Abstract
The 26S proteasome is a multicatalytic complex that acts as primary protease of the ubiquitin-mediated proteolytic pathway in eukaryotes. We provide here the first evidence that the proteasome plays a key role in regulating pollen tube growth. Immunoblotting experiments revealed the presence of high levels of free ubiquitin and ubiquitin conjugates in rehydrated and germinating pollen of kiwifruit [Actinidia deliciosa var. deliciosa (A. Chev) C. F. Liang et A. R. Ferguson]. Proteasome activity, assayed fluorometrically, accompanied the progression of germination. Specific inhibitors of proteasome function such as benzyloxycarbonyl-leucinyl-leucinyl-leucinal (MG-132), clasto-lactacystin β-lactone, and epoxomicin significantly decreased tube growth or altered tube morphology. High-molecular mass, ubiquitinated proteins accumulated in MG-132- and β-lactone-treated pollen, indicating that proteasome function was effectively impaired. The inhibitors were also able to decrease in vitro proteasome activity in pollen extracts. Because MG-132 can inhibit calpains, as well as the proteasome, trans-epoxy succinyl-l-leucylamido-(4-guanidino) butane (E-64), an inhibitor of cysteine proteases, was investigated. Some reduction in tube growth rate was observed, but only at 80 μm E-64, and no abnormal tubes were produced. Furthermore, no inhibition of tube growth was observed when another inhibitor of cysteine proteases, leupeptin, or inhibitors of serine and aspartic proteases (phenylmethylsulfonyl fluoride and pepstatin) were used. Our results indicate that protein turnover during tube organization and elongation in kiwifruit pollen is important, and our results also implicate the ubiquitin/26S proteasome as the major proteolytic pathway involved.
The ubiquitin/proteasome system is a major pathway of proteolysis in eukaryotic cells and may contribute to controlling the intracellular levels of a variety of short-lived regulatory proteins (Viestra, 1993; Ciechanover, 1994; Jentsch and Schlenker, 1995). In this proteolytic pathway, proteins are first modified by covalent conjugation to ubiquitin, which marks them for rapid hydrolysis by the 26S proteasome. Mono-ubiquitination is not sufficient to target a protein for destruction by proteasome. Long polyubiquitin chains must be formed to direct substrates to it. In addition, proteasome-mediated degradation of a few proteins has been shown to occur without ubiquitination (Murakami et al., 1992; Jariel-Encontre et al., 1995). The proteasome is a large multicatalytic complex found in the cytosol and nucleus (Peters, 1994). It is composed of two relatively stable particles, the 20S proteasome, a hollow cylindrical structure that contains the proteolytic active sites in its lumen, and the 19S regulatory particle that binds to either end of the cylinder and provides the ATP dependence and specificity for the ubiquitinated proteins (Coux et al., 1996). A high conservation level has recently been shown in the sequence of α-type proteasome subunits from tobacco, humans, and yeast, adding support to the idea that the proteasome plays an important role in cell growth and development in all eukaryotes, including plants (Bahrami and Gray, 1998).
In plants, the ubiquitin-mediated proteolytic pathway is implicated in a variety of cellular processes, including alkaloid synthesis (Fernandez and DeLuca, 1994), senescence and stress responses (Belknap and Garbarino, 1996; Stephenson and Rubinstein, 1998), hypersensitive and defense responses (Becker et al., 1993, 2000), vascular tissue differentiation (Woffenden et al., 1998), seed formation (Ferreira et al., 1995), and cell cycle regulation (Scoccianti et al., 1997; Genschik et al., 1998).
Pollen is a highly reduced structure comprising only two or three cells when it is released at anthesis. It has a brief existence during which it rapidly extends a pollen tube through which the sperm cells are transported to the embryo sac. Data on levels of free and conjugated ubiquitin in developing pollen from different plant species are contradictory. Callis and Bedinger (1994) found a developmentally related loss of free ubiquitin and ubiquitinated proteins in maize inbred line Ky21, so that the ubiquitin monomer was not detectable at pollen maturity. A reduction in the free ubiquitin level also takes place in the mature pollen stage in olive (Alché et al., 2000). On the contrary, high levels of free ubiquitin and ubiquitin conjugates have been found in mature pollen from several plant species, including some maize inbred lines (Muschietti et al., 1994; Kulikauskas et al., 1995). In our previous work we also observed that pollen maturity is characterized by a large (free and conjugated) ubiquitin content in male kiwifruit (Actinidia deliciosa var. deliciosa). The ubiquitin monomer continuously increases during pollen development and reaches the highest level at the time of pollen release (Scoccianti et al., 1999a, 1999b). It is interesting that the reduction in free ubiquitin observed in maize Ky 21 inbred and olive pollen does not coincide with a parallel reduction in ubiquitin transcripts (Callis and Bedinger, 1994; Alché et al., 2000). This has suggested the possibility of a post-transcriptional regulatory mechanism that adapts a poorly functioning ubiquitin system to a transient slow metabolic stage of these pollen species; in fact, relatively high levels of free ubiquitin are quickly restored upon olive pollen hydration (Alché et al., 2000).
On the basis of these observations and of our previous findings on the presence of high free ubiquitin levels in mature kiwifruit pollen we put forward the hypothesis that all the components of the ubiquitin proteolytic pathway are immediately activated at the onset of pollen germination. In the present paper we provide evidence for the involvement of the ubiquitin proteolytic pathway in pollen germination and tube elongation in kiwifruit. In particular, using inhibitors of the proteasome, we investigated whether this proteolytic pathway has a crucial regulatory role in the re-arrangement of the structural and enzymatic protein population occurring during tube organization and growth. The proteasome inhibitors we used were the reversible, competitive, synthetic tripeptide-aldehyde inhibitor benzyloxycarbonyl-leucinyl-leucinyl-leucinal (MG-132; Tsubuki et al., 1996), which acts as transition-state analog (Lowe et al., 1995) and inhibits calpain and the proteasome (Rock et al., 1994; Tsubuki et al., 1996), and the irreversible specific proteasome inhibitors clasto-lactacystin β-lactone (Dick et al., 1997; Potuschak et al., 1998) and epoxomicin (Meng et al., 1999; Sin et al., 1999). The effects of non-proteasomal protease inhibitors on pollen germination were also tested. The data presented here indicate a direct role of the proteasome in regulating tube emergence and growth in kiwifruit pollen.
