Abstract
The worldwide prevalence of thyroid disease is on the rise, representing a chronic condition that significantly impacts global mortality rates. Machine learning (ML) approaches have demonstrated potential superiority in mitigating the occurrence of this disease by facilitating early detection and treatment. However, there is a growing demand among stakeholders and patients for reliable and credible explanations of the generated predictions in sensitive medical domains. Hence, we propose an interpretable thyroid classification model to illustrate outcome explanations and investigate the contribution of predictive features by utilizing explainable AI. Two real-time thyroid datasets underwent various preprocessing approaches, addressing data imbalance issues using the Synthetic Minority Over-sampling Technique with Edited Nearest Neighbors (SMOTE-ENN). Subsequently, two hybrid classifiers, namely RDKVT and RDKST, were introduced to train the processed and selected features from Univariate and Information Gain feature selection techniques. Following the training phase, the Shapley Additive Explanation (SHAP) was applied to identify the influential characteristics and corresponding values contributing to the outcomes. The conducted experiments ultimately concluded that the presented RDKST classifier achieved the highest performance, demonstrating an accuracy of 98.98 % when trained on Information Gain selected features. Notably, the features T3 (triiodothyronine), TT4 (total thyroxine), TSH (thyroid-stimulating hormone), FTI (free thyroxine index), and T3_measured significantly influenced the generated outcomes. By balancing classification accuracy and outcome explanation ability, this study aims to enhance the clinical decision-making process and improve patient care.
Keywords: Thyroid disease, Machine learning, SMOTE-ENN, Ensemble methods, And explainable AI
1. Introduction
The rising prevalence and high-cost implications of thyroid disorders make it a pressing global public health issue [1]. Insufficient production of thyroid hormones by the thyroid gland leads to abnormal growth and disease development. The disruptions in its functioning can lead to various symptoms, including weight loss, hair loss, irregular heartbeat, hypertension, reduced metabolic rate, weight gain, and a decreased pulse rate [2]. Over time, untreated thyroid disease can lead to various health complications, including joint pain, infertility, obesity, and heart disease. According to data from Ref. [3], there were 43,800 newly reported cases of thyroid disease in the United States in 2022. Of these, 31,940 cases were observed in females, while males accounted for 11,860. In the following year, the American Cancer Society reported 2120 new deaths in the USA due to thyroid cell abnormalities.
The most effective strategy to prevent the serious effects is to detect or anticipate it as early as possible during diagnosis. However, identifying this disorder via a traditional laboratory test is extremely difficult and necessitates a high level of expertise. Additionally, the manual approach might lead to erroneous results and is time-consuming. Machine learning (ML) is the widely accepted method to address these issues and prevent severe consequences by providing early warning of the disease [4,5]. However, the limited ability to explain ML hinders its widespread adoption in healthcare applications, especially when decision-makers need a clear understanding of the underlying rationale [6]. Without explainability, the potential risk of incorrect decisions outweighs the benefits of ML's accuracy and decision-making advantages. Implementing explainable learning methods empowers healthcare professionals to make informed, data-driven decisions and reliable treatments.
Problem Statement and Motivation: Researchers have extensively employed ML approaches to investigate thyroid disease at early stages. These studies encounter notable challenges that necessitate the adoption of state-of-the-art methods. For instance, Gupta [7] and Ahmed [8] have endeavored to propose thyroid classification models using imbalanced datasets, which obstruct the proper capture of underlying patterns and pose difficulties in accurately classifying the minority class. They also faced challenges related to biased learning, poor generalization, and overfitting [9]. To address these concerns, Alyas [10] and Aversano [11] have employed a well-established oversampling method known as the Synthetic Minority Over-sampling Technique (SMOTE). However, it is acknowledged that using SMOTE may introduce noisy and uninformative samples [12,13]. Subsequently, Sonuc [14] and Srivastava [15] have proposed a thyroid classification model retaining the original dimensionality of features, leading to high computation costs and decreased generalization ability.
Chaubey [16] has utilized a single random sampling approach to train and validate the model, which lacks an accurate representation of the sample distribution across classes in the underlying population. Additionally, Savci [17] and Olatunji [18] have employed multiple standalone ML classifiers for training the dataset, limited effectiveness in handling complex and diverse datasets. Recognizing the limitations of individual classifiers, Dharmakar [19] and Yadav [20] have introduced Voting-based hybrid models, combining two baseline classifiers. However, it is important to note that this ensemble method performs best when integrating multiple classifiers as base estimators [21]. Moreover, the black-box behavior of the studies, as mentioned earlier, provided a lack of trust among patients and clinicians. Failure to incorporate interpretable methods while developing ML models may lead to non-compliance with regulatory requirements. Many healthcare regulations mandate transparency and accountability in decision-making.
Novelty and Contribution: Motivated by these concerns, we aim to propose an efficient thyroid classification model. Unlike existing studies, our approach incorporates SMOTE with Edited Nearest Neighbors (ENN) to deal with the imbalanced dataset, where ENN plays a crucial role in eliminating noisy samples generated by SMOTE [22]. Additionally, we have employed two effective feature selection methods to rectify most potential features and reduce the dimensionality of the dataset. To assess the effectiveness of our study, we divided the processed dataset into three distinct subsets (e.g., 70 %, 80 %, and 90 % for training, and 30 %, 20 %, and 10 % for testing), then evaluated the average results for analysis. Then, introduced two hybrid classifiers by integrating three base classifiers, namely Random Forest (RF), Decision Tree (DT), and K-nearest Neighbors (KNN), as popular in the related task [7,10,11,14]. Finally, explainable AI has attracted a lot of attention recently, and explainability has grown to be a heavily researched topic in ML [23]. This is a crucial aspect, especially in the healthcare domain, where incorrect decisions can have severe consequences on patient outcomes. Shapley Additive Explanation (SHAP) is a vital explainable AI tool that assists in clarifying the intricate linkages between multivariate data and computer model predictions. Hence, by integrating SHAP with our proposed model, we ensure the trust and transparency of the system to stakeholders and patients, enabling them to understand the decisions. It also empowers patients with information about the factors influencing their diagnosis, fostering a more patient-centric approach to care. However, the key contributions are as follows.
-
•
We have conducted a series of preprocessing stages encompassing feature dropping, data encoding, imputation missing values, and addressing imbalance issues.
-
•
Introduced two hybrid ML classifiers, namely RDKVT and RDKST, with meticulously optimized parameter configurations.
-
•
The performance of the proposed classifiers was then validated using two thyroid-based datasets with different feature sets. A comprehensive comparison was ensured by evaluating various performance metrics, including two predictive rates, computation time, log loss, and statistical significance test.
-
•
Experimental analysis demonstrates the superiority of the proposed RDKST classifier with a robust accuracy of 98.98 % for the Kaggle dataset, surpassing the performance of existing models.
-
•
Furthermore, incorporating SHAP with the RDKST classifier, we identify that the characteristics T3, TT4, TSH, FTI, and T3_measured exert the most significant influence on the outcome, as their respective values contribute substantially to the prediction.
2. Related studies
Recently, researchers have increasingly focused on developing efficient thyroid classification systems using ML methods. Their efforts have primarily revolved around model comparison, disease detection, and the development of diagnostic models. For example, Naman Gupta et al. [7] employed multiple ML classifiers and applied a modified ant lion optimization algorithm. Their results showed accuracy rates of 95.94 %, 95.66 %, and 92.51 % achieved by RF, DT, and KNN classifiers, respectively. Ahmad et al. [8] proposed a hybrid decision support system based on linear discriminant analysis (LDA), KNN, and adaptive neuro-fuzzy inference (ANFIS). Their LDA-KNN-ANFIS approach demonstrated a remarkable accuracy of 98.5 %. However, it is essential to note that they developed their system using an imbalanced dataset, which introduces several challenges, including biased learning, poor generalization, and overfitting [9].
Tahir et al. [10] compared four ML classifiers by evaluating their performance on both sampled and unsampled datasets. The findings revealed that RF achieved the highest accuracy of 94.8 %. Similarly, with the AOU Federico II Naples hospital dataset, Lerina et al. [11] compared the performance of ten ML classifiers. Before feeding the training model, a range of preprocessing techniques, including SMOTE, were applied to the dataset. Based on the experimental outcome, the Extra Tree (ET) classifier achieved the highest accuracy of 84 %. However, it is necessary to note that utilizing SMOTE in these studies may introduce noisy and irrelevant samples into the dataset [12,13].
Sonuc et al. [14] aimed to classify thyroid disorders utilizing 1250 distinct Iraqi individual samples and achieved a higher accuracy rate of 98.93 % using the RF classifier. Srivastava et al. [15] addressed the data imbalance issues with the BL_SMOTE technique and yielded 98.88 % accuracy by combining RF and DT using a Voting method. Chaubey et al. [16] conducted a classification of thyroid disease using DT, KNN, and logistic regression (LG), where KNN obtained a robust accuracy rate of 96.875 %. Dignata et al. [24] presented a comparison of various ML models, where RF obtained robust accuracy compared to others. Additionally, Awad et al. [25] utilized the Kaggle thyroid dataset and, by training with the SMV classifier, achieved 84.72 % accuracy. However, these studies have limited dimensionality reduction, which can result in high computation costs and decrease the generalizability of the models.
