Abstract
Purpose
We sought to determine the role and retinal cellular location of microRNA-124 (miR-124) in a neuroinflammatory model of retinal degeneration. Further, we explored the anti-inflammatory relationship of miR-124 with a predicted messenger RNA (mRNA) binding partner, chemokine (C-C motif) ligand 2 (Ccl2), which is crucially involved in inflammatory cell recruitment in the damaged retina.
Methods
Human AMD donor eyes and photo-oxidative damaged (PD) mice were labeled for miR-124 expression using in situ hybridization. PDGFRa-cre RFP mice were used for Müller cell isolation from whole retinas. MIO-M1 immortalized cells and rat primary Müller cells were used for in vitro analysis of miR-124 expression and its relationship with Ccl2. Therapeutic efficacy was tested with intravitreal administration of miR-124 mimic in mice, with electroretinography used to determine retinal function. IBA1 immunohistochemistry and photoreceptor row counts were used for assessment of inflammation and cell death.
Results
MiR-124 expression was correlated with progressive retinal damage, inflammation, and cell death in human AMD and PD mice. In addition, miR-124 expression was inversely correlated to Ccl2 expression in mice following PD. MiR-124 was localized to both neuronal-like photoreceptors and glial (Müller) cells in the retina, with a redistribution from neurons to glia occurring as a consequence of PD. Finally, intravitreal administration of miR-124 mimics decreased retinal inflammation and photoreceptor cell death, and improved retinal function.
Conclusions
This study has provided an understanding of the mechanism behind miR-124 in the degenerating retina and demonstrates the usefulness of miR-124 mimics for the modulation of retinal degenerations.
Keywords: retinal degeneration, microRNA, gene therapy, AMD
MicroRNAs (miRNAs) are endogenous short non-coding single-stranded molecules, approximately 18 to 25 nucleotides in length, which play important roles in the regulation of posttranscriptional gene expression.1 MiRNAs bind to the 3′untranslated (3′UTR) regions of specific target messenger RNA (mRNA) and either repress or halt transcription. Due to the similarity in many 3′UTR regions, a single miRNA can regulate multiple genes, often within the same biochemical pathway.2,3 The strong regulatory properties of miRNA have been found in a myriad of biological systems including the eye (reviewed in Ref. 4) and can affect multiple biological processes including development, differentiation, apoptosis, cell proliferation, neuronal cell fates, and inflammation.5,6 As miRNAs are potent regulators of transcription, their dysregulation plays a critical role in the pathogenesis of inflammatory diseases,7–9 cancer,10 neurological disorders,9,11–13 and more recently in the pathogenesis of AMD.14
AMD is a progressive retinal degeneration characterized by the death of photoreceptors and retinal pigment epithelial (RPE) cells, which leads to loss of central vision.15,16 This disease has a multifaceted etiology; however, it is well understood that both activation of the innate immune system and recruitment of inflammatory cells to the retina are significant factors in the development and progression of AMD.17,18 Gene expression studies of AMD donor eyes have shown that a number of chemokines, including chemokine (C-C motif) ligand 2 (CCL2 or MCP-1), are up-regulated in both the neovascular and atrophic forms of AMD.19 CCL2 is a potent microglia/macrophage chemoattractant that is produced and released from Müller cells in response to retinal oxidative damage.20 Down-regulation of CCL2 has been shown previously to reduce photoreceptor cell death in animal models of retinal degeneration.21–23 Therefore, CCL2 represents an ideal therapeutic target to slow the progression of retinal diseases.
Multiple miRNAs have been identified that regulate the immune response,24–26 including miR-124, which has been predicted and reported to modulate Ccl2 expression in rheumatoid arthritis,27 in retinal ganglion cells after Amadori-glycated albumin-induced inflammation,28 and in oral carcinoma tumors.29 MiR-124 is abundant in the neurons of the central nervous system (CNS) and retina30–32 and plays an important role in modulating the inflammatory component of CNS diseases, including Alzheimer's and Parkinson's disease.33,34 The anti-inflammatory properties of miR-124 in neurodegenerative diseases, coupled with its abundance in neurons and validated binding of Ccl2, suggests a role in the progression of retinal degenerations.
In this study, we have demonstrated modulation of miR-124 expression in (1) human AMD tissue, (2) an inflammatory mouse model of photo-oxidative retinal degeneration,35 and (3) immortalized and primary Müller cell cultures. In the degenerating retina, we have identified redistribution in the miR-124 from neurons (photoreceptors and ganglion cells) to the inner nuclear layer (specifically, Müller glia). We also confirmed in retinal cells an immortalized human Müller cell line and primary rodent Müller cell cultures, that miR-124 directly targets Ccl2. In vivo intravitreal delivery of a miR-124 mimic reduced Ccl2 expression levels, microglia/macrophage recruitment and photoreceptor cell death, preserving retinal function. Our findings suggest that in the retina, miR-124 is required for photoreceptor survival and that a balance of miR-124 expression is required to regulate factors such as Ccl2, which contribute to the progression of retinal degenerations.
Methods
Animal Handling and Rodent Photo-Oxidative Damage
All experiments were conducted in accordance with the ARVO Statement for Use of Animals in Ophthalmic and Vision Research. The study was approved by the Australian National University (ANU) Animal Experimentation Ethics Committee (Application ID: 2014/56). Adult (P60) C57BL/6J mice were exposed to photo-oxidative damage (PD) for a period of 1, 3, or 5 days to induce photoreceptor-specific cell death as previously described.35 Adult PDGFRa-cre (platelet derived growth factor receptor, alpha polypeptide) mice (animal #013148; The Jackson Laboratory, Bar Harbor, ME, USA) were used for Müller cell isolation either in dim-reared conditions or after PD. PDGFRa-cre animals were crossed with reporter strain B6.Cg-Gt(ROSA)26Sortm14 (animal #007914; The Jackson Laboratory) to produce PDGFRa-cre RFP mice, which were maintained on a C57BL/6J genetic background. Rodent eyes and retinas were collected and processed as described in previous publications.36 Wistar rats (P8-10) were used for primary Müller cell culture.
Human Tissue
Adult human eyes were obtained through the Lions New South Wales Eye Bank. Research followed the tenets of the Declaration of Helsinki and informed consent was obtained from all human subjects after explanation of the nature and possible consequences of the study. Human tissue collection and processing was approved by the University of Sydney Human Research Ethics Committee (Project No. 2012/218). All collection and processing were performed strictly according to the guidelines set out in the approved protocol mentioned above. Human AMD tissue was extensively catalogued and processed for cryosectioning based on a previously established protocol (n = 3).37
In Situ Hybridization
In situ hybridization was performed on cryosections from both human AMD donor tissue and rodent PD tissue. Ccl2 was cloned from PCR products from rodent retinal cDNA (550 bp amplicon). This template was then synthesized into a digoxigenin (DIG)-labeled riboprobe specific to rodent Ccl2 mRNA according to our published methodology.36 In situ hybridization for Ccl2 mRNA was performed using our previously published protocol.38 The riboprobe was hybridized overnight on retinal sections at 55°C.
To localize expression of miR-124, a double DIG-labeled miR-124-3p miRCURY LNA miRNA Detection Probe (Exiqon, Vedbaek, Denmark) was used on human and rodent retinal cryosections, which were hybridized for 1 hour at 53°C, according to the manufacturer's instructions (Exiqon). The bound probe was visualized with either HNPP/Fast-Red (Roche, Basel, Switzerland) or nitro blue tetrazolium and 5-bromo-4-chloro-3 indoyl phosphate (NBT/BCIP; Sigma-Aldrich Corp., St. Louis, MO, USA).