RESULTS
Kiwifruit Pollen Viability and Germination
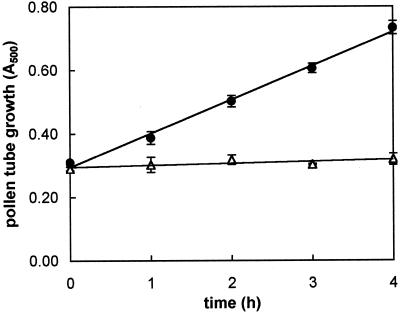
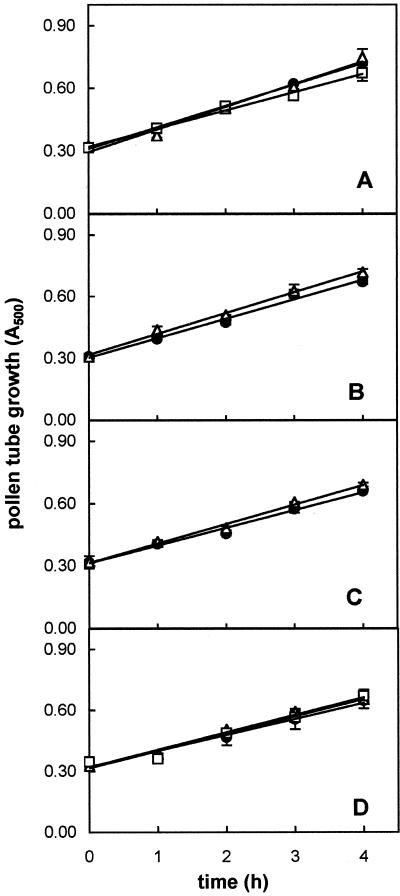
Microscopic counting of the percentage of tube emergence indicated a very good viability of pollen; in fact, close to 85% of pollen grains germinated when cultured in vitro. The time course of kiwifruit pollen tube growth (increase over time of tube mass) was linear over a 6-h incubation period (Fig. 1). To assess the requirement for de novo protein synthesis during pollen tube emergence and elongation, cycloheximide was added to the culture medium at the beginning of incubation. For this experiment we used the dose of 300 μm cycloheximide on the basis of the results obtained by Hoekstra and Bruinsma (1979) and Calzoni and Speranza (1986) on pollen from various plant species. As reported in Figure 1, kiwifruit pollen germination was completely blocked in the presence of cycloheximide.
Figure 1.
Time course of kiwifruit pollen tube growth in standard medium (●) and in the presence of 300 μm cycloheximide (▵). Growth is expressed as A500 of sonicated aqueous suspension of grains/tubes measured at 1-h intervals. Slope of control regression is 0.107 ± 0.005. Data are the means ± sd of three independent experiments.
Protein Pattern, Free Ubiquitin, and Ubiquitin Conjugates during Germination
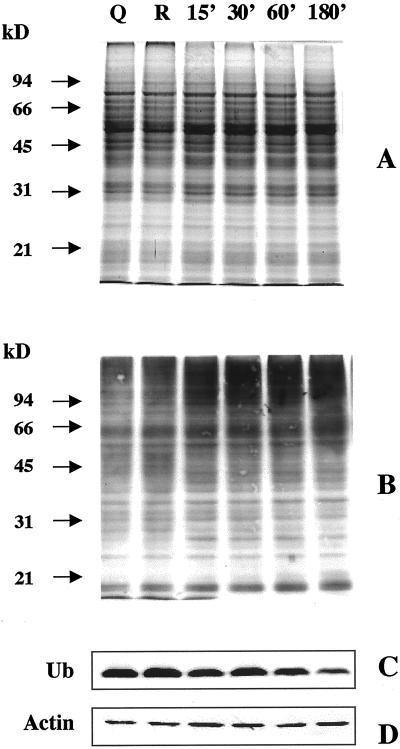
A large number of protein bands were always detectable in germinating kiwifruit pollen (Fig. 2A). The protein pattern was similar in quiescent, rehydrated, and germinating pollen, at least within the limits of resolution of SDS-PAGE and Coomassie staining. Several ubiquitin conjugates of different molecular masses were detected by immunoblotting in all stages tested (Fig. 2B). It is interesting that a time-dependent increase in protein ubiquitination was observed during pollen germination, with the most evident changes in the content of ubiquitin-protein adducts occurring during the first 15 to 30 min of incubation. In comparison with the ubiquitin conjugate banding pattern of quiescent and rehydrated pollen, germinating grains showed an increase in the intensity of several immunoreactive bands and a marked accumulation of high molecular mass (>90 kD) polyubiquitin conjugates (Fig. 2B). The free ubiquitin monomer did not show any significant changes upon hydration, but decreased slightly during pollen germination; only after 180 min of incubation was its level noticeably reduced (Fig. 2C).
Figure 2.
Distribution of free ubiquitin and ubiquitin conjugates in quiescent (Q), rehydrated (R), and germinating (15–180 min of incubation) kiwifruit pollen. A, B, and D, Twenty micrograms of protein was electrophoresed on a 10% (w/v) polyacrylamide gel and stained with Coomassie Blue (A) or electroblotted onto nitrocellulose membrane and probed with an affinity-purified polyclonal anti-ubiquitin antibody (B) or an anti-actin antibody (D). C, Immunoblot detection of free ubiquitin. Five micrograms of protein was electrophoresed on a 14% (w/v) polyacrylamide gel. The nitrocellulose membrane was probed as reported above. The positions of molecular mass markers (in kilodaltons) are shown on the left.
To make sure that cytoplasmic proteins were successfully extracted, we used the detection of actin as a control in all our experiments. No marked differences in the actin band were observed by immunoblot experiments during pollen germination (Fig. 2D).
Ubiquitin-Dependent Proteolytic Activity during Pollen Germination
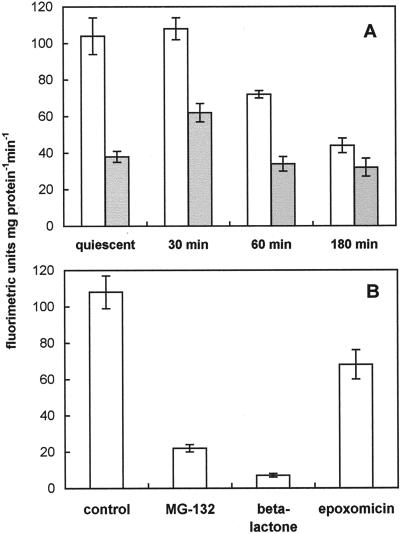
Since proteins tagged with ubiquitin are usually recognized and degraded by the 26S proteasome, the activity of this proteolytic complex was measured in crude pollen extracts by using the fluorogenic peptide N-succinyl-Leu-Leu-Val-Tyr 7-amido-4-methylcoumarin (sLLVY-NH-Mec), which is a substrate for chymotrypsin-like activity of the proteasome. This substrate has been shown to be split by the 26S proteolytic complex, whereas the 20S proteasome, which is not involved in the degradation of ubiquitinated proteins, is known to have latent activity for the peptide breakdown (Kanayama et al., 1992). Since the 26S complex is strictly ATP dependent, whereas the 20S proteasome is not, sLLVY-NH-Mec-hydrolyzing activity was assayed in kiwifruit pollen in the absence or in the presence of exogenous ATP. As shown in Figure 3A, extracts obtained from quiescent and germinating pollen were equally able to hydrolyze the fluorogenic substrate in the absence of exogenous ATP. In fact, with the exception of a peak at 30 min of incubation, no major differences in the proteolytic activity were observed. Upon addition of exogenous ATP, cleavage of sLLVY-NH-Mec was significantly stimulated in extracts obtained from quiescent pollen, as well as from germinating grains, but only during the first 60 min of germination (Fig. 3A). The addition of ATP to extracts from 180-min-germinating pollen did not significantly increase proteasome activity (Fig. 3A). It is interesting that the ATP-dependent hydrolytic activity was similar in quiescent and germinating pollen, whereas it was quite strongly reduced to 70% and 40% of the quiescent value at 60 and 180 min of germination, respectively (Fig. 3A).