Savci et al. [17] have found that ANN achieved a higher accuracy of 98 % from the five classification algorithms. Olatunji et al. [18] developed an ML-based tool using the Saudi Arabian dataset and observed that RF obtained the highest accuracy of 90.91 %. Azrin et al. [26] also introduced a thyroid-based predictive system using an RF classifier. However, relying solely on ML classifiers may have limitations in handling complex and diverse datasets. Thereby, Dharmakar et al. [19] proposed a hybrid classifier called CCTML by integrating C4.5 and RF classifiers using the Voting ensemble approach, which showed an accuracy of 96 %. Moreover, the study [27] presented two hybrid models named Three Stage Hybrid Classifier (3SHC) and Three Stage Hybrid Artificial Neural Network (3SHANN) to forecast the disease. Yadav et al. [20] also presented a hybrid classifier using the Stacking approach, combining DT, and neural networks (NN), and achieved an AUC score of 98.80 %. However, it is worth noting that the Voting ensemble method is considered more suitable for integrating multiple classifiers as base estimators [21]. Finally, Table 1 presents a summarized overview of these studies.
Table 1.
A summary of recent machine learning-based studies on thyroid disease diagnosis.
| Year and reference | Data collection | Number of instances | Output classes | Balancing Technique |
Best Performing classifier |
|---|---|---|---|---|---|
| 2020 [7] | UCI | 7200 | 3 | (−) | RF |
| 2018 [8] | UCI | 3163 | 2 | (−) | LDA-KNN-ANFIS |
| 2022 [10] | UCI | 3163 | 2 | SMOTE | RF |
| 2021 [11] | Naples's hospital | 2784 | 3 | SMOTE | ET |
| 2021 [14] | Hospitals and Labs | 1250 | 3 | (−) | RF |
| 2021 [15] | UCI | 2800 | 2 | BL_SMOTE | Voting (RF, DT) |
| 2020 [16] | UCI | 215 | 3 | (−) | KNN |
| 2022 [17] | UCI | 7200 | 3 | SMOTE | ANN |
| 2021 [18] | Saudi Arabian | 218 | 2 | (−) | RF |
| 2020 [19] | UCI | 7547 | 2 | (−) | CCTML |
| 2019 [20] | Pathologies | 27000 | 3 | (−) | Stacking (DT, NN) |
| 2023 [24] | UCI | 3772 | 2 | (−) | RF |
| 2023 [25] | Kaggle | 3371 | 2 | (−) | SVM |
| 2023 [26] | UCI | 2800 | 2 | SMOTE | RF |
| 2023 [27] | Kaggle | 3773 | 2 | SMOTE | 3SHC |
3. Material and methods
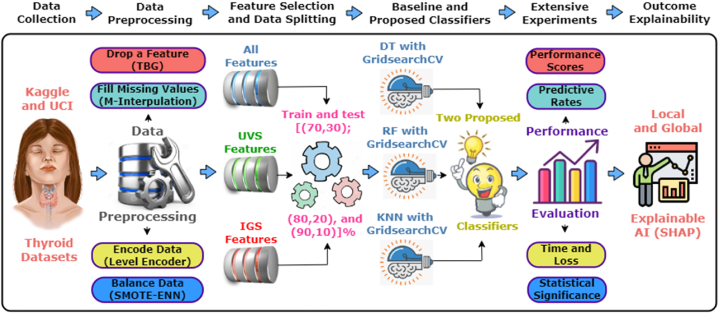
In this section, we have provided a comprehensive discussion of the methods and working procedures employed in the study. The working methodology is divided into six main parts: data collection, data preprocessing, feature selection with data splitting, ML classifiers, performance evaluation matrices, and explainable AI. Fig. 1 shows the overview of the entire working process.
Fig. 1.
The proposed workflow for our study, consisting of the general components.
3.1. Data collection
For this study, we utilized two thyroid-based datasets from well-known data repositories (e.g., Kaggle [28] and UCI [29]). The employed datasets contain different data types, including integers, binaries, categories, and floats. The Kaggle dataset contains a total of 30 features with 3772 different cases. These features included various clinical and laboratory characteristics with a target feature, namely age, sex, on thyroxine (On-T), query on thyroxine (QOT), on antithyroid medication (OAM), sick, pregnant, Thyroid surgery (TS), I131 treatment (I131-T), query hypothyroid (QU-HO), query hyperthyroid (QU-HE), lithium, goiter, tumor, hypopituitary, psych, TSH measured (TSH-M), TSH, T3 measured (T3-M), TT4 measured (TT4-M), TT4, T4U measured (T4U-M), T4U, FTI measured (FTI-M), FTI, TBG, TBG measured (TBG-M), T3, referral source (RS), and class. Upon analyzing the test reports, we found that 3541 patients were unaffected by disease, while only 231 cases were affected. On the other hand, the UCI dataset included 25 clinical and laboratory features and one target feature. The included clinical and laboratory attributes are age, sex, On-T, QOT, OAM, TS, QU-HO, QU-HE, pregnant, sick, tumor, lithium, goiter, TSH-M, TSH, T3-M, T3, TT4-M, TT4, T4U-M, T4U, FTI-M, FTI, TBG-M, and TBG. The dataset comprises a total of 3163 different cases, with 3012 classified as negative and 151 classified as positive cases.
3.2. Data preprocessing
Data preprocessing is essential for an ML model to turn raw data into a useful format. Our datasets have issues with missing values in different features (e.g., age, gender, TSH, T3, TT4, T4U, FTI, TBG), which could impact the model. We solved this issue by eliminating attributes with missing values greater than 50 %, which removed the TBG attribute [30]. The remaining gaps in the Kaggle (29 features) and UCI (24 features) datasets were filled using mean interpolation, thereby maximizing data utilization and maintaining integrity. Non-numeric categorical data gave an additional barrier since classifiers could misinterpret them. We utilized the level-encoder approach to transform these variables into numerical representations while retaining ordinal correlations and not increasing the dataset's dimensionality.
Afterward, it is worth noting that the dataset exhibits a significant class imbalance, with 3541 and 3012 instances belonging to the negative class and only 231 and 151 cases classified as the positive class for Kaggle and UCI datasets, respectively. This class imbalance poses challenges in accurately predicting the minority class. To address this issue, the authors [10,11,17,26,27] have utilized the most popular data balancing technique, SMOTE. However, SMOTE tends to generate synthetic samples that are too close to the original ones, which can introduce noise and potentially lead to overfitting [12,13]. Also, it does not consider the potential overlapping regions between different classes, which can result in the misclassification of samples. Hence, we have introduced an advanced technique named SMOTE-ENN to address the disadvantages of SMOTE. SMOTE-ENN first applies SMOTE to generate the synthetic samples, and then ENN serves to remove the noisy and misclassified samples. By incorporating SMOTE and ENN, we have reduced the chances of overfitting and enhanced the quality of synthetic instances. Initially, SMOTE chose the nearest neighbor in synthesizing new instances from raw samples ; then, the unknown instance is built according to Eq. (1).
| (1) |
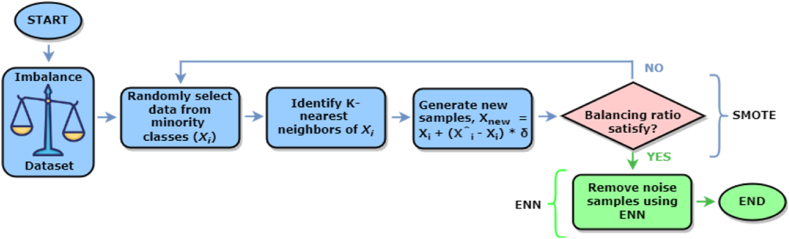
To enhance the quality of new instances, ENN is a powerful technique for eliminating noisy and uninformative samples from the dataset [22]. It examines the samples and compares the corresponding class levels with the majority class neighbors. This combined method is specifically beneficial when the class imbalance rate is severe. Applying the SMOTE-ENN approach resulted in a nearly balanced distribution for both datasets. Specifically, the Kaggle dataset consisted of 2655 cases belonging to the negative class and 3256 instances belonging to the positive class. In the case of the UCI dataset, there were 2879 negative cases and 2973 positive cases reconstructed after the process. These results demonstrate that the approximate balance ratio is 0.82 for the Kaggle dataset and 0.97 for the UCI dataset. The working procedure of SMOTE-ENN is depicted in Fig. 2, illustrating how synthetic instances are generated for the minority class using SMOTE and how ENN is refining the dataset by removing noisy samples.
Fig. 2.
Working diagram of SMOTE-ENN to produce synthetic instances and eliminate the noisy samples.
3.3. Feature selection and data splitting
The feature selection method is crucial in reducing the model dimension and developing a predictive model. It is an essential concept in ML that significantly influences the model's performance and execution time. There are three main categories of feature selection methods: filter, wrapper, and embedded. Among these, filter methods are more scalable and effectively deal with high-dimensional datasets. Several feature selection techniques exist in this type of feature selection method. In our study, we incorporated two widely recognized filter-based feature selection approaches (i.e., Univariate selection (UVS) and Information Gain Selection (IGS)). Numerous thyroid studies have witnessed efforts to utilize the mentioned feature selection techniques and achieved generalizable results [[31], [32], [33], [34]]. These consistent findings and outcomes have motivated us to apply these feature selection techniques in our study.
