ABSTRACT
Ependymoma is the second most common malignant paediatric brain tumour composed of nine methylation‐defined, clinically relevant subgroups. It is unclear if there are sex differences in methylation profiles within these subgroups which could guide future treatment options. We obtained available methylation data from the National Center for Biotechnology Information Gene Expression Omnibus (GEO). Differentially methylated probes (DMPs) between sexes were identified in each ependymoma sample and mapped to genes. Reactome pathways resulting from genes were identified. Survival was estimated for each sex within molecular subgroups. There were 492 cases included in the main analysis: PF‐EPN‐A (n = 238) PF‐EPN‐B (n = 52), PF‐SE (n = 34), SP‐MPE (n = 26), SP‐EPN (n = 21), ST‐EPN‐RELA (n = 87), ST‐EPN‐YAP1 (n = 13) and ST‐SE (n = 21). Females were observed to have better, but statistically nonsignificant, 5‐year overall survival (OS) and better, marginally significant 5‐year progression‐free survival (PFS) than males. One subgroup, ST‐EPN‐RELA, showed significantly better OS in females. There was a difference in immune cell composition within tumour subgroups. One gene, RFTN1, was consistently differentially methylated by sex among all subgroups. There were biologic pathways identified from genes with differential methylation by sex in the following subgroups: PF‐EPN‐B, PF‐SE, ST‐EPN‐RELA and ST‐EPN‐YAP1. Many of the identified pathways may be options for potential therapeutic targets.
1. Introduction
Ependymoma is the third most common paediatric brain tumour and the second most common malignant paediatric brain tumour [1, 2, 3, 4]. On average, it affects 200 children in the United States annually [5] and accounts for around 10% of brain tumours in children [3]. Historically, ependymal tumours were classified by histopathology regardless of location [4], which was of little use in predicting tumour behaviour [3, 6]. More recently, clinically relevant and prognostic classification methods have been prioritised and, now using methylation data, nine subgroups have been identified [3, 6, 7]. These subgroups are found among three different anatomical areas, including supratentorial (ST), posterior fossa (PF) and the spinal cord [3]. Subgroups include three different entities within each anatomic compartment [8]. For example, supratentorial ependymomas are associated with RELA (poor outcome) or YAP fusions (better outcomes) [4], whereas adults generally have spinal tumours associated with changes in the NF2 gene and tumours are of the SP‐MPE subgroup [7]. Overall survival (OS) is around 60% in paediatric patients [7, 8], but progression‐free survival (PFS) is much lower, around 20%–50% depending on the methylation‐defined subgroup [2, 6]. How these outcomes vary by sex has been less frequently explored in the paediatric population.
There are distinct sex differences in ependymoma incidence such that males have an estimated 23% excess in incidence compared to females in the paediatric population [9]. Additionally, we have reported on sex differences in ependymoma survival that were not mediated by the stage of disease at diagnosis, suggesting that male sex itself may be a risk factor for death after ependymoma diagnosis [10]. When looking at ependymoma outcomes generally, males have worse OS when compared to females with 5‐year OS of 71% as opposed to 78% in females [10]. Most epidemiologic studies lack methylation‐based subgroups, but there is evidence of sex differences in the distribution of these subgroups from published clinical case series [6].
DNA methylation helps regulate gene expression, but aberrant methylation can be associated with gene transcription modifications that are pathogenic [11]. Methylation differences based on sex are seen throughout the body in different organ systems [12]. Given the male excess in ependymoma incidence and death, along with our knowledge that methylation plays a role in the development of cancers, particularly in brain tumour formation and progression [12], we sought to identify sex differences in tumour methylation profiles within prognostic subgroups of ependymoma. Using publicly available data from Pajtler et al. (2015) [6], we identified sex differences in survival and methylation profiles and then validated the methylation findings in the Molecular Characterisation Initiative [13] using tumour data from individuals with ependymoma for some subgroups.
2. Materials and Methods
2.1. Data Source
Publicly available DNA methylation data from Pajtler et al. (2015) [6] were used for this analysis (GEO Series accession number GEO: GSE65362, date downloaded: 30 May 2021). There were 492 ependymoma samples from the initial diagnosis. Subgroups were included in the analysis if there were greater than 10 samples within the subgroup. Subgroups included in the analysis were PF‐EPN‐A (n = 238), PF‐EPN‐B (n = 52), PF‐SE (n = 34), SP‐EPN (n = 21), SP‐MPE (n = 26), ST‐SPN‐RELA (n = 87), ST‐TPN‐YAP1 (n = 13) and ST‐SE (n = 21). Already normalised (preprocessIllumina) methylated and unmethylated signal data were available. Clinical data for the Pajtler samples were obtained under Institutional Review Board approval at the University of Minnesota. Additional DNA methylation data (IDAT files, EPIC v1 array) for ependymoma tumours were obtained from Childhood Cancer Data Initiative (CCDI) Molecular Characterisation Inititative (MCI) [13]. The table of diagnosis and patient's included for both Pajtler and MCI can be found in Table 1.
