Abstract
Although in most plant species no more than two annexin genes have been reported to date, seven annexin homologs have been identified in Arabidopsis, Annexin Arabidopsis 1–7 (AnnAt1–AnnAt7). This establishes that annexins can be a diverse, multigene protein family in a single plant species. Here we compare and analyze these seven annexin gene sequences and present the in situ RNA localization patterns of two of these genes, AnnAt1 and AnnAt2, during different stages of Arabidopsis development. Sequence analysis of AnnAt1–AnnAt7 reveals that they contain the characteristic four structural repeats including the more highly conserved 17-amino acid endonexin fold region found in vertebrate annexins. Alignment comparisons show that there are differences within the repeat regions that may have functional importance. To assess the relative level of expression in various tissues, reverse transcription-PCR was carried out using gene-specific primers for each of the Arabidopsis annexin genes. In addition, northern blot analysis using gene-specific probes indicates differences in AnnAt1 and AnnAt2 expression levels in different tissues. AnnAt1 is expressed in all tissues examined and is most abundant in stems, whereas AnnAt2 is expressed mainly in root tissue and to a lesser extent in stems and flowers. In situ RNA localization demonstrates that these two annexin genes display developmentally regulated tissue-specific and cell-specific expression patterns. These patterns are both distinct and overlapping. The developmental expression patterns for both annexins provide further support for the hypothesis that annexins are involved in the Golgi-mediated secretion of polysaccharides.
Annexins are a diverse, multigene family of calcium-dependent, membrane-binding proteins that serve as targets for Ca2+ in most eukaryotic cells. Annexins structurally all have a 60- to 70-amino acid motif that is repeated four to eight times. Annexins have been studied extensively in animal cells and appear to be multifunctional and play a role in essential cellular processes such as membrane trafficking, ion transport, mitotic signaling, cytoskeleton rearrangement, and DNA replication (for review, see Gerke and Moss, 1997; Seaton and Dedman, 1998). Plant annexins share the basic property of Ca2+-dependent membrane binding and are structurally similar to their animal counterparts (for review, see Clark and Roux, 1995; Delmer and Potikha, 1997; Clark et al., 2001). Like animal annexins, plant annexins have also been shown to undergo a cellular redistribution in response to certain stimuli (Thonat et al., 1997; Clark et al., 2000). There is general evidence linking animal annexins with regulated secretion, and recently annexin XIIIb has been demonstrated to directly participate in apical secretion in kidney epithelial cells (Lafont et al., 1998). In plant cells, annexins have been immunolocalized primarily at the cell periphery of highly secretory cell types including developing vascular tissue, epidermal cells, and outer rootcap cells and in the tip region of cells exhibiting polar growth (Blackbourn et al., 1992; Clark et al., 1992, 1994, 1995). Based on these localization results it has been hypothesized that annexins play a role in the Golgi-mediated secretion of newly synthesized plasma membrane and wall materials in plant cells, but other functions have been postulated for them in plants as well.
Thus far, Southern-blot analysis in Arabidopsis (Gidrol et al., 1996) and other plant species (Battey et al., 1996; Proust et al., 1996, 1999) have all indicated that the annexin gene family is relatively simple in plants. Although annexins in plants appear to be a smaller, less diverse gene family than in vertebrates, there are at least two different annexins in most plant species tested, and current sequence data indicate that Arabidopsis has seven distinct annexin genes, Annexin Arabidopsis1–7 (AnnAt1–AnnAt7; Clark and Roux, 1999; Clark et al., 1999).
Interest in plant annexins is growing, and recent data linking annexins to several physiological processes in plants suggest that they are involved in more cellular processes than just Golgi-mediated secretion and are likely to be functionally diverse. Other functions for plant annexins are suggested by evidence of inherent enzyme activity and association with specific cellular functions. For example, nucleotide phosphodiesterase activity has been found in corn, tomato, bell pepper, and cotton fiber annexins (McClung et al., 1993; Calvert et al., 1996; Lim et al., 1998; Shin and Brown, 1999; Hofmann et al., 2000). Another enzyme activity attributed to plant annexins is peroxidase (Gidrol et al., 1996). A celery annexin is vacuolar specific, associated with cell expansion, and may be involved in vacuole biogenesis (Seals and Randall, 1997). In addition, there is a recent report of a spinach annexin which binds to the outer surface of the chloroplast in a Ca2+-dependent manner (Seigneurin-Berny et al., 2000). There is also an annexin in Medicago trunculata, AnnMt1, which is up-regulated by Nod factors and may play a role in nodulation signaling (Niebel et al., 1998). Annexins have also been documented in plant nuclei where they may participate in DNA replication (Clark et al., 1998; Kovacs et al., 1998).
Crystallography studies have indicated that certain animal annexins have a hydrophilic pore, which could act as a channel for Ca2+ ions. Furthermore, Ca2+ channel activity has been demonstrated for some of these annexins in vitro. Structural similarities between Ca2+ channels in the algae Nitellopsis obtusa and those formed by animal annexins have previously been discussed (Ternovsky and Berestovsky, 1998). The first three-dimensional structure of a plant annexin was determined recently by x-ray diffraction of a crystallized bell pepper annexin (Hofmann et al., 2000). This structure shows the presence of the typical annexin fold, but also indicates differences in certain domains of the membrane-binding loops. This bell pepper annexin was also shown to have Ca2+ channel activity in vitro and this activity was higher than any of the animal annexins tested (Hofmann et al., 2000). Another recent paper describes a wheat annexin that accumulates in the plasma membrane in response to cold treatment and may be acting as a Ca2+ channel (Breton et al., 2000). The authors show that this annexin protein associates with the plasma membrane in a Ca2+-independent manner and acts like an intrinsic membrane protein. Thus, another intrinsic activity that may be important functionally for plant annexins is Ca2+ channel activity.
Plant annexin gene expression is influenced by environmental and developmental signals. For example, annexin mRNA levels are up-regulated in response to stress and abscisic acid in alfalfa (Kovacs et al., 1998). During fruit ripening in strawberry and bell pepper there is an annexin mRNA that is also up-regulated (Wilkinson et al., 1995; Proust et al., 1996). There are two tobacco annexins that are cell cycle regulated and expressed mainly in dividing tissues (Proust et al., 1999). Two previous studies have employed in situ hybridization techniques to examine the expression pattern of annexin in plant cells. In situ hybridization experiments have indicated that the maize p35 annexin is expressed in rootcap cells and differentiating vascular tissue in roots (Carroll et al., 1998), and AnnMt1 is expressed in root nodules early in the zone of infection by Rhizobium meliloti (Niebel et al., 1998).
To begin understanding the diverse roles of different annexins, we have confirmed the presence of seven different annexin cDNAs in Arabidopsis and have studied the expression patterns of all seven of these annexins at the tissue level using reverse transcriptase (RT)-PCR. We have also used northern-blot analysis and RNA in situ hybridization to document the expression pattern of two Arabidopsis annexins at the cellular level. Here, we report the results of these studies, and we provide further insight into the potential diversity in annexin function by describing and discussing a comparative sequence alignment of the Arabidopsis annexin gene family.
RESULTS
Isolation and Sequence Analyses of Annexin cDNAs
We have previously reported the isolation of cDNAs encoding four different Arabidopsis annexins, AnnAt1–AnnAt4 (Clark and Roux, 1999; Clark et al., 1999). To complete the molecular study of Arabidopsis annexins, we have isolated partial cDNAs corresponding to three more annexins, AnnAt5–AnnAt7, and have analyzed the main predicted properties of all seven of the annexin cDNA sequences, with special attention to some potentially important motif differences between AnnAt1 and AnnAt2, the two annexins used for the in situ RNA localization study.
