Abstract
It is known that 25%–30% of individuals with cutaneous psoriasis (PsC) will develop psoriatic arthritis (PsA). To date, the reasons for the development of PsA in individuals with PsC have not been identified. Furthermore, there are considerable delays in the diagnosis and treatment of PsA, which lead to joint and bone deformation and chronic pain. It is therefore important to develop more precise diagnostic and screening tools. In this narrative review of the literature, clinical risk factors and novel molecular biomarkers (genetic markers, blood and inflammatory markers, lipid, metabolite and protein biomarkers) have been evaluated. The review included 38 publications that were reported between May 2020 and May 2024. Similar to previous reviews, nail involvement was one of the strongest clinical risk factors for the development of PsA, while molecular biomarkers did not provide a clear and robust differentiation between PsC and PsA groups. The seemingly poor performance of molecular markers may be largely attributed to small study populations and heterogeneity in study designs. Data and sample sharing in large consortia such as HIPPOCRATES (Health initiatives in Psoriasis and PsOriatic arthritis ConsoRTium European States) could help to overcome the limitations of small studies and enable the development of more robust diagnostic and screening tools for PsA.
Keywords: Arthritis, Psoriatic; Psoriasis; Biomarkers
WHAT IS ALREADY KNOWN ON THIS TOPIC
25%–30% of individuals with cutaneous psoriasis will develop psoriatic arthritis (PsA), but to date, the reasons for this have not been identified.
WHAT THIS STUDY ADDS
In this narrative review, we evaluated the known clinical risk factors and novel molecular biomarkers for the development of PsA.
We found nail involvement was one of the strongest clinical risk factors. Molecular biomarkers did not provide a clear and robust differentiation; this is likely due to the small study populations and heterogeneity in study designs.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Data and sample sharing in large consortia such as the HIPPOCRATES consortium could help to overcome these limitations and support the development of more robust diagnostic and screening tools for PsA.
Introduction
Approximately 25%–30% of people with cutaneous-only psoriasis (PsC) attending dermatologists have evidence of an inflammatory musculoskeletal (MSK) disease, psoriatic arthritis (PsA) when examined by rheumatologists.1 2 PsA disease features can be heterogeneous and can include one or more of the following features: peripheral arthritis often affecting just a few joints in early disease with more joints becoming involved as the disease progresses,3 enthesitis, dactylitis or axial inflammation. This heterogeneity contributes to poor recognition of PsA by both general practitioners and dermatologists, but it is not the only reason. In early disease, features may be transient, there are no diagnostic criteria or laboratory diagnostic tests, and as a result, the diagnosis and treatment of PsA may be delayed, leading to a poor outcome.4
There are several clinical or environmental features which may occur more commonly in PsA compared with PsC. These features include nail dystrophic changes, scalp or inverse psoriasis, psoriasis severity, family history of PsA, a history of trauma, smoking or excess alcohol intake.5 The association with PsA is controversial for some (eg, smoking, trauma), while others (eg, nail dystrophic change, obesity) are more consistently associated with PsA development. None of these clinical or environmental features are sufficiently discriminatory to be used in clinical practice. At best, they may be helpful in pointing to those people with PsC who might be screened for emerging MSK disease more carefully but that risks missing people with PsC who develop PsA and who do not have those risk factors. Furthermore, features which occur more commonly in PsA are not necessarily there at baseline when PsC first develops. Data from prospective observational study cohorts are required to confirm that the features are predictive of risk for PsA. There are limited data from such cohorts available with results of multivariate analysis of 464 people with PsC followed up for 8 years, during which time 51 developed a diagnosis of PsA, showing that psoriatic nail pitting (relative risk 2.5, p=0.002) and uveitis (relative risk 31.5, p=0.0002) were associated with the development of PsA.6
There have also been several studies which have sought to identify molecular markers which differentiate patients with PsA from those with PsC. In a systematic literature review (SLR) by Mulder et al,7 these markers were reviewed in detail. Some of the studies were prospective identifying potential markers present in people with PsC before the development of PsA. Some of the studies included significant numbers of subjects but most studies were relatively small. None of the markers to date have been validated in large, independent cohorts. In preparation for studies from the HIPPOCRATES consortium (https://www.hippocrates-imi.eu/), where it is planned to identify and validate clinical/molecular risk factors for the development of PsA in those with PsC, we have conducted a narrative literature review to provide an update since the date of the last SLR. The results of this review are presented here. Further, we discuss the limitations of the current candidate markers as well as strategies to address the issue of prediction of PsA and possible disease prevention. We propose that combined analysis of clinical/molecular features using artificial intelligence approaches may identify algorithms, which initially separate clearly those with PsC from those with PsA, but which can then be applied to prospective cohorts to explore whether they might also operate as identifying those at risk of progression from PsC to PsA.
This literature review summarised new clinical and omic-based biomarkers published in the literature between May 2020 and May 2024. The clinical and molecular biomarkers discussed in this publication have the potential to be used as diagnostic biomarkers to distinguish between PsC and PsA patient populations.
Methods
Search strategy
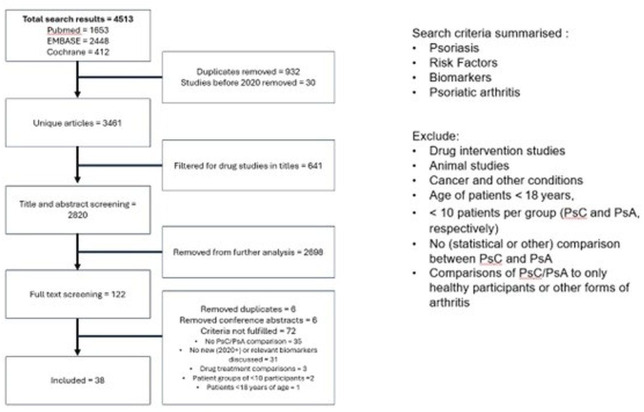
Three bibliographic databases (PubMed, Excerpta Medica dataBASE (EMBASE) and Cochrane) were searched for studies from May 2020 to April 2024. Search terms compromised keywords including PsC, PsA, risk factors and biomarkers. The following types of studies were excluded: drug intervention studies, animal studies, cancer and other conditions, age of patients <18 years, <10 patients per group (PsC and PsA, respectively), no (statistical or other) comparison between PsC and PsA and comparisons of PsC/PsA to only healthy participants or other forms of arthritis. The flow chart in figure 1 summarises the search strategy and results.
Figure 1. Flow chart summarising the search strategy and results. PsA, psoriatic arthritis; PsC, cutaneous psoriasis.
Study selection
Studies were screened for eligibility based on title and abstract by one independent reviewer (TG). Potentially relevant papers were assessed in full text by two independent reviewers (TG and AV). Any disagreement was resolved by consensus or by discussion with a third reviewer (OF).
