Summary
Background
Manually extracted imaging-based body composition measures from a single-slice area (A) have shown associations with clinical outcomes in patients with cardiometabolic disease and cancer. With advances in artificial intelligence, fully automated volumetric (V) segmentation approaches are now possible, but it is unknown whether these measures carry prognostic value to predict mortality in the general population. Here, we developed and tested a deep learning framework to automatically quantify volumetric body composition measures from whole-body magnetic resonance imaging (MRI) and investigated their prognostic value to predict mortality in a large Western population.
Methods
The framework was developed using data from two large Western European population-based cohort studies, the UK Biobank (UKBB) and the German National Cohort (NAKO). Body composition was defined as (i) subcutaneous adipose tissue (SAT), (ii) visceral adipose tissue (VAT), (iii) skeletal muscle (SM), SM fat fraction (SMFF), and (iv) intramuscular adipose tissue (IMAT). The prognostic value of the body composition measures was assessed in the UKBB using Cox regression analysis. Additionally, we extracted body composition areas for every level of the thoracic and lumbar spine (i) to compare the proposed volumetric whole-body approach to the currently established single-slice area approach on the height of the L3 vertebra and (ii) to investigate the correlation between volumetric and single slice area body composition measures on the level of each vertebral body.
Findings
In 36,317 UKBB participants (mean age 65.1 ± 7.8 years, age range 45–84 years; 51.7% female; 1.7% [634/36,471] all-cause deaths; median follow-up 4.8 years), Cox regression revealed an independent association between VSM (adjusted hazard ratio [aHR]: 0.88, 95% confidence interval [CI] [0.81–0.91], p = 0.00023), VSMFF (aHR: 1.06, 95% CI [1.02–1.10], p = 0.0043), and VIMAT (aHR: 1.19, 95% CI [1.05–1.35], p = 0.0056) and mortality after adjustment for demographics (age, sex, BMI, race) and cardiometabolic risk factors (alcohol consumption, smoking status, hypertension, diabetes, history of cancer, blood serum markers). This association was attenuated when using traditional single-slice area measures. Highest correlation coefficients (R) between volumetric and single-slice area body composition measures were located at vertebra L5 for SAT (R = 0.820) and SMFF (R = 0.947), at L3 for VAT (R = 0.892), SM (R = 0.944), and at L4 for IMAT (R = 0.546) (all p < 0.0001). A similar pattern was found in 23,725 NAKO participants (mean age 53.9 ± 8.3 years, age range 40–75; 44.9% female).
Interpretation
Automated volumetric body composition assessment from whole-body MRI predicted mortality in a large Western population beyond traditional clinical risk factors. Single slice areas were highly correlated with volumetric body composition measures but their association with mortality attenuated after multivariable adjustment. As volumetric body composition measures are increasingly accessible using automated techniques, identifying high-risk individuals may help to improve personalised prevention and lifestyle interventions.
Funding
This project was conducted using data from the German National Cohort (NAKO) (www.nako.de). The NAKO is funded by the Federal Ministry of Education and Research (BMBF) [project funding reference numbers: 01ER1301A/B/C, 01ER1511D, and 01ER1801A/B/C/D], federal states of Germany and the Helmholtz Association, the participating universities and the institutes of the Leibniz Association. This research has been conducted using the UK Biobank Resource under Application Number 80337. MJ was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)-518480401. VKR was funded by American Heart Association Career Development Award 935176 and National Heart, Lung, and Blood Institute-K01HL168231.
Keywords: Magnetic resonance imaging, Artificial intelligence, Deep learning, Body composition, Public health, Mortality
Research in context.
Evidence before this study
We searched PubMed for articles between database inception (1946) and June 5, 2024, using the search terms ((MRI [Title/Abstract]) OR (Magnetic Resonance Imaging [Title/Abstract]) OR (CT [Title/Abstract]) OR (Computed Tomography [Title/Abstract])) AND ((Body Composition [Title/Abstract]) AND (Mortality [Title/Abstract])) AND (Humans [Filter]) NOT (Patients [Title/Abstract]) NOT (Systematic Review [Title/Abstract]). We identified 52 articles, of which 46 were original research articles. Of these 46 articles, 40 used cross-sectional imaging studies (computed tomography [CT] and magnetic resonance [MR] imaging) for body composition analysis. Of these, 30 articles focused on disease-specific populations. The remaining ten articles used single-slice CT images to measure body composition and assessed its association with adverse health outcomes in screening populations or population-based cohorts. Five out of these ten studies reported an association between two-dimensional muscle or adipose tissue area/density with mortality. Of the remaining five studies, four used small cohorts (n < 3.300). Only one study was performed in a larger cohort of 9223 adults who underwent routine colorectal cancer screening.
Added value of this study
In contrast, our study proposes a novel deep learning framework for three-dimensional body composition analysis from whole-body MR images and assesses the association with mortality in more than 36,000 individuals from the general population.
We showed that deep learning enables fully automated and robust quantification of volumetric body composition from whole-body MR imaging and predicts all-cause mortality beyond traditional clinical risk factors in more than 36,000 individuals from a large Western population. Although single-slice area measures of body composition, the most widely used approach in research studies, were highly correlated with volumetric measures, the association between area measures and mortality attenuated after multivariable adjustment.
Implications of all the available evidence
As volumetric body composition analysis from medical imaging studies becomes increasingly accessible using automated deep learning techniques, it could be used to opportunistically identify high-risk individuals to improve personalised prevention and lifestyle interventions.
Introduction
Body composition measures, including adipose tissue compartments and skeletal muscle, are associated with clinical outcomes in patients with prevalent cardiometabolic disease and cancer and have been identified as potential imaging biomarkers to improve personalised risk assessment and prognostication.1, 2, 3 Several studies have demonstrated added prognostic value of different body composition measures beyond traditional cardiovascular and oncological risk factors.4, 5, 6 However, routine quantification of body composition measures from cross-sectional imaging studies such as magnetic resonance imaging (MRI) is not performed in clinical radiology workflows due to time and equipment constraints.
