Abstract
Gibberellins (GAs) are plant hormones with diverse roles in plant growth and development. SPINDLY (SPY) is one of several genes identified in Arabidopsis that are involved in GA response and it is thought to encode an O-GlcNAc transferase. Genetic analysis suggests that SPY negatively regulates GA response. To test the hypothesis that SPY acts specifically as a negatively acting component of GA signal transduction, spy mutants and plants containing a 35S:SPY construct have been examined. A detailed investigation of the spy mutant phenotype suggests that SPY may play a role in plant development beyond its role in GA signaling. Consistent with this suggestion, the analysis of spy er plants suggests that the ERECTA (ER) gene, which has not been implicated as having a role in GA signaling, appears to enhance the non-GA spy mutant phenotypes. Arabidopsis plants containing a 35S:SPY construct possess reduced GA response at seed germination, but also possess phenotypes consistent with increased GA response, although not identical to spy mutants, during later vegetative and reproductive development. Based on these results, the hypothesis that SPY is specific for GA signaling is rejected. Instead, it is proposed that SPY is a negative regulator of GA response that has additional roles in plant development.
Gibberellins (GAs) are diterpenoid hormones with multiple roles in plant development. For example, GAs promote seed germination in many, but not all, species and abscisic acid (ABA) can often antagonize the action of GA. A well-studied example is the role of GA and ABA in the germination and reserve mobilization in monocot seeds (Lovegrove and Hooley, 2000). Perhaps the best known physiological role of GAs is the promotion of shoot extension growth across a wide range of species. The importance of GAs in vegetative growth is illustrated by Mendel's dwarf pea (Lester et al., 1997; Martin et al., 1997) and the “green revolution” rht dwarfing alleles (Peng et al., 1999) that cause reduced GA biosynthesis and response, respectively. GAs are also involved in flower induction in some species, although the role of GA appears to be complex and it varies from species to species (Evans, 1999).
The physiological role of GAs has been investigated over many years by a variety of approaches, including the application of active GAs and their precursors, chemical inhibitors of GA biosynthesis, and the analysis of mutants in plants such as maize, pea, and Arabidopsis. In whole plants, GA action involves the coordinated processes of GA metabolism (biosynthesis and catabolism) and GA signal transduction. Over the last several years, many of the enzymes involved in the synthesis and degradation of biologically active GAs such as GA1 and GA4 have been characterized at the molecular and biochemical levels. In addition, the understanding of the controls of GA biosynthesis, particularly the homeostatic control of GA levels within the plant and the regulation of GA biosynthesis by environmental signals such as light, has increased greatly (Kamiya and Garcia-Martinez, 1999). The well-characterized role of GAs in promoting stem elongation, combined with the cloning of several genes encoding GA enzymes, has also opened up the possibility of using genetic engineering to control the growth of crop plants (Hedden and Phillips, 2000).
The last few years have also seen considerable progress in understanding GA signal transduction, largely based on molecular-genetic analysis using Arabidopsis and the aleurone system of monocotyledonous grains. This work has led to the identification of several GA-signaling proteins including RGA (REPRESSOR OF GA1–3), GAI (GA INSENSITITIVE), SPY (SPINDLY), SHI (SHORT INTERNODES), and PKL (PICKLE), in Arabidopsis (Thornton et al., 1999), and GAMyb in barley (Gubler et al., 1999). A role for heterotrimeric G proteins has also be suggested, based on work with inhibitors in wild oat aleurones (Jones et al., 1998) and analysis of the d1 mutant of rice (Ashikari et al., 1999; Fujisawa et al., 1999). Several second messengers that play a role in the process have also been identified (Lovegrove and Hooley, 2000).
The SPY locus was originally identified in a genetic screen for increased GA response mutants able to germinate in the presence of paclobutrazol, a chemical inhibitor that acts early in the GA biosynthesis pathway and prevents germination of wild-type (WT) seeds (Jacobsen and Olszewski, 1993). Additional alleles have subsequently been identified as suppressors of GA deficiency caused by the ga1-3 mutation and as suppressors of the reduced GA response gain-of-function gai dwarf mutant (Wilson and Somerville, 1995; Peng et al., 1997; Silverstone et al., 1997). Based on the loss-of-function spy mutant phenotype, SPY is genetically defined as a negatively acting component of the GA signal transduction pathway. To further address SPY's function, Robertson et al. (1998) used the Arabidopsis SPY gene to identify and then to investigate the role of the barley SPY (HvSPY) in GA response of barley aleurone cells. Expression of HvSPY in Arabidopsis spy mutants partially suppresses the mutant phenotype, suggesting that HvSPY is the barley ortholog of SPY. Consistent with SPY's proposed role as a negative regulator of GA signaling, cobombarding HvSPY driven by a constitutive promoter into aleurone cells with a β-glucuronidase reporter gene under the control of an α-amylase promoter blocked the GA stimulated activation of the reporter. In an unexpected result, HvSPY also induced expression of an ABA-regulated dehydrin gene, suggesting that at least when highly expressed in aleurone cells, HvSPY may also modify the expression of genes that are not regulated by GA. Another unexpected result was that in the absence of exogenous GA, HvSPY caused a small but significant increase in α-amylase reporter activity (Robertson et al., 1998).
The predicted amino acid sequence of SPY and HvSPY exhibit significant similarity, extending through the N-terminal tetratricopeptide repeat (TPR) domain and the C-terminal putative catalytic domain to cytosolic O-linked N-acetyl glucosamine (GlcNAc) transferases (OGTs; Thornton et al., 1999; Roos and Hanover, 2000). Genetic analysis of spy mutants indicates that the N- and C-terminal domains participate in GA signal transduction (Jacobsen et al., 1996). TPR domains in other proteins have been shown to participate in protein-protein interactions, suggesting that SPY is part of a multiprotein complex. Several spy mutants have been found to contain alterations in terminal GlcNAc modification in protein extracts and SPY produced using the baculovirus expression system has GlcNAc transferase activity (Thornton et al., 1999). The OGT enzyme activity was originally identified in mammals, most of the information on OGT function is based on the study of animal systems. Current models of SPY function in plants are based on these studies. OGT is present in the cytosol and nucleus, and O-GlcNAc modification of cytosolic and nuclear proteins is about as common and as readily reversible as Ser/Thr phosphorylation (Snow and Hart, 1998; Comer and Hart, 2000). Animal OGT transfers a single GlcNAc molecule from UDP-GlcNAc to specific Ser and/or Thr residues of target proteins, all of which are phosphoproteins and components of multiprotein complexes. Deletion of the mouse O-GlcNAc transferase gene causes embryo lethality (Shafi et al., 2000), suggesting that O-GlcNAc modification plays a role in essential and diverse signal transduction pathways controlling animal development and physiology.
