Abstract
The binding protein (BiP) is an important component of endoplasmic reticulum stress response of cells. Despite extensive studies in cultured cells, a protective function of BiP against stress has not yet been demonstrated in whole multicellular organisms. Here, we have obtained transgenic tobacco (Nicotiana tabacum L. cv Havana) plants constitutively expressing elevated levels of BiP or its antisense cDNA to analyze the protective role of this endoplasmic reticulum lumenal stress protein at the whole plant level. Elevated levels of BiP in transgenic sense lines conferred tolerance to the glycosylation inhibitor tunicamycin during germination and tolerance to water deficit during plant growth. Under progressive drought, the leaf BiP levels correlated with the maintenance of the shoot turgidity and water content. The protective effect of BiP overexpression against water stress was disrupted by expression of an antisense BiP cDNA construct. Although overexpression of BiP prevented cellular dehydration, the stomatal conductance and transpiration rate in droughted sense leaves were higher than in control and antisense leaves. The rate of photosynthesis under water deficit might have caused a degree of greater osmotic adjustment in sense leaves because it remained unaffected during water deprivation, which was in marked contrast with the severe drought-induced decrease in the CO2 assimilation in control and antisense leaves. In antisense plants, the water stress stimulation of the antioxidative defenses was higher than in control plants, whereas in droughted sense leaves an induction of superoxide dismutase activity was not observed. These results suggest that overexpression of BiP in plants may prevent endogenous oxidative stress.
Protein folding in the endoplasmic reticulum (ER) is facilitated by molecular chaperones, which prevent nonproductive intermolecular interactions of folding intermediates and subsequent misaggregation of proteins within the lumen of the ER (for review, see Hammond and Helenius, 1995). The expression of these ER-molecular chaperones is regulated according to cellular requirements for their functions. Thus, both the increase of secretory activity and accumulation of unfolded proteins within the ER result in the induction of ER-molecular chaperones (for review, see Lee, 1992). This induction is achieved through a signaling pathway named the unfolded protein response (UPR) pathway, which coordinately up-regulates the transcription of a set of ER-resident proteins, including the molecular chaperone binding protein (BiP; Lee, 1992).
In plants, the regulation of BiP gene expression has been examined primarily by the detection of BiP RNA and protein levels under stress conditions and at different developmental stages of the plant organs (for review, see Denecke, 1996). In general, developmental events that are associated with high secretory activity of the cells and exposure of cells to agents that result in accumulation of unfolded proteins in the ER cause induction of plant BiP (for review, see Boston et al., 1996; Galili et al., 1998). Plant BiP expression has also been shown to respond to a variety of abiotic and biotic stress conditions, such as water stress, fungus infestation, insect attack, nutritional stress, cold acclimation, and elicitors of the plant-pathogenesis response (Anderson et al., 1994; Denecke et al., 1995; Kalinski et al., 1995; Fontes et al., 1996; Figueiredo et al., 1997; Fontes et al., 1999). In the endosperm of maize floury-2 mutant, the synthesis of a zein-like storage protein variant, which contains an uncleavable signal sequence, is associated with increased accumulation of BiP (Boston et al., 1991; Fontes et al., 1991; Coleman et al., 1995; Gillinkin et al., 1997). Likewise, the expression of an assembly-defective mutant of the bean storage protein phaseoline induces BiP synthesis in tobacco (Nicotiana tabacum L. cv Havana) leaf protoplasts (Pedrazzini et al., 1994). Furthermore, tunicamycin, apotent activator of the UPR pathway, efficiently induces BiP expression at both mRNA and protein level in several plant systems (Fontes et al., 1991; D'amico et al., 1992; Figueiredo et al., 1997). These results have led to the conclusion that, like mammal and yeast (Saccharomyces cerevisiae) BiP, plant BiP is most likely regulated through a UPR pathway.
Several components of the UPR signaling pathway have been characterized in yeast. The most upstream component characterized is an ER transmembrane kinase protein, Ire1p, which is thought to be responsible for sensing and transmitting the unfolded protein signal to the appropriate downstream components (Mori et al., 1993). Ire1p also exhibits a sequence-specific endonucleolytic activity whose activation seems to be mediated by its kinase activity upon accumulation of unfolded protein within the ER (Shamu and Walter, 1996; Sidrauski and Walter, 1997). The unfolded protein response is mediated at the level of gene expression by the UPR element (UPRE), a 22-bp upstream activating sequence that is necessary and sufficient to activate transcription of a linked promoter in response to accumulation of unfolded proteins within the ER (Mori et al., 1992). Transcriptional activation is mediated by a basic-Leu zipper transcription factor, Hac1p, whose activity is controlled by regulated splicing of its mRNA through a spliciosome-independent pathway, involving tRNA ligase and the endonuclease activity of Ire1p (Cox and Walter, 1996; Sidrauski et al., 1996). Recently, discovery of an Ire1p homolog in mammalian cells has provided direct evidence for the conservation of the ER-stress response mediated by the UPR pathway among eukaryotic cells (Wang et al., 1998). Since these UPR-induced proteins have been shown to act as chaperones, they are believed to function in an ER protective mechanism against protein misaggregation. In fact, overexpression of BiP in mammalian cultured cells (Morris et al., 1997) and tobacco protoplasts (Leborgne-Castel et al., 1999) prevents the induction of UPR-induced genes and increases cell tolerance to stress, suggesting that BiP directly alleviates the ER stress. Furthermore, transfection of mammalian cultured cells with BiP antisense mRNA expression constructs suppressed the induction of BiP without altering basal BiP levels. These cells also showed increased sensitivity to ionophores (Li and Lee, 1991; Li et al., 1992), oxidative stress (Gomer et al., 1991), and cell-mediated toxicity (Sugawara et al., 1993). Although these studies clearly indicated a protective role of BiP against ER stress in cultured cells, they have not been extended to whole multicellular organisms. Here, we have investigated the effect of BiP level variations on protection from ER stress and tolerance to drought in tobacco transgenic lines expressing a soybean (Glycine max) BiP cDNA either in the sense or antisense orientation.
RESULTS
Generation of Tobacco Transgenic Lines
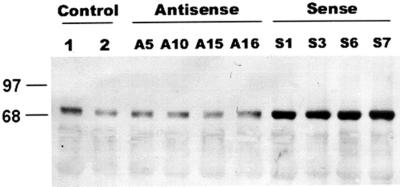
Tobacco was transformed via Agrobacterium tumefaciens with the BiP gene, either in the sense or antisense orientation, under the control of 35S cauliflower mosaic virus (CaMV) promoter and the 3′ nos polyadenylation signal (Fig. 1). T0 primary transformants were selected in tissue culture on the basis of their kanamycin resistance, and most of the rooted plants were tested further for the incorporation of the nptII gene by PCR analysis (data not shown). Several independent transgenic lines were established, transferred into soil, and grown in greenhouse to generate seeds (T1 seeds). The integration and gene copy number of the construct in the transformed plants were further confirmed by Southern blot analysis of genomic DNA digested with BamHI (data not shown) and segregation analysis of the nptII gene in the T0 progenies (T1 plants). Four independent transgenic sense lines (35S-BiPS1, 35S-BiPS3, 35S-BiPS6, and 35S-BiPS7) were selected for further analyses. Under normal, non-stressed conditions, the BiP protein levels detected in their leaves (Fig. 2, lanes S1, S3, S6, and S7) were significantly higher than those of wild type (lane 1) and pBI121-transformed control (lane 2) tobacco leaves. The calibration curves generated with immunoblotting of increasing amounts of purified recombinant soybean protein and tobacco seed protein indicated that our soybean BiP antibody cross-reacted with the endogenous tobacco BiP with similar efficiency (data not shown).
