Abstract
Background
Immune checkpoint blockade is a promising anticancer therapy, whereas the presence of T cells in tumor sites is indispensable for its therapeutic efficacy. To promote the infiltration of T cells and dendritic cells (DCs) into the tumor, we previously proposed a local cell therapy using chemokine (C-C motif) ligand 19 (CCL19)-expressing immortalized syngeneic immortalized mesenchymal stem cells (syn-iMSC/CCL19). However, the preparation of syngeneic/autologous MSC from individual hosts limits the clinical application of this cell therapy.
Methods
In this study, we further developed a new cell therapy using allogeneic iMSC/CCL19 (allo-iMSC/CCL19) using several tumor mice models.
Results
The allo-iMSC/CCL19 therapy exerted drastic antitumor effects, in which the host’s T cells were induced to respond to allogeneic MSC. In addition, the allo-iMSC/CCL19 therapy promoted the infiltration of CD103+ interleukin (IL)-12-producing DCs and priming of CD8+ T cells at tumor sites compared with that using syn-iMSC/CCL19. The antitumor effect of allo-iMSC/CCL19 therapy was not influenced by fingolimod, a sphingosine 1-phosphate receptor modulator, implying no involvement of draining lymph nodes in the priming of tumor-specific T cells.
Conclusion
These results suggest that allo-iMSC/CCL19 therapy exerts dramatic antitumor effects by promoting the infiltration of CD103+ IL-12-producing DCs and thereby priming tumor-specific CD8+ T cells at tumor sites. This local cell therapy could be a promising approach to anticancer therapy, particularly for overcoming dysfunction in the cancer-immunity cycle.
Keywords: Immune modulatory, Immune Checkpoint Inhibitor, Combination therapy, Cytokine
WHAT IS ALREADY KNOWN ON THIS TOPIC
Immune checkpoint blockade (ICB) is a powerful therapy for patients with cancer, which mechanism is to restore T cells from exhaustion. It is known that cell therapy with (C-C motif) ligand 19 (CCL19)-expressing immortalized mesenchymal stem cells (iMSC/CCL19) promotes the infiltration of dendritic cells (DCs) and T cells in the syngeneic tumor mice model. In general, tumor-specific CD8+ T cells could be primed/activated in draining lymph nodes.
WHAT THIS STUDY ADDS
This study demonstrates that cell therapy with allogeneic iMSC/CCL19 (allo-iMSC-CCL19) can exert a robust antitumor effect by increasing CD103+ interleukin-12-producing cells in tumor sites. Notably, allo-iMSC/CCL19 therapy increased tumor-specific CD8+ T cells at tumor sites without involving draining lymph nodes. Allo-iMSC/CCL19 therapy combined with anti-programmed death-ligand 1 antibody shows significant antitumor effects even in ICB-resistant tumor models.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study provides a possibility of allogeneic cell therapy for patients with cancer. Allogeneic cell therapy behaves as an adjuvant and involves not only T cells but also innate immunity such as natural killer cells. Moreover, these findings suggest an important role of tumor-infiltrating CD103+ DCs in the efficacy of cell therapy.
Background
Mesenchymal stem cells (MSC) have been isolated from various tissues, including bone marrow, umbilical cord and adipose tissue.1 Notably, it has been reported that MSC are capable of homing to injured or inflammatory tissues2 and can be engrafted to tumor sites.3 To take advantage of these properties, MSC-based anticancer therapies, as a cell therapy, have been reported using mouse models.4,6 Indeed, third-party (non-autologous) MSC could be used to prevent graft-versus-host disease following allogeneic hematopoietic stem cell transplantation.7 Given that the preparation of syngeneic MSC is cumbersome and time-consuming, using allogeneic MSC might be a useful and powerful approach for anticancer therapy.
Chemokine (C-C motif) ligand 19 (CCL19) is one of the homeostatic chemokines expressed in the thymus and lymph nodes that regulate immune-cell trafficking and adaptive immunity.8 CCL19 attracts dendritic cells (DC) and T cells via its receptor C-C chemokine receptor type 7 (CCR7).9 CCL19 expression correlates positively with the infiltration of CD8+ T cells in tumors and the survival of patients with cancer.10,12 In colorectal cancer, the expression levels of CCL19 messenger RNA and protein correlate negatively with tumor size and invasiveness, and patients positive for CCL19 showed better prognosis than those negative for CCL19.13 These findings suggest that exogenous expression of CCL19 in cells could be useful as an anticancer therapy. Indeed, CCL19-producing chimeric antigen receptor T cells14 and embryonic endothelial cells have been reported to be effective anticancer therapies.15
We previously reported, using a mouse model, that the local injection of CCL19-expressing immortalized MSC (iMSC/CCL19) prepared from syngeneic mice can invoke antitumor effects.16 However, there are practical difficulties in preparing syngeneic (autologous) MSC from individual patients with cancer. In this study, we explored the possibility of using allogeneic iMSC/CCL19 (allo-iMSC/CCL19) for anticancer cell therapy using several mouse tumor models. We observed that allo-iMSC/CCL19 therapy induced antitumor effects by promoting the infiltration of CD103+ interleukin (IL)-12-producing DCs and thereby priming tumor-specific CD8+ T cells at tumor sites, irrespective of the cancer-immunity cycle.
Materials and methods
Mice
BALB/c, C57BL/6 and CB6F1 6-week-old female mice were purchased from CLEA Japan (Tokyo, Japan) and maintained under specific pathogen-free (SPF) conditions. The experiments were carried out according to the Ethical Guidelines for Animal Experiments of the Shimane University Faculty of Medicine (IZ3-79, IZ3-80, IZ3-112, IZ4-34 and IZ5-47). Retired mice were purchased from CLEA Japan (Tokyo, Japan) and maintained under SPF conditions for 53 weeks old (IZ3-122).
Isolation of MSC and Fib
Murine MSC were isolated from the bone marrow of 6 weeks old C57BL/6 female mice as described previously.16 Briefly, crushed bones from femurs and tibias were treated with 0.2% collagenase (Wako) in DMEM for 1 hour at 37°C. Then, the cell suspension was filtered through a cell strainer. The cells were re-suspended in Hank’s Balanced Salt Solution and incubated for 30 min on ice with the following monoclonal antibodies (eBioscience): biotinylated anti-PDGFRα, FITC-conjugated anti-Sca-1, PE-conjugated anti-CD45, and PE-conjugated anti-TER119. Biotinylated antibodies were visualized with APC-conjugated streptavidin (Invitrogen). PDGFRα+ Sca-1+ CD45-TER119 cells were sorted by a triple-laser MoFlo (Dako). Murine primary fibroblasts were isolated from the back skin of C57BL/6 mice; 10 mm diameter skin samples were collected, washed with phosphate-buffered saline and treated with 0.2% collagenase I solution (Wako) for 30 min.
Cell lines
B16-OVA, Renca, Renca-OVA and CT26 were maintained in RPMI-1640 medium (Sigma-Aldrich, St. Louis, Missouri, USA) supplemented with 10% FBS and 20 µg/mL gentamicin (Sigma-Aldrich). Syngeneic and allogeneic iFib, iFib/CCL19, iMSC and iMSC/CCL19 were maintained in DMEM (Nacalai Tesque) supplemented with 10% FBS and 20 µg/mL gentamicin (Nacalai Tesque).
Generation of iFib/CCL19 and iMSC/CCL19
The pWZLneo-CCL19 vector was transfected into Plat-E packaging cells (Cell Biolabs). The supernatant was collected after 48 hours post-transfection, filtered with a 0.45 µm filter (Millipore) and used to infect iFib and iMSC derived from C57BL/6 mice as previously described.16 At 24 hours after retroviral infection, transfectants were cultured in a medium with G418 (400 µg/mL).
Genomic PCR
Tumor DNA was isolated by NucleoSpin Tissue Kit (#740952.50, TAKARA). Template DNA was amplified for 30 cycles of PCR using Ex Taq HS (#RR006A, TAKARA) with the following primers: Egfp, 5′-ACGTAAACGGCCACAAGTTC-3′ (forward) and 5′- AAGTCGTGCTGCTTCATGTG- 3′ (reverse); β-actin, 5′- TGGA ATCC TGTG GCAT CCATG-AAAC- 3′ (forward) and 5′- TAAA ACGC AGCT CAGT AACA GTCCG-3′ (reverse). The PCR products were verified using electrophoresis on 1.8% agarose gels followed by staining with ethidium bromide.
