Abstract
The cloning of two highly homologous chicory (Cichorium intybus var. foliosum cv Flash) fructan 1-exohydrolase cDNAs (1-FEH IIa and 1-FEH IIb) is described. Both isoenzymes could be purified from forced chicory roots as well as from the etiolated “Belgian endive” leaves where the 1-FEH IIa isoform is present in higher concentrations. Full-length cDNAs were obtained by a combination of reverse transcriptase-polymerase chain reaction (PCR), PCR and 5′- and 3′-rapid amplification of cDNA ends using primers based on N-terminal and conserved amino acid sequences. 1-FEH IIa and 1-FEH IIb cDNA-derived amino acid sequences are most homologous to a new group of plant glycosyl hydrolases harboring cell wall-type enzymes with acid isoelectric points. Unlike the observed expression profiles of chicory 1-FEH I, northern analysis revealed that 1-FEH II is expressed when young chicory plants are defoliated, suggesting that this enzyme can be induced at any developmental stage when large energy supplies are necessary (regrowth after defoliation).
The most prominent storage carbohydrate in the plant kingdom is starch, but fructan (a Fru polymer) is used as a storage compound in about 15% of flowering plant species (Hendry, 1993). Inulin-type fructans consists of linear β- (2→1) linked fructofuranosyl units and occur mainly in dicotyledonous species, among which Jerusalem artichoke and chicory (Cichorium intybus) are the most extensively studied species. Levan consists of linear β- (2→6) linked fructofuranosyl units, but more complex and branched fructan types (graminan) are common in monocotyledonous species (Shiomi, 1989; Livingston et al., 1993).
Besides acting as a reserve carbohydrate, fructan might fulfill also other functions such as stress protectant (drought and cold) or osmoregulator (Hendry, 1993; Livingston and Henson, 1998; Pilon-Smits et al., 1995; Hincha et al., 2000), but the molecular mechanism behind these putative roles is still obscure. Unlike starch, fructans are water-soluble and are believed to be localized in the vacuole (Frehner et al., 1984; Wiemken et al., 1986). However, the exclusive vacuolar localization of fructan metabolism has recently been questioned (Livingston and Henson, 1998).
Inulin synthesis involves two distinct enzymes: Suc:Suc 1-fructosyl transferase (1-SST) and fructan:fructan 1-fructosyl transferase (1-FFT; Edelman and Jefford, 1968). Additional enzymes are required for the synthesis of more complex branched-type fructan, e.g. fructan:fructan 6G fructosyl transferase and Suc:fructan 6-fructosyl transferase (Vijn and Smeekens, 1999). Fructan breakdown is catalyzed by fructan exohydrolase (FEH; G − Fn + H2O → G − F(n − 1) + F with n > 1) essentially transferring a Fru moiety to a water molecule as acceptor. Only a few plant FEH enzymes have been purified to electrophoretic homogeneity. They include fructan 1-exohydrolase I (1-FEH I) and 1-FEH II from chicory (Claessens et al., 1990; De Roover et al., 1999a), a 1-FEH from Jerusalem artichoke (Marx et al., 1997a), a fructan 6-exohydrolase from Lolium perenne (Marx et al., 1997b), and a FEH that preferentially hydrolyzes β (2→6) (oat, Henson and Livingston, 1996) or β (2→1) linkages (barley; Henson and Livingston, 1998).
Nowadays, food and non-food industries are eagerly looking for new compounds like fructan and its derivatives or modified starches (Heyer et al., 1999). Inulin is now widely recognized as a health-improving compound when added to various foods as low-calorie sweetener, dietary fiber, or fat replacer (Fuchs, 1991). It is suggested that a daily intake of low amounts already generates a bifidogenic and antitumor effect (Roberfroid et al., 1998; Taper et al., 1999).
We found sharp 1-FEH activity increases in mature chicory roots after cold induction in the field or in cold rooms (Van den Ende and Van Laere, 1996), but also in young fructan-containing chicory roots that were defoliated or received high nitrogen nutrition after a period of nitrogen starvation (De Roover et al., 1999b; Van den Ende et al., 1999). However, it was not clear what particular enzyme (1-FEH I and/or 1-FEH II) was induced at what particular stage since it is not possible to distinguish between 1-FEH I and 1-FEH II activities in crude extracts (De Roover et al., 1999a). To resolve this issue, the cloning of the respective 1-FEH cDNAs would allow a detailed study of the expression of the respective genes throughout several developmental stages and under different stress conditions. Next to chicory 1-FEH I cDNA (Van den Ende et al., 2000a), the 1-FEH II cDNAs are, to our knowledge, the first plant FEH cDNAs described. This is in severe contrast to the cDNAs from plant fructan biosynthetic enzymes of which a large number have been cloned already (for review, see Vijn and Smeekens, 1999). Also, a large number of microbial FEH cDNAs are known (Burne et al., 1999; Kang and Kim, 1999 and refs. therein).
Plant FEHs are expected to be more homologous to plant invertases than to plant fructosyl transferases since invertases and FEHs use water and not fructan or Suc as a fructosyl acceptor. Moreover, for several plant species it was demonstrated that a multigenic family of invertases occurs that contains silent genes or genes that are expressed at very particular places and/or during particular developmental stages (Unger et al., 1994; Godt and Roitsch, 1997; Tymowska-Lalanne and Kreis, 1998; Sturm, 1999). For all these reasons we first derived information at the protein level (purification and N-terminal sequence analysis) and used this information in the first steps of our cloning strategy.
In this paper we focus on the analysis and cloning of two 1-FEH II isoforms from witloof chicory roots, which are used for the production of the vegetable “Belgian endives.”
RESULTS
Purification of 1-FEH IIa and IIb and N-Terminal Analysis
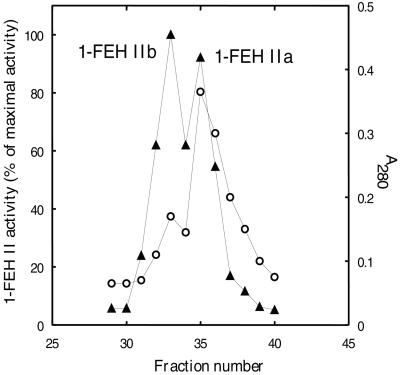
De Roover et al. (1999a) described the purification of 1-FEH II. In their final fraction, a single 64-kD band was found on SDS-PAGE. However, later N-terminal amino acid sequencing on this band was ambiguous, indicating that this fraction was not pure. By performing an additional Uno S cation exchange step at pH 5.2 on this fraction and analyzing the fractions 29 through 40 by activity and A280 measurements (Fig. 1), we demonstrate here the presence of two different isoforms in the fraction obtained by De Roover et al. (1999a). The two independent enzymatically active isoforms were fully separated and were termed 1-FEH IIa and 1-FEH IIb. In etiolated leaves of Belgian endives, 1-FEH IIa is present at a higher concentration, but has a lower specific activity than 1-FEH IIb (Fig. 1). For forced roots, comparable results were obtained except that overall enzyme concentrations were lower. Automated Edman degradation on the two independent FEH II isoforms revealed the following N-terminal sequences: QQIEQPYRTGYHFQP for 1-FEH IIa (Uno S fraction 36) and QQIQQPYRTGYHFQP for 1-FEH IIb (Uno S fraction 32).