3.3.1. Univariate feature selection (UVS)
The UVS technique selects features according to their correlation with the goal variable. Here, we used the chi-square test, a univariate statistical method. It measured the dependency between the features based on the target variable. The higher chi-squared value of a feature indicates that the feature is more likely to be relevant to the target feature. Mathematically, the chi-square statistic is calculated using the formula expressed in Eq. (2). Where is the summation of the classes, is the number of considered feature possible values.
| (2) |
3.3.2. Information gain feature selection (IGS)
IGS utilizes information theory to determine the interdependency between features and the target variable, thereby effectively identifying the most informative attributes. This selection technique commonly uses the tree-based algorithm working procedures. It measures the reduction of entropy in the target variable achieved by splitting the dataset based on the value of a particular feature. Mathematically, entropy is defined as in Eq. (3), where E(Y) is the entropy of target variable Y, c is the number of classes, and is the probability of class i occurring in Y.
| (3) |
When a dataset is split based on a feature X, the information gain IG(X) is calculated as the difference between the entropy and a target variable before and after the split, stated in Eq. (4). Where E(Y|X) is the conditional entropy, calculated in Eq. (5), wherein v refers the number of unique values of feature X, indicates the probability of occurrence of value in feature X, and finally, is the entropy of Y given that feature X takes the value . Features with higher IG are considered more relevant for predicting the target variable and selected for the study.
| (4) |
| (5) |
As shown in Table 2, we identified the 12 most essential characteristics using both methods from the initial feature sets [35]. By exploiting these significant characteristics, we hope to simplify the model's input variables, improve its predictive performance, and maximize the model's overall effectiveness. Therefore, we explore the predictive power of these features in the experimental section.
Table 2.
A list of selected features with rank values using two feature selection techniques for both Kaggle and UCI datasets.
| Kaggle dataset |
UCI dataset |
||||||
|---|---|---|---|---|---|---|---|
| UVS selected features |
IGS selected features |
UVS selected features |
IGS selected features |
||||
| Name | Score | Name | Score | Name | Score | Name | Score |
| T3 | 6184.89 | T3 | 0.5406 | TT4 | 8290.95 | FTI | 0.6328 |
| age | 1474.32 | age | 0.4562 | FTI | 6005.52 | TT4 | 0.4353 |
| TT4 | 626.66 | TT4 | 0.4285 | TSH | 2165.33 | TSH | 0.4314 |
| T3-M | 563.17 | FTI | 0.4218 | T3 | 1958.23 | T3 | 0.4311 |
| On-T | 381.73 | TSH | 0.3418 | TSH-M | 393.49 | T4U | 0.4134 |
| TSH-M | 232.64 | sex | 0.1779 | On-T | 342.71 | age | 0.3173 |
| TT4-M | 232.31 | T3-M | 0.0542 | T4U | 278.40 | sex | 0.1427 |
| QU-HE | 204.56 | TT4-M | 0.0375 | T4U-M | 254.39 | T4U-M | 0.0310 |
| psych | 148.23 | On-T | 0.0373 | TT4-M | 253.87 | TSH-M | 0.0304 |
| T4U | 128.78 | psych | 0.0202 | FTI-M | 253.11 | On-T | 0.0301 |
| T4U-M | 92.59 | TSH-M | 0.0157 | TBG-M | 252.91 | TT4-M | 0.0254 |
| sex | 92.41 | pregnant | 0.0148 | QU-HE | 211.89 | QU-HE | 0.0186 |
3.3.3. Data splitting
To effectively evaluate the performance of our model, we partitioned the processed dataset and the reduced feature sets into different subsets: 70 %, 80 %, and 90 % for training, and correspondingly, 30 %, 20 %, and 10 % for testing. Table 3 shows the representativeness of the sample size after applying these different train-test ratios. By employing multiple training and testing splits, we aim to capture the variability in the data and obtain a more comprehensive understanding of the model's performance across different scenarios. To ensure the reliability of our evaluation, we adopt the practice of averaging the obtained results from multiple testing splits. This approach enables us to get a more accurate and representative measure of the model's performance by mitigating the potential bias introduced by a single train-test split.
Table 3.
The representativeness size of samples used in the experiment from different train-test ratios for both Kaggle and UCI datasets.
| Train-test ratio | Kaggle dataset |
UCI dataset |
||||
|---|---|---|---|---|---|---|
| Training | Testing | All | Training | Testing | All | |
| 70:30 % | 4137 | 1774 | 5911 | 4096 | 1756 | 5852 |
| 80:20 % | 4728 | 1183 | 5911 | 4681 | 1171 | 5852 |
| 90:10 % | 5319 | 592 | 5911 | 5266 | 586 | 5852 |
3.4. Machine learning baseline and proposed classifiers
We initially employ three traditional Machine Learning classifiers—Random Forest (RF), Decision Tree (DT), and K-nearest Neighbor (KNN). During the training of the dataset, the GridSearchCV is utilized to explore the fine-tuned parameters. Subsequently, we present two hybrid classifiers, RDKVT and RDKST, by combining these baseline classifiers with the help of the Voting and Stacking ensemble approaches.
3.4.1. Random Forest
Random Forest (RF) is a pliable supervised ML algorithm widely used in healthcare to analyze patients' medical histories and diagnose diseases. RF creates numerous decision trees and blends them to generate a more accurate and reliable prediction. It uses the Gini Index as a selection criterion for attributes, which assesses an attribute's impurity concerning classes. RFs are robust against overfitting, effectively handle high-dimensional data, and provide feature importance measures.
3.4.2. Decision tree
A decision tree (DT) functions by segmenting the input data into subsets iteratively based on the value of one of its properties. The goal of DT is to build a training model that can forecast the value of the target variable by mastering basic decision rules. The algorithm selects the best feature to split the data at each node, aiming to maximize information gain or minimize impurity. This process continues until a stopping criterion is met, producing leaf nodes representing the final predictions. There are several variants of DT available, and in this study, we used the C4.5 algorithm due to its ease of implementation and interpretability.
3.4.3. K-nearest neighbors
The K-Nearest Neighbor (KNN) algorithm attempts to determine the optimal class for the test data by calculating the distance between the test data and the training points. KNN can be modified in a variety of ways, giving a boost to several KNN types or modifications. This algorithm is robust to training data with such a large amount of noise and will be sufficient if the training data is substantial. KNN primarily operates by determining the Euclidean distance between each raw set of training data and the test data.
3.4.4. Proposed hybrid RDKVT classifier
Lately, there has been a growing emphasis among researchers on developing hybrid classifiers to tackle the challenges associated with using single classifiers. A single traditional classifier may exhibit sensitivity to specific data types, have limited capacity to capture diverse aspects of the dataset, and potentially be biased towards certain scenarios. By leveraging the collective power of multiple classifiers simultaneously, each classifier can capture distinct facets of the data from different viewpoints. The individual strengths of each classifier make the final hybrid classifier achieve more robust and generalized outcomes. Hence, we are motivated to combine the RF, DT, and KNN classifiers using the Voting (VT) ensemble method. Which makes the final prediction based on the highest probability of the chosen class, aiming to enhance the prediction accuracy and robustness of the model. Two different voting schemes, namely hard and soft VT, are available in this approach. In this study, we employ soft Voting, which is compatible with all classifiers and calculates the average probability score for all classes to generate the final prediction. By using soft voting, the developed classifier operates according to Eqs. (6), (7), (8)), where and represent the training and testing data.
| (6) |
| (7) |
| (8) |
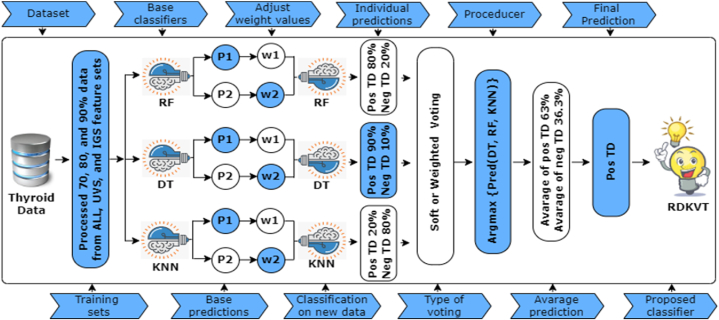
After training and evaluating the individual ML classifiers RF, DT, and KNN, the obtained predictions are derived as , , and , respectively. The final prediction generates the most significant sum of weighted probabilities. The procedure of the RDKVT classifier is shown in Fig. 3. In this ensemble approach, RF brings the benefit of reducing overfitting and handling high-dimensional data by constructing multiple decision trees with random feature selection and bootstrap sampling. Subsequently, DT provides interpretability and the ability to capture complex decision boundaries, and KNN handles non-linear relationships. By combining these models through the VT ensemble method, we can exploit the strengths of each model while mitigating their weaknesses. Thus, it can capture a broader range of patterns and relationships, improving overall prediction performance. Detailed steps of this hybrid model are illustrated in Algorithm 1. This algorithm provides all precious steps, including data processing, data splitting, individual training, evaluating these sets using three baseline classifiers and aggregating their predictions, and finally executing the result for a new sample based on the proposed classifier. The aggregation of different models specialized in handling specific data types can be integrated to create a more versatile classifier. Therefore, it has the greater ability to dynamically adapt to changing conditions or drift in data patterns.