Data extraction
Data extracted included study design, biomarker type, sample type, analysis, list of biomarkers, biomarker association with PsC/PsA, total number of participants, PsC/PsA participant ratio, female/male ratio, PsC/PsA age PsC diagnosis years in PsC group, PsC diagnosis years in PsA group and PsA diagnostic criteria. Extraction was performed by two reviewers (TG and AV).
Narrative synthesis
We descriptively summarised the new clinical and omic-based biomarkers published in the literature.
Clinical markers associated with PSA development
Clinical and anthropometric markers of patients diagnosed with PsC and PsA have been evaluated to identify associations with PsA development. Of the 13 identified studies (table 1), most reported that PsC was diagnosed via dermatologists, while PsA was diagnosed by rheumatologists with participants meeting the ClASsification of Psoriatic ARthritis (CASPAR) Criteria. Statistical models were employed in those 13 studies to investigate what clinical and anthropometric factors could increase the likelihood of progression to PsA. The risk factors reported frequently in the identified literature were nail involvement,8,14 followed by patient age,1114,16 obesity or body mass index (BMI)1015,17 and comorbidities.14 15 18 These findings are supported by previous literature.19 The finding that both nail involvement and obesity are risk factors suggests that microtrauma at entheseal sites may play a role in triggering inflammation locally. A link between nail involvement and distal interphalangeal joint extensor tendon enthesopathy has been proposed,20 and being overweight may contribute to microtrauma at lower limb entheseal sites. Furthermore, three publications reported that taking medication increased the likelihood of PsA diagnosis.13,15 It is likely that this would not be attributed to the use of medications but could be explained by the comorbidities or by PsC being more severe requiring additional treatment. In at least two publications, female sex8 13 or sex in general,17 plaque psoriasis,8 11 psoriasis severity13 17 and duration,9 11 untreated psoriasis skin lesions9 or skin lesions on extremities,8 other forms of arthritis,15 17 back pain17 18 and fatigue14 17 contributed to a higher risk of PsA development. The largest study cohorts analysed by Green et al17 and Merola et al14 indicated fatigue in addition to MSK symptoms are indicative of PsA risk.
Table 1. Clinical markers differentiating PsC and PsA.
| Author | Study type | Sample type | Analysis | Biomarkers and associations | Total participant number | PsC/PsA ratio |
| Alrubaiaan 20238 | Retrospective cohort (8 years) | Demographic, clinical data | Female, plaque psoriasis, psoriatic lesions on extremities, scalp, trunk, dystrophy | 487 | 438/49 | |
| Belman 20219 | Retrospective cohort (12 years) | Cox proportional hazard regression, adjusted for sex | >60 years of PsC, untreated lesions (patients scored the severity of their typical untreated lesions with an induration card (by the National Psoriasis Foundation) and photographs as reference points), nail, pustular, Koebner phenomenon | 627 | 499/128 | |
| El-Garf 202110 | Cross-sectional observational study | Univariate statistics (logistic regression) | Obesity, nail psoriasis, intergluteal cleft site | 200 | 140/60 | |
| Green 202217 | Retrospective cohort (16 years) | Bayesian model of demographics, habitual data, psoriasis and MSK data | Psoriasis severity, BMI category, sex, arthritis, swelling, arthralgia, back pain, fatigue, finger pain, hand pain, hip pain, knee pain, myalgia, non-specific arthritis, unspecified swelling | 90 189 | 88780/1409 | |
| Guldberg-Møller 202254 | Experimental | Ultrasound, MRI, X-ray | No significant differences found between PsC and PsA patients | 75 | 12/50 | |
| Liu 202211 | Observational study | Logistic regression of clinical biomarkers | Duration of having plaque psoriasis, nail involvement, erythematous lunula, onychorrhexis, oil drop, subungual hyperkeratosis, and age at onset | 855 | 746/109 | |
| Liu 202312 | Case-control study | Serum | Blood inflammatory markers and clinical markers | Nail psoriasis | 156 | 109/47 |
| Loo 202413 | Retrospective cohort | Demographic data, clinical data; adjusted for: sex, education, marital status, diabetes mellitus, nail involvement, affected body area, topical therapy and oral systemic therapy use | Female sex, nail involvement, severe skin psoriasis and prior oral systemic therapy | 330 | 247/83 | |
| Merola 202314 | Retrospective cohort (11 years) | Demographic data, clinical data; Cox proportional hazards regression | Patient age (30–39, 40–49, 50–59), high Charlson Comorbidity Index Score, presence of MSK, fatigue, nail dystrophy, prescription medication, procedure/assessment and healthcare | 116 203 | 110118/6085 | |
| Simon 202255 | Prospective cohort study | Demographic, lifestyle, Quantitative Computed Tomography (Q-CT) scan | Structural entheseal lesions | 114 | 90/24 | |
| Lee 202315 | Case-control study | Logistic regression, and neural network analysis of clinical biomarkers | Age, autoimmune connective tissue diseases, rheumatoid arthritis and other inflammatory polyarthritis, anxiety and depression, ankylosing spondylitis and other inflammatory spondylopathies, osteoporosis and pathologic fracture, atopic dermatitis, menopausal and postmenopausal disorders, other chronic comorbidities (renal diseases, obesity and metabolic syndrome, dyslipidaemia, cardiovascular disorders, diabetes and hypertension), medications (Disease-modifying anti-rheumatic drugs (DMARDs), methotrexate, azathioprine, acitretin, aminoquinolines like hydroxychloroquine and chloroquine, calcineurin inhibitors like ciclosporin, selective immunosuppressants like leflunomide and tofacitinib and sulfasalazine), topical medications (tars, corticosteroids, vitamin D analogues, calcipotriol, calcitriol) | 2215 | 1772/443 | |
| Xu 202316 | Retrospective cohort | Logistic regression (LR), and machine learning approaches to find the best model | Age, height, waist-hip ratio (WHR), erythrocyte sedimentation rate, waist, Psoriasis Area and Severity Index (PASI), body surface area (BSA), BMI and Dermatology Life Quality Index | 4539 | 3961/578 | |
| Su 202018 | Retrospective cohort (14 years) | Plasma | MicroRNA, real-time PCR, clinical data | Gout, hyperlipidaemia, axial spondylopathy (inflammatory back pain) and allergic rhinitis | 629 | 527/102 |
Studies often included healthy participants or those diagnosed with other types of arthritis, and these participants were included in the total participant number in the table; however, their analysis was not included in this review.