With recent advances in artificial intelligence (AI), fully automated approaches to quantify volumetric body composition from cross-sectional imaging studies have become feasible. In contrast to other techniques, such as dual-energy X-ray absorptiometry, ultrasound, or superficial estimates (3D surface scanning or waist circumference), MRI may provide a more comprehensive estimate of an individual's body composition. This includes the capability to better characterise different tissue types (e.g., subdivide fat compartments and characterise muscle quality) and reflect their distribution within the body. In addition, an automated volumetric MRI approach may provide a more accurate estimate of an individual's body composition than the currently widely used single-slice area approach at the third lumbar vertebra, as it captures the full three-dimensional structure of the body rather than relying on a single cross-sectional slice.7, 8, 9, 10, 11
In this proof-of-concept study, we developed and tested a fully automated deep learning framework to estimate volumetric body composition defined as subcutaneous and visceral adipose tissue (SAT and VAT), skeletal muscle (SM), SM fat fraction (SMFF), and intramuscular adipose tissue (IMAT) from whole-body MRI studies across different sites, scanner types, and field strengths. We then tested whether these body composition measures were associated with all-cause mortality beyond traditional clinical risk factors in over 30,000 individuals from the UK Biobank (UKBB). Finally, we compared our framework to the commonly used method of estimating body composition from a single-slice area.12,13
Methods
Data sources
This study used data from two large population-based cohort studies: (i) the UK Biobank (UKBB, age range 45–84 years) and (ii) the NAKO Gesundheitsstudie/German National Cohort (NAKO, age range 40–75 years).14,15 Both studies collected detailed clinical information and acquired a comprehensive MR imaging protocol in a subgroup of participants, including an axial oriented whole-body T1-weighted 3D VIBE two-point Dixon sequence, which was used for body composition quantification in this study. Detailed information on the data sources is provided in Supplemental Methods.
Ethics
Informed consent was obtained from all participants in the UK Biobank and the German National Cohort study. In addition, we received local IRB approval (IRB of the University of Freiburg: 23-1316-S1-retro and 24-1099-S1-retro).
Overview of the study design
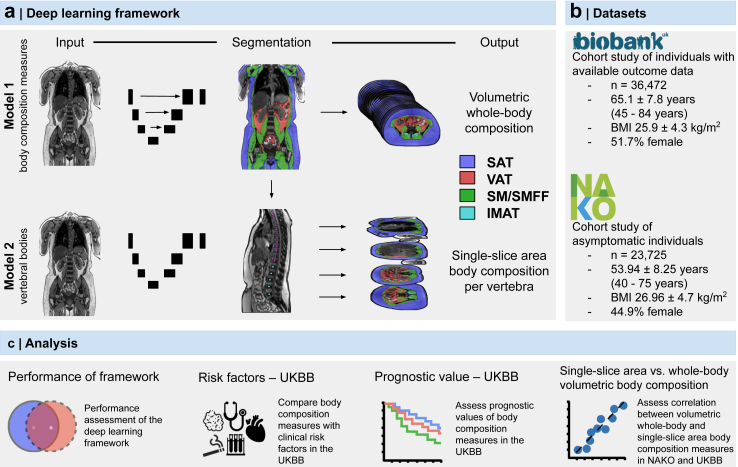
The primary aim of this study was to develop and test a deep learning model to automatically quantify volumetric body composition from whole-body MR imaging and investigate their prognostic value to predict mortality in a large sample of a Western population (UKBB). Additional aims were to compare the proposed volumetric whole-body approach to the currently established single-slice area approach on the height of the L3 vertebra and to investigate the correlation between volumetric whole-body and single-slice area body composition measures on the level of each vertebral body (UKBB and NAKO).9,16,17
For the survival analysis, the continuous body composition measures were categorised into groups (≥upper 10th percentile, middle 10–90th percentile, and low <10th percentile). To reduce overfitting, these thresholds were defined in the NAKO and then applied to the UKBB for all further analysis. The NAKO data was also used to replicate the correlation analysis between volumetric whole-body vs. single-slice body composition areas performed in the UKBB. An overview of the study design is provided in Fig. 1.
Fig. 1.
Overview of the study design. (a) The deep learning framework was developed to quantify body composition (SAT, VAT, SM, SMFF, and IMAT) from MR imaging as volumetric whole-body and single-slice area body composition at each level of the thoracic and lumbar spine. (b) Body composition was quantified in over 60,000 individuals from two large observational cohort studies. (c) After independent testing, the association, correlation, and prognostic value of the body composition measures were investigated. Biobank, UK biobank. IMAT, intramuscular adipose tissue. L, lumbar vertebra. NAKO, German National Cohort. SAT, subcutaneous adipose tissue. SM, skeletal muscle. SMFF, skeletal muscle fat fraction. T, thoracic vertebra. VAT, visceral adipose tissue. MRI, magnetic resonance imaging.
Deep learning framework development and testing
We propose a fully automated deep learning framework for body composition analysis from whole-body T1-weighted Dixon MR imaging that can quantify (i) volumetric whole-body and (ii) single-slice area body composition at the height of each thoracic and lumbar vertebra. Body composition was defined as subcutaneous and visceral adipose tissue (SAT and VAT), skeletal muscle (SM), SM fat fraction (SMFF), and intramuscular adipose tissue (IMAT). The only inputs to the framework were the axially oriented in- and opposed-phase images of the whole-body T1-weighted two-point VIBE Dixon MR sequence; the output of the model were segmentation masks for SAT, VAT, SM, and IMAT that estimated the whole-body volumes (dm3). The framework consists of two independent models (i) volumetric whole-body composition segmentation and (ii) spine labelling to extract single-slice body composition areas at the height of each thoracic and lumbar vertebra. All manual segmentations for model development and testing were generated by an experienced senior year radiology resident (5 years of experience in MR imaging) using all four image contrasts (in-phase, opposed-phase, water, fat) and three imaging planes (axial, as well as coronal and sagittal multiplanar reconstructions) on the T1-weighted Dixon sequences. For quality control, all segmentations were independently validated and adjusted where necessary by a board-certified radiologist (10 years of experience in MR imaging). We used the open-source NORA - medical imaging platform (www.nora-imaging.org, Freiburg, Germany) for all annotations and image-based calculations, model development, and testing. Further detailed information on model development and testing is provided in Supplemental Methods.
Endpoint in the UKBB
The primary endpoint of this study was all-cause mortality in the UKBB. The UKBB receives death notifications regularly through linkage to national death registries. The dates of death were obtained from baseline until the date of data download, May 25, 2023. All survival analyses were performed in the UKBB only, as outcome data were not available for the NAKO.
Covariates
Detailed information on the extraction and definition of covariates from UKBB and NAKO is provided in Supplemental Methods.
Statistical analysis
Data harmonisation
We observed minor distribution shifts between UKBB and NAKO data for SMFF and IMAT, likely due to technical artefacts, as described in Supplemental Methods. Distributions were harmonised before further analyses (Supplemental Methods).
Baseline demographics
Baseline characteristics of the UKBB and NAKO participants were presented as mean ± standard deviation (SD) or median with interquartile ranges (IQR) for continuous variables and absolute counts with percentages for categorical variables. Normality assumptions were informally assessed using Q–Q plots and histograms due to the large sample size in this study. Differences between males and females were assessed using Welch's t-test for normal and Mann–Whitney-U-Test for non-normally distributed variables.