Figure 1.
Purification of chicory 1-FEH IIa and IIb. Protein content profile (○) and 1-FEH II activity profile (▴) of the different fractions obtained during final Uno S chromatography at pH 5.2. 1-FEH activity was measured by the Fru production from 3% (w/v) inulin.
Cloning Strategy
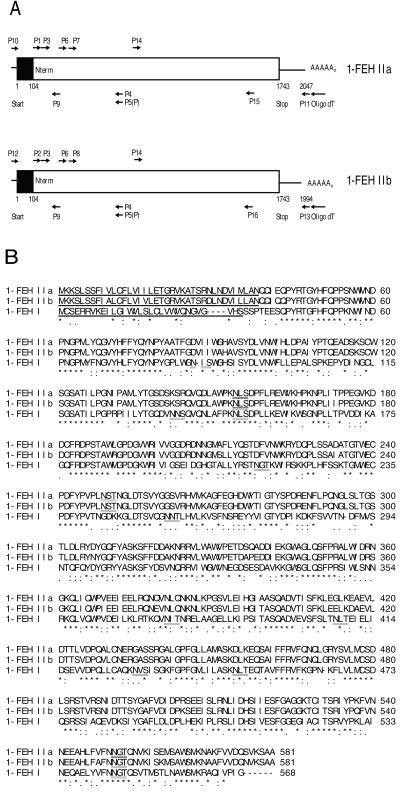
Partial chicory 1-FEH IIa and IIb cDNAs were obtained by performing reverse transcriptase- (RT) PCR using N-terminal sequencing information (P1, P2, and P3) combined with an antisense primer conserved within 1-FEH I and cell wall-type invertases (P5, Van den Ende et al., 2000a). After subcloning, two types of very homologous (but not identical) clones were obtained. Using this sequencing information, new (specific and/or phosphorylated) primers were derived for performing 5′- and 3′-RACE RT-PCR and semi-nested PCR (Fig. 2A, for details, see also “Materials and Methods”). Specific 5′- and 3′-outer primers and a proofreading DNA polymerase were used to amplify the whole cDNAs (2 kb in length). Both cDNAs encode a polypeptide of 581 amino acids. The coding region is preceded by a very short 5′-untranscribed part and followed by longer 3′-untranslated regions (304 bp for 1-FEH IIa and 251 bp for 1-FEH IIb).
Figure 2.
Cloning of chicory 1-FEH IIa and IIb. A, Schematic representation of the 1-FEH IIa and IIb cDNAs from chicory. Both cDNAs contain a single open reading frame (Start-Stop) of 1,743 bp. The first part of this open reading frame is a putative signal sequence (black) of 104 bp. 3′- and 5′-untranslated parts are also present (line). Primers used during RT-PCR, PCR, 5′-, and 3′-RACE RT-PCR are indicated with arrows (for details, see “Materials and Methods”). Probes were prepared by using the primers P14-P15 (1-FEH IIa) or P14-P16 (1-FEH IIb). B, Alignment of the deduced amino acid sequences of chicory root 1-FEH IIa, 1-FEH IIb, and 1-FEH I. The posttranslationally removed signal sequences are indicated (underlined and bold). Potential glycosylation sites are double underlined. For 1-FEH IIa and IIb, tryptic fragments found after quadrupole-time of flight (Q-TOF) mass spectrometry/mass spectroscopy (MS/MS) analysis are indicated (bold). Consensus line: asterisks indicate identical residues, colons indicate conserved subsitutions, and periods indicate semi-conserved substitutions.
The deduced amino acid sequences for chicory 1-FEH IIa, 1-FEH IIb, and 1-FEH I are presented in Figure 2B. Comparison of the cDNA-derived amino acid sequences with the experimentally determined N-terminal sequences (see higher), as well as the unambiguous identification of the amino terminal tryptic fragments by collision-induced dissociation (CID) MS/MS (Tables I and II), demonstrates that the primary translation products have a 38-amino acid signal peptide that is post-translationally removed. The cDNA-derived isoelectric points (pIs) of chicory 1-FEH IIa and IIb are calculated at 5.24, which perfectly fits with isoelectric focusing-PAGE and chromatographic behavior (De Roover et al., 1999a). Furthermore, the mature 1-FEH IIa and IIb proteins contain three potential glycosylation sites (N-X-S/T; see Fig. 2B). The calculated molecular mass of the mature 1-FEH II enzymes (61 kD) is slightly lower than the estimated 64 kD from SDS-PAGE, but this can be explained by the effective glycosylation being present at least on two out of three potential glycosylation sites (Table I).
Table I.
Fragment ions detected in Q-TOF after tryptic digest of 1-FEH IIa, with calculated matches to theoretical digest of virtual cDNA derived protein, and confirmation of identity by tandem MS/MS sequencing
| Observed Mass | Charge State | Calculated Mass (Presumptive 1-FEH IIa Fragment Ion) | MS/MS Sequence (from N to C Terminus) |
|---|---|---|---|
| 415.2 | 2+ | 415.19 [T20+2H]2+ | 278 SFFDDAK 284 |
| 417.25 | 2+ | 417.22 [T40+2H]2+ | 467 SEEISLR 473 |
| 461.3 | 2+ | 461.26 [T47+2H]2+ | 533 FVVDQSVK 540 |
| 481.3 | 2+ | 481.26 [T36+2H]2+ | 427 VFQNQLGR 434 |
| 499.3 | 2+ | 499.26 [T35+2H]2+ | 419 EQSAIFFR 426 |
| 531.3 | 2+ | 531.27 [T1+2H]2+ | 1 QQIEQPYR 8 |
| 542.8 | 2+ | 542.79 [T5+2H]2+ | 107 QVQDLAWPK 115 |
| 559.8 | 2+ | 559.79 [T24+2H]2+ | 306 GWAGLQSFPR 315 |
| 585.3 | 2+ | 585.27 [T45+2H]2+ | 520 ISEMSAWSMK 529 |
| 621.3 | 2+ | 621.28 [T19+2H]2+ | 268 YDYGQFYASK 277 |
| 666.9 | 2+ | 666.87 [T33+2H]2+ | 402 GALGPFGLLAMASK 415 |
| 677.4 | 2+ | 677.36 [T34-35+2H]2+ | 416 DLKEQSAIFFR 426 |
| 829.4 | 2+ | 829.43 [T41+2H]2+ | 474 NLIDHSIIESFGAGGK 489 |
| 935.9 | 2+ | 935.95 [T39+2H]2+ | 450 SNIDTTSYGAFVDIDPR 466 |
| 1,066.5 | 2+ | No match | T6 N-linked glycopeptide:116 NLSDPFLR 123 (Deoxyhexose)1 (Pentose)1 (Man)3 (GlcNAc)2 |
| 691.3 | 3+ | 691.31 [T17+3H]3+ | AGFEGHDWYTLGTYSPDR |
| 892.05 | 3+ | No match | T6-T7 N-linked glycopeptide:116 NLSDPFLREWVK 127 (Deoxyhexose)1 (Pentose)1 (Man)3 (GlcNAc)2 |
| 950.14 | 3+ | 950.17 [T27-28+3H]3+ | QLIQWPVEEIEELRQNQVNLQNK |
| 1,234.25 | 3+ | No match | T44 N-linked glycopeptide:500 FVNNEEAHLFVFNNGTQNVK 519 (Hexose)3 (Man)3 (GlcNAc)2 |
Glycosylation sites NX(S/T) are presented in bold. The position of the fragments in the mature 1-FEH IIa protein are indicated.