TUNEL
Retinal cryosections were stained for apoptotic cells using a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) kit (Roche Applied Science, Penzberg, Germany), following our previous methodology.39 To quantify photoreceptor cell death, TUNEL+ cells in the ONL were counted throughout the full length of each section cut in the parasagittal plane (superoinferior). For each animal, technical duplicates were counted, and these counts were averaged for each experimental group.
Immunohistochemistry
Immunohistochemistry was performed on retinal cryosections according to previously described protocols, with minor modifications.40 Table 1 provides a list of primary and secondary antibodies used. Immunolabeled IBA1+ microglia/macrophages were quantified across the full length of each retinal section. The number of IBA1+ microglia/macrophages in the outer retina was quantified by counting the IBA1+ cells in the ONL and subretinal space.
Table 1.
Immunohistochemistry Antibodies
| Antibody | Raised In | Dilution | Company |
|---|---|---|---|
| IBA1 | Rabbit | 1:500 | Wako, Osaka, Japan |
| Rhodopsin | Rabbit | 1:500 | Millipore, Burlington, MA, USA |
| Vimentin | Mouse | 1:200 | Thermo Fisher Scientific, Waltham, MA, USA |
Confocal Imaging
Immunofluorescence was captured in Z-stacked form, using a laser-scanning A1 confocal microscope (Nikon Instruments, Inc., Tokyo, Japan). Excitation wavelengths of 488 nm (green) and 561 nm (red) were used. Images were obtained using uniform gain settings and processed using Photoshop CS6 software (Adobe Systems, Inc., San Jose, CA, USA).
Densitometry was measured in the relevant regions of interest specified in the results. Regions of staining were measured for relative colorimetric intensity using the Nikon A1 confocal microscope following application of uniform gain settings.
Fluorescence-Activated Cell Sorting (FACS)
Retinas removed from PDGRFa-cre RFP mice eyes (control and PD) were used for Müller cell isolation through fluorescence-activated cell sorting (FACS). Retinas were processed for FACS based on a previously published protocol.40 The resultant samples were sorted using a FACSAria II (BD Biosciences, Franklin Lakes, NJ, USA), gating for RFP+ cells, which indicates an isolated population of Müller cells. The isolated cells were collected in staining buffer and kept chilled on ice until miRNA extraction could be commenced.
Analysis of miRNA and mRNA Expression
RNA extraction and purification was performed on retinas using a combination of TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and an RNAqueous Total RNA Isolation Kit (Thermo Fisher Scientific), as described in our previous publication.39 MiRNA extraction and purification was performed using an acid-phenol:chloroform extraction method with the mirVana miRNA isolation kit (Thermo Fisher Scientific) according to the manufacturer's instructions.
From a miRNA template, cDNA was synthesized using the TaqMan MicroRNA RT kit (Thermo Fisher Scientific) according to the manufacturer's protocol. From an mRNA template, cDNA synthesis was performed using a Tetro cDNA Synthesis Kit (Bioline Reagents, London, UK) as described previously.40 Quantitative real-time PCR (qRT-PCR) was performed on cDNA samples using Taqman hydrolysis probes (Table 2). The hydrolysis probes were utilized according to the manufacturer's instructions in a 10 µl reaction mix with the cDNA and TaqMan Gene Expression Master Mix (Thermo Fisher Scientific). The qRT-PCR reactions were run on a QuantStudio Flex 12 K instrument (Thermo Fisher Scientific). Each biological sample was amplified in a technical replicate, and the average Ct (cycle threshold) value was used to determine the change in expression. Percentage change was calculated using the comparative Ct (ΔΔCt) method, where target miRNAs were normalized to the expression of small nuclear RNA U6 and small nucleolar RNA 234, and target mRNAs were normalized to Gapdh expression as per our previous analyses.20,41
Table 2.
TaqMan Probes*
| Gene Symbol | Name | Catalog |
| Ccl2 | Chemokine (C-C motif) ligand 2 | Mm99999056_m1 |
| Hs00234140_m1 | ||
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | Mm99999915_g1 |
| Ap-1/Jun | Activator protein 1/Jun | Mm00495062_s1 |
| Hs01103582_s1 | ||
| miR-124a | mmu-miR-124-3p | 001182 (assay ID) |
| U6 snRNA | U6 small nuclear RNA | 001973 (assay ID) |
| snoRNA234 | Small nucelolar RNA 234 | 001234 (assay ID) |
Thermo Fisher Scientific (Waltham, MA, USA).
Cell Culture
Cell culture experiments were performed using an immortalized human Müller cell line (MIO-M1) and primary cultured rat (Wistar, P8-10) Müller cells. Cells were used within 10 passages. Cells were authenticated and validated from Cell Bank Australia and were further tested using RT-PCR for expression of cell specific markers (vimentin for MIO-M1 cells). Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum, L-glutamine (3 mM), sodium pyruvate (1 mM), and glucose (25 mM) and grown in flasks in a humidified incubator at 37°C with 5% CO2. Human IL-1β protein (10 ng/ml) was used as a pro-inflammatory stimulus for MIO-M1.
3′UTR CCL2 Luciferase Assay
A luciferase assay was performed on MIO-M1 cells using a commercially available vector containing the luciferase gene fused to the human CCL2 3′UTR (CCL2 LightSwitch 3′UTR Reporter GoClone; Switchgear Genomics, Menlo Park, CA, USA) and a pLightSwitch-3′UTR Luciferase assay system (Switchgear Genomics), following the manufacturer's protocol. MIO-M1 cells were seeded in a 96 well plate (3000 cells per well), cultured for 24 hours and transfected with the vector using Viafect (Promega Corp., Madison, WI, USA) transfection agent. After 36 hours, cells were treated with 10 ng/ml IL-1β protein for a duration of 12 hours, following which the luciferase assay was performed. The pLightSwitch Luciferase assay system was performed according to the manufacturer's protocol and luciferase activity was measured using an Infinite 200 plate reader (TECAN, Grödig, Austria).
In Vitro Transfection of miR-124 Mimic
MIO-M1 cells were seeded in clear plastic 6-well tissue culture plates at a density of 80,000 cells per well, and the medium was replaced with low serum (1% FBS) DMEM after sufficient adhering time (80%–90% confluence). Cells were incubated at 37°C in 5% CO2 for 12 hours and then transfected with 10 nM miR-124-3p mimic, miR-124-3p inhibitor, or negative control miRNA mimic (Thermo Fisher Scientific), using Lipofectamine RNAiMAX (Invitrogen). After 24 hours, cells were exposed to 10 ng/ml IL-1β protein for a duration of 12 hours. The cells were then harvested using TRIzol (Invitrogen) reagent and RNA extracted for qPCR as previously described. CCL2 expression for the different experimental groups was compared to nontransfected/treated MIO-M1 cells.
Transfection of miR-124 Mimic In Vivo
Both miR-124-3p and negative, scrambled control miRNA mimics (Thermo Fisher Scientific) were used for in vivo transfection. Before administration, miRNAs were encapsulated in a cationic lipid-based transfection agent (Invivofectamine 3.0; Thermo Fisher Scientific) according to the manufacturer's protocol. Each encapsulated miRNA was formulated to a final concentration of 1 µg/µl in endotoxin-free 0.1 M PBS solution after being spun at 4000g through an Amicon Ultra-4 Centrifugal Filter Unit (Merck Millipore, Billerica, MA, USA). Intravitreal injections were performed as described in our previous publication.21 One µl of miR-124-3p mimic or negative control miRNA formulation was injected into both eyes of C57BL/6J mice. Animals were recovered for 24 hours in dim conditions prior to the onset of PD.