Cell viability assay
C57BL/6-derived splenocytes were co-cultured with BALB/c-derived splenocytes treated with 50 µg/mL mitomycin C for 3 hours. After 4 days, iFib and iMSC which derived from C57BL/6 and BALB/c mice were co-cultured in flat-bottomed 96-well plates for 24 hours. Then, cell viability was measured by Cell Counting Kit-8 (#341–07761, DOJINDO). After the addition of 10 µL CCK-8 solution, the plates were read at 450 nm using Multiskan FC (Thermo Fisher Scientific).
In vivo treatment protocol
In a co-injection model, 5×105 CT26 cells and 1×105 allogeneic iFib/CCL19 or iMSC/CCL19 cells were co-inoculated subcutaneously (s.c.) into the right flank of BALB/c mice. In a therapeutic model, BALB/c mice were injected s.c. with 5×105 CT26 cells in the right flank, followed by intraperitoneal (i.p.) injection of 200 µg anti-programmed death-ligand 1 (PD-L1) (clone: 10F.9G2, Bio X Cell) antibody on 13 and 16 days with or without intratumoral (i.t.) injection of 5×105 allo-iMSC/CCL19 cells on 10 and 13 days after tumor inoculation. In B16-OVA tumor model, 1×106 B16-OVA cells were s.c. injected into the right flank of C57BL/6 mice. 1×106 allo-iMSC or allo-iMSC/CCL19 was i.t. injected on days 10 and 12. In Renca-OVA tumor model, 1×106 Renca-OVA cells were s.c. injected into BALB/c mice. 1×106 syngeneic iMSC (syn-iMSC), allo-iMSC, syn-iMSC/CCL19 or allo-iMSC/CCL19 were i.t. injected on days 10 and 12. CT26-bearing mice were i.p. administrated 2 mg/kg FTY720 (Cayman Chemical Company) on days 10–20. CT26-bearing mice were i.p. administrated with 100 µg anti-CD103 antibody (clone: M290, Bio X Cell) on days 9 and 11 or 200 µg anti-IL-12p40 antibody (clone: C17.8, Bio X Cell) on days 10 and 13. In vivo depletion, 100 µg anti-asialo GM1 (#014–09801, Fujifilm), anti-CD4 (clone: GK1.5, BioLegend) and anti-CD8 (clone: 53–6.7, BioLegend) antibodies were i.p. administered on days 10 and 13 after CT26 tumor inoculation. For neutralization or blocking, 100 µg anti-interferon (IFN)-γ (clone: XMG1.2, BioLegend), anti-TNF-α (clone: MP6-XT22, BioLegend) or anti-CD40L antibody (clone: MR1, BioLegend) antibodies were i.p. administered 9 and 12 days after CT26 tumor inoculation. Tumor size was calculated by (long diameter × short diameter). All in vivo experiments were performed at least twice.
Flow cytometric analysis
Tumors and draining lymph nodes were harvested and treated with 0.2% collagenase D (Merck) and 0.1% DNase I (Merck) for 1 hour at 37°C. Then, cell suspensions were blocked with anti-mouse CD16//32 antibody (clone: S17011E, BioLegend) and stained with the following fluorescence-conjugated antibodies: anti-CD3ε (clone: 145–2 C11), anti-CD4 (clone: GK1.5), anti-CD8α (clone: 53–6.7), anti-CD11c (clone: N418), anti-NKp46 (clone: 29A1.4), anti-CD45 (clone: 30-F11), anti-IL-12p40 (clone: c15.6), anti-CD103 (clone: #2E7), anti-CD69 (clone: H1.2F3), anti-F4/80 (clone: BM8), anti-CCR7 (clone: 4B12), anti-NKp46 (clone: 29A1.4) purchased from BioLegend, PE-conjugated H-2Ld MuLV gp70 tetramer (TS-M521-1, MBL) and PE-conjugated H-2Kb OVA tetramer (TS-5001–1C, MBL). Analysis was performed with a CytoFLEX (Beckman Coulter).
ELISA
Spleen were harvested from CT26-bearing mice treated with anti-PD-L1 antibody, allo-iMSC/CCL19 or combination therapy. Splenocytes were co-cultured with MMC-treated C57BL/6 splenocyte for 48 hours. Then, the supernatants were collected and the levels of IFN-γ or TNF-α were measured using the murine IFN-γ Standard ELISA Development Kit or TNF-α Standard ELISA Development Kit (900-T98, 900-T54, PeproTech) and Multiskan FC Basic plate-reader at 450 nm (Thermo Fisher Scientific).
RNA sequencing
Total RNA was extracted using ISOSPIN Cell & Tissue RNA (NIPPON GENE). RNA sequencing was conducted on total RNAs extracted from tumor tissues of each mouse group. RNA sequencing and analysis was performed by Azenta Life Sciences (Japan). Differential expression analysis used the DESeq2 Bioconductor package, a model based on the negative binomial distribution. The estimates of dispersion and logarithmic fold changes incorporate data-driven prior distributions, padj of genes were set at <0.05 to detect differentially expressed ones. GO-TermFinder was used to identify Gene Ontology (GO) terms that annotate a list of enriched genes with a significant padj value<0.05.
Statistical analysis
Quantitative data were analyzed with the two-tailed Student’s t-test or one-way analysis of variance followed by the Tukey-Kramer test. The analysis was performed with Excel software. A p value of <0.05 was considered statistically significant.
Results
Allo-iMSC/CCL19 survived longer than CCL19-expressing allogeneic fibroblasts at tumor sites
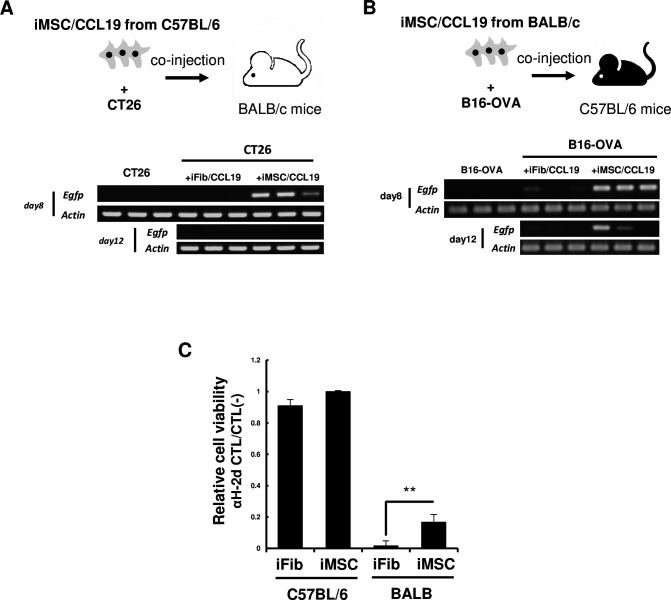
We previously reported that syn-iMSC/CCL19 survived longer than CCL19-expressing fibroblasts (iFib/CCL19).16 To this end, we first compared the survival of allo-iMSC/CCL19 and allo-iFib/CCL19 in vivo. CT26 cells were s.c. injected into syngeneic BALB/c mice with either allogeneic C57BL/6 mouse-derived allo-iMSC/CCL19 or allo-iFib/CCL19 (figure 1A). Genomic PCR analysis capable of detecting the EGFP gene revealed that allo-iMSC/CCL19 survived on day 8 after co-injection, but that allo-iFib/CCL19 were not detected. We also injected B16-OVA cells s.c. into syngeneic C57BL/6 mice with either allogeneic BALB/c mouse-derived allo-iMSC/CCL19 or allo-iFib/CCL19 (figure 1B). Although allo-iFib/CCL19 was detected in only one of three C57BL/6 mice on day 8 after co-injection, allo-iMSC/CCL19 survived in all three mice (figure 1B). Indeed, it was still detectable on day 12 after co-injection. These results indicate that allo-iMSC/CCL19 can survive longer than allo-iFib/CCL19 in allogeneic mice.
Figure 1. Allogeneic MSC/CCL19 retain in tumor longer than fibroblast (A) iMSC/CCL19 derived from C57BL/6 mice were subcutaneously co-injected with CT26 colon carcinoma into BALB/c mice. 8 and 12 days after co-injection, the survivability of iMSC/CCL19 was evaluated by genomic PCR with a primer set which detect the EGFP gene tagged in iMSC/CCL19. (B) iMSC/CCL19 derived from BALB/c mice were co-injected with B16-OVA into C57BL/6 mice. (C) C57BL/6 splenocytes were stimulated with mitomycin C-treated BALB/c splenocytes. After 4 days, iFib and iMSC which derived from C57BL/6 and BALB/c mice were co-cultured for 24 hours. Relative cell viability was shown. **p<0.01 by two-sided t-test. These experiments were performed twice. CCL19, (C-C motif) ligand 19; iMSC, immortalized mesenchymal stem cell.