Overall, the primary sequences for AnnAt1–AnnAt7 are fairly divergent from each other. All seven genes encode proteins with a predicted molecular mass of approximately 36 kD; however, the predicted pIs of these proteins are very divergent, ranging between 5.0 and 9.5. When the deduced amino acid sequences of AnnAt1 and AnnAt2 are compared, they show 64% identity and 72% similarity with no gaps. The cDNA sequences of AnnAt6 and AnnAt7 are both very similar to AnnAt2, showing 76% to 80% identity at the amino acid level and 78% to 80% identity at the nucleotide level in the open reading frame. These two annexins are also the two most closely related annexins because they are 86% identical at the nucleotide level and 83% identical at the amino acid level. The other three cDNAs, AnnAt3–AnnAt5, are more divergent and range from 29% to 39% identity and 37% to 51% similarity allowing for several gaps. There are short insertions found within the structural repeats of two of these annexins. AnnAt4 has an 11-amino acid insertion in its first repeat, and AnnAt3 has a five-amino acid insertion (boxed) in its second repeat that closely resembles an SV40-like nuclear localization signal (Fig. 1).
Figure 1.
An alignment of the deduced amino acid sequences of seven Arabidopsis annexin genes obtained using clustal on Macvector. Identical amino acids are indicated in dark gray, conserved amino acids in light gray. A potential nuclear localization domain in AnnAt3 is boxed. The potential heme-binding domain is underlined and an arrow marks the critical His residue in that domain. Various amino acid motifs discussed in the text are aligned above the annexin sequences, and the numbers found below the corresponding amino acid residue are based on the AnnAt1 protein sequence. GenBank accession nos. are as follows: AnnAt1 (AF083913), AnnAt2 (AF083914), AnnAt3 (AF1888362), AnnAt4 (AF188363), AnnAt5 (AY014797), AnnAt6 (AY014798), and AnnAt7 (AY014799).
The main characteristic of all annexins is their ability to bind acidic phospholipids in a Ca2+-dependent manner. The type II Ca2+-binding sites consists of the sequence GXGTD (marked GXGTD), which is found within the endonexin fold and is followed 42 amino acids downstream of the first Gly residue by a Glu (marked E, 68) or an Asp (marked D, 300; Fig. 1). There is also a Trp residue (marked W, 27) found in the first repeat of most plant annexins, which may be important in phospholipid binding (Fig. 1). Examination of the Arabidopsis annexins for the presence of type II Ca2+-binding sites revealed that AnnAt1, AnnAt2, AnnAt6, and AnnAt7 have fairly well conserved Ca2+-binding sites (one amino acid substitution) in their first and fourth repeat (Fig. 1). AnnAt3 has the entire type II Ca2+-binding site conserved in the first repeat as well as the fourth repeat, while AnnAt5 has the entire Ca2+-binding site conserved only in its fourth repeat (Fig. 1). AnnAt4 has only one potential Ca2+-binding loop located in its first repeat (Fig. 1).
In addition to the Ca2+-binding motif found in annexins, there are other sequences that have been suggested to be important for annexin function. One such sequence is the 30-amino acid heme-binding sequence found in plant peroxidases (marked with solid line) containing the critical conserved His residue needed for heme binding (marked by arrow; Fig. 1). Other potentially important motifs, include the GTP-binding motifs (marked GXXXXGKT and DXXG), and an F-actin-binding motif (marked IRI; Fig. 1).
Northern-Blot and RT-PCR Analysis
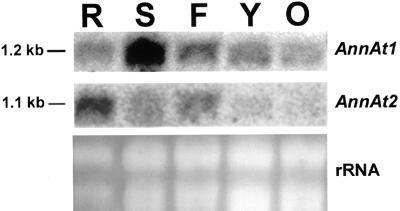
The 3′-untranslated region (UTR) nucleotide sequences for AnnAt1 and AnnAt2 are different in length and are only 45% identical over 120 nucleotides. The 3′ UTRs for these two genes were used as gene-specific probes in northern-blot analysis of various Arabidopsis tissues and revealed distinct expression patterns. AnnAt1 is expressed at varying levels in all tissues examined with the highest level of expression in stem tissue and the lowest level in older leaves. AnnAt2 is expressed mainly in roots and flowers, but there are lower levels of expression in stems and almost no expression in leaves (Fig. 2). The open reading frames of AnnAt1 and AnnAt2 are 67% identical at the nucleotide level and, as expected, are more similar than the 3′ UTRs. However, the open reading frames of AnnAt1 and AnnAt2 also proved to be gene-specific in northern-blot experiments that probed total RNA from Arabidopsis whole plants, and showed hybridization with a 1.2- and 1.1-Kb band, respectively (data not shown).
Figure 2.
Northern-blot analysis of Arabidopsis tissue-specific RNA using gene-specific 3′ UTR probes for AnnAt1 and AnnAt2. R, Roots; S, stems; F, flowers; Y, young leaves; O, old leaves.
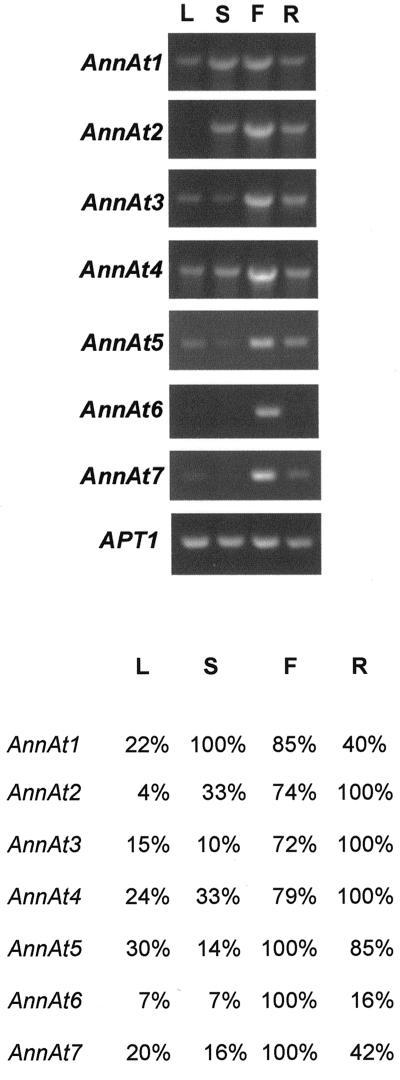
This is the first report to our knowledge describing most of these Arabidopsis annexins; thus, it was informative to analyze their expression patterns. For this purpose, total RNA was extracted from leaves, stems, flowers, and roots and was analyzed by RT-PCR. As shown in Figure 3, this analysis revealed a fairly widespread occurrence for the annexin gene family. Most of the annexin gene transcripts were found at one level or another in all four of the tissues examined. The APT1 gene that codes for adenine phophoribosyltransferase (Moffatt et al., 1994) is constitutively expressed and was used as a control in quantitative RT-PCR (Arenas-Huertero et al., 2000). The RT-PCR results obtained for the AnnAt1 and AnnAt2 transcripts provide further support for the northern-blot analysis of their expression pattern (Fig. 2). The expression profiles for AnnAt3–5 show expression in all four tissues with the highest level of transcript found in flower and root tissue. AnnAt6 and AnnAt7, two closely related annexins, appear to be expressed primarily in flowers.
Figure 3.
RT-PCR analysis of the expression profiles of seven Arabidopsis annexins. Total RNA (2 μg) from leaf (L), stem (S), flower (F), and root (R) tissue was used to synthesize cDNA. A fraction (1/30) of the synthesized cDNA was used to amplify and quantitate AnnAt1–7 (accession nos. AF083913, AF083914, AF1888362, AF188363, AY014797, AY014798, and AY014799, respectively) gene transcripts. The sizes of the annexin PCR products are 954; 954; 966; 960; 951; 1,003; and 1,005 bp, respectively. The RT-PCR product of the APT1 gene (accession no. Y07681; 478 bp) was used as an internal control. A, Ethidium bromide-stained RT-PCR products separated in 1% (w/v) agarose gel. B, A chart depicting the relative amount of each annexin gene transcript. Optical densitometry was performed for each PCR product and normalized against the optical density obtained for the APT1 gene transcript for each tissue type. For each gene transcript, the tissue with the highest normalized optical density was designated 100% and the normalized optical densities for the remaining three tissues were expressed as a percentage.