Measurement of Retinal Function Using Electroretinography (ERG)
Full-field scotopic ERG was performed to assess the retinal function of mice after 5 days of photo-oxidative damage as described previously.35 Briefly, mice were dark-adapted overnight, anesthetized by intraperitoneal injection of xylazil (10 mg/kg) and ketamine (100 mg/kg), and the pupils dilated with 1% atropine sulfate. A single- or twin-flash paradigm was used to elicit mixed (rod and cone) or isolated cone responses, respectively. Flash stimuli for mixed responses were provided by an LED-based system (FS-250A Enhanced Ganzfeld; Photometric Solutions International, Huntingdale VIC, Australia) over a stimulus intensity range of 6.3 log cd s m−2 (range, −4.4 to 1.9 log cd s m−2).
Statistical Analyses
For experiments comparing two groups, unpaired Student's t-tests were used. Multiple group comparisons were made using a 1-way ANOVA with Tukey's post hoc test for multiple comparisons, and 2-way ANOVA with Sidak's post hoc test for multiple comparisons for the ERG data sets. Statistical analyses were performed using GraphPad Prism V5 (GraphPad Software, La Jolla, CA, USA). Statistical significance was defined by P < 0.05.
Results
MiR-124 Expression in Human AMD Tissue
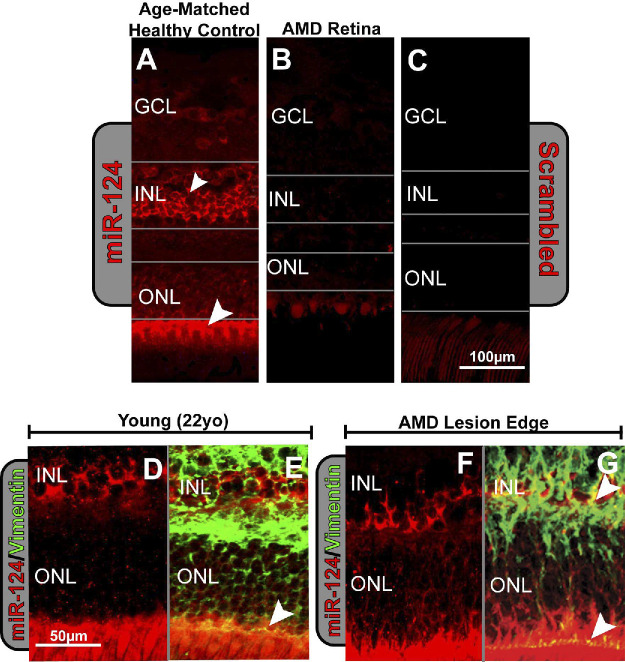
Using in situ hybridization, we localized miR-124 labeling to the inner nuclear layer (INL) and outer limiting membrane (OLM) of the retina with some low-level labeling in the ganglion cell layer (GCL) in healthy human retinal tissue (Fig. 1A). In central, foveal AMD retinal sections, the degenerating macular region was found to be devoid of miR-124 labeling with gray lines outlining the relative sizes of the nuclear layers, showing a heavily degenerated outer nuclear layer (ONL; Fig. 1B). No background labeling was seen in the human retinal sections when labeled with a scrambled miRNA probe (Fig. 1C). The cellular location of miR-124 was compared to both normal, aged human retinas (Fig. 1A) and young adult retinas (Figs. 1D, 1E), in which high levels of miR-124 expression were seen uniformly across the retina in the INL and OLM. At the lesion edge of AMD patients, a similar distribution of miR-124 (Figs. 1F, 1G) was exhibited for both the young adult and age-matched controls, indicating that only the central retina of AMD patients was devoid of miR-124 expression.
Figure 1.
In situ hybridization of miR-124 in human retinas. (A) A normal age-matched human retina (96 years old, female) shows strong labeling of miR-124 in the inner nuclear layer (INL) and outer limiting membrane (OLM) compared with (B), an AMD-affected retina (97 years old, female) where staining of miR-124 is depleted (gray lines indicate the layers of the retina). (C) A scrambled control probe showed no labeling in the human retina. (D) Healthy 22-year-old (female) human retina shows staining of miR-124 in the INL and OLM. (E) Colocalization with the Müller cell marker vimentin is evident in the OLM (arrowhead). (F) 97-year-old, AMD, human retina at the lesion edge also shows staining of miR-124 in the INL and outer limiting membrane as well as colocalization with (G) vimentin INL and OLM (arrowheads).
Sections double-labeled with miR-124 and vimentin, a Müller cell marker, showed low-level colocalization in the INL in the young adult retinas and double-labeling in the OLM (Fig. 1E). By comparison, at the lesion edge of AMD patient sections (Figs. 1F, 1G), increased colocalization with vimentin was seen in the INL with the Müller cell end feet still showing evidence of staining (Fig. 1G). This indicates an increase in vimentin colocalization in the INL with miR-124 in AMD peripheral retinas.
MiR-124 Expression in Müller Cells
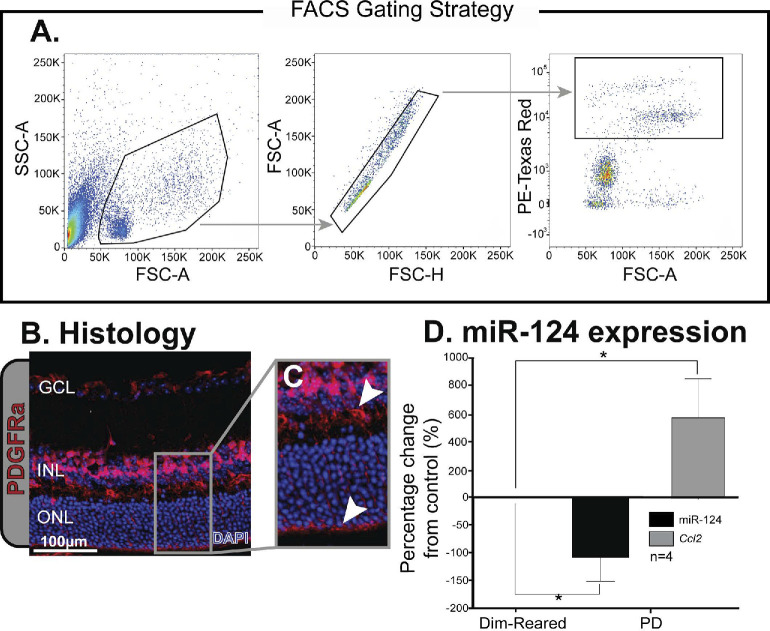
The FACS gating strategy for isolating PDGFRa-cre RFP cells is shown in Figure 2A. Histology of retinal sections from PDGFRa-cre RFP reporter animals showed Müller cell-specific fluorescence in the GCL, INL, and OLM as well as the Müller cells processes spanning across the retinal layers (Figs. 2B, 2C). FACS-isolated Müller cells from PDGFRa-cre RFP reporter mice (Fig. 2D) showed decreased expression of miR-124 following 5 days PD when compared to dim-reared animals (P < 0.05), while Ccl2 expression was significantly increased at 5 days PD (P < 0.05, Fig. 2D).
Figure 2.
Isolation of Müller cells from PDGFRa-cre RFP mice. (A) Gating strategy used to isolate cells expressing RFP from PDGFRa-cre positive animals. (B, C) Histology of retinal tissue from PDGFRa-cre RFP animals shows fluorescence in the ganglion cell layer (GCL), inner nuclear layer (INL), and outer limiting membrane (OLM) as well as the Müller cell processes spanning across the retinal layers (B). (D) Expression levels of miR-124 decreased, while Ccl2 increased in Müller cells isolated from PDGFRa-cre RFP mice that were subjected to 5 days of PD. *Significance using a Student's t-test, P < 0.05 and error bars indicate SEM, n = 4.