Figure 3.
Proteasome activity in quiescent and germinating kiwifruit pollen. Extracts of ungerminated or germinated pollen were incubated in the presence of 200 μm sLLVY-NH-Mec at 30°C. The breakdown of the fluorogenic peptide was monitored by a fluorescence spectrophotometer for 5 min. A, Proteolytic activity in the presence (white columns) or absence (gray columns) of 2 mm ATP. B, Proteolytic activity in 30-min-germinated pollen extract as affected by the proteasome inhibitors MG-132 (50 μm), clasto-lactacystin β-lactone (10 μm), or epoxomicin (5 μm). Data are means ± sd of three measurements.
To demonstrate that the proteolytic activity observed in pollen extracts was supported by the proteasome, the breakdown of the fluorogenic peptide sLLVY-NH-Mec was also assayed in the presence of well-known proteasome inhibitors such as MG-132, clasto-lactacystin β-lactone, and epoxomicin. For this purpose, extracts of 30-min-cultured pollen were used. All the inhibitors tested reduced sLLVY-NH-Mec-hydrolyzing activity, causing an inhibition of 82%, 95%, and 64%, respectively, compared with the control (Fig. 3B).
Proteasome Inhibitors Reduce Pollen Tube Emergence and Growth
To determine whether the ubiquitin/proteasome system plays a role in pollen germination, MG-132, clasto-lactacystin β-lactone, and epoxomicin were added to the culture medium at the start of incubation. The concentrations of the different inhibitors tested were comparable with those used in animal (Palombella et al., 1994; Meng et al., 1999), yeast, and plant systems (Lee and Goldberg, 1996; Genschik et al., 1998; Woffenden et al., 1998).
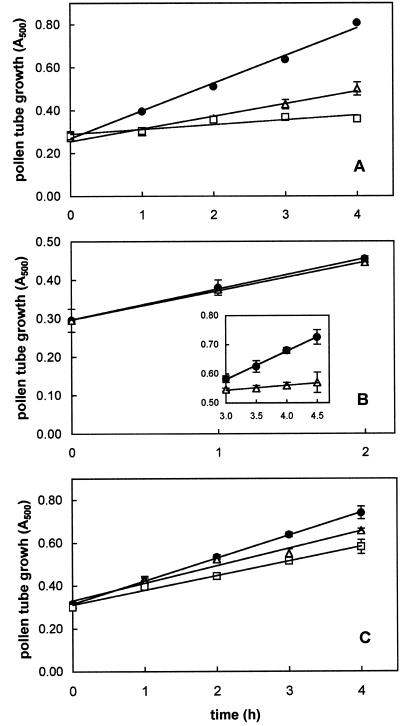
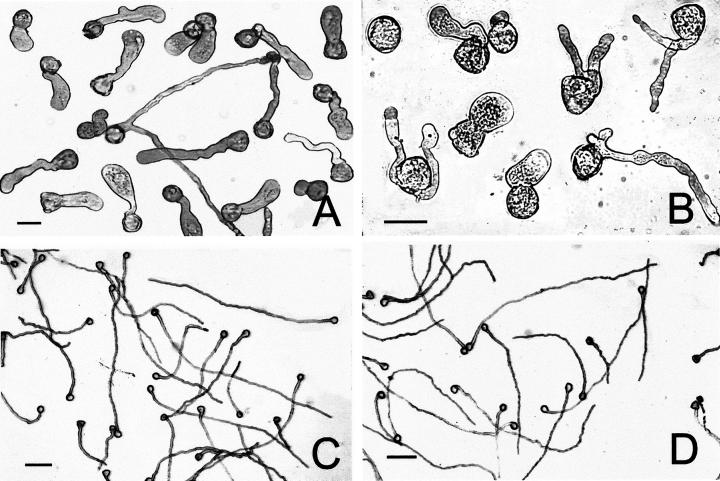
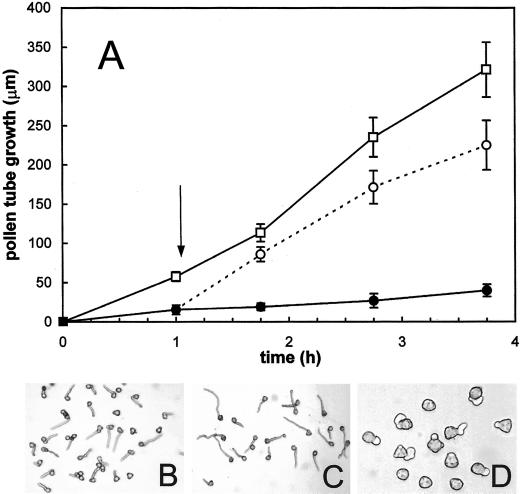
All the proteasome inhibitors tested produced significant effects on tube growth (Figs. 4 and 5). As reported in Figure 4A, MG-132 treatment drastically affected kiwifruit pollen germination. Microscopic counting indicated that only 11.4% and 3.3% of pollen grains had germinated after 60 min of incubation in the presence of 40 and 80 μm MG-132, respectively. After 180 min, the percentage of tube emergence was 54.1% (40 μm) and 31.7% (80 μm), whereas at this time, more than 85% of pollen tubes had emerged in the control. Slopes of the linear regressions of MG-132-treated pollen tubes showed an extremely significant (P < 0.0001) difference with respect to controls. In particular, tube elongation rate was reduced to about 45% (40 μm) and 17% (80 μm) of that of controls. As a consequence, the total mass of tubes produced after 4 h of incubation (expressed as the increase over time in A500 of pollen suspensions) fell to about 43% (40 μm) and 15% (80 μm) of that of controls. Furthermore, tube morphology of MG-132-treated pollen appeared altered, as shown by diffuse tube tip swelling, tube branching, and multisiphonous pollen grains (Fig. 5). To exclude the possibility that reduction by MG-132 of pollen germination and tube growth was due to a toxic effect, the inhibitor was removed from the culture medium after 1 h of incubation, when a significant inhibition was already detectable (Fig. 4A). As shown in Figure 6, the removal of 80 μm MG-132 completely restored growth. In fact, tube growth from pollen incubated for 1 h in the presence of the inhibitor, washed, and then transferred to fresh medium containing dimethyl sulfoxide (DMSO), but not MG-132, was comparable with that of DMSO controls handled in the same way (Fig. 6A). Observations carried out 45 min after transfer showed that tube shape of pollen pre-treated for 1 h with MG-132 (Fig. 6B) was also comparable with that of DMSO controls (Fig. 6C). By contrast, after the same incubation period (105 min), pollen treated with MG-132 throughout exhibited extremely poor and altered tube growth (Fig. 6D).