Detailed analysis of mutant plant phenotypes is one of the most powerful ways to determine the biochemical function and physiological role of individual genes in plant development. The phenotype of spy mutants has, therefore, been examined in more detail, particularly in regard to SPY's proposed role in negatively regulating GA signal transduction. We have also overexpressed the SPY mRNA as an additional approach to understanding the role of SPY in GA signaling and plant development.
RESULTS
spy Mutants Are Not Complete Phenocopies of GA-Treated WT Plants
Previous work with spy mutants had led to the conclusion that the action of SPY is restricted to the GA signal transduction pathway. This hypothesis was tested by a more detailed examination of the spy mutant phenotype, including two severe mutants, spy-2 and spy-4. The spy-2 mutation alters RNA splicing, whereas the spy-4 allele possesses a T-DNA insertion just upstream of the SPY coding region that results in reduced SPY mRNA levels (Jacobsen et al., 1996). In contrast to the previously reported observations that loss-of-function spy mutants in the Columbia and Wassilewskija (WS) backgrounds resemble GA-treated WT plants (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996), in the Landsberg–erecta (La–er) background spy-2 and spy-4 mutants display novel whole plant phenotypes not expected for plants with a specific increase in GA response. For example, although GAs promote internode elongation in Arabidopsis, in this genetic background, spy-2 (0.53 ± 0.03 cm) and spy-4 (0.31 ± 0.03 cm) possess significantly (P < 0.001) shorter internodes than WT La-er plants (2.26 ± 0.18 cm). These mutants are also smaller than WT La-er plants in terms of rosette size, they have abnormal phylotaxy of flowers on the inflorescence, and they do not obviously resemble GA-treated La-er plants. Thus, the phenotype displayed by severe spy mutants is modified by the genetic background. One possibility is that the different alleles of ER in Columbia and WS versus La-er is responsible for these differences, with the reduced ER activity in La-er modifying the spy phenotype. This hypothesis was confirmed when a similar effect on the spy-4 phenotype was observed by combining spy-4 with the loss-of-function er-102 allele (Torii et al., 1996) in the Columbia background (data not shown). Despite the dramatic effect of er mutations on the spy mutant phenotype, spy mutations in the La-er background are still able to increase GA response. For example, La-er spy alleles partially suppress the dwarf phenotype caused by chemical inhibitors of GA biosynthesis, and by the ga1 or gai mutations (Carol et al., 1995; Peng et al., 1997; Silverstone et al., 1997).
The phenotype of spy-4 in the La-er genetic background prompted us to more carefully examine this allele in the Columbia background (Table I). This analysis revealed that even in a background where spy-4 does display the expected “spindly” phenotype, mutant plants are only a partial phenocopy of WT plants treated with GA. Although some mutant phenotypes are similar to the effect of treating WT plants with repeated and high doses of GA3, others are opposite to the observed effect of applied GA or the predicted effect of increased GA response. For example, spy-4 plants and WT treated with GA3 flower with fewer rosette leaves than untreated WT plants, whereas loss of SPY function, at least for severe alleles, and GA3 treatment have opposite effects on rosette leaf length.
Table I.
spy-4 plants are not phenocopies of WT plants treated with GA3
| WT (Columbia)
|
spy-4 (Columbia)
|
spy-4 Phenocopies GA Treatment of WT? | |||
|---|---|---|---|---|---|
| −GA | +GAa | −GA | +GA | ||
| No. of rosette leaves | 19.2 ± 1.2 | 11.7 ± 0.4 | 8.5 ± 0.6 | 6.3 ± 0.6 | Yes |
| No. of cauline leaves | 5.0 ± 0.5 | 6.7 ± 0.4 | 3.5 ± 0.2 | 3.9 ± 0.3 | No |
| No. of total leaves | 24.2 ± 1.7 | 18.4 ± 0.7 | 12.0 ± 0.7 | 9.1 ± 0.6 | Yes |
| Height (cm)b | 11.2 ± 0.1 | 15.1 ± 0.7 | 8.3 ± 0.4 | 9.3 ± 0.4 | No |
| Internode length (cm)c | 2.0 ± 0.2 | 2.0 ± 0.1 | 1.7 ± 0.1 | 1.9 ± 0.1 | – |
| Length of longest leaf in rosette (cm) | 4.2 ± 0.2 | 4.8 ± 0.1 | 2.5 ± 0.1 | 2.3 ± 0.2 | No |
| Width of first internode (mm) | 0.99 ± 0.04 | 0.97 ± 0.03 | 0.65 ± 0.04 | 0.49 ± 0.03 | No |
| Fertilityd | 8.8 ± 0.3 | 5.6 ± 1.0 | 4.5 ± 0.6 | 0.3 ± 0.2 | Yes |
| Phylotaxye | 0 | 0 | 100 | 100 | No |
Plants were grown in standard long day (LD) conditions.
Seeds were imbibed in 3 × 10−5 M GA3 for 3 d at 4°C and were then treated three times with 5 μL of 3 × 10−2 M GA3 in ethanol during seedling development. Control plants were imbibed in water and were treated with ethanol only. A single GA treatment of this type early in seedling development is sufficient to rescue the ga1 dwarf phenotype and restore flower fertility.
Distance between base of inflorescence and pedicel of first flower.
Average internode length for internodes between the rosette and first flower.
No. of siliques with at least one developed seed from the first 10 flowers on the main inflorescence stem.
Percentage of plants with inflorescence phylotaxy clearly deviating from the spiral arrangement exhibited by untreated WT plants.
Constructs Designed to Overexpress SPY Are Not Equally Effective at Preventing Germination of spy Seeds
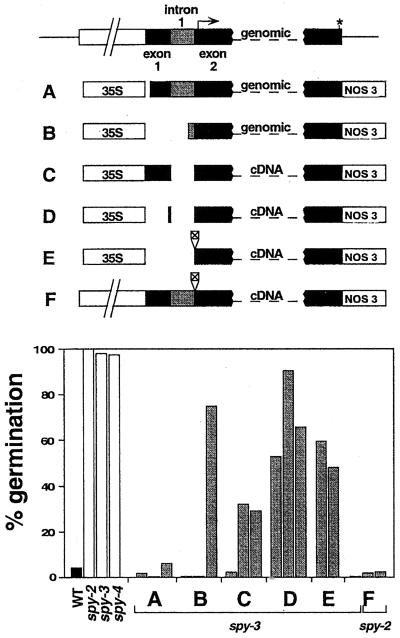
In an attempt to further test the hypothesis that SPY acts to inhibit GA signaling and to explore the role of SPY in plant development, a number of constructs were made in which the 35S promoter from cauliflower mosaic virus was used to drive expression of SPY (Fig. 1). Constructs containing the genomic and cDNA sequence were used. In addition, a construct was prepared in which expression of the SPY cDNA was under the control of the presumed SPY promoter (Fig. 1; data not shown). The functionality of the different constructs was determined by introducing them into spy-2 or spy-3 plants and determining if the sensitivity of germination to paclobutrazol was restored.