CCL2 as a miR-124 Target
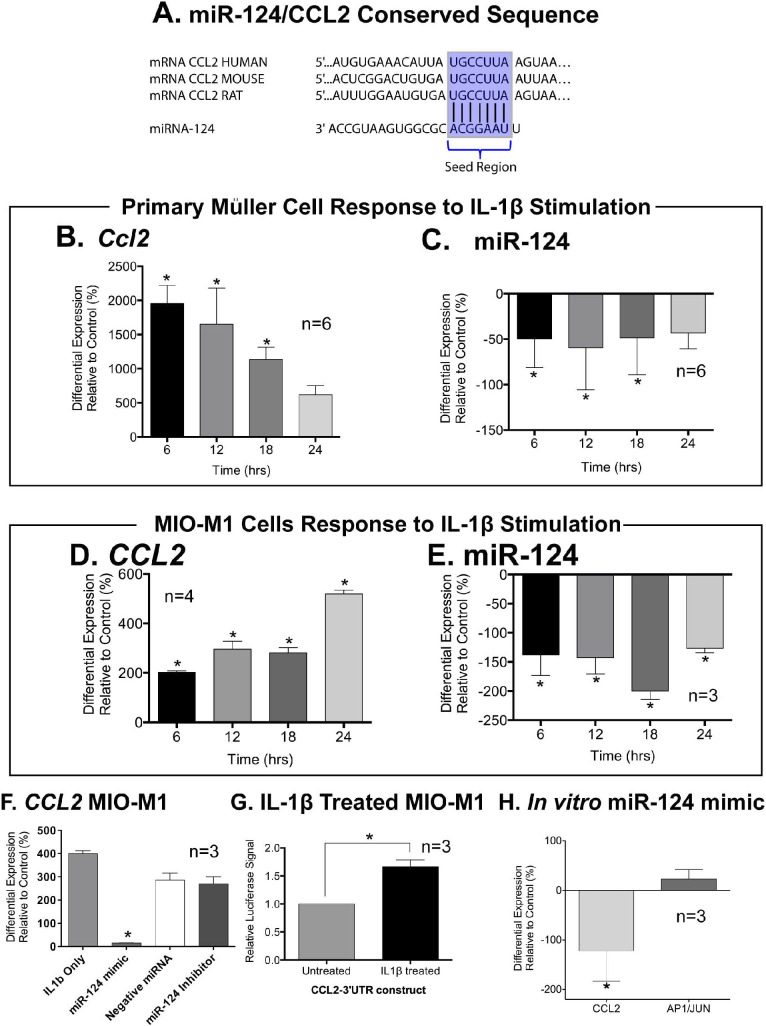
Binding of the miR-124 mimic 8-mer seed region to CCL2 is conserved across human, mouse, and rat tissue (Fig. 3A). Rat primary Müller cells stimulated with IL-1β showed an increase of CCL2 expression at 6, 12, and 18 hours after the introduction of IL-1β (P < 0.05, Fig. 3B), with a concurrent decrease in miR-124 at the same time points (P < 0.05, Fig. 3C). Cells of the immortalized Müller cell line MIO-M1, stimulated with IL-1β over a 24-hour period, showed a similar expression profile with increased Ccl2 expression (P < 0.05, Fig. 3D) at each time point, and decreased miR-124 expression (P < 0.05, Fig. 3E).
Figure 3.
MiR-124 expression in primary and immortalized Müller cells. (A) Schematic showing the conserved sequence of the 3′UTR of CCL2 across human, mouse, and rat and its perfect complementarity with the seed region of miR-124. (B) In primary Müller cells, Ccl2 significantly increased up to 18 hours, and (C) and miR-124 levels down-regulated at all corresponding time points. (D) CCL2 expression increased after stimulating MIO-M1 cells with IL-1β over a 24-hour period while (E) levels of miR-124 expression were found to significantly decrease in MIO-M1 after stimulation. (F) A significant decrease in CCL2 expression levels was detected in IL-1β-treated cells after transfection with a miR-124 mimic, compared with cells stimulated only with IL-1β and cells transfected with negative miRNA and miR-124 inhibitor. (G) Further, using a luciferase assay for interactions between CCL2 and miR-124, IL-1β stimulation of MIO-M1 cells produced an increase in relative luciferase signal compared to non-treated control cells. (H) In the absence of IL-1β, transfection of miR-124 mimics in MIOM-1 cells decreased Ccl2 expression with Ap-1/Jun is unchanged. *Significance using 1-way ANOVA with Tukey's post hoc test or a Student's t-test, P < 0.05 and error bars indicate SEM, n = 3–4.
Using a luciferase assay, we transfected IL-1β-treated MIO-M1 cells with a miR-124 mimic, a negative control mimic, or a miR-124 inhibitor. The resulting data (Fig. 3F) indicated that CCL2 expression was downregulated in the presence of the miR-124 mimic but not in the presence of the negative control mimic or the miR-124 inhibitor (P < 0.05). MIO-M1 cells stimulated with IL-1β showed an increased luciferase signal (P < 0.05, Fig. 3G), indicating decreased miRNA binding to the 3′UTR. MiR-124-transfected MIO-M1 cells displayed a downregulation of CCL2 expression, while a marker of cell death, AP1/JUN, remained unchanged (P < 0.05, Fig. 3H).
Cellular Localization of miR-124 in Retinal Degenerations
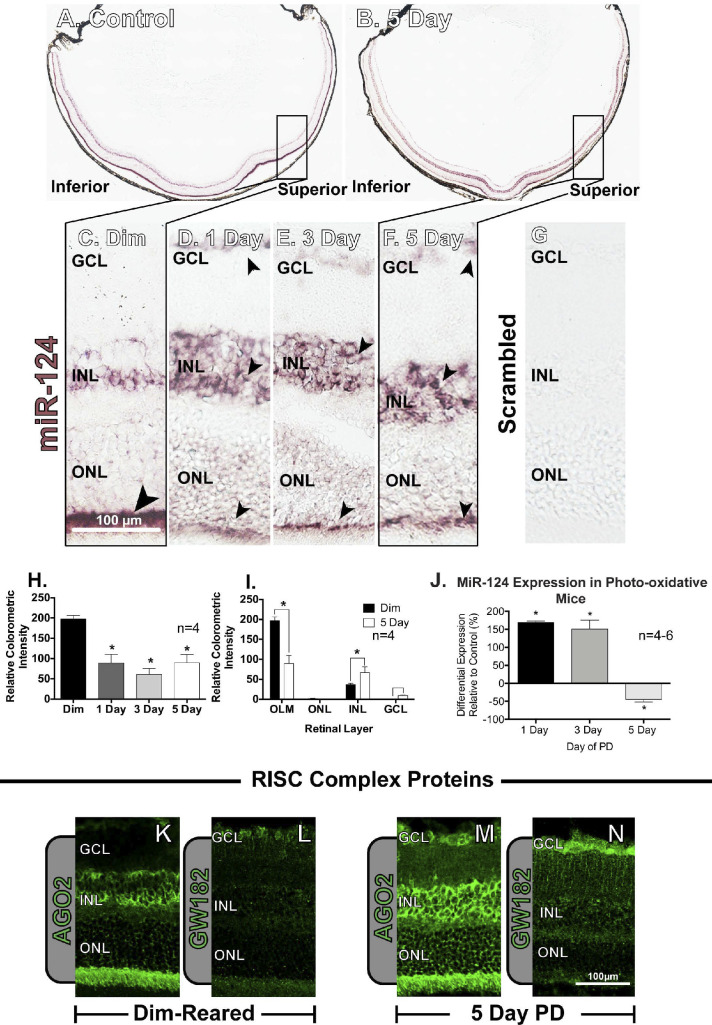
In situ hybridization labeling for miR-124 in dim-reared mice was heavily congregated in the superior retina (“hotspot” region of focal damage35) in the OLM (Fig. 4A). This labeling pattern in the superior retina was more intense in the INL following 5 days of PD (Fig. 4B). In dim-reared control animals, miR-124 labeling was strongly present in the OLM (Fig. 4C), whereas high-resolution images showed increasing intensity of labeling in the INL between 1 and 5 days of PD (Fig. 4D–F). The scrambled control showed no background labeling (Fig. 4G). Colorimetric quantification of miR-124 labeling in the central retinal region showed a decline in labeling over the 5 days of PD (P < 0.05, Fig. 4H), matching the visualization of the images as the intense staining in OLM seen in dim-reared animals decreased after PD. However, analysis of the distribution of miR-124 between the outer and inner retina identified a decrease in staining intensity in the OLM with an increase in the INL (P < 0.05, Fig. 4I). A small but significant increase in expression was shown in the GCL at 5 days (P < 0.05). While a redistribution of miR-124 was observed following PD, levels of retinal miR-124 show a global expression decrease after 5 days PD (P < 0.05, Fig. 4J). Overall, we showed that the retinal location of miR-124 expression shifted from primarily ONL prior to damage to the INL after the onset of injury.