Table II.
Fragment ions detected in Q-TOF after tryptic digest of 1-FEH IIb, with calculated matches to theoretical digest of virtual cDNA derived protein, and confirmation of identity by tandem MS/MS sequencing
| Observed Mass | Charge State | Calculated Mass (Presumptive1-FEH IIb Fragment Ion) | MS/MS Sequence (from N to C Terminus) |
|---|---|---|---|
| 415.2 | 2+ | 415.19 [T21+2H]2+ | 278 SFFDDAK 284 |
| 417.25 | 2+ | 417.22 [T41+2H]2+ | 467 SEEISLR 473 |
| 481.3 | 2+ | 481.26 [T37+2H]2+ | 427 VFQNQLGR 434 |
| 499.3 | 2+ | 499.26 [T36+2H]2+ | 419 EQSAIFFR 426 |
| 530.8 | 2+ | 530.78 [T1+2H]2+ | 1 QQIQQPYR 8 |
| 542.8 | 2+ | 542.79 [T5+2H]2+ | 107 QVQDLAWPK 115 |
| 559.8 | 2+ | 559.79 [T25+2H]2+ | 306 GWAGLQSFPR 315 |
| 602.8 | 2+ | 602.82 [T47+2H]2+ | 530 NAAFVVDQNVK 540 |
| 829.4 | 2+ | 829.43 [T42+2H]2+ | 474 NLIDHSIIESFGAGGK 489 |
| 921.95 | 2+ | 921.94 [T40+2H]2+ | 450 SNIDTTSYGAFVDIDPK 466 |
| 1,066.5 | 2+ | No match | T6 N-linked glycopeptide:116 NLSDPFLR 123 (Deoxyhexose)1 (Pentose)1 (Man)3 (GlcNAc)2 |
Glycosylation sites NX(S/T) are presented in bold. The position of the fragments in the mature 1-FEH IIb protein are indicated.
The 1-FEH IIa Gene also Contains the Typical Invertase Mini-Exon
Most invertases (Sturm, 1999) contain a mini-exon (exon 2) that encodes for the tripeptide DPN of the conserved motif W(I/M) NDPNG (Pons et al., 1998). For chicory 1-FEH IIa we performed PCR on genomic DNA by using a combination of undegenerated versions of primers P1 and P5, followed by nested PCR with primers P3 and P5. This resulted in a genomic fragment that was subcloned and sequenced at its 5′ part. The fragment contained the 3′ part of the first exon, followed by a first AT-rich (62%) intron of 264 bp, the 9-bp mini-exon 2, and finally, part of a second intron (not fully sequenced). Similar data were obtained for chicory 1-FEH I (not shown).
Homology to Other Glycosyl Hydrolases
As observed for chicory 1-FEH I (Van den Ende et al., 2000a), the cDNA-derived amino acid sequences of 1-FEH IIa and IIb are more homologous to cell wall invertases (43%–59% identical amino acids) than to vacuolar invertases (40%–41% identical) and fructan biosynthetic enzymes (33%–39% identical). Homologies to microbial fructan hydrolases are much lower (13%–24% identical). 1-FEH IIa and IIb are 94% homologous. Homology to chicory 1-FEH I is much lower: 51% for 1-FEH IIa and 50% for 1-FEH IIb.
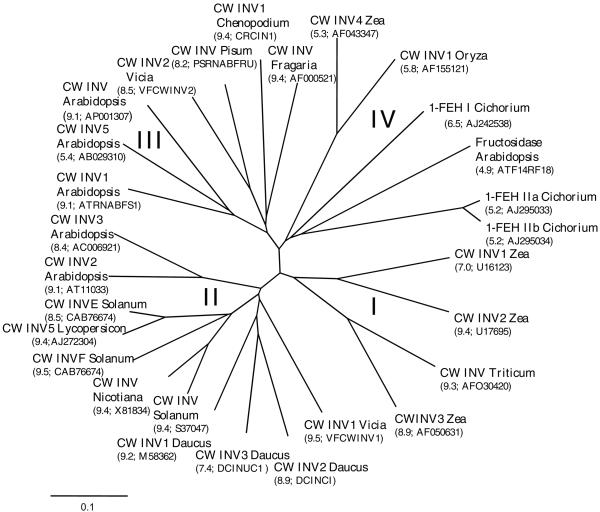
An unrooted radial tree of some members of cell wall-type glycosyl hydrolases is presented in Figure 3. Four distinct groups can be discerned: the first group (I) contains monocotyledonous and mainly basic cell wall invertases. The second group (II) contains dicotyledonous, basic cell wall-type invertases. A third group (III) also contains dicotyledonous and mainly basic cell wall-type invertases, but there is one exception: an acid cell wall invertase from Arabidopsis (CW INV5). A fourth group (IV) contains monocotyledonous and dicotyledonous enzymes, and harbors the three chicory 1-FEH cDNAs. Within this group, all members have an acidic pI.
Figure 3.