Association between body composition measures and all-cause mortality in the UKBB
Survival analyses were only performed in the UKBB. The association between body composition measures and all-cause mortality was explored for continuous and categorised body composition measures. To avoid overfitting, we did not define the thresholds to categorise the continuous body composition measures into groups in the UKBB itself. Instead, we used body composition data from the NAKO to define cutoffs for risk categories in an independent dataset. Cutoffs were defined as high (≥10th percentile of NAKO participants), middle (10–90th percentile of NAKO participants), and low (<10th percentile of NAKO participants), which were calculated for males/females and volumetric/single-slice area body composition measures separately and are summarised in Supplemental Table S1. Subsequently, these cutoffs were applied to the UKBB.
To investigate time to death in the UKBB, Kaplan–Meier survival estimates and log-rank tests were computed using the above-defined body composition categories. The association between the continuous body composition measures and mortality was evaluated via univariable and multivariable Cox proportional hazards regression analysis adjusted for age, sex, BMI, race, alcohol consumption, smoking status, hypertension, diabetes, history of cancer, stroke, or myocardial infarction, total cholesterol, HDL cholesterol, LDL cholesterol, triglycerides, glucose, and HbA1c. These confounders were identified using a modified disjunctive cause criterion.
Follow-up time in the UKBB was calculated from the date of the MRI examination (start time and origin time for survival analysis) to the date of death, the date of loss to follow-up (0.02% [7/36,317] leaving the UK), or May 25, 2023 (date of UKBB data download), whichever came first. The proportional hazards assumption was tested by computing scaled Schoenfeld residuals, and linearity was assessed using Martingale residuals. Both assumptions were satisfied for all models. All results of Cox proportional hazards regression analyses were reported as Hazard Ratios (HR) per 1-unit change and 95% (2.5th and 97.5th percentiles) confidence intervals (CI). All analyses were performed using the volumetric whole-body composition measures. In addition, we present the same analyses for the currently established approach estimating body composition measures from a single-slice area at the height of the L3 vertebra to allow for a better comparison to the currently published literature. Furthermore, we calculated the continuous net reclassification improvement (NRI) for the volumetric vs single-slice area measures based on the above-defined risk categories using the nricens R package.18,19 95% (2.5th and 97.5th percentiles) CIs were estimated by using 5000 nonparametric bootstrap samples.
Correlation between volumetric whole-body and single-slice area body composition measures
To investigate the correlation between volumetric whole-body and single-slice area body composition measures at the height of each vertebra (thoracic and lumbar spine), Pearson's correlation coefficients were calculated for normally distributed body composition measures (SM, SMFF) and Kendall's rank correlation coefficient for non-normally distributed body composition measures (SAT, VAT, and IMAT) in the UKBB for the entire cohort and stratified by sex and BMI. Subsequently, these analyses were repeated with the NAKO data using a similar approach.
All statistical analyses were performed using R V4.2.1 (R Core Team, www.r-project.org, 2022). Statistical significance was indicated by p-values < 0.05.
Role of funders
The funders of this study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Study cohorts
The UKBB cohort consisted of 36,317 individuals (18,777 females and 17,540 males) with a mean age of 65.1 ± 7.8 years (age range 45–84 years) and a mean BMI of 25.9 ± 4.3 kg/m2. VSAT, VSMFF, and VIMAT were higher in females than in males (all p < 0.0001; Table 1). In contrast, males had higher VVAT and VSM (all p < 0.0001; Table 1).
Table 1.
Baseline characteristics – UKBB.
| Characteristic | Overall, N = 36,317a | Female, N = 18,777a | Male, N = 17,540a |
|---|---|---|---|
| Age (y) | 65.1 ± 7.8 | 64.4 ± 7.7 | 65.7 ± 7.9 |
| BMI (kg/m2) | 25.9 ± 4.3 | 25.4 ± 4.6 | 26.4 ± 3.8 |
| Race (white) | 35,124/36,221 (97%) | 18,170/18,735 (97%) | 16,954/17,486 (97%) |
| SAT (dm3) | 15.00 (IQR 11.62–19.38) | 17.10 (IQR 13.44–21.72) | 13.10 (IQR 10.43–16.45) |
| L3 SAT (dm2) | 2.03 (IQR 1.54–2.7) | 2.29 (IQR 1.67–3.05) | 1.83 (IQR 1.45–2.33) |
| VAT (dm3) | 3.50 (IQR 2.01–5.35) | 2.43 (IQR 1.44–3.65) | 5.02 (IQR 3.43–6.70) |
| L3 VAT (dm2) | 1.4 (IQR 0.78–2.26) | 0.94 (IQR 0.54–1.48) | 2.08 (IQR 1.37–2.89) |
| SM (dm3) | 11.44 ± 3.01 | 9.06 ± 1.34 | 13.99 ± 2.08 |
| L3 SM (dm2) | 1.30 ± 0.35 | 1.04 ± 0.17 | 1.59 ± 0.25 |
| SMFF (%) | 16.04 ± 3.25 | 17.20 ± 3.05 | 14.79 ± 2.99 |
| L3 SMFF (%) | 24.59 ± 5.63 | 26.36 ± 5.47 | 22.70 ± 5.16 |
| IMAT (10−1 dm3) | 1.42 (IQR 1.07–1.85) | 1.51 (IQR 1.18–1.92) | 1.31 (IQR 0.96–1.76) |
| L3 IMAT (10−1 dm2) | 0.40 (IQR 0.26–0.60) | 0.48 (IQR 0.33–0.68) | 0.32 (IQR 0.2–0.49) |
| Regular alcohol consumptionb | 25,679/36,033 (71%) | 12,120/18,612 (65%) | 13,559/17,421 (78%) |
| Ever smokersc | 14,804/36,154 (41%) | 6993/18,677 (37%) | 7811/17,477 (45%) |
| History of hypertension | 19,793/33,350 (59%) | 9001/17,083 (53%) | 10,792/16,267 (66%) |
| History of diabetes | 2057/35,985 (5.7%) | 745/18,593 (4.0%) | 1312/17,392 (7.5%) |
| History of stroke | 407/36,317 (1.1%) | 140/18,777 (0.7%) | 267/17,540 (1.5%) |
| History of myocardial infarction | 857/36,317 (2.4%) | 161/18,777 (0.9%) | 696/17,540 (4.0%) |
| History of cancer | 4345/35,981 (12%) | 2347/18,588 (13%) | 1998/17,393 (11%) |
| All-cause death | 633/36,317 (1.7%) | 222/18,777 (1.2%) | 411/17,540 (2.3%) |
| Follow up time (y) | 4.77 (IQR 3.92–6.12) | 4.78 (IQR 3.92–6.14) | 4.76 (IQR 3.90–6.09) |
IMAT, intramuscular adipose tissue. L3, 3rd lumbar vertebra. SAT, subcutaneous adipose tissue. SM, skeletal muscle. SMFF, skeletal muscle fat fraction. UKBB, UK biobank. VAT, visceral adipose tissue. Y, years.