Figure 1.
Constructs designed to overexpress SPY mRNA and their effects on germination. Top, Schematic representation of the SPY locus and various constructs designed for SPY expression in transgenic plants. Exon 1 is not translated and the apparent start codon is represented by an arrow in exon 2. The stop codon is represented by an asterisk. Exon 1 is 324 bp and intron 1 is 320 bp. The size of the promoter used in construct F is about 2.8 kb from the 5′ end to the start codon. The square in constructs E and F represents an N-terminal epitope tag from the pRSET vector (Kroll et al., 1993). Bottom, Seed germination on 1.2 × 10−4 M paclobutrazol after 10 d for WT and spy mutant seeds, and for seeds homozygous for spy-2 or spy-3 and a single transgene locus. A through F correspond to the constructs shown in the top panel, and each gray vertical bar represents an independent transgenic line.
The two 35S:genomic SPY constructs (Fig. 1, A and B) were capable of fully restoring paclobutrazol sensitivity to spy-3 seeds, confirming that these constructs produce biologically active SPY protein in transgenic plants. By contrast, constructs with 35S driving expression of the cDNA (Fig. 1, C–E) were generally not fully effective at preventing germination of spy-3 on paclobutrazol, although one line containing construct C was able to prevent spy-3 seeds from germinating (Fig. 1). It appears that this result is not due to mutated or missing translated cDNA sequence, since the same SPY coding region driven by the SPY promoter (construct F) was completely effective at functionally complementing the spy-2 and spy-3 mutations at germination (Fig. 1) and throughout plant development (data not shown). These results suggest that the choice of promoter and the presence or absence of the first intron and exon influence the functionality of the construct. Further analysis was restricted to construct A, which was most effective of the 35S-driven constructs. Although we originally examined construct A in the spy-3 background (see above), most of the analysis of older plants was done in a WT SPY background because of the nature of the observed 35S:SPY phenotypes (see below). Nevertheless, the 35S:SPY construct appears to cause identical phenotypes in the WT SPY and spy− genetic backgrounds (e.g. for germination and hypocotyl length, see below).
35S:SPY Plants Have Elevated SPY mRNA Levels
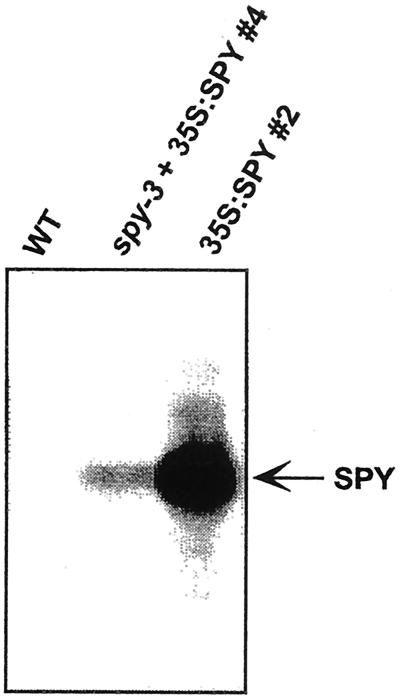
Northern-blot analysis confirmed that construct A caused overexpression of SPY in young seedlings (Fig. 2) in the spy-3 and WT SPY backgrounds. The line spy-3 + 35S:SPY #4 rescues the germination phenotype of spy-3 (Fig. 1) and contains elevated levels of SPY mRNA that is the same size as the endogenous SPY transcript. This result was confirmed when poly(A)+ mRNA was examined (data not shown).
Figure 2.
35S:SPY plants possess elevated levels of SPY mRNA. Northern-blot analysis of total RNA from 11-d-old WT seedlings and two independent 35S:SPY lines hybridized with a SPY cDNA probe. SPY mRNA of the same size as the major band observed in 35S:SPY lines was detected from WT seedlings if the film was exposed for a longer period of time. Equal loading of RNA was confirmed by visualizing the RNA with ethidium bromide. Line number 4 is also homozygous for the spy-3 mutation. Lines #2 and #4 display phenotypes typical of 35S:SPY plants.
35S:SPY Reduces GA Response during Seed Germination
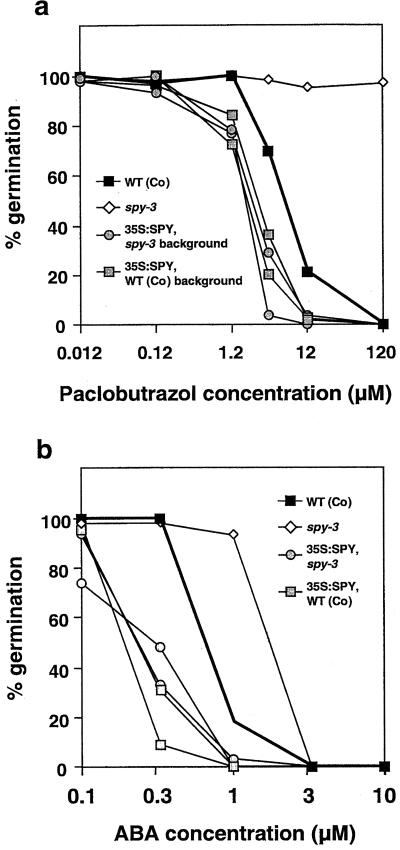
One prediction of the hypothesis that SPY is a negative regulator of GA responses is that 35S:SPY seeds will have decreased GA response and, as a consequence, an increased response to ABA. To test this hypothesis seeds were collected from plants grown together under identical conditions and germinated on various concentrations of paclobutrazol, to prevent de novo GA biosynthesis, or ABA (Fig. 3). As has been shown previously, spy-3 seeds were less sensitive than WT seeds to paclobutrazol (Jacobsen et al., 1993) and ABA (Steber et al., 1998), whereas 35S:SPY seeds were more sensitive to these growth regulators. This result was observed for multiple independent lines in the WT and spy-3 genetic backgrounds, including the lines 35S:SPY #2 and spy-3 + 35S:SPY #4. As expected, if SPY is a negatively acting component of the GA signaling pathway, these results suggest that GA sensitivity is increased by reducing SPY activity and decreased by increasing SPY activity. On media without growth regulators, 35S:SPY seeds germinate more slowly (1–2 d later) than WT seeds, but after several days, a similar proportion (typically about 95%) of seeds germinate (data not shown).
