Abstract
Cholesterol oxidase represents a novel type of insecticidal protein with potent activity against the cotton boll weevil (Anthonomus grandis grandis Boheman). We transformed tobacco (Nicotiana tabacum) plants with the cholesterol oxidase choM gene and expressed cytosolic and chloroplast-targeted versions of the ChoM protein. Transgenic leaf tissues expressing cholesterol oxidase exerted insecticidal activity against boll weevil larvae. Our results indicate that cholesterol oxidase can metabolize phytosterols in vivo when produced cytosolically or when targeted to chloroplasts. The transgenic plants exhibiting cytosolic expression accumulated low levels of saturated sterols known as stanols, and displayed severe developmental aberrations. In contrast, the transgenic plants expressing chloroplast-targeted cholesterol oxidase maintained a greater accumulation of stanols, and appeared phenotypically and developmentally normal. These results are discussed within the context of plant sterol distribution and metabolism.
Cholesterol oxidase is a bacterial enzyme that has potent insecticidal activity against the cotton boll weevil (Anthonomus grandis grandis Boheman). Upon ingestion, this protein causes developmental arrest and death of boll weevil larvae (Purcell et al., 1993), and a marked decrease in fecundity of female adult boll weevils (Greenplate et al., 1995). The enzyme also exhibits more moderate insecticidal effects when ingested by several species of lepidopteran cotton insect pests including tobacco budworm (Heliothis virescens), corn earworm (Helicoverpa zea), and pink bollworm (Pectinophora gossypiella; Greenplate et al., 1997). These lepidopteran pests can be effectively controlled using transgenic cotton plants that express the Cry1Ac δ-endotoxin from Bacillus thuringiensis (B.t.; Perlak et al., 1990) or by chemical insecticide sprays. Boll weevil populations are more difficult to control with chemical sprays due to the fact that eggs are deposited within immature flower buds (squares) or cotton fruit (bolls). The larvae feed and develop enclosed within these organs, protected from chemical spray applications. Furthermore, neither the Cry1Ac δ-endotoxin, nor any other currently known B.t. toxin, has sufficient insecticidal activity against boll weevil to achieve commercial control using transgenic plant strategies. Because of the particularly potent effects of cholesterol oxidase on boll weevil larvae and adults, cholesterol oxidase genes may be useful in the production of genetically modified cotton plants that are self-protected against boll weevil infestation. Toward this end, we previously cloned, sequenced, and expressed in Escherichia coli and tobacco (Nicotiana tabacum) plant protoplasts an insecticidally active cholesterol oxidase gene, choM (Corbin et al., 1994).
The lethal effect of cholesterol oxidase on boll weevil larvae has been attributed to the oxidation of cholesterol in the midgut epithelial membrane, resulting in physical and functional disruption of the membrane (Purcell et al., 1993). In vitro, cholesterol oxidase can catalyze the oxidation of cholesterol and other 3-hydroxysterols, resulting in production of the corresponding 3-ketosteroids and hydrogen peroxide (Smith and Brooks, 1976; Fig. 1). We have demonstrated that the enzyme also oxidized cholesterol in isolated boll weevil midgut brush border membrane vesicles, with concomitant alterations in activity of brush border membrane vesicles marker enzymes (Shen et al., 1997). These studies support a direct mode of action for cholesterol oxidase that involves enzymatic oxidation of midgut epithelial membrane cholesterol.
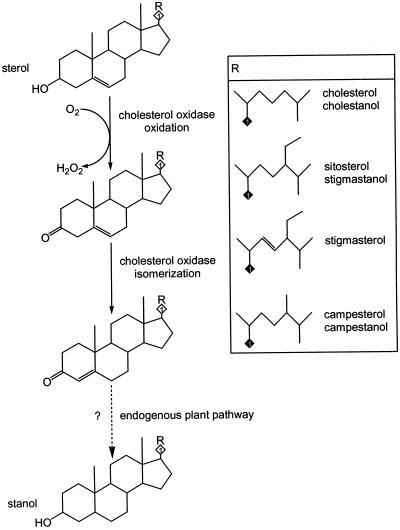
Figure 1.
Cholesterol oxidase reaction and proposed pathway for cholesterol oxidase-dependent sterol metabolism in transgenic tobacco plants. Oxidation and isomerization steps are catalyzed directly by cholesterol oxidase. Reduction of the 3-ketosteroids to stanols occurs by an uncharacterized endogenous plant pathway. The aliphatic groups in the side box show the side chain structures of the four corresponding 3-hydroxysterols detected in this study. Stanol derivatives with side chain structures corresponding to cholesterol (cholestanol), sitosterol (stigmastanol), and campesterol (campestanol) were observed in transgenic tobacco expressing cholesterol oxidase. No steroid was observed having both the unsaturated stigmasterol side chain and a saturated ring nucleus. Complete reduction of stigmasterol leads to a structure with a side chain identical to that found in sitosterol and stigmastanol.
In contrast to the δ-endotoxin insecticidal proteins from B. thuringiensis, which have no known enzymatic activity, cholesterol oxidase is an insecticidal protein that has a well-known enzymatic property. For cholesterol oxidase to be successfully deployed in transgenic plants, its effects on plant sterols in vivo need to be evaluated and appropriate expression strategies need to be devised. Plants have a characteristically complex sterol mixture, with as many as 61 sterols and pentacyclictriterpenes having been identified in maize (Zea mays; Guo et al., 1995). The common phytosterols sitosterol, stigmasterol, and campesterol (24-methyl-cholesterol) predominate within most higher plant species. Sitosterol and campesterol are proposed to regulate membrane fluidity and permeability, similar to cholesterol in mammalian cell membranes, whereas stigmasterol may be required for cell proliferation (Goad, 1990; Schuler, 1990 and 1991). Recent in vitro evidence suggests that plant sterols are able to modulate the activity of plasma membrane H+-ATPase from maize roots (Grandmougin-Ferjani et al., 1997). Cholesterol was found to stimulate proton pumping, whereas a variety of other sterols tested behaved as inhibitors. The requirement for sterols within higher plant membranes is obvious; however, little information exists regarding the levels of sterol required for maintenance of physiological function or if particular mixtures of sterols are required for successful growth and reproduction of higher plants. Because cholesterol oxidase can oxidize 3-hydroxysterols, including the common plant sterols sitosterol, stigmasterol, campesterol, and cholesterol (Fig. 1), the production of cholesterol oxidase in plant cells may result in oxidation of plant sterols and may have detrimental effects on plant growth and development.
In this paper, we report the stable transformation of tobacco plants with a cholesterol oxidase gene and demonstrate expression of insecticidally active cholesterol oxidase in plants using several transcriptional promoters. Expression of cytosolic and chloroplast-targeted forms of cholesterol oxidase is described, and we compare their effects on plant sterol metabolism and phenotype.
RESULTS
Construction of Cholesterol Oxidase Plant Transformation Vectors
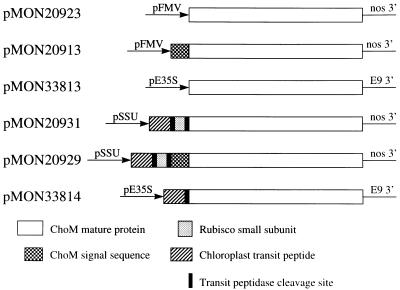
Figure 2 summarizes the composition of the choM plant gene expression cassettes contained in the plant transformation vectors. Plant transformation vectors pMON20923 and pMON20913 carried the mature and full-length forms of the choM gene, respectively, under control of the figwort mosaic virus promoter. Vector pMON33813 carried the mature choM gene under control of the e35S promoter. Plant transformation vectors pMON20931 and pMON20929 carried the mature and full-length forms of the gene, respectively, translationally fused at their N termini to the chloroplast-targeting peptide (CTP) sequence CTP1. In pMON20931 and pMON20929, the chloroplast-targeted versions of the gene were placed under transcriptional control of the Arabidopsis SSU promoter. In transformation vector pMON33814, the mature choM gene was translationally fused at its N terminus to CTP1Δ and placed under transcriptional control of the e35S promoter.
