Summary
Evaluating the invasiveness of lung adenocarcinoma is crucial for determining the appropriate surgical strategy, impacting postoperative outcomes. This study developed a multimodality model combining radiomics, intraoperative frozen section (FS) pathology, and clinical indicators to predict invasion status. The study enrolled 1,424 patients from two hospitals, divided into multimodal training, radiology testing, and pathology testing cohorts. A prospective validation cohort of 114 patients was selected between March and May 2023. The radiomics + pathology + clinical indicators multimodality model (multi-RPC model) achieved an area under the curve (AUC) of 0.921 (95% confidence interval [CI] 0.899–0.939) in the multimodal training cohort and 0.939 (95% CI 0.878–0.975) in the validation cohort, outperforming single- and dual-modality models. The multi-RPC model’s predictive accuracy of 0.860 (95% CI 0.782–0.918) suggests that it could significantly reduce inappropriate surgical procedures, enhancing precision oncology by integrating multimodal information to guide surgical decisions.
Subject areas: Oncology, Medical imaging, Artificial intelligence applications
Graphical abstract
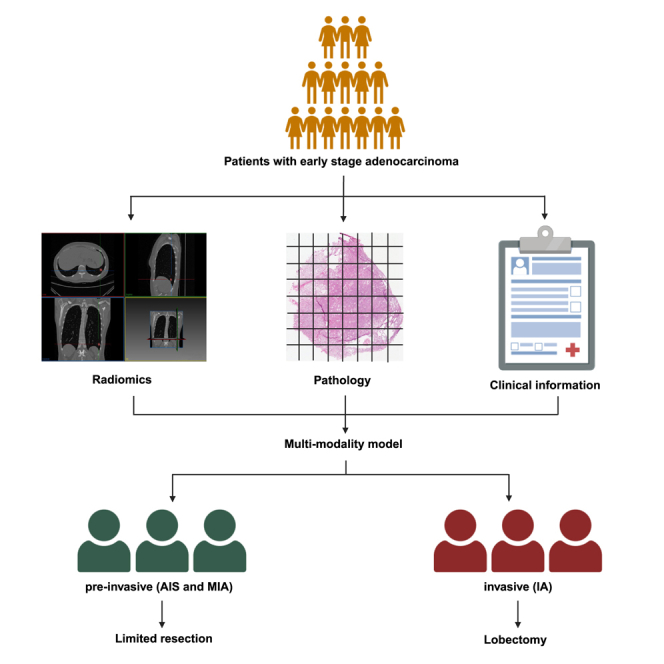
Highlights
-
•
A high-efficient pipeline was put forward by combining multi-modal information
-
•
The EfficientNet B5 model was used to analyze pathologic images
-
•
The performance of the multimodality model was tested in an independent cohort
Oncology; Medical imaging; Artificial intelligence applications
Introduction
The diagnosis of small-sized non-small-cell lung cancer (NSCLC) has increased with the increasing utilization of computed tomography (CT) scanning of the thorax as the primary lung cancer screening program. Two new categories of lung adenocarcinoma (LUAD), proposed by the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society,1 adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA), have garnered the interest of thoracic surgeons and pathologists. These two categories are often referred to as the pre-invasive stages of LUAD.2 Multiple studies have revealed that patients with AIS and MIA have a 100% 5-year disease-free survival rate after resection3,4,5; however, patients with invasive adenocarcinoma (IA) have a worse 5-year disease-specific survival rate with a higher risk of recurrence.6 Many surgeons currently propose an appropriate technique for limited resection, including wedge resection and segmentectomy, as an effective treatment for AIS/MIA. Furthermore, compared to lobectomy, sublobar resection can better maintain pulmonary function and enable another resection in case of subsequent primary lung cancer.7,8 However, lobectomy remains the standard treatment for most IA, owing to improved prognosis.
Intraoperative frozen section (FS) diagnosis may be an effective method to determine AIS, MIA, and IA and to guide surgical strategy for small-sized LUAD. According to Liu et al., the total concordance rate between FS and final pathology (FP) is 84.4%.9 In some investigations, the concordance rate among patients diagnosed with AIS or MIA using FS was only 63.24%.10 Pre-invasive stages and nodules smaller than 1 cm are more challenging to diagnose, and errors are more prevalent in these instances.11 Additionally, an overestimated level of invasiveness is frequently observed.12 Severe distortion of the architecture and other sampling problems may compromise the accuracy and reliability of the interpretation.13 The potential for limited resection is mostly based on intraoperative FS diagnosis; hence, there is an increasing demand for timely preoperative or intraoperative diagnosis with high accuracy.
Many methods have been developed to aid pathological classification preoperatively or intraoperatively, particularly using features based on CT imaging.14,15,16 Matthew et al. developed highly accurate machine learning models based on radiomic features extracted from low-dose CT images that can assess pulmonary nodules for risk of malignancy.17 Contrary to previous studies, we attempted to develop a model that integrates multimodal information, including clinical and laboratory data, radiomics, and intraoperative FS analysis, to improve the diagnosis accuracy for identifying AIS, MIA, and IA intraoperatively, in particular focusing on the small-sized (≤3 cm) lung nodule cases, which have a high probability for incorrect diagnosis or with uncertainty during intraoperative FS diagnosis. Artificial intelligence-based image analysis of radiologic and pathologic images enables the development of an automated method for processing and analyzing multimodal data that is efficient and labor-saving with satisfactory performance.
Results
Clinical characteristics of patients with stage Ⅰ LUAD who received surgery
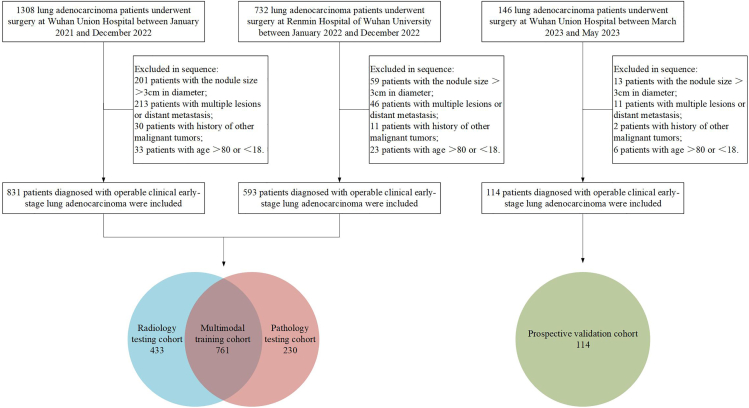
The process of patient selection for four cohorts was presented in Figure 1, and the research protocol of this study was shown in Figure 2. Table 1 presents the clinical characteristics of the four cohorts. No significant differences were found between multimodal training cohort and other cohorts in terms of age, sex, CT location, and smoking history (p > 0.05). The median sizes of the nodules were 14 mm (interquartile range [IQR], 10–19 mm), 12 mm (IQR, 9–16 mm), 10 mm (IQR, 7–15 mm), and 15 mm (IQR, 11–20 mm) for largest diameter in the multimodal training, radiology testing, pathology testing, and prospective validation cohorts, respectively. The numbers of patients identified with AIS were 84 (11.1%), 92 (21.3%), 67 (29.1%), and 17 (14.9%) in the multimodal training, radiology testing, pathology testing, and prospective validation cohorts, respectively. The number of patients identified with MIA was 170 (22.3%), 194 (44.8%), 63 (27.4%), and 20 (17.5%) in the four cohorts, respectively. The proportions of patients identified with IA were 66.6%, 33.9%, 43.5%, and 67.5% in the four cohorts, respectively. The total concordance rates between FS and FP were 78.3%, 71.2%, and 68.4% in the multimodal training, pathology testing, and prospective validation cohorts, respectively.
Figure 1.