MSKmusculoskeletalPsApsoriatic arthritisPsCcutaneous psoriasis
Genetic markers of PsA
Genetic markers associated with higher risks of PsA development have been described in previous literature,7 21 but often these genetic markers could not be confirmed by multiple studies. Two studies reported hypermethylation of DNA sites in study participants with PsC who converted to PsA22 and in participants with PsA23 (table 2). The characterisation of miRNAs inside extracellular vesicles (EVs) is a new avenue for potential disease biomarkers, and more studies are needed to investigate the impact and diagnostic capacity of these miRNAs. The work of Soomro et al 24 investigated genetic markers that were associated with either PsC or PsA on population data from the Biomarkers and Stratification To Optimise outcomes in Psoriasis study (PsA-BSTOP) cohort and UK Biobank data. In addition to two previously identified loci (Major Histocompatibility Complex (MHC) region (rs1050414) and the IL23R gene (rs72676069)) that were associated with PsA, four additional loci were found in both datasets that could potentially be linked to PsA (CNTN4 (rs17194140), no associated gene (rs11665266, rs76800961) and PAXIP1 (rs306281)).24 The association with these new loci will need confirmation in other, independent datasets.
Table 2. Genetic markers to differentiate PsC and PsA.
| Author | Study type | Sample type | Analysis | Biomarkers and associations | Total participant number | PsC/PsA ratio |
| Genetic studies | ||||||
| Caputo 202056 | Experimental | Whole blood sample | Extraction, PCR, Sanger sequencing | COL10A1 associated with PsA, plays a role in maintaining bone homeostasis and cartilage metabolism, and organisation and remodelling of the extracellular matrix | 1417 | 393/424 |
| Coto-Segura 202357 | Experimental | Whole blood sample | Real-time PCR and sequencing | No genetic markers were significantly different between PsC and PsA | 572 | 401/171 |
| Soomro 202224 | Experimental | Whole blood sample | Sanger sequencing for SNPs | MHC region (rs1050414) and the IL23R gene (rs72676069) associated with PsA development | 10 771 | 6431/4340 |
| Epigenetic studies | ||||||
| Cruz-Correa 202322 | Prospective, longitudinal study | Whole blood sample | Bisulphite converted using the EZ DNA methylation kit; genome-wide methylation profile, CpG sites and CpG island were measured using Infinium Methylation EPIC BeadChips (Illumina) | PsC patients converting to PsA show hypermethylation of 630 sites compared with those who have PsC and did not convert to PsA;pathways affected (n=298) were interleukin signalling, T-cell differentiation, cytoskeleton remodelling and included non-canonical Wnt, PI3K-AKT-mTOR, receptor tyrosine kinases, RAS, GPVI-mediated platelet activation and CXCR4 signalling pathways | 117 | 59/58 |
| Deng 202223 | Experimental | Whole blood sample;CD4+ T-cells, CD8+ T-cells, CD19+ B cells and CD14+monocytes (validation) | Illumina 850K array, and validated by pyrosequencing | 135 CpG sites are significantly different between PsC and PsA;associated with phosphatidylglycerol metabolism, Cytidine diphosphate diacylglycerol (CDP)–diacylglycerol metabolism and biosynthesis, cardiolipin metabolism; genes were enriched in glycerophospholipid and glycerolipid metabolism;chr12: cg16459382 and chr2: cg16348668—two differentially methylated CpG sites were consistently hypermethylated in PsA in whole blood and in immune regulatory cells | 64 (screening), 259 (validation) | 20/25 (screening), 48/60 (validation) |
| MiRNA studies | ||||||
| Lättekivi 202258 | Experimental | Serum EVs | EV purification with size exclusion chromatography, small RNA sequencing for miRNA analysis | No genetic markers were significantly different between PsC and PsA | 36 | 12/12 |
| Pasquali 202059 | Experimental | Plasma EVs | miRNA next-generation sequencing (discovery), Locked Nucleic Acid (LNA) miRNA Pick & Mix qPCR (validation) | Nine miRNAs in EVs were upregulated in PsA (has-miR-23a-3p, −379–5 p, −98–5 p, 29 a-3p, 27b-3p, 27 a-3p, 26 a-5p, 146 a-5p, has-let-7e-5p);10 miRNAs in EVs were downregulated in PsA (has-miR-92a-3p, −139–3 p, −92b-3p, −486–5 p, −1180–3 p, −3158–3 p, −4732–3 p, −203 a, has-let-7b-5p, −7b-3p);Downregulation of let-7b-5p and miR-30e-5p in PsA confirmed in the validation cohort | 29 (discovery), 57 (validation) | 14/15 (discovery), 29/28 (validation) |
| Su 202018 | Retrospective cohort (14 years) | Plasma | microRNA, real-time PCR, clinical data | miR-210–3 p significantly lower in PsA | 629 | 527/102 |
Studies often included healthy participants or those diagnosed with other types of arthritis, and these participants were included in the total participant number in the table; however, their analysis was not included in this review.
EVsextracellular vesiclesPsApsoriatic arthritisPsCcutaneous psoriasis
Genetic markers that have previously been reported in a literature review by Mulder et al,7 with conflicting or moderate evidence for being associated with PsA, are HLA-B*27 and HLA-B*38. These genetic markers have also been identified in the review by Caputo et al.25 To confirm genetic markers that clearly differentiate between PsC and PsA populations, larger, international cohort studies are needed.
Cellular and soluble biomarkers
Multiple studies have investigated the potential of immune cell counts and respective genetic markers that were upregulated or downregulated within immune cells as markers of progression from PsC to PsA (table 3). Four studies investigated immune cell counts and markers in Peripheral Blood Mononuclear Cells (PBMCs).26,29 Leijten et al,26 Liu et al 27 and Mulder et al 28 reported that certain immune cells (CD8+CD45RO+CCR10+; CD16 monocytes and CD4+CD196+CD183 CD194+ Th17-like and CD4+CD196 CD183-CD194+T-cells, respectively) are significantly increased in PsA populations as compared with PsC, while other cells were reduced in PsA populations as compared with PsC (B memory and a CD4 T effector memory (TEM) cells; CD196+and CD197+ monocytes, memory T-cells, CD4+CD197+CD45RA- T-cells, CD8+CD45RA-CD27- T-cells, Treg, and CD19+IgD+ CD5++B cells; respectively). However, the resulting associations with PsC and PsA are conflicting, perhaps relating to differences in study inclusion criteria. For example, Mulder et al’s 28 study reported the proportion of PsC/PsA participants that were taking conventional synthetic and biologic disease-modifying anti-rheumatic drugs, but this was not reported in Leijten et al’s 26 and Liu et al’s 27 studies. Therefore, larger study populations, with clear reporting of the study inclusion, are required to confirm these results.