Figure 3.
35S:SPY seeds exhibit altered paclobutrazol and ABA sensitivity. Dose-response curves for seed germination were used to compare the sensitivity with paclobutrazol (a), an inhibitor of GA biosynthesis, and ABA (b) of WT (Columbia), spy-3, and four independent 35S:SPY lines in a WT SPY or mutant spy-3 genetic background. The original spy mutants were isolated at the highest paclobutrazol concentration shown, which is 1.2 × 10−4 M.
Hypocotyl Elongation Is Altered in 35S:SPY Lines and in spy Mutants
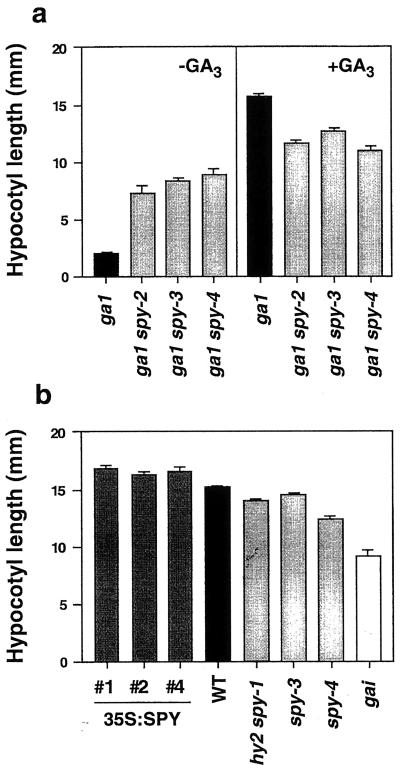
Since GAs are one of the many factors known to influence hypocotyl length, the effects of several spy mutations and the 35S:SPY construct on this phenotype were examined. To eliminate possible complicating effects caused by altered photoreceptor levels or activity, final hypocotyl length was measured for dark-grown seedlings (Fig. 4). As previously shown for light-grown seedlings (Silverstone et al., 1997), spy mutations are able to partially suppress the short hypocotyl phenotype of ga1. In contrast, when grown on a saturating dose of 3 × 10−4 M GA3, ga1 spy double mutants possessed shorter hypocotyls than similarly treated ga1 plants. A similar observation was made by Silverstone et al. (1997) in the La-er background for light-grown seedlings.
Figure 4.
Hypocotyl length of dark-grown mutants and transgenic plants overexpressing SPY. Final hypocotyl length of seedlings grown in the dark on filter paper saturated with 1× Murashige and Skoog salts and 1% (w/v) Suc. In a, the “−GA” treatment consisted of imbibing the seeds for 3 d in 3 × 10−5 M GA3, rinsing, and transferring to a solution without GA. The “+GA” seedlings were allowed to germinate and grow in the presence of 3 × 10−4 M GA3. In b, no exogenous GA was added. Data from three independent 35S:SPY lines are shown. Line #4 is also homozygous for the spy-3 mutation.
The effect of several spy mutations and the 35S:SPY construct on dark-grown hypocotyl length was also examined in the GA1 (i.e. otherwise WT) background. To varying degrees, different spy mutants were found to have short hypocotyls (P < 0.01) compared with WT Columbia plants (Fig. 4b). In white or red light, spy-1 hy2 double mutant seedlings (Jacobsen et al., 1996) possess long hypocotyls due to the hy2 mutation-dependent and light-dependent decrease in phytochrome function. In contrast, and similar to the other spy alleles examined, dark-grown spy-1 hy2 seedlings possess short hypocotyls. In the La-er background, dark-grown WT plants (13.16 ± 0.30 mm) also displayed significantly (P < 0.02) longer hypocotyls than spy-5 plants (10.80 ± 0.90 mm). The short hypocotyl length phenotype was also observed for spy-4 in the light and is opposite to the predicted phenotype for plants with increased GA response or those treated with GA. Treatment of ga1 spy double mutants (Fig. 4a) or spy-4 plants (data not shown) with a saturating GA dose did not restore hypocotyl length to WT values.
Consistent with increased SPY gene expression, 35S:SPY lines exhibited a subtle but significant (P < 0.01) long hypocotyl phenotype opposite to that displayed by loss-of-function spy mutants and gai plants (Fig. 4b). 35S:SPY plants also possessed significantly (P < 0.001) longer hypocotyls than WT plants in white light (data not shown). Line #4, which is homozygous for the spy-3 mutation and contains elevated levels of SPY mRNA (Fig. 2), also possessed long hypocotyls in the dark (Fig. 4), demonstrating that the 35S:SPY construct can functionally complement the germination (Fig. 1) and short hypocotyl length spy mutant phenotypes.
Vegetative Growth of Plants with Altered GA Levels or Response
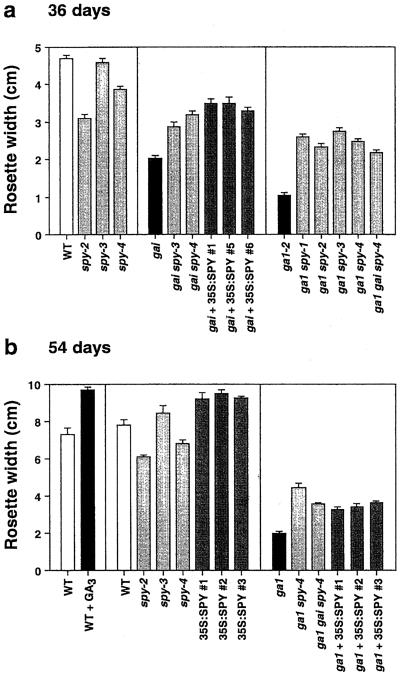
One of the most obvious effects of GA deficiency in Arabidopsis, the decreased diameter of the vegetative rosette, demonstrates that GA is required for normal leaf growth in LD and short day (SD) photoperiods (Fig. 5). Repeated treatment of WT plants with GA3 produced paler, larger plants with longer leaves and larger rosettes. By contrast, severe spy mutants such as spy-2 and spy-4, although paler than WT plants, possess smaller rosettes. This phenotype was not due to differing angles of the leaf above the horizontal for different genotypes, since rosettes were flattened when measured. Furthermore, under SD conditions, spy-2 and spy-4 plants were clearly smaller than WT plants several weeks before flower buds were visible, suggesting that the reduced plant size is not a consequence of increased assimilate distribution to reproductive growth in the early-flowering spy mutants.