Figure 4.
Effect of proteasome inhibitors on kiwifruit pollen tube growth over time. Growth is expressed as A500 of a sonicated aqueous suspension of grains/tubes measured at 1-h intervals. A, Effect of MG-132 on tube growth. Slope of DMSO control (●) regression is 0.129 ± 0.007; slope of 40 μm MG-132-treated (▵) regression is 0.058 ± 0.005; and slope of 80 μm MG-132-treated (□) regression is 0.022 ± 0.006. B, Effect of 10 μm clasto-lactacystin β-lactone on tube growth during the first 2 h and during the following 3 to 4.5 h of incubation (inset). Slope of DMSO control (●) regression over total 4.5 h of incubation is 0.099 ± 0.003 and slope of β-lactone-treated (▵) regression during 3 to 4.5 h of incubation is 0.016 ± 0.002. C, Effect of epoxomicin on tube growth. Slope of DMSO control (●) regression is 0.107 ± 0.001; slope of 1 μm epoxomicin-treated (▵) regression is 0.081 ± 0.008; and slope of 5 μm epoxomicin-treated (□) regression is 0.068 ± 0.004. Data are the means ± sd of three independent experiments.
Figure 5.
Abnormal morphology of kiwifruit pollen tubes induced by MG-132 treatment. A, Three hours of incubation in the presence of 40 μm MG-132; bar = 30 μm. B, Threee hours of incubation in the presence of 80 μm MG-132; bar = 30 μm. C, Untreated pollen tubes after 3 h of incubation in standard medium; bar = 100 μm. D, Untreated pollen tubes (controls) after 3 h of incubation in standard medium containing DMSO solvent; bar = 100 μm.
Figure 6.
Reversion of the inhibitory effect of MG-132 on kiwifruit pollen tube growth. A, Pollen tube growth over time. DMSO controls (□) and 80 μm MG-132-treated pollen (○) were collected and washed after 1 h of incubation (arrow) and were transferred to fresh standard medium containing DMSO. Another lot of 80 μm MG-132-treated pollen (●) was incubated in the presence of the inhibitor throughout. Growth was monitored by measuring tube length (micrometers) as described in “Materials and Methods.” Data are the means ± sd (n = 300). B, Pollen treated for 1 h with 80 μm MG-132, 45 min after transfer to inhibitor-free medium. C, DMSO controls, treated as described above, 45 min after transfer to fresh medium containing DMSO. D, Pollen after 105 min of incubation in the presence of 80 μm MG-132.
Because MG-132 can inhibit calpains as well as the proteasome (Rock et al., 1994), the effect of trans-epoxy succinyl-l-leucylamido-(4-guanidino) butane (E-64) ester, a cell permeable inhibitor of Cys proteases, was also investigated. As reported in Figure 7A, 40 μm E-64 did not affect pollen tube growth (no significant difference between the slopes at P > 0.5). At the higher concentration (80 μm), the elongation rate was reduced to 85% of that of controls. The difference between the slopes of the linear regressions was significant (P < 0.05); however, the production of abnormal pollen tubes and a decrease in percent tube emergence did not occur after treatment with E-64 (data not shown).
Figure 7.
Effect of non-proteasomal protease inhibitors on kiwifruit pollen tube growth over time. Growth is expressed as A500 of a sonicated aqueous suspension of grains/tubes measured at 1-h intervals. A, Effect of E-64 on tube growth. Slope of control (●) regression is 0.102 ± 0.002; slope of 40 μm E-64-treated (▵) regression is 0.108 ± 0.009; and slope of 80 μm E-64-treated (□) regression is 0.087 ± 0.005. B, Effect of leupeptin on tube growth. Slope of control (●) regression is 0.099 ± 0.006 and slope of 50 μm leupeptin-treated (▵) regression is 0.101 ± 0.005. C, Effect of pepstatin on tube growth. Slope of ethanol control (●) regression is 0.089 ± 0.006 and slope of 50 μm pepstatin-treated (▵) regression is 0.093 ± 0.005. D, Effect of PMSF on tube growth. Slope of ethanol control (●) regression is 0.084 ± 0.007; slope of 50 μm PMSF-treated (▵) regression is 0.086 ± 0.006; and slope of 100 μm PMSF-treated (□) regression is 0.085 ± 0.009. Data are the means ± sd of three independent experiments.
When 10 μm clasto-lactacystin β-lactone was added to the culture medium, tubes elongated at the same rate as controls during the first 2 h of incubation. A dramatic inhibition of growth subsequently occurred and tubes nearly ceased to elongate; the differences between the slopes of the linear regressions were extremely significant (P < 0.0001; Fig. 4B). At this time, the growth rate was reduced to about 16% of that of controls.
Epoxomicin caused an appreciable inhibition at both the concentrations tested, causing a reduction of pollen tube growth rate of 25% (1 μm) and 36% (5 μm) compared with the control (P < 0.01; Fig. 4C).
Non-proteasomal protease inhibitors phenylmethylsulphonyl fluoride (PMSF), pepstatin, and leupeptin, which inhibit Ser-proteases, aspartic-proteases, and Ser/Cys-proteases, respectively, did not affect tube emergence and growth rate at the concentrations tested (Fig. 7, B–D). In fact, no significant differences between the slopes of control and treated tube linear regressions were found (P > 0.1).
Proteasome Inhibitors Increase the Level of High-Molecular Mass Ubiquitin Conjugates
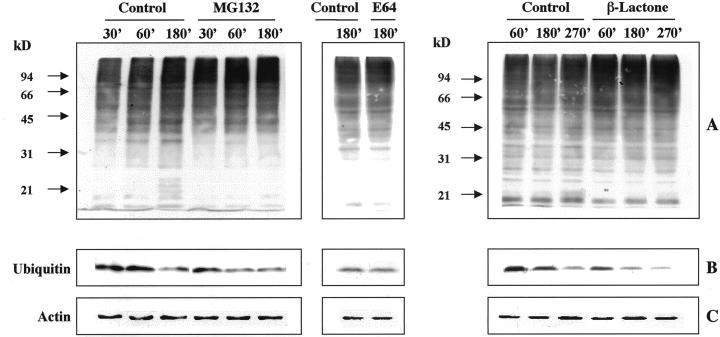
Because inhibition of proteasome function should result in the accumulation of ubiquitinated proteins, the effect of MG-132 on the levels of ubiquitin-protein conjugates was analyzed by immunoblot. The addition of the inhibitor (40 μm) to the culture medium resulted in the accumulation of multiple, high-molecular mass bands recognized by an anti-ubiquitin antibody (Fig. 8A). The conjugates already were detectable after 30 min of incubation and their level increased with time. In parallel, a more pronounced decrease in the levels of free ubiquitin monomer compared with the control was observed (Fig. 8B). Similar results were obtained when β-lactone was added to the culture, although the effects produced by this inhibitor were evident only later, starting from 60 min of incubation (Fig. 8A).