Table 3. Blood and inflammatory cell markers to differentiate PsC and PsA.
| Author | Study type | Sample type | Analysis | Biomarkers and associations | Total participant number | PsC/PsA ratio |
| Cellular studies | ||||||
| Leijten 202126 | Experimental | PBMCs from blood, and skin biopsy from plaque and healthy skin | Flow cytometry;RNA sequencing was performed on small subset of <10 PsC/PsA (not included) | CD8+CD45RO+CCR10+ cells significantly higher expressed in PsA;Within CD8+CCR10+ T-cells high expression of DNAM-1 in PsA;TIGIT coexpression was reduced in patients with PsA; | 78 | 21/21 |
| Lembo 202129 | Experimental | PBMCs | Osteo assay | No significant biomarkers detected between PsC and PsA | 30 (and rheumatoid arthritis, and healthy controls but not reported how many) | 20/10 |
| Liu 202227 | Experimental | PBMCs | PBMC extraction, RNAseq | Differentially expressed proteins and genes detected between PsC and PsA;CD16 monocytes are highly expressed in PsA;B memory (cluster 1) and a CD4 TEM (cluster 2) cells reduced in PsA;Top 20 differentially expressed genes (DEG) or proteins (DEP) identified in the model (out of 257 DEGs and 258 DEPs) can be used to identify PsC and PsA;DEGs alone showed more accuracy in the Area Under the Curve (AUC) analysis compared with DEPs alone; | 95 | 24/28 |
| Mulder 202128 | Experimental | Whole blood | Flow cytometry, random forest model to distinguish between PsC/PsA | PsA patients showed but increased fractions of CD4+CD196+CD183 CD194+ Th17-like and CD4+CD196 CD183-CD194+ T-cells;PsA patients had reduced levels of CD196+and CD197+ monocytes, memory T-cells, CD4+CD197+CD45RA- T-cells, CD8+CD45RA-CD27- T-cells, Treg and CD19+IgD+ CD5++ B cells; | 86 | 45/41 |
| Soluble studies | ||||||
| Li 202260 | Experimental | Serum | ELISA of MPO-DNA complex | Myeloperoxidase (MPO)-DNA complex significantly increased in PsA | 152 | 58/74 |
| Liu 202312 | Case-control study | Serum | Blood inflammatory markers and clinical markers | IL-6, platelet to lymphocyte ration (PLR), and Systemic Immune-inflammation Index (SII) significantly higher in PsA | 156 | 109/47 |
Studies often included healthy participants or those diagnosed with other types of arthritis, and these participants were included in the total participant number; however, their exact numbers were not reported in this table.
PsApsoriatic arthritisPsCcutaneous psoriasis
Lipid, metabolite, protein and peptide markers
Omic technologies provide a comprehensive picture of disease-specific biomarkers. In table 4, lipidomic, metabolomic and proteomic studies are listed (n=12 in total). Only a single study investigated lipid biomarkers which differentiate PsC and PsA and found 18 distinct features that were classified as fatty acids, carboxylic acids, glycerophospholipids, steroids and sphingolipids.30 Three metabolomic studies used LC-MS/MS-based methodologies and identified very different metabolites that were associated with either PsC or PsA (14,15 diHETrE, tyramine, glycoursodeoxycholic acid sulphate, glycodeoxycholate 3-sulphate and deoxycholic acid 12-sulphate significantly lower in PsA populations; mucic acid, 12-HHTrE and guanine significantly higher in PsA populations).31,33 The proteomic studies identified in this literature review were targeted and mostly measured specific proteins by ELISA (gelsolin, calprotectin, type I collagen degradation, IL-17, CXCL10 and HMGB1).34,39 Two proteomic studies reported an association of CXCL10 with PsA;38 40 the study of Abji et al 40 reported a dramatic increase in CXCL10 levels just prior to PsA diagnosis, which then after diagnosis fell to PsC levels.
Table 4. Lipidomic, metabolomic and proteomic markers to differentiate PsC and PsA.
| Author | Omics type | Study type | Sample type | Analysis | Biomarkers and associations | Total participant number | PsC/PsA ratio |
| Koussiouris 202330 | Lipidomics | Experimental | Serum | Liquid Chromatography - Mass Spectrometry (LC-MS/MS) | Valerylcarnitine,(tentatively annotated features: tryptophyl-histidine, Lipoprotein A (LPA) (18:2(9Z,12Z)/0:0), sofalcone, cymorcin diglucoside, dolichyl b-D-glucosyl phosphate, phenylalanyl-tryptophan, aldosterone, arginyl-histidine, (3′-sulpho)Galbeta-Cer(d18:1/16:0), SM(d18:1/24:0), acetyl-D-carnitine, 19R-hydroxy-PGE2, 20-hydroxy-PGE2, isoleucyl-isoleucine, bilirubin diglucuronide, sialyllacto-N-tetraose a, tryptophyl-phenylalanine, 2,3-dihydroxy-2-methylbutanoic acid)Significantly differentiated between PsC and PsA | 78 | 27/26 |
| Coras 202131 | Metabolomic | Experimental | Serum | LC-MS/MS, oxylipins separated by reverse-phase chromatography and measured using electrospray ionisation in negative ion mode and MRM | 14,15 diHETrE (14,15-dihydroxyeicosatrienoate)—significantly lower in PsA | 39 | 20/19 |
| Kishikawa 202132 | Metabolomic | Experimental | Plasma | Capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) and Liquid chromatography time-of-flight mass spectrometry (LC-TOFMS) | Tyramine significantly decreased in PsA;Mucic acid significantly increased in PsA | 131 | 50/43 |
| Paine 202233 | Metabolomic | Experimental | Serum | Untargeted metabolome analysis; reverse-phase and hydrophilic interaction liquid chromatography coupled to Q-exactive mass spectrometry | 12-hydroxyheptadecatrenoic acid (12-HHTrE) and guanine significantly higher in PsA;three bile acids significantly lower in PsA (glycoursodeoxycholic acid sulphate, glycodeoxycholate 3-sulphate and deoxycholic acid 12-sulphate) | 71 | 21/34 |
| Abji 202040 | Proteomic | Longitudinal (10 years) | Serum | Milliplex MAP human magnetic bead panels | CXCL10 levels significantly elevated and declining to PsC levels prior to PsA development | 81 | 52/29 |
| Esawy 202034 | Proteomic | Experimental | Whole blood | ESR, ELISA for gelsolin | Gelsolin significantly lower in PsA | 156 | 40/76 |
| Leijten 202161 | Proteomic | Experimental | Serum | Olink proteomics | No significant differences between PsC and PsA | 79 | 18/20 |
| Li 202335 | Proteomic | Experimental | Serum | ELISA for calprotectin | Calprotectin significantly higher in PsA | 176 | 55/71 |
| Holm Nielsen 202336 | Proteomic | Experimental | Serum | ELISA for collagen and inflammation biomarkers | Type I collagen degradation (C1M) significantly higher in PsA | 101 | 30/30 |
| Ruscitti 202362 | Proteomic | Experimental | Serum | Luminex (19 Plex) of inflammatory cytokines, chemokine, cell adhesion and cellular mediators | E-selectin, IL-8 significantly increased in PsC;ICAM-1 significantly increased in PsA | 80 | 19/41 |
| Saif 202037 | Proteomic | Experimental | Serum | IL-17 ELISA and MRI | IL-17 significantly increased in PsA | 200 | 62/38 |
| Soliman 202238 | Proteomic | Experimental | Serum | Ultrasound, MRI, ELISA for cytokines | CXCL10 levels significantly increased in PsA | 80 | 50 |
| Yildirim 202339 | Proteomic | Experimental | Serum | ELISA for HMGB1 | HMGB1 significantly increased in PsA | 91 | 43/48 |
Studies often included healthy participants or those diagnosed with other types of arthritis, and these participants were included in the total participant number; however, their exact numbers were not reported in this table.