Figure 4.
Regulation of miR-124 expression in the rodent retina. (A, B) In situ hybridization showing the difference in distribution of miR-124 dim-reared control retinas compared to photo-oxidative damage (PD) retinas. The boxed regions of the superior side of the retina (rectangles in A, B) show the location for greatest susceptibility to cell death in the photo-oxidative model. (C–G) Dim-reared animals show labeling predominantly below the ONL in the OLM, while damaged retinas have higher levels in the INL and depletion of labeling in the OLM, comparatively. This change is seen as early as 1 day, with consistent labeling at 3 days and 5 days of PD. (H, I) Quantification of in situ hybridization using relative colorimetric intensity displayed a decreased expression of miR-124 at each of the experimental time points, with an increase in INL and GCL and a decrease in OLM expression at 5 days relative to controls. (J) Gene expression revealed a significant decrease of miR-124 levels at 5 days of PD. In dim-reared animals, RISC complex protein, (K) AGO2 was labeled in the INL and OLM with (L) GW182 faintly expressed in the GCL. Both proteins were both strongly labeled after PD with (M) AGO-2 expression in the GCL, INL, and OLM, and (N) GW182 more evident in the GCL and in the Müller cell processes. *Significance using 1-way ANOVA with Tukey's post hoc test or a Student's t-test, P < 0.05 and error bars indicate SEM, n = 3–6.
This shift in cellular localization of miR-124 from the OLM to the INL is correlated with a change in labeling of two RNA-induced silencing complex (RISC) proteins involved with miRNA-mediated gene silencing in mammalian cells AGO2 and GW182 (Figs. 4K–N). AGO2 labeling initially is dispersed throughout the retina with intense labeling in the OLM, photoreceptor inner and outer segments and the INL (Fig. 4K), while GW182 is more diffusely spread throughout the retinal layers (Fig. 4L). At 5 days PD, both AGO2 and GW182 labeling is more pronounced throughout the retina, particularly in the INL for AGO2 (Fig. 4M) and in the INL and GCL for GW182 (Fig. 4N).
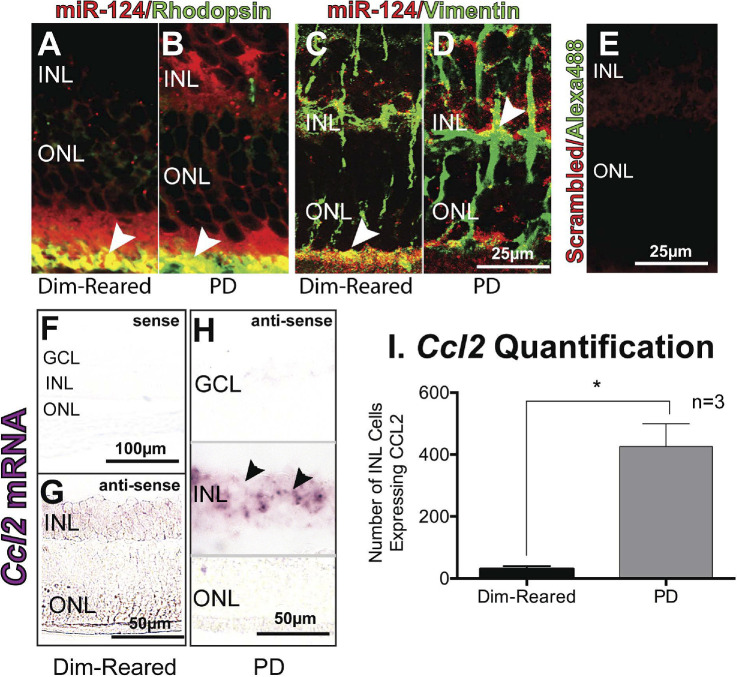
Labeling with the rod photoreceptor marker, rhodopsin, revealed colocalization with miR-124 in the photoreceptors in dim-reared and PD animals (Figs. 5A, 5B). Labeling with Müller cell marker, vimentin (Figs. 5C, 5D), also indicated colocalization with miR-124 in the OLM as well as the INL with the majority of vimentin labeling localized to the middle of the INL. Little to no background staining was seen in the scrambled miRNA and Alexa-488 double-labeled control (Fig. 5E). Minimal Ccl2 in situ hybridization labeling was detected using the sense probe in control retina (Fig. 5F), as well as in the antisense in the dim-reared control retina (Fig. 5G). However, intense Ccl2 mRNA labeling staining was detected 24 hours post-PD in the INL (Fig. 5H, gray lines). Ccl2 mRNA was localized to cells in the INL, most likely Müller cells based on our previous studies20,21 (Fig. 5H), with an expression pattern in the INL similar to miR-124 at 24 hours PD, including an increased number of Ccl2-expressing cells (Fig. 5I).
Figure 5.
Colocalization of miR-124 with photoreceptors and Müller cells. (A, B) Immunohistochemistry using a photoreceptor specific marker, rhodopsin (green), showed colocalization with miR-124 staining (red) deep to the ONL (arrowheads, yellow) in both dim-reared and PD rodents. (C, D) Immunohistochemistry with the Müller cell marker, vimentin (green), also showed colocalization with miR-124 (red) in the deep ONL for dim-reared rodents, and in the INL for PD rodents (arrowheads, yellow). (E) No labeling was seen in the scrambled control. (F–H) In situ hybridization for Ccl2 shows increased expression in the INL in PD rodents, compared to dim-reared (data from Natoli R, Fernando N, Madigan M, et al. Microglia-derived IL-1β promotes chemokine expression by Müller cells and RPE in focal retinal degeneration. Mol Neurodegener. 2017;12(1):31.) (I) Quantification of Ccl2-expressing cells in the INL of dim-reared vs PD rodents demonstrates a significant difference. *Student t-test, P < 0.05; error bars indicate SEM.