Unrooted phylogenetic tree containing cell wall-type invertase-like cDNA-derived amino acid sequences. Four groups can be discerned. I, Maize cell wall invertase (CW INV) 1, 2, and 3; and wheat CW INV. II, Broad bean CW INV1; carrot CW INV 1, 2, and 3; potato CW INV1, E, and F; tomato CW INV5; and Arabidopsis CW INV2 and 3. III, Arabidopsis CW INV, CW INV1, and 5; broad bean CW INV2; pea CW INV; and Chenopodium rubrum CWINV 1 and Fragaria × ananassa CW INV; IV, Maize CW INV4; rice CW INV; Arabidopsis fructosidase; and chicory 1-FEH I, IIa, and IIb. pIs, as well as cDNA names or accession numbers, are given between brackets in the figure. The scale bar indicates a distance value of 0.1.
Q-TOF Analyses on 1-FEH IIa and IIb Tryptic Fragments
Theoretical tryptic digests on the cDNA-derived 1-FEH IIa and IIb protein sequences yielded 48 peptides for both isoforms (designated T1–T48 from N to C terminus). The two independent 1-FEH IIa (Uno S fraction 36) and 1-FEH IIb (Uno S fraction 32) isoforms were gel purified. Masses of ZipTip-eluted tryptic peptides were determined by Q-TOF and were compared (Tables I and II) with the masses of theoretical peptides (with the consideration of one possible missed cleavage site). All except four masses detected matched, within the acceptable mass measurement error of ±1 D, with one of the theoretical fragments (Tables I and II). CID MS/MS analysis yielded a number of sequence tags (Mann and Wilm, 1994), which proved the identity of the tryptic peptides (Tables I and II). Special attention was given to the unexplained 1,066.5 [2+], 892.05 [3+], and 1,234.25 [3+] fragments. After fragmentation they all proved to be glycosylated peptides. One of these fragments (892.05 [3+]) contains one missed cleavage site (Table I). These fragments fit perfectly well with two out of three potential N-glycosylation sites (see Fig. 2B). Further detailed analysis revealed the carbohydrate composition of the respective glycosyl rests (Tables I and II).
1-FEH II Expression
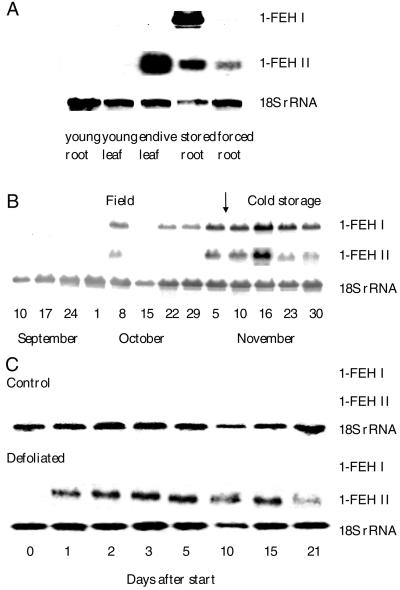
1-FEH IIa and IIb cDNAs are very homologous (92% identity at the DNA level) and hence, no specific probes could be developed to differentiate between the two isoforms (cross-hybridization of probes during Southern analysis at 68°C, data not shown). Therefore, during northern analysis we only used a 1-FEH IIa probe. We verified that 1-FEH IIa and 1-FEH I probes did not cross-hybridize (not shown). From Figure 4A, it is clear that chicory 1-FEH II is abundantly expressed in leaves of Belgian endives, but also in the cold-stored (vernalized) and forced chicory roots, whereas no signals were found in leaves and roots of very young chicory plants. The pattern is different from 1-FEH I, which is mainly expressed in cold-stored roots and not at all in Belgian endive leaves.
Figure 4.
Expression of chicory 1-FEH genes. A, Northern blot containing RNA from a 7-week-old chicory root, a 7-week-old chicory leaf, a mature chicory root after 1 week of cold storage, and a forced chicory root after 3 weeks of forcing. B, Northern blot containing RNA from mature field-grown chicory roots between September 10 and November 5 and subsequent storage for 3 weeks in cold room (arrow indicates start of storage). Sampling dates are presented on the figure. C, Northern blot containing 10 μg RNA from the roots of control and defoliated young plantlets. Chicory root 1-FEH I, IIa, and 18S rRNA were used as probes.
In field-grown roots throughout the growing season, the expression pattern of chicory 1-FEH II is similar to that of 1-FEH I, although expression seems to be induced at a later stage of development. The strongest signals for 1-FEH I and 1-FEH II mRNA were obtained when plants were defoliated and roots were stored in a cold room for 1 week (Fig. 4B).
Figure 4C shows the expression of chicory 1-FEH I and II after defoliation of young chicory plants. No expression of 1-FEH I and 1-FEH II could be detected in roots of control plants. However, in roots of defoliated plants, expression of 1-FEH IIa was rapidly initiated (after 1 d), whereas no expression of 1-FEH I became apparent. Because plant regrowth after defoliation was slow, 1-FEH IIa expression remained high until d 15 after start and only declined on the last sampling date.
DISCUSSION
In this paper we describe the isolation of two highly homologous cDNAs encoding chicory root 1-FEH IIa and IIb. We purified the two independent isoforms from Belgian endive leaves (Fig. 1) and from forced chicory roots to deduce their N-terminal protein sequences and perform molecular mass analyses on their tryptic fragments. For cloning, the use of N-terminal and conserved degenerate primers proved a successful strategy to isolate partial 1-FEH II cDNAs. 5′- and 3′-RACE RT-PCR were used to obtain the complete 1-FEH II cDNAs (Fig. 2). Q-TOF MS/MS analyses on the two independent 1-FEH II isoforms (Tables I and II) convincingly demonstrated the link between the cDNAs obtained and the biologically active 1-FEH II enzymes (Fig. 1) by approving 35% (FEH IIa) or 21% (FEH IIb) of the cDNA-derived sequence information (Tables I and II; Fig. 2B).
Chicory 1-FEH cDNAs group together with cell wall-type enzymes (Fig. 3) and not with vacuolar invertases and fructosyl transferases (Van den Ende et al., 2000a). Genes encoding plant fructan biosynthetic enzymes (1-SST, 1-FFT, Suc:fructan 6-fructosyl transferase, and fructan:fructan 6G fructosyl transferase) seem to have evolved from ancestral vacuolar invertase genes by relatively few mutational changes (Wiemken et al., 1995). On the contrary, genes encoding plant 1-FEHs seem to have evolved from an ancestral cell wall invertase gene. Four subgroups of cell wall invertases become apparent (Fig. 3), suggesting multiple duplication of the ancestral cell wall invertase gene. One subgroup containing the chicory 1-FEHs (IV, Fig. 3) can clearly be discerned from the other groups by their acidic pIs and the presence of putative C-terminal vacuolar-targeting signals (see Van den Ende et al., 2000a). This suggests an evolutionary pressure toward the development of “cell wall-like” vacuolar FEHs and/or invertases. The reasons why chicory 1-FEHs have evolved from a cell wall-type ancestral invertase and not from a vacuolar invertase gene are far from clear. It can be speculated that cell wall invertase genes might have been better candidates for the development of genuine 1-FEHs, being capable of using oligo- or polysaccharides instead of Suc as fructosyl donors. In this regard, it has been demonstrated that native purified extracellular invertases from carrot show a much higher cleavage rate for raffinose than carrot vacuolar enzymes (Unger et al., 1994 and refs. therein). Moreover, it was recently demonstrated that the different substrate specificity between vacuolar invertases (containing the conserved WECVD motif) and extracellular invertases (containing the conserved WECPD motif) was fully determined by a single amino acid substitution (P to V; Goetz and Roitsch, 1999). All members of group IV (Fig. 3) contain a P in this conserved motif.