Figure 2.
Cholesterol oxidase plant gene expression cassettes. pMON numbers adjacent to each cassette designate binary plant transformation vectors carrying that particular choM gene cassette. Identity of gene segments and gene expression elements are indicated. White segments represent the coding region of the mature, secreted ChoM protein. Checkered segments represent the 43-amino acid secretory signal sequence of ChoM. Slashed segments represent the chloroplast-targeting sequence from the Arabidopsis 1A Rubisco small subunit (SSU) gene. Dotted segments represent the N-terminal 24-amino acid sequence from the Arabidopsis 1A Rubisco SSU gene. Solid segments represent the chloroplast transit peptide peptidase cleavage sequence from the Arabidopsis 1A Rubisco SSU gene. pFMV, Figwort mosaic virus promoter; pE35S, enhanced cauliflower mosaic virus promoter; pSSU, Rubisco SSU promoter.
Characterization of Nontargeted ChoM Expression in Transgenic Tobacco Plants
Cholesterol oxidase-expressing R0 tobacco plants that had been transformed with nontargeted choM vectors pMON20913 and pMON20923 were initially identified by qualitative western-blot analysis using anti-ChoM antibodies. Expression levels in leaves of these plants were estimated to range from 5 to 15 μg ChoM g−1 fresh weight using the cholesterol oxidase enzyme activity assay. Cholesterol oxidase-expressing R0 tobacco plants transformed with the nontargeted choM vector pMON33813 were identified by quantitative ELISA. In a R0 population of 27 plants transformed using pMON33813, ChoM expression levels in leaves ranged from 0 to 24 μg ChoM g−1 fresh weight, with an average level of 12.42 ± 6.49 μg ChoM g−1 fresh weight among the 19 expressing plants. Because cholesterol oxidase-expressing plants derived from the nontargeted vectors pMON20913, pMON20923, and pMON33813 were all sterile (see below), analysis of expression in these cases was limited to the R0 generation.
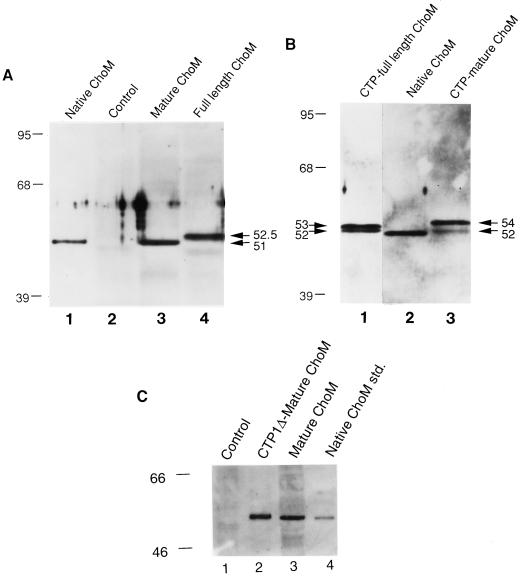
Western-blot analysis was performed to determine the sizes of the ChoM proteins produced in the nontargeted transgenic tobacco plants. Plants derived from pMON20923 and pMON33813, which each encode the nontargeted, mature form of ChoM, produced a band that migrated on SDS-PAGE gels with an Mr of 51,000, very similar to the mobility of native, secreted cholesterol oxidase (Fig. 3A, lane 3, and Fig. 3C, lane 3). Plants derived from pMON20913, the vector encoding the nontargeted, full-length form of ChoM, produced a band that migrated with an Mr of 52,500 (Fig. 3A, lane 4). Thus, the cleavage of the 43-amino acid secretory signal, which has a predicted molecular mass of approximately 5,000 D, appears to have occurred at a site upstream of the native cleavage site and was not completely removed from the primary translation product of the full-length gene in plant cells. The subcellular location of the processed form of the nontargeted, full-length protein was not determined in this study.
Figure 3.
Characterization of ChoM proteins produced in transgenic tobacco plants by western-blot analysis. A, Nontargeted ChoM proteins in crude leaf extracts. Mature ChoM was encoded by pMON20923. Full-length ChoM was encoded by pMON20913. B, Chloroplast-targeted ChoM proteins in crude leaf extracts. CTP-mature ChoM was encoded by pMON20931. CTP full-length ChoM was encoded by pMON20929. C, Chloroplast-targeted and nontargeted ChoM proteins in crude leaf extracts. Mature ChoM was encoded by pMON33813. CTP1Δ mature ChoM was encoded by pMON33814. Lanes designated Native ChoM contain purified, secreted bacterial cholesterol oxidase as a standard. Lanes designated Control contain extracts from plants transformed with a vector that lacks the choM gene. Numbers on the left without arrows indicate positions of protein Mr size standards in kilodaltons. Numbers with arrows indicate relative mobilities, in kilodaltons, of specific ChoM protein bands.
Characterization of Chloroplast-Targeted ChoM Expression in Transgenic Tobacco Plants
Cholesterol oxidase-expressing R0 transgenic plants transformed with the chloroplast-targeted vectors pMON20929, pMON20931, and pMON33814 were identified by western blot or ELISA. Most of these plants were fertile, and expression levels were quantitated by ELISA in the R1 generation. Leaf expression levels in R1 plants derived from two lines each of pMON20929 and pMON20931 ranged from 6 to 30 μg ChoM g−1 fresh weight. Three independent plant lines derived from transformation with pMON33814 that exhibited normal Mendelian segregation of cholesterol oxidase expression were also analyzed. In a population of 15 R1 plants, five from each of the three independent lines, expression levels ranged from 38 to 69 μg ChoM g−1 fresh weight, with an average of 50.8 ± 8.8 μg ChoM g−1 fresh weight.
Western-blot analysis was performed to characterize the sizes of the ChoM proteins produced in plants using the chloroplast-targeted versions of the choM gene. The apparent sizes of the ChoM proteins produced in the chloroplast-targeted plants depended upon which chloroplast-targeting peptide was used and whether the mature or full-length choM gene was used. In the cases of pMON20929 and pMON20931, where CTP1 was used, two bands were consistently observed (Fig. 3B, lanes 1 and 3). In each case, one band migrated with an Mr of 52,000, migrating only slightly slower than native, mature cholesterol oxidase (Fig. 3B, lane 2). The relative mobility of the second band differed between plants transformed with the two vectors, and thus depended upon whether the mature (pMON20931) or full-length (pMON20929) choM gene was used. With the full-length gene the upper band migrated with an Mr of 53,000 (Fig. 3B, lane 1). With the mature gene (pMON20931) the upper band migrated with an Mr of 54,000 (Fig. 3B, lane 3). In the case of pMON33814, which used CTP1Δ and the mature choM gene, only one ChoM band was observed in western blots (Fig. 3C, lane 2). This band migrated only slightly slower than native, mature cholesterol oxidase (Fig. 3C, lane 4) and the nontargeted mature ChoM from pMON33813 (Fig. 3C, lane 3).