In Situ Hybridization Analysis
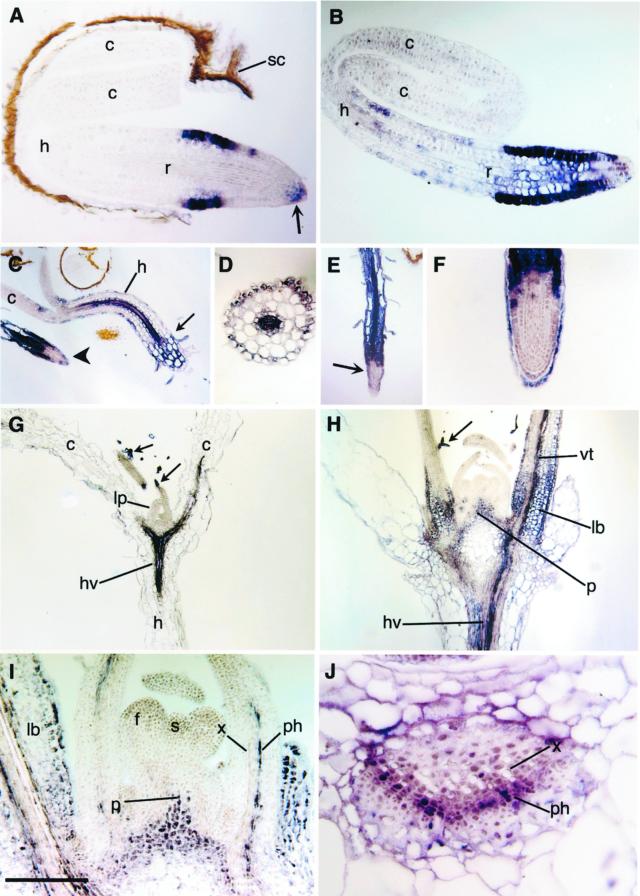
The open reading frames for AnnAt1 and AnnAt2 served as templates for the probes used in the in situ hybridization experiments. As controls, AnnAt1 and AnnAt2 sense probes were routinely used in parallel experiments and gave no signal (data not shown). Using the antisense annexin probes, we followed cell-specific expression in a developmental sequence from germinating seedlings to the transition between vegetative to reproductive growth. As the seedling emerges from the seed coat, AnnAt1 expression is limited to epidermal cells of the root in the zone of cell elongation, to the distal rootcap, and to a few adjacent cells of the root apex (Fig. 4A). As germination continues, expression persists in the same cells, but expands to also include interior cells of the root, as shown in Figure 4B. With further growth, cells in the hypocotyl begin to express AnnAt1. This expression is most noticeable in the vascular tissue and in cells of the epidermis (Fig. 4C). In roots at this stage, expression is seen throughout the interior cells except at the tip (Fig. 4C; see also Fig. 4, E and F). A cross-section confirms the staining pattern seen in the hypocotyl in Figure 4C and also shows low levels of staining in the innermost layer of the cortex (endodermis) (Fig. 4D). The staining pattern seen in the root in Figure 4C remains the same as the root elongates; expression at the root tip is restricted to the rootcap cells (Fig. 4, E and F). Root hairs also show expression of AnnAt1 (Fig. 4, C and E).
Figure 4.
AnnAt1 expression. A, Early expression in the epidermis in the zone of elongation in a root (r) tip, and in the root cap (arrow) of the germinating seedling; cotyledon (c), hypocotyl (h), and seedcoat (sc). B, Later expression includes internal cells of the root, and hypocotyl (h) epidermal cells. C, Expression in the seedling hypocotyl (h) occurs in the epidermis and vascular tissue. A small arrow shows the hypocotyl-root junction, and the large arrow indicates the root tip of another seedling. D, Cross section of a hypocotyl showing staining in the epidermis and vascular tissue. E, Root from a germinating seedling showing expression throughout the root base, including the hair cells, and excluded from the center of the root tip (arrow). F, Close-up of a root tip showing expression restricted to the epidermis of the root tip and outer rootcap cells. G, Longitudinal section through d 7 seedling showing expression in the trichomes (arrows), the hypocotyl vasculature (hv), and cotyledon (c) vasculature; lp, leaf primordia. H, Longitudinal section through d 14 plant showing expression in trichomes (arrow), pith meristem (p), leaf bases (lb), hypocotyl vasculature (hv), and leaf vascular tissue (vt). I, Close-up of d 14 plant showing vascular expression occurs in the phloem (ph) and not the xylem (x); floral meristem (f), shoot meristem (s), and pith meristem (p). J, Close-up of a cross section through a leaf base showing expression in the phloem (ph) and not the xylem (x). Scale bar lower left: A, 60 μm; B, 75 μm; C and E, 500 μm; D, 140 μm; F, 130 μm; G and H, 400 μm; I, 180 μm; and J, 50 μm.
By d 7 expression can be observed in the initiating trichomes (arrows) on leaf primordia and in the vasculature in the hypocotyl and cotyledon (Fig. 4G). At d 14 with the transition to reproductive growth, expression expands to include the vasculature, epidermis, and some parenchyma cells in intermediate aged leaves, mesophyll cells at the leaf bases, and the pith meristem (Fig. 4, H and I). Higher magnification shows that the vascular expression is primarily in the phloem, in what appear to be phloem and phloem-associated cells (Fig. 4, I and J).
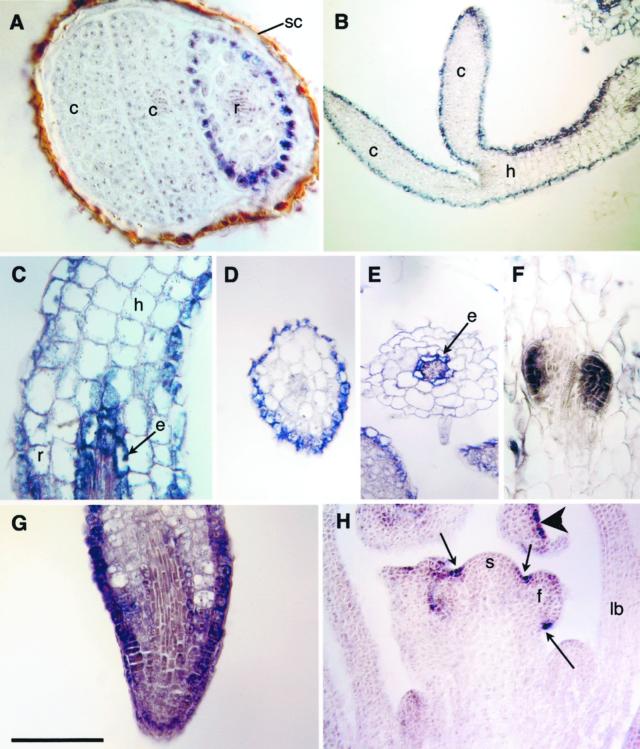
AnnAt2 expression is first observed in root epidermal cells in germinating seedlings (Fig. 5A) and subsequently expands to include the epidermal cells of the hypocotyl and cotyledons (Fig. 5B). By d 4, expression in the root expands to include cells of the endodermis near the root-hypocotyl junction (Fig. 5C). A cross-section of the hypocotyl confirms that expression at this stage is restricted to the epidermal cells as suggested by Figure 5, B and C (Fig. 5D). By contrast a cross-section of the root adjacent to the hypocotyl shows staining of the endodermis and no staining of the epidermis (Fig. 5E). AnnAt2 is also expressed throughout initiating lateral roots (Fig. 5F). In the root tip, staining is seen in epidermal cells basipetal to the apex and light staining is seen in some interior cells (Fig. 5G). Expression in the epidermis of the hypocotyl and cotyledons decreases as the seedling matures. By d 14, during the transition from vegetative to reproductive growth, expression of AnnAt2 is restricted to the epidermis in a spotty manner and in creases between the shoot meristem and its lateral primordia (Fig. 5H).