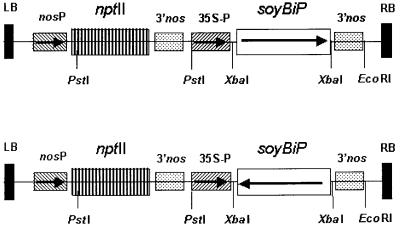
Figure 1.
Schematic diagram of the chimeric BiP constructs transformed into tobacco via pBI121-derived binary vector. The soy BiP gene in the sense (A) and antisense (B) orientation was placed under the control of the constitutive CaMV 35S promoter (35S-P) and the 3′ nos polyadenylation signal (3′nos). The nptII gene expression is driven by the nos promoter (nos-P). LB and RB correspond to the T-DNA left and right borders, respectively. The positions of some restriction enzyme sites are indicated.
Figure 2.
Enhanced levels of BiP in transgenic tobacco plants. Equivalent amounts of total protein (30 μg per lane) extracted from the fully expanded third leaf of untransformed, wild-type plant (lane 1), pBI121-trasnformed control plant (lane 2), four independent transgenic 35S-BiPAS (antisense) tobacco plants (lanes A5, A10, A15, and A16), and four independent transgenic 35S-BiPS (sense) tobacco plants (lanes S1, S3, S6 and S7) were fractionated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with an anti-soybean BiP antibody. Different numbers after S and A symbols indicate that the transgenic plants were originated from independent events of transformation. The positions of prestained molecular markers are indicated on the left in kilodaltons.
Segregation analysis suggested that the lines 35S-BiPS1, 35S-BiPS3, and 35S-BiPS7 T0 plants appeared to have an integrated T-DNA locus on a single chromosome, since 75% of their T1 segregating seedlings were resistant to kanamycin (Table I). Homozygous lines of the 35S-BiPS3 and 35S-BiPS7 transgenic plants were established by selecting T1 plants that had exclusively produced kanamycin-resistant T2 plants after self-crossing.
Table I.
Expression of kanamycin resistance and tunicamycin tolerance in the T1 generation of transgenic tobacco plants
| Plant Lines Tested | Kanamycin-Resistant Seedlings | Ratio | χ2 | Tunicamycin-Tolerant Seedlings | Ratio | χ2 |
|---|---|---|---|---|---|---|
| 35S-BiPS1 | 780+ /282− | 3:1 | 0.81 | 174+ /054− | 3:1 | 0.21 |
| 35S-BiPS3 | 305+ /093− | 3:1 | 0.28 | 305+ /093− | 3:1 | 0.56 |
| 35S-BiPS6 | 400+ /250− | – | – | NDa | ND | ND |
| 35S-BiPS7 | 739+ /271− | 3:1 | 1.72 | 175+ /054− | 3:1 | 0.25 |
| ControlAb | 000+ /105− | – | – | 003+ /301− | – | – |
| ControlBc | 640+ /41− | 15:1 | 0.18 | 004+ /208− | – | – |
χ2 Tests indicate good agreement with the segregation ratio indicated.
ND, Not determined.
Untransformed, wild-type plants.
pBI121-transformed plants.
Overexpression of BiP Attenuates the Lethality Caused by the Toxic Effect of Tunicamycin during Seed Germination
The BiP protective properties against ER stressors have been investigated in engineered mammalian cell lines (Morris et al., 1997; Laitusis et al., 1999) and also in protoplasm from transgenic cell lines (Leborgne-Castel et al., 1999). Here we analyzed, at the whole plant level, the effect of BiP overexpression on a typical ER stress response using a germination/survival assay in the presence of tunicamycin, a potent activator of the UPR pathway. For this assay, T1 seeds were allowed to germinate for 5 d in a solid Murashige and Skoog-based medium supplemented with 5 μg mL−1 of tunicamycin and then transferred to a tunicamycin-free medium. The seeds expressing the soybean BiP gene recovered and germinated into seedlings, whereas those lacking the transgene failed to germinate and eventually died (less than 1% of the wild type seedlings recovered after removing tunicamycin). For the 35S-BiPS1, 35S-BiPS3, and 35S-BiPS7 independent transgenic lines analyzed, the tunicamycin-tolerant germination phenotype was found to be linked to the nptII gene because it segregated with the same ratio as the kanamycin-resistant phenotype (Table I). Since transformation of tobacco with the nptII gene alone did not confer resistance to tunicamycin during seed germination, we concluded that the tunicamycin-tolerant germination phenotype was caused by overexpression of the BiP gene.
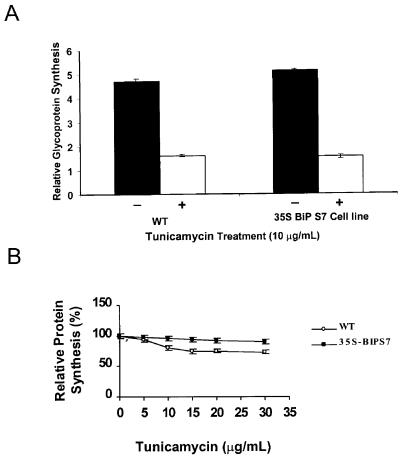
Tunicamycin inhibits glycosylation of N-linked glycoproteins and, as a consequence, promotes the accumulation of unfolded proteins in the ER. BiP did not interfere directly in the toxicity mechanism of tunicamycin because elevated levels of BiP did not prevent the glycosylation block in the tunicamycin-treated 35S-BiPS7 sense cell line. In fact, tunicamycin treatment reduced accumulation of glycoproteins in the 35S-BiPS7 cell line to the same extent as in control cells (Fig. 3A). Tunicamycin treatment has been shown to inhibit the synthesis of secretory proteins in tobacco cell cultures (Leborgne-Castel et al., 1999). The BiP-mediated protection against ER stressors has been previously shown to be due to restoration of the protein synthetic capability under ER stress conditions (Morris et al., 1997; Laitusis et al., 1999; Leborgne-Castel et al., 1999). We asked whether overexpression of BiP in the 35S-BiPS7 cell line could affect the protein synthesis inhibition mediated by tunicamycin (Fig. 3B). Treatment with 15 μg mL−1 tunicamycin inhibited protein synthesis in control cells to 74% of untreated cells. In contrast, tunicamycin treatment of 35S-BiPS7-overexpressing BiP cells only inhibited protein synthesis to 93%, demonstrating that they were partially resistant to this inhibition.
Figure 3.
Total protein synthesis and glycoprotein accumulation in tunicamycin-treated cell line overexpressing BiP. Wild-type (WT) and 35S-BiPS7 suspension cells were treated with the indicated concentrations of tunicamycin for 12 h and then were labeled for 3 h with [35S]Met and [35S]Cys. Incorporation of radiolabeled amino acids was measured by monitoring trichloroacetic acid (TCA)-precipitable radiolabeled proteins from [35S]Met and [35S]Cys-labeled cell lysates. A, Labeled glycoproteins were affinity purified using concanavalin-A-Sepharose resin and determined by liquid scintillation counting (Beckman Instruments, Fullerton, CA). Relative synthesis of glycoprotein was calculated by normalizing the TCA-precipitable activity (100%) in labeled cell lysates. Values are the mean ± sd from three replicates. B, Protein synthetic rates were calculated as the radioactivity incorporation per microgram of protein. Values are the mean ± sd from three replicates.