PsApsoriatic arthritisPsCcutaneous psoriasis
Limitations of current molecular markers
It is evident that a relatively large number of clinical and molecular markers have emerged as candidates for the early identification of PsA in those with PsC. However, in most studies, the cohorts are retrospective and of modest size. As noted before, there are very few examples of biomarkers that have been consistently identified in independent studies (using different cohorts). Notably, two proteomic studies identified changes in CXCL10 with PsA,38 40 but one of these highlighted the importance of longitudinal data.40 In this study, low CXCL10 levels were observed in PsC and these increased just before the diagnosis of PsA and subsequently returned to the level found in PsC. This indicates that the temporal profile of potential biomarkers is important, and comparing levels of a biomarker in PsC with those diagnosed with PsA may not reveal new (and important) biomarkers. It is perhaps not surprising given the modest numbers of samples and complexity of the analyses that there is very little overlap in the candidate molecular markers identified in individual (lipidomic, metabolomic or proteomic) studies. This issue must be addressed if reliable markers are to be identified and arguably this needs to be done before large-scale validation studies are performed. Even once such studies have been performed, the subsequent development of diagnostic tests presents many challenges. These challenges are well-known and have been highlighted in review articles.41,43
So, good progress is being made in the identification of candidate biomarkers, but their continued development will likely require international collaboration to share samples, harmonise methodologies, apply sophisticated statistical methods and agree on the markers that may have potential clinical utility and are worthy of continued development.
Strategies to address the issue of prediction of PsA
Prediction of PsA is important to guide interventions and management in clinical practice. Currently, there is a marked delay in diagnosis of PsA that persists despite increasing focus on this disease.44 While one in three people with psoriasis will develop PsA, the majority of people living with psoriasis are not aware of this risk and educating those at higher risk is key to improving the current diagnostic delay. Screening tools for PsA are available but not routinely implemented. A recent UK study of 2225 people with psoriasis did not indicate a significant benefit to routine screening in primary care for PsA but focusing on those at higher risk would likely make this more cost-effective.45
As summarised above, there are a number of clinical and molecular biomarkers that can be used to address the prediction of PsA development. Currently, they are not routinely used to identify those at higher risk. Clinicians consider all patients with psoriasis to be at potential risk of PsA, and often well-recognised clinical markers like the presence of nail psoriasis are used in practice to identify those at high risk.
The current limitation is creating a more definitive risk prediction model that could be applied to individuals. Previous work done with routine primary care data in the UK46 has shown that routinely collected data on demographics, symptoms and blood tests can be used to identify those at risk. At the simplest level, using this kind of routinely collected data within primary or secondary care settings could be used to implement a risk stratification tool and guide interventions. With the advent of exciting new research in biomarkers, more accurate risk prediction may be possible, and research is needed to see the added benefit of using such biomarkers alongside clinical data. These data can guide further interventions including screening questionnaires, referrals to rheumatology and consideration of interventions to reduce or prevent the risk of PsA.
Disease prevention
In recent years, there has been an increasing interest in disease interception in rheumatology including in PsA. If we can accurately identify people at high risk of PsA, then potential strategies can be tested to reduce this risk. To successfully take this idea forward, we need to design optimal trials considering the population, intervention, control and outcomes required. A risk prediction model, that works for individual people, is key to identifying who should be included in a trial. A model needs to indicate a significant risk to ensure that the trial is statistically powered and that interventions have a reasonable risk/benefit balance.
There are likely two key types of interventions that could be used: lifestyle or drugs. Research has suggested that maintaining a healthy body weight and regular physical activity are associated with a lower risk and interventions supporting lifestyle changes towards these goals may be beneficial. Some observational studies have suggested that biologic therapies used for psoriasis may be associated with a lower risk of PsA development.47,49 This seems biologically plausible, despite many biases that will affect such retrospective data. However, we cannot be sure if these drugs are really preventing or changing the risk of PsA or whether they are only delaying clinical evidence of disease. Studies are currently ongoing investigating drug treatment for PsA prevention including the Preventing Arthritis in a Multi-Center Psoriasis At-Risk Cohort (PAMPA) study which is testing guselkumab versus placebo in patients with psoriasis and subclinical ultrasound inflammation.50 However, the control group in these studies is potentially problematic. It is ideal to include patients with severe psoriasis as they are known to be at increased risk of arthritis, but then systemic treatment for psoriasis is indicated and it would be unethical to withhold this.
The key consideration throughout the design is that a future trial needs to be acceptable to potential participants. Here, risk prediction is key as this influences patient acceptability for interventions, particularly medications with potential side effects. Surveys of people with psoriasis have suggested that a higher baseline risk and a likely reduction in PsA risk of 30%–40% is required for them to consider preventative drug treatment, even when considering potential moderate side effects.51 The level of risk that would justify interventions needs to be considered from both the patient’s and the researcher’s perspectives. This risk level may vary for different interventions though. If the intervention was a healthy lifestyle intervention (eg, weight reduction, healthy diet, increased exercise) that has clear general health benefits and a very low risk of side effects, those at lower risk could be considered. If the intervention is an expensive or risky medication with significant side effects, then it is likely that a higher baseline risk would be appropriate to make the intervention acceptable to patients and ethical reviewers. For us to communicate and test potential prevention strategies, accurate individual patient risk prediction is required.
Summary and conclusions
In summary, this literature review confirms that nail involvement, patient age, being overweight or obese and comorbidities are the most predictive clinical risk factors for PsA development. The studies reporting clinical risk factors generally included larger population groups compared with those investigating novel molecular biomarkers. It should be pointed out that the study by Eder et al on the Psoriatic Arthritis Risk Estimation Tool (PRESTO) did not come up in the literature search, perhaps because it describes a model or tool, rather than individual clinical features.52 The PRESTO model identifies a combination of clinical features, different at 1 year and 5 years, which are moderately effective (both AUCs <75) in predicting the development of PsA. It is possible that PRESTO could be applied in routine clinical settings but it will require validation. The addition of other blood-based biomarkers could well improve performance.
As the diversity of molecular biomarkers identified in this literature review corroborates the findings of previous literature reviews, this strongly emphasises the need for large cohort studies and the use of advanced statistical modelling to identify relevant biomarkers suitable for PsA diagnosis. Current PsA risk prediction and risk screening tools are not accurate enough to facilitate early diagnosis and treatment, while this is of utmost importance to prevent loss of function in affected body parts. Once molecular biomarkers are reliably measured and confirmed by multiple studies to differentiate PsA from PsC patients, they could be included in routine blood screenings for PsA. Disease prevention can only be targeted once there are more accurate diagnostic tools available to medical professionals.