By vacuole isolation experiments from protoplasts and enzymatic activity measurements, it has long been demonstrated that fructan and fructan metabolizing enzymes, including FEH, are located in the vacuolar sap (Wiemken et al., 1986). However, in a recent report, fructan and FEH activity were also found in the crown apoplast of winter oat (Livingston and Henson, 1998), suggesting the presence of FEH outside the cell under certain circumstances. We have purified chicory 1-FEH IIa and IIb in roughly the same way as the soluble, vacuolar 1-SST and 1-FFT enzymes (same amount of starting material, same extraction buffer, removal of cell wall fraction by centrifugation, and a similar purification strategy) and we found about the same amount of pure protein. In a similar manner as described for chicory 1-FEH I (Van den Ende et al., 2000a), we attempted to purify the 1-FEH II enzymes from apoplastic fluid (prepared as in Boller and Métraux, 1988; Isla et al., 1999) as well as from total root extracts. No significant amounts higher than could be explained by cellular leakage were detected in the apoplastic fluid (data not shown), suggesting that the 1-FEH II enzymes are localized in the vacuole.
These observations are in conflict with the data obtained for INCW4 of maize, another representative of group IV (Fig. 3) of acidic cell wall-type enzymes (Kim et al., 2000). The authors suggested that INCW4 could be a new type of cell wall invertase present in a free form in the apoplast, although their results do not fully exclude a vacuolar localization. Immunocytochemical methods might help corroborating in the cellular localization the chicory 1-FEH enzymes or other representatives of group IV (Fig. 3).
The widely used criteria to discriminate between vacuolar and extracellular enzymes (extracellular enzymes have a basic pI and vacuolar enzymes have an acid pI; extracellular enzymes have WECPD and vacuolar enzymes have WECVD; Goetz and Roitsch, 1999) perhaps need to be reconsidered. Besides the different pI and putative vacuolar localization of group IV glycosyl hydrolases, their genomic organization resembles that of typical invertases (presence of a mini-exon in 1-FEH IIa and 1-FEH I, data not shown).
Chicory 1-FEH II is highly expressed in Belgian endive leaves and in forced roots (Fig. 4A). Throughout the 1998 growing season (Fig. 4B), 1-FEH I was expressed earlier than 1-FEH II in field-grown chicory roots, but both 1-FEH genes were heavily expressed when roots were harvested and cold stored. As a consequence, manipulation of fructan catabolism in commercial chicory roots would not only imply the control of chicory 1-FEH I, but also of 1-FEH IIa and IIb. Increased FEH activity results in massive breakdown of fructan and production of Fru and inulo-n-oses (Van den Ende et al., 1996a, 1996b). The latter phenomena are to be avoided for industrial fructan production.
We earlier demonstrated that a sudden 1-FEH activity increase and fructan breakdown could be induced in young chicory roots after defoliation (De Roover et al., 1999b). Due to the loss of photosynthate (Suc) entering the root, the root has to switch abruptly from a sink to a source organ since substrates and energy are needed for general maintenance and for sustaining the leaf meristematic tissues that remain a strong sink for carbohydrate. We show here that chicory 1-FEH II and not chicory 1-FEH I is responsible for the observed 1-FEH activity after defoliation (Fig. 4C). This indicates that 1-FEH II very likely can be considered as a “survival” enzyme that can be induced at any physiological stage or whenever energy demands greatly increase. This can for instance be the case in periods when an increased nitrogen uptake, reduction and metabolism of nitrogen occur (Van den Ende et al., 1999), or when a fast regrowth is necessary (early spring regrowth, flowering, or after grazing, mowing, or other physical damage to the photosynthetic apparatus).
From Figure 4, it can be concluded that chicory 1-FEH I and IIa are regulated in a different way. The 1-FEH II genes might be induced by a sudden drop of photosynthate (Suc?) entering the root (see also Mino et al., 1978). It is apparent that the same trigger is not able to induce chicory 1-FEH I, at least not in young roots. It was previously suggested that cold is an essential trigger for 1-FEH I induction (Van den Ende et al., 1996b, 2000a). Although chicory 1-FEH II now has been shown to provide the energy for leaf regrowth after defoliation, the role of 1-FEH I induction in mature field-grown chicory roots during autumn is still obscure since no large energy supplies are necessary at this moment. In this case it can be speculated that the shift from higher to lower degree of polymerization fructan by the concerted action of 1-FEH I and 1-FFT might in some way protect the plants against frost damage.
Further research is necessary to understand how and why fructan hydrolases have evolved from cell wall invertases, where they are localized, and how they are regulated throughout different developmental stages. Other types of FEH, controlled in a different way, may be present in nature, e.g. the FEH that is responsible for the rapid breakdown of fructan in flower petals driving flower opening by osmotic forces (Bieleski, 1993; Vergauwen et al., 2000).
In conclusion, we have cloned two different 1-FEH II cDNAs that encode two independent enzymatically active 1-FEH II isoforms. The cloning of chicory 1-FEH II cDNAs is interesting from a scientific and an agro-economical point of view. Like chicory 1-FEH I, 1-FEH IIa and IIb are unexpectedly related to cell wall invertases, although a vacuolar localization is proposed. Our data indicate that chicory 1-FEH I and 1-FEH II are expressed under different conditions and are regulated in a different way, but further research is necessary to understand the fine-tuning of the gene expression and regulation. 1-FEH II is strongly induced in roots after defoliation, suggesting that 1-FEH II can be considered a “survival” enzyme that is induced at times when energy demands largely increase.
MATERIALS AND METHODS
Plant Material
Chicory (Cichorium intybus var. foliosum cv Flash) was sown in a local field with sandy, loamy soil on May 18, 1998. Since the induction of 1-FEH activity in field-grown chicory roots coincides with low temperatures around mid-October (Van den Ende and Van Laere, 1996; Van den Ende et al., 1996b), field-grown chicory roots were collected weekly between September 10 and November 5. Afterward, on November 8, plants were uprooted, defoliated, and stored for 3 weeks at 1°C. Throughout storage, root samples were taken on November 10, 16, 23, and 30. Roots were subsequently forced (start December 1), as described in Van den Ende et al. (1996b). After 3 weeks, samples were taken from Belgian endive leaves, as well as from the forced roots on which they were grown. Similar material was used for enzyme purification purposes.