Mean ± SD; median (IQR); n/N (%).
Regular alcohol consumption was defined as (“Daily or almost daily”, “Three or four times a week”, “Once or twice a week”).
“Ever smokers” includes former and current smokers.
Similar results were found for NAKO participants (23,725 individuals; mean age of 53.9 ± 8.3 years [age range 40–75 years] and a mean BMI of 27 ± 4.7 kg/m2; Table 2) and for single-slice area body composition measures at L3 in both cohorts.
Table 2.
Baseline characteristics – NAKO.
| Characteristic | Overall, N = 23,725 | Female, N = 10,651 | Male, N = 13,074 |
|---|---|---|---|
| Age (y) | 53.9 ± 8.3 | 54.1 ± 8.2 | 53.8 ± 8.3 |
| BMI (kg/m2) | 27 ± 4.7 | 26.4 ± 5.3 | 27.4 ± 4.1 |
| SAT (dm3) | 14.41 (IQR 10.96–19.10) | 16.71 (IQR 12.71–21.98) | 12.99 (IQR 10.05–16.62) |
| L3 SAT (dm2) | 2.01 (IQR 1.47–2.71) | 2.24 (IQR 1.57–3.07) | 1.88 (IQR 1.43–2.45) |
| VAT (dm3) | 3.35 (IQR 1.82–5.15) | 2.02 (IQR 1.12–3.29) | 4.62 (IQR 3.10–6.19) |
| L3 VAT (dm2) | 1.41 (IQR 0.72–2.26) | 0.81 (IQR 0.41–1.41) | 2.00 (IQR 1.27–2.73) |
| SM (dm3) | 12.62 ± 3.28 | 9.62 ± 1.45 | 15.05 ± 2.14 |
| L3 SM (dm2) | 1.43 ± 0.37 | 1.11 ± 0.17 | 1.70 ± 0.25 |
| SMFF (%) | 15.87 ± 3.25 | 17.20 ± 3.05 | 14.79 ± 2.99 |
| L3 SMFF (%) | 24.34 ± 5.61 | 26.36 ± 5.47 | 22.70 ± 5.16 |
| IMAT (10−1dm3) | 1.41 (IQR 1.05–1.85) | 1.51 (IQR 1.17–1.92) | 1.31 (IQR 0.96–1.76) |
| L3 IMAT (10−1dm2) | 0.40 (IQR 0.26–0.58) | 0.48 (IQR 0.34–0.68) | 0.33 (IQR 0.21–0.50) |
IMAT, intramuscular adipose tissue. L3, 3rd lumbar vertebra. NAKO, German National Cohort. SAT, subcutaneous adipose tissue. SM, skeletal muscle. SMFF, skeletal muscle fat fraction. VAT, visceral adipose tissue. Y, years.
Mean ± SD; median (IQR).
Association between body composition measures and all-cause mortality in the UKBB
Survival analysis using volumetric whole-body composition measures
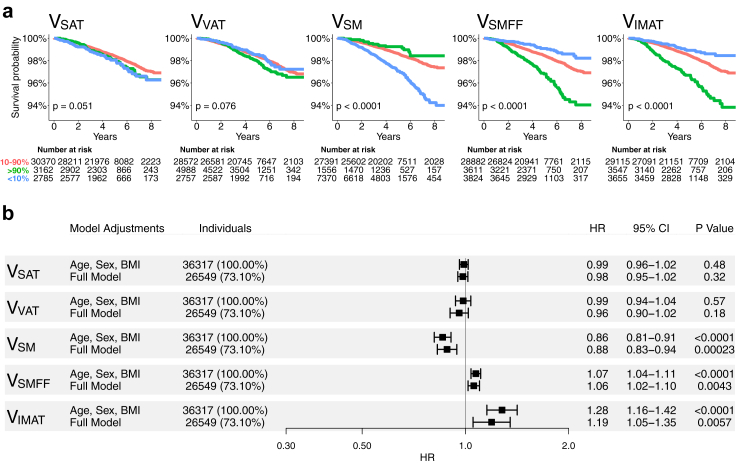
Over a median follow-up of 4.77 years (IQR 3.92–6.12 years), 634 deaths (1.7%) of all causes were observed. Detailed information on the UKBB covariates is shown in Table 1. Kaplan–Meier survival curves showed that the whole-body lower 10th percentile VSM category and the upper 10th percentile VSMFF and VIMAT category had a 6% mortality rate over 8 years, 3-fold higher than the middle category (Log-rank p < 0.0001; Fig. 2a).
Fig. 2.
Kaplan Meier survival estimates and forest plots for volumetric whole-body composition measures in the UKBB. (a) Overall survival in the UKBB for volumetric body composition according to 10th percentile (blue, <10%), mid 80th percentile (red, 10%–90%), and upper 10th percentile (green, >90%) of VSAT, VVAT, VSM, VSMFF, and VIMAT. Cutoffs were derived from the NAKO cohort to avoid overfitting. Kaplan Meier survival estimates show substantial differences between VSM, VSMFF, and VIMAT categories. (b) Forest plots show hazard ratios per 1-unit change with 95% confidence intervals of Cox proportional hazards regression analysis with endpoint all-cause mortality in the UKBB. Models were adjusted for age, sex, and BMI (upper row), and additional adjustment for traditional risk factors (lower row) for each body composition measure. VSM, VSMFF, and VIMAT showed robust associations with all-cause mortality beyond baseline demographics and traditional risk factors. Outcome data was only available in the UKBB (median follow-up 4.8 years). Full models were adjusted for age, sex, BMI, race, alcohol consumption, smoking status, hypertension, diabetes, history of stroke, history of myocardial infarction, history of cancer, and blood serum markers (TC, HDL, LDL, triglycerides, glucose, and HbA1c). IMAT, intramuscular adipose tissue. L, lumbar vertebra. MACE, major adverse cardiovascular event. NAKO, German National Cohort. SAT, subcutaneous adipose tissue. SM, skeletal muscle. SMFF, skeletal muscle fat fraction. UKBB, UK biobank. VAT, visceral adipose tissue.