The Leaf BiP Levels Correlated with the Maintenance of the Shoot Turgidity under Water Deficit Conditions
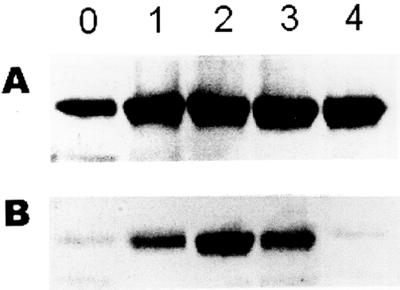
Because plant BiP has also been shown to be induced by a variety of environmental stresses, including water stress (Figueiredo et al., 1997; Cascardo et al., 2000), we next analyzed the response of the transgenic lines to drought stress. The water stress condition was gradually established by ceasing irrigation from young seedlings grown in a greenhouse. After 4 weeks (30 d) under progressive dehydration, a water stress-tolerant phenotype was clearly developed by the transgenic plants overexpressing a BiP gene (Fig. 4). Whereas the untransformed, wild-type (data not shown) and pBI121-transformed control leaves (bottom) were completely wilted, the transgenic sense leaves kept the turgidity to a normal level (top). Prolonged drought inhibited growth completely, and eventually the wild-type and pBI121-transformed control plants died (Fig. 4). Whereas nearly all 35S-BiPS7 homozygous plants were tolerant to water stress, less than 5% of control plants survived the drought stress treatment. During the period of water deprivation, the level of BiP accumulation in transgenic and pBI121-transformed control leaves was monitored by immunoblotting analysis (Fig. 5). In the control leaves (Fig. 5B), as a direct result of water deficit, the endogenous BiP was induced (lane 2), but as the stress condition persisted, accumulation of BiP declined to undetectable levels (lane 4). The decrease of BiP levels coincided with the appearance of the wilting phenotype in the control leaves, whereas in the transgenic sense leaves the BiP levels were kept high during the period of stress (Fig. 5A). So, under progressive drought, the BiP levels in the leaves correlated with the maintenance of the shoot turgidity. In addition, the water stress-tolerant phenotype segregated with the BiP transgene in the T0 progenies (T1 plants). These results represent the first demonstration that elevated levels of BiP increase tolerance of plants to water deficit.
Figure 4.
Elevated levels of BiP confer water stress tolerance to transgenic plants. Water stress was induced in 6-week-old seedlings (at the same developmental stage) of pBI121-transformed control and transformed 35S-BiPS7 tobacco plants by withholding irrigation for 4 weeks. The stress condition was prolonged until death of control (pBI121) plants (bottom). At the top, 35S-BiPS7 transgenic plants (kanamycin-resistant T1 generation) were submitted to the same water stress conditions as control plants.
Figure 5.
BiP accumulation in transgenic and control leaves during progressive water deficit. Leaf soluble proteins from the transgenic 35S-BiPS7 (A) and pBI121-tranformed control (B) plants grown under water deprivation for 1 week (lane 1), 2 weeks (lane 2), 3 weeks (lane 3), and 4 weeks (lane 4) were fractionated by SDS-PAGE and immunoblotted using an anti-soybean BiP serum. Lane 0 shows the BiP levels in plants before withholding watering.
Decreased Expression of Endogenous BiP Disrupts Water Stress Tolerance
We predicated that if overexpression of BiP was the basis for the water stress-tolerant phenotype, antisense repression should further debilitate the plant under water deficit condition. In fact, selective target of BiP with antisense cDNA expression interferes with induction of BiP and other ER stress proteins and disrupts the ER stress response (Little and Lee, 1995; Liu et al., 1997; data not shown). We target endogenous tobacco BiP using the entire antisense-coding region of the soyBiPD clone under the control of the S35 promoter. Nevertheless, under unstressed conditions, the antisense plants accumulated BiP to normal levels (Fig. 2, lanes A5, A10, A15, and A16), suggesting that the inhibitor effect of the mRNA antisense on BiP levels may be compensated by activation of the UPR pathway and up-regulation of endogenous BiP mRNA. The lack of coregulation between the antisense 35S promoter and endogenous UPR-regulated BiP promoter together with the essential nature of BiP may explain the normal level of BiP in the selected antisense plants. In fact, in yeast and mammalian cells, a minimum basal level of BiP expression is required for cell viability (Rose et al., 1989; Li et al., 1992). Likewise, antisense repression studies in plant cells indicate that a complete knockout of BiP function may result in cell lethality (Leborgne-Castel et al., 1999). Nevertheless, antisense BiP gene expression prevents BiP induction in response to ER stressors (Little and Lee, 1995; Liu et al., 1997; data not shown).
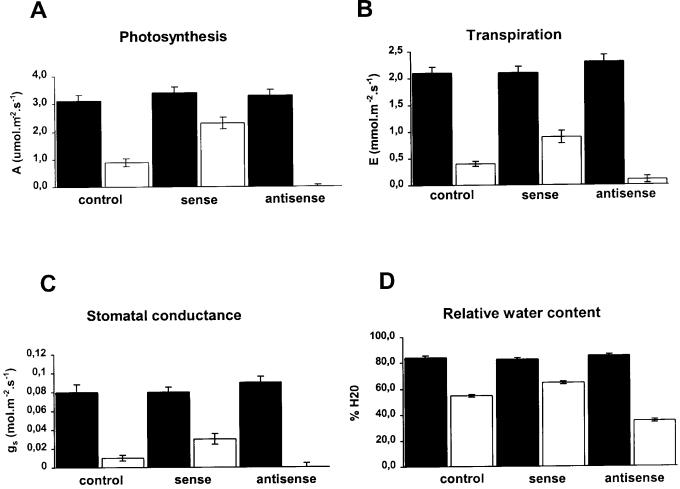
In view of these observations, the response of the antisense transgenic lines to water stress conditions was examined. For this experiment, the plants were transferred to a growth chamber where one-half of the plants received no irrigation and the remaining ones were irrigated throughout of the experiment. The pBI121-transformed control plants, which showed normal induction of BiP (Fig. 5), tolerated the stress treatment relatively well during the period of the experiment (15 d, Fig. 6). In contrast, the antisense plants with impaired BiP induction showed increased sensitivity to water deficit, as their leaves were completely wilted at 15 d after stress induction. Therefore, a deficiency in stress induction of BiP in antisense plants correlated with an increase in water stress sensitivity over that of control plants. An inverse correlation of effects was evident in sense plants exposed to water deficit and confirmed the previous results. Although the transgenic sense leaves kept the turgidity to a normal level under water deprivation, their stomatal conductance and transpiration were higher than in control plants (Fig. 7B, 7C). Thus, neither the water stress-tolerant phenotype of BiP transgenic plants nor the turgidity of transgenic plants was a result of stimulation of stomatal closure aperture. The maintenance of turgidity and water content in stressed sense plants (Fig. 7D) may suggest that a degree of osmotic adjustment in these plants prevented cellular dehydration.
Figure 6.
Antisense expression of BiP genes disrupts the water stress tolerance. Water stress condition was induced in 6-week-old seedlings grown in growth chamber by withholding irrigation for 15 d (from left to right in duplicates): 35S-BiPS3 sense transgenic tobacco plant, pBI121-transformed control plant, and 35S-BiPAS5 antisense plant.