To explore the underlying mechanisms, we evaluated the expression of PD-L1, CD80 and major histocompatibility complex (MHC) class I by flow cytometry, and we observed no significant differences between iFib and iMSC (online supplemental figure S1). We then examined the susceptibility of iFib and iMSC to anti-allo H-2 CTLs. To prepare anti-H-2d CTLs, C57BL/6 spleen cells were in vitro cultured with inactivated BALB/c spleen cells for 4 days. These cultured cells, containing anti-H-2d CTLs, were co-cultured with iFib and iMSC derived from either C57BL/6 or BALB/c mice, and evaluated their survival by cell viability assays (figure 1C). Although there were no differences in survival between C57BL/6 mouse-derived iFib and iMSC, BALB/c mice-derived iMSC were more resistant to anti-H-2d CTLs than BALB/c mice-derived iFib. These increased resistance of iMSC to anti-allo CTLs may explain the longer survival of iMSC in allogeneic mice.
Therapeutic efficacy of allo-iMSC/CCL19 in treating OVA-expressing tumor models
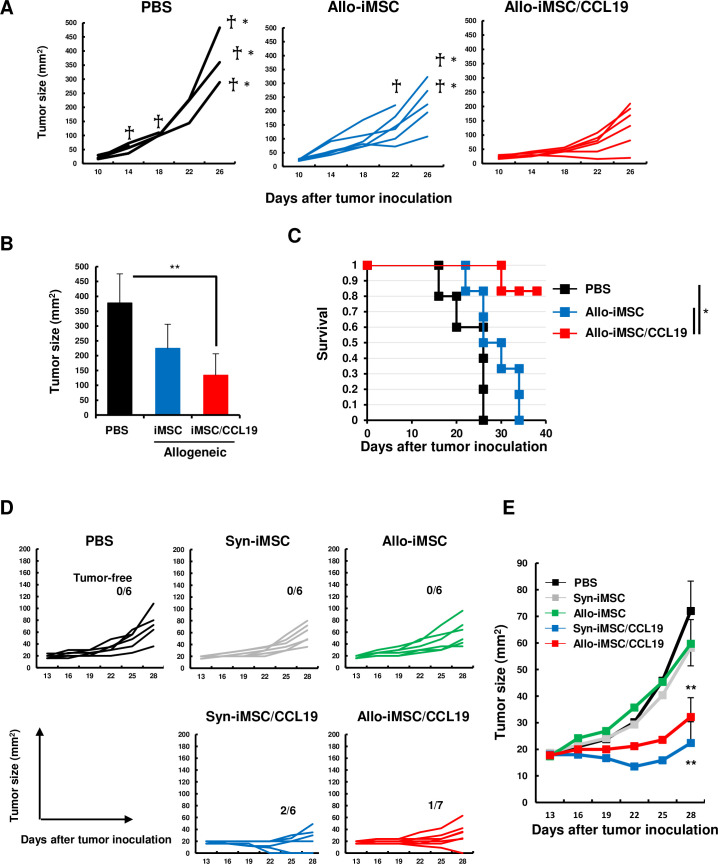
To examine the therapeutic efficacy of allo-iMSC/CCL19, B16-OVA-bearing mice were injected i.t. with allo-iMSC or allo-iMSC/CCL19. Although some mice treated with or without allo-iMSC died during the experimental period, all mice treated with allo-iMSC/CCL19 survived, and they had significantly smaller tumor sizes and significantly longer survival than the other two groups (figure 2A–C). We also performed a similar experiment using murine kidney carcinoma Renca, but we observed no definite antitumor effect with either syn-iMSC/CCL19 or allo-iMSC/CCL19 (online supplemental figure S2). Therefore, to increase the immunogenicity of Renca cells, we generated OVA-expressing Renca (Renca-OVA) (online supplemental figure S3). When Renca-OVA-bearing BALB/c mice were treated, tumor growth was significantly suppressed by the local injection of either syn-iMSC/CCL19 or allo-iMSC/CCL19 (figure 2D,E), but not by that of syn-iMSC or allo-iMSC. These results indicate that CCL19 expression was critical in the therapeutic effect, and that the therapeutic efficacy of the syn-iMSC/CCL19 or allo-iMSC/CCL19 was dependent mainly on the immunogenicity of tumor cells.
Figure 2. Allo-iMSC/CCL19 therapy delayed B16-OVA and Renca-OVA tumor growth in vivo (A–C) B16-OVA cells were s.c. injected into C57BL/6 mice. PBS, allo-iMSC or allo-iMSC/CCL19 were intratumorally (i.t.) injected 10 and 12 days after tumor inoculation. Individual tumor size (A) and mean±SD on days 26 (B) and survival (C) are shown. (D, E) Renca-OVA cells were s.c. injected into BALB/c mice. PBS, syn-iMSC, allo-iMSC, syn-iMSC/CCL19 or allo-iMSC/CCL19 were i.t. injected 10 and 12 days after tumor inoculation. Individual tumor size (D) and mean±SD (E) are shown. Dead † and euthanized †* mice were indicated. *p<0.05, **p<0.01 by Tukey (analysis of variance). These experiments were performed at least twice. CCL19, (C-C motif) ligand 19; iMSC, immortalized mesenchymal stem cell; PBS, phosphate-buffered saline; s.c., subcutaneously; syn-iMSC, syngeneic iMSC.
Allo-iMSC/CCL19 therapy can induce complete regression of established CT26 tumor
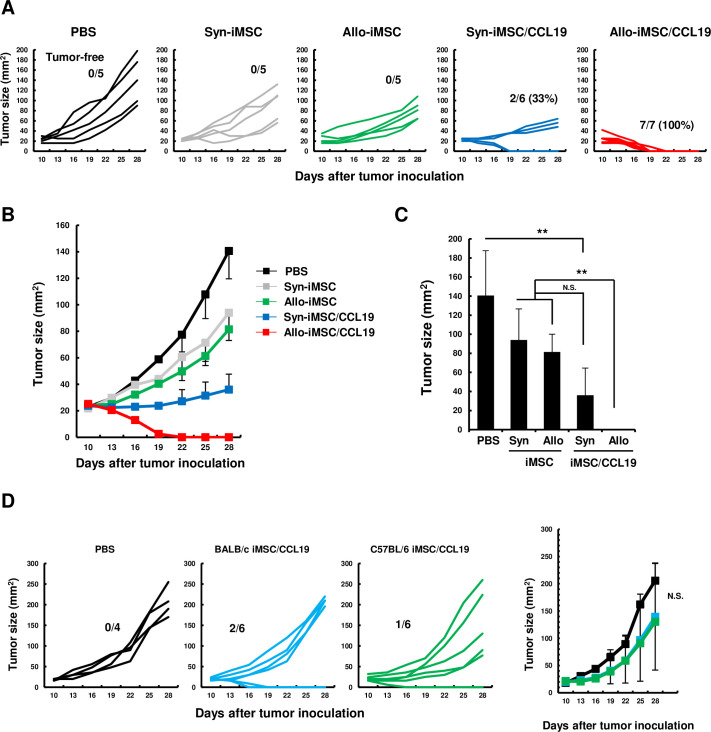
Because CT26 colon carcinoma is an immunogenic tumor and we previously reported the therapeutic efficacy of syn-iMSC/CCL19 in CT26-bearing BALB/c mice,16 we mainly used CT26 in the following experiments. As shown in figure 3A–C, syn-iMSC/CCL19 therapy significantly suppressed the growth of the CT26 tumor, and two out of six mice (33%) were cured. However, allo-iMSC/CCL19 therapy induced complete regression in all seven mice. Next, we tested the possibility that anti-allo T-cell response was involved in the antitumor effect. For this purpose, we used CB6F1 (H-2d/b) mice, which are crossbred between BALB/c and C57BL/6 mice. CB6F1 mice were s.c. injected with CT26 and subsequently treated with BALB/c-derived or C57BL/6-derived iMSC/CCL19. In this model, T cells of CB6F1 mice showed no anti-allo T-cell response to BALB/c-derived or C57BL/6-derived iMSC/CCL19. As a result, although therapy with these types of iMSC/CCL19 tended to suppress CT26 tumor growth compared with the untreated group, there was no definite antitumor effect (figure 3D). These results indirectly suggest that the anti-allo T-cell response of CT26-bearing host mice toward allo-iMSC/CCL19, that is, an allogeneic effect, contributed to the antitumor effects of allo-iMSC/CCL19 therapy.