Plants were also grown in a controlled growth chamber as described in De Roover et al. (2000). Seven weeks after sowing, a number of young plants were defoliated and root samples were taken at 0, 1, 2, 3, 5, 10, 15, and 21 d after defoliation. Intact chicory seedlings were used as control.
Purification of 1-FEH IIa and IIb and N-Terminal Sequencing
1-FEH IIa and IIb were purified from Belgian endive leaves and also from forced roots as described (De Roover et al., 1999a) except that an additional Uno S step at pH 5.2 was performed to fully separate the two different isoforms. 1-FEH IIa (Uno S fraction 36) and IIb (Uno S fraction 32) were subjected to SDS-PAGE, blotted on a polyvinylidene difluoride membrane, and their N-termini were sequenced by automated Edman degradation. FEH activities were measured as described in Van den Ende et al. (1999).
Q-TOF Analyses on FEH II Tryptic Fragments
The two different SDS-PAGE protein bands of 1-FEH IIa (Uno S fraction 36) and IIb (Uno S fraction 32) exhibiting 1-FEH activity were also subjected to MS identification. The Coomassie Brilliant Blue-stained proteins were excised from the gel, thoroughly washed in water and 50% (v/v) acetonitrile, respectively, and dried by vacuum centrifugation. The gel pieces were subsequently reswollen and incubated for 16 h at 37°C in 30 μL of 25 mm NH4HCO3 (pH 8.0) containing 10% (v/v) acetonitrile and 0.1 μg of trypsin (sequencing grade-modified porcine trypsin, Promega, Madison, WI). The resulting tryptic fragments were extracted twice with 50 μL of 7.5% (v/v) acetonitrile/0.1% (v/v) trifluoracetic acid (TFA) and were desalted using ZipTip C18 pipette tips (Millipore, Bedford, MA). The ZipTips were subsequently washed in 100% (v/v) acetonitrile (HPLC grade, Riedel-De Haën, Seelze, Germany) and 50% (v/v) acetonitrile with 0.1% (v/v) TFA in water. The tips were then equilibrated in 0.1% (v/v) TFA. The peptide extract was bound to the C18 silica by repeated pipetting. Repeated washing and desalting of the peptides was done in 0.1% (v/v) TFA followed by 0.1% (v/v) formic acid in water. The peptides were eluted in 3 μL of 60% (v/v) acetonitrile-1% (v/v) formic acid in water. This solution was deposited into a gold-coated borosilicate nanoflow needle (Protana, Odense, Denmark) that was fitted in the nano-electrospray source of a hybrid quadrupole-TOF electron spray ionization instrument (Q-TOF by Micromass, Wythenshawe, UK). An MS survey spectrum was acquired, after which individual peptides were selected in the instrument's quadrupole and subjected to CID, yielding MS/MS spectra of the fragmented peptides. Sequence information was derived from the MS/MS spectra with the aid of the MaxEnt 3 software (deconvoluting and de-isotoping of data) as well as the PepSeq component of the Biolynx software package (Micromass).
RNA Isolation, RT-PCR, and Subcloning
Total RNA was isolated from forced roots and from Belgian endive leaves by using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). Based on the N-terminal amino acid sequence of the purified 1-FEH IIa (QQIEQPYRTGYHFQP) and 1-FEH IIb (QQIQQPYRTGYHFQP), we constructed three degenerated sense primers: P1 (5′-CARCARATHGARCARCC-3′), P2 (5′- CARCARATHCARCARCC-3′), and P3 (5′-GGNTAYCAYTTYCARCC-3′). The conserved amino acid sequence MWECVD in vacuolar invertases and fructosyl transferases was used to make the antisense primer, P4 (5′-GTANARRTCNACRCAYTCCCACAT-3′). The conserved amino acid sequence ECPDF in cell wall invertases was used to create the antisense primer, P5 (5′- GGGWAMARRTCNGGRCAYTCC-3′). One-step RT-PCR was performed (Access RT-PCR System, Promega) by using the combinations P1-P4, P1-P5, P2-P4, and P2-P5. It was surprising that only weak bands of about 600 bp were produced for the P1-P5 and P2-P5 combinations. “Partial touchdown RT-PCR” on total RNA was used under the following conditions: 94°C for 3 min followed by 10 cycles at 94°C for 40 s and 52°C for 40 s (with temperature decrease of 0.5°C each cycle), and 72°C for 1 min, and then 25 more cycles at 94°C for 40 s, 47°C for 40 s (constant), and 72°C for 1 min. Final extension was at 72°C for 10 min (PCR Access kit, Promega). Semi-nested PCR was subsequently performed on these PCR products with primers P3 and P5. PCR conditions were identical. The resulting PCR fragments were ligated in the Topo-TA vector and transformed to Escherichia coli (Topo-TA cloning kit, Invitrogen, Groningen, The Netherlands). Plasmid was extracted using Wizard Plus SV Minipreps (Promega). Partial sequencing yielded two types of clones (named pFEH IIa and pFEH IIb).
5′- and 3′-RACE
The 3′ parts of both cDNAs were found by first performing one-step RT-PCR (see above) with P6 (5′- ACCAATACAATCCGTATGCAG -3′) and an oligo dT-based antisense primer (5′-CTCGCTCGCCCAT27–3′). Two specific primers were subsequently chosen for each clone: P7 (5′- ACAATCCGTATGCAGCAACG -3′) for FEH IIa and P8 (5′-ACAATCCGTATGCAGCAACC-3′) for FEH IIb. Semi-nested PCR was then performed (P7-oligo dT; P8-oligo dT), resulting in PCR products of about 1,800 bp that were subcloned and from which the 3′ parts were sequenced. A P5-derived 5′-phosphorylated antisense primer was then constructed (5′-P-GGGTAAAARTCCGGGCACTCC-3′). Total RNA was reverse-transcribed with this primer and was ligated to obtain single-stranded circular cDNA (5′-Full RACE Core Set, TaKaRa, Otsu, Shiga, Japan). This mixture was used in a PCR reaction (LA PCR TaKaRa) with the primers P6 and P9 (5′-GTCCATTGGGATCGTTCATC-3′). A second semi-nested PCR with P7-P9 (1-FEH IIa) or P8-P9 (1-FEH IIb) resulted in a few bands. The largest ones were excised from the gel and PCR products were recovered as described in the Wizard PCR preps protocol (Promega). After subcloning and sequencing, the 5′ part of 1-FEH IIa and IIb cDNAs were determined.