Figure 7.
Physiological measurements of transgenic tobacco lines continuously irrigated (black bars) or exposed to 15 d of water deprivation (white bars). On the 15th d of the experiment, the relative water content (D) of well-watered and droughted pBI121-transformed control, 35S-BiPS sense, and 35S-BiPAS antisense transgenic leaves was measured. Photosynthetic rate (A), transpiration rate (B), and stomatal conductance (C) of the third leaf of control, sense, and antisense transgenic plants were measured by the LCA-2 infrared (IR) gas analyzer at 600 μmol m−2 s−1 irradiance. Each value represents the mean ± sd of five replicates from three independent experiments.
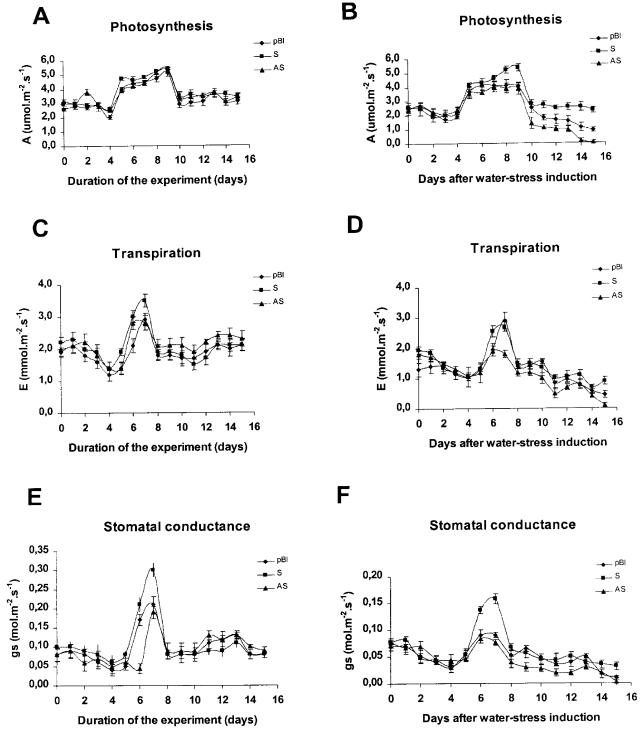
The photosynthetic rate, transpiration rate, and stomatal conductance did not differ significantly in well-watered PBI121-transformed control, sense, and antisense plants throughout the period of the experiment (Fig. 8, A, C, and E). However, under a water deprivation regime, the photosynthetic CO2 assimilation decreased to a different extent in antisense, control, and sense plants (Figs. 7A and 8B). After 10 d of water deprivation, the rate of CO2 assimilation rapidly decreased, reaching a minimum on the 15th d of stress in antisense (0% of the initial value), in control (30% of the initial value), and in sense lines (80% of the initial value). The loss of CO2 assimilation capacity in the droughted antisense leaves was higher than in pBI121-transformed control leaves, whereas in the droughted sense leaves it was much less affected than in control leaves. Thus, under progressive drought conditions, the photosynthetic rate in sense leaves decreased to a much less extent than control and antisense lines.
Figure 8.
Physiological measurements of sense and antisense transgenic tobacco plants during progressive water deficit. Sense (S), antisense (AS), and PBI121-transformed control (pBI) young seedlings were exposed to the follow water regime: one-half received normal water supply (A, C, and E) and the other half received no irrigation (B, D, and F) during 15 d. Photosynthetic rate (A and B), transpiration rate (C and D), and stomatal conductance (E and F) of the third leaf of pBI121-transformed control, 35S-BiPS, and 35S-BiPAS transgenic plants were measured by the LCA-2 IR gas analyzer at 600 μmol m−2 s−1 irradiance during the period of the experiment. Each value represents the mean ± sd of five replicates from three independent experiments.
Alterations of photosynthesis rate may be a result of stomatal closure or degradation of the photosynthetic apparatus by the stress conditions. The effect of water stress on the integrity of the photosynthetic apparatus was analyzed by determining the fluorescence ratio (Fv/Fm) as a measure for the photochemical efficiency of photosystem II. Under progressive drought stress, the photochemical efficiency (Fv/Fm ratio) of sense, control, and antisense leaves remained unaltered and was similar to that of the well-watered control plants (data not shown). In contrast, stomatal conductance varied to different extents in antisense, sense, and control leaves and correlated well with the photosynthesis rate of these plants (Figs. 7C and 8F). Consistent with the higher rate of photosynthesis in sense lines, the stomatal conductance was higher in sense leaves than in control and antisense leaves (Figs. 7B and 8C). Variation on transpiration rate followed the same pattern as stomatal conductance, although the differences observed among sense, antisense, and control plants were lower (Fig. 8C). Taken together, these results indicate that the decrease in photosynthetic rate of antisense plants under water deficit was more likely a result of stomatal closure than of water stress-induced damage of the photosynthetic apparatus.
Fluctuations of BiP Levels Correlate Inversely with the Activity of Oxidative Stress-Induced Enzymes
Despite the observation that enhanced accumulation of BiP seems to prevent dehydration, our results indicated that the maintenance of normal turgidity of sense leaves under water deficit may not be a consequence of stomatal functioning. In this case, BiP may be acting directly to alleviate intracellular stress caused by water deprivation. One of the effects of severe water stress is the enhancement of the production of reactive oxygen species, leading to oxidative stress, which in turn stimulates enhanced levels or activities of antioxidant enzymes, such as superoxide dismutase (SOD). In view of this observation, we measured SOD activity in sense, antisense, and control droughted plants as a means to monitor intracellular stress (Table II). In fact, water deficit caused an increase of SOD activity in control plants. The activity of SOD remained practically unaltered in droughted sense plants as compared to their irrigated counterparts, whereas in antisense plants the extent of the increase in SOD activity caused by water deprivation was higher than in control plants. Taken together, these results are consistent with a BiP-mediated mechanism that alleviates endogenous oxidative stress.
Table II.
Specific activity of SOD in transgenic tobacco leaves continuously irrigated (well-watered plants) or exposed to 15 d of water deprivation (droughted plants)
| Water Regime | SOD Activity
|
||
|---|---|---|---|
| Controla | Senseb | Antisensec | |
| unit mg−1 | |||
| Well-watered plants | 13.656 ± 2.702 | 12.384 ± 0.541 | 11.810 ± 0.806 |
| Droughted plants | 17.302 ± 1.354 | 13.470 ± 1.641 | 24.423 ± 0.538 |
Values represent means ± se (n = 5).
pBI121-transformed control plants.
BiP-overexpressing plants (35S-BiPS).
Antisense BiP cDNA-transformed control plants (35S-BiPAS).