Figure 5.
AnnAt2 expression. A, Expression in the germinating embryo begins in epidermal cells of the root (r); cotyledon (c) and seed coat (sc). B, Seedling showing epidermal expression in the epidermis of the hypocotyl (h) and cotyledons (c). C, Oblique longitudinal section through the hypocotyl (h)-root (r) junction showing expression in the hypocotyl epidermis and the endodermis (e) of the root. D, Cross section through a seedling hypocotyl showing epidermal expression. E, Cross section through seedling root showing expression in the endodermis (e). F, Oblique longitudinal section through a root showing expression in two lateral root primordia. G, Oblique section through a root tip showing enhanced expression in the epidermis. H, Longitudinal section through d 14 plant showing epidermal expression in the creases (arrows) between floral primordia (f) and the shoot meristem (sm). An arrowhead shows patchy epidermal expression in a flower bud; leaf base (lb). Scale bar lower left: A and C, 60 μm; B, 70 μm; D and E, 130 μm; F and G, 100 μm; and H, 200 μm.
DISCUSSION
As a first step toward addressing the question of whether plant annexins are functionally diverse, we have examined the expression patterns and analyzed the deduced amino acid sequences of seven different annexins in Arabidopsis. The annexin gene family in Arabidopsis appears to have seven members, which have been named AnnAt1–AnnAt7 according to the nomenclature for plant annexins suggested by Delmer and Potikha (1997). A survey through the complete Arabidopsis genomic sequence does not reveal any other obvious annexin genes. However, there is a sequence located on chromosome IV that is annotated as a putative annexin (accession no. AL161559) based on its similarity with the N-terminal region of annexin VII from Dictyostelium discoideum. However, this sequence is lacking the characteristic annexin structural repeats and thus the putative gene containing this sequence is unlikely to be a member of the annexin family.
cDNAs almost identical with AnnAt1 have been found and sequenced previously by two other groups (accession nos. U28415 and X99224). Our sequence for AnnAt1 is 99% identical at the nucleotide level with the sequence obtained by Gidrol et al. (1996) (accession no. U28415), and the slight differences in primary nucleotide sequence result in four amino acid changes at the end of the third repeat of the deduced amino acid sequences. It is unclear whether the differences in these three AnnAt1 sequences represent minor variations in the same gene or are due to minor errors in sequencing. AnnAt3 and AnnAt4 genes are located adjacent to each other and are transcribed in divergent orientation. Thus, they could be coregulated by sharing upstream enhancer sequences that are not orientation specific. It is also likely that one of these annexins arose as a result of a gene duplication event.
A consistent difference between animal and plant annexin sequences is that plant annexins appear to typically have a well-conserved endonexin fold in only one of their repeats, usually the first repeat, whereas animal annexins usually have a conserved endonexin fold in three of the repeats. AnnAt3 has two well-conserved endonexin fold regions and is the only Arabidopsis annexin that has the classical Ca2+-binding loop in more than one of its repeats. There are only two other plant annexins thus far that also have conserved endonexin folds in two repeats (Kovacs et al., 1998; Lim et al., 1998). AnnAt4, the most divergent of the Arabidopsis annexins, has a modified sequence within the Ca2+-binding site in its first repeat and is completely divergent in the Ca2+-binding region in the fourth repeat. It remains to be tested whether the modified sequence found in AnnAt4 can serve as a Ca2+-binding site. An annexin sequence that also lacks the classical conserved Ca2+-binding loop has been reported in Giardia lamblia. Instead, this annexin has a modified loop that has been demonstrated to bind phosphatidyl-Ser in a Ca2+-dependent manner (Bauer et al., 1999).
It is worth noting that the Glu (E, 68) or Asp (D, 300) located at the C-terminal region of the Ca2+-binding site is conserved in the first repeat and in the fourth repeat of all seven Arabidopsis annexins except for the fourth repeat of AnnAt4. Lim et al. (1998) tested the Ca2+-dependent phospholipid binding activity of an overexpressed tomato annexin, p35, in which substitutions were made in the C-terminal Asp and Glu residues that help to stabilize the type II Ca2+-binding sites found in the first and fourth repeats. Based on their results, they suggested that repeat IV in plant annexins plays a more critical role in Ca2+-binding. They proposed that binding of Ca2+ to repeat IV may result in a conformational change which exposes a Trp residue (Trp-27) found in repeat I, leading to a favored state in this repeat for Ca2+ and phospholipid binding (Lim et al., 1998). This Trp residue, which is conserved in almost all other plant annexins, can be found in all of the Arabidopsis annexins except AnnAt4 and AnnAt5. In the three-dimensional structure of the bell pepper annexin this Trp residue is shown to be on the surface of the molecule and is suggested to be an important residue for membrane binding and/or annexin aggregation (Hofmann et al., 2000).
The main source of functional diversity in vertebrate annexins is thought to be the N-terminal region of the proteins, which differs in length and sequence and serves as a site for phosphorylation and other posttranslational modifications. Translations of all plant annexin cDNA sequences to date, including all seven Arabidopsis annexins, reveal that they have only very short N-terminal regions preceding the first structural repeat of the proteins. Data from the x-ray structure of the bell pepper annexin show that the short N-terminal domain associates with the protein core and the authors suggest that this domain may be an important regulatory element in plant annexins (Hofmann et al., 2000). However, the short N-terminal domains found in plant annexins do not show the same variation in length and sequence found in mammalian annexins and thus functional differences that exist between various plant annexins may be due to variation found within the structural repeats.
The 70-amino acid repeat regions that are characteristic of the annexin gene family are present in each of the seven Arabidopsis annexins. Within these repeat sequences, the positions with conserved amino acid properties for all annexins (Barton et al., 1991) are most often conserved. However, comparison of the structural repeats from all seven Arabidopsis annexins shows that there are significant sequence differences within the repeat regions. Sequence analysis of AnnAt1–AnnAt7 reveals the presence of different motifs within the repeat regions. These motifs could be responsible for specialized, unique functions for each of these plant annexins. As an example of one of these motifs, AnnAt3 has a potential nuclear localization signal inserted in its second repeat. The existence of nuclear annexins in pea and alfalfa cells has been documented recently (Clark et al., 1998; Kovacs et al., 1998), and it will be of interest to determine if the putative NLS in AnnAt3 is functional; i.e. whether AnnAt3 is a nuclear protein.
AnnAt1 has previously been reported to have inherent peroxidase activity, and it has a sequence in the first repeat that is similar to a heme-binding domain found in plant peroxidases (Gidrol et al., 1996). This potential heme-binding domain is most highly conserved in AnnAt1; however, all of the Arabidopsis annexins show similarity with this heme-binding region and contain the critical conserved His residue needed except for AnnAt3 and AnnAt4. This potential heme-binding domain overlaps the potential Ca2+-binding site. This raises the possibility that Ca2+ could regulate the peroxidase activity observed in AnnAt1 (and which may also be present in the other annexins), and it will be informative to test this possibility experimentally.
Several plant annexins have also been found to have nucleotide phosphatase activity in vitro, although they appear to lack the classical Walker ATPase motif. A recent paper describing this activity in a cotton fiber annexin, AnnGh1, shows that this annexin binds to and hydrolyzes GTP more efficiently than ATP and the GTPase activity is inhibited by Ca2+ (Shin and Brown, 1999). By deletion mapping, GTP binding was demonstrated to be dependent on the presence of the fourth repeat in AnnGh1. There is a tripartite consensus sequence GXXXXGKS(T), DXXG, N(T) KXD which is present in many GTP-binding proteins (Kjeldgaard et al., 1996). In examining the fourth repeat of the cotton annexin we found two sequences similar with two of these GTP-binding motifs, GXXXXGKT and DXXG.