Characterization of Chloroplast-Targeted ChoM in Isolated Tobacco Chloroplasts
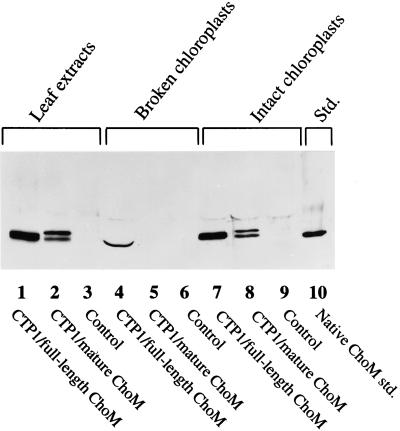
Intact and broken (stripped of the outer membrane and stromal contents) chloroplast fractions from tobacco plants transformed with pMON20929 (full length) and pMON20931 (mature) were subjected to western-blot analysis (Fig. 4). Two ChoM bands were observed in the intact chloroplast fractions that comigrated with the two ChoM bands observed in crude leaf extracts (Fig. 4, lanes 1, 2, 7, and 8). The broken chloroplast fraction from plants transformed with pMON20929 was found to contain only one ChoM protein band, which migrated closely with the lower of the two bands observed in crude leaf extracts (Fig. 4, lane 4). ChoM protein was not detected in the broken chloroplast fraction from plants transformed with pMON20931 (Fig. 4, lane 5). ChoM proteins were not detected in leaf tissue or chloroplasts from control plants that had been transformed with a control vector lacking the choM gene (Fig. 4, lanes 3, 6, and 9). These results demonstrate that the CTP1 element used in these vectors does result in localization of cholesterol oxidase to chloroplasts in tobacco. Without the bacterial secretory signal sequence, ChoM is localized to the stroma and thus is not detected in the broken chloroplast preparation. The presence of the bacterial secretory signal sequence in the full-length, chloroplast-targeted gene causes cholesterol oxidase to accumulate in the broken chloroplast fraction, suggesting that this element functions to transport ChoM across the thylakoid membrane.
Figure 4.
Characterization of chloroplast-targeted ChoM proteins in chloroplast fractions of transgenic tobacco plants by western-blot analysis. Proteins in whole leaf extracts are included for comparison. Lanes designated Control represent samples that had been transformed with a control vector that lacks the choM gene. Lane 10 labeled Native ChoM std contains purified, secreted bacterial cholesterol oxidase. CTP1/full-length ChoM was encoded by pMON20929. CTP1/mature ChoM was encoded by pMON20931.
Phenotypic Characterization of Transgenic ChoM Tobacco Plants
As mentioned above, tobacco plants expressing cholesterol oxidase that had been transformed with vectors carrying nontargeted choM genes, pMON20913, pMON20923, and pMON33813, were sterile and did not develop floral buds. Nontargeted plants also consistently exhibited severe vegetative abnormalities. The plants were severely stunted, and when compared with control plants were characterized by thick, wrinkled leaves and shortened internodal regions.
Tobacco plants expressing cholesterol oxidase that had been transformed with vectors carrying chloroplast-targeted choM genes, pMON20929, pMON20931, and pMON33814, were indistinguishable from control plants that were not transformed or that had been transformed with an empty transformation vector lacking the choM gene. The chloroplast-targeted cholesterol oxidase plants appeared morphologically and developmentally normal and they were fertile. Figure 5 shows representative ChoM-expressing plants transformed with a nontargeted choM vector (pMON33813) and a chloroplast-targeted choM vector (pMON33814), and compares them with a plant derived from a control vector lacking the choM gene.
Figure 5.
Phenotype of R0 tobacco plants transformed with nontargeted and chloroplast-targeted choM genes. The nontargeted plant on the left was transformed with the choM gene from pMON33813. The chloroplast-targeted plant on the right was transformed with the choM gene from pMON33814. The control plant in the center was transformed with a vector lacking a choM gene.
Insecticidal Activity of ChoM-Expressing Transgenic Plants
Because boll weevil larvae do not feed on whole tobacco plants or fresh tobacco tissues, leaf tissue from ChoM-expressing tobacco plants was incorporated in insect artificial diet bioassays to demonstrate that the ChoM protein produced in plants was insecticidally active. Table I shows that the transformed plant tissue expressing chloroplast targeted mature (pMON20931) and chloroplast targeted full-length (pMON20929) choM genes had potent insecticidal activity against boll weevil larvae. Levels of ChoM in these plant lines ranged from 5 to 15 μg ChoM g−1 fresh weight, giving a final dietary concentration of ChoM of approximately 0.5 μg ChoM g−1 fresh weight when sample lyophilization, resuspension, and dilution are taken into account. The ChoM-producing tobacco tissue caused mortality and severe developmental stunting of neonate boll weevil larvae compared with leaf tissue from control plants that had been transformed with a vector lacking the choM gene.
Table I.
Insecticidal activity of R1 transgenic tobacco leaf tissue on boll weevil larvae
| Plant Sample | Mortality | No. of Survivors | Mean Larval Wt | Surviving Biomass |
|---|---|---|---|---|
| % | mg | |||
| Control 1 | 14 | 13 | 12.5 ± 2.9 | 175.0 |
| Control 2 | 1 | 15 | 13.7 ± 3.2 | 205.5 |
| Control 3 | 0 | 15 | 13.0 ± 3.0 | 208.0 |
| Control 4 | 0 | 16 | 7.3 ± 2.7 | 116.8 |
| Control 5 | 21 | 12 | 11.5 ± 3.0 | 138.0 |
| Control 6 | 0 | 18 | 11.3 ± 2.8 | 203.4 |
| pMON20929-1 | 74 | 4 | 0.4 ± 0.1 | 1.6 |
| pMON20929-2 | 87 | 2 | 0.3 ± 0.2 | 0.6 |
| pMON20929-3 | 67 | 4 | 0.8 ± 0.2 | 4.0 |
| pMON20931-1 | 54 | 7 | 0.3 ± 0.1 | 2.1 |
| pMON20931-2 | 61 | 6 | 0.4 ± 0.1 | 2.4 |
| pMON20931-3 | 67 | 5 | 0.7 ± 0.3 | 3.5 |
Neonate boll weevil larvae were allowed to feed on each tobacco leaf tissue sample that had been applied to the surface of artificial insect diet. After 6 d, the no. of surviving larvae and their wts were determined.
Chemical Characterization of Transgenic Tobacco Plants
The cholesterol oxidase enzyme catalyzes the oxidation of 3-hydroxysterols in vitro, resulting in production of the corresponding 3-ketosteroids and hydrogen peroxide (Fig. 1). Because we observed differences in the plant phenotype conferred by the chloroplast targeted and cytosolic versions of ChoM, we conducted an analysis of the steroid composition of the transgenic tobacco to determine if the phenotypic differences could be correlated with possible differences in sterol metabolism. We found that the ChoM-producing tobacco tissues did not accumulate detectable levels of 3-ketosteroids. However, stanols, which are possible products of 3-ketosteroid metabolism, did accumulate in all ChoM producing tobacco tissue. It appears likely that a proportion of the tobacco sterol pool, including sitosterol, stigmasterol, campesterol, and cholesterol, is oxidized to the 3-ketosteroid metabolite and reduced by endogenous enzymes to produce the observed stanols (Fig. 1). The reduction of the 3-ketosteroids derived from the oxidation of sitosterol and stigmasterol by ChoM result in the accumulation of the same product, stigmastanol. Therefore, within the context of the results and discussion of this study, the level of stigmastanol will be compared with the levels of both sitosterol and stigmasterol.
Plants expressing nontargeted ChoM contained the expression cassette from pMON33813. The nontargeted analysis group consisted of three samples designated group A for statistical purposes. These samples were taken from three different R0 plants, each produced by an independent transformation event.
Plants expressing chloroplast-targeted ChoM contained the expression cassette from pMON33814. Three independent transgenic tobacco lines expressing chloroplast-targeted ChoM were separated into groups B, C, and D for statistical purposes. The chloroplast-targeted transgenic lines were separated into ChoM-positive and ChoM-negative R1 plant groups. The ChoM-negative plants segregating in the R1 generation served as a comparison group to the ChoM-positive groups. Control tissues discussed throughout the study were untransformed wild-type tobacco.
