Abstract
Increasing atmospheric carbon dioxide (CO2) concentrations are impacting the global climate, resulting in significant interest in soil carbon sequestration as a mitigation strategy. While recognized that mineral-associated organic matter (MAOM) in soils is mainly formed through microbial activity, our understanding of microbial-derived MAOM formation processes remains limited due to the complexity of the soil environment. To gain insights into this issue, we incubated fresh soil samples for 45 days with one of three mineral additions: Sand, Kaolinite+Sand, or Illite+Sand. 16S rRNA V3/V4 gene amplicon sequencing was then conducted on samples using an Illumina NextSeq 2000 flow cell. The reads were analyzed and taxonomically assigned with QIIME2 v2023.5.1 and SILVA 138. The dataset has been made publicly available through NCBI GenBank under BioProject ID PRJNA1124235. This dataset is important and useful as it provides valuable insights into the interactions between soil minerals and microbial communities, which can inform strategies for enhancing soil carbon sequestration and mitigating climate change. Moreover, it serves as a crucial reference for future studies, offering a foundational understanding of microbial dynamics in soil systems and guiding further research in microbial ecology and carbon cycling.
Keywords: Mineral-associated organic matter, Soil mineral mesocosms, Microbiome, Climate change
Specifications Table
| Subject | Agricultural Microbiology, Environmental Engineering, Microbiome. |
| Specific subject area | Soil Metagenomics. |
| Type of data | Table, Figure Raw, Analyzed, Filtered, Processed |
| Data collection | Soil samples from a depth of 0–10 cm were collected from a long-term cultivated agricultural field at Colorado State University. Amplicon sequencing of the 16S rRNA V3/V4 gene region was performed using the Quick-16S kit (Zymo Research). The 341F (CCTACGGGDGGCWGCAGCCTAYGGGGYGCWGCAG) and 806R (GACTACNVGGGTMTCTAATCC) primers were used on a P1 600-cycle NextSeq2000 Flowcell (Illumina), producing 2 × 301 bp paired-end reads. |
| Data source location | Soil samples were collected from Fort Collins, Colorado, USA (40°44′ N; 104°59′ W). Sequencing data were generated by SeqCenter (91 43rd Street, Ste. 250, Pittsburgh, PA 15,201). |
| Data accessibility | Repository name: National Center for Biotechnology Information Sequence Read Archive (NCBI SRA). Data identification number: BioProject ID PRJNA1124235 [Accession Numbers: SRR29422095 - SRR29422109] Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1124235 Instructions for accessing these data: Click the above URL |
| Related research article | None |
1. Value of the Data
-
•
The data provides insights into the microbial diversity of mineral-treated soils, offering valuable baseline information for studies on soil carbon dynamics and microbial ecology.
-
•
Significant shifts in microbial composition were observed in kaolinite and illite-treated soils, compared with control samples.
-
•
Despite the importance of understanding microbial roles in MAOM formation, there is a scarcity of such studies, and this dataset contributes to addressing that gap.
2. Background
Soil organic matter (SOM), the largest terrestrial carbon reservoir than the atmosphere and vegetation combined, is crucial for soil fertility, climate change mitigation, and human well-being [1,2]. Recent studies found that 34–47 % of the mineral-associated organic matter (MAOM) pool originates from microbial inputs, highlighting the need to clarify the poorly understood molecular mechanisms and specific microbial characteristics that derive MAOM formation [1,3].
3. Data Description
Microbial composition in various types of mineral-treated soil was identified using V3/V4 16S rRNA amplicon sequencing. The links and accession numbers to the raw reads can be found in Table 1. A total of 6,309,552 raw sequencing reads were obtained with 341F (CCTACGGGDGGCWGCAGCCTAYGGGGYGCWGCAG) as the forward primer and 806R (GACTACNVGGGTMTCTAATCC) as the reverse primer across 15 samples. Denoised sequences were assigned operational taxonomic units (OTUs) using the SILVA 138 database and the VSEARCH4 utility within QIIME2’s feature-classifier plugin. The OTUs were then collapsed to their lowest taxonomic level, and their counts were adjusted to reflect their relative frequency within each sample. In total, 815 species were identified across all samples.
Table 1.