Delivery of miR-124 Mimic in Retinal Degenerations
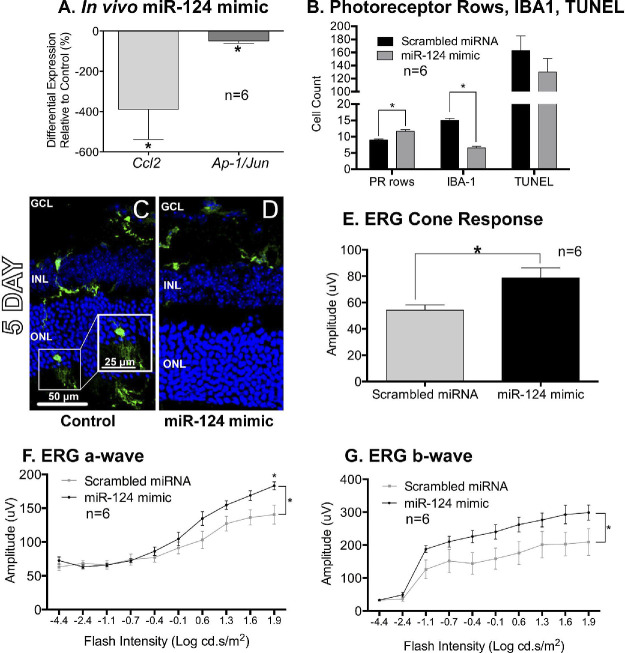
Intravitreal delivery of miR-124 mimic to mouse eyes targeting the central retina was performed prior to PD. miR-124 mimics resulted in a significant decrease in both Ccl2 and Ap1/Jun (P < 0.05, Fig. 6A) expression in the retinas after 5 days of PD. Analysis of ONL thickness in retinal sections (Fig. 6B) indicate increased survival of photoreceptors (PR rows) at 5 days PD in the animals treated with miR-124 mimics, compared with animals injected with scrambled miRNA (P < 0.05). No differences in TUNEL+ cells in the ONL were observed (Fig. 6B, P > 0.05). Furthermore, increased photoreceptor survival was accompanied by decreased numbers of IBA1+ microglia/macrophage cells infiltrating into the ONL in the mimic-treated retinas at 5 days compared to scrambled controls (P < 0.05, Figs. 6B–D).
Figure 6.
Intravitreal delivery of miR-124 mimics in rodent photo-oxidative damage. (A) Intravitreal injections of miR-124 mimic decreased the expression of Ccl2 as well as the apoptotic marker Ap-1/Jun after 5 days of PD. (B) Animals injected with miR-124 mimic had a higher amount of photoreceptor rows (PR rows) compared to control animals (scrambled miRNA injection). No differences were seen in TUNEL+ cell count. (B–D) Inflammatory cells labeled with IBA1 were significantly reduced in number in the ONL 5 days after the miR-124 mimic injection compared to scrambled controls. (E–G) Electroretinography (ERG) showed a larger response amplitude in a-wave, b-wave, and the cone response in animals receiving a miR-124 mimic compared to controls. *Indicates significance using a Student's t-test or 2-way ANOVA with Sidak's post hoc test for multiple comparisons, P < 0.05 error bars indicate SEM, n = 6.
Finally, electroretinography (ERG) of 5-day PD animals treated with miR-124 mimic injections demonstrated a higher a-wave, b-wave, and cone response compared to scrambled control-injected animals (P < 0.05, Figs. 6E–G). Overall, the delivery of miR-124 mimic led to decreased photoreceptor loss, reduced inflammation and an improved retinal function.
Discussion
It is well established that activation of the innate immune system plays a critical role in the pathogenesis of many retinal degenerations, including AMD.42–45 For this reason, improving understanding of the molecular mechanisms involved in regulating inflammatory pathways is likely to result in novel treatment strategies and improved patient outcomes (reviewed in Ref. 18). In this study, we explored the role of miR-124 in retinal degenerations and demonstrated the potential use of miRNA, a potential new class of therapeutics for the treatment of retinal degenerations. We demonstrate that in both human and rodent tissues, miR-124 expression and cellular location were altered in the degenerating retina, with overall expression of miR-124 decreasing in both human AMD retinas and in a mouse model of retinal degeneration.35 In addition to a global decrease in expression, the cellular location of miR-124 was found to change from predominantly OLM in the normal retina, to the INL during degeneration. We identified Ccl2, a chemokine produced in Müller glia, as a target of miR-124 in retina cells and demonstrated that miR-124 mimics can reduce levels of retinal inflammation and photoreceptor cell death, thereby improving overall retinal function.
Cellular Redistribution of miR-124 in Retinal Degenerations
Our findings showed that miR-124 is constitutively expressed in the normal human and rodent retina and undergoes both a decrease in total retinal expression and a change in cellular location during retinal degeneration. miR-124 is largely considered to be neuronal-specific46,47 and is enriched in the retina with high levels in photoreceptors.30,47,48 Additionally, in vitro, it has been reported that there is an absence of miR-124 in Müller cells cultured from young animals (p12),30 or that it is expressed at very low levels,46 suggesting that Müller cells produce little to no miR-124 in the absence of neurons.46
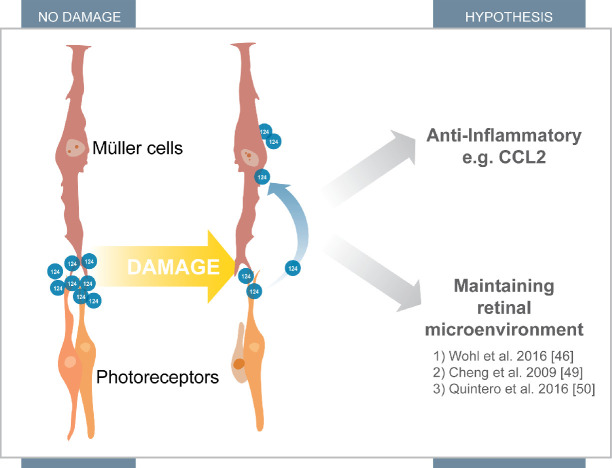
The current study is consistent with previous findings, showing little to no expression of miR-124 in primary Müller cell cultures from similar aged animals,47,49,50 and our results indicate that miR-124 levels decrease with each passage, suggesting that in the absence of neuronal-glial interactions Müller cell expression of miR-124 is eventually lost (Supplementary Fig. 1). Despite this low level of miR-124 expression, we have demonstrated that in response to an inflammatory stimulus, miR-124 levels are significantly decreased in both primary and immortalized Müller cells. PDGFRa-cre RFP Müller cells isolated after retinal degeneration showed a decrease in miR-124 expression in response to photoreceptor cell death, consistent with our observations in AMD tissue. These results suggest that miR-124 is either lowly or non-endogenously produced by Müller cells, but we have speculated that it is likely to be translocated from the photoreceptors to the Müller cells during retinal damage (Fig. 7).46,49,50
Figure 7.
Proposed mechanism of action for miR-124. MiR-124 accumulates at the end feet of the Müller cells and in the photoreceptors under homeostasis. However, after damage, a large population of miR-124 translocates toward the Müller cell bodies. This translocation helps attribute to its anti-inflammatory nature through regulation of CCL2. It may also be implicated in its role in maintaining the retinal microenvironment.46,49,50
In support of the hypothesis that miR-124 is redistributed to the Müller cell bodies during retinal degeneration, we found increased localization of RISC complex proteins (AGO2 and GW182) in the inner retina following retinal degeneration. These proteins are part of the RISC complex, which facilitates binding of miRNA with mRNA1 and are required for miRNA functionality. Their changed expression profile under PD-induced degeneration corresponds with changes in miR-124 distribution, supporting the hypothesis that miR-124 is constitutively expressed in the retina, and mobilized in stress conditions to the cell bodies of Ccl2-expressing Müller cells in the INL. In the macula of AMD retinas, the absence of miR-124 is consistent with a highly pro-inflammatory environment and a dysregulated expression of CCL2, along with other pro-inflammatory cytokines.