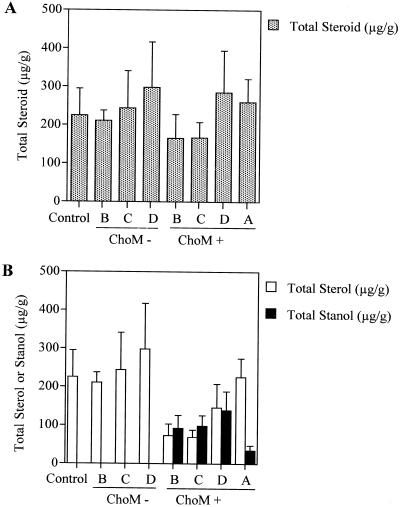
Within all transgenic lines analyzed, the average concentration of steroid assessed on a total micrograms of steroid per gram fresh weight did not differ significantly from wild-type controls (Student's t test, P > 0.3; Fig. 6A). The designation of steroid refers to the total sterol accumulation combined with the total stanol accumulation within a given tissue sample. Control tissues and ChoM-negative transgenic plant lines contain undetectable levels of stanol within our system; therefore, the combined accumulation of stanol and sterol within transgenic tissues was compared with the accumulation of sterol in control and ChoM-negative tissue. The average level of steroid per group varied from a high of 298 (sem 118) μg steroid g−1 fresh weight within group D ChoM-negative to a low of 165 (sem 62) μg steroid g−1 fresh weight within group B ChoM-positive. Wild-type controls accumulate on average 224 (sem 70) μg steroid g−1 fresh weight (Fig. 6A). Group A tissue accumulated on average a greater amount of total steroid than control tissues, whereas groups B, C, and D demonstrated variability in total steroid accumulation when compared with controls (Fig. 6A). On average, stanol production was greatest in groups B, C, and D ChoM-positive plants, whereas group A demonstrated an approximately 3-fold lower accumulation of stanol than that observed for groups B, C, and D ChoM-positive (Fig. 6B). Groups B, C, and D ChoM-negative and control tissues produced undetectable levels of stanol (Fig. 6B).
Figure 6.
Total steroid content of transgenic and control tobacco. All data is represented as average micrograms of steroid per gram fresh weight with calculated ses of the mean. A, Total steroid content for control and groups A, B, C, and D. B, Total sterol and stanol content for control and groups A, B, C, and D. Group A plants express the nontargeted choM gene from pMON33813. Group B, C, and D ChoM+ represent R1 transgenic plant lines, which express the chloroplast-targeted choM gene from pMON33814. Group B, C, and D ChoM represent segregating, non-ChoM-expressing R1 transgenic plant lines derived from R0 plants originally transformed using pMON33814. Control represents non-transformed tobacco of the same developmental age as experimental lines.
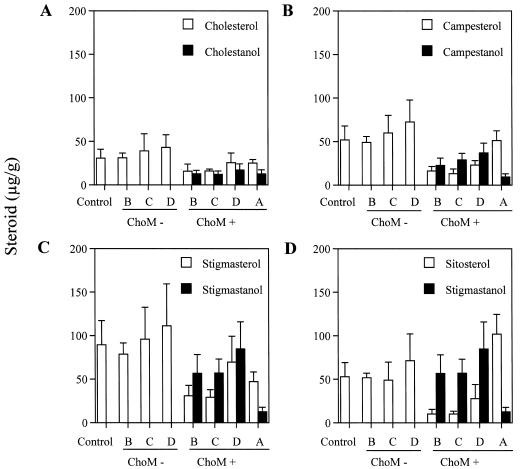
Stanol accumulation was obvious in groups B, C, and D ChoM-positive and group A (Fig. 7, A–D). Comparisons of individual sterol and stanol levels on a micrograms of steroid per gram fresh weight basis within the stanol producing lines demonstrates a consistent pattern of stanol accumulation. Group A tissues, in each instance, accumulate a greater level of sterol than the corresponding stanol (Fig. 7, A–D). However, group B, C, and D ChoM-positive tissues accumulate higher levels of campestanol and stigmastanol than the corresponding sterols campesterol, stigmasterol, or sitosterol. Cholesterol levels are equal to or exceed cholestanol levels in these same tissues (Fig. 7A). Individual sterol accumulations within group B, C, and D ChoM-negative tissue in all instances exceed the level of corresponding sterol accumulated in group B, C, and D ChoM-positive lines (Fig. 7, A–D).
Figure 7.
Individual sterol and stanol contents of transgenic and control tobacco lines. All data is represented as average micrograms of steroid per gram fresh weight with calculated ses of the mean. A, Total cholesterol and cholestanol levels for control and groups A, B, C, and D. B, Total campesterol and campestanol levels for control and groups A, B, C, and D. C, Total stigmasterol and stigmastanol levels for control and groups A, B, C, and D. D, Total sitosterol levels compared to stigmastanol for control and groups A, B, C, and D. The complete reduction of sitosterol and stigmasterol produce stigmastanol; therefore, stigmastanol will be used in comparison to both sterols. Group A plants express the nontargeted choM gene from pMON33813. Group B, C, and D ChoM+ represent R1 transgenic plant lines which express chloroplast-targeted choM gene from pMON33814. Group B, C, and D ChoM- represent segregating R1 transgenic plant lines derived from R0 plants transformed using pMON33814, but that do not express ChoM. Control represents nontransformed tobacco of the same developmental age as the experimental lines.
DISCUSSION
This report demonstrates that a cholesterol oxidase gene originally isolated from an actinomycete can be expressed in transgenic tobacco plants. The structural gene used in these studies had been minimally modified from its native sequence to contain restriction enzyme sites at the N and C termini to facilitate cloning and, in the case of the chloroplast-targeted cholesterol oxidase, a chloroplast-targeting peptide sequence has been added. It was shown previously that when native genes encoding insecticidal proteins such as the B.t. toxins from B. thuringiensis were expressed in plants, relatively low levels of expression were achieved (Fischhoff et al., 1987; Vaeck et al., 1987). Weak expression of native B.t. genes was attributed to their high A + T content (approximately 63%), which resulted in an under-representation of plant preferred codons and the occurrence of mRNA destabilization and polyA addition recognition sequence motifs. Modified and synthetic versions of B.t. genes with a lower A + T content were used to increase expression levels and improve effectiveness of insect control in transgenic plant tissues (Perlak et al., 1990, 1991, 1993). In contrast to B.t. genes, the A + T content in the native actinomycete cholesterol oxidase choM gene is only 31%. As a result, this gene in its native form was more amenable to expression in plants, and expression levels in leaf tissues routinely ranged from approximately 5 to 50 μg ChoM g−1 fresh weight.
In the absence of a chloroplast-targeting sequence, cholesterol oxidase expression resulted in severe abnormalities in plant development and fertility. Similar phenotypic effects were observed with either the mature or the full-length cholesterol oxidase. When expressed as a fusion with a chloroplast-targeting peptide, expression of the mature and the full-length cholesterol oxidases did not cause the deleterious phenotypic effects observed with untargeted cholesterol oxidase. Amelioration of the abnormal morphological phenotype was observed in every instance in which cholesterol oxidase was targeted to chloroplasts. The inclusion within CTP1 of 24 amino acids from the N terminus of the SSU of Rubisco resulted in the production of two discernible forms of cholesterol oxidase. The smaller and larger forms were detected in isolated intact chloroplast fractions, indicating that both forms resulted from chloroplast import and processing (Fig. 4). In contrast, CTP1Δ, which lacked the N-terminal 24 amino acids of the SSU of Rubisco, resulted in a single discernible form of cholesterol oxidase when it was used to target either the mature or full-length cholesterol oxidase. Thus, presence of a secretory signal sequence and an SSU Rubisco sequence at the N terminus of the protein affected the nature of the final protein product. The inclusion of the Rubisco SSU peptide has been reported to improve expression and import of some fusion proteins into chloroplasts (Kuntz et al., 1986; Wasmann et al., 1986; Kavanagh et al., 1988; Russell et al., 1993). Although we did not make a quantitative comparison between the use of CTP1 and CTP1Δ in the present study, our results indicate that the Rubisco SSU peptide is not necessary for high ChoM expression levels and that its presence causes some aberrant processing of a significant fraction of the precursor protein. Although leaf tissues expressing chloroplast-targeted mature and full-length cholesterol oxidase exerted insecticidal activity in insect diet overlay bioassays, it was not determined if the fully processed products, the partially processed products, or both, contributed the insecticidal activity.