DISCUSSION
Overexpression studies of BiP genes in cultured mammalian cells and tobacco leaf protoplasts demonstrated that BiP attenuates ER stress caused by ionophore or tunicamycin (Morris et al., 1997; Laitusis et al., 1999; Leborgne-Castel et al., 1999). In both cases, overexpression of BiP has been shown to protect against inhibition of protein synthesis caused by these ER stressors. Here, we showed that the effectiveness of BiP protection against ER stress caused by accumulation of unfolded proteins into the ER could be extended to the whole organism level, as judged by the results of our germination/survival assay in the presence of tunicamycin. The blockage of protein glycosylation by tunicamycin is expected to result in the accumulation of underglycosylated protein, which cannot fold properly, raising a pool of unfolded or nonfunctional proteins into the ER. In such a highly concentrated protein environment, hydrophobic side chains exposed by denatured proteins will interact with other unfolded proteins or with hydrophobic surfaces on proteins in process of synthesis or assembly, triggering misaggregation of proteins. At high concentrations of BiP, unfolded protein domains would be more likely to interact with BiP than with other reactive surfaces. Under this circumstances, enhanced accumulation of BiP would raise the ER protein processing capacity, which in turn would allow high rates of protein synthesis. A dynamic relationship has been shown to exist between the rate of protein processing and mRNA translation. Compelling evidence in the literature supports the hypothesis that BiP functions in part to coordinate the respective rates of ER protein processing and mRNA translation. In mammalian cells, alterations in the availability of BiP affected translational initiation and the degree of eukaryotic initiation factor 2 phosphorylation (Morris et al., 1997; Laitusis et al., 1999). Likewise, in protoplasts of tobacco (Leborgne-Castel et al., 1999) and in our 35S-BiPS7 cell line, overexpression of BiP confers tolerance to translational inhibition in response to ER stress.
We have also demonstrated that constitutive overexpression of BiP in tobacco is enough to confer tolerance to water stress. At the cellular level, plant responses to dehydration may result from membrane and protein structure damage, whereas other responses correspond to adaptive processes (for review, see Shinozaki and Yamaguchi-Shinozaki, 1997). Based on BiP function as molecular chaperone and regulation of its expression in response to stresses, BiP may act in both mechanisms. The protective role of BiP against water stress may be associated with preservation of protein structure and membrane integrity as well as with the maintenance of high secretory activity mediated by the water stress adaptive cellular response (Ingram and Bartels, 1996). BiP might facilitate proper folding and maturation of a selected group of water stress-induced secretory proteins, which are probably involved in the osmotic response mechanism. In fact, drought-induced proteins, which are targeted to the secretory pathway, have been identified in a wide range of plant species (Ingram and Bartels, 1996; Riccardi et al., 1998) and BiP has been shown to associate with water-stress induced proteins (Cascardo et al., 2000). Overexpression of BiP may provide an indirect effect in stress tolerance by allowing the cells to control more efficiently the concentration of specific defense proteins before the stress condition has reached its maximum deleterious effect. Investigation of the induction kinetics of secretory water stress-induced proteins in BiP overproducers will allow us to address this possibility, providing insight into the mechanism by which BiP-dependent increases in water stress tolerance are achieved.
Based on the phenotype of the drought-tolerant transgenic lines, overexpression of BiP was associated with a water stress tolerance, which led to the maintenance of turgidity and water content. This water stress-tolerant phenotype suggests that BiP might also be involved in preventing cellular dehydration. Stomatal closure in response to water stress constitutes one of the well-characterized drought adaptation mechanisms. Nevertheless, under water deficit, the stomatal conductance and transpiration rate in the BiP-overexpressing leaves were higher than in control and antisense leaves. More likely, a degree of greater osmotic adjustment prevented turgidity loss and dehydration in sense plants. This possibility is consistent with the observation that under progressive drought conditions the photosynthesis rate in sense leaves was much less affected than in control and antisense leaves. Thus, while CO2 assimilation is maintained in droughted sense leaves, their expansion growth is inhibited by water stress (data not shown). This leads to a reduced utilization of photoassimilates that could account at least in part for the increase in solute concentration in sense leaf tissues. Compelling evidence has implicated photosynthesis as the major source of organic solute accumulation under water stress. Centrosema brasilianum, Centrosema pascuorum, and Centrosema pubescens, stomatal closure causes the cessation of osmotic adjustment (Ludlow et al., 1983). Likewise, the excision of photosynthetic cotyledons from soybean and sunflower seedlings prevents solute accumulation and osmotic adjustment, causing a significant decrease in turgor (Meyer and Boyer, 1972, 1981; Kutschera and Köhler, 1994). The higher rates of CO2 assimilation in water-stressed sense leaves may be correlated with their stomatal conductance under water deficit. An inverse correlation of effects was observed in antisense plants. Since we did not detect water stress-induced damage of the photosynthetic apparatus in transgenic plants, the decrease in photosynthetic rate of antisense plants under water deficit was more likely a consequence of stomatal closure.
The cytoplasmic Ca2+ signal transduction pathway is thought to regulate turgor pressure of plant cells and to coordinate stomatal responses to leaf dehydration (for review, see Shinozaki and Yamaguchi-Shinozaki, 1997). During stomatal closure, the level of cytoplasmic Ca2+ increases as a result of inositol 1,4,5-triphosphate-mediated Ca2+ mobilization from intracellular stores. In fact, increases in the concentration of cytosolic Ca2+ can initiate closure of the stomatal aperture in tobacco. Although little is yet known to evaluate a direct connection among BiP function, Ca2+ release from the ER, and cell turgor, it is largely known that the gradual depletion of intracellular Ca2+ stores promotes up-regulation of BiP genes by a distinct pathway from the tunicamycin-mediated BiP induction (Roy et al., 1996; McCormick et al., 1997; Morris et al., 1997). BiP has also been demonstrated to play a direct and relevant role in the storage of a rapidly exchanging pool of Ca2+ within the ER lumen, and variations on the protein level induced adjustment of the cellular Ca2+ homeostasis (Lièvremont et al., 1997). Thus, overexpression of BiP in sense plants may increase the ER Ca2+ storage capacity affecting stomatal functioning.
The present investigation shows that the magnitude of stimulation of the antioxidative protection system by water stress correlated inversely with the levels of BiP. A component of the antioxidative defense system, SOD, was investigated, and the water stress-induced increase in its activity was higher in antisense water-stressed leaves than in pBI121-transformed control, whereas in sense plants the SOD activity remains practically unaltered during water deprivation. As the antisense plants exhibit an increased sensitivity to water deficit and SOD contributes to the removal of O2−, the increase of SOD activity in antisense droughted leaves may reflect the magnitude of the water stress-induced oxidative stress rather than the intensity of a protective response mechanism in antisense leaves.
In contrast to the antisense plants, overexpression of BiP was apparently associated with a decreased responsiveness of antioxidative enzymes under water deficit. The presumed oxidative stress may be limited in sense plants because overexpression of BiP prevented endogenous stress. In mammalian cells, the control of Ca2+ release from the ER by BiP prevented oxidative stress (Liu et al., 1997, 1998). Although further studies will be necessary to discern the precise mechanism of BiP-mediated water stress tolerance, the potential BiP control on ER Ca2+ homeostasis may provide a link for the apparently pleiotropic physiological effects of BiP protective properties under water stress. However, our data did not rule out the possibility that the O2− production rate was reduced in sense leaves by activation of alternative dissipative pathways.
Whole plants respond to water stress through morphological, physiological, and metabolic adjustments, and some of the adaptive cellular responses are clearly interconnected with other environmental stress responses. In natural conditions, water stress alone is unlikely to occur, since it intrinsically affects the uptake of essential nutrients leading to nutritional stress. Likewise, cell water deficit results in osmotic stress, which also can be caused by salt stress. Consequently, in situations of water stress, nutritional deprivation and osmotic and salt stresses are likely to debilitate the plant. In this complex interplay of physiological stresses, general stress-related proteins that permit coordinate adaptive cellular responses under a large array of stress conditions are more likely to provide protective functions and physiological advantage to plants under field conditions. Thus, the effectiveness of BiP overexpression on plant protection against stresses may be related to its induction by a large variety of physiological stress conditions (Galili et al., 1998). The fundamental bases for these acquired physiological advantages can be exploited on the agronomic level and extended to other economically important crops.