TABLE 1.
Clinical data and predicted molecular subtypes of ependymoma by predicted sex in Pajtler and COG MCI.
| Pajtler | N = 492 | Male, N = 302 | Female, N = 190 |
|---|---|---|---|
| Predicted subtypes, n (%) | |||
| PF‐EPN‐A | 238 | 154 (65) | 84 (35) |
| PF‐EPN‐B | 52 | 21 (40) | 31 (60) |
| PF‐SE | 34 | 26 (76) | 8 (24) |
| SP‐EPN | 21 | 13 (62) | 8 (38) |
| SP‐MPE | 26 | 14 (54) | 12 (46) |
| ST‐EPN‐RELA | 87 | 56 (64) | 31 (36) |
| ST‐EPN‐YAP1 | 13 | 3 (23) | 10 (77) |
| ST‐SE | 21 | 15 (71) | 6 (29) |
| Vital status, n (%) | |||
| Alive | 273 | 154 (56) | 119 (44) |
| Death | 83 | 56 (67) | 27 (33) |
| Missing | 136 | 92 | 44 |
| Progression, n (%) | |||
| Not progression | 168 | 87 (52) | 81 (48) |
| Progression | 198 | 127 (64) | 71 (36) |
| Missing | 126 | 88 | 38 |
| COG MCI | N = 103 | Male, N = 58 | Female, N = 45 |
| Predicted subtypes, n (%) | |||
| PF‐EPN‐A | 45 | 23 (51) | 22 (49) |
| PF‐EPN‐B | 34 | 24 (71) | 10 (29) |
| SP‐MPE | 14 | 5 (36) | 9 (64) |
| ST‐EPN‐RELA | 10 | 6 (60) | 4 (40) |
Note: One case diagnosed with astrocytoma was excluded. Distributions of molecular subtypes of ependymoma in Pajtler and COG APEC14B1‐MCI were statistically significantly different by sex according to Fisher's exact test for count data with simulated p value (based on 2000 replicates).
2.2. Survival Analysis
Five‐year OS and PFS were estimated for males and females in all subgroups combined and within molecular subgroups of ependymoma. We defined OS as the time from the date of diagnosis of ependymoma to the date of the last follow‐up or date of death in months (n = 359), and PFS as the time from the date of diagnosis of ependymoma to the date of cancer progression in months (n = 365). We estimated 5‐year OS and PFS (95% confidence intervals) by sex in all subgroups combined and within molecular subgroups of ependymoma. Kaplan–Meier (KM) curves were constructed to identify sex differences in the 5‐year OS and PFS by sex in all subgroups combined and within molecular subgroups of ependymoma. The log‐rank p values were estimated to identify sex differences in 5‐year OS and PFS. We used R version 4.2.2 (2022‐10‐31 ucrt) and survival package survminor to implement survival analysis.
2.3. Quality Control and Sex Prediction
For each dataset, badly performing arrays (samples) were identified and removed using minfi's get QC function. Prenormalised methylated and unmethylated signals were used for the Pajtler dataset, while the MCI dataset was normalised using preprocess Funnorm. Probes containing known SNPs and probes with detection p values > 0.01 were removed from the dataset. Sex prediction was performed using minfi getSex on each dataset. As the Pajtler and MCI datasets came from different arrays, overlapping probes were selected for each dataset. Beta values for arrays (samples) and probes that passed QC and are present on both arrays were included in the final dataset.
2.4. Subgroup Prediction
Beta values for each dataset were combined, and study‐related batch effects were removed using limma removeBatchEffects(). The Pjalter dataset included subgroup information for each tumour and was used to create a random forest model to predict subgroups for the MCI dataset. Batch‐corrected beta values for the Pjalter dataset were divided into a training set (70% of the dataset) and a test set (30% of the dataset). Principal components (PCs) were calculated from the top 10,000 most variable probes. The top 10 PCs were used to train a random forest model (randomForest()) to predict subgroups from the training set. This model was then used to predict subgroups for the training set resulting in an OOB (out of bag) error of 0.75% (1 wrong call). The beta values for the probes included in the model were then isolated from the MCI dataset, and the top 10 PCs were calculated. These PCs were used as input for the model to predict subgroups for the MCI dataset.