Microbiome amplicon data summary. Soil T0, Soils at beginning of incubation; Soil T1, Soils at 45 days of incubation (control); Sand, Soils incubated with sand for 45 days; Kaolinite+Sand, Soils incubated with a mixture (1:1) of kaolinite and sand for 45 days; Illite+Sand, Soils incubated with a mixture (1:1) of illite and sand for 45 days. Raw Sequencing data is available on the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under PRJNA1124235.
| Sample ID | Sample type | Collection Date (mo/day/yr) | No. of Reads | SRA Accession |
|---|---|---|---|---|
| a48c0 | Soil T0 | 07/24/2023 | 270,546 | SRR29422109 |
| a5f6a | Soil T0 | 07/24/2023 | 233,566 | SRR29422108 |
| a608d | Soil T0 | 07/24/2023 | 377,244 | SRR29422102 |
| a65e1 | Soil T1 | 9/14/2023 | 354,654 | SRR29422101 |
| a67ba | Soil T1 | 9/14/2023 | 282,982 | SRR29422100 |
| a6a79 | Soil T1 | 9/14/2023 | 301,058 | SRR29422099 |
| a6cc3 | Sand | 9/14/2023 | 393,770 | SRR29422098 |
| a6dd4 | Sand | 9/14/2023 | 464,496 | SRR29422097 |
| a6e47 | Sand | 9/14/2023 | 450,618 | SRR29422096 |
| a7805 | Kaolinite+Sand | 9/14/2023 | 519,446 | SRR29422095 |
| a88eb | Kaolinite+Sand | 9/14/2023 | 499,708 | SRR29422107 |
| a8e39 | Kaolinite+Sand | 9/14/2023 | 515,168 | SRR29422106 |
| a9171 | Illite+Sand | 9/14/2023 | 583,830 | SRR29422105 |
| a92b0 | Illite+Sand | 9/14/2023 | 561,644 | SRR29422104 |
| a9956 | Illite+Sand | 9/14/2023 | 500,822 | SRR29422103 |
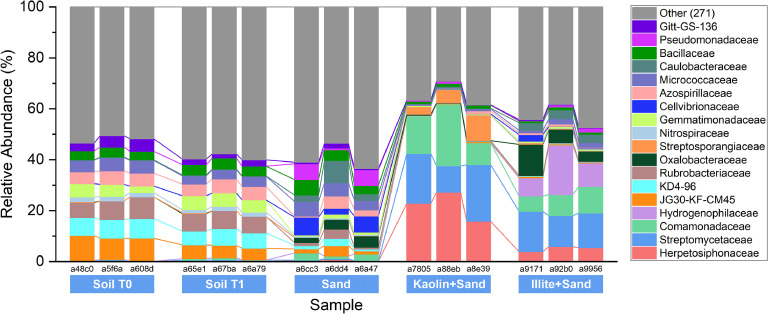
Microbiome amplicon data analyzed in this study (Table 1 and Fig. 1) reveal no single dominant family (> 10 % relative abundance) in source soil samples (i.e., Soil T0), control soil incubation samples (i.e., Soil T1), and sand only incubated samples. Kaolinite+Sand samples were dominated by Herpetosiphonaceae (15.81–27.24 %), Streptomycetaceae (15.81–24.26 %), and Comamonadaceae (8.63–24.26 %), while Illite+Sand samples were dominated by the Streptomycetaceae (12.06–15.81 %) and Hydrogenophilaceae (7.11–19.49 %) (Fig. 1). These samples exhibited lower alpha diversity, indicated by the Shannon index (median values of 6.0 vs. 7.5; P value of < 0.01, Kruskal–Wallis Test). For beta-diversity, we utilized permutational multivariate analysis of variance (PERMANOVA) based on Bray Curtis distance metrics to evaluate the differentiation of microbial composition among soil samples. Samples were grouped by two categorical factors: sampling time (i.e., 0 days & 45 days) and mineral addition (i.e., binary information where 0 indicates non-added and 1 indicates added). Both factors significantly affected the beta-diversity of the bacterial communities (R² = 0.1911 for different time points and R² = 0.2856 for mineral addition, p < 0.005, PERMANOVA). These data serve as a robust baseline for future research, providing a detailed understanding of microbial composition in mineral-treated soils. These data are a valuable resource for researchers exploring the complex interactions between soil microbes and MAOM formation, and they offer a foundation for long-term studies on microbial ecology.
Fig. 1.
Relative abundance of the taxonomic composition of the microbiomes derived from soil samples and minerals. Extraction from soil samples was performed at the beginning (T0) and the end of incubation (T1 after 45 days for all other samples). Sample IDs of Soil T0 (a48c0, a5f6a, a608d), Soil T1 (a65e1, a67ba, a6a79), Sand (a6cc3, a6dd4, a6a47), Kaolinite+Sand (a7805, a88eb, a8e39), and Illite+Sand (a9171, a92b0, a9956).