In Cox regression adjusted for age, sex, and BMI, VSM was associated with a lower risk (aHR: 0.86, 95% CI [0.81–0.91], p < 0.0001), and VSMFF (aHR: 1.07, 95% CI [1.04–1.11], p < 0.0001) and VIMAT (aHR: 1.28, 95% CI [1.05–1.35], p < 0.0001) with a higher risk of all-cause mortality (Fig. 2b). These associations remained robust after further adjustment for race, alcohol consumption, smoking status, hypertension, diabetes, history of stroke, history of myocardial infarction, history of cancer, and blood serum markers (TC, HDL, LDL, triglycerides, glucose, HbA1c) (VSM, aHR: 0.88, 95% CI [0.83–0.94], p < 0.0001; VSMFF, aHR: 1.06, 95% CI [1.02–1.10], p = 0.004; VIMAT, aHR: 1.19, 95% CI [1.05–1.35], p = 0.0057; Fig. 2b). There was no substantial association between VSAT or VVAT and all-cause mortality (VSAT, aHR: 0.98, 95% CI [0.95–1.02], p = 0.32; VVAT, aHR: 0.96, 95% CI [0.90–1.02], p = 0.18; Fig. 2b).
Survival analysis using single-slice area measures at L3
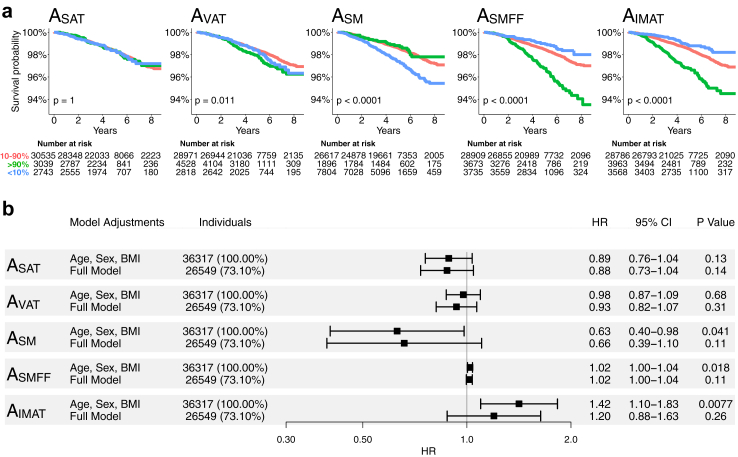
The Kaplan–Meier plots for the body composition risk categories for the single-slice areas showed results that were largely similar to those for the volumetric risk categories (Fig. 3a).
Fig. 3.
Kaplan Meier survival estimates and forest plots for single-slice area body composition measures in the UKBB. (a) Overall survival in the UKBB for single-slice area body composition according to 10th percentile (blue, <10%), mid 80th percentile (red, 10%–90%), and upper 10th percentile (green, >90%). Cutoffs were derived from the asymptomatic NAKO cohort to avoid overfitting. Kaplan Meier survival estimates showed graded associations for AVAT, ASM, ASMFF, and AIMAT categories. (b) Forest plots show hazard ratios per 1-unit change with 95% confidence intervals of Cox proportional hazards regression analysis with endpoint all-cause mortality in the UKBB. Models were adjusted for age, sex, and BMI (upper row), and additional adjustment for traditional risk factors (lower row) for each body composition measure. After full adjustment for baseline demographics and traditional risk factors, the association between single-slice area body composition measures and all-cause mortality could not be reliably estimated. Outcome data was only available in the UKBB (median follow-up 4.8 years). Full models were adjusted for age, sex, BMI, race, alcohol consumption, smoking status, hypertension, diabetes, history of stroke, history of myocardial infarction, history of cancer, and blood serum markers (TC, HDL, LDL, triglycerides, glucose, and HbA1c). IMAT, intramuscular adipose tissue. L, lumbar vertebra. MACE, major adverse cardiovascular event. NAKO, German National Cohort. SAT, subcutaneous adipose tissue. SM, skeletal muscle. SMFF, skeletal muscle fat fraction. UKBB, UK biobank. VAT, visceral adipose tissue.
In Cox proportional hazards regression analysis adjusted for age, sex, and BMI, ASM was associated with a lower risk (aHR: 0.63, 95% CI [0.40–0.98], p = 0.041), while ASMFF (aHR: 1.02, 95% CI [1.00–1.04], p = 0.018) and AIMAT (aHR: 1.42, 95% CI [1.10–1.83], p = 0.0077; Fig. 3b) were associated with a higher risk of all-cause mortality, similar to the volumetric whole-body measures. Conversely to the volumetric measures, however, these associations could not be reliably estimated after further adjustment for the risk factors listed above for single-slice area SM and IMAT (ASM, aHR: 0.66, 95% CI [0.39–1.10], p = 0.11; AIMAT, aHR: 1.20, 95% CI [0.88–1.63], p = 0.26; Fig. 3b). There was no substantial association between ASAT, AVAT, and ASMFF with all-cause mortality after adjustment (ASAT, aHR: 0.88, 95% CI [0.73–1.04], p = 0.14; AVAT, aHR: 0.93, 95% CI [0.82–1.07], p = 0.31; ASMFF, aHR: 1.02, 95% CI [1.00–1.04], p = 0.11; Fig. 3b).
In reclassification analysis based on the volumetric whole-body vs. single-slice area body composition measure categories (≥10th; 10–90th; <10th percentile) for each investigated measure, we found that the volumetric whole-body composition categories better identified high-risk individuals for all-cause mortality than the single-slice area categories with the highest NRI for SM (NRI = 0.053, 95% CI [0.016–0.089]; Supplemental Figure S3).
Correlation between volumetric whole-body and single-slice area body composition at different vertebral heights
The highest correlation coefficients between whole-body volumes and single-slice areas were found at L5 for SAT (R = 0.820, 95% CI [0.818–0.822]), at L3 for VAT (R = 0.892, 95% CI [0.891–0.893]), SM (R = 0.944, 95% CI [0.943–0.945]), L4 at IMAT (R = 0.546, 95% CI [0.541–0.550]), and at L5 for SMFF (R = 0.947, 95% CI [0.946–0.948] all p < 0.0001, Supplemental Table S2).
A largely similar pattern was observed in the NAKO (Supplemental Table S3). Additional stratified analyses by sex and BMI are provided in Supplemental Tables S4–S6 for the UKBB and in Supplemental Tables S7–S9 for the NAKO, which revealed substantial variability for the vertebral level with the highest correlation between volumetric whole-body and single-slice areas for the different body composition measures, which was more pronounced for BMI strata than for sex strata.