MATERIALS AND METHODS
Construction of Plasmids
DNA manipulations were performed essentially as described by Sambrook et al. (1989). The cDNA encoding BiP protein, designated soyBiPD cDNA (GenBank accession no. AF031241), was isolated from a soybean (Glycine max) seed expression library and has been previously described (Figueiredo et al., 1997). The BiP coding region was amplified by PCR from the λgt11 recombinant DNA with the forward primer A (5′-ctcgagagagcatatggctggctcgtg-3′), the reverse primer B (5′-ggatccgactaatctagagctcatcgt-3′), and Pfu DNA polymerase. The underlined sequences of the forward A and reverse B primers correspond to the XhoI and BamHI restriction sites, respectively, which were introduced in the BiP cDNA by PCR-based mutagenesis. The 2-kb amplified sequence, spanning the entire protein-coding region and lacking the 3′ untranslated sequences, was subcloned into the SmaI site of pUC118 in the antisense orientation to obtain the clone pUFV41. The BiP coding region was released from pUFV41 with XhoI/BamHI double digestion and subcloned into pGEM7zf (−) previously digested with the same enzymes and dephosphorylated. The resulting clone, pUFV42, harbors the BiP coding sequence in the right orientation. To construct the chimeric BiP gene under the control of the CaMV 35S promoter, the 2.0-kb XbaI cDNA insert of pUFV42 was subcloned into an XbaI site of a pBI121-derived binary vector, which had been obtained by releasing the gene gus with SacI and SmaI, repairing SacI site with T4 DNA ligase, and then circularizing it in vitro. The resulting binary plant transformation plasmids, pUFVBiPS and pUFVBiPAS, harbor the BiP cDNA in the sense and antisense orientation, respectively, under the control of the CaMV 35S promoter and the polyadenylation signal of the T-DNA nopaline synthase (nos) gene.
Plant Transformation
Leaf discs from in vitro-grown tobacco (Nicotiana tabacum L. cv Havana) plants were cocultivated for 15 min with Agrobacterium tumefaciens strain LBA4404 containing the binary plasmid pUFVBiPS or pUFVBiPAS or the binary vector pBI121. Transformed shoots were selected on Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 6-benzylaminopurine (500 μg mL−1), cefotaxime (400 μg mL−1), and kanamycin sulfate (100 μg mL−1) (Pedra et al., 2000). Regenerated shoots were rooted on phytohormone-free medium containing kanamycin (100 μg mL−1), transferred into soil, and grown in standardized greenhouse conditions (T0 plants) to generate seeds. Four independently regenerated kanamycin-resistant plants harboring the BiP sense construct and three antisense plants were grown for further analyses. Two kanamycin-resistant plants for the pBI121-incorporated binary vector were used as control.
Analysis of Transgenic Plants
The presence of nptII and BiP transgenes was analyzed by PCR from leaf tissue samples. PCR was carried out on 20 ng of genomic DNA isolated from 4-week-old greenhouse-grown transgenic plants, using 1.5 μm each of nptII primers or soyBiPD gene-specific primers (GenBank accession no. AF031241) and 1 unit of Taq polymerase in a final volume of 25 μL. The soybean BiP-specific primers were 5′-atctggaggagccctaggcggtgg-3′ (coordinates 1966–1990, upstream) and 5′-cttgaagaagcttcgtcgtaaactaag-3′ (positions 2157–2184, downstream). The primers specific for the nptII gene were 5′-tcgacgttgtcactgaagcgcg-3′ (positions 627–648, upstream) and 5′-gcggtcagcccattcgccgcc-3′ (coordinates 1082–1102, downstream). The PCR reactions were conducted for 30 cycles (50 s at 94°C, 75 s at 47°C, and 120 s at 72°C) with a final extension at 72°C, for 10 min. Transgene copy number was determined by Southern blot and segregation analyses.
For genomic DNA gel blot analysis, DNA was extracted from young leaves, digested overnight with BamHI, precipitated with 70% (v/v) ethanol and separated on a 1% (w/v) agarose gel. The gel was washed with 250 mm HCl followed by alkaline denaturation (Sambrook et al., 1989). After neutralization, the DNA was transferred to nylon membranes and UV fixed (Stratalinker, Stratagene, La Jolla, CA). The nptII DNA fragment was radiolabeled with [α-32P]dCTP by random primed labeling (Amersham Pharmacia Biotech, Barra Funda, SP, Brazil). Hybridization and washing of the blots were performed using standard procedures (Sambrook et al., 1989). Autoradiography was performed at −80°C using a Lightning-Plus intensifying screen (Sigma, St. Louis).
For segregation analysis, seeds were germinated on Murashige and Skoog medium containing 150 μg mL−1 kanamycin. Homozygous T1 lines with respect to the T-DNA loci were selected by determining the frequency of their antibiotic-resistant T2 seeds after self-pollination. Accumulation of BiP was monitored in each generation by immunoblotting analysis.
Protein Extraction and Immunoblotting Analysis
Total protein was extracted from an acetone dry powder, using a protocol adapted from Görg et al. (1988). Briefly, plant tissues (cells, leaves, and seeds) were crushed in liquid nitrogen, and 2 g of the powder were homogenized with 10% (w/v) TCA in acetone containing 0.07% (v/v) 2-mercaptoethanol. Total protein was precipitated for 40 min at −20°C, recovered by centrifugation at 16,000g for 15 min, and washed two to three times with acetone containing 0.07% (v/v) 2-mercaptoethanol. The pellet was dried under vacuum, and 100 mg of the acetone dry powder was homogenized in 1 mL of 50 mm Tris-HCl (pH 7.5), 1% (w/v) SDS, and 25 mm EDTA. Cell debris was removed by centrifugation at 25,000g for 20 min and protein concentration was determined as described by Hill and Straka (1988).
Equivalent amounts of total protein (30 μg) were resolved by SDS-PAGE (Laemmli, 1970) and transferred to nitrocellulose membrane using a blot apparatus (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. The membrane was blocked with 3% (w/v) bovine serum albumin in TBST [100 mm Tris-HCl (pH 8), 150 mm NaCl, 0.05% (v/v) Tween 20]. BiP was detected using a polyclonal antibody raised against the carboxyl region of soybean BiP at a 1:1,000 dilution (Figueiredo et al., 1997), followed by a goat anti-rabbit IgG conjugated to alkaline phosphatase (Sigma) at a 1:5,000 dilution. The activity of alkaline phosphatase was assayed using 5-bromo-4-chloro-3-indolyl phosphate (Life Technologies do Brasil Ltda, São Paulo, Brazil) and p-nitroblue tetrazolium (Life Technologies).
Water Stress Tolerance of Transgenic Plants
Untransformed, transformed control (pBI121 vector alone), transformed sense (35S-BiPS lines), and antisense (35S-BiPAS lines) tobacco plants were grown in standardized greenhouse conditions. Transgenic plants used for analysis of water stress tolerance were primary transformant and homozygous, selfed T1 progeny of the primary transformant (T0 plants). Transgenic T1 seeds were germinated in kanamycin-containing medium for 3 weeks before transplantation. Plants were grown in a mixture of soil, sand, and dung (3:1:1) for 2 weeks in greenhouse conditions under natural conditions of light, relative humidity 70%, and controlled temperature, 18°C and 30°C (night and day). Water stress condition was induced by withholding watering for the period of time as indicated in the figure legends. The water content of the soil was monitored throughout the experiment by the gravimetric method (Slavik, 1974). The soil water content of all samples was progressively recorded as a function of time to ensure that the extent of soil drying or the severity of plant water stress was similar for all samples analyzed. This procedure ruled out the possibility that differential rates of soil water depletion among the lines could account for the observed phenotypes.