Flowchart of patient selection
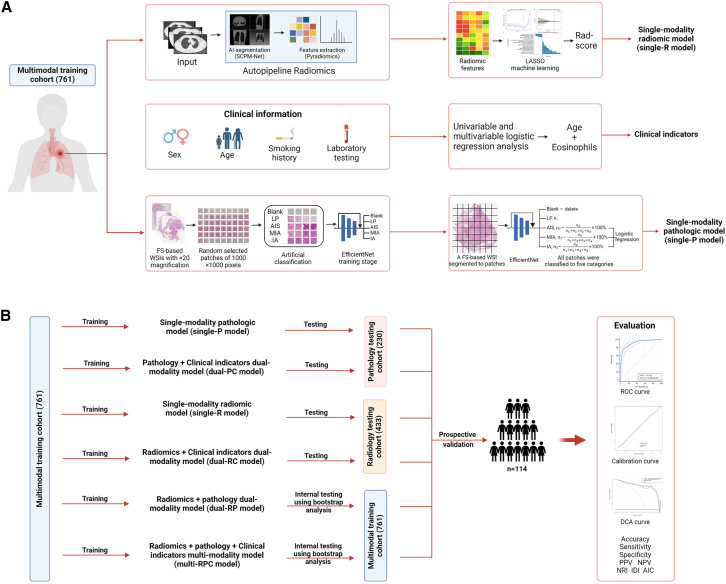
Figure 2.
Workflow of the study
(A) Images of non-enhanced thoracic CT and FS-based hematoxylin and eosin-stained WSIs with 20× magnification were collected and segmented for radiomic feature extraction and pathologic parameter generation, respectively. The classification of five pathologic categories (Blank, LP, AIS, MIS, and IA) for WSI patches was accomplished through pretrained EfficientNet model. Single-R model was developed with Rad-score, and single-P model was developed with AIS%, MIA%, and IA%. Clinical indicators, including age and eosinophils, were selected by using univariable and multivariable logistic regression analysis. All dual-/multimodality models were developed by logistic regression.
(B) All models were trained in the multimodality training cohort, while single-P and dual-PC models were tested in the pathology testing cohort, single-R and dual-RC models were tested in the radiology testing cohort, and dual-RP and multi-RPC models underwent internal testing by bootstrap analysis in the multimodality training cohort. All models were further validated in the prospective validation cohort. FS: frozen section; WSI: whole-slide image; LP: lung parenchyma; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma; IA, invasive adenocarcinoma. AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; NRI, net reclassification improvement; IDI, integrated discrimination improvement; AIC, Akaike information criterion.
Table 1.
Patient characteristics
| Characteristics | Multimodal training cohort (761) | Radiology testing cohort (433) | Pathology testing cohort (230) | Prospective validation cohort (114) |
|---|---|---|---|---|
| Age, years | ||||
| Median | 58 | 56 | 57.5 | 58 |
| IQR | (51–65) | (48–64) | (50–65) | (52–65) |
| Sex | ||||
| Male | 279 (36.7) | 147 (33.9) | 88 (38.3) | 39 (34.2) |
| Female | 482 (63.3) | 286 (66.1) | 142 (61.7) | 75 (65.8) |
| Asian | 761 (100) | 433 (100) | 230 (100) | 114 (100) |
| CT location | ||||
| Left upper lobe | 192 (25.3) | 125 (28.9) | 65 (33.5) | 33 (28.9) |
| Left lower lobe | 112 (14.8) | 50 (11.5) | 21 (10.8) | 20 (17.5) |
| Right upper lobe | 274 (36.1) | 162 (37.4) | 70 (36.1) | 34 (29.8) |
| Right middle lobe | 46 (6.1) | 33 (7.6) | 10 (5.2) | 8 (7.0) |
| Right lower lobe | 134 (17.7) | 63 (14.5) | 28 (14.4) | 19 (16.7) |
| CT size, mm | ||||
| Median | 14 | 12a | 10a | 15 |
| IQR | (10–19) | (9–16) | (7–15) | (11–20) |
| Smoking history | ||||
| No | 620 (81.5) | 361 (83.4) | 189 (82.2) | 92 (80.7) |
| Yes | 141 (18.5) | 72 (16.6) | 41 (17.8) | 22 (19.3) |
| Invasion status diagnosed by FFPE | ||||
| AIS | 84 (11.1) | 92 (21.3)a | 67 (29.1)a | 17 (14.9) |
| MIA | 170 (22.3) | 194 (44.8)a | 63 (27.4)a | 20 (17.5) |
| IA | 507 (66.6) | 147 (33.9)a | 100 (43.5)a | 77 (67.5) |
| Concordance rate between FS and FP (%) | ||||
| AIS | 53.8% | – | 70.1% | 88.2% |
| MIA | 63.8% | – | 61.9% | 70.0% |
| IA | 85.2% | – | 78.0% | 63.6% |
| Total | 78.3% | – | 71.2% | 68.4% |
Data presented as number (percentage, %), with the exception of age and CT size (median [IQR]).
IQR, interquartile range; CT, computed tomography; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma; IA, invasive adenocarcinoma; FS, frozen section; FP, final pathology.
p value (<0.05) is statistically significant for the radiology testing cohort, pathology testing cohort, and prospective validation cohort, respectively, when compared with the multimodal training cohort.
Preoperative clinical indicators and CT radiomic feature selection
Univariable and multivariable analyses were used to select clinical indicators in the multimodal training cohort, with 254 pre-invasive LUAD cases and 507 invasive LUAD cases (Table S2). Univariable and multivariable logistic regression analyses demonstrated that age (hazard ratio [HR], 1.10 [95% confidence interval (CI), 1.04–1.17]; p = 0.001) and eosinophil count (HR, 20.96 [95% CI, 1.18–373.04]; p = 0.038) were associated with an increasing probability of invasive LUAD. Subsequently, age and eosinophil count were combined to predict the invasive status in the multimodal training cohort, with an area under the curve (AUC) of 0.672 (95% CI 0.637–0.705), indicating the discriminative capacity (Figure S1). Characteristics of demographic information and clinical indicators of participants in radiology testing, pathology testing, and prospective validation cohorts were shown in Table S3. Overall, 100 radiomic features were extracted from each patient’s CT images. Seventeen features with nonzero coefficients were selected to establish the Rad-score using a least absolute shrinkage and selection operator logistic regression (Figure S2).
Intraoperative FS pathologic classification
The AUCs for the EfficientNet B5 model to distinguish “blank,” “lung parenchyma,” “AIS,” “MIA,” or “IA” patches from the other four classes were 1.00, 1.00, 0.95, 0.89, and 0.96, respectively. In addition, the overall accuracy for identifying the five pathologic classes was 0.82 (Figure S3). After blank patches were deleted, AIS%, MIA%, and IA% were calculated for each whole-slide image (WSI) collected in the multimodal training, pathology testing, and prospective validation cohorts (Table S4). The IA% was extremely low (median <4%) in the AIS and MIA groups of all three cohorts, whereas the median AIS% was the highest in the MIA groups of all three cohorts. However, the three proportions were not as diverse in IA groups as in the AIS and MIA groups in all three cohorts.
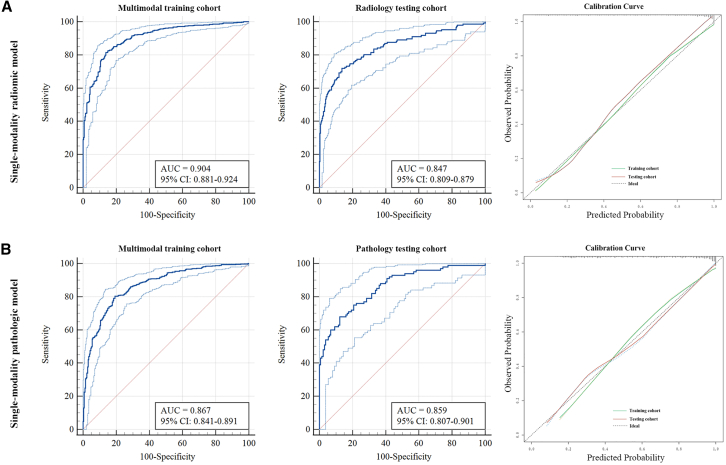
Predictive performance of single-R and single-P models
Based on the selected radiomic features, the CT-based single-modality radiomic model (single-R model) achieved an AUC of 0.904 (95% CI 0.881–0.924) in the multimodal training cohort. Subsequently, the single-R model was tested in the radiology testing cohort, achieving an AUC of 0.847 (95% CI 0.809–0.879) (Figure 3A). The single-modality pathologic model (single-P model) achieved an AUC of 0.867 (95% CI 0.841–0.891) in the multimodal training cohort and attained a similar AUC of 0.859 (95% CI 0.807–0.901) when tested in the pathology testing cohort (Figure 3B). The single-P model demonstrated a higher level of generalizability, with a more proximate AUC of the training and testing cohorts than the single-R model. Furthermore, the calibration curve demonstrated the single-R and single-P models’ high accuracy in predicting the invasiveness of LUAD. The accuracy of the single-R model was higher than that of the single-P model in the multimodal training cohort (0.838 vs. 0.810) (Table 2).