Scientific collaborations and consortia are ideal solutions for tackling the limitations of individual studies. The HIPPOCRATES consortium is a collaboration of scientists, clinicians, patient partners, small and medium-sized enterprises and pharmaceutical industry partners, who are collaborating to share data and samples from previously conducted studies on PsC and PsA cohorts, to detect novel biomarkers in these samples via omics technologies and to analyse these large dataset via statistical models, to overcome the biases of small study populations.53 Furthermore, the risks for PsA development are evaluated in the HIPPOCRATES Prospective Observational Study, which is aiming to recruit 25 000 PsC patients across Europe to monitor their risk factors over a period of 3 years. The work outlined in the HIPPOCRATES consortium aims to identify molecular biomarkers for the early diagnosis of PsA, to identify risk factors for PsA development in PsC patients, to find preventative measures for joint and bone damage in PsA and to understand what medical interventions work best for the individual. Similarly, other consortia have formed to tackle the difficulties of PsA diagnosis, for example, by monitoring PsA risk via an app (iPROLEPSIS, https://www.iprolepsis.eu/), or identifying biomarkers that could explain endotypes in psoriasis and atopic dermatitis (Biomarkers in Atopic Dermatitis and Psoriasis (BIOMAP), https://www.biomap-imi.eu/overview). The future of PsA research arguably depends on such collaborative efforts to find robust biomarkers, diagnostic tools and preventative strategies.
Footnotes
Funding: This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement number 101007757 (HIPPOCRATES). The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. Any dissemination of results must indicate that it reflects only the author’s view and that the JU is not responsible for any use that may be made of the information it contains. AV is funded by an NIHR Doctoral Research Fellowship (Award ID: NIHR302335).
Patient consent for publication: Not applicable.
Ethics approval: Not applicable.
Provenance and peer review: Commissioned; externally peer reviewed.
Contributor Information
Teresa Grohmann, Email: teresa.grohmann@ucd.ie.
Arani Vivekanantham, Email: arani.vivekanantham@ndorms.ox.ac.uk.
Laura C Coates, Email: laura.coates@ndorms.ox.ac.uk.
Stephen Pennington, Email: stephen.pennington@ucd.ie.
Oliver FitzGerald, Email: oliver.fitzgerald@ucd.ie.
Data availability statement
All data relevant to the study are included in the article.
References
- 1.Haroon M, Kirby B, FitzGerald O. High prevalence of psoriatic arthritis in patients with severe psoriasis with suboptimal performance of screening questionnaires. Ann Rheum Dis. 2013;72:736–40. doi: 10.1136/annrheumdis-2012-201706. [DOI] [PubMed] [Google Scholar]
- 2.Alinaghi F, Calov M, Kristensen LE, et al. Prevalence of psoriatic arthritis in patients with psoriasis: A systematic review and meta-analysis of observational and clinical studies. J Am Acad Dermatol. 2019;80:251–65. doi: 10.1016/j.jaad.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 3.Kane D, Stafford L, Bresnihan B. A classification study of clinical subsets in an inception cohort of early psoriatic peripheral arthritis--’DIP or not DIP revisited’. Rheumatol Sunnyvale. 2023;42:1469–76. doi: 10.1093/rheumatology/keg445. [DOI] [PubMed] [Google Scholar]
- 4.Haroon M, Gallagher P, FitzGerald O. Diagnostic delay of more than 6 months contributes to poor radiographic and functional outcome in psoriatic arthritis. Ann Rheum Dis. 2015;74:1045–50. doi: 10.1136/annrheumdis-2013-204858. [DOI] [PubMed] [Google Scholar]
- 5.Scher JU, Ogdie A, Merola JF, et al. Preventing psoriatic arthritis: focusing on patients with psoriasis at increased risk of transition. Nat Rev Rheumatol. 2019;15:153–66. doi: 10.1038/s41584-019-0175-0. [DOI] [PubMed] [Google Scholar]
- 6.Eder L, Haddad A, Rosen CF, et al. The Incidence and Risk Factors for Psoriatic Arthritis in Patients With Psoriasis: A Prospective Cohort Study. Arthritis Rheumatol. 2016;68:915–23. doi: 10.1002/art.39494. [DOI] [PubMed] [Google Scholar]
- 7.Mulder MLM, van Hal TW, Wenink MH, et al. Clinical, laboratory, and genetic markers for the development or presence of psoriatic arthritis in psoriasis patients: a systematic review. Arthritis Res Ther. 2021;23:168. doi: 10.1186/s13075-021-02545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alrubaiaan MT, Alsulaiman SA, Alqahtani A, et al. Prevalence and Clinical Predictors of Psoriatic Arthritis in Saudi Patients With Psoriasis: A Single-Center Retrospective Cohort Study. Cureus. 2023;15:e46632. doi: 10.7759/cureus.46632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belman S, Walsh JA, Carroll C, et al. Psoriasis Characteristics for the Early Detection of Psoriatic Arthritis. J Rheumatol. 2021;48:1559–65. doi: 10.3899/jrheum.201123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Garf A, Teleb DA, Said ER, et al. Psoriatic arthritis among Egyptian patients with psoriasis attending the dermatology clinic: prevalence, comorbidities, and clinical predictors. Reumatologia. 2021;59:394–401. doi: 10.5114/reum.2021.112238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Kuang Y, Ye L, et al. Predicting the Risk of Psoriatic Arthritis in Plaque Psoriasis Patients: Development and Assessment of a New Predictive Nomogram. Front Immunol. 2021;12:740968. doi: 10.3389/fimmu.2021.740968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Zhao Y, Mu Z, et al. The Combination of IL-6, PLR and Nail Psoriasis: Screen for the Early Diagnosis of Psoriatic Arthritis. Clin Cosmet Investig Dermatol. 