2.5. Differential Methylation Analysis
All samples and probes that passed QC in the Pajtler dataset were used for differential methylation analysis using dmpFinder. DMP analysis was done between males and females within each subgroup. Significant DMPs had False discovery rate (FDR) < 0.05. DMPs were mapped to the nearest gene using the annotation information that comes with the array. Heatmaps were generated using the normalised beta values for significant DMPs.
2.6. Reactome Pathway Analysis
We identified biological pathways that had significant sex‐DMP in each subgroup of ependymoma. This was done using reactome pathway analysis. Pathways of interest were chosen if their FDR was 10% or less.
2.7. Immune Cell Profiling Based on Methylation Values
To assess immune cell composition within each tumour subgroup by sex and sex overall, we used the MethylCIBERTSORT package (version 0.2.0) [14] and CIBERSORTx (https://cibersortx.stanford.edu/) [15]. Beta values for reference probes were uploaded as percentages to CIBERSORTx. Using 1000 permutations without quartile normalisation, the data were analysed, and boxplots were created in R.
2.8. Validation Analysis
We validated the DMP results from Pajtler data in a linked dataset from the CCDI: MCI [16]. MCI samples were run on the Illumina Human Methylation EPIC array. Methylation data for cases were pulled on (date: 12 June 2024). The R package minfi was used to perform quality control (minfi plotQC) and sex prediction (minfi getSex) following the methods described in the section Quality Control and Sex Prediction. According to the bad sample cutoff equal to 10.5, samples with problematic probes that had lower median intensities and did not cluster were removed. Subgroups were removed if there were not at least 10 samples within the subgroup. Subgroups that remained in the analysis included: PF‐EPN‐A, PF‐EPN‐B, SP‐MPE and ST‐EPN‐RELA. A total of 103 samples were retained in the analysis (male: 58 (56%) vs. female: 45 (44%)). Two‐sample t‐tests were used to examine statistically significant differences in DNA methylation within molecular subgroups of ependymoma by sex, as we had done in the Pajtler data. FDR correction was performed for adjusting multiple comparisons, FDR cutoff used was < 0.05. Furthermore, an agreement between the results regarding sex differences in DNA methylation within molecular subgroups of ependymoma in the training and validation datasets was defined as the same direction of mean differences between the two sexes.
3. Results
There were 492 cases included in the discovery analysis: PF‐EPN‐A (n = 238) PF‐EPN‐B (n = 52), PF‐SE (n = 34), SP‐MPE (n = 26), SP‐EPN (n = 21), ST‐EPN‐RELA (n = 87), ST‐EPN‐YAP1 (n = 13) and ST‐SE (n = 21).
Females were detected as having better, but statistically insignificant 5‐year (82% vs. 73%) OS compared to males. Females also showed better and marginally significant 5‐year PFS overall than males (49% vs. 37%). One subgroup, ST‐EPN‐RELA, also showed significantly better 5‐year OS of females when compared to males (100% vs. 63%). Both females and males were reported 100% 5‐year OS in PF‐SE, SP‐EPN, SP‐MPE, SP‐SE and ST‐EPN‐YAP1. These results are found in Table 2 and Figure 1.
TABLE 2.
5‐Year overall survival and progression‐free survival estimates by sex in molecular subtypes of ependymoma.