Figure 3.
Performance of single-modality models in training and testing cohorts
(A) Performance of single-modality radiomic model showed by ROC curves in multimodal training and radiology testing cohort and calibration curve.
(B) Performance of single-modality pathologic model showed by ROC curves in multimodal training and pathology testing cohort and calibration curve.
Table 2.
Predictive performance of six types of models
| Multimodal training cohort | Radiology testing cohort | Pathology testing cohort | Prospective validation cohort | |
|---|---|---|---|---|
| Single-R model | ||||
| AUC | 0.904 (0.880–0.924) | 0.847 (0.809–0.879) | – | 0.936 (0.874–0.973) |
| Accuracy | 0.838 (0.810–0.864) | 0.822 (0.782–0.857) | – | 0.860 (0.782–0.918) |
| Sensitivity | 0.814 (0.778–0.847) | 0.719 (0.639–0.790) | – | 0.818 (0.714–0.897) |
| Specificity | 0.862 (0.813–0.902) | 0.864 (0.818–0.901) | – | 0.946 (0.818–0.993) |
| PPV | 0.922 (0.896–0.941) | 0.729 (0.664–0.786) | – | 0.969 (0.891–0.992) |
| NPV | 0.699 (0.658–0.737) | 0.858 (0.822–0.887) | – | 0.714 (0.607–0.802) |
| Single-P model | ||||
| AUC | 0.867 (0.841–0.891) | – | 0.859 (0.807–0.901) | 0.883 (0.809–0.935) |
| Accuracy | 0.810 (0.781–0.838) | – | 0.787 (0.728–0.838) | 0.798 (0.713–0.868) |
| Sensitivity | 0.804 (0.767–0.838) | – | 0.680 (0.579–0.770) | 0.766 (0.656–0.855) |
| Specificity | 0.806 (0.752–0.853) | – | 0.877 (0.808–0.928) | 0.892 (0.746–0.970) |
| PPV | 0.893 (0.866–0.915) | – | 0.810 (0.725–0.873) | 0.937 (0.853–0.974) |
| NPV | 0.673 (0.631–0.713) | – | 0.781 (0.727–0.827) | 0.647 (0.546–0.736) |
| Dual-RC model | ||||
| AUC | 0.905 (0.882–0.925) | 0.846 (0.808–0.878) | – | 0.929 (0.865–0.968) |
| Accuracy | 0.839 (0.811–0.865) | 0.810 (0.770–0.846) | – | 0.860 (0.782–0.918) |
| Sensitivity | 0.844 (0.809–0.874) | 0.706 (0.624–0.778) | – | 0.818 (0.714–0.897) |
| Specificity | 0.850 (0.800–0.891) | 0.864 (0.818–0.901) | – | 0.946 (0.818–0.993) |
| PPV | 0.918 (0.893–0.938) | 0.725 (0.660–0.783) | – | 0.969 (0.891–0.992) |
| NPV | 0.731 (0.688–0.770) | 0.852 (0.817–0.881) | – | 0.714 (0.607–0.802) |
| Dual-PC model | ||||
| AUC | 0.874 (0.848–0.896) | – | 0.869 (0.819–0.910) | 0.876 (0.802–0.931) |
| Accuracy | 0.812 (0.782–0.839) | – | 0.783 (0.724–0.834) | 0.781 (0.694–0.853) |
| Sensitivity | 0.804 (0.767–0.838) | – | 0.870 (0.788–0.929) | 0.701 (0.586–0.800) |
| Specificity | 0.806 (0.752–0.853) | – | 0.739 (0.654–0.812) | 0.946 (0.818–0.993) |
| PPV | 0.893 (0.866–0.915) | – | 0.719 (0.655–0.775) | 0.964 (0.874–0.991) |
| NPV | 0.673 (0.631–0.713) | – | 0.881 (0.815–0.925) | 0.603 (0.517–0.684) |
| Dual-RP model | ||||
| AUC | 0.919 (0.898–0.938) | – | – | 0.942 (0.883–0.977) |
| Accuracy | 0.835 (0.807–0.861) | – | – | 0.868 (0.792–0.924) |
| Sensitivity | 0.765 (0.725–0.801) | – | – | 0.818 (0.714–0.897) |
| Specificity | 0.937 (0.899–0.963) | – | – | 0.973 (0.858–0.999) |
| PPV | 0.960 (0.938–0.975) | – | – | 0.984 (0.901–0.998) |
| NPV | 0.666 (0.629–0.700) | – | – | 0.720 (0.615–0.806) |
| Multi-RPC model | ||||
| AUC | 0.921 (0.899–0.939) | – | – | 0.939 (0.878–0.975) |
| Accuracy | 0.842 (0.814–0.867) | – | – | 0.860 (0.782–0.918) |
| Sensitivity | 0.796 (0.759–0.831) | – | – | 0.831 (0.729–0.907) |
| Specificity | 0.897 (0.853–0.932) | – | – | 0.973 (0.858–0.999) |
| PPV | 0.939 (0.915–0.957) | – | – | 0.985 (0.902–0.998) |
| NPV | 0.688 (0.649–0.725) | – | – | 0.735 (0.627–0.820) |
Data are presented as mean (95% CI).
AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; Single-R model, single-modality radiomic model; Single-P model, single-modality pathologic model; Dual-RC model, radiomics + clinical indicators dual-modality model; Dual-PC model, pathology + clinical indicators dual-modality model; Dual-RP model, radiomics + pathology dual-modality model; Multi-RPC model, radiomics + pathology + clinical indicators multimodality model.
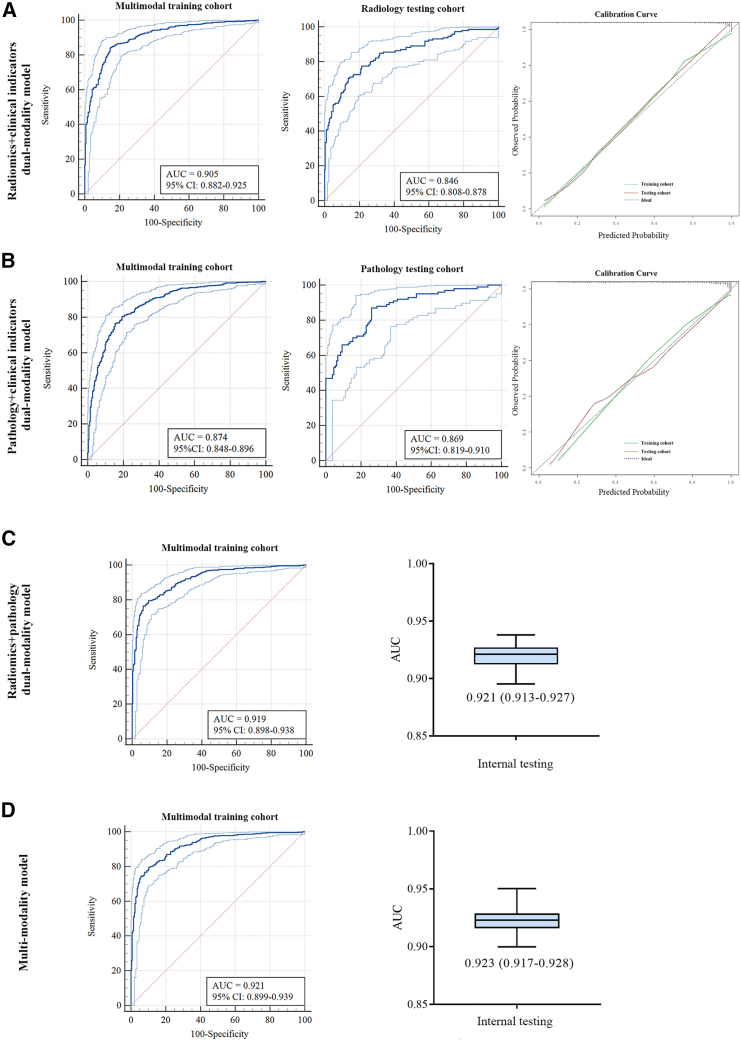
Different kinds of integration based on multimodal information in the multimodal training cohort
We systematically compared various combinations of dual-modality and multimodal features after assessing the predictive performance of single-modality features. Combining clinical indicators and Rad-score (dual-RC model) resulted in a minor improvement in predictive performance in both the multimodal training (from AUC = 0.904, 95% CI 0.880–0.924 to AUC = 0.905, 95% CI 0.882–0.825) and radiology testing cohort (form AUC = 0.847, 95% CI 0.809–0.879 to AUC = 0.846, 95% CI 0.808–0.878) (Figure 4A). Combining clinical and pathologic indicators (dual-PC model) also resulted in a minor improvement in predictive performance in both the multimodal training (from AUC = 0.867, 95% CI 0.841–0.891 to AUC = 0.874, 95% CI 0.848–0.896) and pathology testing cohort (from AUC = 0.859, 95% CI 0.807–0.901 to AUC = 0.869, 95% CI 0.819–0.910) (Figure 4B). A dual-modality model integrating Rad-score and pathologic indicators (dual-RP model) resulted in an AUC of 0.919 (95% CI 0.898–0.938) in the multimodal training cohort with bootstrap internal testing analysis, obtaining a median AUC of 0.921 (IQR 0.913–0.927) (Figure 4C). The integration of the three modes of information to develop the radiomics + pathology + clinical indicators multimodality model (multi-RPC model) resulted in the highest accuracy of 0.842 (95% CI 0.814–0.867) with an AUC of 0.921 (95% CI 0.899–0.939) (Table 2). The median AUC of the bootstrap internal testing analysis was 0.923 (IQR 0.917–0.928) (Figure 4D). Generally, combining different feature sets enhances the performance of a single modality set.