Figure 5.
Rosette diameter of plants with altered GA levels or response. Rosette width (at the widest point of the plant) was determined at 36 (a) and 54 (b) days of age in SD conditions. At 36 d, WT and 35S:SPY plants were of similar size as were ga1 mutants and ga1 35S:SPY plants (not shown). For treatment of WT with GA, seeds were germinated on 3 × 10−5 M GA3, and were then transferred to soil and after 12 d were treated weekly with 0.5 μL of 3 × 10−2 M GA3 in ethanol until flowering. Control plants received ethanol only. The suppression of the gai dwarf phenotype by spy mutations or by 35S:SPY is not due to altered rates of leaf initiation because all three genotypes possessed similar numbers of leaves.
Although severe spy mutants possess smaller rosettes in a GA1 GAI background, loss of SPY activity partially suppressed the dwarf phenotype of ga1 and gai plants (Fig. 5; Jacobsen et al., 1993, 1996). Careful examination revealed that whereas gai spy-4 and GAI spy-4 plants are very similar in appearance, spy-4 is not truly epistatic to gai since gai spy-4 double mutants are slightly smaller (Fig. 5) and possess darker leaves and shorter inflorescence internodes than spy-4 mutants (data not shown). A similar lack of epistasis of spy-4 over gai was also observed in the ga1 background for rosette size (Fig. 5) and leaf color (data not shown).
Similar to the case for dark-grown hypocotyl elongation, 35S:SPY plants are larger than WT plants at 54 d of age in SD, a phenotype opposite to that exhibited by severe spy mutants and consistent with increased SPY protein levels in 35S:SPY plants. Despite the difference between severe spy mutants and 35S:SPY plants in terms of rosette size, the 35S:SPY construct and spy mutations partially suppress the vegetative dwarf phenotype of the ga1 and gain-of-function gai mutants.
The suppression of the ga1 phenotype has been used as a major criterion for increased GA response in several genetic screens. The partial suppression of the ga1 dwarf phenotype by loss of SPY function and the 35S:SPY construct led us to re-examine how diagnostic this phenotype is of changed GA response. The suppression of the ga1 phenotype by several other mutants known to alter plant growth, but not necessarily alter GA signal transduction, was also examined (Table II). In addition to spy and rga, mutants thought to possess increased GA response, the mutants tested possess impaired phytochrome activity caused by reduced chromophore biosynthesis (hy1; Davis et al., 1999; Muramoto et al., 1999), reduced phytochrome B levels (hy3; Reed et al., 1993), reduced phytochrome action (hy5; Ang et al., 1994), and loss of ELONGATED (ELG) (Halliday et al., 1996) activity. Of the range of mutants examined, only the spy and rga mutations can visibly suppress the vegetative defects of ga1 and only spy mutations, and to a lesser extent the 35S:SPY construct, can suppress the defects in flower development caused by severe GA deficiency.
Table II.
Suppression of severe GA deficiency by various mutations and 35S∶SPY in LD
| Genotype | Background | Heighta | Visible Petals and Mature Pollen | Fertile Fruit |
|---|---|---|---|---|
| mm | ||||
| WT | La-er | 66.7 ± 4.6 | Yes | Yes |
| gal-2 | La-er | <1 | No | No |
| gal-2 hy3 | La-er | <1 | No | No |
| gal-2 hy5 | La-er | <1 | No | No |
| gal-3 elg | La-er | <1 | No | No |
| gal-3 rga-24 | La-er | 6.3 ± 1.0 | No | No |
| WT | Columbia | 97.1 ± 3.6 | Yes | Yes |
| gal-2 | Columbia | <1 | No | No |
| gal-2 hy1-100 | Columbia | <1 | No | No |
| gal-2 spy-3 | Columbia | 18.8 ± 1.7 | Yes | Yesb |
| gal-2 spy-4 | Columbia | 20.3 ± 2.2 | Yes | Yesb |
| gal-2 35S∶SPY #1 | Columbia | 5.6 ± 0.6 | Yes | No |
| gal-2 35S∶SPY #2 | Columbia | 25.5 ± 2.0 | Yes | Noc |
| gal-2 35S∶SPY #3 | Columbia | 16.5 ± 2.5 | Yes | Noc |
Distance between base of inflorescence and pedicel of first flower.
Some fruit sterile, particularly those from earliest flowers.
Occasionally fertile fruit present, but in most flowers pollen can not reach stigmatic surface because anthers are not sufficiently long.
Flowering Time of Plants with Altered GA Levels or Response
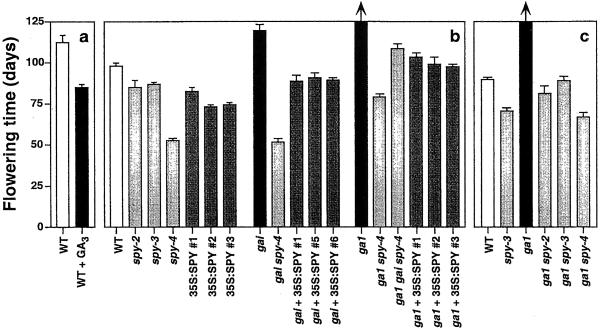
GAs are known to be involved in flower induction in many, but not all, species. In Arabidopsis, GAs act to promote flowering, and under laboratory SD conditions with light provided by standard white-light fluorescent tubes, severe ga1 mutants do not flower. The flowering time of spy mutants and 35S:SPY lines was examined under LD and SD conditions (Fig. 6). In agreement with previous results, treatment of WT plants with GA3 induced earlier flowering under SD conditions. Mutant spy plants also flower earlier (P < 0.001) than WT plants, consistent with an increase in GA response and previous reports (Jacobsen et al., 1993; Kania et al., 1997). Homozygous spy-2 and spy-3 plants flowered at a similar time, whereas spy-4 flowered significantly (P < 0.001) earlier than the other mutants. The spy-4 mutation restored the flowering time of ga1 spy-4 plants to that of WT plants, demonstrating that in this regard loss of spy function can completely suppress phenotypes caused by GA deficiency. In agreement with the phenotypes for vegetative growth (Fig. 5), ga1 gai spy-4 triple mutants flowered later (measured by the day of the first open flower) than ga1 spy-4 double mutants, confirming that spy-4 cannot completely suppress the gain-of-function gai phenotype.
Figure 6.