The chloroplast fractionation experiment showed that the chloroplast-targeted full-length cholesterol oxidase had different chloroplast localization characteristics than did the chloroplast-targeted mature cholesterol oxidase. The smaller of the processed products from the targeted full-length protein, but not from the targeted mature protein, was found to be associated with the broken chloroplast fraction, indicating that the native secretory signal sequence of the full-length protein functioned to transport chloroplast-targeted cholesterol oxidase across the thylakoid membrane. The structural similarity of bacterial protein secretory signal sequences and plant thylakoid-targeting sequences has been noted (von Heijne et al., 1989). The leader domain from plant thylakoid proteins can function to target proteins to the periplasm in E. coli, allowing processing of the secreted precursor proteins (Seidler and Michel, 1990; Meadows and Robinson, 1991). Furthermore, leader peptides of prokaryotic origin can be cleaved by the thylakoid signal peptidase (Halpin et al., 1989). The data we present here shows that a native bacterial signal sequence can function in planta to target a foreign protein to the chloroplast thylakoid compartment.
Stanols have been reported in many groups of organisms, including higher plants (Kanazawa et al., 1978; Applequist et al., 1981; Iida et al., 1981; Teshima and Patterson, 1981). The stanol content ranges from less than 1% of the total sterol pool in most organisms to 100% of the accumulated sterol pool in the slime molds (Nes and McKean, 1977). Stanol metabolism has been described in marine invertebrates and in mammals (Smith and Goad, 1975; Sheikh and Djerassi, 1977; Ballantine and Lavis, 1978). A proposed pathway for the biosynthesis of stanols includes a 3-ketosteroid intermediate, which is similar to the product of cholesterol oxidase (Smith and Goad, 1975; Smith and Brooks, 1976; Salt, 1984). In our analysis of tobacco plants, stanols were not detected in untransformed control plants and ChoM-negative segregating progeny of plants transformed with the choM gene. However, in all plants expressing ChoM, significant accumulation of stanol was observed, demonstrating that a product of cholesterol oxidase was endogenously converted to stanol. Transgenic lines expressing chloroplast-targeted ChoM produce on average 3-fold higher levels of stanols than transgenic tissues expressing nontargeted ChoM (Fig. 6B). The expression levels of chloroplast-targeted ChoM are approximately 4-fold higher than that observed with nontargeted ChoM (50.8 versus 12.4 μg ChoM g−1 fresh weight), suggesting that the differences in the observed stanol levels may be primarily due to greater ChoM accumulation in the chloroplast-targeted plants. The present study did not address the levels of conjugated sterol or stanol and therefore we will not discuss the potential contributions made by these pools to the overall steroid budget within the plant or the observed morphologies.
Average sterol accumulation within group B, C, and D ChoM-negative plants exceeded the level of corresponding sterol accumulated in group B, C, and D ChoM-positive plants (Fig. 7). These data demonstrate that chloroplast-targeted cholesterol oxidase is promoting significant conversion of sterols to corresponding stanols, thereby reducing the total pool of free sterol within the cell.
Chloroplast-targeted ChoM was associated with higher levels of stanols, and these plants maintained a phenotype and ontogeny similar to wild-type tobacco plants. In contrast, cytosolic ChoM resulted in lower levels of stanols, and these plants were characterized by severe stunting and infertility. The major sterol accumulated within wild-type tobacco is stigmasterol, followed by sitosterol, campesterol, and cholesterol in descending order of mass accumulated. The cytolsolic ChoM plant lines maintained a total level of sterol comparable to that observed for control tissues (Figs. 6 and 7). The only difference appears to be a shift in the relative abundance of specific sterols accumulated within the transgenic lines. The lines expressing ChoM cytosolically have an altered ratio of stigmasterol to sitosterol in which sitosterol is now more abundant (Fig. 7). The transgenic lines expressing chloroplast-targeted ChoM exhibit no effects on ontogeny even though these tissues maintain on average 55% of their steroid pool as stanol. Within these chloroplast-targeted ChoM-positive plant lines the relative proportions of free sterols which remained was similar to wild type, suggesting that the ratios of certain free sterols, and not the total level of sterol, contained within tissues may be important to maintain normal growth and development. In another study, transgenic Arabidopsis demonstrated reduced growth in direct correlation with the alteration in the ratio of campesterol (24-methyl cholesterol) to sitosterol (Schaller et al., 1998). The results of the present study suggest that at least 50% of the accumulated sterol pool is not essential for normal plant growth and development, as long as the ratio of the remaining free sterol is comparable to wild type. The mechanism by which altered sterol ratio affects plant growth and development in transgenic tobacco is unclear; however, it has been suggested recently that altered sterol ratios influence cell division in Arabidopsis (Schaller et al., 1998).
It is also possible that cholesterol oxidase may be affecting an additional biochemical pool that was not addressed in this study and the phytotoxic effects observed are not related to the common sterol pool already discussed. Steroid hormones play important roles in growth and development of various organisms. The plant steroid hormones, known collectively as brassinosteroids, elicit a variety of responses during plant growth and development (Mandava, 1988; Sasse, 1997). Several studies of dwarf mutants have provided strong evidence that brassinosteroids are required for cell elongation (Clouse et al., 1996). Brassinosteroids structurally are derived from campestanol with modifications occurring to the A and B rings and the side chain (Fujioka and Sakurai, 1997). Results from the present study suggest that accumulation of stanols in general does not influence the ontogeny of the transgenic tobacco. Elevated levels of campestanol exist in all ChoM-positive transgenic lines when compared with wild type. Transgenic tobacco expressing chloroplast-targeted ChoM maintains a normal phenotype, whereas lines expressing ChoM cytosolically maintain a dwarf phenotype. The dwarf phenotype observed when expressing cytosolic cholesterol oxidase is interesting because many brassinosteroid mutants display a similar phenotype. However, a connection between cholesterol oxidase activity, increased stanol synthesis, and alteration of brassinosteroid synthesis cannot be concluded from this data. Our data does not address the subcellular localization of campestanol within wild-type or transgenic tissues. However, the suggestion can be made that localized cytosolic concentrations of campestanol may influence brassinosteroid synthesis, whereas chloroplastic concentrations of campestanol have little effect. In addition, cholesterol oxidase should not directly recognize and alter brassinosteroid structure due to the saturated steroid nucleus (Fujioka and Sakurai, 1997).
The sterol biosynthetic pathway historically has been described as a cytosolic pathway in higher plant cells. However, the occurrence of two distinct isoprene pathways in higher plants has been described recently (Lichtenthaler, 1999; Rohmer, 1999); the classical mevalonate pathway and the 2C-methyl erythritol-4-phosphate pathway. The results of our present study in tobacco also suggest the existence of two distinct sterol biosynthetic pathways within tobacco or a highly regulated exchange of sterol between the cytosol and the chloroplast. If multiple pathways for sterol biosynthesis exist in tobacco, then one pathway appears cytosolic in nature, whereas the second pathway is chloroplastic. For this conclusion to be valid it has to be assumed that ChoM targeted to the chloroplast is not active during transit from the cytosol to the chloroplast. If this dual pathway proposal is correct, the levels of sterol and stanol observed within the transgenic lines predict that both pathways contribute considerably to the overall accumulation of sterol within the cell. Several transgenic plant lines expressing chloroplast-targeted ChoM demonstrate on average 55% of their total sterol pool converted to stanol, suggesting that a substantial amount of carbon is moving through this pathway (Fig. 6B). The similarity in total steroid accumulation between the control and transgenic tissues suggests that some form of coordinate regulation is occurring between the two pathways to limit carbon moving into end product sterols or that the stanol end product is assessed similarly to sterol within plant cell physiology (Fig. 6A).