Figure 4.
Performance of dual-modality and multimodality models in training and testing cohorts
(A) Performance of radiomics + clinical indicators dual-modality model showed by ROC curve in multimodal training and radiology testing cohort and calibration curve.
(B) Performance of pathology + clinical indicators dual-modality model showed by ROC curve in multimodal training and pathology testing cohort and calibration curve.
(C) Performance of radiomics + pathology dual-modality model showed by ROC curve in multimodal training cohort and AUC through internal testing by bootstrap analysis.
(D) Performance of multimodality model showed by ROC curve in multimodal training cohort and AUC through internal testing by bootstrap analysis.
The improvements in predicting the invasion status of dual-/multimodality models over the single-R or single-P models were further evaluated using net reclassification improvement tests (all p < 0.05, except in comparisons between single-R and dual-RC or multi-RPC models, and between single-P and dual-PC models, Table S5) and integrated discrimination improvement tests (all p < 0.05, Table S6) in the multimodal training cohort. Additionally, the multi-RPC model yielded the lowest Akaike information criterion value, suggesting that the multimodality information integration provided the complementary capacity to improve predictive performance without increasing the risk of overfitting (Table S7).
Prospective evaluation of dual-modality and multimodality models in the clinical identification of invasiveness of LUAD
The AUCs of the dual-RC, the dual-RP, and multi-RPC models were 0.929 (95% CI 0.865–0.968), 0.942 (95% CI 0.883–0.977), and 0.939 (95% CI 0.878–0.975), respectively, which were higher than the AUC of the dual-PC model (0.874, 95% CI 0.848–0.896) in the prospective validation cohort. The overall accuracy of the dual-RP model was the highest (0.868, 95% CI 0.792–0.924) in the prospective validation cohort (Table 2; Figure 5A). Decision curve analysis was performed to demonstrate the clinical utility of the dual-/multimodality models (Figures 5B and 5C).
Figure 5.
Performance of models in prospective validation cohort
(A) Comparison of ROC curves for dual-/multimodality models in prospective validation cohort.
(B) Decision curve analysis demonstrating the clinical utility in predicting invasiveness of the five models in multimodal training cohort.
(C) Decision curve analysis demonstrating the clinical utility in predicting invasiveness of the five models in prospective validation cohort.
(D) Comparison of final pathology, intraoperative frozen section diagnosis and multi-RPC model predictive results in the prospective validation cohort. Dual-RC model, radiomics + clinical indicators dual-modality model; dual-PC model, pathology + clinical indicators dual-modality model; dual-RP model, radiomics + pathology dual-modality model; multi-RPC model, radiomics + pathology + clinical indicators multimodality model; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma; IA, invasive adenocarcinoma.
If the predictive results of the multi-RPC model were applied to guide the resection strategy, the number of pre-invasive cases that underwent lobectomy owing to misdiagnosis as IA or ambiguous diagnosis of LUAD by intraoperative FS would be reduced from 8 to 2; the number of invasive cases that underwent limited resection owing to misdiagnosis as AIS/MIA by intraoperative FS would be reduced from 23 to 13 (Figure 5D). The overall accuracy can be improved from 68.4% (78/114) of intraoperative FS diagnosis to 86.8% (99/114) of multi-RPC model. The compute performance measures on the basis of one intraoperative process are showed in STAR Methods, and it takes less than 20 min from making up the glass slide to gaining the predictive results. The correspondent predictive results of the single-R and dual-RP models in the prospective validation cohort are displayed in Figure S5. The overall accuracies for the prediction of single-R and dual-RP models were 86.0% (98/114) and 86.8% (99/114), respectively.
Discussion
Multimodal data integration based on artificial intelligence technologies has presented significant potential to guide decision-making on precision oncology.18 Complex diagnostic tasks, such as lung cancer classification, can improve cancer management; moreover, the clinical outcomes for patients can be improved if the accuracy of the diagnosis can be improved.19 This study successfully established a remarkable multimodal predictive model by integrating clinical information, radiomics, and FS pathologic features to determinate the invasiveness of ≤3 cm early LUAD, which is crucial for selecting an appropriate resection method. The robustness and generalizability of this multimodal predictive model indicate that it is a reliable and propagable tool for assisting in the invasive diagnosis of operable early LUAD. However, the predictive performance of the models to distinguish AIS and MIA was far from satisfactory (all AUC < 0.75, Table S8), which suggested the indistinctive differences of features between AIS and MIA, and more detailed feature analysis is required in the future.
Several large-scale multicenter clinical trials have indicated that the overall survival of patients with T1aN0 peripheral NSCLC showed no significant difference between sublobar resection and lobectomy.20,21,22 Studies have demonstrated that limited resection is curative for atypical adenomatous hyperplasia (AAH), AIS, and MIA, while its survival and recurrence rates are non-inferior to lobectomy.23,24 Conversely, IA often suggests a high risk of metastasis and recurrence, which is more reasonable to treat with lobectomy.25 Determining the invasion status of early LUAD before or during the surgery is crucial, particularly with the increasing use of sublobar resection, which has the advantage of preserving lung function. Since AAH is not included in TNM staging and the occurrence of AAH in patients without concurrent cancer is uncommon,26 AAH was not included as a separate histologic type analyzed in our research. Additionally, the prognosis of LUADs with AAH has not been shown to be different from that of those without AAH.27 Undoubtedly, CT, the most popular screening tool for lung diseases, has been widely used to explore several aspects for lung cancer, including invasiveness. Masahiro et al. used high-spatial-resolution CT findings to predict the invasiveness of LUAD and presented high diagnostic performance; however, this study collected some of the features using subjective image analysis via the radiologists.28 Nobuyuki et al. and Sun et al. used radiomics for invasive prediction: one combined radiomic features and radiographic features to construct a nomogram,14 and the other investigated the voxel-based histogram of 3D CT images.29 Wang et al. proposed a C-Lung-RADS system for precise risk stratification of pulmonary nodules by integrating CT imaging information, demographic characteristics, and follow-up data.30 Even though the results of these studies are encouraging to some degree, the accuracy is variable, and the parameters lack uniform standards.