Sequence analysis of the Arabidopsis annexin cDNAs indicates that there are sequences in the fourth repeat of AnnAt1, AnnAt2, and AnnAt7 that are similar to the first two GTP-binding motifs. AnnAt2 and AnnAt7 appear to be the most likely candidates for GTP binding because they both contain a modified GXXXXGLT sequence that has only one amino acid substitution and the conserved DXXG sequence in its fourth repeat. However, AnnAt1 has a much more modified motif, which has only three amino acids between the glycines and an extra Ala residue following the second Gly GXXXGALT and a conserved DXXG sequence in the fourth repeat. There are 32 amino acids between these two motifs, a distance similar to that found in other GTP-binding proteins. The third motif N(T) KXD that cannot be found in either of these annexins is believed to be important for determining guanine nucleotide specificity and is missing in proteins which are able to bind GMP in addition to GDP and GTP (Cheng et al., 1991). These potential GTP-binding motifs overlap with the Ca2+-binding site in this repeat and could explain the inhibition of GTPase activity by Ca2+ ions. It is interesting that GTP was shown to inhibit the annexin/ Ca2+-dependent secretion in maize rootcap cells. Taken together, these results suggest a model in which Ca2+ and GTP are able to affect “regulated” secretion in plant cells directly through their interaction with the fourth repeat of annexin proteins. In animal cells, the membrane fusion activity of annexin VII is regulated by both Ca2+ and GTP (Caohuy et al., 1996).
Annexin V has been shown to bind filamentous actin in a Ca2+-dependent manner in vitro, and it binds to the actin cytoskeleton at the plasma membrane of activated platelets (Tzima et al., 1999). Certain plant annexins can bind F actin in vitro, and a potential F-actin-binding sequence, IRI, has been previously noted in some plant annexin sequences (Lim et al., 1998). This IRI motif is part of a heptapeptide and a nonapeptide sequence shown to be critical for myosin and annexin interaction with F actin, respectively (Eto et al., 1990; Jones et al., 1992). All of the Arabidopsis annexins except for AnnAt3 and AnnAt4 contain an IRI motif, indicating that these annexins are more likely to associate with the actin cytoskeleton. The IRI motif in AnnAt5 overlaps with the Ca2+-binding site and this could have structural implications for Ca2+-annexin-actin interactions for this annexin.
The sequence analyses of these Arabidopsis annexins provide further insight into the potential diversity of this gene family in plants. In comparing the internal repeats, it is apparent that overall these annexins are structurally conserved, but that there are important sequence differences. These sequence differences may be indicative of unique activities and regulatory mechanisms for each annexin. We are now testing whether the various sequence motifs found in these genes are functional motifs by overexpressing and purifying each annexin and then testing them for various activities in vitro.
In general, annexins are relatively abundant proteins in animal and plant cells and have been estimated to be 0.1% of the total extractable protein content of plant cells (Delmer and Potikha, 1997). As of February 2001, AnnAt1 had over 40 entries in the Arabidopsis Expressed Sequence Tag database, compared with only four entries for AnnAt2. Assuming theses numbers reflect the relative abundance of the mRNAs expressed, AnnAt1 appears to be the major annexin in Arabidopsis. The northern-blot results (Fig. 2) clearly indicate that the mRNA for AnnAt1 is certainly more abundant than AnnAt2 overall.
Northern-blot analysis of annexins from various plant species has revealed that most annexins have a fairly widespread expression (Gidrol et al., 1996; Proust et al., 1999), except for the Medicago truncatula annexin, AnnMt1, involved in nodulation signaling, which is expressed almost exclusively in roots (Niebel et al., 1998). In this study, the northern-blot results show expression levels of AnnAt1 and AnnAt2 in various tissues taken from 2.5-week-old plants. Northern-blot analysis reveals that AnnAt1 and AnnAt2 clearly have some overlapping tissue expression, but that there are also major differences in their levels of expression in these tissues. For example, AnnAt1 is most abundant in stems, whereas AnnAt2 is most abundant in roots. Another obvious difference is seen in leaves; AnnAt2 is not expressed, whereas AnnAt1 is expressed especially in younger leaves and to a lesser extent in older leaves. The northern results that we obtained for AnnAt1 are similar to those obtained by Gidrol et al. (1996) for the same annexin, except that our results indicated that the highest level of expression occurs in stems and their results showed the highest level of expression in roots. This discrepancy between their results and ours could be attributable to the difference in the age of the plants used (4-week-old plants versus 2.5-week-old plants). Northern-blot results for AnnAt1 and AnnAt2 were confirmed by RT-PCR. The RT-PCR results also showed that most of the Arabidopsis annexins are not expressed in a tissue-specific manner with the possible exception of AnnAt6, which appears to be expressed mainly in flowers.
The in situ hybridization experiments follow the cellular expression patterns for AnnAt1 and AnnAt2 during different developmental stages. They also address the question of whether these two annexins are expressed in the same cell types in certain tissues. The results show that the expression of both AnnAt1 and AnnAt2 is turned on early in development and is dynamic throughout development, and they reveal both similarities and differences in the tissue-specific expression patterns of these two annexins. Expression pattern differences and similarities are particularly evident in epidermal cells. Both AnnAt1 and AnnAt2 are first expressed in the epidermis of the roots of developing seedlings. At this early stage, AnnAt1 expression appears to be restricted to only the epidermal cells of the elongation zone of the root, whereas AnnAt2 expression is observed in epidermal cells of the root, hypocotyl, and cotyledons. Later in development, expression of AnnAt1 is also found in the epidermis of the hypocotyl and thus may overlap with AnnAt2 expression there.
In roots at a later stage of development, AnnAt1 and AnnAt2 display different expression patterns. AnnAt2 is expressed in endodermal cells near the root-hypocotyl junction, in epidermal cells near the apex, and in initiating lateral roots. In contrast, AnnAt1 is expressed in all cells of the root, except at the tip, where expression is restricted to the rootcap. The in situ results obtained for maize annexin p35 in maize roots (Carroll et al., 1998) is very similar to the expression pattern for AnnAt1 in Arabidopsis roots. Both show expression in the rootcap and in the internal layers of the root. If AnnAt1 is functionally homologous to maize p35, then it is possible that AnnAt1 is involved in the directed secretion of polysaccharides by rootcap cells. Only AnnAt1 is expressed in root hairs, providing evidence that this may be the annexin involved in polar growth of root hair cells.
Expression of AnnAt1 and AnnAt2 occurs in distinct tissues and cell types in more mature shoots. AnnAt1 is primarily expressed in vascular tissues, and ground tissues of the stem and leaf bases, and in developing trichomes. AnnAt2 expression is very restricted and is found only in epidermal cells in the creases between meristems and primordia, possibly for secretion of wall-softening enzymes.
Although northern blots using the AnnAt2 in situ probe showed hybridization specifically with a 1.1-Kb band, it is possible that in certain cases this pattern may actually represent expression of AnnAt 6 and/or AnnAt7 because these two sequences show a high degree of sequence identity with AnnAt2. However, although it is more likely that the expression pattern obtained for AnnAt2 truly represents this annexin, even if some part of this pattern can be attributed to AnnAt6 and/or AnnAt7, the pattern is clearly distinct from the one obtained using the AnnAt1 in situ probe. Thus, the conclusion that different annexins are expressed in different cell types during development is still valid.
Overall the expression data in this study matches well with previous analyses of the distribution of annexin proteins in plants using immunodetection methods. Pea annexins have been immunolocalized in highly secretory cell types, such as epidermal cells, developing vascular cells, and rootcap cells (Clark et al., 1992). All of these cell types at some stage of development also showed strong expression of either AnnAt1 or AnnAt2 in Arabidopsis. In particular, the previously documented immunostaining of annexins in young developing phloem cells (Clark et al., 1992) and in mature sieve elements (Clark et al., 2000) is paralleled by the strong expression of AnnAt1 mRNA in phloem cells and in associated phloem parenchyma cells within the vasculature. Thus, the in situ expression data for these two Arabidopsis annexins provides another line of evidence consistent with the hypothesis that annexins are involved in Golgi-mediated secretion of polysaccharides and other materials in plant cells.