2023;16:1703–13. doi: 10.2147/CCID.S413853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo WY, Tee YC, Han WH, et al. Predictive factors of psoriatic arthritis in a diverse population with psoriasis. J Int Med Res. 2024;52:03000605231221014. doi: 10.1177/03000605231221014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merola JF, Patil D, Egana A, et al. Prevalence of Musculoskeletal Symptoms in Patients with Psoriasis and Predictors Associated with the Development of Psoriatic Arthritis: Retrospective Analysis of a US Claims Database. Dermatol Ther (Heidelb) 2023;13:2635–48. doi: 10.1007/s13555-023-01025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee LT-J, Yang H-C, Nguyen PA, et al. Machine Learning Approaches for Predicting Psoriatic Arthritis Risk Using Electronic Medical Records: Population-Based Study. J Med Internet Res. 2023;25:e39972. doi: 10.2196/39972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Ou J, Li C, et al. Multi-modality data-driven analysis of diagnosis and treatment of psoriatic arthritis. NPJ Digit Med. 2023;6:13. doi: 10.1038/s41746-023-00757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green A, Tillett W, McHugh N, et al. Using Bayesian networks to identify musculoskeletal symptoms influencing the risk of developing psoriatic arthritis in people with psoriasis. Rheumatology (Sunnyvale) 2022;61:581–90. doi: 10.1093/rheumatology/keab310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su YJ. Early diagnosis of psoriatic arthritis among psoriasis patients: clinical experience sharing. Clin Rheumatol. 2020;39:3677–84. doi: 10.1007/s10067-020-05132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pennington SR, FitzGerald O. Early Origins of Psoriatic Arthritis: Clinical, Genetic and Molecular Biomarkers of Progression From Psoriasis to Psoriatic Arthritis. Front Med (Lausanne) 2021;8:723944. doi: 10.3389/fmed.2021.723944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aydin SZ, Castillo-Gallego C, Ash ZR, et al. Ultrasonographic assessment of nail in psoriatic disease shows a link between onychopathy and distal interphalangeal joint extensor tendon enthesopathy. Dermatology . 2012;225:231–5. doi: 10.1159/000343607. [DOI] [PubMed] [Google Scholar]
- 21.Gurke R, Bendes A, Bowes J, et al. Omics and Multi-Omics Analysis for the Early Identification and Improved Outcome of Patients with Psoriatic Arthritis. Biomedicines. 2022;10:2387. doi: 10.3390/biomedicines10102387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruz‐Correa OF, Pollock RA, Machhar R, et al. Prediction of Psoriatic Arthritis in Patients With Psoriasis Using DNA Methylation Profiles. Arthritis Rheumatol. 2023;75:2178–84. doi: 10.1002/art.42654. [DOI] [PubMed] [Google Scholar]
- 23.Deng M, Su Y, Wu R, et al. DNA methylation markers in peripheral blood for psoriatic arthritis. J Dermatol Sci. 2022;108:39–47. doi: 10.1016/j.jdermsci.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Soomro M, Stadler M, Dand N, et al. Comparative Genetic Analysis of Psoriatic Arthritis and Psoriasis for the Discovery of Genetic Risk Factors and Risk Prediction Modeling. Arthritis Rheumatol. 2022;74:1535–43. doi: 10.1002/art.42154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caputo V, Strafella C, Termine A, et al. Overview of the molecular determinants contributing to the expression of Psoriasis and Psoriatic Arthritis phenotypes. J Cell Mol Med. 2020;24:13554–63. doi: 10.1111/jcmm.15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leijten EF, van Kempen TS, Olde Nordkamp MA, et al. Tissue-Resident Memory CD8+ T Cells From Skin Differentiate Psoriatic Arthritis From Psoriasis. Arthritis Rheumatol. 2021;73:1220–32. doi: 10.1002/art.41652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Kumar S, Hong J, et al. Combined Single Cell Transcriptome and Surface Epitope Profiling Identifies Potential Biomarkers of Psoriatic Arthritis and Facilitates Diagnosis via Machine Learning. Front Immunol. 2022;13:835760. doi: 10.3389/fimmu.2022.835760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulder MLM, He X, van den Reek JMPA, et al. Blood-Based Immune Profiling Combined with Machine Learning Discriminates Psoriatic Arthritis from Psoriasis Patients. Int J Mol Sci . 2021;22:10990. doi: 10.3390/ijms222010990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lembo C, Raimondo A, de Paulis A, et al. Clinical predictors of psoriatic arthritis and osteoclast differentiation. Exp Dermatol. 2021;30:1834–7. doi: 10.1111/exd.14424. [DOI] [PubMed] [Google Scholar]
- 30.Koussiouris J, Looby N, Kulasingam V, et al. A Solid-Phase Microextraction-Liquid Chromatography-Mass Spectrometry Method for Analyzing Serum Lipids in Psoriatic Disease. Metabolites. 2023;13:963. doi: 10.3390/metabo13080963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coras R, Kavanaugh A, Kluzniak A, et al. Differences in oxylipin profile in psoriasis versus psoriatic arthritis. Arthritis Res Ther. 2021;23:200. doi: 10.1186/s13075-021-02575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kishikawa T, Arase N, Tsuji S, et al. Large-scale plasma-metabolome analysis identifies potential biomarkers of psoriasis and its clinical subtypes. J Dermatol Sci. 2021;102:78–84. doi: 10.1016/j.jdermsci.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Paine A, Brookes PS, Bhattacharya S, et al. Dysregulation of Bile Acids, Lipids, and Nucleotides in Psoriatic Arthritis Revealed by Unbiased Profiling of Serum Metabolites. Arthritis Rheumatol. 2023;75:53–63. doi: 10.1002/art.42288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esawy MM, Makram WK, Albalat W, et al. Plasma gelsolin levels in patients with psoriatic arthritis: a possible novel marker. Clin Rheumatol. 2020;39:1881–8. doi: 10.1007/s10067-020-04959-y. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Li G, Song Z, et al. Serum Calprotectin as a Promising Inflammatory Biomarker in Psoriatic Arthritis: a 1-Year Longitudinal Study. Rheumatol Ther. 2023;10:149–60. doi: 10.1007/s40744-022-00501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm Nielsen S, Magee C, Groen SS, et al. Differentiating patients with psoriasis from psoriatic arthritis using collagen biomarkers. Clin Exp Rheumatol. 2023;41:574–80. doi: 10.55563/clinexprheumatol/jmt9jv. [DOI] [PubMed] [Google Scholar]
- 37.Saif DS, El Tabl MA, Afifi N, et al. Interleukin-17A biomarker as a predictor for detection of early axial spondyloarthritis changes in patients with psoriasis. Int J Rheum Dis. 2020;23:1664–9. doi: 10.1111/1756-185X.13997. [DOI] [PubMed] [Google Scholar]
- 38.Soliman SG, Gaber MA, Labeeb AA, et al. Role of musculoskeletal ultrasound, magnetic resonance imaging, and serum chemokine C-X-C motif ligand 10 in early detection of arthritis in patients with psoriasis. The Egyp Rheum. 2022;44:219–24. doi: 10.1016/j.ejr.2021.11.006. [DOI] [Google Scholar]
- 39.Yildirim D, Baykul M, Edek YC, et al. Could serum HMGB1 levels be a predictor associated with psoriatic arthritis? Biomark Med. 2023;17:871–80. doi: 10.2217/bmm-2023-0490. [DOI] [PubMed] [Google Scholar]