Intraoperative FS can be considered a superior alternative because locating and sampling small lesions is a challenge for the diagnostic performance of preoperative biopsy (transbronchial or transthoracic fine-needle biopsies). However, the concordant rate of pathologic diagnosis between FS and FP relies heavily on the experience of the pathologists and specimen quality. Although H&E-stained WSIs collected from biopsy and paraffin-embedded sections (final pathology) have been widely explored to assist in disease diagnosis and prognosis prediction, such as distinguish luminal and non-luminal breast cancers31 and predict NSCLC prognosis,32 H&E-stained WSIs derived from intraoperative FS have not garnered much attention. Our team brought experts from specific domains to address various features to facilitate this study. Experienced radiologists carefully verified the segmentation of the targeted lung nodules, and PyRadiomics, a standardized and open-source radiomics pipeline, was used to extract the image biomarker standardization initiative compliant features.33 This platform renders radiomics studies more reproducible and comparable. A senior thoracic pathologist annotated the digitized H&E-stained WSIs of the intraoperative FS, including the classification of 1,220 randomly selected images to train the EfficientNet B5 classifier and recheck the invasion status of patients from all cohorts. The EfficientNet B5 model achieved high accuracy with fewer parameters, and thus was able to rapidly analyze WSIs.34
We integrated age, eosinophil count, radiomics, and FS pathologic features to develop a multi-RPC model for predicting the intraoperative invasion status. Although the multi-RPC model exhibited the best predictive performance in the multimodal training cohort, clinical information contributed little to the multimodal integration improvement compared with radiomics and pathologic information. In the prospective validation cohort, the dual-RP model demonstrated the highest AUC and accuracy; nevertheless, the complementary information provided by clinical indicators cannot be neglected. The method we used to analyze FS WSIs, on the one hand, focused on the most representative and informative image parts to train the classifier; on the other hand, the classification of small patches spared the manual delineation of tumor regions of interest, which is a time-consuming procedure, and improved the accuracy without losing holistic features. More importantly, the outcomes of the three modalities can be easily integrated using logistic regression, a classic and widely used machine learning method. The workflows and software provided in this study can stimulate interest in deploying a standardized and automated collection of multimodal data and have broad applications for other diseases. Various data have been combined to develop predictive models for various purposes. Wang et al. combined machine learning pathomics, radiomics features, immunoscore, and clinical factors to develop a nomogram to predict the outcomes of patients with colorectal cancer.35
In conclusion, our results demonstrated that the integration of radiomics, intraoperative FS pathologic information, and clinical indicators can effectively predict the invasiveness of small-sized early LUAD by developing a multimodality model based on computational and machine learning methods. The multi-RPC model is a promising approach to intraoperatively guide surgical strategies and promote the development of precision oncology practices in decision-making.
Limitations of the study
Although the advantages of the proposed approach are evident, this study has some limitations. First, the data collected from the two medical centers may be insufficient to train or test the multimodality models. And the present study was implemented only in a Chinese population, which might restrict the generalizability of our findings to other ethnic populations. Comprehensive training and testing strategies require datasets from multiple sites with consistent and stringent workflows to adjust for the heterogeneity properly. Second, the H&E-stained WSIs used in our study were at 20× magnification; however, a widespread criterion for digitalizing histopathology images has not yet been established, and retraining of prediction models is required for WSIs with different magnification levels. Third, long-term survival information was not collected to explore the difference in prognosis between patients with pre-invasive and invasive LUAD in this study, which deserves further research.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Yang Jin (whuhjy@126.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. All original code has been deposited at GitHub and is publicly available as of the date of publication. DOI is listed in the key resources table. Any additional information that supports the findings of this study is available from the lead contact upon reasonable request.
Acknowledgments
This work was supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (no. 2022YFF1203300), National Natural Science Foundation of China (no. 82330003, 82172034, 81900095, and 82001988), Major Project of the Department of Science and Technology of Hubei (no. 2022BCA016), Major Program of Special Project for Technology Innovation of Hubei Province (no. 2023BCB014), Natural Science Foundation of Hubei Province (2022CFB220), and Research Fund of Hubei Provincial Health Commission (WJ2021M226). We are grateful to Hangzhou Deepwise & League of PHD Technology Co., Ltd. for providing technical support about the segmentation of the targeted lung nodules. We would like to thank Editage (www.editage.cn) for English language editing.
Author contributions
Conceptualization, X.T., G.Y., and Y.J.; methodology, F.P., N.Z., S.W., Z.D., Y.L., G.Y., Y.D., H.Y., Y.C., M.Z., S.T., and L.Y.; software, F.P. and Z.D.; validation, X.T. and F.P.; formal analysis, X.T., F.P., and N.Z.; investigation, F.P., Y.L., B.H., and S.T.; resources, X.T., F.P., N.Z., Y.L., L.Y., and Y.J.; data curation, X.T., S.W., Y.L., G.Y., B.H., H.X., Y.C., M.Z., Z.L., Q.H., L.Z., M.Z., and L.Y.; writing – original draft, X.T., F.P., and N.Z.; writing – review and editing, S.W., Z.D., and Y.J.; visualization, X.T., F.P., and N.Z.; supervision, Y.D. and Y.J.; project administration, X.T., S.W., and L.Y.; funding acquisition, Z.L., Q.H., L.Y., and Y.J.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw CT | This paper | N/A |
| Raw WSI | This paper | N/A |
| Code for models | This paper | https://github.com/tanxueyun-code/invasiveness |
| Software and algorithms | ||
| Python (version 3.6.5) | Python Software Foundation | https://www.python.org/ |
| R (version 4.2.0) | R software | http://www.R-project.org |
| MITK (version 2022.10) | github | https://www.mitk.org/wiki/The_Medical_Imaging_Interaction_Toolkit_(MITK) |
| PyRadiomics (version 3.0.1) | github | http://www.radiomics.io/pyradiomics.html |
| EfficientNet B5 | github | https://github.com/tensorflow/tpu/tree/master/models/official/efficientnet |
| MedCalc (version 11.4.2.0) | MedCalc software | https://www.medcalc.org/ |
Experimental model and study participant details
Study design and participants
A retrospective multimodal training cohort was selected from the clinical database of the Wuhan Union Hospital and Renmin Hospital of Wuhan University between January 2021 and December 2022. The enrollment criteria for patients with multimodal information were as follows: (1) patients were diagnosed with operable clinical early-stage lung adenocarcinoma, (2) thoracic CT was performed within three months before surgery, and the nodule size was ≤3cm in diameter on CT, (3) no therapy history was discovered for lung cancer, and (4) an intraoperative FS examination was performed during the operation. The exclusion criteria were as follows: (1) tumor size >3cm, (2) multiple lesions or distant metastasis, (3) history of additional malignant tumors, and (4) age < 18 or > 80. Overall, 761 participants with complete multimodal data were included (Figure 1). Additionally, 433 participants were included in the radiology testing cohort as they had undergone preoperative diagnostic thoracentesis with insufficient intraoperative FS evaluation. Due to the unavailability of preoperative thoracic CT scans, 230 patients were included in the pathology testing cohort because they had undergone positron emission tomography (PET) examinations before surgery (Figure 1). The prospective validation cohort (n=114) was selected consecutively from Wuhan Union Hospital between March 2023 and May 2023 according to the same enrollment criteria. The gold standard for the pathological diagnosis of invasion status (AIS, MIA, IA) for all patients included in the four cohorts is a postoperative Formalin-Fixed Paraffin-Embedded section analysis (FFPE), defined as the final pathology (FP). Three physicians, YC, HX, and MZ, obtained clinical information from electronic medical records of the enrolled patients, including sex, age, race, smoking history, CT features and laboratory test results.
Ethical statement
This study was performed according to the International Council for Harmonization Guidelines for Good Clinical Practice and the Declaration of Helsinki. The Wuhan Union Hospital (No. S0802) and the Renmin Hospital of Wuhan University (No. WDRY2023-K004) Institutional Ethics Committee approved the study protocol and exempted the study from the necessity of obtaining informed consent owing to its observational design.