In regard to its potential role in secretion control, annexins have also been previously immunolocalized at the extreme tips of cell types exhibiting polar growth such as pollen tubes and fern rhizoids (Blackbourn et al., 1992; Clark et al., 1995). These are rapidly growing cells with active, directed secretion that requires Ca2+ (Thiel and Battey, 1998). This raises the question of which Arabidopsis annexin(s) are involved in polar growth. In this study, the only result that addresses this question is the observation that AnnAt1 is expressed in root hairs. It will be important to determine if this Arabidopsis annexin is also found in other tip growing cells such as pollen tubes.
In the immunolocalization studies discussed above, annexin typically has a strong cell peripheral localization at the light microscope level and is plasma membrane associated at the electron microscope level. There are reports that annexins are among the major proteins found in isolated plasma membrane fractions of soybeans and Arabidopsis (Shi et al., 1995; Santoni et al., 1998). In their Arabidopsis study, Santoni et al. (1998) showed that AnnAt1 is enriched in leaf plasma membrane preparations, where it appears as two isoforms at 34 and 39 kD, and that the 34-kD isoform appears to behave more like an integral membrane protein because it remains in the membrane fraction after sodium carbonate and detergent treatment. In a recent paper, Breton et al. (2000) describe a wheat annexin that associates with the plasma membrane in a Ca2+-independent manner and also acts like an intrinsic membrane protein when membranes are treated with sodium carbonate followed by proteinase K. The proteome data of Santoni et al. (1998) may be another indication that AnnAt1 is a multifunctional protein. The abundance of the AnnAt1 transcript and its widespread expression pattern also suggest that this annexin could be functionally diverse. The demonstration that AnnAt1 exhibits peroxidase activity and can rescue an oxyR mutant in Escherichia coli certainly supports a function for this annexin in H202 stress responses (Gidrol et al., 1996).
To our knowledge, a comparative study of the cell-specific expression of two different annexins in the same plant species has not been reported previously. Our finding that the expression patterns for these two annexin cDNAs are mainly nonoverlapping suggests distinct tissue-specific functions for each annexin. However, in the case of epidermal cells we observed an overlapping expression pattern during development. The expression of these two annexins in the same cell type at the same time could be just an example of genetic redundancy. However, recent evidence indicates that multigene protein families, such as the plant actin gene family, may co-express members of a gene family that differ in at least one activity, leading to a more robust and buffered response system, termed isovariant dynamics (Meagher et al., 1999). Although as of yet different activities have not been demonstrated for AnnAt1 and AnnAt2, there are several potentially important sequence differences between these two annexins, and so the possibility that the co-expression of these two annexins in epidermal cells may be an example of isovariant dynamics should be considered.
MATERIALS AND METHODS
Plant Material
Plants used for in situ experiments were grown in Sunshine Mix soil, in 22°C growth chambers, under 18-h-light days, and watered with Miracle Grow plant food. Plants used for RT-PCR experiments and northern analyses of the stems, leaves, and flowers were grown in the same conditions except under continuous light. Plants used for northern analyses of the roots were grown on 0.8% (w/v) Murashige and Skoog basal salt mixture agar plates in 22°C growth chambers under continuous light.
Isolation of cDNA Clones Encoding the Annexin cDNAs
The first two Arabidopsis annexin genes, AnnAt1 and AnnAt2, were identified by the sequencing of several annexin-like Arabidopsis expressed sequence tags (GenBank accession nos. AF083913 and AF083914; Newman et al., 1994; Clark and Roux, 1999). A genomic version of AnnAt1 has been sequenced from a chromosome I bacterial artificial chromosome (AC021198) and matches our AnnAt1 sequence. The genomic version of AnnAt2 has been found more recently on chromosome V (GenBank accession no. AB019236). Five other putative Arabidopsis annexin genes were first discovered through genome sequencing efforts. The third and fourth Arabidopsis annexins were found by sequencing a chromosome II bacterial artificial chromosome (GenBank accession no. AC005499). We have confirmed that these putative annexin genes are expressed in Arabidopsis plants by using RT-PCR techniques to obtain the corresponding cDNAs, AnnAt3 and AnnAt4, from leaf tissue (GenBank accession nos. AF188362 and AF188363; Clark et al., 1999).
Another putative annexin gene was found and sequenced on chromosome 1 (GenBank accession no. AC012563) and we have also isolated the partial cDNA for this annexin, AnnAt5, from stem tissue by using RT-PCR techniques (GenBank accession no. AY014797). The genome sequencing effort more recently has yielded two more novel Arabidopsis annexin genes which are aligned next to each other on chromosome V (GenBank accession nos. AL356332 and AL360334) and we have isolated the partial cDNAs for these two annexins from leaf tissue using RT-PCR techniques (GenBank accession nos. AY014798 and AY014799).
RT-PCR
TRIzol reagent (Gibco, Gaithersburg, MD) was used to extract mRNA from whole plants, leaves, stems, flowers, and roots of 2.5-week-old Arabidopsis using a modified procedure as recommended by the manufacturer for purification of plant RNA. For RT, RNA samples (2 μg) were treated with Dnase I (amp grade, Gibco) for 15 min, annealed to the primer 5′-TTCTAGAATTCAGCGGCCGC(T)30-3′ for 10 min, and then the cDNAs were synthesized using 200 units of RT (Superscript, Gibco) in buffer containing 10 mm dithiothreitol and 1.25 mm dNTPs. The genomic sequences for AnnAt5–AnnAt7 were used to design primers for RT-PCR. For obtaining the partial cDNA for AnnAt5, gene-specific primers were designed based on the N- and C-terminal sequences of the open reading frame. For obtaining the partial cDNAs for AnnAt6 and AnnAt7, gene-specific primers were designed based on UTR sequences. The primers used and the corresponding size products are as follows: AnnAt5, 5′-ATGGCAACAATGAAAATACCA-3′ and 5′-TCAAACGTTGGGGCCTAAAAGAGA-3′, 951bp; AnnAt6, 5′-GAGAAATATTCAGT GGTCGGAGA-3′ and 5′GACTATGAAACGATGATGTTGTT-3′, 1003 bp; and AnnAt7 5′-CATACAGAAATTTCACTTGTTCG-3′ and 5′-TCTTAAACAAAACTTGCAAATGT-3′, 1,005 bp. The reaction mixture contained 1 μL of first-strand cDNA, 0.25 mm dNTPs, 1.5 mm MgCl2, 0.2 μm primers, and 1.25 units of Taq polymerase (Gibco) in a total volume of 50 μL. The PCR cycling conditions were as follows: an initial denaturation step at 94°C for 6 min, 30 cycles (94°C for 1 min, 55°C for 30 s, and 72°C for 1 min), and a final elongation step at 72°C for 10 min. For quantitative RT-PCR, linearity for each amplification was confirmed and the APT1 (accession no. Y07681) transcript was used as an internal control. The same primers used for obtaining the partial cDNAs for AnnAt5, AnnAt6, and AnnAt7 were used for quantitiative RT-PCR. The other primers used and the corresponding size products are as follows: AnnAt1, 5′-ATGGCGACTCTTAAGGTTTCTGAT-3′ and 5′-TTAAGCATCATCTTCACCGAGAAGTGC-3′, 954 bp; AnnAt2, 5′-ATGGCGTCTCTCAAAGTC CCAAGC-3′ and 5′-TCAAGCATCGCCATGTCCGAGAAGAGC-3′, 954 bp; AnnAt3, 5′-ATGGCCACCATTAGAGTACCAAAC-3′ and 5′-TCAGATTTTGGATCCAAGTAAGGTG AT-3′, 966 bp; AnnAt4, 5′-ATGGCTCTTCCTCTCGAGCTCGAA-3′ and 5′-TCAATCG GATTTGGAGAGAAGTGTGAG-3′, 960 bp; and APT1, 5′TCCCAGAATCGCTAAGATT GCC-3′ and 5′-CCTTTCCCTTAAGCTCTG-3′, 478 bp.