Testing of the deep learning framework
Overall performance of the volumetric whole-body composition segmentation model was high, with Dice coefficients ≥0.88 in the NAKO testing dataset and ≥0.86 in the UKBB testing dataset. Pearson's correlation coefficients assessing the linear relationship between manual and automatically generated volumetric segmentation masks were r > 0.99 in the NAKO testing dataset and r > 0.97 in the UKBB testing set. The spine labelling model's performance was highly accurate, with mean distance errors of the automated labels in the craniocaudal direction of −1 ± 7 mm in the NAKO testing dataset and 4 ± 8 mm in the UKBB testing dataset. The Dixon swap correction model correctly changed the swapped fat contrast to a water contrast and vice versa when a Dixon swap artefact was present in the original images and left the correct contrasts unchanged on a per-scan basis.
Detailed testing results of the proposed deep learning framework are provided in Supplemental Results and Supplemental Figures S4 and S5 and Supplemental Table S10.
Discussion
In this study, we developed and tested a fully automated deep learning framework to estimate body composition from whole-body MRI and investigated the prognostic value to predict mortality in over 30,000 individuals from a Western population. Our major findings were (i) that deep learning enables robust and reliable volumetric body composition analysis from whole-body MRI, (ii) that only volumetric whole-body measures were independent predictors for all-cause mortality in a large sample of a Western population beyond traditional demographic and cardiometabolic risk factors with a 12% lower (SM), 6% higher (SMFF), and 19% higher (IMAT) hazard per 1-unit change for all-cause mortality, and (iii) that single-slice body composition areas showed strong but highly sex- and BMI-dependent correlations with volumetric whole-body composition at varying levels.
These results are clinically relevant as medical imaging-based body composition quantification is increasingly recognised as a strong and independent prognostic factor in patients with prevalent cardiometabolic and oncologic diseases that can be quantified from different cross-sectional imaging studies and could complement currently established strategies to improve personalised risk assessment and clinical decision making in screening and hospital populations.4,20, 21, 22, 23 Our study demonstrated that body composition is an independent prognostic marker beyond traditional clinical risk factors not only in diseased patients but also in a large Western population, which could be integrated into clinical care as an opportunistic screening tool to help identify high-risk individuals to initiate personalised risk discussion and intensified prevention.24 This opportunistic body composition assessment could be performed regardless of the initial indication of an imaging test and could serve as an easy to obtain instrument to improve patient care.
However, to date, the use of body composition measures is limited to retrospective research studies because manual, expert-level segmentation is costly and time-consuming, which hinders seamless integration into current clinical workflows.25,26 To ease the labour-intensive and time-consuming manual segmentation process, most studies have measured body composition areas on a single axial slice12,13 as multiple analyses have reported high correlations between single-slice areas and whole-body composition volumes.9,16,17,27 While several height levels have been suggested for optimal single-slice area body composition estimation, the most widely used approach is a single-slice area at the level of the L3 vertebra.16,17,28 However, body composition areas, including height- and/or BMI-adjusted measures, are known to have substantial age-, sex-, and race-related variations and are strongly associated with standard anthropometric measures.2,29,30 In line with these results, we found strong correlations between single-slice and volumetric whole-body composition measures in over 60,000 individuals, but the vertebral level with the highest correlation coefficient showed substantial variation between the different body composition measures and was highly dependent on sex and BMI strata.31 This variability is an important limitation that reduces the accuracy, generalisability, and comparability between different studies and clinical settings with the risk that readily available and prognostically important information from routine medical imaging scans may go unnoticed.7,8
Advances in artificial intelligence have made automated analyses of large imaging cohorts feasible. Previously, Glasser et al.32,33 found that artificial intelligence could estimate all-cause mortality risk in older adults using 2D dual-energy absorptiometry images. In addition, Langner et al.33 showed that artificial intelligence could estimate body composition metrics from a two-dimensional representation of a whole-body MR image. To our knowledge, ours is the first study to test whether body composition measures derived from three-dimensional imaging are associated with clinical outcomes and the first to comprehensively estimate volumetric muscle and fat compartments using a manual segmentation gold standard. Our results in over 30,000 individuals showed that SM, SMFF, and IMAT were the body composition measures with the greatest predictive value, highlighting the advantage of MRI-based body composition assessment over other techniques used, such as dual-energy X-ray absorptiometry or surface scanning, which lack the ability to subdivide different fat compartments and assess SMFF. Our study further showed that the associations between single-slice body composition areas and mortality attenuated after multivariable adjustment for demographic and cardiometabolic risk factors, whereas the association for volumetric whole-body measures remained robust beyond these risk factors. In addition, reclassification analysis based on single-slice area vs. volumetric whole-body composition analysis showed that especially the VSM measure more frequently identified individuals at high risk for all-cause mortality than ASM measures. Based on these results and increasingly available fully automated tools, we consider volumetric body composition estimation to be the preferred method for clinical integration over the traditional single-slice area approach because it has the potential to (i) more accurately quantify an individual's true body composition, (ii) serve as a more robust prognostic factor, and (iii) facilitate comparability across studies and patient groups.
This study has limitations. First, the two investigated population-based cohort studies consist of predominantly white Western Europeans with a limited age range (40–84 years). Whether our results are generalisable to more heterogeneous groups needs to be investigated in future studies. Second, data for outcome analysis were only available in the UKBB with a median follow-up of 4.8 years; the adjusted hazard ratios reported in this study may not generalise to longer follow-up periods and are limited for proper causal inference due to differential depletion of the groups over time. This reduced our data set for assessing the association between body composition and all-cause mortality. Third, despite the possibility of integrating the fully automated deep learning framework for volumetric whole-body composition segmentation into clinical practice without disrupting routine radiology workflows, clinical use cases may currently be limited because whole-body MRI is not a commonly performed examination in daily routine, and it remains unclear whether typically imaged body regions (such as the liver or the pelvis) carry the same prognostic information as embedded in the volumetric whole-body measures. In addition, our model was developed for a T1-weighted VIBE Dixon sequence, which is widely used in clinical radiology but may not be part of every protocol. Furthermore, although our model provided robust results across sites, different scanner types, and field strengths, future studies will need to investigate whether the model also generalises to non-Siemens scans. Also, further studies are needed to assess whether our findings are reproducible in computed tomography (CT), where whole-body scans are routinely performed, e.g., for cancer staging. Finally, our study has methodological limitations, including potential unmeasured confounding and residual confounding due to measurement error in confounding variables such as smoking status and alcohol consumption. In addition, we did not perform a sample size estimation, as we used fixed sample sizes based on the two available cohorts. However, given the precise confidence intervals of the adjusted hazard ratios for the volumetric whole-body measures, the sample size seemed adequate for our analysis.
In conclusion, deep learning allowed for automated and robust quantification of body composition from whole-body MR imaging and predicted mortality in a large Western population beyond traditional clinical risk factors. Single-slice body composition measures were highly correlated with volumetric measures but the association with mortality attenuated after multivariable adjustment. As volumetric body composition measures are increasingly accessible using automated techniques, identifying high-risk individuals may help to improve personalised prevention and lifestyle interventions.