Role of miR-124 in Regulating Ccl2-Mediated Inflammation in the Retina
MiR-124 has been found to modulate the inflammatory response in various disorders, including those of the central nervous system.33,34 It is highly abundant in the brain and has been shown to be up-regulated in AMD.30,31 Our rodent and human data suggest that miR-124 is mobilized toward the site of Ccl2 expression, the Müller cell bodies in the INL20 during retinal degeneration. We have previously shown up-regulation of Ccl2 and other chemokines by microarray analyses in a rodent PD model of retinal degeneration.39 We have also previously established a key role for Ccl2 in the progression of degeneration by showing that small-interfering RNA (siRNA)-mediated down-regulation of Ccl2 attenuates the inflammatory profile of the retina and reduces photoreceptor death.21 Ccl2 has been demonstrated to be upregulated in a number of retinal diseases, including retinitis pigmentosa,51 diabetic retinopathy,52 and AMD.19 As Müller cells produce high levels of Ccl2 during PD,20 we have postulated that miR-124 is redistributing to the Müller cell bodies during PD to inhibit the production of Ccl2 and dampen the inflammatory cascade. In our study, we showed that miR-124 dampens pro-inflammatory signaling in vitro through down-regulation of CCL2 expression in MIO-M1 cells. Further, we demonstrated an inverse correlation in miR-124 and Ccl2 expression levels in isolated Müller cells.
Taken together, this data suggests a possible novel mechanistic role of miR-124 in stabilizing the retina through a neuronal-glial molecular interaction and regulation of Ccl2 expression, as illustrated in Figure 7. The lack of expression of miR-124 in the human AMD retina, coupled with the decrease in miR-124 expression in Müller cells following PD, suggests that this regulation becomes unbalanced, which would inevitably lead to an increased inflammatory response and cell loss.
It has been postulated that therapeutic approaches employing miR-124 may control neuroinflammation (reviewed in Ref.53), with the majority of studies concentrating on the activity in microglia/macrophages.54,55 Their potential as therapeutics for neurodegenerative diseases such as AMD, however, has not been widely investigated, with research focusing more on miRNAs in eye development and the use of circulating miRNA as early diagnostics of disease.56 In this study, we investigated the use of miR-124 mimics in vivo as a potential therapeutic for retinal degenerations. Our data showed that the intravitreal delivery of miR-124 mimics decreased infiltration of macrophages into the photoreceptor layer—a process correlated with photoreceptor cell death in retinal degenerations.57–61 Most importantly, the strong anti-inflammatory effects of miR-124 mimics in the retina, coupled with a reduction in photoreceptor loss, results in a preservation of retinal function.
Due to many other predicted binding partners of miR-124, it is unlikely that the proposed redistribution from photoreceptors is targeting only Ccl2. MiR-124 has been shown to interact with a number of other mRNA involved in neuronal diseases, including Rab3a,62 an essential component of the inner and outer plexiform layers of the retina.63 Further, alteration of miR-124 in Müller cells has also been shown to trigger neuronal differentiation through expression of the proneural transcription factor Asc11,46 indicating a possible uncharacterized function of mammalian retinal neuronal-glial interactions. Further investigations into the interplay between neurons, Müller cells, and miR-124 are needed to improve our understanding in this area.
Conclusions
This study provides an understanding of the role of miR-124 in the degenerating retina. By demonstrating its cellular relocation and anti-inflammatory properties, our current findings pave the way for further investigation of the therapeutic potential of miRNA mimics in neurodegenerative diseases, including AMD, Parkinson's disease, and Alzheimer's disease. However, to fully assess the therapeutic efficacy of miR-124 in neurodegeneration, identification of the full spectrum of regulatory targets of miR-124 is required.
Supplementary Material
Acknowledgments
The authors thank the New South Wales Tissue and Organ Donation Service and the Lions New South Wales Eye Bank for the human donor eyes.
Supported by the National Health and Medical Research Council in Australia (APP1127705, 2017-2019), the Australian Government Research Training Program, the Gretel and Gordon Bootes Foundation (2013), and the Ophthalmic Research Institute of Australia/Eye Surgeons' Foundation (2015).
Disclosure: J.A. Chu-Tan, None; M. Rutar, None; K. Saxena, None; R. Aggio-Bruce, None; R.W. Essex, None; K. Valter, None; H. Jiao, None; N. Fernando, None; Y. Wooff, None; M.C. Madigan, None; J. Provis, None; R. Natoli, None
References
- 1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 2. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011; 12: 861–874. [DOI] [PubMed] [Google Scholar]
- 3. Lewis BP, Burge CB, Bartel DP.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 4. Sundermeier TR, Palczewski K.. The impact of microRNA gene regulation on the survival and function of mature cell types in the eye. FASEB J. 2016; 30: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel DP, Chen CZ.. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004; 5: 396–400. [DOI] [PubMed] [Google Scholar]
- 6. Ardekani AM, Naeini MM.. The role of MicroRNAs in human diseases. Avicenna J Med Biotechnol. 2010; 2: 161–179. [PMC free article] [PubMed] [Google Scholar]
- 7. Ortega FJ, Moreno-Navarrete JM, Pardo G, et al.. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One. 2010; 5: e9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kantharidis P, Wang B, Carew RM, Lan HY.. Diabetes complications: the microRNA perspective. Diabetes. 2011; 60: 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alexandrov PN, Dua P, Lukiw WJ.. Up-regulation of miRNA-146a in progressive, age-related inflammatory neurodegenerative disorders of the human CNS. Front Neurol. 2014; 5: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naidu S, Magee P, Garofalo M.. MiRNA-based therapeutic intervention of cancer. J Hemat Oncol. 2015; 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shafi G, Aliya N, Munshi A.. MicroRNA signatures in neurological disorders. Can J Neurol Sci. 2010; 37: 177–185. [DOI] [PubMed] [Google Scholar]
- 12. Maciotta S, Meregalli M, Torrente Y.. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci. 2013; 7: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rao P, Benito E, Fischer A.. MicroRNAs as biomarkers for CNS disease. Front Mol Neurosci. 2013; 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S, Koster KM, He Y, Zhou Q.. miRNAs as potential therapeutic targets for age-related macular degeneration. Future Med Chem. 2012; 4: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP.. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol. 2003; 48: 257–293. [DOI] [PubMed] [Google Scholar]
- 16. Green WR, Key SN III. Senile macular degeneration: a histopathologic study. Trans Am Ophthalmol Soc. 1977; 75: 180–254. [PMC free article] [PubMed] [Google Scholar]
- 17. Penfold PL, Madigan MC, Gillies MC, Provis JM.. Immunological and aetiological aspects of macular degeneration. Prog Retin Eye Res. 2001; 20: 385–414. [DOI] [PubMed] [Google Scholar]
- 18. Ambati J, Atkinson JP, Gelfand BD.. Immunology of age-related macular degeneration. Nat Rev Immunol. 2013; 13: 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newman AM, Gallo NB, Hancox LS, et al.. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012; 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rutar M, Natoli R, Valter K, Provis JM.. Early focal expression of the chemokine Ccl2 by Muller cells during exposure to damage-inducing bright continuous light. Invest Ophthalmol Vis Sci. 2011; 52: 2379–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rutar M, Natoli R, Provis JM.. Small interfering RNA-mediated suppression of Ccl2 in Muller cells attenuates microglial recruitment and photoreceptor death following retinal degeneration. J Neuroinflamm. 2012; 9:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ambati J, Anand A, Fernandez S, et al.. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat Rev. 2003; 9: 1390–1397. [DOI] [PubMed] [Google Scholar]