Generation of Full-Length cDNA
Based on the sequences of 5′- and 3′-RACE products, full-length cDNAs were obtained by choosing specific primers at the extreme 5′ and 3′ parts. For 1-FEH IIa, P10 (5′-CACACACTCATCATGAAGAAATCA-3′) and P11 (5′-TTTTTTTGCTCA-AATATTGTAGTTGTA-3′) were chosen. For 1-FEH IIb, P12 (5′-CACACACTCATC-ATGAAGAAATCT-3′) and P13 (5′-TTTAATGAAACAATAATTTGTTACAATG-3′) were chosen. First, total RNA was reverse-transcribed with AMV Reverse Transcriptase XL (TaKaRa) using P12 and P13, respectively, at 50°C for 1 h. A number of independent PCRs were then performed with P10-P11 and P12-P13, respectively. Proofreading Pfu DNA polymerase was used (Promega). PCR conditions included 35 cycles at 94°C for 30 s; 59°C for 40 s, and 72°C for 5 min. After A-tailing with dATP, fragments were ligated in TOPO-XL vector (Invitrogen). The sequences were deposited in the EMBL sequence library (accession no. AJ295033 for 1-FEH IIa and AJ295034 for 1-FEH IIb).
Sequencing and Computer Analysis
From a number of clones, the full-length 1-FEH IIa and 1-FEH IIb fragments were fully sequenced on both DNA strands by Eurogentec (Seraing, Belgium) using the BigDye Terminator technology (Applied Biosystems, Foster City, CA). General DNA and protein sequence analyses were carried out with the DNASIS 1.2 software (Hitachi, Alameda, CA ). For the construction of a phylogenetic tree and multiple alignments, sequences were extracted from EMBL, PIR, and SWISSPROT databases and were sent to CLUSTAL W from which the results were exported to the TREEVIEW program for construction of a phylogenetic tree.
Probe Preparation and Northern Analysis
PCR with primer combinations P14 (5′-GGTGTACGGCGGCAGTG-3′) and P15 (5′-TTTTGAGTCCCATTATTGAACA-3′) for 1-FEH IIa or P14 and P16 (5′-TTTTGAGTCCCGTTATTGAACG-3′) for 1-FEH IIb was performed for probe preparation. The 850-bp PCR products were further purified on a PCR-Wizard column (Promega) and were then labeled by a random-primed method using the DNA-labeling T7 QuickPrime Kit (Pharmacia Biotech, Piscataway, NJ) and [α-32P] dCTP, as described in Feinberg and Vogelstein (1984). 18S ribosomal probe was prepared as described (De Roover et al., 2000).
RNA from 1 g of frozen plant material was extracted using an RNA extraction kit (TR-118, Euromedex, France). Total RNA (10 μg) was pretreated, blotted, and hybridized as earlier described (Van den Ende et al., 2000b). To check whether equal amounts of RNA were loaded in each well, the membrane was also hybridized with the radiolabeled chicory 18S rRNA probe.
ACKNOWLEDGMENT
The authors want to thank E. Nackaerts for his technical assistance.
Footnotes
This work was supported by the Fund for Scientific Research, Flanders (grant to W.V.d.E., S.P.C., and J.D.R.).
LITERATURE CITED
- Bieleski RL. Fructan hydrolysis drives petal expansion in the ephemeral daylily flower. Plant Physiol. 1993;103:213–219. doi: 10.1104/pp.103.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Métraux JP. Extracellular localization of chitinase in cucumber. Physiol Mol Plant Pathol. 1988;33:11–16. [Google Scholar]
- Burne RA, Wen ZT, Chen YY, Penders JE. Regulation of expression of the fructan hydrolase gene of streptococcus mutans GS-5 by induction and carbon catabolite repression. J Bacteriol. 1999;181:2863–2871. doi: 10.1128/jb.181.9.2863-2871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens G, Van Laere A, De Proft M. Purification and properties of an inulinase from chicory roots (Cichorium intybus L.) J Plant Physiol. 1990;136:35–39. [Google Scholar]
- De Roover J, De Winter M, Van Laere A, Timmermans JW, Van den Ende W. Purification and properties of a second fructan exohydrolase from the roots of Cichorium intybus L. Physiol Plant. 1999a;106:28–34. [Google Scholar]
- De Roover J, Van den Branden K, Van Laere A, Van den Ende W. Drought induces fructan synthesis and 1-SST (sucrose:sucrose 1-fructosyl transferase) in roots and leaves of chicory seedlings (Cichorium intybus L) Planta. 2000;210:808–814. doi: 10.1007/s004250050683. [DOI] [PubMed] [Google Scholar]
- De Roover J, Van Laere A, Van den Ende W. Effect of defoliation on fructan pattern and fructan metabolizing enzymes in chicory roots (Cichorium intybus) Physiol Plant. 1999b;106:158–163. [Google Scholar]
- Edelman J, Jefford TG. The mechanism of fructosan metabolism in higher plants as exemplified in Helianthus tuberosus. New Phytol. 1968;67:517–531. [Google Scholar]
- Feinberg AP, Vogelstein B. Addendum: a technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Frehner M, Keller F, Wiemken A. Localization of fructan metabolism in the vacuoles isolated from protoplasts of Jerusalem artichoke. J Plant Physiol. 1984;116:197–208. doi: 10.1016/S0176-1617(84)80089-9. [DOI] [PubMed] [Google Scholar]
- Fuchs A. Current and potential food and non-food applications of fructans. Biochem Soc Trans. 1991;19:555–560. doi: 10.1042/bst0190555. [DOI] [PubMed] [Google Scholar]
- Godt DE, Roitsch T. Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol. 1997;115:273–282. doi: 10.1104/pp.115.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Roitsch T. The different pH optima and substrate specificities of extracellular and vacuolar invertases from plants are determined by a single amino acid substitution. Plant J. 1999;20:707–711. doi: 10.1046/j.1365-313x.1999.00628.x. [DOI] [PubMed] [Google Scholar]
- Hendry G. Evolutionary origins and natural functions of fructans: a climatological, biogeographic and mechanistic appraisal. New Phytol. 1993;123:3–14. [Google Scholar]
- Henson CA, Livingston DP. Purification and characterization of an oat fructan exohydrolase that preferentially hydrolyzes β-2,6 fructans. Plant Physiol. 1996;110:639–644. doi: 10.1104/pp.110.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson CA, Livingston DP. Characterization of a fructan exohydrolase purified from barley stems that hydrolyzes multiple fructofuranosidic linkages. Plant Physiol Biochem Paris. 1998;36:715–720. [Google Scholar]
