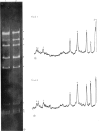
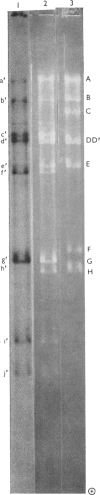
Abstract
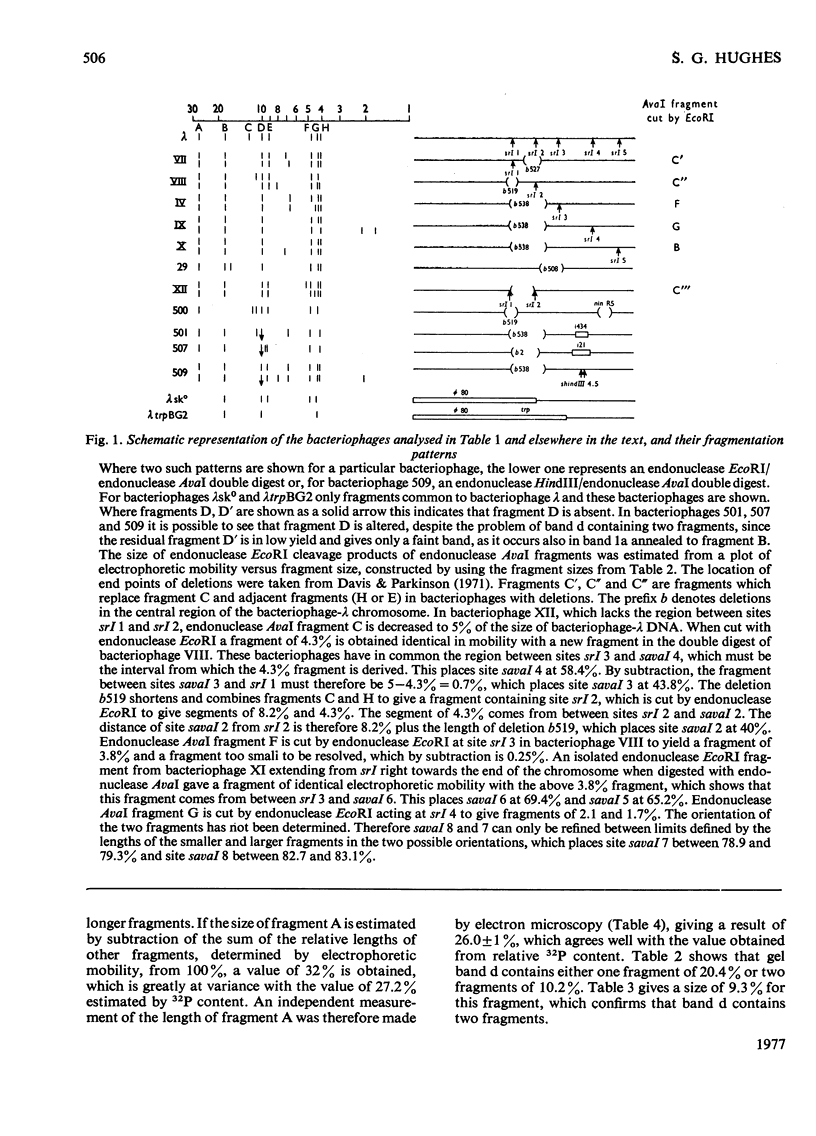
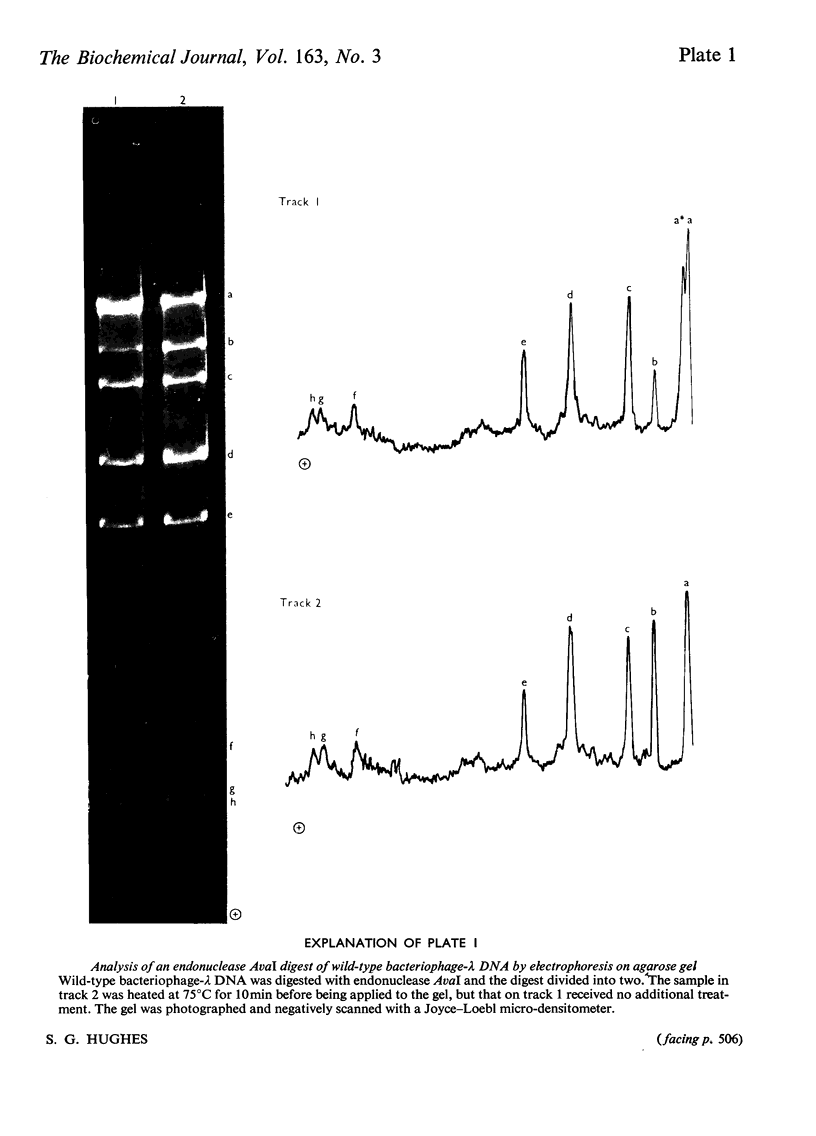
The linear order of nine fragments generated by the action of endonuclease AvaI on the DNA of bacteriophage lambda was determined from the altered fragmentation patterns of bacteriophages containing known deletions and of hybrids of bacteriophages lambda and phi80. Digestion of 5'-terminally 32P-labelled bacteriophage-lambda DNA was used to identify the terminal fragments. Measurement of relative fragment lengths permitted rough mapping of the endonuclease-AvaI cleavage sites relative to the ends of the bacteriophage-lambda chromosome. The fragment order was confirmed and the map refined by analysis of the fragmentation of derivative phages containing single cleavage sites for endonuclease EcoRI.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Jeppesen P. G., Katagiri K. J., Delius H. Mapping the DNA fragments produced by cleavage by lambda DNA with endonuclease RI. Nature. 1973 Jan 12;241(5385):120–123. doi: 10.1038/241120a0. [DOI] [PubMed] [Google Scholar]
- Allet B., Solem R. Separation and analysis of promoter sites in bacteriophage lambda DNA by specific endonucleases. J Mol Biol. 1974 Jan 5;85(4):475–484. doi: 10.1016/0022-2836(74)90310-6. [DOI] [PubMed] [Google Scholar]
- Arber W., Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Brammar W. J., Murray N. E., Winton S. Restriction of lambda trp bacteriophages by Escherichia coli K. J Mol Biol. 1974 Dec 25;90(4):633–647. doi: 10.1016/0022-2836(74)90529-4. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Parkinson J. S. Deletion mutants of bacteriophage lambda. 3. Physical structure of att-phi. J Mol Biol. 1971 Mar 14;56(2):403–423. doi: 10.1016/0022-2836(71)90473-6. [DOI] [PubMed] [Google Scholar]