Cloning and Sequencing
The cDNA bands of the expected sizes were gel purified (Wizard PCR preps DNA purification system, Promega, Madison, WI) and subcloned into pCR2.1 (Invitrogen, Carlsbad, CA). All sequencing was conducted on both strands of cDNA and was performed at the Sequencing Center of the University of Texas (Austin) using the dRhodamine Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA) for an ABIPrism 377. Sequence alignments and homology analyses were performed using the PileUp and Gap programs, respectively, on SeqWeb (Version 1.1, Wisconsin package Version 10.0, Genetics Computer Group, Madison, WI). The deduced amino acid sequences for the annexin cDNAs were also aligned using Mac Vector (Version 6.5.3, Oxford Molecular Ltd., Oxford).
Northern-Blot Analysis
Ecotype Wassilewskija (WsO) tissue was harvested and frozen immediately in liquid nitrogen. Total RNA was isolated from 2.5-week-old Arabidopsis plants using the standard TRIzol (Gibco) RNA extraction procedure (extra polysaccharide precipitation steps were performed as described in the protocol). Tissue samples included: roots, stems, flowers and a small amount of peduncle, young leaves, and old leaves. A total of 20 μg of RNA was run on a 1.4% (w/v) denaturing formaldehyde gel. The RNA was subsequently transferred via standard northern-blot procedure to a Bio-Rad Zetaprobe membrane and UV cross-linked. Probes were generated using PCR in the presence of a 1:10 ratio of hot (P32):cold dCTP. Following PCR, probes were cleaned from unincorporated nucleotides using a sephadex G-50 column. Probe AnnAt2 was hybridized to the membrane at 55°C for 24 h in hybridization solution (1 mm EDTA, 0.5 m Na2HPO4, 7% [w/v] SDS, and 1× Denhardt's solution). Hybridization of probe AnnAt1 was done as above but at 60°C. High stringency washes were carried out at the corresponding hybridization temperatures using 1 mm EDTA, 40 mm Na2HPO4, 5% (w/v) SDS, 1 mm EDTA, 40 mm Na2HPO4, and 1% (w/v) SDS wash solutions as stated in Bio-Rad Zetaprobe Instruction manual. Membranes were subsequently exposed to a Molecular Dynamics Phosphorimager screen for 4 d before development.
In Situ Hybridizations
Ecotype Columbia (ColO) tissue was fixed in FAA (3.7% [w/v] formaldehyde, 5% [w/v] acetic acid, and 50% [w/v] ethanol) under vacuum for 15 min, followed by fresh FAA for 2 h. Fixed tissue was dehydrated through ethanol and Histoclear, embedded in Paraplast Plus (Fisher, Pittsburgh), sectioned at 8 μm, and affixed to Probe On Plus slides (Fisher). Slides were dewaxed in Histoclear (National Diagnostics) twice for 10 min, and rehydrated through an ethanol series. Slides were treated for 20 min with 0.2 m HCl, followed by 5 min distilled, deionized water; 5 min 5× SSC; 5 min distilled, deionized water; 20 min 1 μg mL−1 proteinase K (Roche) in 0.1 m Tris/0.05 m EDTA (pH 8.0); 2 min phosphate-buffered saline (PBS); 10 min 4% (w/v) formaldehyde in PBS; twice for 5 min in PBS; and dehydrated through an ethanol series, and dried under vacuum.
Digoxigenin-labeled antisense RNA probes of the open reading frames of AnnAt1 and AnnAt2 were transcribed from pBluescript subclones of AF083913 (pAnnAt1) and AF083914 (pAnnAt2) by linearizing with BamHI and SacI, and transcribing with T3 and T7 RNA polymerase respectively, and hybridized overnight at 55°C to slides in 6× SSC, 3% (w/v) SDS, 50% (w/v) formamide, and 100 μg mL−1 tRNA at empirically determined probe concentrations. Digoxigenin sense probes (pAnnAt1 NotI/T7 RNA polymerase and pAnnAt2 SalI/T3 RNA polymerase) were also constructed and used in parallel experiments. Alkaline phosphatase detection was performed according to the method of Ferrandiz et al. (2000) and according to the manufacturer's (Boehringer Mannheim, Indianapolis) directions.
ACKNOWLEDGMENTS
We thank Marty Yanofsky for help with in situ hybridizations. We thank Marianne Dauwalder for helpful discussions during the preparation of this manuscript. We also thank Iris Steinbrunner, Stuart Reichler, and Mari Salmi for assistance with this project.
Footnotes
This work was supported by the National Aeronautics and Space Administration (grant no. NAGW 1519 to S.J.R.). A.S. was funded by a Department of Energy fellowship from the Life Sciences Research Foundation.
LITERATURE CITED
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Barton GJ, Newman RH, Freemont PS, Crumpton MJ. Amino acid sequence analysis of the annexin super-gene family of proteins. Eur J Biochem. 1991;198:749–760. doi: 10.1111/j.1432-1033.1991.tb16076.x. [DOI] [PubMed] [Google Scholar]
- Battey NH, James NC, Greenland AJ. cDNA isolation and gene expression of the maize annexins p33 and p35. Plant Physiol. 1996;112:1391–1396. doi: 10.1104/pp.112.3.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer B, Engelbrecht S, Bakker-Grunwald T, Scholze H. Functional identification of α1-giardin as an annexin of Giardia lamblia. FEMS Microb Lett. 1999;173:147–153. doi: 10.1111/j.1574-6968.1999.tb13496.x. [DOI] [PubMed] [Google Scholar]
- Blackbourn HD, Barker PJ, Huskisson NS, Battey NH. Properties and partial sequence of plant annexins. Plant Physiol. 1992;99:864–871. doi: 10.1104/pp.99.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G, Vasquez-Tello A, Danyluk J, Sarhan F. Two novel intrinsic annexins accumulate in wheat membranes in response to low temperature. Plant Cell Physiol. 2000;41:177–184. doi: 10.1093/pcp/41.2.177. [DOI] [PubMed] [Google Scholar]
- Calvert MC, Gant SJ, Bowles DJ. Tomato annexins p34 and p35 bind to F-actin and display nucleotide phosphodiesterase activity inhibited by phospholipid binding. Plant Cell. 1996;8:333–342. doi: 10.1105/tpc.8.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caohuy H, Srivastava M, Pollard HB. Membrane fusion protein synexin (annexin VII) as a Ca2+/GTP sensor in exocytotic secretion. Proc Natl Acad Sci USA. 1996;93:10797–10802. doi: 10.1073/pnas.93.20.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AD, Moyen C, Van Kesteren P, Tooke F, Battey NH, Brownlee C. Ca2+, annexins, and GTP modulate exocytosis from maize root cap protoplasts. Plant Cell. 1998;10:1267–1276. doi: 10.1105/tpc.10.8.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y-SE, Patterson CE, Staeheli P. Interferon-induced guanylate-binding proteins lack an N(T) KXD consensus motif and bind GMP in addition to GDP and GTP. Mol Cell Biol. 1991;11:4717–4725. doi: 10.1128/mcb.11.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GB, Dauwalder M, Roux SJ. Purification and immunolocalization of annexin-like protein in pea seedlings. Planta. 1992;187:1–9. doi: 10.1007/BF00201617. [DOI] [PubMed] [Google Scholar]
- Clark GB, Dauwalder M, Roux SJ. Immunolocalization of an annexin-like protein in corn. Adv Space Res. 1994;14:341–346. doi: 10.1016/0273-1177(94)90421-9. [DOI] [PubMed] [Google Scholar]
- Clark GB, Rafati DS, Bolton RJ, Dauwalder M, Roux SJ. Redistribution of annexin in gravistimulated pea plumules. Plant Physiol Biochem. 2000;38:937–947. doi: 10.1016/s0981-9428(00)01206-7. [DOI] [PubMed] [Google Scholar]
- Clark GB, Rives AE, Beauchamp LM, Roux SJ. Isolation and characterization of two novel Arabidopsis annexin cDNAs (accession nos. AF188362 and AF188363) Plant Physiol. 1999;121:1054. [Google Scholar]