Figure 3. Allo-iMSC/CCL19 therapy regressed CT26 tumor growth in vivo (A–C) PBS, syn-iMSC, allo-iMSC, syn-iMSC/CCL19 or allo-iMSC/CCL19 were i.t. injected into CT26-bearing BALB/c mice 10 and 13 days after tumor inoculation. Individual tumor size (A) and mean (B) are shown. The percentage of tumor-free mice is indicated. (C) The mean±SD of tumor size on day 28 are shown. (D) PBS, BALB/c-derived iMSC/CCL19 or C57BL/6-derived iMSC/CCL19 were i.t. injected into CT26-bearing CB6F1 mice 10 and 13 days after tumor inoculation. Individual tumor size and the number of tumor-free mice are shown (left). The mean±SD are shown (right). *p<0.05, **p<0.01 by Tukey (analysis of variance). N.S.: not significant. These experiments were performed twice. CCL19, (C-C motif) ligand 19; iMSC, immortalized mesenchymal stem cell; i.t., intratumoral; PBS, phosphate-buffered saline; syn-iMSC, syngeneic iMSC.
Allo-iMSC/CCL19 therapy upregulates cDC1-related and Th1-related genes at tumor sites
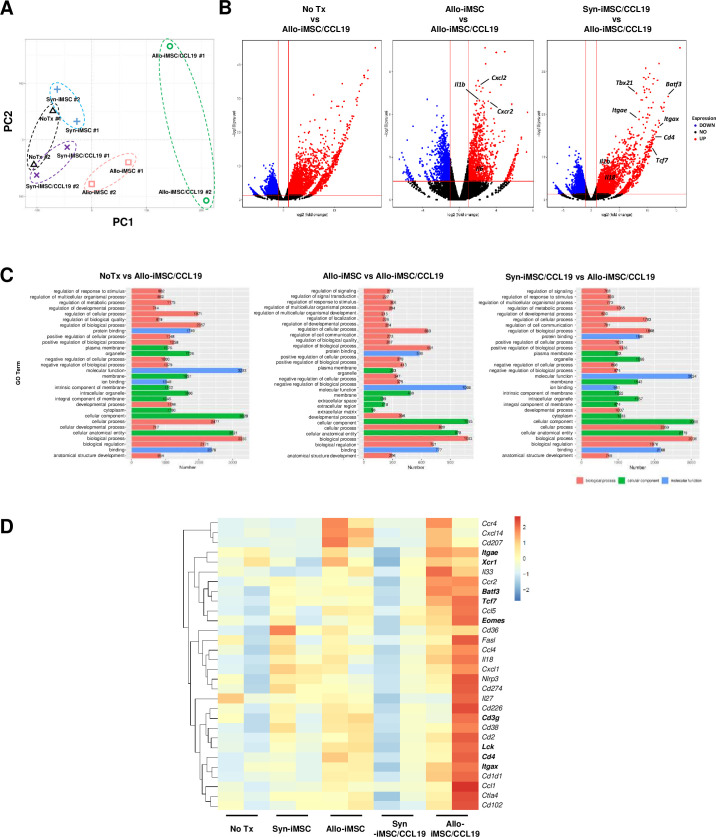
To investigate the underlying mechanisms by which allo-iMSC/CCL19 therapy induced drastic antitumor effects on CT26, we performed RNA sequencing transcriptome analyses of tumor samples from five groups (no treatment (No Tx), syn-iMSC, syn-iMSC/CCL19, allo-iMSC and allo-iMSC/CCL19). Principal component analysis revealed that each group was distinct in terms of its RNA expression, especially the allo-iMSC/CCL19 group (figure 4A). Allo-iMSC/CCL19 therapy upregulated >200 genes compared with the No Tx group. Inflammation-related cytokines such as Il6, Il1b and Cxcl2 were upregulated by allo-iMSC/CCL19 therapy compared with allo-iMSC therapy. Allo-iMSC/CCL19 therapy also upregulated inflammation-related genes such as Il1b and Il18 compared with syn-iMSC/CCL19 therapy (figure 4B). GO analysis revealed that allo-iMSC/CCL19 treatment modulated the tumor microenvironment, especially in terms of molecular functions, cellular components, and biological processes (figure 4C). We also investigated which genes were involved in anti-allo T-cell reactivity. Compared with the allo-iMSC and allo-iMSC/CCL19 therapy groups, we found that several immune-related genes were upregulated, including Xcr1, Itgae, Itgax and Batf3, representing conventional type 1 dendritic cells (cDC1) feature. In addition, the upregulation of genes such as Cd3g, Cd4, Eomes, Tcf7 and Lck was specific to Th1 cells (figure 4D). Consistent with the RNA sequencing results, quantitative RT-PCR analysis revealed that the gene expression of Ccl19, Ccr7, Itgax, and Batf3 was upregulated by allo-iMSC/CCL19 therapy, and that of both p35 and p40, which are IL-12 components, was also upregulated (online supplemental figure S4). These results suggest that allo-iMSC/CCL19 therapy effectively upregulates cDC1-related and Th1-related genes compared with syn-iMSC/CCL19 and allo-iMSC therapy, indicating that both the use of allogeneic cells and the expression of CCL19 were required for therapeutic efficacy.
Figure 4. Allo-iMSC/CCL19 therapy modifies tumor microenvironment CT26-bearing mice were treated with PBS, syn-iMSC, allo-iMSC, syn-iMSC/CCL19 or allo-iMSC/CCL19 10 days after tumor inoculation. Tumors were harvested on 12 days and analyzed by RNA sequencing. (A) Principal component of each treatment group is shown. (B) Volcano plot showing log2 (fold change) and −log10(p value) for DEGs in No Tx (PBS) versus allo-iMSC/CCL19, allo-iMSC versus allo-iMSC/CCL19 and Syn-iMSC/CCL19 versus allo-iMSC/CCL19. (C) Gene Ontology analysis in No Tx (PBS) versus allo-iMSC/CCL19, allo-iMSC versus allo-iMSC/CCL19 and syn-iMSC/CCL19 versus allo-iMSC/CCL19 are shown. (D) Upregulated genes in syn-iMSC/CCL19 versus allo-iMSC/CCL19 group are shown in heatmap. RNA sequencing was performed once. CCL19, (C-C motif) ligand 19; iMSC, immortalized mesenchymal stem cell; NO Tx, no treatment, PBS, phosphate-buffered saline; syn-iMSC, syngeneic iMSC.
CCR7+ CD103+ IL-12-producing DCs play crucial roles in antitumor effects induced by allo-iMSC/CCL19 therapy
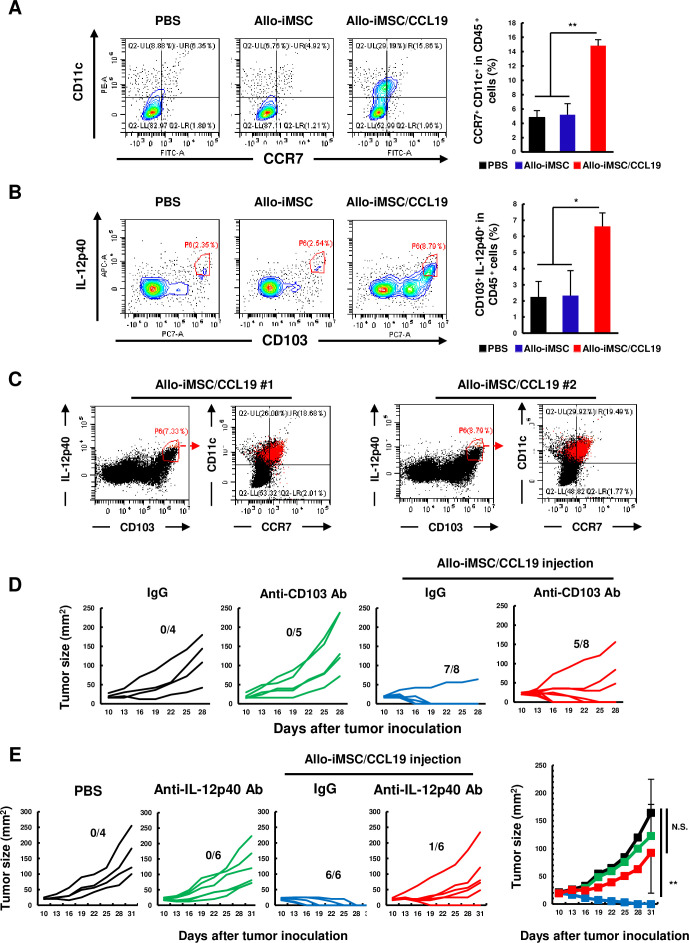
We previously reported that CCR7+ DCs were increased by syn-iMSC/CCL19 therapy.16 Based on the results of RNA sequencing, we evaluated the infiltration of DCs into tumor sites after allo-iMSC/CCL19 therapy. Because IL-12 plays an important role in the induction of the antitumor Th1 response,17,20 we also assessed the proportions of IL-12-producing DCs. Allo-iMSC/CCL19 therapy increased the proportion of CCR7+ CD11c+ DCs and CD103+ IL-12-producing DCs (figure 5A,B), indicating that increased DCs were cDC1 phenotype. In addition, we examined the relationship of CCR7+ CD11c+ DCs and CD103+ IL-12-producing DCs, and found that CCR7+ CD11c+ DCs and CD103+ IL-12-producing DCs, both of which increased by allo-iMSC/CCL19 therapy, were almost the same population (figure 5C). When CD103+ cells were depleted in vivo by anti-CD103 antibody, the antitumor effects of allo-iMSC/CCL19 therapy were partially attenuated (figure 5D). This reduced effect of anti-CD103 antibody could be due to the incomplete depletion of CD103+ cells in vivo.21 In addition, the antitumor effect of allo-iMSC/CCL19 therapy was negated by neutralization of IL-12p40 (figure 5E). Taken together, these results suggest that CCR7+ CD11c+ and CD103+ IL-12-producing DCs were almost the same population and that CCR7+ CD103+ IL-12-producing DCs play crucial roles in the antitumor effects induced by allo-iMSC/CCL19 therapy.