A second possible explanation for the observed biochemical phenotypes is that dynamic cycling of sterols between subcellular compartments includes the transient movement of sterols through the chloroplasts. The results of this study demonstrate that plants expressing chloroplast-targeted ChoM accumulate significant levels of stanol. If sterol biosynthesis is localized solely to the cytosol, movement of sterol to the chloroplast must be possible based solely on the amount of stanol accumulated in transgenic plants in which ChoM is targeted to the chloroplast. Assuming the cytosolic and chloroplastic ChoM enzymes are equally active in their respective compartments, it appears that a substantial portion of the cellular sterol must at some point be exposed to the interior of the chloroplast. Sterol accumulation patterns in higher plants demonstrate that the microsomal and mitochondrial fractions contain the bulk of the accumulated sterols, suggesting that exposure of sterols to the chloroplast may be transient.
The present study clearly demonstrates that enzymatically and insecticidally active cholesterol oxidase can be produced in transgenic plants. The enzyme effectively oxidized the major sterols in tobacco causing the accumulation of elevated levels of stanols. Although the physiological effect of stanol accumulation was not fully delineated in this study, it is clear that differential subcellular localization of cholesterol oxidase results in tremendous differences in plant development. Further biochemical analysis and characterization of the subcellular localization of cholesterol oxidase and any subsequent reactions leading to the accumulation of stanol products will address important questions regarding the metabolism and function of sterols in higher plant ontogeny.
MATERIALS AND METHODS
Cholesterol Oxidase Genes
Cloning of a functional cholesterol oxidase gene from Actinomyces sp. A19249 and construction of N terminally modified recombinant genes was previously described (Corbin et al., 1994). The native cholesterol oxidase gene, choM, encodes a protein of 547 amino acids. The N-terminal 43 amino acids constitute a signal sequence that is removed from the primary translation product during secretion. The recombinant full-length choM gene includes the N-terminal secretory signal sequence. The recombinant mature choM gene encodes only the processed, mature form of the enzyme. These two recombinant choM genes were used to construct all the plant gene expression cassettes used in the present study.
Plant Gene Expression Elements
The chloroplast-targeting sequence CTP1, from the Arabidopsis Rubisco SSU 1A gene, was previously described (Wong et al., 1992). The CTP1 element consisted of the 55-amino acid SSU chloroplast-targeting sequence plus the N-terminal 24 amino acids of the Rubisco SSU protein. The native transit peptidase cleavage site at the junction of the targeting sequence and the SSU sequence was preserved, and a second peptidase cleavage site and a NcoI restriction site were introduced at the 3′ end of the Rubisco coding sequence.
The chloroplast-targeting sequence CTP1Δ was derived from CTP1 by deleting the 24 amino acids of Rubisco and one of the two transit peptidase cleavage sites. This deletion was accomplished using two SphI restriction enzyme sites, one contained within each of the duplicated transit peptidase cleavage site sequences.
Transcriptional promoters used for plant expression were the figwort mosaic virus promoter (Richins et al., 1987), the enhanced cauliflower mosaic virus 35S (e35S) promoter (Kay et al., 1987), and the Arabidopsis Rubisco SSU 1A promoter (Wong et al., 1992).
Plant polyadenylation signals used were the nopaline synthase 3′-UTR region from the Agrobacterium tumefaciens Ti plasmid (Bevan et al., 1983) and the 3′-UTR region from the E9 SSU Rubisco gene from Pisum sativa (Hunt and MacDonald, 1989).
Tobacco (Nicotiana tabacum cv Samsum) Transformation
Tobacco was transformed using the A. tumefaciens leaf disc transformation method (Horsch et al., 1985). All plant transformation vectors carried a 5-enolpyruvylshikimate-3-phosphate synthase gene and transformed plants were selected using glyphosate selection (Zhou et al., 1995).
Detection of Cholesterol Oxidase in Transgenic Plants
Native, secreted ChoM for use as a protein standard in western blot experiments was purified from Actinomyces strain A19249 culture filtrates as previously described (Purcell et al., 1993).
Cholesterol oxidase enzyme activity was determined using the colorimetric method of Gallo (1981). In some experiments, cholesterol oxidase expression levels were estimated based upon observed enzyme activities and the specific activity of a purified cholesterol oxidase preparation. Extracts were prepared by homogenizing fresh tobacco leaf tissue in phosphate-buffered saline-Tween (PBST) extraction buffer consisting of 0.07% (w/v) Tween 20 in phosphate-buffered saline (8 g L−1 NaCl, 1.15 g L−1 Na2HPO4, 0.2 g L−1 KH2PO4, and 0.2 g L−1 KCl). Ten milligrams of leaf tissue was homogenized in 0.5 mL of PBST. Tissue debris was removed by centrifugation at 10,000g for 10 min at 4°C.
Protein samples for western-blot analysis were prepared by pulverizing tobacco leaf tissue that had been frozen on dry ice in a 1.5-mL microcentrifuge tube. One-half milliliter of boiling SDS/PAGE extraction/loading buffer was added to 10 mg pulverized tissue and the samples were immediately placed in a boiling water bath for 10 min. Tissue debris was removed by centrifugation at 10,000g for 10 min. Proteins were subjected to SDS-PAGE electrophoresis in 10% (w/v) polyacrylamide gels and transferred to nitrocellulose membranes. Cholesterol oxidase protein bands were detected by using anti-ChoM antibodies as previously described (Corbin et al., 1994).
Protein samples for quantitative ELISA analysis were prepared from fresh leaf tissue by homogenizing tissue in PBST followed by centrifugation at 10,000g for 10 min to remove tissue debris. Five to 20 μl of extract was analyzed in a direct ELISA that used purified rabbit anti-cholesterol oxidase IgG as a coating antibody and horseradish peroxidase-conjugated purified rabbit anti-cholesterol oxidase IgG as a detection antibody. Detection was accomplished by reaction with 3,3′,5,5′-tetramethyl-benzidine peroxidase substrate and hydrogen peroxide followed by determination of optical density at 450 nm with a reference wavelength of 655 nm using a model 3550 plate reader (Bio-Rad, Hercules, CA).
Steroid Analysis
Reverse phase-HPLC was performed in conjunction with gas chromatography (GC)-mass spectroscopy (MS) analysis. Sterols and stanols were quantified by reverse phase-HPLC using previously produced standard curves for all compounds (Grebenok et al., 1991). Previous analysis of control tissue and transgenic tissue expressing ChoM prior to and after base saponification produced no appreciable difference in steroid titer or distribution (data not shown). Therefore, samples used in this study were not subjected to base saponification.
Lyophilized plant material was extracted with ethanol by previously published methods (Grebenok et al., 1991). At the time of extraction, 40 μg of lathosterol was added as an internal standard for quantification purposes. The steroid fraction was evaporated to dryness under nitrogen and subsequently solubilized in 200 μL of methylene chloride. Sterols and stanols were identified through gas chromatography-mass spectroscopy (GC-MS) using a 5890 gas chromatograph and a 5970 mass spectrometer (Hewlett-Packard, Palo Alto, CA). GC separation of isolated sterols and stanols was achieved on a one-column (15 m × 0.3 mm; 0.25-mm film thickness; Hewlett-Packard). Oven temperature was programmed from 200°C to 280°C at 20°C per min and the carrier gas was helium at a velocity of 30 cm s−1. The MS was operated at an ionizing potential of 70 eV, the ion source was maintained at 280°C, and the injector port was maintained at 250°C. Cholesterol, campesterol, stigmasterol, sitosterol, stigmastanol, campestanol, and cholestanol were identified through cochromatography with authentic standards (Sigma Chemical, St. Louis) on the gas chromatograph, and by characteristic mass spectrum (Lenfant et al., 1970; Iida et al., 1981; Rahier and Benveniste, 1989). The parent ion of a stanol contains two additional mass units when compared with the corresponding sterol, with the most abundant ion being m/e 215. Cholestanol displayed ions at m/e: M+ 388, 373, 355, 273, 257, 248, 233, and 215 (100%). The two additional mass units present in the parent ion (m/e 388) are in agreement with a saturated sterol nucleus when compared with cholesterol. The most abundant ion (m/e 215) is in agreement with previously published stanol fragmentation. Campestanol displayed similar characteristics with ions at m/e: M+ 402, 387, 369, 257, 248, 233, and 215 (100%). Sitostanol/stigmastanol displayed ions at m/e: M+ 416, 401, 398, 383, 344, 275, 257, 248, 233, and 215 (100%; Lenfant et al., 1970; Iida et al., 1981).