- Clark GB, Roux SJ. Annexins of plant cells. Plant Physiol. 1995;109:1133–1139. doi: 10.1104/pp.109.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GB, Dauwalder M, Roux SJ. Immunological and biochemical evidence for nuclear localization of annexin in peas. Plant Physiol Biochem. 1998;36:621–627. doi: 10.1016/s0981-9428(98)80010-7. [DOI] [PubMed] [Google Scholar]
- Clark GB, Roux SJ. Isolation and characterization of two different Arabidopsis annexin cDNAs (accession Nos. AF083913 and AF083914) Plant Physiol. 1999;120:340. [Google Scholar]
- Clark GB, Thompson G, Roux SJ. Signal transduction mechanisms in plants: an overview. Curr Sci. 2001;80:170–177. [PubMed] [Google Scholar]
- Clark GB, Turnwald S, Tirlapur UK, Haas CJ, von der Mark K, Roux SJ, Scheuerlein R. Polar distribution of annexin-like proteins during phytochrome-mediated initiation and growth of rhizoids in the ferns Dryopteris and Anemia. Planta. 1995;197:376–384. doi: 10.1007/BF00202660. [DOI] [PubMed] [Google Scholar]
- Delmer DP, Potikha TS. Structures and functions of annexins in plants. Cell Mol Life Sci. 1997;53:546–553. doi: 10.1007/s000180050070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto M, Suzuki R, Morita F, Kuwayama H, Nishi N, Tokura S. Roles of the amino acid side chains in the actin-binding S-site of myosin heavy chain. J Biochem. 1990;108:499–504. doi: 10.1093/oxfordjournals.jbchem.a123228. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky M. Redundant regulation of meristem identity and plant architecture by FRUITFULL APETALA1, and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Gerke V, Moss SE. Annexins and membrane dynamics. Biochim Biophys Acta. 1997;1357:129–154. doi: 10.1016/s0167-4889(97)00038-4. [DOI] [PubMed] [Google Scholar]
- Gidrol X, Sabelli PA, Fern YS, Kush AK. Annexin-like protein from Arabidopsis thaliana rescues delta oxyR mutant of Escherichia coli from H2O2 stress. Proc Natl Acad Sci USA. 1996;93:11268–11273. doi: 10.1073/pnas.93.20.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Proust J, Dorowski A, Schantz R, Huber R. Annexin 24 from Capsicum annuum: x-ray structure and biochemical characterization. J Biol Chem. 2000;275:8072–8082. doi: 10.1074/jbc.275.11.8072. [DOI] [PubMed] [Google Scholar]
- Jones PG, Moore GJ, Waisman DM. A nonapeptide to the putative F-actin binding site of annexin II tetramer inhibits its calcium-dependent activation of actin filament bundling. J Biol Chem. 1992;267:13993–13997. [PubMed] [Google Scholar]
- Kjeldgaard M, Nyborg J, Clark BFC. The GTP binding motif: variations on a theme. FASEB J. 1996;10:1347–1368. [PubMed] [Google Scholar]
- Kovacs I, Ayaydin F, Oberschall A, Ipacs I, Bottka S, Dudits D, Toth EC. Immunolocalization of a novel annexin-like protein encode by a stress and abscisic acid responsive gene in alfalfa. Plant J. 1998;15:185–197. doi: 10.1046/j.1365-313x.1998.00194.x. [DOI] [PubMed] [Google Scholar]
- Lafont F, Lecat S, Verkade P, Simons K. Annexin XIIIb associates with lipid microdomains to function in apical delivery. J Cell Biol. 1998;142:1413–1427. doi: 10.1083/jcb.142.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E-K, Roberts MR, Bowles DJ. Biochemical characterization of tomato annexin p35, independence of calcium binding and phosphatase activities. J Biol Chem. 1998;273:34920–34925. doi: 10.1074/jbc.273.52.34920. [DOI] [PubMed] [Google Scholar]
- McClung AD, Carroll AD, Battey NH. Identification and characterization of ATPase activity associated with maize (Zea mays) annexins. Biochem J. 1993;303:709–712. doi: 10.1042/bj3030709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, McKinney EC, Kandasamy MK. Isovariant dynamics expand and buffer the responses of complex systems: the diverse plant actin gene family. Plant Cell. 1999;11:995–1005. doi: 10.1105/tpc.11.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, McWhinnie EA, Agarwal SK, Schaff DA. The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene. 1994;143:211–216. doi: 10.1016/0378-1119(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Newman T, deBruijn EJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel FC, Lescure N, Cullimore JV, Gamas P. The Medicago trunculata MtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Mol Plant-Microbe Interact. 1998;11:504–513. doi: 10.1094/MPMI.1998.11.6.504. [DOI] [PubMed] [Google Scholar]
- Proust J, Houlne G, Schantz M-L, Schantz R. Characterization and gene expression of an annexin during fruit development in Capsicum annum. FEBS Lett. 1996;383:208–212. doi: 10.1016/0014-5793(96)00252-9. [DOI] [PubMed] [Google Scholar]
- Proust J, Houlne G, Schantz M-L, Shen W-H, Schantz R. Regulation of biosynthesis and cellular localization of Sp32 annexins in tobacco BY2 cells. Plant Mol Biol. 1999;39:361–372. doi: 10.1023/a:1006199814795. [DOI] [PubMed] [Google Scholar]
- Santoni V, Rouquie D, Doumas P, Mansion M, Boutry M, Degand H, Dupree P, Packman L, Sherrier J, Prime T. Use of a proteome strategy for tagging proteins present at the plasma membrane. Plant J. 1998;16:633–641. doi: 10.1046/j.1365-313x.1998.00335.x. [DOI] [PubMed] [Google Scholar]
- Seals DF, Randall SK. A vacuole associated annexin protein, VcaB42, correlates with the expansion of tobacco cells. Plant Physiol. 1997;115:753–761. doi: 10.1104/pp.115.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton BA, Dedman JR. Annexins. Biometals. 1998;11:399–404. doi: 10.1023/a:1009205925714. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Rolland N, Dorne A-J, Joyard J. Sulfolipid is a potential candidate for annexin binding to the outer surface of chloroplast. Biochem Biophys Res Commun. 2000;272:519–524. doi: 10.1006/bbrc.2000.2805. [DOI] [PubMed] [Google Scholar]
- Shi J, Dixon RA, Gonzales RA, Kjellbom P, Bhattacharyya MK. Identification of cDNA clones encoding valosin-containing protein and other plant membrane-associated proteins by a general immunoscreening strategy. Proc Natl Acad Sci USA. 1995;92:4457–4461. doi: 10.1073/pnas.92.10.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Brown M. GTPase activity and biochemical characterization of a recombinant cotton fiber annexin. Plant Physiol. 1999;119:925–934. doi: 10.1104/pp.119.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternovsky VI, Berestovsky GN. Effective diameter and structural organization of reconstituted calcium channels from the Characeae algae Nitellopsis. Membr Cell Biol. 1998;12:79–88. [PubMed] [Google Scholar]
- Thiel G, Battey N. Exocytosis in plants. Plant Mol Biol. 1998;38:111–125. [PubMed] [Google Scholar]
- Thonat C, Mathieu C, Crevecoeur M, Penel C, Gaspar T, Boyer N. Effects of a mechanical stimulation on localization of annexin-like proteins in Bryonia dioica internodes. Plant Physiol. 1997;114:981–988. doi: 10.1104/pp.114.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzima E, Trotter PJ, Orchard MA, Walker JH. Annexin V binds to the actin-based cytoskeleton at the plasma membrane of activated platelets. Exp Cell Res. 1999;251:185–193. doi: 10.1006/excr.1999.4553. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Conner TW, Klee HJ. Identification of mRNAs with enhanced expression in ripening strawberry fruit using polymerase chain reaction differential display. Plant Mol Biol. 1995;27:1097–1108. doi: 10.1007/BF00020883. [DOI] [PubMed] [Google Scholar]