Figure 5. CCR7+ CD103+ IL-12-producing dendritic cells play an important role in antitumor immunity evoked by allo-iMSC/CCL19 therapy CT26-bearing mice were treated with PBS, allo-iMSC or allo-iMSC/CCL19 10 days after tumor inoculation. 2 days after treatment, tumors were collected and analyzed by flow cytometry. (A) Representative dot plot of CCR7+ CD11c+ cells, gated on CD45+ cells (left) and mean±SD (right) are shown. (B) Representative dot plot and histogram of CD103+ IL-12p40+ cells, gated on CD45+ cells (left) and mean±SD (right) are shown. (C) Two representative dot plots of CD103+ IL-12p40+ cells and CCR7+ and CD11c+ cells are shown in red dot. (D) CT26-bearing mice were treated with or without allo-iMSC/CCL19 (on days 10 and 12) followed by intraperitoneally (i.p.) administration of PBS, control IgG or anti-CD103 antibody (100 µg, on days 9 and 11). Individual tumor size and the number of tumor-free mice are indicated. (E) PBS, control IgG or anti-IL-12p40 neutralization antibody (200 µg) was i.p. administrated on days 10 and 13 after CT26 tumor inoculation. PBS or allo-iMSC/CCL19 was intratumoral injected on days 10 and 13. Individual tumor size and number of tumor-free mice (left) and mean±SD (right) were shown. *p<0.05, **p<0.01 by Tukey (analysis of variance). N.S.: not significant. These experiments were performed twice. CCL19, (C-C motif) ligand 19; iMSC, CCR7, C-C chemokine receptor type 7; IL, interleukin; iMSC, immortalized mesenchymal stem cell; PBS, phosphate-buffered saline.
Allo-iMSC/CCL19 therapy activates CD4+ T and natural killer cells and primes/activates CD8+ T cells without involving draining lymph nodes
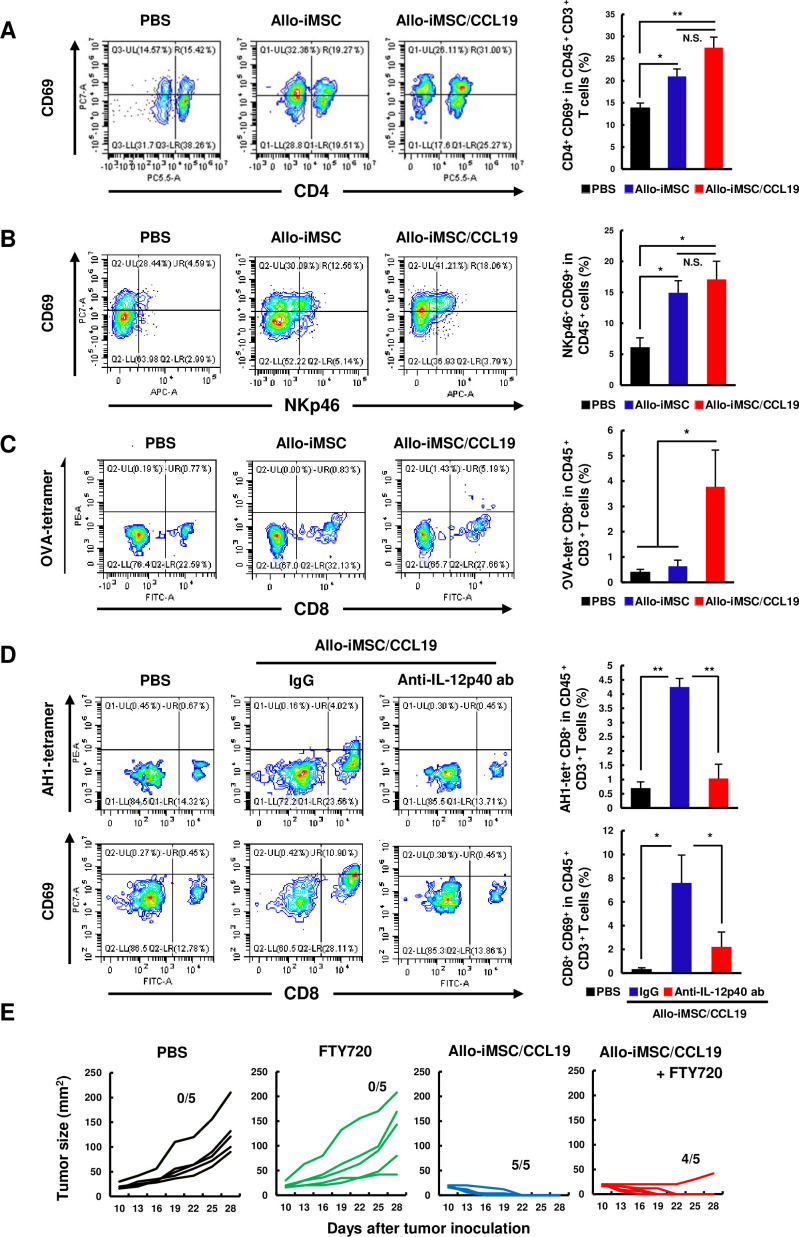
Next, we tested the activation levels of CD4+ T, natural killer (NK) and CD8+ T cells at tumor sites after the treatment with either allo-iMSC therapy or allo-iMSC/CCL19 therapy. We found that both therapies increased the level of CD69 on CD4+ T cells and NKp46+ cells (figure 6A,B). Although there was no statistical difference, the level of CD69 was slightly higher in the case of allo-iMSC/CCL19 therapy. Nevertheless, these results suggest that allo-iMSC with or without CCL19 secretion induces the activation of CD4+ T and NK cells in tumor sites, probably via anti-allo response.
Figure 6. Allo-iMSC/CCL19 therapy promotes the activation of CD4+ T, NK and tumor-specific CD8+ T cells in tumor sites CT26-bearing mice were treated with PBS, allo-iMSC or allo-iMSC/CCL19 10 days after tumor inoculation. On day 12, tumors were harvested and analyzed by flow cytometry. (A) Representative dot plots of CD69+ CD4+ cells, gated on CD45+ CD3+ cells (left) and mean±SD (right) in tumors are shown. (B) Representative dot plot of CD69+ NKp46+ cells, gated on CD45+ cells (left) and mean±SD (right) are shown. (C) B16-OVA-bearing mice were treated with PBS, allo-iMSC or allo-iMSC/CCL19 10 days after tumor inoculation. On day 12, tumors were harvested and analyzed by flow cytometry. Representative dot plot of OVA-tetramer+ CD8+ gated on CD45+ CD3+ cells (left) and mean±SD (right) are shown. (D) CT26-bearing mice were treated with PBS of allo-iMSC/CCL19 on day 10 followed by i.p. administration of 100 µg anti-IL-12p40 antibody. On day 12, tumors were harvested and analyzed by flow cytometry. Representative dot plot of AH1 (MuLV gp70)-tetramer+ CD8+ cells, gated on CD45+ CD3+ cells and mean±SD are shown (top). Representative dot plot of CD69+ CD8+ cells, gated on CD45+ CD3+ cells and mean±SD are shown (down). (E) CT26-bearing mice were treated with PBS or allo-iMSC/CCL19 on days 10 and 13 followed by i.p. administration of 2 mg/kg FTY720 (Fingolimod) on days 10–20. Tumor size and the number of tumor-free mice are shown. **p<0.01 by Tukey (analysis of variance). N.S.: not significant. These experiments were performed twice. CCL19, (C-C motif) ligand 19; iMSC, immortalized mesenchymal stem cell; i.p., intraperitoneal; NK, natural killer; PBS, phosphate-buffered saline.