Figure 8.
Effect of proteasome inhibitors on accumulation of high-molecular mass ubiquitin-conjugated proteins in germinating kiwifruit pollen. A and C, Immunoblotting of total protein (20 μg per lane) extracted from pollen incubated with 40 μm MG-132, 80 μm E-64, or 10 μm β-lactone for different times and from pollen incubated in the medium without the respective inhibitor. Total protein was electrophoresed on 10% (w/v) polyacrylamide gels and was immunoblotted using polyclonal anti-ubiquitin antibody (A) or an anti-actin antibody (C). B, Immunoblot detection of free ubiquitin (each lane was loaded with 5 μg of protein). Molecular mass of standard proteins are indicated on the left (in kilodaltons).
Accumulation of high-molecular mass ubiquitin conjugates and a decrease in free ubiquitin level were not detectable in pollen germinated for 180 min in the presence of 80 μm E-64 (Fig. 8, A and B).
Quantitative evaluation of ubiquitin conjugates performed with a solid-phase dot-blot immunoassay showed a 44% increase in ubiquitin conjugate levels after 180 min of incubation in MG-132-treated pollen, compared with the amount found in the control (Table I). A 29% increase was induced by β-lactone treatment after 270 min of incubation. No differences from controls were observed at 180 min in the E-64-treated tubes.
Table I.
Content of ubiquitin-protein conjugates in extracts from kiwifruit pollen incubated in the presence of different inhibitors
| Treatment | Time | Ubiquitin-Protein Conjugates |
|---|---|---|
| min | pmol mg protein−1 | |
| Control | 180 | 189 ± 3 |
| 40 μm MG-132 | 180 | 273 ± 12a |
| Control | 180 | 217 ± 15 |
| 80 μm E-64 | 180 | 231 ± 17b |
| Control | 270 | 181 ± 16 |
| 10 μm β-Lactone | 270 | 233 ± 15c |
Ubiquitin-protein conjugates were quantified by solid-phase immunoassay. Data are means ± sd (n = 3).
P < 0.001 compared with controls.
Not significantly different from controls (P > 0.1).
P < 0.005 compared with controls.
DISCUSSION
Tube emergence and growth in kiwifruit pollen is strongly dependent upon de novo protein synthesis, as demonstrated by complete inhibition of germination in the presence of cycloheximide. Plants show wide differences in the ability of their pollen to germinate and grow in the presence of protein synthesis inhibitors. Some pollen species are completely insensitive to cycloheximide. In other cases, mature pollen already possesses the proteins required for germination, but new protein synthesis is needed for tube elongation; in yet others, germination and pollen tube growth require new protein synthesis (Hoekstra and Bruinsma, 1979; Mascarenhas, 1993; Taylor and Hepler, 1997). It has been hypothesized that the same genes that were active during the terminal stages of pollen maturation continue to be expressed during germination. Most of the late gene products seem to have primary functions in germination and tube growth (Mascarenhas, 1992, 1993). It is generally accepted that the mRNA required for germination is already present in mature pollen, although the dependence of protein synthesis on presynthesised mRNA varies with the plant species; mRNA synthesis occurs during the first moments of germination and tube growth (Mascarenhas and Mermelstein, 1981; Mascarenhas, 1993). It was recently reported that pollen-specific sequences encoding proteins with regulatory or signaling functions are transcribed during the earliest moments of pollen germination in petunia (Guyon et al., 2000).
Besides a transcriptional and translational control, we also hypothesized a key role for protein turnover during pollen germination. Ubiquitin and ubiquitin-protein conjugates have been found in virtually all gymnosperm and angiosperm pollen examined, suggesting a role for the ubiquitin proteolytic system in pollen development and maturation (Kulikauskas et al., 1995; Scoccianti et al., 1999a, 1999b). Here we provide the first evidence, based not only on the detection of free ubiquitin and ubiquitin conjugates, but mostly on a functional analysis of the 26S proteolytic complex responsible for the turnover of ubiquitinated proteins, that the ubiquitin system is strongly involved in pollen germination and tube growth in kiwifruit. A marked accumulation of high-molecular mass polyubiquitin conjugates occurs at early stages of germination and the levels of ubiquitinated proteins remain high during pollen tube growth. The 26S proteolytic complex, responsible for the degradation of ubiquitinated substrates, seems to already be present in quiescent pollen, as demonstrated by the ability of pollen extracts obtained from quiescent grains to hydrolyze the fluorogenic substrate sLLVY-NH-Mec in an ATP-dependent manner. It is well known that ATP is required for the association of the 20S particle with the 19S regulatory complex to generate the 26S proteasome; this interaction results in the stimulation of different peptidase activities associated with the 20S catalytic core (Chu-Ping et al., 1994; Coux et al., 1996). The multicatalytic protease complex is also active in the first stages of germination (30 min of incubation), coincident with a high availability of ubiquitinated substrates, whereas it becomes less active at later stages (i.e. 180 min) when the levels of the ubiquitin monomer also markedly decrease. This strongly suggests a major role for tightly regulated protein turnover mediated by the ubiquitin/proteasome system in the very early phases of pollen germination. The loss of ubiquitin monomer at 180 min of incubation could be due to its incorporation into polyubiquitinated substrates that, in the presence of lower levels of the 26S proteasome, could not be degraded, allowing the recycling of free ubiquitin. Although at the moment we do not have direct evidence to support this hypothesis, it is interesting to note that in the presence of proteasome inhibitors, the decline in free monomer levels occurs earlier, in parallel with accumulation of ubiquitinated proteins.
The key role played by the proteasome during pollen germination is clearly demonstrated by the evidence that proteasome inhibitors markedly inhibited pollen tube growth. MG-132 was responsible for a wide range of inhibitory effects regarding percent tube emergence, as well as growth rate and shape. The predominant aberrant morphology induced by MG-132 treatment consisted of expanded tubes with swollen tips. Disruption of the regular cylindrical shape means that cells lose their polarity, delocalizing the growth site otherwise confined to the very tip of the tube. Thus, we assume that MG-132 was rapidly taken up by pollen grains and then it strongly affected proteasome-mediated degradation of crucial factor(s), leading to isotropic growth of tubes. It is interesting that when we removed MG-132 from the medium, reversion of the inhibitory effect on tube growth was observed.