- Heyer AG, Lloyd JR, Kossmann J. Production of modified polymeric carbohydrates. Curr Opin Biotechnol. 1999;10:169–174. doi: 10.1016/s0958-1669(99)80030-5. [DOI] [PubMed] [Google Scholar]
- Hincha DK, Hellwege EM, Heyer AG, Crowe JH. Plant fructans stabilize phosphatidylcholine liposomes during freeze-drying. Eur J Biochem. 2000;267:535–540. doi: 10.1046/j.1432-1327.2000.01028.x. [DOI] [PubMed] [Google Scholar]
- Isla MI, Vattuone MA, Ordóñez RM, Sampietro AR. Invertase activity associated with the walls of Solanum tuberosum tubers. Phytochemistry. 1999;50:525–534. doi: 10.1016/s0031-9422(98)00474-9. [DOI] [PubMed] [Google Scholar]
- Kang SI, Kim SI. Molecular cloning and sequence analysis of an endo-inulinase gene from Arthrobacter sp. Biotechnol Lett. 1999;21:569–574. [Google Scholar]
- Kim J-Y, Mahé A, Guy S, Brangeon J, Roche O, Chourey PS, Prioul J-L. Characterization of two members of the maize gene family, Incw3 and Incw4, encoding cell-wall invertases. Gene. 2000;245:89–102. doi: 10.1016/s0378-1119(00)00034-2. [DOI] [PubMed] [Google Scholar]
- Livingston DP, Chatterton NJ, Harrison PA. Structure and quantity of fructan oligomers in oat (Avena spp.) New Phytol. 1993;123:725–734. [Google Scholar]
- Livingston DP, Henson CA. Apoplastic sugars, fructans, fructan exohydrolase, and invertase in winter oat: responses to second-phase cold hardening. Plant Physiol. 1998;116:403–408. [Google Scholar]
- Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- Marx SP, Nösberger J, Frehner M. Seasonal variation of fructan-β-fructosidase (FEH) activity and characterization of a β-(2, 1)-linkage specific FEH. New Phytol. 1997a;135:267–277. [Google Scholar]
- Marx SP, Nösberger J, Frehner M. Hydrolysis of fructan in grasses: a β-(2, 6)-linkage specific fructan-β-fructosidase from stubble of Lolium perenne. New Phytol. 1997b;135:279–290. [Google Scholar]
- Mino Y, Shimada A, Yamatoto S. Effect of cutting height on the carbohydrate metabolism in the haplocorm of timothy (Phleum pratense L.) J Jap Grass Sci. 1978;24:34–39. [Google Scholar]
- Pilon-Smits EAH, Ebskamp MJM, Jeuken MJW, Weisbeek PJ, Smeekens SCM. Improved performance of transgenic fructan-accumulating tobacco under drought stress. Plant Physiol. 1995;107:125–130. doi: 10.1104/pp.107.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons T, Olmea O, Chinea G, Beldarraín A, Márquez G, Acosta N, Rodríguez L, Valencia A. Structural model for family 32 of glycosyl-hydrolase enzymes. PROTEINS. 1998;33:383–395. doi: 10.1002/(sici)1097-0134(19981115)33:3<383::aid-prot7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Roberfroid MB, Van Loo JAE, Gibson GR. The bifidogenic nature of chicory inulin and its hydrolysis products. J Nutr. 1998;128:11–19. doi: 10.1093/jn/128.1.11. [DOI] [PubMed] [Google Scholar]
- Shiomi N. Properties of fructosyltransferases involved in the synthesis of fructan in Liliaceous plants. J Plant Physiol. 1989;134:151–155. [Google Scholar]
- Sturm A. Invertases: primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121:1–7. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taper H, Lemort C, Roberfroid MB. Inhibition of dietary inulin and oligofructose on the growth of transplantable mouse tumor. Anticancer Res. 1999;18:4123–4126. [PubMed] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M. Expression of the Arabidopsis thaliana invertase gene family. Planta. 1998;207:259–265. doi: 10.1007/s004250050481. [DOI] [PubMed] [Google Scholar]
- Unger C, Hardegger M, Lienhardt S, Sturm A. cDNA cloning of carrot (Daucus carota) soluble acid β-fructofuranosidases and comparison with the cell wall isoenzyme. Plant Physiol. 1994;104:1351–1357. doi: 10.1104/pp.104.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W, De Roover J, Van Laere A. In vitro synthesis of fructofuranosyl-only oligosaccharides from inulin and fructose by purified chicory root fructan:fructan fructosyl transferase. Physiol Plant. 1996a;97:346–352. [Google Scholar]
- Van den Ende W, De Roover J, Van Laere A. Effect of nitrogen concentration on fructan and fructan metabolizing enzymes in young chicory plants (Cichorium intybus) Physiol Plant. 1999;105:2–8. [Google Scholar]
- Van den Ende W, Michiels A, De Roover J, Verhaert P, Van Laere A. Cloning and functional analysis of chicory root fructan 1-exohydrolase I (1-FEH I): a vacuolar enzyme derived from a cell-wall invertase ancestor? Mass fingerprint of the 1-FEH I enzyme. Plant J. 2000a;24:447–456. doi: 10.1046/j.1365-313x.2000.00890.x. [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, Van Wonterghem D, Vergauwen R, Van Laere A. Cloning, developmental and tissue-specific expression of 1-SST (sucrose:sucrose 1-fructosyl transferase) from Taraxacum officinale: fructan localization in roots. Plant Physiol. 2000b;123:71–80. doi: 10.1104/pp.123.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W, Mintiens A, Speleers H, Onuoha A, Van Laere A. The metabolism of fructans in roots of Cichorium intybus L. during growth, storage and forcing. New Phytol. 1996b;132:555–563. doi: 10.1111/j.1469-8137.1996.tb01874.x. [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Van Laere A. Fructan synthesizing and degrading activities in chicory roots (Cichorium intybus L.) during growth, storage and forcing. J Plant Physiol. 1996;149:43–50. [Google Scholar]
- Vergauwen R, Van den Ende W, Van Laere A. The role of fructans in flowering of Campanula rapunculoides. J Exp Bot. 2000;51:1261–1266. [PubMed] [Google Scholar]
- Vijn I, Smeekens SCM. Fructan: more than a reserve carbohydrate? Plant Physiol. 1999;120:351–359. doi: 10.1104/pp.120.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemken A, Frehner M, Keller F, Wagner W. Fructan metabolism, enzymology and compartmentation. Curr Topics Plant Biochem Physiol. 1986;5:17–37. [Google Scholar]
- Wiemken A, Sprenger N, Boller T. Fructans: an extension of sucrose by sucrose. In: Pontis HG, Salerno GL, Echeverria EJ, editors. Current Topics in Plant Physiology. Vol. 14. Rockville, MD: American Society of Plant Physiologists; 1995. pp. 179–189. [Google Scholar]