- 40.Abji F, Lee K-A, Pollock RA, et al. Declining levels of serum chemokine (C-X-C motif) ligand 10 over time are associated with new onset of psoriatic arthritis in patients with psoriasis: a new biomarker? Br J Dermatol. 2020;183:920–7. doi: 10.1111/bjd.18940. [DOI] [PubMed] [Google Scholar]
- 41.Füzéry AK, Levin J, Chan MM, et al. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin Proteomics. 2013;10:13. doi: 10.1186/1559-0275-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drucker E, Krapfenbauer K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J. 2013;4:7. doi: 10.1186/1878-5085-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savva K-V, Kawka M, Vadhwana B, et al. The Biomarker Toolkit - an evidence-based guideline to predict cancer biomarker success and guide development. BMC Med. 2023;21:383. doi: 10.1186/s12916-023-03075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charlton R, Coates L, Galloway J, et al. Diagnostic delay and less intensive therapy for people with psoriatic arthritis compared with rheumatoid arthritis: a nested matched cohort study from within the uk national early inflammatory arthritis audit. ACR Meeting Abstracts. American College of Rheumatology; 2022. p. 74 (Suppl 9). Available. [Google Scholar]
- 45.McHugh N, Tillett W, Helliwell P, et al. Enhanced surveillance for the detection of psoriatic arthritis in a UK primary care psoriasis population: results from the TUDOR trial. Rheumatol (Oxford) 2024:keae374. doi: 10.1093/rheumatology/keae374. [DOI] [PubMed] [Google Scholar]
- 46.Rudge A, Mchugh N, Tillett W, et al. POS0964 DYNAMIC prediction of psoriatic arthritis in a cohort of psoriasis patients using uk primary care electronic health records. EULAR. 2024;83:699. [Google Scholar]
- 47.Singla S, Putman M, Liew J, et al. Association between biological immunotherapy for psoriasis and time to incident inflammatory arthritis: a retrospective cohort study. Lancet Rheumatol. 2023;5:e200–7. doi: 10.1016/S2665-9913(23)00034-6. [DOI] [PubMed] [Google Scholar]
- 48.Gisondi P, Bellinato F, Targher G, et al. Biological disease-modifying antirheumatic drugs may mitigate the risk of psoriatic arthritis in patients with chronic plaque psoriasis. Ann Rheum Dis. 2022;81:68–73. doi: 10.1136/annrheumdis-2021-219961. [DOI] [PubMed] [Google Scholar]
- 49.Rosenthal YS, Schwartz N, Sagy I, et al. Incidence of Psoriatic Arthritis Among Patients Receiving Biologic Treatments for Psoriasis: A Nested Case–Control Study. Arthritis Rheumatol. 2022;74:237–43. doi: 10.1002/art.41946. [DOI] [PubMed] [Google Scholar]
- 50.Haberman RH, MacFarlane KA, Catron S, et al. Efficacy of guselkumab, a selective IL-23 inhibitor, in Preventing Arthritis in a Multicentre Psoriasis At-Risk cohort (PAMPA): protocol of a randomised, double-blind, placebo controlled multicentre trial. BMJ Open. 2022;12:e063650. doi: 10.1136/bmjopen-2022-063650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Groothuizen S, Bolt JW, Veldwijk J, et al. AB1643-pare assessment of psoriasis patients’ preferences for interventions to prevent psoriatic arthritis using a probabilistic threshold technique. Ann Rheum Dis. 2024;83:2196–7. [Google Scholar]
- 52.Eder L, Lee K-A, Chandran V, et al. Derivation of a Multivariable Psoriatic Arthritis Risk Estimation Tool (PRESTO): A Step Towards Prevention. Arthritis Rheumatol . 2023 doi: 10.1002/art.42661. [DOI] [PubMed] [Google Scholar]
- 53.FitzGerald O, Pennington SR. HIPPOCRATES: improving diagnosis and outcomes in psoriatic arthritis. Nat Rev Rheumatol. 2022;18:123–4. doi: 10.1038/s41584-022-00748-w. [DOI] [PubMed] [Google Scholar]
- 54.Guldberg-Møller J, Mogensen M, Ellegaard K, et al. Multimodal imaging of the distal interphalangeal-joint synovio-entheseal complex in psoriatic arthritis (MIDAS): a cross-sectional study on the diagnostic accuracy of different imaging modalities comparing psoriatic arthritis to psoriasis and osteoarthritis. RMD Open. 2022;8:e002109. doi: 10.1136/rmdopen-2021-002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon D, Tascilar K, Kleyer A, et al. Association of Structural Entheseal Lesions With an Increased Risk of Progression From Psoriasis to Psoriatic Arthritis. Arthritis Rheumatol . 2022;74:253–62. doi: 10.1002/art.41239. [DOI] [PubMed] [Google Scholar]
- 56.Caputo V, Strafella C, Termine A, et al. RNAseq-Based Prioritization Revealed COL6A5, COL8A1, COL10A1 and MIR146A as Common and Differential Susceptibility Biomarkers for Psoriasis and Psoriatic Arthritis: Confirmation from Genotyping Analysis of 1417 Italian Subjects. Int J Mol Sci . 2020;21:2740. doi: 10.3390/ijms21082740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coto-Segura P, Vázquez-Coto D, Velázquez-Cuervo L, et al. The IFIH1/MDA5 rs1990760 Gene Variant (946Thr) Differentiates Early- vs. Late-Onset Skin Disease and Increases the Risk of Arthritis in a Spanish Cohort of Psoriasis. Int J Mol Sci . 2023;24:14803. doi: 10.3390/ijms241914803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lättekivi F, Guljavina I, Midekessa G, et al. Profiling Blood Serum Extracellular Vesicles in Plaque Psoriasis and Psoriatic Arthritis Patients Reveals Potential Disease Biomarkers. Int J Mol Sci. 2022;23:4005. doi: 10.3390/ijms23074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pasquali L, Svedbom A, Srivastava A, et al. Circulating micro rna s in extracellular vesicles as potential biomarkers for psoriatic arthritis in patients with psoriasis. Acad Dermatol Venereol . 2020;34:1248–56. doi: 10.1111/jdv.16203. [DOI] [PubMed] [Google Scholar]
- 60.Li B, Li G, Yang X, et al. NETosis in Psoriatic Arthritis: Serum MPO-DNA Complex Level Correlates With Its Disease Activity. Front Immunol. 2022;13:911347. doi: 10.3389/fimmu.2022.911347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leijten E, Tao W, Pouw J, et al. Broad proteomic screen reveals shared serum proteomic signature in patients with psoriatic arthritis and psoriasis without arthritis. Rheumatology (Oxford) 2021;60:751–61. doi: 10.1093/rheumatology/keaa405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruscitti P, Esposito M, Di Cola I, et al. Cytokine profile characterization of naïve patients with psoriasis and psoriatic arthritis: implications for a pathogenic disease continuum. Front Immunol. 2023;14:1229516. doi: 10.3389/fimmu.2023.1229516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data relevant to the study are included in the article.