Flowering time of plants with altered GA levels or response. Flowering time was recorded as the day the first flower fully opened and the petals were completely reflexed. Plants were grown under SD conditions to maximize the differences in flowering time. a through c show data from three separate experiments. Treatment of WT plants with GA was as described for Figure 5. Homozygous ga1 plants did not flower during these experiments, each of which lasted over 5 months. Combining ga1 with spy mutations or with 35S:SPY allowed flowering, as did treatment with GA (data not shown).
Although several of the phenotypes displayed by 35S:SPY plants are opposite to those observed in spy mutants (see above), plants overexpressing SPY mRNA also flower early (Fig. 6). This phenotype was observed in LD and SD for multiple independent lines, and also occurred if total or rosette leaf number, rather than time, were used to measure flower induction (data not shown). In addition, early flowering due to the 35S:SPY transgene under LD and SD photoperiods was shown to cosegregate as a dominant trait with the kanamycin resistance phenotype in a population in which the 35S:SPY transgene (line 35S:SPY #2) was segregating (data not shown). The late flowering phenotype of the ga1 and gai mutants was suppressed by spy mutations and the 35S:SPY construct. This result is similar to the suppression of the dwarf phenotype of these mutants (Fig. 5) and is consistent with the early flowering of 35S:SPY lines (Fig. 6).
Other spy Mutant and 35S:SPY Phenotypes
An additional spy mutant phenotype is a reduction in the size and number of leaf serrations, particularly in SD-grown plants: spy mutants and gai spy double mutants possess essentially unserrated smooth-edged leaves (Table III). By contrast, WT and ga1 plants with or without GA3 treatments and gai plants all possess serrated leaves. Leaf morphology, therefore, represents another example of a spy mutant phenotype that cannot be duplicated by repeated treatment of WT plants with GA. The leaf serration phenotype also provides a criterion to distinguish between the suppression of gai caused by loss of SPY activity or the 35S:SPY construct. Double-mutant gai spy plants and gai 35S:SPY lines are larger than gai dwarf plants, and the 35S:SPY construct leads to suppression at least as strong as that caused by spy-3 or spy-4 in terms of rosette diameter (Fig. 5). Nevertheless, the overall appearance of gai spy− and gai 35S:SPY plants is not identical since unlike gai spy− plants, gai 35S:SPY plants possess serrated leaves (Table III).
Table III.
Leaf serration and stem death phenotypes under SD conditions
| Genotype (All in Columbia) | Leaves Serrated | Proportion of Plants Exhibiting Stem Death |
|---|---|---|
| WT | Yes | 1/23 |
| WT + GA3a | Yes | 0/6 |
| gal-2 | Yes | No inflorescence stem |
| gal-2+ GA3b | Yes | 0/7 |
| gai | Yes | 0/12 |
| spy-2 | No | 5/5 |
| spy-3 | No | 0/8 |
| spy-4 | No | 10/10 |
| gai spy-2 | No | 3/3 |
| gai spy-3 | No | 0/8 |
| gai spy-4 | No | 7/18 |
| gai+ 35S∶SPY #1 | Yes | 0/10 |
| gai+ 35S∶SPY #5 | Yes | 0/9 |
| gai+ 35S∶SPY #6 | Yes | 0/10 |
| 35S∶SPY #1 | No | 1/8 |
| 35S∶SPY #2 | No | 4/8 |
| 35S∶SPY #3 | No | 5/10 |
One cotyledon or leaf treated weekly with 5 μg of GA3 in 0.5 μL of ethanol.
Single leaf treated once with 20 μg of GA3 in 2 μL of ethanol to induce flowering.
A novel and unexplained phenotype was observed in SD-grown plants after flowering (Table III). Depending on the experiment, variable numbers of the severe spy-2 and spy-4 mutants would exhibit cell death approximately 3 cm below the apical meristem, which subsequently led to death of the apical tissue above this point. Lateral meristems from lower nodes continued to grow and eventually set seeds. This phenotype was also observed in gai spy-2 and gai spy-4 plants and in 35S:SPY plants, but not in gai 35S:SPY plants. Although one out of 23 WT plants exhibited a similar phenotype, the frequency did not markedly increase following GA treatment.
DISCUSSION
spy mutants and 35S:SPY lines display a range of phenotypes, suggesting that GA response is altered. In addition, severe spy mutants in the Columbia and La-er backgrounds exhibit phenotypes not expected for plants with increased GA response (e.g. Table I). This is particularly evident in the La-er background where the presence of a loss-of-function er allele leads to stunted spy mutant plants that possess reduced internode lengths. Although this phenotype suggests impaired growth responses, spy mutants in the La-er background can still partially suppress the ga1 and gai dwarf phenotype. This demonstrates that, regardless of other growth defects, GA response is increased in er− spy− plants. By contrast, a comparable effect on the phenotype of 35S:SPY lines has not been observed in the La-er background.
For final hypocotyl length in the La-er (Silverstone et al., 1997) and Columbia backgrounds (Fig. 4), loss of SPY activity increases GA response at low endogenous GA concentrations, but surprisingly, it decreases growth in plants with a normal or elevated amount of GA. It is not clear why spy mutants display this phenotype, although it cannot be due to decreased levels of endogenous GAs since treatment of ga1 spy double mutants (Fig. 4) or spy-4 plants (data not shown) with a saturating GA dose cannot restore hypocotyl length to WT values. In this case, the apparent change in GA response, as determined by final hypocotyl length, for normal or high exogenous GA levels is likely to reflect roles for SPY in a pathway(s) other than GA signaling (see below).
Introduction of the 35S:SPY construct into a range of Arabidopsis genotypes reduces GA response at seed germination, but causes changes in growth consistent with increased GA response throughout the above-ground part of the plant. Several lines of evidence suggest that the effects of the 35S:SPY construct on the above-ground portion of the plant are not likely to be due to cosuppression. The 35S:SPY lines possess greatly elevated SPY mRNA levels (Fig. 2), and the ability of the 35S:SPY construct to prevent germination of spy mutant seeds on paclobutrazol (Fig. 1) demonstrates that this construct encodes a functional protein. Although we have not been able to determine the level of SPY protein in plant tissue, 35S:SPY plants possess a range of phenotypes that are opposite to, or different from, those shown by spy mutants (Figs. 4 and 5; Table III). The 35S:SPY transgene also delays flowering in a spy-4 background (data not shown) consistent with increased SPY protein levels.