4. Experimental Design, Materials and Methods
4.1. Sample collection and processing
Soil samples were collected from a cornfield at Colorado State University (40°44′ N, 104°59′ W) at 0–10 cm depth, transported on ice, and refrigerated. Samples were sieved (2 mm) to remove plant debris and refrigerated. Sieved soils had moisture and water holding capacity (SWHC) determined. 250 µm mesh bags were soaked in methanol, rinsed with water, and dried to kill potential live organisms. Minerals sand (SiO2), Kaolinite, and Illite were grounded to <53 µm. A pH=4 artificial root exudate solution was made with a 2.16:1 glucose ratio (6.8 M) and citric acid (3.2 M). Treatments were composed of 2 soil controls, 1 undergoing 45-day incubation (Soil T1) and the other one sampled on the day of experimental setup (Soil T0). The other 3 treatments were composed of Sand only, Kaolinite+Sand, or Illite+Sand, with the Kaolinite or Illite mixed with Sand (1:1 ratio) to prevent clumping. Minerals were wetted with 1 mL of calcium chloride (0.01 M) and 6 ml of root exudate, then, 12 g of each mineral were added to the washed mesh bags. The soil mesocosms were set up in 250 mL mason jars and contained 1 mineral mesh bag in the middle of two layers of 25g of 60 % SWHC-wetted soil. Control samples used 50g of 60 % SWHC-wetted soil. Each treatment was replicated three times. Jars were sealed with parafilm and incubated at 25 °C for 45 days.
4.2. DNA extraction and sequencing
Samples were sent to SeqCenter (Pittsburgh, PA, USA) following the incubation period; except for the no-incubation control samples (Soil T0), which were sent at the experimental setup. DNA was extracted using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research, Irvine, CA, USA). DNA concentrations were determined with Qubit 1X dsDNA assays (Invitrogen, Thermo Fisher Scientific, Wilmington, DE, USA). Amplicon sequencing of the 16S rRNA V3/V4 gene was conducted with the Quick-16S kit (Zymo Research) using the 341F (CCTACGGGDGGCWGCAGCCTAYGGGGYGCWGCAG) and 806R (GACTACNVGGGTMTCTAATCC) primers on a P1 600cyc NextSeq2000 Flowcell (Illumina), generating 2 × 301 bp paired end reads. Quality control and adapter trimming were performed with bcl-convert1 v4.2.4 (Illumina).
4.3. Data processing
Data analysis were conducted using QIIME2 v2023.5.1 [4]. The Cutadapt plugin was used to remove primer sequences (forward trim sequence: CCTAYGGGNBGCWGCAG, reverse trim sequence GACTACNVGGGTMTCTAATCC) [5], and the DADA2 plugin was used to denoise the data [6]. Denoised sequences were assigned operational taxonomic units (OTUs) using the SILVA 138 99 % OTUs full-length sequence database and the VSERACH utility within QIIME2’s feature-classifier plugin [7]. OTUs were then collapsed to their lowest taxonomic units, and their counts were converted to reflect their relative frequency within each sample. Microbial composition and alpha diversity (Shannon's diversity index, observed features, Faith's Phylogenic Diversity, and Pielou's Evenness) analyses were performed with QIIME2. Differences in alpha diversity among different types of samples were identified using the Kruskal–Wallis test in R version 4.3.2. Permutational multivariate analysis of variance (PERMANOVA) was applied to assess the effects of measured variables, such as time points and mineral addition, on the Bray Curtis distances. This was done using the Adonis2 function of the vegan package in R version 4.3.2.
Limitations
Not applicable.
Ethics Statement
The study follows the ethical requirements for publication in Data in Brief. It does not involve human subjects, animal experiments, or any data collected from social media platforms.
Credit Author Statement
Daniel Lee: writing—original draft, data analysis, visualization; Fernanda C C Oliveira: sample collection, writing—review and editing; Richard T. Conant: conceptualization, supervision, writing—review and editing; Minjae Kim: supervision, conceptualization, writing—review and editing.
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The author (Daniel Lee) is also affiliated with Paul Laurence Dunbar High School.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Richard T. Conant, Email: rich.conant@colostate.edu.
Minjae Kim, Email: minjae.kim@uky.edu.
Data Availability
References
- 1.Chang Y., Sokol N.W., van Groenigen K.J., Bradford M.A., Ji D., Crowther T.W., Liang C., Luo Y., Kuzyakov Y., Wang J. A stoichiometric approach to estimate sources of mineral-associated soil organic matter. Glob. Change Biol. 2024;30(1):e17092. doi: 10.1111/gcb.17092. [DOI] [PubMed] [Google Scholar]
- 2.Angst G., Mueller K.E., Nierop K.G., Simpson M.J. Plant-or microbial-derived? A review on the molecular composition of stabilized soil organic matter. Soil Biol. Biochem. 2021;156 [Google Scholar]
- 3.Kleber M., Bourg I.C., Coward E.K., Hansel C.M., Myneni S.C., Nunan N. Dynamic interactions at the mineral–organic matter interface. Nat. Rev. Earth Environ. 2021;2(6):402–421. [Google Scholar]
- 4.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 6.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.