Insect Bioassays
Cotton boll weevil (Anthonomus grandis grandis Boheman) larval bioassays were performed in 96-well microtiter plates using a modification of the methods of MacIntosh et al. (1990) and Purcell et al. (1992) using an agar-based artificial diet (Marrone et al., 1985). Tobacco leaf tissues to be assayed were harvested, frozen on dry ice, lyophilized to dryness, and then pulverized to a fine powder. The lyophilized powder was suspended as a 2% (w/v) slurry in sterile distilled water. Fifty microliters of the slurry was applied to each well containing 200 μL of diet and allowed to air dry. Each plant sample was tested in 16 wells. Each well received at least one boll weevil egg. Boll weevil eggs were obtained from the Robert T. Gast Rearing Laboratory (U.S. Department of Agriculture, Starkville, MS). After egg hatch, neonate larvae were allowed to feed and develop for 6 d. Mortality for each plant sample was scored as the percentage of infested wells that did not contain any surviving larva after 6 d. Surviving larvae were weighed.
Chloroplast Isolation
Chloroplast isolation was accomplished using a modification of the Percoll step gradient protocol described by Orozco et al. (1986) in which polyethylene glycol was omitted from the homogenization buffer. Tobacco plants used for chloroplast isolation were maintained in a greenhouse with supplemental lighting to provide 16 h of light and 8 h of dark. The plants were maintained in the dark for 48 h prior to leaf harvesting in order to reduce chloroplast starch accumulation. Forty grams of fresh leaf tissue, with mid-veins removed, was homogenized in 200 mL ice-cold buffer {50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH8.0, 2 mm EDTA, 1 mm MgCl2, 1 mm MnCl2, 0.33 m sorbitol, 0.1% [w/v] ascorbic acid, and 0.1% [w/v] bovine serum albumin} using three 5-s cycles in a blender (Waring, New Hartford, CT) set at high speed. The crude homogenate was filtered through four layers of Miracloth and the filtrate was centrifuged at 5,000g for 10 min. The supernatant was discarded and the crude organelle pellet was resuspended in 10 mL of homogenization buffer and layered onto the tops of 40% to 85% (v/v) Percoll step gradients. Loaded gradients were centrifuged at 14,800g in a swinging bucket rotor for 10 min. Two green chloroplast-enriched bands were observed and collected. The lower chloroplast band at the 40%/85% interface consisted of intact chloroplasts and the upper chloroplast band at the buffer/40% interface consisted of broken chloroplasts depleted of chloroplast envelope and stroma. Light microscopy of the two fractions substantiated their identification as intact and broken chloroplast fractions. Samples were washed twice by 5 to 10-fold dilution in homogenization buffer followed by centrifugation.
ACKNOWLEDGMENTS
The authors thank Barbara Reich and Aundrea Warren for conducting the ELISA analysis, Nancy Mathis and Deborah Stone for performing tobacco transformations, Dr. Ganesh Kishore and Dr. Fred Perlak for insightful suggestions, and Dr. Murtaza Alibhai and Dr. Kenneth Gruys for critical reading of the manuscript.
LITERATURE CITED
- Applequist LD, Kornfeldt AK, Wennerholm JE. Sterols and steryl esters in some Brassica and Sinapsis seeds. Phytochemistry. 1981;20:207–210. [Google Scholar]
- Ballantine JA, Lavis A. Marine sterols: VIII. The sterol compositions of two marine sponges: occurrence of new C26 and C30 stanols in an oceanic sponge. Comp Biochem Physiol. 1978;63:119–123. [Google Scholar]
- Bevan M, Barnes WM, Chilton M-D. Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 1983;11:369–385. doi: 10.1093/nar/11.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin DR, Greenplate JT, Wong EY, Purcell JP. Cloning of an insecticidal cholesterol oxidase gene and its expression in bacteria and plant protoplasts. Appl Environ Microbiol. 1994;60:4239–4244. doi: 10.1128/aem.60.12.4239-4244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischhoff DA, Bowdish KS, Perlak FJ, Marrone PG, McCormick SM, Niedermeyer JG, Dean DA, Kusano-Kretzmer K, Mayer EJ, Rochester DE. Insect tolerant transgenic tomato plants. Bio/Technology. 1987;5:807–813. [Google Scholar]
- Fujioka S, Sakurai A. Biosynthesis and metabolism of brassinosteroids. Plant Physiol. 1997;100:710–715. [Google Scholar]
- Gallo LL. Sterol ester hydrolase from rat pancreas. Methods Enzymol. 1981;71:664–674. doi: 10.1016/0076-6879(81)71079-6. [DOI] [PubMed] [Google Scholar]
- Goad LJ. Applications of sterol synthesis inhibitors to investigate the sterol requirements of protozoa and plants. Biochem Soc Trans. 1990;18:63–65. doi: 10.1042/bst0180063. [DOI] [PubMed] [Google Scholar]
- Grandmougin-Ferjani A, Schuler-Muller I, Hartmann M-A. Sterol modulation of the plasma membrane H+-ATPase activity from corn root reconstituted into soybean lipids. Plant Physiol. 1997;113:163–174. doi: 10.1104/pp.113.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenok RJ, Ripa PV, Adler JH. Occurrence and levels of ecdysteroids in spinach. Lipids. 1991;26:666–668. [Google Scholar]
- Greenplate JT, Corbin DR, Purcell JP. Proceedings of the Beltwide Cotton Conferences. National Cotton Council of America, New Orleans. 1997. Cholesterol oxidase: potent boll weevil larvicidal and oostatic agent suitable for transgenic cotton development; pp. 877–880. [Google Scholar]
- Greenplate JT, Duck NB, Pershing JC, Purcell JP. Cholesterol oxidase: an oostatic and larvicidal agent active against the cotton boll weevil, Anthonomus grandis. Entomol Exp Appl. 1995;74:253–258. [Google Scholar]
- Guo DA, Venkatramesh M, Nes WD. Developmental regulation of sterol biosynthesis in Zea mays. Lipids. 1995;30:203–219. doi: 10.1007/BF02537823. [DOI] [PubMed] [Google Scholar]
- Halpin C, Elderfield PD, James HE, Zimmermann R, Dunbar B, Robinson C. The reaction specificities of the thylakoid processing peptidase and Escherichia coli leader peptidase are identical. EMBO J. 1989;8:3917–3921. doi: 10.1002/j.1460-2075.1989.tb08572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Hunt AG, MacDonald MH. Deletion analysis of the polyadenylation signal of a pea ribulose-1,5-bisphosphate carboxylase small-subunit gene. Plant Mol Biol. 1989;13:125–138. doi: 10.1007/BF00016132. [DOI] [PubMed] [Google Scholar]
- Iida T, Tamura T, Matsumoto T. Stigmastanol in the seeds of Trichosanthes cucumeroide. Phytochem. 1981;20:857. [Google Scholar]
- Kanazawa A, Teshima S, Hyodo S. Sterols of the sponges (Porifera, Class Demospongiae) Comp Biochem Physiol. 1978;62:521–525. [Google Scholar]
- Kavanagh TA, Jefferson RA, Bevan MW. Targeting a foreign protein to chloroplasts using fusions to the transit peptide of a chlorophyll a/b protein. Mol Gen Genet. 1988;215:38–45. doi: 10.1007/BF00331300. [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1303. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Kuntz M, Simons A, Schell J, Schreier PH. Targeting of protein to chloroplasts in transgenic tobacco by fusion to mutated transit peptide. Mol Gen Genet. 1986;205:454–460. [Google Scholar]