Fig. 3.
Outlining the procedures (including processed training set, fitting base classifiers, and functional stages of soft Voting) of the proposed RDKVT classifier (TD refers to Thyroid).
Algorithm 1.
Showcasing the Major Working Steps of RDKVT.
| 1: Inputs: Dataset, |
| 2: Outputs: Classify whether the thyroid is affected or not |
| 3: D(a) ← D.drop ([T BG], axis = columns) |
| 4: D(b) ← MedianImpute {} |
| 5: D(c) ← LabelEncoder {} |
| 6: D(d) ← SMOTEENN {} |
| 7: ← input {(N × M matrix)}, output (N × 1 vector) |
| 8: ← TrainTestSplit () |
| 9: ← TrainTestSplit () |
| 10: ← TrainTestSplit () |
| 11: while (execute − different − TrainTestSets) do |
| 12: ← RandomForest () |
| 13: ← DecisionTree () |
| 14: ← KNearestNeighbors () |
| 15: end while |
| 16: procedure RDKVT () |
| 17: RDKVT ← argmax () |
| 18: while (fitting – different − TrainTestSets) |
| 19: RDKVT ← RDKVT.fit () |
| 20: end while |
| 21: Result ← RDKVT. predict (New−sample) |
| 22: ReturnResult |
3.4.5. Proposed hybrid RDKST classifier
We have introduced another hybrid classifier to identify the strengths of different ensemble methods and provide flexibility in model deployment. In the proposed RDKST classifier, we again combined the RF, DT, and KNN classifiers using the Stacking (ST) ensemble method. By incorporating the ST ensemble method, we aim to further explore and leverage the advantages of ensemble modeling for our specific problem. The ST ensemble method is a powerful technique that combines multiple models to achieve improved accuracy and robustness. In the RDKST classifier, we train a meta-model on the predictions made by the base classifiers. The base classifiers (RF, DT, and KNN) generate predictions on the input data, and these predictions are subsequently used as inputs for the meta-model. Here we used Logistic Regression (LG) as a meta-model to train the base predictions from the base estimators. The working functions of our proposed RDKST classifier are stated in Eqs. (9), (10), (11)).
| (9) |
| (10) |
| (11) |
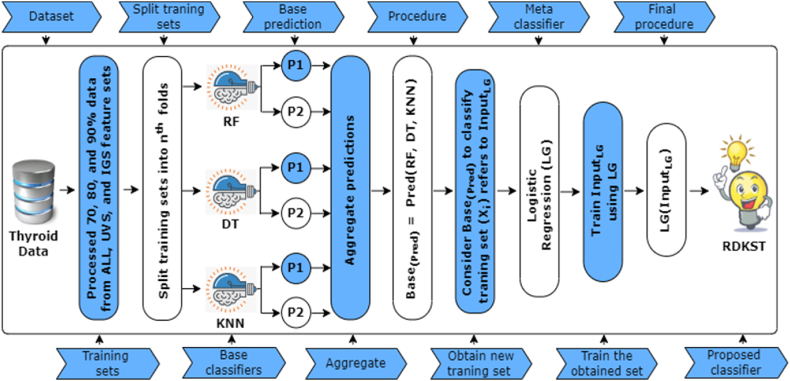
where, refers to the predictions from base classifiers, which are further used in Eq. (10) to classify the training instances , these classified training instances are used as inputs for a meta classifier, namely LG. Finally, Eq. (11) states the procedure of the meta classifier, is the intercept, and to is the coefficient of the generated input features to . By combining the RF, DT, and KNN classifiers using the ST ensemble method, the RDKST classifier takes advantage of the complementary strengths of each classifier. Where RF is known for its robustness and ability to handle high-dimensional data, DT is adept at capturing complex decision boundaries, and KNN excels in identifying local patterns and similarities. Through the ST ensemble approach, the RDKST classifiers aim to improve the overall predictive performance by leveraging the collective knowledge and expertise of the base classifiers. Fig. 4 represents the working process of the RDKST classifier. Furthermore, to address diverse learning preferences, we have shown an inclusive strategy of this method in Algorithm 2. Compared to the RDKDT, the step employed in this model is identical for data processing, splitting, and execution processes. The main difference is instead of evaluating the outcome based on first-level prediction (), it used the first-level prediction as a new training set for LG. LG is trained on the outputs of , allowing it to capture complex relationships and dependencies. The potential superiority of RVKST lies in its ability to adaptively learn and optimize the combination of diverse base models based on the characteristics of the data.
Fig. 4.
Outlining the procedures (includes processed training set, fitting base classifiers, and functional stages of Stacking) of the proposed RDKST classifier.
Algorithm 2.
Showcasing the Major Working Steps of RDKST.
| 1: Inputs: Dataset, , Meta classifiers = LG |
| 2: Outputs: Classify whether the thyroid is affected or not |
| 3: ← D.drop ([TBG], axis = columns) |
| 4: ← MedianImpute {} |
| 5: ← LabelEncoder {} |
| 6: ← SMOTEENN {} |
| 7: ← input{(N × M matrix)}, output (N × 1 vector) |
| 8: ← TrainTestSplit () |
| 9: ← TrainTestSplit () |
| 10: ← TrainTestSplit () |
| 11: while (execute − different − TrainTestSets) do |
| 12: ← RandomForest () |
| 13: ← DecisionTree () |
| 14: ← KNearestNeighbors () |
| 15: end while |
| 16: ← concatenate () |
| 17: fordo |
| 18: Apply to classify training instances |
| 19: ← () |
| 20: ← (, ), where ← () |
| 21: end for |
| 22: RDKST ← LG {} |
| 23: Result ← RDKST. predict (New−sample) |
| 24: ReturnResult |
3.5. Prevention overfitting from the proposed classifiers
In this study, we proposed two hybrid ML classifiers for effective thyroid disease classification. However, atypical data conditions can affect their generalizability, possibly leading to overfitting during classification. To address this, we used several preprocessing techniques to clean the dataset, removing missing values and minimizing noise. We also tackled class imbalance for balanced distribution and selected relevant features to improve model performance. These steps aim to lower overfitting risk and enhance classifier generalizability. Then, we developed the proposed classifiers by integrating multiple baseline classifiers using ensemble methods to reduce individual biases and capture diverse perspectives, thereby mitigating overfitting. Furthermore, the integrated baseline classifiers were trained with a fine-tuned set of parameters to control the learning process. In this regard, we used the GridSearchCV, which automates the hyperparameter tuning process by systematically exploring the hyperparameter space and identifying the optimal configuration for the model. Table 4 illustrates the best parameter of our employed traditional classifiers for different feature sets. By utilizing these aforementioned methodologies, we can hypothesize that our proposed classifiers are less susceptible to overfitting and capable of producing more generalized results.
Table 4.
Utilized the set of fine-tuned parameters for employed baseline classifiers in different feature sets by using GridsearchCV.
| Feature set | RF | DT | KNN |
|---|---|---|---|
| ALL Features |
n_estimator = 10, random_state = 10, max_depth = 7, max_features = ‘sqrt’ | max_depth = 5, random_state = 5, | n_neighbors = 10, leaf_size = 25, |
| criterion = ‘entropy’, | metric = ‘minkowski’, | ||
| splitter = ‘best’ | algorithm = ‘brute’ | ||
| UVS Features |
n_estimator = 8, random_state = 15, max_depth = 5, max_features = ‘sqrt’ | max_depth = 8, random_state = 15, | n_neighbors = 15, leaf_size = 30, |
| criterion = ‘gini’, | metric = ‘minkowski’, | ||
| splitter = ‘best’ | algorithm = ‘auto’ | ||
| IGS Features |
n_estimator = 7, random_state = 10, max_depth = 10, max_features = ‘sqrt’ | max_depth = 5, random_state = 20, | n_neighbors = 5, leaf_size = 30, |
| criterion = ‘gini’, | metric = ‘minkowski’, | ||
| splitter = ‘best’ | algorithm = ‘auto’ |
3.6. Performance evaluation metrics
To validate the effectiveness of our proposed classifiers, we conducted a comprehensive evaluation using various performance evaluation scores such as accuracy (ACC), precision (PRE), recall (REC), f1-score (F1S), the area under the curve (AUC), and cohen kappa score (CKS). The ACC calculates the ratio of correctly identified samples to the total number of instances. PRE quantifies the model's ability to identify positive cases accurately. REC assesses the model's ability to capture all positive samples. F1S comprehensively evaluates the model's prediction ability by combining PRE and REC. The AUC evaluates a model's ability to differentiate between positive and negative classes, and CKS assesses inter-rater reliability for qualitative items.
Additionally, we analyzed two predictive rates: false positive rate (FPR) and false negative rate (FNR). The FPR measures the rate at which the classifier incorrectly predicts the positive class, while the FNR measures the rate at which the classifier fails to classify positive cases accurately. Subsequently, to assess the computational efficiency of the classifiers, we analyzed the required computation time for accessing individual predictions, which aids in determining the practical applicability of our proposed classifiers. We also evaluated the performance of the classifiers using the log loss metric, which measures the logarithmic loss between the predicted probabilities and the actual labels.