- Goldberg A. R., Howe M. New mutations in the S cistron of bacteriophage lambda affecting host cell lysis. Virology. 1969 May;38(1):200–202. doi: 10.1016/0042-6822(69)90148-2. [DOI] [PubMed] [Google Scholar]
- Haggerty D. M., Scheif R. F. Location in bacteriophage lamdba DNA of cleavage sites of the site-specific endonuclease from Bacillus amyloliquefaciens H. J Virol. 1976 May;18(2):659–663. doi: 10.1128/jvi.18.2.659-663.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. G., Brown P. R. Improved preparation of bacteriophage T4-infected Escherichia coli for the isolation of polynucleotide kinase. Biochem J. 1973 Mar;131(3):583–583. doi: 10.1042/bj1310583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D., HOGNESS D. S. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J Mol Biol. 1960 Dec;2:392–415. doi: 10.1016/s0022-2836(60)80050-2. [DOI] [PubMed] [Google Scholar]
- Lang D., Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9(3):373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin D. A., Schaller H. The topology of DNA from the small filamentous bacteriophage fd. J Mol Biol. 1966 Jan;15(1):1–7. doi: 10.1016/s0022-2836(66)80204-8. [DOI] [PubMed] [Google Scholar]
- McParland R. H., Brown L. R., Pearson G. D. Cleavage of lambda DNA by a site-specific endonuclease from Serratia marcescens. J Virol. 1976 Sep;19(3):1006–1011. doi: 10.1128/jvi.19.3.1006-1011.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Hughes S. G., Brown J. S., Bruce S. A. Isolation and characterization of two sequence-specific endonucleases from Anabaena variabilis. Biochem J. 1976 Nov;159(2):317–322. doi: 10.1042/bj1590317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Murray K. Nucleotide sequence analysis with polynucleotide kinase and nucleotide "mapping" methods. 