- Lenfant M, Lecompte MF, Farrugia G. Identification des sterols de Physarum polycephalum. Phytochemistry. 1970;9:2529–2535. [Google Scholar]
- Lichtenthaler HK. The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Ann Rev Plant Physiol Plant Mol Bio. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- MacIntosh SC, Stone TB, Sims SR, Hunst PL, Greenplate JT, Marrone PG, Perlak FJ, Fischhoff DA, Fuchs RL. Specificity and efficacy of purified Bacillus thuringiensis proteins against agronomically important insects. J Invertebr Pathol. 1990;56:258–266. doi: 10.1016/0022-2011(90)90109-j. [DOI] [PubMed] [Google Scholar]
- Mandava NB. Plant growth promoting brassinosteroids. Ann Rev Plant Physiol Plant Mol Biol. 1988;39:23–52. [Google Scholar]
- Marrone PG, Ferri FD, Mosley TR, Meinke LJ. Improvements in laboratory rearing of the southern corn rootworm. J Econ Entomol. 1985;78:290–293. [Google Scholar]
- Meadows JW, Robinson C. The full precursor of the 33 kDa oxygen-evolving complex protein of wheat is exported by Escherichia coli and processed to the mature size. Plant Mol Biol. 1991;17:1241–1243. doi: 10.1007/BF00028739. [DOI] [PubMed] [Google Scholar]
- Nes WR, McKean ML. W.R. Nes and M.L. McKean, eds, Biochemistry of Steroids and Other Isopentenoids. Baltimore: University Park Press; 1977. Occurrence, physiology, and ecology of sterols; pp. 411–533. [Google Scholar]
- Orozco EM, Jr, Mullet JE, Hanley-Bowdoin L, Chua N-H. In vitro transcription of chloroplast protein genes. Methods Enzymol. 1986;118:232–235. doi: 10.1016/0076-6879(86)18076-1. [DOI] [PubMed] [Google Scholar]
- Perlak FJ, Deaton RW, Armstrong TA, Fuchs RL, Sims SR, Greenplate JT, Fischhoff DA. Insect resistant cotton plants. Bio/Technology. 1990;8:939–943. doi: 10.1038/nbt1090-939. [DOI] [PubMed] [Google Scholar]
- Perlak FJ, Fuchs RL, Dean DA, McPherson SL, Fischhoff DA. Modification of the coding sequence enhances plant expression of insect control protein genes. Proc Natl Acad Sci USA. 1991;88:3324–3328. doi: 10.1073/pnas.88.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlak FJ, Stone TB, Muskopf YM, Petersen LJ, Parker GB, McPherson SA, Wyman J, Love S, Reed G, Biever D. Genetically improved potatoes: protection from damage by Colorado potato beetles. Plant Mol Biol. 1993;22:313–321. doi: 10.1007/BF00014938. [DOI] [PubMed] [Google Scholar]
- Purcell JP, Greenplate JT, Jennings MG, Ryerse JS, Pershing JC, Sims SR, Prinsen MJ, Corbin DR, Tran M, Sammons RD. Cholesterol oxidase: a potent insecticidal protein active against boll weevil larvae. Biochem Biophys Res Commun. 1993;196:1406–1413. doi: 10.1006/bbrc.1993.2409. [DOI] [PubMed] [Google Scholar]
- Purcell JP, Greenplate JT, Sammons RD. Examination of midgut luminal proteinase activities in six economically important insects. Insect Biochem Mol Biol. 1992;22:41–47. [Google Scholar]
- Rahier A, Benveniste P. Mass spectral identification of phytosterols. In: Nes WD, Parish EJ, editors. Analysis of Sterols and Other Biologically Significant Steroids. New York: Academic Press; 1989. pp. 223–250. [Google Scholar]
- Richins R, Scholthof H, Shepard R. Sequence of the figwort mosaic virus DNA (caulimovirus group) Nucleic Acids Res. 1987;15:8451–8466. doi: 10.1093/nar/15.20.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- Russell DA, DeBoer DL, Stark DM, Preiss J, Fromm ME. Plastid targeting of E. coli β-glucuronidase and ADP-glucose pyrophosphorylase in maize (Zea mays L.) cells. Plant Cell Rep. 1993;13:24–27. doi: 10.1007/BF00232309. [DOI] [PubMed] [Google Scholar]
- Salt TA. Sterol chemosystematics in the order Caryophyllales. PhD thesis. Philadelphia: Drexel University; 1984. [Google Scholar]
- Sasse JM. Recent progress in brassinosteroid research. Plant Physiol. 1997;100:696–701. [Google Scholar]
- Schaller H, Bouvier-Naveí P, Benveniste P. Overexpression of an Arabidopsis cDNA encoding a sterol-C24-methyltransferase in tobacco modifies the ratio of 24-methylcholesterol to sitosterol and is associated with growth reduction. Plant Physiol. 1998;118:461–469. doi: 10.1104/pp.118.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler I. Soybean phosphatidylcholine vesicles containing plant sterols: a fluorescence anisotropy study. Biochim Biophys Acta. 1990;1028:82–88. doi: 10.1016/0005-2736(90)90268-s. [DOI] [PubMed] [Google Scholar]
- Schuler I. Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phophatidylcholine bilayers. Proc Natl Acad Sci USA. 1991;88:6926–6930. doi: 10.1073/pnas.88.16.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler A, Michel H. Expression in Escherichia coli of the psbO gene encoding the 33 kd protein of the oxygen-evolving complex from spinach. EMBO J. 1990;9:1743–1748. doi: 10.1002/j.1460-2075.1990.tb08298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh YM, Djerassi C. Biosynthesis of sterols in the sea cucumber Strichopus californicus. Tetrahedron Lett. 1977;36:3111–3114. [Google Scholar]
- Shen Z, Corbin DR, Greenplate JT, Grebenok RJ, Galbraith DW, Purcell JP. Studies on the mode of action of cholesterol oxidase on insect midgut membranes. Arch Insect Biochem Physiol. 1997;34:429–442. [Google Scholar]
- Smith A, Goad JL. The conversion of cholest-5-en-3β-ol into cholest-7-en-3β-ol by the echinoderms Asterias rubens and Solaster papposus. Biochem J. 1975;146:35–40. doi: 10.1042/bj1460035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Brooks CJW. Cholesterol oxidase: properties and applications. J Steroid Biochem. 1976;7:705–713. doi: 10.1016/0022-4731(76)90071-6. [DOI] [PubMed] [Google Scholar]
- Teshima S, Patterson GW. Identification of 4α-methylsterols in the oyster, Crassostera virginica. Comp Biochem Physiol. 1981;69:175–181. [Google Scholar]
- Vaeck M, Reynaerts A, Hofte H, Jansens S, De Beuckeleer M, Dean C, Zabeau M, Van Montagu M, Leemans J. Transgenic plants protected from insect attack. Nature. 1987;328:33–37. [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wasmann CC, Reiss B, Bartlett SG, Bohnert HJ. The importance of the transit peptide and the transported protein for protein import into chloroplasts. Mol Gen Genet. 1986;205:446–453. [Google Scholar]
- Wong EY, Hironaka CM, Fischhoff DA. Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thuringiensis proteins in transgenic plants. Plant Mol Biol. 1992;20:81–93. doi: 10.1007/BF00029151. [DOI] [PubMed] [Google Scholar]
- Zhou H, Fry JE, Arrowsmith JW, Fromm ME. A glyphosate-tolerant EPSPS gene as a selectable marker in wheat transformation. Plant Cell Rep. 1995;15:159–163. doi: 10.1007/BF00193711. [DOI] [PubMed] [Google Scholar]