Moreover, the Mann-Whitney U statistic is a non-parametric test employed to determine whether there is a significant difference between two independent groups in terms of variable distribution. It assesses whether two independent samples come from the same population or have similar distributions. Mathematically illustrated in Eq. (12), the statistical value U can be calculated using the following formula, where and are the sum of ranks; and are the sizes of the first and second samples, respectively. The total number of possible pairs is used to calculate the maximum possible value of U. After that, it can be compared to the critical value from the Mann-Whitney U distribution or used to calculate the P-value to decide on the null hypothesis.
| (12) |
Furthermore, we used another non-parametric statistical test named Mood's Median Test to determine whether the medians of the two groups are significantly different. It extends the median test for two independent samples to multiple groups. The test counts the number of observations in each group that are both above and below the total group median of the aggregated data. The working formula of this test is stated in Eq. (13). Where and are the observed frequencies of samples above and below the median, respectively; and are the expected frequencies of samples above and below the median, respectively.
| (13) |
3.7. Explainable AI
In ML, explainability refers to elucidating a model's inner workings, specifically understanding the relationship between its input and output and the underlying reasons for such connections. This concept aims to address the black box problem by enhancing the interpretability of models, allowing users to gain insights into their decision-making processes. There are multiple popular methodologies used for model interpretability, such as Local Interpretable Model-agnostic Explanation (LIME), Shapley Additive Explanation (SHAP), Permutation Feature Importance (PFI), and Anchor. However, the LIME and Anchor-based techniques can be sensitive, leading to potential inconsistencies for similar samples [36,37]. These techniques are primarily designed to explain local behaviors [38,39], meaning they provide insights into individual instances without considering the interaction at the global scale. Moreover, PFI values can vary between different runs of the permutation process, leading to instability in feature rankings and interpretations [40]. In contrast, SHAP provides consistency and stable explanations, ensuring the reliability of local behaviors and offers both local and global explanations [41]. Originating from the concepts of cooperative game theory, SHAP values offer a comprehensive way to evaluate the relative relevance of distinct characteristics in prediction models. This is achieved by evaluating each feature's influence on predictions while considering all potential feature combinations and their contributions. By using a feature importance plot that incorporates SHAP values to rank the input features according to their impacts, the prediction outcomes of the model are evaluated. The contribution of each input characteristic to the final estimate for the data instance is quantified by Eq. (14). Where f refers to a feature, x is a data instance, is the calculated SHAP value, F indicates the subset of f, refers the size of F, xF is the outcome of predicted model for x with F, and refers to the outcome of predicted model for x with F excluding f.
| (14) |
Although SHAP has numerous advantages over other interpretable techniques, it has some drawbacks. Specifically, it can be computationally expensive and may not always provide intuitive explanations in all cases. Additionally, interpreting the influential characteristics and their corresponding values can be challenging. We employed carefully optimized parameters to address such limitations while developing our proposed classifiers. This optimization aimed to reduce the computational complexity of our models calculating SHAP values. Regarding the other limitations, we provided additional context and domain-specific knowledge in the subsection dedicated to developing an interpretable model. By incorporating relevant information and explanations, we aimed to bridge the gap between the SHAP explanations and their practical understanding. Also, we employed interactive plots that effectively represent the SHAP values and highlight the influential characteristics. Through this visualization, we aimed to make complex information more accessible and facilitate user interpretation. Furthermore, we provided comparative visualizations of two different examples, which contribute to a better understanding of the model's behavior and provide valuable insights into the factors driving the model's predictions.
4. Experiment and results
This section has conducted a comprehensive comparative analysis to evaluate the models that were used using various performance indicators across three distinct feature sets. This analysis aims to identify the classifier with the feature set that yields the highest performance. Subsequently, an interpretable thyroid classification model was developed by incorporating SHAP and the highest-performing classifier with the feature set.
4.1. Experimental Setup
Modeling experiments were performed on computer hardware featuring an Intel Core i3 processor, 10th generation, running at 3.3 GHz with 4 GB of RAM to evaluate the performance of both the proposed and baseline classifiers. Colab Notebook, a cloud-based Jupyter Notebook environment, provided the framework for creating and testing the approaches. The availability of several libraries—such as Scikit-learn, Matplotlib, Pandas, and NumPy—that are necessary for machine learning models made this decision easier.
4.2. Analysis of the various performance scores
Several classification scores were measured, including ACC, PRE, REC, F1S, AUC, and CKS to compare the different feature sets. Table 5 presents a comparative representation of these performed scores for the Kaggle dataset. This table demonstrates both RDKVT and RDKST are able to obtain high and generalized scores. Specifically, the RDKST achieved the highest ACC of 0.9898 when utilizing the IGS-selected features. Similarly, the RDKST achieved a robust PRE score of 0.9799 with the IGS-selected features, closely followed by the RF classifier with a score of 0.9761. The RDKVT attained the highest REC score of 0.9986 when utilizing the UVS features. Considering the F1S, the KNN obtained the lowest score of 0.8836 with the UVS features. In contrast, the RDKST achieved the highest F1S of 0.9886 with the IGS features. Regarding the AUC score, both hybrid classifiers, RDKVT and RDKST, demonstrated high and generalized scores. Specifically, for the IGS-selected features, the RDKVT classifier achieved an AUC score of 0.9990, while the RDKST achieved an even higher score of 0.9995. On the contrary, the UVS-based features have obtained 0.9958 and 0.9978 AUC scores from RDKVT and RDKST, respectively. Furthermore, when evaluating the CKS, the proposed RDKST outperformed other classifiers with a score of 0.9794 for IGS-selected features, indicating excellent agreement between the model predictions and the actual class values.
Table 5.
The performed accuracy (ACC), precision (PRE), recall (REC), f1-score (F1S), the area under the curve (AUC), and Cohen's Kappa score (CKS) for the Kaggle dataset.
| Feature sets | Classifiers | ACC | PRE | REC | F1S | AUC | CKS |
|---|---|---|---|---|---|---|---|
| All features |
RF | 0.9561 | 0.9649 | 0.9390 | 0.9517 | 0.9866 | 0.9115 |
| DT | 0.9465 | 0.9323 | 0.9477 | 0.9399 | 0.9704 | 0.8917 | |
| KNN | 0.9375 | 0.9047 | 0.9537 | 0.9286 | 0.9834 | 0.8713 | |
| RDKVT | 0.9618 | 0.9348 | 0.9790 | 0.9564 | 0.9952 | 0.9223 | |
| RDKST |
0.9774 |
0.9624 |
0.9896 |
0.9758 |
0.9957 |
0.9566 |
|
| UVS features |
RF | 0.9662 | 0.9292 | 0.9946 | 0.9611 | 0.9940 | 0.9313 |
| DT | 0.9622 | 0.9523 | 0.9632 | 0.9577 | 0.9867 | 0.9237 | |
| KNN | 0.8947 | 0.8797 | 0.8776 | 0.8836 | 0.8986 | 0.7875 | |
| RDKVT | 0.9741 | 0.9436 | 0.9986 | 0.9703 | 0.9958 | 0.9474 | |
| RDKST |
0.9774 |
0.9719 |
0.9749 |
0.9765 |
0.9979 |
0.9545 |
|
| IGS features | RF | 0.9746 | 0.9761 | 0.9677 | 0.9719 | 0.9939 | 0.9761 |
| DT | 0.9718 | 0.9423 | 0.9947 | 0.9678 | 0.9889 | 0.9428 | |
| KNN | 0.9634 | 0.9260 | 0.9919 | 0.9578 | 0.9936 | 0.9256 | |
| RDKVT | 0.9831 | 0.9674 | 0.9948 | 0.9809 | 0.9990 | 0.9657 | |
| RDKST | 0.9898 | 0.9799 | 0.9974 | 0.9886 | 0.9995 | 0.9794 |
Afterward, Table 6 represents a comparative overview of these performance indicators for the UCI dataset. Regarding ACC, the RDKST again obtained robust scores of 0.9861 with IGS-selected features. The attained rate is nearly comparable to the highest-performing outcome recorded in the Kaggle dataset. Regarding PRE, another proposed RDKVT yields remarkable scores of 0.9888 when employing the same feature set. When concentrating on the REC and F1S scores, it is visible that the RDKST significantly has 0.9894 and 0.9843 scores, respectively. Regarding the AUC score, most of the performing classifiers have demonstrated high and generalized scores with IGS-based selected features. Specifically, the RDKVT and RDKST classifiers achieved 0.9968 and 0.9980 AUC scores, respectively. Furthermore, the proposed RDKST outperformed with a 0.9732 CKS score for IGS-selected features, indicating excellent agreement between the model predictions and the actual class values. These results demonstrate the effectiveness of the RDKST using the IGS features, as indicated by its high ACC, PRE, REC, F1S, AUC, and CKS. The outcomes from different performance metrics suggest that the IGS-based features possess significantly more potential than the UVS-based features.
Table 6.