| Characteristic | 5‐Year overall survival | 5‐Year progression‐free survival | |||||
|---|---|---|---|---|---|---|---|
| N | Event N | Probability (95% CI) | N | Event N | Probability (95% CI) | ||
| Total | |||||||
| Overall | 359 | 60 | 76% (71%, 82%) | 359 | 78 | 69% (63%, 76%) | |
| Predicted sex | 359 | 60 | 359 | 78 | |||
| Male | 212 | 42 | 73% (66%, 80%) | 212 | 54 | 64% (56%, 73%) | |
| Female | 147 | 18 | 82% (74%, 90%) | 147 | 24 | 76% (68%, 86%) | |
| PF‐EPN‐A | |||||||
| Overall | 199 | 48 | 67% (59%, 75%) | 199 | 57 | 60% (51%, 69%) | |
| Predicted sex | 199 | 48 | 199 | 57 | |||
| Male | 129 | 31 | 68% (59%, 78%) | 129 | 39 | 57% (47%, 69%) | |
| Female | 70 | 17 | 65% (53%, 81%) | 70 | 18 | 64% (51%, 80%) | |
| PF‐EPN‐B | |||||||
| Overall | 48 | 1 | 96% (89%, 100%) | 48 | 3 | 93% (85%, 100%) | |
| Predicted sex | 48 | 1 | 48 | 3 | |||
| Male | 19 | 0 | 100% (100%, 100%) | 19 | 1 | 94% (83%, 100%) | |
| Female | 29 | 1 | 93% (82%, 100%) | 29 | 2 | 92% (83%, 100%) | |
| PF‐SE | |||||||
| Overall | 9 | 0 | 100% (100%, 100%) | 9 | 0 | 100% (100%, 100%) | |
| Predicted sex | 9 | 0 | 9 | 0 | |||
| Male | 4 | 0 | 100% (100%, 100%) | 4 | 0 | 100% (100%, 100%) | |
| Female | 5 | 0 | 100% (100%, 100%) | 5 | 0 | 100% (100%, 100%) | |
| SP‐EPN | |||||||
| Overall | 9 | 0 | 100% (100%, 100%) | 9 | 0 | 100% (100%, 100%) | |
| Predicted sex | 9 | 0 | 9 | 0 | |||
| Male | 4 | 0 | 100% (100%, 100%) | 4 | 0 | 100% (100%, 100%) | |
| Female | 5 | 0 | 100% (100%, 100%) | 5 | 0 | 100% (100%, 100%) | |
| SP‐MPE | |||||||
| Overall | 1 | 0 | 100% (100%, 100%) | 1 | 0 | 100% (100%, 100%) | |
| Predicted sex | 1 | 0 | 1 | 0 | |||
| Male | 1 | 0 | 100% (100%, 100%) | 1 | 0 | 100% (100%, 100%) | |
| Female | 0 | NA | NA | 0 | NA | NA | |
| ST‐EPN‐RELA | |||||||
| Overall | 77 | 11 | 76% (65%, 90%) | 77 | 18 | 54% (39%, 76%) | |
| Predicted sex | 77 | 11 | 77 | 18 | |||
| Male | 49 | 11 | 63% (47%, 84%) | 49 | 14 | 51% (34%, 76%) | |
| Female | 28 | 0 | 100% (100%, 100%) | 28 | 4 | 64% (38%, 100%) | |
| ST‐EPN‐YAP1 | |||||||
| Overall | 10 | 0 | 100% (100%, 100%) | 10 | 0 | 100% (100%, 100%) | |
| Predicted sex | 10 | 0 | 10 | 0 | |||
| Male | 1 | 0 | 100% (100%, 100%) | 1 | 0 | NA | |
| Female | 9 | 0 | 100% (100%, 100%) | 9 | 0 | 100% (100%, 100%) | |
| ST‐SE | |||||||
| Overall | 6 | 0 | 100% (100%, 100%) | 6 | 0 | 100% (100%, 100%) | |
| Predicted sex | 6 | 0 | 6 | 0 | |||
| Male | 5 | 0 | 100% (100%, 100%) | 5 | 0 | 100% (100%, 100%) | |
| Female | 1 | 0 | NA | 1 | 0 | NA | |
Abbreviation: CI, confidence interval.
FIGURE 1.
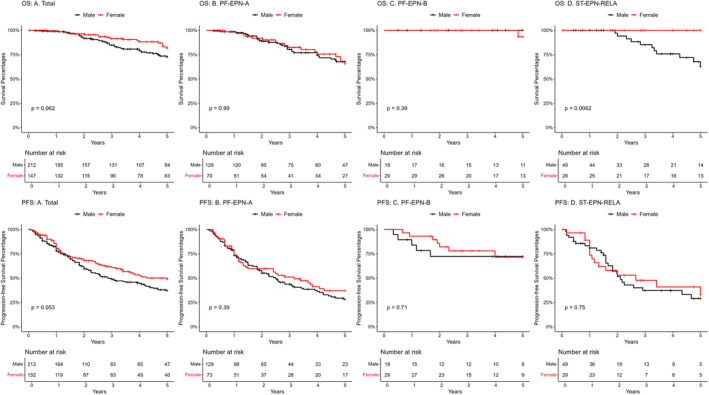
Kaplan–Meier survival curves showing five‐year overall survival (OS) and five‐year progression‐free survival (PFS) for those subgroups with less than 100% survival at 5 years by sex from the Pajtler data. (OS: A) Total, (OS: B) PF‐EPN‐A, (OS: C) PF‐EPN‐B, (OS: D) ST‐EPN‐RELA, (PFS: A) Total, (PFS: B) PF‐EPN‐A, (PFS: C) PF‐EPN‐B and (PFS: D) ST‐EPN‐RELA.