As mentioned above, allo-iMSC/CCL19 therapy increased CCR7+ CD103+ IL-12-producing DCs at tumor sites. In general, DCs prime the tumor-specific CD8+ T cells in draining lymph nodes.22,24 To this end, we next attempted to determine the sites where DCs primed tumor-specific CD8+ T cells using B16-OVA model. 2 days after allo-iMSC/CCL19 therapy, OVA-tetramer+ CD8+ T cells were increased in the tumor sites (figure 6C). Given that CD69 is an activation marker of T cells, such CD8+ T cells could be activated in tumor sites. Therefore, we tested this possibility using CT26 tumor model. As a result, CT26 tumor-specific AH1-tetramer+ CD8+ T cells and CD69+ CD8+ T cells were increased by allo-iMSC/CCL19 therapy. Importantly, administration of anti-IL-12p40 antibody negated increases in these cell population (figure 6D), suggesting that IL-12, produced by tumor-infiltrating CD103+ IL-12-producing DCs, plays key roles in antitumor effect by allo-iMSC/CCL19 therapy. In contrast, at the same time point, the percentages of tumor-peptide-specific CD8+ T cells were not changed in the draining lymph nodes (online supplemental figure S5). Therefore, we further performed experiments using FTY720 (Fingolimod), which is a sphingosine 1-phosphate receptor modulator that inhibits T cells to exit from draining lymph nodes.25 26 The administration of FTY720 showed a reduction of CD3+ T cells at the tumor sites but not in the spleen (online supplemental figure S6). Interestingly, FTY720 treatment showed no effect on the antitumor effect of allo-iMSC/CCL19 therapy (figure 6E). These data suggest that allo-iMSC/CCL19 therapy helped tumor-specific CD8+ T cells to be primed or activated at tumor sites without involving draining lymph nodes.
NK, CD4+ T and CD8+ T cells participated in the antitumor effects of allo-iMSC/CCL19 therapy
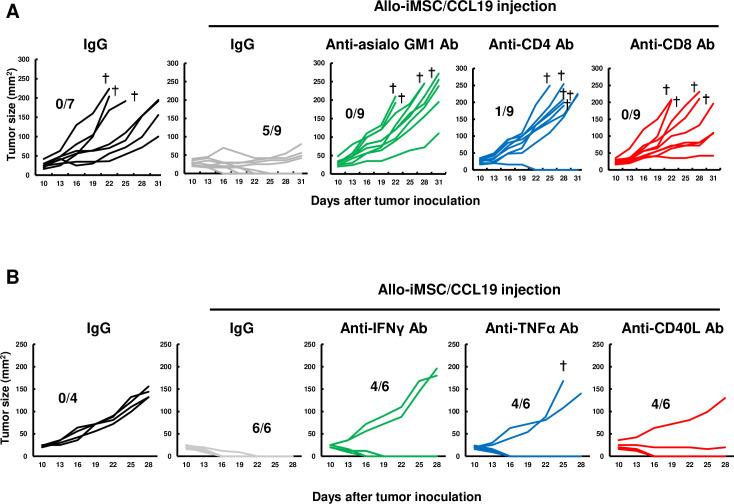
Next, we explored the types of cells contributing to allo-iMSC/CCL19 therapy by selectively depleting NK, CD4+ T, or CD8+ T cells in vivo. The antitumor effects induced by allo-iMSC/CCL19 therapy were negated by either the anti-CD4 or anti-CD8 antibodies (figure 7A). The anti-asialo GM1 antibody also negated the antitumor effects. In this regard, several groups have shown that asialo GM1 is expressed on a part of CD8+ T cells.27,30 However, in our experimental model, the expression of asialo GM1 was observed on the NK and NKT cells, but not on CD8+ T cells (online supplemental figure S7). We further determined whether inflammatory cytokine/molecules were involved in the antitumor effect or not. As shown in figure 7B, either of anti-IFN-γ, anti-TNF-α or anti-CD40 ligand (CD40L) antibody attenuated the antitumor effect induced by allo-iMSC/CCL19. These results indicate that NK, CD4+ T and CD8+ T cells collaborated to exert the antitumor effects of allo-iMSC/CCL19 therapy, and that IFN-γ and TNF-α also participate in the antitumor effects. In addition, given that the interaction between CD40 and CD40L leads to IL-12 production by DCs, the inhibitory effect of anti-CD40L antibody on antitumor effects could result from a negative effect on IL-12 production by DCs.
Figure 7. Depletion of NK, CD4+, or CD8+ cells attenuates antitumor effect of allo-iMSC/CCL19 therapy (A) CT26-bearing mice were treated with allo-iMSC/CCL19 on days 10 and 13 after tumor injection. Control IgG, anti-asialo GM1, anti-CD4 or anti-CD8 antibody (100 µg) was i.p. administrated on days 10 and 13. Individual tumor size and the number of tumor-free mice are shown. (B) CT26-bearing mice were treated with allo-iMSC/CCL19 on days 10 and 13 after tumor injection. Control IgG, anti-IFN-γ, anti-TNF-α or anti-CD40L antibody (100 µg) was i.p. administrated on days 9 and 12. Individual tumor size and the number of tumor-free mice are shown. †Mice were euthanized due to tumor size. **p<0.01 by Tukey (analysis of variance). N.S.: not significant. These experiments were performed twice. CCL19, (C-C motif) ligand 19; IFN, interferon; iMSC, immortalized mesenchymal stem cell; i.p., intraperitoneal.
Combination therapy with allo-iMSC/CCL19 and anti-PD-L1 antibody in CT26-bearing young and aged mice
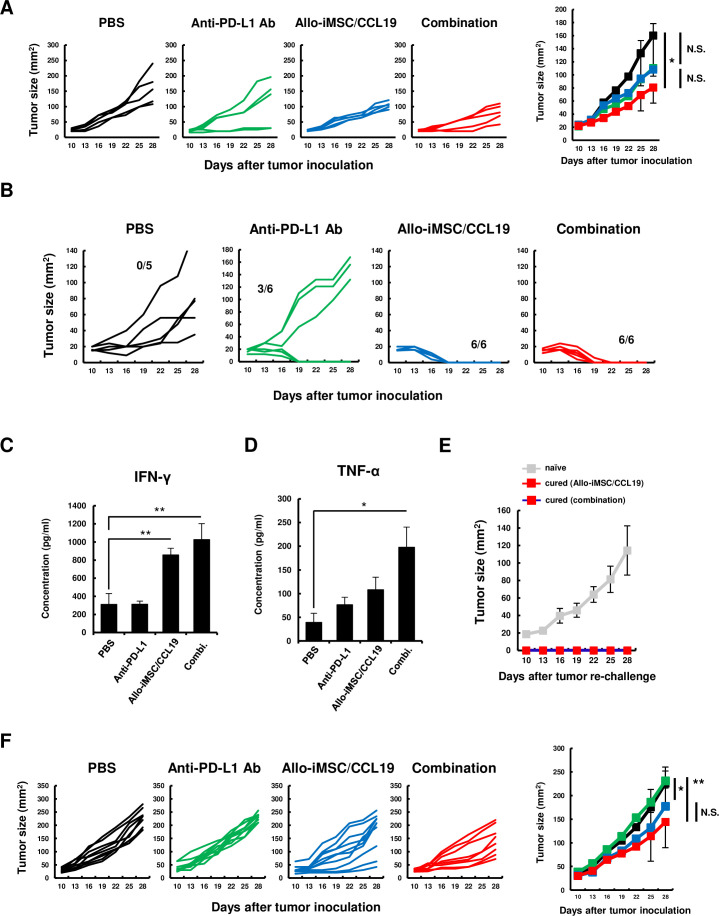
To represent a clinical situation, we next examined the antitumor effects of allo-iMSC/CCL19 therapy combined with anti-PD-L1 antibody. Although the allo-iMSC/CCL19 therapy or anti-PD-L1 antibody monotherapy showed no definite effect on the tumor growth in the Renca tumor model, combination therapy significantly reduced the tumor growth (figure 8A). We examined the therapeutic efficacy of this combination therapy in the CT26 tumor model. Anti-PD-L1 antibody cured the established CT26 tumor in three out of six young BALB/c mice. When CT26-bearing mice were treated with allo-iMSC/CCL19 therapy, complete regression was observed even without the anti-PD-L1 antibody (figure 8B). To evaluate the levels of anti-allo T-cell responses, the spleen cells of each group were harvested and cultured for 48 hours in vitro with splenocytes from allogeneic C57BL/6 mice. As a result, allo-iMSC/CCL19 therapy, with or without anti-PD-L1 antibody treatment, showed increased production of IFN-γ (figure 8C). On the other hand, the levels of TNF-α were higher than in the other groups only when allo-iMSC/CCL19 therapy was combined with anti-PD-L1 antibody treatment (figure 8D). Cured mice that were treated with allo-iMSC/CCL19 therapy, with or without anti-PD-L1 antibody treatment, rejected the CT26 re-challenge (figure 8E).