The performed accuracy (ACC), precision (PRE), recall (REC), f1-score (F1S), the area under the curve (AUC), and Cohen's Kappa score (CKS) for the UCI dataset.
| Feature sets | Classifiers | ACC | PRE | REC | F1S | AUC | CKS |
|---|---|---|---|---|---|---|---|
| All features |
RF | 0.9406 | 0.9523 | 0.9317 | 0.9443 | 0.9802 | 0.9074 |
| DT | 0.9358 | 0.9357 | 0.9334 | 0.9341 | 0.9732 | 0.9005 | |
| KNN | 0.9467 | 0.9494 | 0.9447 | 0.9479 | 0.9870 | 0.9131 | |
| RDKVT | 0.9594 | 0.9618 | 0.9510 | 0.9588 | 0.9936 | 0.9192 | |
| RDKST |
0.9656 |
0.9624 |
0.9695 |
0.9674 |
0.9910 |
0.9526 |
|
| UVS features |
RF | 0.9620 | 0.9704 | 0.9611 | 0.9658 | 0.9883 | 0.9293 |
| DT | 0.9612 | 0.9573 | 0.9661 | 0.9633 | 0.9878 | 0.9210 | |
| KNN | 0.9646 | 0.9707 | 0.9611 | 0.9679 | 0.9886 | 0.9263 | |
| RDKVT | 0.9715 | 0.9823 | 0.9696 | 0.9777 | 0.9905 | 0.9394 | |
| RDKST |
0.9742 |
0.9749 |
0.9725 |
0.9765 |
0.9938 |
0.9486 |
|
| IGS features | RF | 0.9703 | 0.9781 | 0.9691 | 0.9710 | 0.9920 | 0.9526 |
| DT | 0.9667 | 0.9614 | 0.9717 | 0.9686 | 0.9893 | 0.9488 | |
| KNN | 0.9711 | 0.9635 | 0.9820 | 0.9743 | 0.9911 | 0.9542 | |
| RDKVT | 0.9807 | 0.9888 | 0.9776 | 0.9839 | 0.9968 | 0.9610 | |
| RDKST | 0.9861 | 0.9839 | 0.9894 | 0.9843 | 0.9980 | 0.9732 |
4.3. Analysis of misclassified false predictive rates
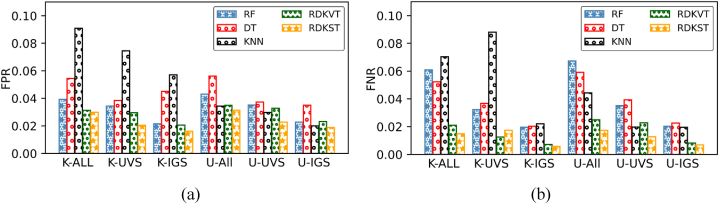
Fig. 5(a) and (b) provide a comparative representation of the FPR and FNR for all performing classifiers. In the case of FPR, KNN exhibits the highest FPR value of 0.0909 with UVS features for the Kaggle dataset. On the other hand, RDKST achieves the lowest FPR value of 0.016. with IGS features. Notably, the RDKST classifier demonstrates effective FPR rates for the UCI dataset, precisely 0.0187 with IGS features. Meanwhile, the DT classifier yields the highest FPR values for all feature sets. Regarding the FNR, RDKVT, and RDKST classifiers attain remarkable values for the Kaggle dataset, 0.0071 and 0.0058, respectively, for the IGS features set. With an identical set of features, these proposed classifiers achieve an admirable FNR rate of 0.0082 and 0.0068 for the UCI dataset. Conversely, the KNN and DT classifiers exhibit the highest FNR rates compared to others.
Fig. 5.
Performed false positive rate (FPR (a)) and false negative rate (FNR (b)) rates for used classifiers on both Kaggle (K) and UCI (U) datasets.
4.4. Analysis of computation time and log loss
Table 7 provides insights into both datasets' computation time and log loss. In the Kaggle dataset, the DT exhibits the lowest computation time of 20.9 ms (ms) when using UVS features. On the other hand, the KNN and RDKVT require a significantly longer time, 508 and 520 ms for ALL features. RDKST shows relatively similar ranges from 250 to 550 ms across the different feature sets. Considering log loss, the KNN shows the highest loss of 3.63 %. In contrast, the RDKST has the lowest losses for all different feature sets. Regarding the UCI dataset, the RDKST demands the highest compilation time of 504 ms for ALL features, while DT takes relatively less time. In the case of the log loss, the RDKST significantly produces only a 0.48 % loss with the IGS-selected features.
Table 7.
Analysis of the computation time (in ms) and log loss (%) for Kaggle and UCI datasets over various feature sets.
| Per-formed classifiers |
Kaggle dataset |
UCI dataset |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compilation time |
Log loss |
Compilation time |
Log loss |
|||||||||
| ALL | UVS | IGS | ALL | UVS | IGS | ALL | UVS | IGS | ALL | UVS | IGS | |
| RF | 36.7 | 32.7 | 26.3 | 1.51 | 1.16 | 0.87 | 33.6 | 29.3 | 24.9 | 1.72 | 1.32 | 0.94 |
| DT | 24 | 20.9 | 21.5 | 1.84 | 1.30 | 0.97 | 30 | 21.7 | 22.3 | 1.97 | 1.39 | 1.09 |
| KNN | 508 | 217 | 224 | 2.15 | 3.63 | 1.26 | 445 | 198 | 202 | 1.63 | 1.22 | 0.88 |
| RDKVT | 520 | 282 | 277 | 1.32 | 0.89 | 0.58 | 478 | 267 | 221 | 1.47 | 0.91 | 0.67 |
| RDKST | 470 | 243 | 257 | 0.73 | 0.77 | 0.34 | 504 | 232 | 249 | 1.25 | 0.83 | 0.48 |
4.5. Analysis of the statistical significance
We have evaluated the Mann-Whitney U statistic to determine whether there is a difference between two independent groups. The P-value is determined by comparing the computed test statistic with a critical value or an approximation derived from the normal distribution. If the P-value falls below a pre-selected significance level (α), we reject the null hypothesis in favor of the alternative hypothesis, suggesting a difference between the paired measurements. The U Test represents the sum of ranks for one of the groups in the comparison. A larger U Test value indicates that the distribution of values for the first group is generally higher than the distribution of values for the second group. Here, we set the α to 0.05 for all different feature subsets on both datasets. It represents the probability of rejecting a true null hypothesis, also known as a Type I error. In other words, the significance level is the threshold used to determine whether the observed results are statistically significant (SGNF). Table 8 represents the P-value, U Test, and SGNF for three different types of features on both datasets. This table demonstrates the sum of ranks is very high for all different classifiers (e.g., the proposed RDKST given 326459 and 275559 for the IGS feature set on both Kaggle and UCI datasets, respectively. The higher the U statistic, the more evidence there is that one group's values tend to be larger than the other. The p-value is close to zero, indicating strong evidence against the null hypothesis. The extremely small P-value (0.0015) suggests a highly significant result, indicating strong evidence to reject the null hypothesis that there is no difference between the two groups. As the P-value is lower than the significance level or α for all subsets with different classifiers, it indicates a significant difference between the two groups and is statistically significant (✓).
Table 8.
Evaluate the statistical significance between two independent groups (thyroid and non-thyroid) by using the Mann-Whitney U Statistical Test.
| Kaggle Dataset | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Employed classifiers | ALL features (α = 0.05) |
UVS features (α = 0.05) |
IGS features (α = 0.05) |
||||||
| P-Value | U Test | SGNF | P-Value | U Test | SGNF | P-Value | U Test | SGNF | |
| RF | 0.0747 | 246124 | (✗) | 0.0416 | 278421 | (✓) | 0.0461 | 318499 | (✓) |
| DT | 0.7638 | 167246 | (✗) | 0.0718 | 229867 | (✗) | 0.0519 | 263345 | (✗) |
| KNN | 1.3413 | 135678 | (✗) | 0.2461 | 197781 | (✗) | 0.0524 | 226578 | (✗) |
| RDKVT | 0.0057 | 195278 | (✓) | 0.0403 | 308754 | (✓) | 0.0497 | 325142 | (✓) |
| RDKST |
0.0023 |
225982 |
(✓) |
0.0021 |
293454 |
(✓) |
0.0015 |
326459 |
(✓) |
|
UCI Dataset | |||||||||
| RF | 0.0675 | 184151 | (✗) | 0.1156 | 198713 | (✗) | 0.0413 | 245638 | (✓) |
| DT | 1.3413 | 107031 | (✗) | 1.1835 | 184907 | (✗) | 0.0575 | 175645 | (✗) |
| KNN | 1.5678 | 118109 | (✗) | 0.8961 | 179337 | (✗) | 1.5876 | 176324 | (✗) |
| RDKVT | 0.0945 | 173192 | (✗) | 0.0509 | 215095 | (✗) | 0.0477 | 246144 | (✓) |
| RDKST | 0.0021 | 167523 | (✓) | 0.0016 | 227552 | (✓) | 0.0025 | 275559 | (✓) |
Moreover, we performed another statistical test called the Mood's Median Test to assess the difference between two paired groups. Table 9 showcases the P-values of different sets of features for both the Kaggle and UCI datasets, where the first value in the P-value column indicates the value for the Kaggle dataset and the last value is for the UCI dataset. In the following column, the significance level is determined based on their corresponding P-values, whether the observed results are statistically significant (SGNF) or not. The sign (✓) indicates the pair of models has statistically significant differences, and the sign (✗) represents the mentioned pair with no significant differences. This table illustrates the very small P-value for our proposed RDKVT and RDKST, indicating strong evidence against the null hypothesis in both Kaggle and UCI datasets.