- 23. Sennlaub F, Auvynet C, Calippe B, et al.. CCR2(+) monocytes infiltrate atrophic lesions in age-related macular disease and mediate photoreceptor degeneration in experimental subretinal inflammation in Cx3cr1 deficient mice. EMBO Mol Med. 2013; 5: 1775–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marques-Rocha JL, Samblas M, Milagro FI, et al.. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015; 29: 3595–3611. [DOI] [PubMed] [Google Scholar]
- 25. O'Connell RM, Rao DS. Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012; 30: 295–312. [DOI] [PubMed] [Google Scholar]
- 26. Zhu J, Chen T, Yang L, et al.. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS One. 2012; 7: e46551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kawano S, Nakamachi Y.. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann Rheum Dis. 2011; 70(suppl 1): i88–i91. [DOI] [PubMed] [Google Scholar]
- 28. Dong N, Xu B, Shi H, Tang X.. Baicalein inhibits amadori-glycated albumin-induced MCP-1 expression in retinal ganglion cells via a MicroRNA-124-dependent mechanism. Invest Ophthalmol Vis Sci. 2015; 56: 5844–5853. [DOI] [PubMed] [Google Scholar]
- 29. Li X, Fan Q, Li J, Song J, Gu Y.. MiR-124 down-regulation is critical for cancer associated fibroblasts-enhanced tumor growth of oral carcinoma. Exp Cell Res. 2017; 351: 100–108. [DOI] [PubMed] [Google Scholar]
- 30. Sanuki R, Onishi A, Koike C, et al.. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011; 14: 1125–1134. [DOI] [PubMed] [Google Scholar]
- 31. Lagos-Quintana M, Rauhut R, Yalcin A, et al.. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002; 12: 735–739. [DOI] [PubMed] [Google Scholar]
- 32. Karali M, Persico M, Mutarelli M, et al.. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016; 44: 1525–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun Y, Luo ZM, Guo XM, Su DF, Liu X.. An updated role of microRNA-124 in central nervous system disorders: a review. Front Mol Neurosci. 2015; 9: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang D, Zhang H, Li M, et al.. MicroRNA-124 controls the proliferative, migratory, and inflammatory phenotype of pulmonary vascular fibroblasts. Circ Res. 2014; 114: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Natoli R, Jiao H, Barnett NL, et al.. A model of progressive photo-oxidative degeneration and inflammation in the pigmented C57BL/6J mouse retina. Exp Eye Res. 2016; 147: 114–127. [DOI] [PubMed] [Google Scholar]
- 36. Rutar M, Natoli R, Kozulin P, et al.. Analysis of complement expression in light-induced retinal degeneration: synthesis and deposition of C3 by microglia/macrophages is associated with focal photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2011; 52: 5347–5358. [DOI] [PubMed] [Google Scholar]
- 37. Shelley EJ, Madigan MC, Natoli R, Penfold PL, Provis JM.. Cone degeneration in aging and age-related macular degeneration. Arch Ophthalmol. 2009; 127: 483–492. [DOI] [PubMed] [Google Scholar]
- 38. Cornish EE, Madigan MC, Natoli R, et al.. Gradients of cone differentiation and FGF expression during development of the foveal depression in macaque retina. Vis Neurosci. 2005; 22: 447–459. [DOI] [PubMed] [Google Scholar]
- 39. Natoli R, Zhu Y, Valter K, et al.. Gene and noncoding RNA regulation underlying photoreceptor protection: microarray study of dietary antioxidant saffron and photobiomodulation in rat retina. Mol Vis. 2010; 16: 1801–1822. [PMC free article] [PubMed] [Google Scholar]
- 40. Rutar M, Natoli R, Chia RX, Valter K, Provis JM.. Chemokine-mediated inflammation in the degenerating retina is coordinated by Muller cells, activated microglia, and retinal pigment epithelium. J Neuroinflammation. 2015; 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rutar M, Valter K, Natoli R, Provis JM.. Synthesis and propagation of complement C3 by microglia/monocytes in the aging retina. PLoS One. 2014; 9: e93343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edwards AORitter R IIIAbel KJ, et al.. Complement factor H polymorphism and age-related macular degeneration. Science. 2005; 308: 421–424. [DOI] [PubMed] [Google Scholar]
- 43. Hageman GS, Anderson DH, Johnson LV, et al.. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005; 102: 7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haines JL, Hauser MA, Schmidt S, et al.. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005; 308: 419–421. [DOI] [PubMed] [Google Scholar]
- 45. Klein RJ, Zeiss C, Chew EY, et al.. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wohl SG, Reh TA.. miR-124-9-9* potentiates Ascl1-induced reprogramming of cultured Muller glia. Glia. 2016; 64: 743–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wohl SG, Reh TA.. The microRNA expression profile of mouse Müller glia in vivo and in vitro. Sci Rep. 2016; 6: 35423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karali M, Manfredi A, Puppo A, et al.. MicroRNA-restricted transgene expression in the retina. PLoS One. 2011; 6: e22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng LC, Pastrana E, Tavazoie M, Doetsch F.. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009; 12: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quintero H, Gomez-Montalvo AI, Lamas M.. MicroRNA changes through Muller glia dedifferentiation and early/late rod photoreceptor differentiation. Neurosci. 2016; 316: 109–121. [DOI] [PubMed] [Google Scholar]
- 51. Guo C, Otani A, Oishi A, et al.. Knockout of ccr2 alleviates photoreceptor cell death in a model of retinitis pigmentosa. Exp Eye Res. 2012; 104: 39–47. [DOI] [PubMed] [Google Scholar]
- 52. Yoshimura T, Sonoda KH, Sugahara M, et al.. Comprehensive analysis of inflammatory immune mediators in vitreoretinal diseases. PLoS One. 2009; 4: e8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guedes J, Cardoso AL, Pedroso de Lima MC.. Involvement of microRNA in microglia-mediated immune response. Clin Dev Immunol. 2013; 2013: 186872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL.. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011; 17: 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ponomarev ED, Veremeyko T, Weiner HL.. MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia. 2013; 61: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Szemraj M, Bielecka-Kowalska A, Oszajca K, et al.. Serum MicroRNAs as Potential Biomarkers of AMD. Med Sci Monit. 2015; 21: 2734–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cherepanoff S, McMenamin P, Gillies MC, Kettle E, Sarks SH.. Bruch's membrane and choroidal macrophages in early and advanced age-related macular degeneration. Br J Ophthalmol. 2010; 94: 918–925. [DOI] [PubMed] [Google Scholar]
- 58. Ezzat MK, Hann CR, Vuk-Pavlovic S, Pulido JS.. Immune cells in the human choroid. Br J Ophthalmol. 2008; 92: 976–980. [DOI] [PubMed] [Google Scholar]
- 59. Gupta N, Brown KE, Milam AH.. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp Eye Res. 2003; 76: 463–471. [DOI] [PubMed] [Google Scholar]
- 60. Roque RS, Rosales AA, Jingjing L, Agarwal N, Al-Ubaidi MR.. Retina-derived microglial cells induce photoreceptor cell death in vitro. Brain Res. 1999; 836: 110–119. [DOI] [PubMed] [Google Scholar]
- 61. Rutar M, Provis JM, Valter K.. Brief exposure to damaging light causes focal recruitment of macrophages, and long-term destabilization of photoreceptors in the albino rat retina. Curr Eye Res. 2010; 35: 631–643. [DOI] [PubMed] [Google Scholar]
- 62. Kye MJ, Goncalves Ido C.. The role of miRNA in motor neuron disease. Front Cell Neurosci. 2014; 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grabs D, Bergmann M, Urban M, Post A, Gratzl M.. Rab3 proteins and SNAP-25, essential components of the exocytosis machinery in conventional synapses, are absent from ribbon synapses of the mouse retina. Eur J Neurosci. 1996; 8: 162–168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.