Contributors
Matthias Jung: conceptualisation, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualisation, writing – original draft.
Vineet K. Raghu: conceptualisation, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualisation, writing – original draft.
Marco Reisert: conceptualisation, data curation, formal analysis, methodology, resources, software, supervision, validation, writing – review & editing.
Hanna Rieder: data curation, formal analysis, methodology, resources, software, writing – review & editing.
Susanne Rospleszcz: conceptualisation, data curation, formal analysis, investigation, methodology, project administration, resources, supervision, validation, writing – review & editing.
Tobias Pischon: conceptualisation, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing – original draft, writing – review & editing.
Thoralf Niendorf: conceptualisation, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Hans-Ulrich Kauczor: conceptualisation, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Henry Völzke: conceptualisation, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Robin Bülow: conceptualisation, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Maximilian F. Russe: conceptualisation, data curation, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Christopher L. Schlett: conceptualisation, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Michael T. Lu: conceptualisation, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Fabian Bamberg: conceptualisation, data curation, funding acquisition, methodology, project administration, resources, software, supervision, validation, writing – review & editing.
Jakob Weiss: conceptualisation, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualisation, writing – original draft.
All authors read and approved the final version of the manuscript.
Data sharing statement
Due to the restrictions imposed by the NAKO ethics committee, the data are not publicly available. UKBB data is available by application to access the UK Biobank research resource.
Code sharing
The model will be made available via bitbucket (https://bitbucket.org/reisert/bodycompmodel).
Declaration of interests
Vineet K. Raghu: Grants or contracts from any entity: Norn Group, Johnson and Johnson Innovation, National Academy of Medicine.
Tobias Pischon: Leadership in board: Member of the board of directors of the NAKO e.V., who are leading the NAKO study (unpaid position).
Hans-Ulrich Kauczor: Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events: Siemens Healthineers, Philips, Boehringer Ingelheim Participation on a Data Safety Monitoring Board or Advisory Board: Median, contextflow.
Christopher L. Schlett: Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events: Siemens Healthineers, Bayer Healthcare.
Michael T. Lu: Grants or contracts from any entity: American Heart Association, AstraZeneca, Ionis, Johnson & Johnson Innovation, Kowa Pharmaceuticals America, MedImmune, National Academy of Medicine, National Heart, Lung, and Blood Institute, and Risk Management Foundation of the Harvard Medical Institutions.
Fabian Bamberg: Grants or contracts from any entity: Siemens Healthineers, Bayer Healthcare Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events: Siemens Healthineers, Bayer Healthcare.
Leadership or fiduciary role in other board, society, committee or advocacy group, paid or unpaid: German Roentgen Society.
Jakob Weiss: Consulting fees: Onc.AI.
Acknowledgements
This project was conducted using data from the German National Cohort (NAKO) (www.nako.de). The NAKO is funded by the Federal Ministry of Education and Research (BMBF) [project funding reference numbers: 01ER1301A/B/C, 01ER1511D and 01ER1801A/B/C/D], federal states of Germany and the Helmholtz Association, the participating universities and the institutes of the Leibniz Association. This research has been conducted using the UK Biobank Resource under Application Number 80337. We thank all participants who took part in the NAKO and UKBB study and the staff of these research initiatives. MJ was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)-518480401. VKR was funded by American Heart Association Career Development Award 935176 and National Heart, Lung, and Blood Institute-K01HL168231. Open Access funding enabled and organized by Projekt DEAL.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105467.
Contributor Information
Matthias Jung, Email: mjung6@mgh.harvard.edu, matthias.jung@uniklinik-freiburg.de.
Vineet K. Raghu, Email: vraghu@mgh.harvard.edu.
Marco Reisert, Email: marco.reisert@uniklinik-freiburg.de.
Hanna Rieder, Email: hanna.rieder@uniklinik-freiburg.de.
Susanne Rospleszcz, Email: susanne.rospleszcz@uniklinik-freiburg.de.
Tobias Pischon, Email: tobias.pischon@mdc-berlin.de.
Thoralf Niendorf, Email: thoralf.niendorf@mdc-berlin.de.
Hans-Ulrich Kauczor, Email: Hans-Ulrich.Kauczor@med.uni-heidelberg.de.
Henry Völzke, Email: voelzke@uni-greifswald.de.
Robin Bülow, Email: robin.buelow@med.uni-greifswald.de.
Maximilian F. Russe, Email: maximilian.russe@uniklinik-freiburg.de.
Christopher L. Schlett, Email: christopher.schlett@uniklinik-freiburg.de.
Michael T. Lu, Email: mlu@mgh.harvard.edu.
Fabian Bamberg, Email: fabian.bamberg@uniklinik-freiburg.de.
Jakob Weiss, Email: jakob.benedikt.weiss@uniklinik-freiburg.de.