Table 9.
Evaluate the statistical significance between two independent groups (thyroid and non-thyroid) by using the Mood's Median Test.
| (Kaggle Dataset, UCI Dataset) | ||||||
|---|---|---|---|---|---|---|
| Employed classifiers | ALL features (α = 0.05) |
UVS features (α = 0.05) |
IGS features (α = 0.05) |
|||
| P-Value | SGNF | P-Value | SGNF | P-Value | SGNF | |
| RF | (0.0347, 1.5678) | (✓, ✗) | (0.0716, 1.8182) | (✗, ✗) | (0.0377, 0.1001) | (✓, ✗) |
| DT | (0.7638, 1.3413) | (✗, ✗) | (0.0418, 0.1231) | (✓, ✗) | (0.0661, 1.4555) | (✗, ✗) |
| KNN | (1.4657, 0.9675) | (✗, ✗) | (0.2461, 0.3417) | (✗, ✗) | (0.0519, 0.0891) | (✗, ✗) |
| RDKVT | (0.0513, 0.0425) | (✗, ✓) | (0.0403, 0.0345) | (✓, ✓) | (0.0204, 0.0321) | (✓, ✓) |
| RDKST | (0.0123, 0.0321) | (✓, ✓) | (0.0112, 0.0214) | (✓, ✓) | (0.0041, 0.0035) | (✓, ✓) |
Upon examining the overall results, we can infer that the RDKST classifier demonstrates superior performance for the Kaggle dataset when using IGS-selected features. Hence, our study suggests that the SMOTE-ENN, IGS, and RDKST approaches can be utilized for efficient thyroid detection. Therefore, the proposed interpretable model has been developed using the Kaggle IGS features set and RDKST classifier.
4.6. Developing an interpretable model
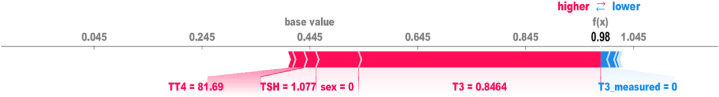
To develop an interpretable model, we have employed the SHAP in conjunction with the RDKST classifier trained in the Kaggle IGS-based selected features. SHAP provides valuable insights into the reasoning behind individual predictions, allowing us to understand why a particular instance is classified in a specific manner. To illustrate this, we have randomly selected two cases, one representing a patient with thyroid disease and the other without it. Fig. 6 presents the most influential features with their values that affect the model's positive prediction. These features include T3, TSH, TT4, and sex, with their corresponding values of 0.8464, 1.077, 81.69, and 0 (female), respectively.
Fig. 6.
Local Explanations generated by SHAP force plot, highlighting the most influential features for a random positive case.
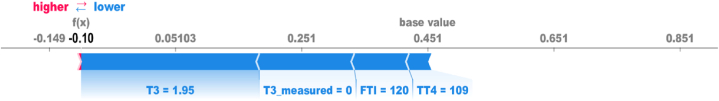
In Fig. 7, the model determines that the patient does not have thyroid disease, influenced by specific feature values. These include a T3 value of 1.95, a T3_measure of 0 (false), an FTI value of 120, and a TT4 value of 109. These feature values play a crucial role in the model's classification process, indicating that they contribute to predicting a negative outcome for thyroid disease. By providing explanations for the model's decisions and identifying the specific features that drive the predictions, we improve the transparency and interpretability of our model. Thus, it strengthens the overall confidence in the model's performance and increases the credibility of our findings.
Fig. 7.
Local Explanations generated by SHAP force plot, highlighting the most influential features for a random negative case.
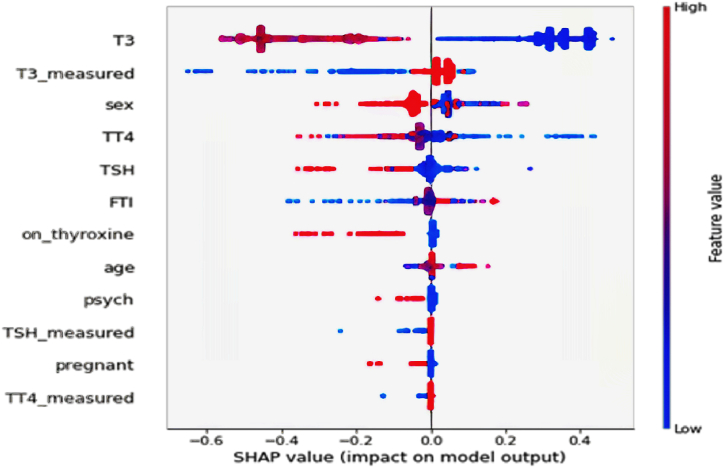
In addition, we utilize a summary plot, such as Fig. 8, to demonstrate the global impact of features on predictions. Higher contributing features are positioned at the top of the plot, with the colors blue, purple, and red signifying low, moderate, and high feature values, respectively. A clear pattern implies that red or purple dots (higher feature values) are related to a decreased risk of thyroid disease, as seen by primarily negative SHAP values. Blue dots representing lower feature values generally indicate a higher disease risk, as positive SHAP values demonstrate. As with local explanations, this figure shows that T3, T3_measured, sex, TT4, TSH, and FTI features significantly impact the dataset's global behaviors.
Fig. 8.
Global Explanations generated by SHAP summary plot, highlighting the overall implications of used features in the diagnosis model.
5. Conclusion
This study signifies a reliable machine-learning model for thyroid disease that will be useful in the medical and healthcare sectors. Multiple preprocessing methods are used to clean the raw dataset and address the imbalance issues with SMOTE-ENN. The results section showed that the proposed RDKST outperformed other classifiers in identifying the disease with the IGS-based selected features. Additionally, different statistical tests were used to emphasize and strengthen overall outcomes. Moreover, we anticipated the local and global factors behind the outcomes using an explainable AI, SHAP. This attempt improves the status of thyroid patient categorization in terms of classification metrics and makes the model's outputs easier for healthcare practitioners to comprehend and interpret. Researchers should conduct further research to test other ensemble methods for classification and utilize different explainable approaches to explore the hidden factors of the disease more thoroughly.
Data availability statement
Two publicly available datasets were utilized in this study. The first dataset was sourced from Kaggle, and the second dataset was obtained from the UCI repository.
Dataset 1 (Kaggle repository): https://www.kaggle.com/datasets/bidemiayinde/thyroid-sickness-determination
Dataset 2 (UCI repository): https://archive.ics.uci.edu/dataset/102/thyroid+disease
CRediT authorship contribution statement
Ananda Sutradhar: Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation. Sharmin Akter: Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation. F M Javed Mehedi Shamrat: Writing – review & editing, Writing – original draft, Visualization, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Pronab Ghosh: Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation. Xujuan Zhou: Writing – review & editing, Validation, Supervision, Project administration, Funding acquisition. Mohd Yamani Idna Bin Idris: Writing – review & editing, Validation, Supervision, Project administration. Kawsar Ahmed: Writing – review & editing, Validation, Supervision, Project administration, Conceptualization. Mohammad Ali Moni: Writing – review & editing, Validation, Supervision, Project administration, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Ananda Sutradhar, Email: anandasutradhar819@gmail.com.
Sharmin Akter, Email: sharmin.happy.25@gmail.com.
F M Javed Mehedi Shamrat, Email: javedmehedicom@gmail.com.
Pronab Ghosh, Email: pghosh1@lakeheadu.ca.
Xujuan Zhou, Email: xujuan.zhou@usq.edu.au.
Mohd Yamani Idna Bin Idris, Email: yamani@um.edu.my.
Kawsar Ahmed, Email: k.ahmed@usask.ca, kawsar.ict@mbstu.ac.bd, k.ahmed.bd@ieee.org.
Mohammad Ali Moni, Email: m.moni@uq.edu.au.