Although 35S:SPY lines #2 and #4 differ in the level of SPY overexpression (Fig. 1), no obvious differences in the phenotypes of these 35S:SPY lines was observed (e.g. Fig. 4). No correlation between SPY mRNA levels and the magnitude of the 35S:SPY phenotypes, which are similar for all of the construct A lines examined, have been observed. Doubling SPY gene copy number by transforming WT plants with a genomic clone containing the entire SPY gene or using construct F (Fig. 1) functionally complements all spy mutant phenotypes (Fig. 1; data not shown), but does cause detectable changes in GA response or plant development.
The suppression of the gai dwarf phenotype by spy mutations potentially allows the gene order of WT GAI and SPY to be determined. To define gene order in a genetic pathway, epistasis must be defined as the inability, based on plant phenotype, to determine which allele (in this case GAI or gai) is present in a spy mutant background. Because of its severe nature and the fact that it may be a null allele, spy-4 was used for this experiment. Our results demonstrate that although spy-4 can partially suppress gai, spy-4 is not epistatic to gai. In other words, gai spy-4 and GAI spy-4 plants are not identical. As a consequence, the gene order of the GAI and SPY loci in GA signal transduction cannot be determined from these genetic studies. Nevertheless, the fact that the spy-4 mutation is able to substantially suppress the combined effects of GA deficiency and the mutant gai protein in ga1 gai spy-4 plants (Fig. 5) strongly supports a role for SPY in regulating GA response. The near-complete masking of the gai phenotype by spy-4 also demonstrates that the ability of the mutant gai protein to impair GA response is largely dependent on normal SPY activity.
The results presented above confirm some previous hypotheses and support several new ideas about the role of SPY in GA signal transduction and plant development. It is clear that some aspects of the spy mutant phenotype indicate that SPY is a negative regulator of GA signaling. The ability of HvSPY to block GA-induced α-amylase expression in barley aleurone cells also supports a role for SPY in the GA-signaling pathway. A range of genetic evidence (Jacobsen et al., 1993; this paper) also strongly suggests that SPY is a negatively acting component of the GA signal transduction pathway in intact plants. By contrast, other phenotypes displayed by some of the spy mutants are difficult to reconcile with known roles for GAs in plant development. This raises two, mutually nonexclusive, possibilities: GAs have additional, previously unestablished, physiological roles during growth and development, and/or SPY also acts on other, as yet undefined, signaling pathways. Although it would seem reasonable to suggest that some spy mutant phenotypes define novel GA-requiring responses, these phenotypes have not been reported in other mutants with altered GA levels or response. Although it is not possible to resolve this issue at present, it is possible that GA response genes such as RGA and GAI, in addition to the known GA biosynthesis genes, only affect a subset of GA responses. If this model is correct, then it appears that treatment of WT plants with exogenous GAs or chemical inhibitors of GAs also does not reveal the full extent of physiological processes requiring or regulated by GAs. The analysis of the lh mutants of pea supports this suggestion. These mutants have been used to demonstrate that GAs are required for normal seed development (Swain et al., 1997), a role not apparent from the analysis of other GA-deficient mutants or from treatment of plants with GAs or GA inhibitors.
Although the spy mutant phenotype is more complex than originally suggested, 35S:SPY plants possess a relatively simple phenotype after seed germination, consistent with an increased GA response: longer hypocotyls, larger rosettes, and earlier flowering. Why overexpression of a negative regulator should have this effect is not known, but several explanations are possible. One is that overexpression of the SPY protein causes titration of other proteins that form a complex with SPY. For example, higher levels of SPY might cause the formation of partial complexes or complexes with reduced activity. Possible SPY partners include RGA and GAI, and consistent with this idea, some 35S:SPY phenotypes such as partial suppression of the ga1 and gai dwarf phenotypes could be explained by reduced GAI or RGA activity.
Another hypothesis is that overexpression of the SPY protein activates GA response, perhaps by inappropriately O-GlcNAc modifying target proteins. In this regard, overexpression of SPY in intact plants may be analogous to the slight induction of an α-amylase reporter in barley aleurone cells by HvSPY in the absence of exogenous GA (Robertson et al., 1998). The first model predicts that since SPY is likely to interact with other proteins via its TPR domain, overexpression of this domain alone should be sufficient to cause dominant-negative phenotypes such as increased GA response. Preliminary results suggest that this is the case (T.S. Tseng, S.M. Swain, and N.E. Olszewski, unpublished data). For example, 35S:SPY and 35S:SPY-TPR lines exhibit early flowering and are able to partially suppress the ga1 and gai dwarf phenotypes. By contrast, for seed germination and hypocotyl elongation, 35S:SPY plants exhibit phenotypes opposite to those observed in spy mutants and 35S:SPY-TPR lines. These observations can be reconciled by the hypothesis that some 35S:SPY phenotypes are due to increased SPY activity, whereas others are caused by dominant-negative effects, perhaps due to accumulation of abnormal SPY proteins. Testing of this hypothesis will require the identification of SPY partners and substrates, and a better understanding of whether developmental processes besides GA signaling require SPY.
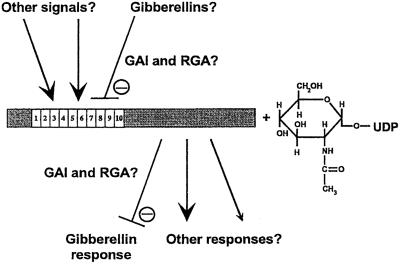
Because SPY was originally identified genetically (Jacobsen et al., 1993) and the similarity to OGT was recognized (Thornton et al., 1999), hypotheses regarding the role of SPY in GA response, and possibly other aspects of plant development, have had to become more complex. It now appears likely that SPY is an OGT that interacts with unknown protein partners via its TPR domain to modify as yet unidentified substrates to regulate plant development in several different ways, one of which is to inhibit the GA signal transduction pathway (Fig. 7). A number of different molecular-genetic and biochemical approaches are presently under way to address these questions.
Figure 7.
Model for SPY's role in GA signaling and plant development. The SPY protein is a putative O-GlcNAc transferase and is represented by the rectangle. The reaction involves the transfer of a GlcNAc moiety from UDP-GlcNAc to Ser or Thr residues of target proteins. The 10 TPR domains may interact with other proteins that modify SPY's activity and substrate specificity. Two other GA response components, RGA and GAI, may be upstream or downstream of SPY. RGA and GAI could interact with the TPR domain and/or serve as substrates for SPY. GAs may act via SPY (and RGA and GAI) or SPY could modify GA response, possibly by regulating RGA and GAI activity. Based on the phenotypes of spy mutants, SPY may also respond to other signals or be involved in other responses, in addition to its role in GA response.