Method details
Radiomics feature analysis for the development of a single-modality radiomic model
Non-enhanced thoracic CT data were obtained from Wuhan Union Hospital and Renmin Hospital of Wuhan University. Thoracic CT examinations were performed at Wuhan Union Hospital on two commercial multi-detector CT scanners: Philips Ingenuity Core128 (Philips Medical Systems) and SOMATOM Definition AS (Siemens Healthineers), with images reconstructed at a section thickness of 1.25–5 mm. CT scans were performed at the Renmin Hospital of Wuhan University on four commercial multi-detector CT scanners: GE Optima CT680 CT, 64-slice LightSpeed VCT, BrightSpeed Elite CT, and Revolution CT (GE Medical Systems). The images were reconstructed at a section thickness of 0.75-5 mm. The targeted lung nodules were segmented using the pulmonary nodule CT imaging AI-assisted diagnosis system (Dr.Wise@PulmonaryNodule) (Hangzhou Deepwise & League of PHD Technology Co., Ltd).36,37 After CT images transmitted to the Dr. Wise system, automatic detection and segmentation of lung nodules were performed at the lung window setting (level, − 600 HU, and width, 1600 HU). A qualified radiologist (F.P., with 14 years of thoracic radiology experience) corrected the segmentation slice-by-slice using open-source Medical Imaging Interaction Toolkit (MITK) software (version 2022.10).
Radiomics features were extracted using PyRadiomics (version 3.0.1),38 a widely used software package which is implemented in Python for radiomics feature extraction. After CT image preprocessing, five feature classes, including a class for first-order statistics, a class for shape descriptors, and texture classes Gray Level Co-occurrence Matrix, Gray Level Run Length Matrix, and Gray Level Size Zone Matrix, were used for feature extraction from whole volume (3D) segmentations. Further information regarding the detailed formulas of the radiomic features can be discovered at https://pyradiomics.readthedocs.io/en/latest/features.html. A hundred radiomic features were extracted from the segmented data. Details about these features are provided in Table S1.
After extracting the 100 radiomic features, z-score normalization was applied to ensure that all features were on a comparable scale and to eliminate any potential bias resulting from their varied ranges. To further refine the feature selection process, a t-test feature selection was performed to assess the statistical significance of each feature in distinguishing between different types of lung nodules. A threshold P value of 0.01 was selected to identify the most informative features. Least absolute shrinkage and selection operator (LASSO) logistic regression with L1 regularization was employed to develop a single-modality radiomic (single-R) model utilizing the selected features. Five-fold cross-validation was performed to ensure the robustness and generalizability of the model.
LASSO regression yielded a linear equation involving the radiomic signatures considered the most relevant for differential diagnosis to obtain the Rad-score.
Rad-Score = 1.3229 + -0.1551 × original_shape_Flatness + 0.5883 × original_shape_LeastAxisLength + 0.2976 × original_shape_Maximum2DDiameterSlice + -0.0124 × original_shape_Sphericity + 0.7493 × original_firstorder_90Percentile + 0.0189 × original_firstorder_InterquartileRange + 0.4060 × original_firstorder_Median + 0.0905 × original_firstorder_RootMeanSquared + -0.2789 × original_firstorder_Skewness + -0.0126 × original_glcm_ClusterProminence + -0.2726 × original_glcm_DifferenceVariance + 0.1688 × original_glcm_JointEnergy + 0.0687 × original_glcm_JointEntropy + 0.0513 × original_glszm_SizeZoneNonUniformityNormalized + 0.1848 × original_glszm_ZoneEntropy + 0.2546 × original_gldm_DependenceEntropy + -0.0120 × original_gldm_LargeDependenceLowGrayLevelEmphasis.
FS pathologic image analysis for the development of a single-modality pathologic model
At first, among FS-based haematoxylin and eosin (H&E) stained whole-slide images (WSIs) with 20× magnification collected in 2020 from Wuhan Union Hospital and Renmin Hospital of Wuhan University, a random selected dataset comprising 1220 pathologic image tiles (resolution: 1000×1000 pixels), selected by an experienced pathologist (N.Z.) and classified into five categories (including “Blank”, “lung parenchyma (LP)”, “AIS”, “MIA”, and “IA”). We adopted the EfficientNet B5 model as our network backbone to exploit the potential of deep learning and to achieve the classification of five pathologic categories for WSI tiles with a high accuracy.39 This dataset was divided into training and validation sets using a 7:3 split. EfficientNet, being among the most advanced convolutional neural network (CNN) architectures currently available, is known for its efficiency and efficacy due to the joint application of depthwise separable convolution (MBConv) and squeeze and excitation (SE) modules.39,40,41 It achieves an optimal balance between model size and performance by utilizing a novel compound scaling algorithm that optimizes depth, width, and resolution.40,41 We applied transfer learning to leverage the pre-trained weights from IMAGENET1K_V1, further fine-tuning the network for the pathologic diagnosis task.42,43 Data augmentation strategies included resizing to 224×224, auto-segment policies, random horizontal flips, and patch erasing to enhance generalization.44 A dropout strategy with a 0.5 threshold probability was employed to determine the final classifier layer.39,45,46 Cross-entropy loss was utilized, and optimization was performed with Stochastic Gradient Descent (SGD) and a specific learning rate of 0.001, a batch size of 30, and training for 100 epochs. Learning rate scheduling utilized OneCycleLR.1 The model was trained on a cloud GPU platform (gpuhub.com/home) with an Nvidia A5000 24G GPU, a 60-core Intel(R) Xeon(R) Platinum 8358P CPU, and 360GB of RAM, utilizing PyTorch 1.11.0 + CUDA 11.3. The model selection criterion prioritized minimizing the loss of the validation set to prevent overfitting.
Then, we obtained 399 and 592 FS-based WSIs between January 2021 and December 2022 from Wuhan Union Hospital and Renmin Hospital of Wuhan University, respectively, each containing WSIs with dimensions of up to 100,000×100,000 pixels at a 20× magnification, corresponding to a resolution of 0.25 μm per pixel. To ensure interoperability with our trained network, these considerable WSIs were segmented into small contiguous patches, each with a uniform resolution of 1000×1000 pixels. These patches were subsequently input into the pre-trained and fine-tuned EfficientNet B5 model. The model then generated probability distributions for all patches, categorizing them into the five pathologic classes, including “Blank”, “LP”, “AIS”, “MIA”, and “IA”. Blank patches were deleted from the entire count of one WSI, and the numbers of LP, AIS, MIA, and IA patches were denoted by , , and , respectively. Three proportions were calculated to develop the single-modality pathologic (single-P) model in the multimodal training cohort using logistic regression:
Development and validation of dual-modality and multimodality predictive models
In this study, clinical indicators were selected based on age, sex, smoking history, and laboratory testing results using univariable and multivariable logistic regression analyses to predict the invasiveness of lung adenocarcinoma. In the multimodal training cohort (Figure 2), the Rad-score and clinical indicators were combined to develop the radiomics + clinical indicators dual-modality (dual-RC) model. Three pathological proportions (AIS%, MIA%, and IA%) calculated in the single-P model and clinical indicators were combined to develop the pathology + clinical indicators dual-modality (dual-PC) model. The Rad-score calculated in the single-R model and three pathological proportions (AIS%, MIA%, and IA%) calculated in the single-P model were combined to develop the radiomics + pathology dual-modality (dual-RP) model. The multi-RPC model was developed from the integration of the Rad-score, three pathological proportions (AIS%, MIA%, and IA%), and two clinical indicators (age and eosinophils). All dual-modal and multimodal models were developed using logistic regression to predict binary outcomes.
The radiology testing cohort was used to test the single-R and dual-RC models, and the pathology testing cohort was used to test the single-P and dual-PC model. A prospective validation cohort was used to validate the models. For all models developed in this study, the binary outcome was “pre-invasive” (AIS and MIA) and “invasive” (IA).
Compute performance measures on the basis of one intraoperative process
First step: The gross specimen of tumor is cut into thin slices; the tissue sections are cut and picked up on glass slide and stained in haematoxylin and eosin. (about 15 min).
Second step: The FS-based haematoxylin and eosin (H&E) stained section is scanned into whole-slide images (WSIs) with 20× magnification. (about 1 min).
Third step: A sliding window approach is used to segment one WSI into small contiguous patches, each with a uniform resolution of 1000×1000 pixels; these patches are subsequently input into the EfficientNet B5 model and categorized into five pathologic classes, including “Blank,” “LP,” “AIS,” “MIA,” and “IA”. (about 30 s).