5'-Terminal sequences of deoxyribonucleic acid from bacteriophages lambda and 424. Biochem J. 1973 Mar;131(3):569–582. doi: 10.1042/bj1310569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Batten P. L., Murray K. Restriction of bacteriophage lambda by Escherichia coli K. J Mol Biol. 1973 Dec 15;81(3):395–407. doi: 10.1016/0022-2836(73)90149-6. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Murray K. Manipulation of restriction targets in phage lambda to form receptor chromosomes for DNA fragments. Nature. 1974 Oct 11;251(5475):476–481. doi: 10.1038/251476a0. [DOI] [PubMed] [Google Scholar]
- Nathans D., Smith H. O. Restriction endonucleases in the analysis and restructuring of dna molecules. Annu Rev Biochem. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- Old R., Murray K., Boizes G. Recognition sequence of restriction endonuclease III from Hemophilus influenzae. J Mol Biol. 1975 Feb 25;92(2):331–339. doi: 10.1016/0022-2836(75)90232-6. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Deletion mutants of bacteriophage lambda. II. Genetic properties of att-defective mutants. J Mol Biol. 1971 Mar 14;56(2):385–401. doi: 10.1016/0022-2836(71)90472-4. [DOI] [PubMed] [Google Scholar]
- Perricaudet M., Tiollais P. Defective bacteriophage lambda chromosome, potential vector for DNA fragments obtained after cleavage by Bacillus amyloliquefaciens endonuclease (BamI). FEBS Lett. 1975 Aug 1;56(1):7–11. doi: 10.1016/0014-5793(75)80099-8. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Sequence of the OR operator of phage lambda. Nature. 1975 Mar 13;254(5496):114–117. doi: 10.1038/254114a0. [DOI] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Thompson R., Hughes S. G., Broda P. Plasmid identification using specific endonucleases. Mol Gen Genet. 1974;133(2):141–149. doi: 10.1007/BF00264835. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]