MATERIALS AND METHODS
All seeds were stratified for 3 d at 4°C under dim light to aid germination. Plants were routinely grown under an 18-h LD photoperiod of 120 μmol m−2 s−1 consisting of white fluorescent light with a temperature of 22°C (day) and 20°C (night). For plants grown under SD conditions, the photoperiod consisted of 8 h of light (same source as LD) and 16 h of darkness. Unless otherwise mentioned, all Arabidopsis genotypes used are in the Columbia background or were backcrossed three or six times into Columbia from La-er or WS. The ga1-2 mutation is a presumed null allele similar to ga1-3 (Sun et al., 1992) and was backcrossed into Columbia three times from La-er. To allow comparison of the spy mutant phenotypes in different ecotypes, the spy-4 mutation, originally generated by a T-DNA insertion in the WS background, was backcrossed into the Columbia (six times) and La-er (three times) genetic backgrounds. The ethyl methanesulfonate-generated allele, spy-2, was also backcrossed from Columbia into La-er three times.
All constructs were generated using standard molecular techniques. Constructs A and B contained an approximately 6-kbp genomic fragment containing the entire SPY coding region downstream from exon 2. Construct A also included most of the first untranslated exon (3′ of a unique XhoI site) and the first intron. Construct B differs from A in that it lacks the first exon and includes only 79 bp of the first intron (3′ of a HindIII site in this intron). Constructs C to F contain the SPY cDNA with or without the first untranslated exon (Fig. 1; Jacobsen et al., 1996). The vectors used for plant transformation were based on derivatives of pOCA18 as described in Robertson et al. (1998).
Plant transformation was essentially as described in Robertson et al. (1998), except that WT (Columbia), spy-3, and gai (backcrossed three times into Columbia and without the La-er er allele) were infiltrated. To introduce construct F (Fig. 1) into spy-2 plants, heterozygous SPYspy-2 plants were initially transformed and progeny homozygous for spy-2 and a single transgene insert (based on kanamycin segregation) subsequently isolated. Independent transgenic lines #1, #2, and #3, generated by transforming WT (Columbia), were crossed with the ga1-2 mutant (backcrossed three times into Columbia and without the La-er er allele) to generate ga1 plants containing 35S:SPY. Line #1 was also combined with the gai mutation by crossing. Line #4 (spy-3 transformation) and lines #5 and #6 (gai transformation) all represent independent transformation events. All lines described in detail here contain a single locus, based on segregation of kanamycin resistance, containing construct A (Fig. 1). All of the 35S:SPY phenotypes described in this paper were consistently observed for multiple independent lines over several generations in at least two experiments. Values are shown as the means ± se, and Student's t test was used to determine the statistical significance of differences between genotypes.
ACKNOWLEDGMENTS
We thank Tina Thornton and Lynn Hartweck for helpful discussions and critical comments on this manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–9604126 to N.E.O.).
LITERATURE CITED
- Ang LH, Deng XW. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol P, Peng J, Harberd NP. Isolation and preliminary characterization of gas1-1, a mutation causing partial suppression of the phenotype conferred by the gibberellin-insensitive (gai) mutation in Arabidopsis thaliana (L.) Heyhn. Planta. 1995;197:414–417. doi: 10.1007/BF00202665. [DOI] [PubMed] [Google Scholar]
- Comer FI, Hart GW. O-Glycosylation of nuclear and cytosolic proteins: dynamic interplay between O-GlcNAc and O-phosphate. J Biol Chem. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA. 1999;96:6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LT. Gibberellins and flowering in long day plants, with special reference to Lolium temulentum. Aust J Plant Physiol. 1999;26:1–8. [Google Scholar]
- Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA. 1999;96:7575–7580. doi: 10.1073/pnas.96.13.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;17:1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Halliday K, Devlin PF, Whitelam GC, Hanhart C, Koornneef M. The ELONGATED gene of Arabidopsis acts independently of light and gibberellins in the control of elongation growth. Plant J. 1996;9:305–312. doi: 10.1046/j.1365-313x.1996.09030305.x. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Manipulation of hormone biosynthetic genes in transgenic plants. Curr Opin Biotechnol. 2000;11:130–137. doi: 10.1016/s0958-1669(00)00071-9. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HD, Smith SJ, Desikan R, Plakidou-Dymock S, Lovegrove A, Hooley R. Heterotrimeric G proteins are implicated in gibberellin induction of α-amylase gene expression in wild oat aleurone. Plant Cell. 1998;10:245–254. doi: 10.1105/tpc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y, Garcia-Martinez JL. Regulation of gibberellin biosynthesis by light. Curr Opin Plant Biol. 1999;2:398–403. doi: 10.1016/s1369-5266(99)00012-6. [DOI] [PubMed] [Google Scholar]
- Kania T, Russenberger D, Peng S, Apel K, Melzer S. FPF1 promotes flowering in Arabidopsis. Plant Cell. 1997;9:1327–1338. doi: 10.1105/tpc.9.8.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll DJ, Abdel-Malek Abdel-Hafiz H, Marcell T, Simpson S, Chen CY, Gutierrez-Hartmann A, Lustbader JW, Hoeffler JP. A multifunctional prokaryotic protein expression system: overproduction, affinity purification, and selective detection. DNA Cell Biol. 1993;12:441–453. doi: 10.1089/dna.1993.12.441. [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3 β-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R. Gibberellin and abscisic acid signaling in aleurone. Trends Plant Sci. 2000;5:102–110. doi: 10.1016/s1360-1385(00)01571-5. [DOI] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F. “Green revolution” genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE. Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell. 1998;10:995–1007. doi: 10.1105/tpc.10.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos MD, Hanover JA. Structure of O-linked GlcNAc transferase: mediator of glycan-dependent signaling. Biochem Biophys Res Commun. 2000;271:275–280. doi: 10.1006/bbrc.2000.2600. [DOI] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martinez ES, Sun T-p. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997;146:1087–1090. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow DM, Hart GW. Nuclear and cytoplasmic glycosylation. Int Rev Cytol. 1998;181:43–74. doi: 10.1016/s0074-7696(08)60416-7. [DOI] [PubMed] [Google Scholar]
- Steber CM, Cooney SE, McCourt P. Isolation of the GA response mutant sly1, a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149:509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Goodman HM, Ausubel FM. Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell. 1992;4:119–128. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Reid JB, Kamiya Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997;12:1329–1338. [Google Scholar]
- Thornton T, Swain SM, Olszewski N. Gibberellin signal transduction presents: the SPY who O-GlcNAc'd me. Trends Plant Sci. 1999;4:424–428. doi: 10.1016/s1360-1385(99)01485-5. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Somerville CR. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 1995;108:495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]