Figure 8. Allo-iMSC/CCL19 therapy combined with anti-PD-L1 antibody exerts anti-tumor effect in aged mice (A) Renca-bearing mice were i.t. injected with allo-iMSC/CCL19 on days 10 and 13 after tumor inoculation. Control IgG or anti-PD-L1 antibody (200 µg) were i.p. administrated on days 13 and 16. The tumor growth in individual mice (left) and the mean±SD (right) are shown. (B) CT26-bearing mice were i.t. injected with allo-iMSC/CCL19 on days 10 and 13 after tumor inoculation. Control IgG or anti-PD-L1 antibody (200 µg) were i.p. injected on days 13 and 16. The tumor growth in individual mice is shown. (C, D) On day 28, the spleen was harvested from each group in (A) and stimulated mitomycin C-treated C57BL/6-derived splenocyte. 48 hours after co-culture, secretion of IFN-γ (C) and TNF-α (D) was measured by ELISA. (E) Cured mice treated with allo-iMSC/CCL19 or combination were s.c. inoculated with CT26. The mean±SD of tumor growth is shown. (F) 53 weeks old aged BALB/c mice were s.c. inoculated with CT26. PBS or allo-iMSC/CCL19 were i.t. injected on days 10 and 13 and control IgG or anti-PD-L1 (200 µg) antibody were i.p. injected on days 13 and 16. The tumor growth in individual mice (left) and the mean±SD (right) are shown. *p<0.05, **p<0.01 by Tukey (analysis of variance). These experiments were performed twice. CCL19, (C-C motif) ligand 19; IFN, interferon; iMSC, immortalized mesenchymal stem cell; i.p., intraperitoneal; i.t., intratumoral; N.S., not significant; PBS, phosphate-buffered saline; PD-L1, programmed death-ligand 1; s.c., subcutaneously.
Given that many patients with cancer are middle-aged or elderly and their immunological potency is attenuated, we next examined the antitumor effects of our combination therapy in aged mice (53 weeks old). In contrast to the results seen in young mice, anti-PD-L1 antibody treatment showed no antitumor effects in the aged mice (figure 8F). Allo-iMSC/CCL19 therapy alone inhibited the tumor growth slightly but significantly compared with the untreated and anti-PD-L1 therapy groups (p<0.05). Although no significant difference was observed between allo-iMSC/CCL19 monotherapy and combination therapy, combination therapy reduced the tumor size significantly compared with the untreated and anti-PD-L1 therapy groups (p<0.01). These results indicate that allo-iMSC/CCL19 therapy could be therapeutically effective in tumor-bearing young mice, and suggest that the additional combination with anti-PD-L1 treatment could induce therapeutic efficacy even in immune checkpoint blockade (ICB)-resistant model and in aged mice.
Discussion
Although MSC have been considered to contribute to tumor progression through their inhibitory effect on immune cells, we did not observe any tumor-promoting effects in our model.16 In this study, we showed that iMSC/CCL19 survived at tumor sites in allogeneic mice for at least 8 days after s.c. inoculation with tumor cells. We also found that iMSC tended to be more resistant to anti-allo CTLs than to iFib, whereas the expression of MHC class I, PD-L1 and CD80 did not differ between them. This difference could account for the longer survival of iMSC in allogeneic mice.
In therapeutic models, allo-iMSC/CCL19 therapy suppressed the in vivo tumor growth of B16-OVA and Renca-OVA. Considering that allo-iMSC/CCL19 therapy failed to control the tumor growth of parental Renca, the immunogenicity of tumor cells is a key to the therapeutic efficacy of this treatment. In addition, we showed that, although anti-PD-L1 antibody alone was ineffective, allo-iMSC/CCL19 therapy in combination with anti-PD-L1 antibody induced antitumor effects even in tumor-bearing aged mice. Taken together, our results indicate that allo-iMSC/CCL19 therapy, with or without anti-PD-L1 antibody could be a promising therapy protocol for aged patients with cancer.
Allo-iMSC/CCL19 therapy markedly regressed CT26 tumor growth (figure 3). In experiments using CB6F1 mice, the therapeutic efficacy of allo-iMSC/CCL19 was significantly attenuated. This result implies that the anti-allo T-cell response of CT26-bearing host mice toward allo-iMSC/CCL19, that is, an allogeneic effect, contributed to the antitumor effects of allo-iMSC/CCL19 therapy. On the other hand, in CB6F1 mice, BALB/c- and C57BL/6-derived iMSC/CCL19 can be under the same condition, in which BALB/c mice were treated by syn-iMSC/CCL19. BALB/c-derived and C57BL/6-derived iMSC/CCL19 regressed the tumor growth two out of six, and one out of six mice, respectively (figure 3D), while syn-iMSC/CCL19 regressed the tumor growth two out of six mice (figure 3A). However, antitumor effects were more apparent in mice treated with syn-iMSC/CCL19 compared with the untreated group. Although we have no clear answer to this issue, the following possibility could explain it. That is, BALB/c-derived and C57BL/6-derived iMSC/CCL19 cells have a higher susceptibility to NK cells of CB6F1 mice. Because BALB/c-iMSC/CCL19 cells lack H-2d and C57BL/6-derived iMSC/CCL19 cells lack H-2b, respectively, the “missing-self” in CB6F1 (H2b/d) mice could lead to shorter survivability of these iMSC/CCL19 in tumor sites.
Transcriptome analysis revealed that allo-iMSC/CCL19 therapy upregulated innate immune-response-related genes, especially those encoding cDC1-related molecules, such as Xcr1, Itgae, Itgax and Batf3 (figure 4D). Consistent with RNA sequencing and quantitative RT-PCR results, we found a high level of infiltration of CD103 (Itgae)+ IL-12-producing DCs into tumor sites (figure 5B and online supplemental figure S4). We also found that CD103+ IL-12-producing cells and CCR7+ DCs are almost the same population, suggesting that allo-iMSC/CCL19 therapy traffic these cells to tumor sites. Since anti-IL-12p40 antibody attenuates the antitumor effect of allo-iMSC/CCL19 and activation of CD8+ T cells in tumor sites, IL-12 secretion by CCR7+ CD103+ DCs might be essential factor (figures5E 6D).
We showed that allo-iMSC/CCL19 therapy increased CD69+ CD8 T cells in tumor sites, whereas not all CD69+ CD8 T cells were tumor-specific and some of them must be anti-allo T cells. How did anti-allo T-cell response induce tumor-specific CD8+ T cells? In this regard, we suppose the following mechanisms. Locally injected allo-iMSC/CCL19 cells accumulated T cells and NK cells into tumor sites and activated them. In addition, activated anti-allo T cells express CD40L on their cell surface and activate DCs via CD40, leading to their IL-12 production. IL-12 increased the cytotoxicity of NK cells and promoted the priming of tumor-specific CD8+ T cells in tumor sites. Locally produced IL-12 probably played crucial roles in antitumor effects of allo-iMSC/CCL19 therapy because neutralization of IL-12 abolished the antitumor effects (figure 5E). In addition, locally produced IFN-γ and TNF-α also contribute to the antitumor effects (figure 7B). This is just a scenario that we assume at present and further studies are needed.