Nevertheless, it should be considered that MG-132 is able to affect calpains as well as the proteasome. No inhibitory effect on pollen growth was observed by using the Cys-protease inhibitor E-64 (40 μm). Some reduction in tube growth rate was induced only with 80 μm E-64, but the percentage of tube emergence was not reduced, and abnormal tube morphology was not observed. From this it may be argued that the effects observed after MG-132 treatment were largely due to inhibition exerted at the proteasome level. The ability of MG-132 to affect pollen proteasome was clearly indicated by results obtained from in vitro assays, and from the accumulation of high-molecular mass ubiquitin-conjugated proteins, as evidenced by immunoblotting experiments. Taken together, this evidence supports the hypothesis that the ubiquitin/proteasome-mediated pathway of protein degradation is a major determinant of tube growth in kiwifruit.
The clasto-lactacystin β-lactone is a derivative, through spontaneous lactonization, of the microbial metabolite lactacystin (Omura et al., 1991). Cells are relatively impermeable to lactacystin, but highly permeable to β-lactone (Dick et al., 1997). The effective β-lactone concentration with kiwifruit pollen tubes, which almost ceased to elongate after 2 h of treatment, was close to double that (4 μm) which prevented tracheary element differentiation in zinnia mesophyll cell cultures after a 96-h exposure (Woffenden et al., 1998). Due to the fact that the inhibitory effect on kiwifruit pollen appeared after a certain time, the percentage of tube emergence was unaffected, and total tube mass was affected to a lesser, though significant, degree than growth rate. No morphological alterations of tubes appeared. As regards the delay of the inhibitory effect, a mechanism of preliminary concentration of the compound could have occurred in the β-lactone-treated tubes. It was reported that once inside cells, β-lactone can hydrolyze to the inactive dihydroxy acid, it can react with the proteasome, or it can reversibly react with glutathione to form lactathione (Dick et al., 1997). Lactathione is able to spontaneously regenerate the β-lactone, thus it can be considered as a reservoir for the prolonged release of the active species (Dick et al., 1997). The β-lactone actually exerted its effects in kiwifruit pollen via inhibition of proteasome; in fact, immunoblot analyses revealed an accumulation of high-molecular mass ubiquitin-conjugated proteins in treated pollen. The inhibitor's ability to bind to pollen proteasome subunits was further confirmed by in vitro activity assays. At 1 h of incubation, some ubiquitin conjugate accumulation had already occurred in β-lactone-treated pollen, that is, before any effect on tube growth became clearly evident. This is not easy to explain, although the same was observed in yeast, where, despite the large reduction in proteolysis, the inhibitor did not reduce growth for several hours (Lee and Goldberg, 1996; Goldberg et al., 1997).
Epoxomicin is a natural product that belongs to a small family of linear peptides sharing a Thr or Ser residue and an α′, β′-epoxyketone. It was isolated based on its potent anti-tumor activity against solid tumors derived from B16 melanome (Meng et al., 1999). Epoxomicin potently inhibits the chymotrypsin-like activity of the proteasome, and also the trypsin-like and peptidyl-glutamyl peptide-hydrolyzing activities. It does not inhibit proteases such as trypsin, chymotrypsin, papain, calpain, and cathepsin B (Meng et al., 1999). Epoxomicin treatment caused a significant inhibition of tube growth rate in kiwifruit pollen, at least in relation to the concentrations tested here. To the best of our knowledge this is the first report on the use of epoxomicin as a tool to explore the role of proteasome in plant cells.
The lack of any inhibition after treatment with different protease inhibitors (apart from E-64), at least at the concentrations we used, suggests that the respective protease activities may not be crucial to sustain growth of pollen tubes.
Growth of pollen tubes is different from that of most other plant cells. In fact, it does not take place over the entire surface of the cell, but is restricted to the tip region. It involves several coordinated processes (vesicle transport and fusion and expansion of the cell wall and plasma membrane) supported by cytoskeletal elements. It requires spatial control (determination of growth site) and temporal regulation; that is, the rate of growth site propagation to which Golgi-derived vesicles are targeted and fused (Li et al., 1999). However, the molecular mechanisms governing tip growth are not fully understood, nor is much known about how many and what sort of enzymes are involved in this process. It has recently been proposed that a pollen-specific Rop GTPase may act as a key molecular switch controlling tip growth (Li et al., 1999). A number of factors, for example, ATPases, Ca2+ gradients, ion channels or regulators of them, and actin-binding proteins are critical for pollen germination (Obermeyer and Bentrup, 1996; Taylor and Hepler, 1997; Franklin-Tong, 1999). Based on this evidence, the inhibition of pollen tube growth observed in the presence of proteasome inhibitors could affect the levels of protein(s) that directly or indirectly regulates the growth mechanism.
The factors that control pollen germination and tube growth are of considerable interest for the comprehension of the metabolic processes involved in fertility and reproduction in plants, and also for the development of molecular tools aimed at manipulating pollen tube growth for practical purposes. The results presented here not only provide the first demonstration that the ubiquitin/proteasome system plays a crucial role in growing pollen tubes, but also that proteasome inhibitors can be used as tools to study pollen tube growth.
MATERIALS AND METHODS
Plant Material
Pollen of male kiwifruit [Actinidia deliciosa var. deliciosa (A. Chev), C. F. Liang et A. R. Ferguson] was obtained from plants of cv Tomuri growing in experimental plots near Faenza (Italy). Anthers collected from central flowers were allowed to dehisce under controlled conditions as described by Calzoni et al. (1979). Pollen was stored at −20°C under NaOH pellets until use.
In Vitro Pollen Germination
After 30 min of rehydration at 30°C under 100% relative humidity, pollen (final concentration of 1 mg mL−1) was suspended in liquid germination medium containing 0.29 m Suc, 1 mm calcium nitrate, and 0.4 mm boric acid, according to Bomben et al. (1999), with some modifications. Aliquots were withdrawn under stirring, transferred to Petri dishes, and incubated at 30°C in the dark. Pollen tube growth over time was quantified photometrically (A500 of sonicated, aqueous suspension of pollen grains and tubes) according to Kappler and Kristen (1987). Increases over time in A500 of kiwifruit pollen cultures were strictly correlated with increases in tube length (Table II). In some cases, tube length and the percentage of tube emergence were determined with an image analyzer-Axioplan microscope (Zeiss, Jena, Germany) combination by scoring at least 300 (length) or 1,000 (emergence) randomly chosen pollen grains per sample, resulting from the sum of several nonoverlapping fields.
Table II.
Tube growth over time of kiwifruit pollen in standard medium
| Time | Photometric Assay | Tube Length |
|---|---|---|
| h | A500 | μm |
| 0 | 0.290 ± 0.003 | 0 |
| 1 | 0.393 ± 0.005 | 76.6 ± 7.5 |
| 2 | 0.497 ± 0.012 | 211.2 ± 25.9 |
| 3 | 0.611 ± 0.014 | 400.0 ± 52.5 |
| 4 | 0.721 ± 0.024 | 720.4 ± 97.0 |
Growth was monitored by photometric assay according to Kappler and Kristen (1987) or by tube length measured with an image analyzer-Zeiss Axioplan microscope combination. Data are means ± sd (n = 5 independent samples for A500 determination; n = 300 pollen grains for tube length measure).