Fourth step: The clinical indicators, radiomics and pathologic results are input into the RPCMM model to get the final prediction of invasion status of the tumor. (about 3 s).
Quantification and statistical analysis
The area under the curve (AUC), accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were used to evaluate the models’ performance in predicting invasion status. Moreover, a calibration plot was performed to assess the discrimination capacity of the predictive models. Additionally, we used net reclassification improvement (NRI) and integrated discrimination improvement (IDI) tests to estimate the model performance in identifying pre-invasive and invasive lung adenocarcinoma. The Akaike information criterion (AIC) test was performed to assess the risk of model overfitting. Decision curve analysis (DCA) was performed to estimate the clinical utility of the models by evaluating their net advantages when applied to a test set at different threshold probabilities. All tests were two-sided, and P < 0.05 indicated statistical significance. MedCalc (version 11.4.2.0), R studio (version 4.2.0), and Python (version 3.6.5) were used for statistical analyses.
Additional resources
This study was registered on the Clinical Trials website (No. NCT05830812). Description: https://clinicaltrials.gov/study/NCT05830812?term=NCT05830812&rank=1.
Published: November 19, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111421.
Contributor Information
Lian Yang, Email: yanglian@hust.edu.cn.
Yang Jin, Email: whuhjy@126.com.
Supplemental information
References
- 1.Travis W.D., Brambilla E., Noguchi M., Nicholson A.G., Geisinger K.R., Yatabe Y., Beer D.G., Powell C.A., Riely G.J., Van Schil P.E., et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H., Carrot-Zhang J., Zhao Y., Hu H., Freeman S.S., Yu S., Ha G., Taylor A.M., Berger A.C., Westlake L., et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat. Commun. 2019;10:5472. doi: 10.1038/s41467-019-13460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu J., Lu C., Guo J., Chen L., Chu Y., Ji Y., Ge D. Prognostic significance of the IASLC/ATS/ERS classification in Chinese patients-A single institution retrospective study of 292 lung adenocarcinoma. J. Surg. Oncol. 2013;107:474–480. doi: 10.1002/jso.23259. [DOI] [PubMed] [Google Scholar]
- 4.Kadota K., Villena-Vargas J., Yoshizawa A., Motoi N., Sima C.S., Riely G.J., Rusch V.W., Adusumilli P.S., Travis W.D. Prognostic significance of adenocarcinoma in situ, minimally invasive adenocarcinoma, and nonmucinous lepidic predominant invasive adenocarcinoma of the lung in patients with stage I disease. Am. J. Surg. Pathol. 2014;38:448–460. doi: 10.1097/PAS.0000000000000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yanagawa N., Shiono S., Abiko M., Ogata S.Y., Sato T., Tamura G. New IASLC/ATS/ERS classification and invasive tumor size are predictive of disease recurrence in stage I lung adenocarcinoma. J. Thorac. Oncol. 2013;8:612–618. doi: 10.1097/JTO.0b013e318287c3eb. [DOI] [PubMed] [Google Scholar]
- 6.Ye T., Deng L., Xiang J., Zhang Y., Hu H., Sun Y., Li Y., Shen L., Wang S., Xie L., Chen H. Predictors of Pathologic Tumor Invasion and Prognosis for Ground Glass Opacity Featured Lung Adenocarcinoma. Ann. Thorac. Surg. 2018;106:1682–1690. doi: 10.1016/j.athoracsur.2018.06.058. [DOI] [PubMed] [Google Scholar]
- 7.Blasberg J.D., Pass H.I., Donington J.S. Sublobar resection: a movement from the Lung Cancer Study Group. J. Thorac. Oncol. 2010;5:1583–1593. doi: 10.1097/jto.0b013e3181e77604. [DOI] [PubMed] [Google Scholar]
- 8.Keenan R.J., Landreneau R.J., Maley R.H., Jr., Singh D., Macherey R., Bartley S., Santucci T. Segmental resection spares pulmonary function in patients with stage I lung cancer. Ann. Thorac. Surg. 2004;78:228–233. doi: 10.1016/j.athoracsur.2004.01.024. discussion 228-33. [DOI] [PubMed] [Google Scholar]
- 9.Liu S., Wang R., Zhang Y., Li Y., Cheng C., Pan Y., Xiang J., Zhang Y., Chen H., Sun Y. Precise Diagnosis of Intraoperative Frozen Section Is an Effective Method to Guide Resection Strategy for Peripheral Small-Sized Lung Adenocarcinoma. J. Clin. Oncol. 2016;34:307–313. doi: 10.1200/JCO.2015.63.4907. [DOI] [PubMed] [Google Scholar]
- 10.He P., Yao G., Guan Y., Lin Y., He J. Diagnosis of lung adenocarcinoma in situ and minimally invasive adenocarcinoma from intraoperative frozen sections: an analysis of 136 cases. J. Clin. Pathol. 2016;69:1076–1080. doi: 10.1136/jclinpath-2016-203619. [DOI] [PubMed] [Google Scholar]
- 11.Walts A.E., Marchevsky A.M. Root Cause Analysis of Problems in the Frozen Section Diagnosis of In Situ, Minimally Invasive, and Invasive Adenocarcinoma of the Lung. Arch. Pathol. Lab Med. 2012;136:1515–1521. doi: 10.5858/arpa.2012-0042-OA. [DOI] [PubMed] [Google Scholar]
- 12.Donington J.S. An Additional Step Toward Personalization of Surgical Care for Early-Stage Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2016;34:295–296. doi: 10.1200/JCO.2015.64.7578. [DOI] [PubMed] [Google Scholar]
- 13.Xu X., Chung J.H., Jheon S., Sung S.W., Lee C.T., Lee J.H., Choe G. The accuracy of frozen section diagnosis of pulmonary nodules: evaluation of inflation method during intraoperative pathology consultation with cryosection. J. Thorac. Oncol. 2010;5:39–44. doi: 10.1097/JTO.0b013e3181c09f9c. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y., Li C., Jin L., Gao P., Zhao W., Ma W., Tan M., Wu W., Duan S., Shan Y., Li M. Radiomics for lung adenocarcinoma manifesting as pure ground-glass nodules: invasive prediction. Eur. Radiol. 2020;30:3650–3659. doi: 10.1007/s00330-020-06776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nitadori J.I., Bograd A.J., Morales E.A., Rizk N.P., Dunphy M.P.S., Sima C.S., Rusch V.W., Adusumilli P.S. Preoperative consolidation-to-tumor ratio and SUVmax stratify the risk of recurrence in patients undergoing limited resection for lung adenocarcinoma ≤2 cm. Ann. Surg Oncol. 2013;20:4282–4288. doi: 10.1245/s10434-013-3212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu E., Xie H., Dai C., Zhang L., Huang Y., Dong Z., Guo J., Su H., Ren Y., Shi P., et al. Intraoperatively measured tumor size and frozen section results should be considered jointly to predict the final pathology for lung adenocarcinoma. Mod. Pathol. 2018;31:1391–1399. doi: 10.1038/s41379-018-0056-0. [DOI] [PubMed] [Google Scholar]
- 17.Warkentin M.T., Al-Sawaihey H., Lam S., Liu G., Diergaarde B., Yuan J.M., Wilson D.O., Atkar-Khattra S., Grant B., Brhane Y., et al. Radiomics analysis to predict pulmonary nodule malignancy using machine learning approaches. Thorax. 2024;79:307–315. doi: 10.1136/thorax-2023-220226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehm K.M., Khosravi P., Vanguri R., Gao J., Shah S.P. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer. 2022;22:114–126. doi: 10.1038/s41568-021-00408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo J., Steuten L., Aftimos P., André F., Davies M., Garralda E., Geissler J., Husereau D., Martinez-Lopez I., Normanno N., et al. Delivering precision oncology to patients with cancer. Nat. Med. 2022;28:658–665. doi: 10.1038/s41591-022-01717-2. [DOI] [PubMed] [Google Scholar]