Water stress experiments were also conducted in controlled environment cabinets. In this case, 2-week-old seedlings were transplanted individually to pots and grown in a growth chamber with a 12-h photoperiod at a 23°C day/18°C night temperature cycle, 240 μmol m−2 s−1 irradiance, and a relative air humidity of 60%. After 30 d of growth with normal water supply, drought stress was conducted by withholding water for 2 weeks from one-half of the sense and antisense plants. The remaining transgenic plants received normal water supply continuously. In control experiments, the same conditions of water availability were applied in transformed pBI121 control plants at the same developmental stage as transgenic sense and antisense BiP plants. All the experiments were conducted with five clones from at least three independently transformed lines for each DNA construct.
Physiological Measurements
Photosynthetic CO2 assimilation, transpiration rate, and stomatal conductance of the third leaf were measured by IR gas analysis using a portable analyzer (model LCA-2, Analytical Development Co., Hoddeston, UK) at 600 μmol m−2 s−1 irradiance during the period of the water stress experiment. Relative water content of leaves was determined by the relative turgidity technique (Catský, 1974).
SOD Activity
For determination of total SOD activity, 1 g of leaves were homogenized with 3 mL of 200 mm sodium phosphate, pH 7.8, containing 2 mm EDTA and 80 mm l-ascorbic acid. After centrifugation at 10,000 g for 25 min, total protein was determined according to Bradford (1976). Total SOD activity was assayed by its ability to inhibit the photochemical reduction of nitroblue tetrazolium (NBT), determined at 560 nm. The reaction mixture contained 50 mm sodium phosphate, pH 7.8, 13 mm l-Met, 2 μm riboflavin, 0.1 μm EDTA, and 75 μm NBT. One unit of the enzyme was defined as the amount of the enzyme required to inhibit NBT reduction by 50% (Giannopolitis and Ries, 1977).
Cell Culture and BiP Induction Assays
Callus cultures were initiated from the pith of untransformed and 35S-BiPS7 (sense) tobacco plants as described by Delú-Filho et al. (2000). Cell culture lines were generated by transferring 2 g of friable calli to 25 mL of medium NT-1 [Murashige and Skoog salts supplemented with 3% (w/v) Suc, 0.0001% (w/v) thiamine-HCl, 0.01% (w/v) inositol, 0.2 μg mL−1 2,4-d, 1.32 mm KH2PO4]. The cell culture was established after four subcultures in the liquid medium prior to the BiP induction assays. Tunicamycin was added to cultures at 4 d after passage by dilution of a 5 mg mL−1 stock in dimethyl sulfoxide into normal growth medium to increasing concentrations (as indicated in the figure legends) and incubated for 12 h.
Control and tunicamycin-treated cells were labeled for 3 h with 50 μCim L−1 Trans [35S]-label (Amersham Pharmacia Biotech, UK). Incorporation of radiolabeled amino acids was determined by measuring TCA-precipitable activity in labeled cell lysates and protein concentration was determined (Bradford, 1976). Glycoproteins were affinity purified from cell lysates using a concanavalin-A-Sepharose (Amersham-Pharmacia Biotech, Uppsala) batch method as described by Thompson et al. (1987), and the affinity purified labeled glycoproteins were quantified by liquid scintillation counting (Beckman).
ACKNOWLEDGMENTS
The authors thank Dr. Becky Boston and Jeff Gillikin for helpful discussions and critical reading of the manuscript and Luís Contim for his technical assistance.
Footnotes
This research was supported by the Brazilian government agencies Financiadora de Estudas e Projetos (grant no. 64.94.0113.00 to E.P.B.F.), Programa de Apoio e Desenvolvimento Científico e Tecnológico (grant no. 62.0272/97.0 to E.P.B.F.), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (grant no. CBB 2598/98 to E.P.B.F.). F.C.A. was supported by a Fundação de Amparo à Pesquisa do Estado de Minas Gerais graduate fellowship from the Minas Gerais State (Brazil). S.M.B.C. and J.C.M.C. received Conselho Nacional de Desenvolvimento Científico e Tecnológico graduate fellowships from the Brazilian Government.
LITERATURE CITED
- Anderson JV, Li QB, Haskell DW, Guy CL. Structural organization of the spinach endoplasmic reticulum-lumenal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994;104:1395–1370. doi: 10.1104/pp.104.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston RS, Fontes EPB, Shank BB, Wrobel RL. Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell. 1991;3:497–505. doi: 10.1105/tpc.3.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in the plants. Plant Mol Biol. 1996;34:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cascardo JCM, Almeida RS, Buzeli RAA, Carolino SMB, Otoni WC, Fontes EPB. The phosphorylation state and expression of soybean BiP isoforms are differentially regulated following abiotic stresses. J Biol Chem. 2000;275:14494–14500. doi: 10.1074/jbc.275.19.14494. [DOI] [PubMed] [Google Scholar]
- Catský J. Water saturation deficit (relative water content) In: Slavik N, editor. Methods of Studying Plant Water Relations. New York: Springer Verlag; 1974. pp. 136–154. [Google Scholar]
- Coleman CE, Lopes MA, Gillikin JW, Boston RS, Larkins BA. A defective signal peptide in the maize high-lysine mutant floury-2. Proc Natl Acad Sci USA. 1995;92:6828–6831. doi: 10.1073/pnas.92.15.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- D'amico L, Valsania B, Daminati MG, Fabrini MS, Nitti G, Bollini R, Ceriotti A, Vitale A. Bean homologues of the mammalian glucose regulated proteins: induction by tunicamycin and interaction with newly-synthesized storage proteins in the endoplasmic reticulum. Plant J. 1992;2:443–445. doi: 10.1111/j.1365-313x.1992.00443.x. [DOI] [PubMed] [Google Scholar]
- Delú-Filho N, Pirovani CP, Pedra JHF, Matrangolo FSV, Macêdo JNA, Otoni WC, Fontes EPB. A sucrose binding protein homologue from soybean affects sucrose uptake in transgenic tobacco suspension-cultured cells. Plant Physiol Biochem. 2000;38:353–361. [Google Scholar]
- Denecke J. Soluble endoplasmic reticulum resident proteins and their function in protein synthesis and transport. Plant Physiol Biochem. 1996;34:197–205. [Google Scholar]
- Denecke J, Carlsson LE, Vidal S, Höglund A-S, Ek B, van Zeiji MJ, Sinjorgo KMC, Palva ET. The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell. 1995;7:391–406. doi: 10.1105/tpc.7.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo JEF, Cascardo JCM, Carolino SMB, Alvim FC, Fontes EPB. Water-stress regulation and molecular analysis of the soybean BiP gene family. Braz J Plant Physiol. 1997;9:103–110. [Google Scholar]
- Fontes EPB, Shank BB, Wrobel RL, Moose SP, O'Brian GR, Wurtzel ET, Boston RS. Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell. 1991;3:483–496. doi: 10.1105/tpc.3.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EPB, Silva CJ, Carolino SMB, Figueiredo JEF, Batista DPO. A soybean binding protein (BiP) homolog is temporally regulated in soybean seeds and associates detectably with normal storage proteins in vitro. Braz J Genet. 1996;19:306–312. [Google Scholar]