Appendix A. Supplementary data
References
- 1.Neeland I.J., Ross R., Despres J.P., et al. Visceral and ectopic fat, atherosclerosis, and cardiometabolic disease: a position statement. Lancet Diabetes Endocrinol. 2019;7(9):715–725. doi: 10.1016/S2213-8587(19)30084-1. [DOI] [PubMed] [Google Scholar]
- 2.Bates D.D.B., Pickhardt P.J. CT-derived body composition assessment as a prognostic tool in oncologic patients: from opportunistic research to artificial intelligence-based clinical implementation. AJR Am J Roentgenol. 2022;219(4):671–680. doi: 10.2214/AJR.22.27749. [DOI] [PubMed] [Google Scholar]
- 3.Bradshaw P.T. Body composition and cancer survival: a narrative review. Br J Cancer. 2024;130(2):176–183. doi: 10.1038/s41416-023-02470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J.C., Caan B.J., Prado C.M., et al. Body composition and cardiovascular events in patients with colorectal cancer: a population-based retrospective cohort study. JAMA Oncol. 2019;5(7):967–972. doi: 10.1001/jamaoncol.2019.0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caan B.J., Cespedes Feliciano E.M., Prado C.M., et al. Association of muscle and adiposity measured by computed tomography with survival in patients with nonmetastatic breast cancer. JAMA Oncol. 2018;4(6):798–804. doi: 10.1001/jamaoncol.2018.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neeland I.J., Poirier P., Despres J.P. Cardiovascular and metabolic heterogeneity of obesity: clinical challenges and implications for management. Circulation. 2018;137(13):1391–1406. doi: 10.1161/CIRCULATIONAHA.117.029617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas E.L., Bell J.D. Influence of undersampling on magnetic resonance imaging measurements of intra-abdominal adipose tissue. Int J Obes Relat Metab Disord. 2003;27(2):211–218. doi: 10.1038/sj.ijo.802229. [DOI] [PubMed] [Google Scholar]
- 8.Shen W., Chen J., Gantz M., Velasquez G., Punyanitya M., Heymsfield S.B. A single MRI slice does not accurately predict visceral and subcutaneous adipose tissue changes during weight loss. Obesity (Silver Spring) 2012;20(12):2458–2463. doi: 10.1038/oby.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faron A., Luetkens J.A., Schmeel F.C., Kuetting D.L.R., Thomas D., Sprinkart A.M. Quantification of fat and skeletal muscle tissue at abdominal computed tomography: associations between single-slice measurements and total compartment volumes. Abdom Radiol (NY) 2019;44(5):1907–1916. doi: 10.1007/s00261-019-01912-9. [DOI] [PubMed] [Google Scholar]
- 10.Koitka S., Kroll L., Malamutmann E., Oezcelik A., Nensa F. Fully automated body composition analysis in routine CT imaging using 3D semantic segmentation convolutional neural networks. Eur Radiol. 2021;31(4):1795–1804. doi: 10.1007/s00330-020-07147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shepherd J.A., Ng B.K., Sommer M.J., Heymsfield S.B. Body composition by DXA. Bone. 2017;104:101–105. doi: 10.1016/j.bone.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prado C.M., Lieffers J.R., McCargar L.J., et al. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: a population-based study. Lancet Oncol. 2008;9(7):629–635. doi: 10.1016/S1470-2045(08)70153-0. [DOI] [PubMed] [Google Scholar]
- 13.Cao K., Yeung J., Arafat Y., Wei M.Y.K., Yeung J.M.C., Baird P.N. Identification of differences in body composition measures using 3D-derived artificial intelligence from multiple CT scans across the L3 vertebra compared to a single mid-point L3 CT scan. Radiol Res Pract. 2023;2023 doi: 10.1155/2023/1047314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudlow C., Gallacher J., Allen N., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlett C.L., Hendel T., Weckbach S., et al. Population-based imaging and radiomics: rationale and perspective of the German National Cohort MRI Study. Röfo. 2016;188(7):652–661. doi: 10.1055/s-0042-104510. [DOI] [PubMed] [Google Scholar]
- 16.Shen W., Punyanitya M., Wang Z., et al. Visceral adipose tissue: relations between single-slice areas and total volume. Am J Clin Nutr. 2004;80(2):271–278. doi: 10.1093/ajcn/80.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irlbeck T., Massaro J.M., Bamberg F., O'Donnell C.J., Hoffmann U., Fox C.S. Association between single-slice measurements of visceral and abdominal subcutaneous adipose tissue with volumetric measurements: the Framingham Heart Study. Int J Obes (Lond) 2010;34(4):781–787. doi: 10.1038/ijo.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pencina M.J., D'Agostino R.B., Sr., Steyerberg E.W. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30(1):11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue E. R Package ‘nricens’. 2018. https://cran.r-project.org/web/packages/nricens/nricens.pdf
- 20.Pickhardt P.J., Graffy P.M., Zea R., et al. Automated CT biomarkers for opportunistic prediction of future cardiovascular events and mortality in an asymptomatic screening population: a retrospective cohort study. Lancet Digit Health. 2020;2(4):e192–e200. doi: 10.1016/S2589-7500(20)30025-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nachit M., Horsmans Y., Summers R.M., Leclercq I.A., Pickhardt P.J. AI-Based CT body composition identifies myosteatosis as key mortality predictor in asymptomatic adults. Radiology. 2023;307(5) doi: 10.1148/radiol.222008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd M.A., Amin J., Mallon P.W., et al. Body composition and metabolic outcomes after 96 weeks of treatment with ritonavir-boosted lopinavir plus either nucleoside or nucleotide reverse transcriptase inhibitors or raltegravir in patients with HIV with virological failure of a standard first-line antiretroviral therapy regimen: a substudy of the randomised, open-label, non-inferiority SECOND-LINE study. Lancet HIV. 2017;4(1):e13–e20. doi: 10.1016/S2352-3018(16)30189-8. [DOI] [PubMed] [Google Scholar]
- 23.Bazzocchi A., Diano D., Battista G. How fat is fat? Lancet. 2012;380(9837):e1. doi: 10.1016/S0140-6736(11)61925-9. [DOI] [PubMed] [Google Scholar]
- 24.Pickhardt P.J. Value-added opportunistic CT screening: state of the art. Radiology. 2022;303(2):241–254. doi: 10.1148/radiol.211561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigiroli F., Zhang D., Molinger J., et al. Automated versus manual analysis of body composition measures on computed tomography in patients with bladder cancer. Eur J Radiol. 2022;154 doi: 10.1016/j.ejrad.2022.110413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paris M.T. Body composition analysis of computed tomography scans in clinical populations: the role of deep learning. Lifestyle Genom. 2020;13(1):28–31. doi: 10.1159/000503996. [DOI] [PubMed] [Google Scholar]
- 27.Derstine B.A., Holcombe S.A., Ross B.E., Wang N.C., Su G.L., Wang S.C. Optimal body size adjustment of L3 CT skeletal muscle area for sarcopenia assessment. Sci Rep. 2021;11(1):279. doi: 10.1038/s41598-020-79471-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maislin G., Ahmed M.M., Gooneratne N., et al. Single slice vs. volumetric MR assessment of visceral adipose tissue: reliability and validity among the overweight and obese. Obesity (Silver Spring) 2012;20(10):2124–2132. doi: 10.1038/oby.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz-Jentoft A.J., Bahat G., Bauer J., et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing. 2019;48(1):16–31. doi: 10.1093/ageing/afy169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng X., Zhang Y., Wang C., et al. The optimal anatomic site for a single slice to estimate the total volume of visceral adipose tissue by using the quantitative computed tomography (QCT) in Chinese population. Eur J Clin Nutr. 2018;72(11):1567–1575. doi: 10.1038/s41430-018-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koster A., Murphy R.A., Eiriksdottir G., et al. Fat distribution and mortality: the AGES-Reykjavik Study. Obesity (Silver Spring) 2015;23(4):893–897. doi: 10.1002/oby.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaser Y., Shepherd J., Leong L., et al. Deep learning predicts all-cause mortality from longitudinal total-body DXA imaging. Commun Med (Lond) 2022;2:102. doi: 10.1038/s43856-022-00166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langner T., Strand R., Ahlstrom H., Kullberg J. Large-scale biometry with interpretable neural network regression on UK Biobank body MRI. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-74633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.