- 20.Altorki N., Wang X., Kozono D., Watt C., Landrenau R., Wigle D., Port J., Jones D.R., Conti M., Ashrafi A.S., et al. Lobar or Sublobar Resection for Peripheral Stage IA Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2023;388:489–498. doi: 10.1056/NEJMoa2212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altorki N.K., Wang X., Wigle D., Gu L., Darling G., Ashrafi A.S., Landrenau R., Miller D., Liberman M., Jones D.R., et al. Perioperative mortality and morbidity after sublobar versus lobar resection for early-stage non-small-cell lung cancer: post-hoc analysis of an international, randomised, phase 3 trial (CALGB/Alliance 140503) Lancet Respir. Med. 2018;6:915–924. doi: 10.1016/s2213-2600(18)30411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saji H., Okada M., Tsuboi M., Nakajima R., Suzuki K., Aokage K., Aoki T., Okami J., Yoshino I., Ito H., et al. Segmentectomy versus lobectomy in small-sized peripheral non-small-cell lung cancer (JCOG0802/WJOG4607L): a multicentre, open-label, phase 3, randomised, controlled, non-inferiority trial. Lancet (London, England) 2022;399:1607–1617. doi: 10.1016/s0140-6736(21)02333-3. [DOI] [PubMed] [Google Scholar]
- 23.Li D., Deng C., Wang S., Li Y., Zhang Y., Chen H. Ten-year follow-up of lung cancer patients with resected adenocarcinoma in situ or minimally invasive adenocarcinoma: Wedge resection is curative. J. Thorac. Cardiovasc. Surg. 2022;164:1614–1622.e1. doi: 10.1016/j.jtcvs.2022.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Li F., Yang L., Zhao Y., Yuan L., Wang S., Mao Y. Intraoperative frozen section for identifying the invasion status of lung adenocarcinoma: A systematic review and meta-analysis. Int. J. Surg. 2019;72:175–184. doi: 10.1016/j.ijsu.2019.10.047. [DOI] [PubMed] [Google Scholar]
- 25.Nitadori J.i., Bograd A.J., Kadota K., Sima C.S., Rizk N.P., Morales E.A., Rusch V.W., Travis W.D., Adusumilli P.S. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. J. Natl. Cancer Inst. 2013;105:1212–1220. doi: 10.1093/jnci/djt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO Classification of Tumours Editorial Board . International Agency for Research on Cancer; 2021. Thoracic tumours.https://publications.iarc.fr/595 WHO Classification of Tumours Series, 5th ed.; vol. 5. [Google Scholar]
- 27.Suzuki K., Nagai K., Yoshida J., Yokose T., Kodama T., Takahashi K., Nishimura M., Kawasaki H., Yokozaki M., Nishiwaki Y. The prognosis of resected lung carcinoma associated with atypical adenomatous hyperplasia: a comparison of the prognosis of well-differentiated adenocarcinoma associated with atypical adenomatous hyperplasia and intrapulmonary metastasis. Cancer. 1997;79:1521–1526. doi: 10.1002/(sici)1097-0142(19970415)79:8<1521::aid-cncr12>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 28.Yanagawa M., Tsubamoto M., Satoh Y., Hata A., Miyata T., Yoshida Y., Kikuchi N., Kurakami H., Tomiyama N. Lung Adenocarcinoma at CT with 0.25-mm Section Thickness and a 2048 Matrix: High-Spatial-Resolution Imaging for Predicting Invasiveness. Radiology. 2020;297:462–471. doi: 10.1148/radiol.2020201911. [DOI] [PubMed] [Google Scholar]
- 29.Yoshiyasu N., Kojima F., Hayashi K., Bando T. Radiomics technology for identifying early-stage lung adenocarcinomas suitable for sublobar resection. J. Thorac. Cardiovasc. Surg. 2021;162:477–485.e1. doi: 10.1016/j.jtcvs.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Wang C., Shao J., He Y., Wu J., Liu X., Yang L., Wei Y., Zhou X.S., Zhan Y., Shi F., et al. Data-driven risk stratification and precision management of pulmonary nodules detected on chest computed tomography. Nat. Med. 2024;30:3184–3195. doi: 10.1038/s41591-024-03211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., Yao Z., Li L., Mao R., Huang W., Hu Z., Hu Y., Wang Y., Guo R., Tang X., et al. Deep learning radiopathomics based on preoperative US images and biopsy whole slide images can distinguish between luminal and non-luminal tumors in early-stage breast cancers. EBioMedicine. 2023;94 doi: 10.1016/j.ebiom.2023.104706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu K.H., Zhang C., Berry G.J., Altman R.B., Ré C., Rubin D.L., Snyder M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016;7 doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mu W., Schabath M.B., Gillies R.J. Images Are Data: Challenges and Opportunities in the Clinical Translation of Radiomics. Cancer Res. 2022;82:2066–2068. doi: 10.1158/0008-5472.Can-22-1183. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z., Ren Y., Zhang Z., Guan T., Wang Z., Chen W., Luo T., Li G. Digital histopathological images of biopsy predict response to neoadjuvant chemotherapy for locally advanced gastric cancer. Gastric Cancer. 2023;26:734–742. doi: 10.1007/s10120-023-01407-z. [DOI] [PubMed] [Google Scholar]
- 35.Wang R., Dai W., Gong J., Huang M., Hu T., Li H., Lin K., Tan C., Hu H., Tong T., Cai G. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J. Hematol. Oncol. 2022;15:11. doi: 10.1186/s13045-022-01225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi L.L., Wu B.T., Tang W., Zhou L.N., Huang Y., Zhao S.J., Liu L., Li M., Zhang L., Feng S.C., et al. Long-term follow-up of persistent pulmonary pure ground-glass nodules with deep learning-assisted nodule segmentation. Eur. Radiol. 2020;30:744–755. doi: 10.1007/s00330-019-06344-z. [DOI] [PubMed] [Google Scholar]
- 37.Qi L.L., Wang J.W., Yang L., Huang Y., Zhao S.J., Tang W., Jin Y.J., Zhang Z.W., Zhou Z., Yu Y.Z., et al. Natural history of pathologically confirmed pulmonary subsolid nodules with deep learning-assisted nodule segmentation. Eur. Radiol. 2021;31:3884–3897. doi: 10.1007/s00330-020-07450-z. [DOI] [PubMed] [Google Scholar]
- 38.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J.C., Pieper S., Aerts H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.Can-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Wan Y., Pan F. Enhancing Multi-disease Diagnosis of Chest X-rays with Advanced Deep-learning Networks in Real-world Data. J. Digit. Imaging. 2023;36:1332–1347. doi: 10.1007/s10278-023-00801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandler M., Howard A., Zhu M.L., Zhmoginov A., Chen L.C. 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)Salt Lake City, UT. 2018. Ieee, MobileNetV2: Inverted Residuals and Linear Bottlenecks; pp. 4510–4520. [DOI] [Google Scholar]
- 41.Hu J., Shen L., Albanie S., Sun G., Wu E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020;42:2011–2023. doi: 10.1109/TPAMI.2019.2913372. [DOI] [PubMed] [Google Scholar]
- 42.Albahli S., Rauf H.T., Algosaibi A., Balas V.E. AI-driven deep CNN approach for multi-label pathology classification using chest X-Rays. PeerJ. Comput. Sci. 2021;7:e495. doi: 10.7717/peerj-cs.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X.S., Peng Y.F., Lu L., Lu Z.Y., Bagheri M., Summers R.M. 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)Honolulu, HI. 2017. Summers, Ieee, ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases; pp. 3462–3471. [DOI] [Google Scholar]
- 44.Cubuk E.D., Zoph B., Mané D., Vasudevan V., Le Q.V., Soc I.C. 32nd IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)Long Beach, CA. 2019. AutoAugment: Learning Augmentation Strategies from Data; pp. 113–123. [DOI] [Google Scholar]
- 45.Ying X., IOP . International Conference on Computer Information Science and Application Technology (CISAT)NE Petr Univ, Daqing. PEOPLES R CHINA; 2018. An Overview of Overfitting and its Solutions. [DOI] [Google Scholar]
- 46.Barrow E., Eastwood M., Jayne C. 23rd International Conference on Neural Information Processing (ICONIP)Kyoto, JAPAN. 2016. Selective Dropout for Deep Neural Networks; pp. 519–528. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. All original code has been deposited at GitHub and is publicly available as of the date of publication. DOI is listed in the key resources table. Any additional information that supports the findings of this study is available from the lead contact upon reasonable request.