According to the concept of the cancer-immunity cycle, antigen-presenting cells uptake tumor antigens in tumors, homing to draining lymph nodes and priming or activating T cells. In this study, although activated CD69+ CD8+ T cells and tumor-specific CD8+ T cells increased in number at tumor sites, the number of tumor-specific CD8+ T cells in draining lymph nodes did not increase (figure 6C,D and online supplemental figure S5). Moreover, when the exit of lymphocytes from draining lymph nodes was inhibited by FTY720, the antitumor effect of allo-iMSC/CCL19 therapy was not negated (figure 6E). These results indicate that the mechanism by which allo-iMSC/CCL19 exerted its antitumor effects was elicited at tumor sites without involving draining lymph nodes. In addition, when anti-asialo GM1, CD4 or CD8 antibodies were administered to CT26-bearing mice and allo-iMSC/CCL19 therapy was applied, the antitumor effects were clearly attenuated. These results indicate that NK, CD4 and CD8 T cells contributed to the antitumor effects of allo-iMSC/CCL19 therapy. These results indicate that allo-iMSC/CCL19 therapy can overcome dysfunction in the cancer-immunity cycle and activate both innate and adoptive immunity. For clinical application, allo-iMSC/CCL19 treatment combined with ICB treatment could be a powerful therapy in ICB-resistant patients with cancer.
supplementary material
Acknowledgements
We thank Dr Taisuke Kondo and Dr Akihiko Yoshimura for kindly gift of B16-OVA cells and Dr Akihiko Murata for the fruitful discussion.
Footnotes
Funding: This study was supported in part by JSPS KAKENHI Grant (no. JP23K06636 to YI).
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not applicable.
Ethics approval: All experimental procedures were approved by the Animal Center of Shimane University
Data availability free text: Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as online supplemental information.
Contributor Information
Yuichi Iida, Email: yiida@med.shimane-u.ac.jp.
Mamoru Harada, Email: haramamo@med.shimane-u.ac.jp.
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request.
References
- 1.Lan T, Luo M, Wei X. Mesenchymal stem/stromal cells in cancer therapy. J Hematol Oncol. 2021;14:195. doi: 10.1186/s13045-021-01208-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rustad KC, Gurtner GC. Mesenchymal Stem Cells Home to Sites of Injury and Inflammation. Adv Wound Care (New Rochelle) 2012;1:147–52. doi: 10.1089/wound.2011.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houghton J, Stoicov C, Nomura S, et al. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–71. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]
- 4.Chawla-Sarkar M, Leaman DW, Borden EC. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin Cancer Res. 2001;7:1821–31. [PubMed] [Google Scholar]
- 5.Okada H, Pollack IF. Cytokine gene therapy for malignant glioma. Expert Opin Biol Ther. 2004;4:1609–20. doi: 10.1517/14712598.4.10.1609. [DOI] [PubMed] [Google Scholar]
- 6.Uchibori R, Okada T, Ito T, et al. Retroviral vector-producing mesenchymal stem cells for targeted suicide cancer gene therapy. J Gene Med. 2009;11:373–81. doi: 10.1002/jgm.1313. [DOI] [PubMed] [Google Scholar]
- 7.Jitschin R, Mougiakakos D, Von Bahr L, et al. Alterations in the cellular immune compartment of patients treated with third-party mesenchymal stromal cells following allogeneic hematopoietic stem cell transplantation. Stem Cells. 2013;31:1715–25. doi: 10.1002/stem.1386. [DOI] [PubMed] [Google Scholar]
- 8.Comerford I, Harata-Lee Y, Bunting MD, et al. A myriad of functions and complex regulation of the CCR7/CCL19/CCL21 chemokine axis in the adaptive immune system. Cytokine Growth Factor Rev. 2013;24:269–83. doi: 10.1016/j.cytogfr.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–71. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 10.Cheng H-W, Onder L, Cupovic J, et al. CCL19-producing fibroblastic stromal cells restrain lung carcinoma growth by promoting local antitumor T-cell responses. J Allergy Clin Immunol. 2018;142:1257–71. doi: 10.1016/j.jaci.2017.12.998. [DOI] [PubMed] [Google Scholar]
- 11.Itakura M, Terashima Y, Shingyoji M, et al. High CC chemokine receptor 7 expression improves postoperative prognosis of lung adenocarcinoma patients. Br J Cancer. 2013;109:1100–8. doi: 10.1038/bjc.2013.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gowhari Shabgah A, Al-Obaidi ZMJ, Sulaiman Rahman H, et al. Does CCL19 act as a double-edged sword in cancer development? Clin Exp Immunol. 2022;207:164–75. doi: 10.1093/cei/uxab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Zhao J, Feng H, et al. Antitumor efficacy of CC motif chemokine ligand 19 in colorectal cancer. Dig Dis Sci. 2014;59:2153–62. doi: 10.1007/s10620-014-3138-y. [DOI] [PubMed] [Google Scholar]
- 14.Adachi K, Kano Y, Nagai T, et al. IL-7 and CCL19 expression in CAR-T cells improves immune cell infiltration and CAR-T cell survival in the tumor. Nat Biotechnol. 2018;36:346–51. doi: 10.1038/nbt.4086. [DOI] [PubMed] [Google Scholar]
- 15.Hamanishi J, Mandai M, Matsumura N, et al. Activated local immunity by CC chemokine ligand 19-transduced embryonic endothelial progenitor cells suppresses metastasis of murine ovarian cancer. Stem Cells. 2010;28:164–73. doi: 10.1002/stem.256. [DOI] [PubMed] [Google Scholar]
- 16.Iida Y, Yoshikawa R, Murata A, et al. Local injection of CCL19-expressing mesenchymal stem cells augments the therapeutic efficacy of anti-PD-L1 antibody by promoting infiltration of immune cells. J Immunother Cancer. 2020;8:e000582. doi: 10.1136/jitc-2020-000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–46. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 18.Landoni E, Woodcock MG, Barragan G, et al. IL-12 reprograms CAR-expressing natural killer T cells to long-lived Th1-polarized cells with potent antitumor activity. Nat Commun. 2024;15:89. doi: 10.1038/s41467-023-44310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dredge K, Marriott JB, Todryk SM, et al. Adjuvants and the promotion of Th1-type cytokines in tumour immunotherapy. Cancer Immunol Immunother. 2002;51:521–31. doi: 10.1007/s00262-002-0309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haicheur N, Escudier B, Dorval T, et al. Cytokines and soluble cytokine receptor induction after IL-12 administration in cancer patients. Clin Exp Immunol. 2000;119:28–37. doi: 10.1046/j.1365-2249.2000.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mang Y, Zhao Z, Zeng Z, et al. Efficient elimination of CD103-expressing cells by anti-CD103 antibody drug conjugates in immunocompetent mice. Int Immunopharmacol. 2015;24:119–27. doi: 10.1016/j.intimp.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 23.Giles JR, Globig A-M, Kaech SM, et al. CD8+ T cells in the cancer-immunity cycle. Immunity. 2023;56:2231–53. doi: 10.1016/j.immuni.2023.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellman I, Chen DS, Powles T, et al. The cancer-immunity cycle: Indication, genotype, and immunotype. Immunity. 2023;56:2188–205. doi: 10.1016/j.immuni.2023.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Rosen H, Sanna G, Alfonso C. Egress: a receptor-regulated step in lymphocyte trafficking. Immunol Rev. 2003;195:160–77. doi: 10.1034/j.1600-065x.2003.00068.x. [DOI] [PubMed] [Google Scholar]
- 26.Matloubian M, Lo CG, Cinamon G, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature New Biol. 2004;427:355–60. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 27.Trambley J, Bingaman AW, Lin A, et al. Asialo GM1(+) CD8(+) T cells play a critical role in costimulation blockade-resistant allograft rejection. J Clin Invest. 1999;104:1715–22. doi: 10.1172/JCI8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosaka A, Lee U, Wakita D, et al. Interleukin-12-responding asialoGM1+CD8+ central memory-type T cells as precursor cells for interferon-gamma-producing killer T cells. Cancer Sci. 2006;97:1236–41. doi: 10.1111/j.1349-7006.2006.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosaka A, Wakita D, Matsubara N, et al. AsialoGM1+CD8+ central memory-type T cells in unimmunized mice as novel immunomodulator of IFN-gamma-dependent type 1 immunity. Int Immunol. 2007;19:249–56. doi: 10.1093/intimm/dxl140. [DOI] [PubMed] [Google Scholar]
- 30.Sung CC, Horng JH, Siao SH, et al. Asialo GM1-positive liver-resident CD8 T cells that express CD44 and LFA-1 are essential for immune clearance of hepatitis B virus. Cell Mol Immunol. 2021;18:1772–82. doi: 10.1038/s41423-020-0376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a public, open access repository. Data are available upon reasonable request.