Pollen Extraction
Extracts from ungerminated or germinated (controls and treated) pollen were obtained by four freeze-thaw cycles in 50 mm Tris-HCl buffer, pH 8.0, containing 0.25 m Suc, 1% (w/v) SDS, 5 mm Na-EDTA, 5 mm N-ethylmaleimide, and protease inhibitors such as 1 μg mL−1 pepstatin, 2 μg mL−1 leupeptin, and 2 mm PMSF. The lysed cells were sonicated three times for 30 s, with a 30-s interval, and were then boiled for 10 min. The supernatants obtained after a 15-min centrifugation at 14,000g were used for protein determination (Lowry et al., 1951) or electrophoresis and immunoblotting.
Electrophoresis and Western Blotting
Twenty micrograms of protein from each sample was fractionated on 10% (w/v) polyacrylamide gels according to Laemmli (1970). Protein bands were visualized by staining with Coomassie Blue R-250 or the gels were electroblotted according to Towbin et al. (1979). Blots were probed with an affinity-purified anti-ubiquitin antibody kindly provided by Prof. A.L. Haas (Medical College of Wisconsin, Milwaukee) or with an anti-actin antibody (Sigma, St. Louis). A goat anti-rabbit horseradish peroxidase-conjugated IgG (Bio-Rad, Hercules, CA) was used as secondary antibody. Enhanced chemiluminescence (Amersham International, Little Chalfont, UK) was used as the detection system.
For immunoblot detection of free ubiquitin, protein extracts (5 μg per sample) were fractionated on 14% (w/v) polyacrylamide gels, blotted onto a nitrocellulose membrane with 0.2-μm pores, and probed as described above. Purified bovine ubiquitin (Sigma) was used as a standard.
Quantification of Conjugated Ubiquitin Pools
Conjugated ubiquitin was quantified by a solid-phase immunochemical method with some modifications from the previously described procedure (Haas and Bright, 1985). Samples were dot-blotted onto polyvinylidene difluoride membranes (Bio-Rad). The membranes were probed sequentially with an affinity-purified rabbit polyclonal antibody specific for conjugated ubiquitin, kindly provided by Prof. A.L. Haas, and horseradish-peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad). Detection was by the enhanced chemiluminescence system, and films were quantified by densitometry in an enhanced laser densitometer (Ultroscan XL, Pharmacia LKB, Uppsala). Student's t test was used to compare means ± sd of control and treated pollen.
Proteasome Activity Assay
The activity of the 20S and 26S proteasome was assayed spectrofluorometrically using the fluorogenic substrate sLLVY-NH-Mec (Sigma). Ungerminated or germinated pollen samples were extracted in cold 50 mm HEPES- [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] KOH buffer, pH 7.8, containing 2 mm dithiothreitol and 0.25 m Suc using a Potter-Elvehjem apparatus, and were then centrifuged for 10 min at 500g. Aliquots of the supernatants corresponding to 500 μg of protein were incubated at 30°C for 5 min in 100 mm HEPES-KOH buffer, pH 7.8, containing 5 mm MgCl2 and 10 mm KCl, in the presence or absence of 2 mm ATP. The reaction was initiated by adding the fluorogenic substrate to a final concentration of 0.2 mm. The breakdown of the peptide was continuously monitored for 5 min using a fluorescence spectrophotometer (FP-770, Jasco, Tokyo), with an excitation wavelength of 370 nm and an emission wavelength of 430 nm.
Proteasome activity was also assayed in vitro in the presence of the proteasome inhibitors MG-132 (Biomol Research Laboratory, Plymouth Meeting, PA), clasto-lactacystin β-lactone (Calbiochem-Novabiochem, La Jolla, CA), or epoxomicin, in part kindly provided by Prof. C.M. Crews and L. Meng (Yale University, New Haven, CT), and in part purchased from Affiniti Research Products (Exeter, UK). In these cases, the extracts were pre-incubated in ice for 5 min in the presence of the inhibitor.
In Vivo Proteasome or Protease Inhibitor Treatments
The proteasome inhibitors MG-132, epoxomicin, and clasto-lactacystin β-lactone, the irreversible inhibitor of calpains and other Cys-proteases E-64 (Biomol), or the Ser-protease, aspartic-protease, and Cys/Ser-protease inhibitors PMSF, pepstatin, and leupeptin (all from Sigma), were separately added to the medium at the time of pollen culture initiation (time 0) at the concentration indicated in figure legends. Controls were set up with DMSO solvent for MG-132, clasto-lactacystin β-lactone, and epoxomicin treatments, or ethanol solvent for PMSF and pepstatin treatments. Solvent concentrations were never higher than 0.2% (w/v). A linear regression test was used to compare the slopes of tube growth over time.
ACKNOWLEDGMENTS
We are thankful to Prof. Arthur L. Haas for providing the anti-ubiquitin antibody and to Prof. Craig M. Crews for providing part of the epoxomicin used in this work.
Footnotes
This work was supported by the Italian Ministero per l'Università e la Ricerca Scientifica e Tecnologica.