References
- 1.An Y., Hu S., Liu S., Zhao J., Zhang Y.D. The research of automatic classification of ultrasound thyroid nodules. Comput. Model. Eng. Sci. 2021;128(1):203–222. [Google Scholar]
- 2.Abbad Ur Rehman H., Lin C.Y., Mushtaq Z. Effective K-nearest neighbor algorithms performance analysis of thyroid disease. J. Chin. Inst. Eng. 2021;44(1):77–87. [Google Scholar]
- 3.Bloxsom J. Treatment of thyroid cancer with stem cells. Microreviews in Cell and Molecular Biology. 2022;9(4) [Google Scholar]
- 4.Ahmed M.T., Imtiaz M.N., Karmakar A. Analysis of Wisconsin breast cancer original dataset using data mining and machine learning algorithms for breast cancer prediction. Journal of Science Technology and Environment Informatics. 2020;9(2):665–672. [Google Scholar]
- 5.Sutradhar A., Al Rafi M., Shamrat F.J.M., Ghosh P., Das S., Islam M.A., Ahmed K., Zhou X., Azad A.K.M., Alyami S.A., Moni M.A. BOO-ST and CBCEC: two novel hybrid machine learning methods aim to reduce the mortality of heart failure patients. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-48486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dave D., Naik H., Singhal S., Patel P. Explainable ai meets healthcare: a study on heart disease dataset. arXiv preprint arXiv:2011.03195. 2020 [Google Scholar]
- 7.Gupta N., Jain R., Gupta D., Khanna A., Khamparia A. Cognitive Informatics and Soft Computing. Springer; Singapore: 2020. Modified ant lion optimization algorithm for improved diagnosis of thyroid disease; pp. 599–610. [Google Scholar]
- 8.Ahmad W., Ahmad A., Lu C., Khoso B.A., Huang L. A novel hybrid decision support system for thyroid disease forecasting. Soft Comput. 2018;22(16):5377–5383. [Google Scholar]
- 9.Susan S., Kumar A. The balancing trick: optimized sampling of imbalanced datasets—a brief survey of the recent State of the Art. Engineering Reports. 2021;3(4) [Google Scholar]
- 10.Alyas T., Hamid M., Alissa K., Faiz T., Tabassum N., Ahmad A. Empirical method for thyroid disease classification using a machine learning approach. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/9809932. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Aversano L., Bernardi M.L., Cimitile M., Iammarino M., Macchia P.E., Nettore I.C., Verdone C. Thyroid disease treatment prediction with machine learning approaches. Procedia Comput. Sci. 2021;192:1031–1040. [Google Scholar]
- 12.Lin M., Zhu X., Hua T., Tang X., Tu G., Chen X. Detection of ionospheric scintillation based on xgboost model improved by smote-enn technique. Rem. Sens. 2021;13(13):2577. [Google Scholar]
- 13.Sutradhar A., Al Rafi M., Alam M.J., Islam S. An early warning system of heart failure mortality with combined machine learning methods. Indonesian Journal of Electrical Engineering and Computer Science. 2023;32(2):1115–1122. [Google Scholar]
- 14.Sonuç E. vol. 1963. IOP Publishing; 2021, July. Thyroid disease classification using machine learning algorithms. (Journal of Physics: Conference Series). 1. [Google Scholar]
- 15.Srivastava R., Kumar P. Proceedings of Second International Conference on Computing, Communications, and Cyber-Security. Springer; Singapore: 2021. BL_SMOTE ensemble method for prediction of thyroid disease on imbalanced classification problem; pp. 731–741. [Google Scholar]
- 16.Chaubey G., Bisen D., Arjaria S., Yadav V. Thyroid disease prediction using machine learning approaches. Natl. Acad. Sci. Lett. 2021;44(3):233–238. [Google Scholar]
- 17.Savcı E., Nuriyeva F. Diagnosis of thyroid disease using machine learning techniques. Journal of Modern Technology and Engineering. 2022;7(2):134–145. [Google Scholar]
- 18.Olatunji S.O., Alotaibi S., Almutairi E., Alrabae Z., Almajid Y., Altabee R., Altassan M., Ahmed M.I.B., Farooqui M., Alhiyafi J. Early diagnosis of thyroid cancer diseases using computational intelligence techniques: a case study of a Saudi Arabian dataset. Comput. Biol. Med. 2021;131 doi: 10.1016/j.compbiomed.2021.104267. [DOI] [PubMed] [Google Scholar]
- 19.Dharamkar B., Saurabh P., Prasad R., Mewada P. Progress in Computing, Analytics and Networking. Springer; Singapore: 2020. An ensemble approach for classification of thyroid using machine learning; pp. 13–22. [Google Scholar]
- 20.Yadav D.C., Pal S. To generate an ensemble model for women thyroid prediction using data mining techniques. Asian Pac. J. Cancer Prev. APJCP: Asian Pac. J. Cancer Prev. APJCP. 2019;20(4):1275. doi: 10.31557/APJCP.2019.20.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra S., Mallick P.K., Tripathy H.K., Jena L., Chae G.S. Stacked KNN with hard voting predictive approach to assist hiring process in IT organizations. Int. J. Electr. Eng. Educ. 2021 [Google Scholar]
- 22.Xu Z., Shen D., Nie T., Kou Y. A hybrid sampling algorithm combining M-SMOTE and ENN based on random forest for medical imbalanced data. J. Biomed. Inf. 2020;107 doi: 10.1016/j.jbi.2020.103465. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed T., Wijewardane N.K., Lu Y., Jones D.S., Kudenov M., Williams C., Villordon A., Kamruzzaman M. Advancing sweetpotato quality assessment with hyperspectral imaging and explainable artificial intelligence. Comput. Electron. Agric. 2024;220 [Google Scholar]
- 24.Sengupta D., Mondal S., Raj A., Anand A. Frontiers of ICT in Healthcare: Proceedings of EAIT 2022. Springer Nature Singapore; Singapore: 2023. Binary classification of thyroid using comprehensive set of machine learning algorithms; pp. 265–276. [Google Scholar]
- 25.Naeem A.B., Senapati B., Chauhan A.S., Makhija M., Singh A., Gupta M., Tiwari P.K., Abdel-Rehim W.M. Hypothyroidism disease diagnosis by using machine learning algorithms. International Journal of Intelligent Systems and Applications in Engineering. 2023;11(3):368–373. [Google Scholar]
- 26.Sultana A., Islam R. Machine learning framework with feature selection approaches for thyroid disease classification and associated risk factors identification. Journal of Electrical Systems and Information Technology. 2023;10(1):32. [Google Scholar]
- 27.Sutradhar A., et al. An Intelligent Thyroid Diagnosis System Utilizing Multiple Ensemble and Explainable Algorithms With Medical Supported Attributes. IEEE Trans. Artific. Intel. 2024;5(6):2840–2855. doi: 10.1109/TAI.2023.3327981. [DOI] [Google Scholar]
- 28.Thyroid dataset 2023. https://www.kaggle.com/datasets/bidemiayinde/thyroid-sickness-determination
- 29.Thyroid dataset 2023. https://archive.ics.uci.edu/dataset/102/thyroid+disease
- 30.Plati D.K., Tripoliti E.E., Bechlioulis A., Rammos A., Dimou I., Lakkas L., Watson C., McDonald K., Ledwidge M., Pharithi R., Gallagher J. A machine learning approach for chronic heart failure diagnosis. Diagnostics. 2021;11(10):1863. doi: 10.3390/diagnostics11101863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y., Rao K., Liu J., Han C., Gong L., Chong Y., Liu Z., Xu X. Machine learning algorithms for the prediction of central lymph node metastasis in patients with papillary thyroid cancer. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.577537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talachian M., Fathian M. A model for diagnosis of thyroid disease based on rules extraction using tree algorithms and feature selection. International Journal of Hospital Research. 2022;11(3) [Google Scholar]
- 33.Ahmad W., Huang L., Ahmad A., Shah F., Iqbal A., Saeed A. Thyroid diseases forecasting using a hybrid decision support system based on ANFIS, k-NN and information gain method. J Appl Environ Biol Sci. 2017;7(10):78–85. [Google Scholar]
- 34.Liu W., Wang S., Xia X., Guo M. A proposed heterogeneous ensemble algorithm model for predicting central lymph node metastasis in papillary thyroid cancer. Int. J. Gen. Med. 2022:4717–4732. doi: 10.2147/IJGM.S365725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dissanayake K., Md Johar M.G. Comparative study on heart disease prediction using feature selection techniques on classification algorithms. Applied Computational Intelligence and Soft Computing. 2021:1–17. 2021. [Google Scholar]
- 36.Bansal N., Agarwal C., Nguyen A. Proceedings of the Ieee/cvf Conference on Computer Vision and Pattern Recognition. 2020. Sam: the sensitivity of attribution methods to hyperparameters; pp. 8673–8683. [Google Scholar]
- 37.Ma W., Li K., Wang G. Location-aware box reasoning for anchor-based single-shot object detection. IEEE Access. 2020;8:129300–129309. [Google Scholar]
- 38.Alikhademi K., Richardson B., Drobina E., Gilbert J.E. Can explainable AI explain unfairness? A framework for evaluating explainable AI. arXiv preprint arXiv:2106.07483. 2021 [Google Scholar]
- 39.Markus A.F., Kors J.A., Rijnbeek P.R. The role of explainability in creating trustworthy artificial intelligence for health care: a comprehensive survey of the terminology, design choices, and evaluation strategies. J. Biomed. Inf. 2021;113 doi: 10.1016/j.jbi.2020.103655. [DOI] [PubMed] [Google Scholar]
- 40.Strobl C., Boulesteix A.L., Zeileis A., Hothorn T. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinf. 2007;8:1–21. doi: 10.1186/1471-2105-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W., Suominen H., Liu T., Rice G., Salomon C., Barnard A.S. Explainable discovery of disease biomarkers: the case of ovarian cancer to illustrate the best practice in machine learning and Shapley analysis. J. Biomed. Inf. 2023;141 doi: 10.1016/j.jbi.2023.104365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Two publicly available datasets were utilized in this study. The first dataset was sourced from Kaggle, and the second dataset was obtained from the UCI repository.
Dataset 1 (Kaggle repository): https://www.kaggle.com/datasets/bidemiayinde/thyroid-sickness-determination
Dataset 2 (UCI repository): https://archive.ics.uci.edu/dataset/102/thyroid+disease