- Fontes MA, Otoni WC, Carolino SMB, Brommonschenkel SH, Fontes EPB, Fári M, Louro RP. Hyperhydricity in pepper plants regenerated in vitro: involvement of BiP (binding protein) and ultrastructural aspects. Plant Cell Rep. 1999;19:81–87. doi: 10.1007/s002990050714. [DOI] [PubMed] [Google Scholar]
- Galili G, Sengupta-Gopalam C, Ceriotti A. The endoplasmic reticulum of plant cells and its role in protein maturation and biogenesis of oil bodies. Plant Mol Biol. 1998;38:1–29. [PubMed] [Google Scholar]
- Giannopolitis CN, Ries SK. Superoxide dismutase: I. Occurrence in higher plants. Plant Physiol. 1977;59:309–314. doi: 10.1104/pp.59.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillinkin JW, Zhang F, Coleman CE, Bass HW, Larkins BA, Boston RS. A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane. Plant Physiol. 1997;114:345–352. doi: 10.1104/pp.114.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomer CJ, Ferrario A, Rucker N, Wong S, Lee AS. Glucose regulated protein induction and cellular resistance to oxidative stress mediated by porphyrin photosensitization. Cancer Res. 1991;15:6574–6579. [PubMed] [Google Scholar]
- Görg A, Postel W, Günther S. Two-dimensional electrophoresis: the current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1988;9:531–546. doi: 10.1002/elps.1150090913. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Hill HD, Straka JG. Protein determination using bicinchonic acid. Anal Biochem. 1988;170:203–208. doi: 10.1016/0003-2697(88)90109-1. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Herman EM. Binding-protein expression is subject to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta. 1995;195:611–621. doi: 10.1007/BF00195722. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Köhler K. Cell elongation, turgor and osmotic pressure in developing sunflower hypocotyls. J Exp Bot. 1994;45:591–595. [Google Scholar]
- Laemmli UK. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laitusis AL, Brostrom MA, Brostrom CO. The dynamic role of GRP78/BiP in the coordination of mRNA translation with protein processing. J Biol Chem. 1999;274:486–493. doi: 10.1074/jbc.274.1.486. [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N, Jelitto-Van Dooren EPWM, Crofts AJ, Denecke J. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell. 1999;11:459–469. doi: 10.1105/tpc.11.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS. Mammalian stress response: induction of the glucose-regulated protein family. Curr Opin Cell Biol. 1992;4:267–273. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- Li LJ, Li XA, Ferrario A, Rucker N, Liu ES, Wong S, Gomer CJ, Lee AS. Establishment of a Chinese hamster ovary cell line that expresses grp78 antisense transcripts and suppresses A23187 induction of both GRP78 and GRP94. J Cell Physiol. 1992;153:575–582. doi: 10.1002/jcp.1041530319. [DOI] [PubMed] [Google Scholar]
- Li XA, Lee AS. Competitive inhibition of a set of endoplasmic reticulum protein genes (GRP78, GRP94 and Erp72) retards cell growth and lowers viability after ionophore treatment. Mol Cell Biol. 1991;11:3446–3453. doi: 10.1128/mcb.11.7.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lièvremont J-P, Rizzuto R, Hendeershot L, Meldolesi J. BiP, a major chaperone protein of the endoplasmic reticulum lumen, plays a direct and important role in the storage of the rapidly exchanging pool of Ca2+ J Biol Chem. 1997;272:30873–30879. doi: 10.1074/jbc.272.49.30873. [DOI] [PubMed] [Google Scholar]
- Little E, Lee AS. Generation of a mammalian cell line deficient in glucose-regulated protein stress induction through targeted ribozyme driven by a stress-inducible promoter. J Biol Chem. 1995;270:9526–9534. [PubMed] [Google Scholar]
- Liu H, Bowes RC, III, van de Water B, Sillence C, Nagelkerke JF, Stevens JL. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J Biol Chem. 1997;272:21751–21759. doi: 10.1074/jbc.272.35.21751. [DOI] [PubMed] [Google Scholar]
- Liu H, Miller E, van de Water B, Steven JL. Endoplasmic reticulum stress proteins block oxidant-induced Ca2+ increases and cell death. J Biol Chem. 1998;273:12858–12862. doi: 10.1074/jbc.273.21.12858. [DOI] [PubMed] [Google Scholar]
- Ludlow MM, Chu ACP, Clements RJ, Kerslake RG. Adaptation of species of Centrosema to water stress. Aust J Plant Physiol. 1983;10:119–130. [Google Scholar]
- McCormick TS, McColl KS, Distelhorst CM. Mouse lymphoma cells destined to undergo apoptosis in response of thapsigargin fail to generate a calcium-mediated grp78/grp98 stress response. J Biol Chem. 1997;272:6087–6092. doi: 10.1074/jbc.272.9.6087. [DOI] [PubMed] [Google Scholar]
- Meyer RF, Boyer JS. Sensitivity of cell division and cell elongation to water potentials in soybean hypocotyls. Planta. 1972;108:77–87. doi: 10.1007/BF00386508. [DOI] [PubMed] [Google Scholar]
- Meyer RF, Boyer JS. Osmoregulation, soluble distribution and growth in soybean seedlings having low water potentials. Planta. 1981;151:482–489. doi: 10.1007/BF00386543. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething M-J, Sambrook JF. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K, Sant A, Khono K, Normington K, Gething M-J, Sambrook JF. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Dorner AJ, Edwards CA, Hendershot LM, Kaufman J. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J Biol Chem. 1997;272:4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Pedra JHF, Delú-Filho N, Pirovani CP, Contim LAS, Dewey RE, Otoni WC, Fontes EPB. Antisense and sense expression of a sucrose binding protein homologue gene from soybean in transgenic tobacco affects plant growth and carbohydrate partitioning in leaves. Plant Sci. 2000;152:87–98. [Google Scholar]
- Pedrazzini E, Giovinazzo G, Bollini R, Ceriotti A, Vitale A. Binding of BiP to an assembly-defective protein in plant cells. Plant J. 1994;5:103–110. [Google Scholar]
- Riccardi F, Gazeau P, Vienne D, Zivy M. Protein changes in response to progressive water deficit in maize. Plant Physiol. 1998;117:1253–1263. doi: 10.1104/pp.117.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. Kar2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Roy B, Li WW, Lee AS. Calcium-sensitive tanscriptional activation of the proximal CCAAT regulatory element of the grp78/BiP promoter by human nuclear factor CBF/NF-Y. J Biol Chem. 1996;271:28995–29002. doi: 10.1074/jbc.271.46.28995. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Slavik N. Brief review of methods used to determine the soil water content. In: Slavik N, editor. Methods of Studying Plant Water Relations. New York: Springer Verlag; 1974. pp. 160–163. [Google Scholar]
- Sugawara S, Takeda K, Lee AS, Dennert G. Suppression of stress protein GRP78 induction in tumor B/C10ME eliminates resistance to cell mediated cytotoxicity. Cancer Res. 1993;53:6001–6005. [PubMed] [Google Scholar]
- Thompson S, Latham JAE, Graham AT. A simple, reproducible and cheap batch method for the analysis of serum glycoproteins using Sepharose-coupled lectins and silver staining. Clin Chem Acta. 1987;167:217–223. doi: 10.1016/0009-8981(87)90374-3. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur HP, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;19:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]