LITERATURE CITED
- Alché JD, Butowt R, Castro AJ, Rodriguez-Garcia MI. Ubiquitin and ubiquitin-conjugated proteins in the olive (Olea europea L.) pollen. Sex Plant Reprod. 2000;12:285–291. [Google Scholar]
- Bahrami AR, Gray JE. Conservation of proteasome structure and activity between plants and other eukaryotes. Biochem Soc Trans. 1998;26:395. doi: 10.1042/bst026s395. [DOI] [PubMed] [Google Scholar]
- Becker F, Buschfeld E, Schell E, Bachmair A. Altered response to viral infection by tobacco plants perturbed in ubiquitin system. Plant J. 1993;3:875–881. [Google Scholar]
- Becker J, Kempf R, Jeblick W, Kauss H. Induction of competence for elicitation of defense responses in cucumber hypocotyls requires proteasome activity. Plant J. 2000;21:311–316. doi: 10.1046/j.1365-313x.2000.00677.x. [DOI] [PubMed] [Google Scholar]
- Belknap WR, Garbarino JE. The role of ubiquitin in plant senescence and stress response. Trends Plant Sci. 1996;1:331–335. [Google Scholar]
- Bomben C, Malossini C, Cipriani G, Testolin R (1999) Long term storage of kiwifruit pollen. Acta Hortic (in press)
- Callis J, Bedinger P. Developmentally regulated loss of ubiquitin and ubiquitinated proteins during pollen maturation in maize. Proc Natl Acad Sci USA. 1994;91:6074–6077. doi: 10.1073/pnas.91.13.6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzoni GL, Speranza A. Developmental state of mature apple pollen: study of RNA and protein synthesis during in vitro germination. Physiol Vég. 1986;24:53–62. [Google Scholar]
- Calzoni GL, Speranza A, Bagni N. In vitro germination of apple pollen. Sci Hortic. 1979;10:49–55. [Google Scholar]
- Chu-Ping M, Vu JH, Proske RJ, Slaughter CA, De Martino GN. Identification, purification and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20S proteasome. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dick LR, Cruikshank AA, Destree AT, Grenier L, McCormack TA, Melandri FD, Nunes SL, Palombella VJ, Parent LA, Plamondon L. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- Fernandez JA, DeLuca V. Ubiquitin-mediated degradation of tryptophan decarboxylase from Catharanthus roseus. Phytochemistry. 1994;36:1123–1128. [Google Scholar]
- Ferreira RMB, Rodrigues Ramos PC, Franco E, Ricardo CPP, Teixeira ARN. Changes in ubiquitin and ubiquitin-protein conjugates during seed formation and germination. J Exp Bot. 1995;46:211–219. [Google Scholar]
- Franklin-Tong VE. Signaling and the modulation of pollen tube growth. Plant Cell. 1999;11:727–738. doi: 10.1105/tpc.11.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J. Cell cycle-dependent proteolysis in plants: identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG-132. Plant Cell. 1998;10:2063–2076. doi: 10.1105/tpc.10.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL, Akopian TN, Kisselev AF, Lee DH. Protein degradation by the proteasome and dissection of its in vivo importance with synthetic inhibitors. Mol Biol Rep. 1997;24:69–75. doi: 10.1023/a:1006860828265. [DOI] [PubMed] [Google Scholar]
- Guyon VN, Astwood JD, Garner EC, Dunker AK, Taylor LP. Isolation and characterization of cDNAs expressed in the early stages of flavonol-induced pollen germination in petunia. Plant Physiol. 2000;123:699–710. doi: 10.1104/pp.123.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Bright PM. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem. 1985;260:12464–12473. [PubMed] [Google Scholar]
- Hoekstra FA, Bruinsma J. Protein synthesis of binucleate and trinucleate pollen and its relationship to tube emergence and growth. Planta. 1979;146:559–566. doi: 10.1007/BF00388832. [DOI] [PubMed] [Google Scholar]
- Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Peichaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Schlenker S. Selective protein degradation: a journey's end within the proteasome. Cell. 1995;82:881–884. doi: 10.1016/0092-8674(95)90021-7. [DOI] [PubMed] [Google Scholar]
- Kanayama H, Tamura T, Ugai S, Kagawa S, Tanahashi N, Yoshimura T, Tanaka K, Ichihara A. Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by closely linked mechanisms. Eur J Biochem. 1992;206:567–578. doi: 10.1111/j.1432-1033.1992.tb16961.x. [DOI] [PubMed] [Google Scholar]
- Kappler R, Kristen U. Photometric quantification of in vitro pollen tube growth: a new method suited to determine the cytotoxicity of various environmental substances. Environ Exp Bot. 1987;27:305–309. [Google Scholar]
- Kulikauskas R, Hou A, Muschietti J, McCormick S. Comparison of diverse plant species reveal that only grasses show drastically reduced levels of ubiquitin monomer in mature pollen. Sex Plant Reprod. 1995;8:326–332. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophylum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mascarenhas JP. Pollen gene expression: molecular evidence. Int Rev Cytol. 1992;140:3–18. doi: 10.1016/s0074-7696(08)61091-8. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP. Molecular mechanisms of pollen tube growth and differentiation. Plant Cell. 1993;5:1303–1314. doi: 10.1105/tpc.5.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP, Mermelstein J. Messenger RNAs: their utilization and degradation during pollen germination and tube growth. Acta Soc Bot Pol. 1981;50:13–20. [Google Scholar]
- Meng L, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo anti-inflammatory activity. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26 S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S. LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 1994;6:321–338. doi: 10.1046/j.1365-313x.1994.06030321.x. [DOI] [PubMed] [Google Scholar]
- Obermeyer G, Bentrup FW. Regulation of polar cell growth and morphogenesis. Progr Bot. 1996;57:54–67. [Google Scholar]
- Omura S, Fujimoto T, Otoguro K, Matsuzaki K, Moriguchi R, Tanaka H, Sasaki Y. Lactacystin, a novel microbial metabolite, induces neuritogenesis of neuroblastoma cells. J Antibiot (Tokyo) 1991;44:113–118. doi: 10.7164/antibiotics.44.113. [DOI] [PubMed] [Google Scholar]
- Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kB1 precursor protein and the activation of NF-kB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Peters JM. Proteasomes: protein degradation machines of the cell. Trends Biochem Sci. 1994;19:377–382. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Stary S, Schlögelhofer P, Becker F, Nejinskaia V, Bachmair A. PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:7904–7908. doi: 10.1073/pnas.95.14.7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Scoccianti V, Corsi D, Magnani M. Ubiquitin conjugation to endogenous proteins in the dormant tuber of Helianthus tuberosus and during the first cell cycle. Physiol Plant. 1997;101:77–85. [Google Scholar]
- Scoccianti V, Speranza A, Crinelli R, Calzoni GL, Biasi R, Altamura MM, Bagni N. Development-related changes of protein ubiquitination in pollen from male and female kiwifruit (Actinidia deliciosa) Physiol Plant. 1999b;107:128–135. [Google Scholar]
- Scoccianti V, Speranza A, Crinelli R, Calzoni GL, Teti G, Bagni N. Protein targeting by ubiquitin during anther and pollen development in male and female flowers of kiwifruit (Actinidia deliciosa) In: Clément C, Pacini E, Audran JC, editors. Anther and Pollen. Berlin: Springer-Verlag; 1999a. pp. 45–53. [Google Scholar]
- Sin N, Kim KB, Elofsson M, Meng L, Auth H, Kwok BHB, Crews CM. Total synthesis of the potent proteasome inhibitor epoxomicin: a useful tool for understanding proteasome biology. Bioorg Med Chem Lett. 1999;9:2283–2288. doi: 10.1016/s0960-894x(99)00376-5. [DOI] [PubMed] [Google Scholar]
- Stephenson P, Rubinstein B. Characterization of proteolytic activity during senescence in daylilies. Physiol Plant. 1998;104:463–473. [Google Scholar]
- Taylor LP, Hepler PK. Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- Towbin H, Stachelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubuki S, Saito Y, Tomioka M, Ito H, Kawashima S. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J Biochem. 1996;119:572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- Viestra RD. Protein degradation in plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:385–410. [Google Scholar]
- Woffenden BJ, Freeman TB, Beers EP. Proteasome inhibitors prevent tracheary element differentiation in zinnia mesophyll cell cultures. Plant Physiol. 1998;118:419–430. doi: 10.1104/pp.118.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]