We observed statistically significant sex differences in DNA methylation within each included subgroup, except for SP‐EPN. PF‐SE had the highest number of DMPs by sex (n = 199), followed by PF‐EPN‐A (n = 164), SP‐MPE (n = 44), ST‐EPN‐RELA (n = 18), PF‐EPN‐B (n = 24), ST‐EPN‐YAP1 (n = 15) and ST‐SE (n = 7). Statistically significant DMPs by sex mapped to the gene RFTN1 in all the subgroups. Unsupervised hierarchical clustering of significant DMPs by sex within each subgroup was done. We found clustering was relatively strong by sex, but not by other characteristics as shown in the heat maps (Figure 2). PF‐EPN‐B, SP‐MPE, ST‐EPN‐YAP1 and ST‐SE each showed complete clustering by sex‐DMPs.
FIGURE 2.
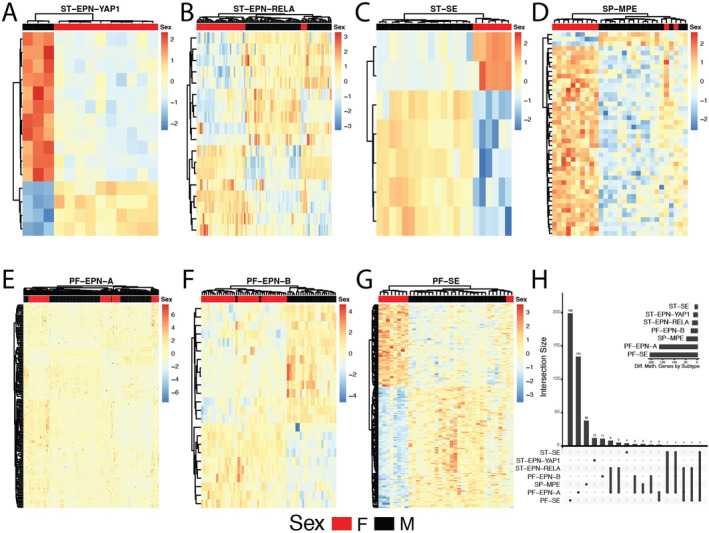
Heatmaps showing methylation levels (row‐scaled b‐values) of statistically significant differentially methylated positions (DMPs) by sex within ependymoma subgroups: (A) ST‐EPN‐YAP1, (B) ST‐EPN‐RELA, (C)ST‐SE, (D) SP‐MPE, (E) PF‐EPN‐A, (F) PF‐EPN‐B, (G) PF‐SE and (H) Upset plot showing unique and shared differentially methylated genes across ependymoma subgroups from the Pajtler data.
Pathways resulting from sex‐DMPs mapped to nearest gene for each subgroup are presented in Table 3. There were specific pathways identified from genes that were differentially methylated by sex in the following subgroups: PF‐EPN‐B, PF‐ SE, ST‐EPN‐RELA, ST‐EPN‐YAP1. Top pathways in PF‐EPN‐B include RNA polymerase transcription and promoter clearance as well as SMAD domain mutants in cancer. The top pathways in PF‐SE include FGFR2 signalling and activation. The top pathways in ST‐EPN‐RELA include mitochondrial and keratan sulfate biosynthesis. Many significant pathways from genes with sex DMPs were found in ST‐EPN‐YAP1 and include ERBB2 signalling, PI3K signalling, Interferon‐gamma and other cytokine signalling.
TABLE 3.
Reactome pathway analysis (FDR < 10%) using genes that had sex‐DMPs identified within each subgroup from the Pajtler data. No pathways were identified for PF‐EPN‐A, ST‐SE and SP‐MPE.
| Reactome pathway name | FDR |
|---|---|
| PF‐EPN‐B | |
| Mitochondrial biogenesis | 0.09443772 |
| RNA polymerase transcription | 0.09443772 |
| HIV transcription initiation | 0.09443772 |
| RNA polymerase II transcription initiation | 0.09443772 |
| RNA polymerase II HIV promoter escape | 0.09443772 |
| RNA polymerase II promoter escape | 0.09443772 |
| RNA polymerase II transcription initiation and promoter clearance | 0.09443772 |
| Loss of function SMAD4 in cancer | 0.09443772 |
| SMAD4 MH2 domain mutants in cancer | 0.09443772 |
| SMAD2/3 MH2 domain mutants in cancer | 0.09443772 |
| PF‐SE | |
| Signalling by FGFR2 IIIa TM | 0.02478109 |
| FGFR2 mutant receptor activation | 0.02478109 |
| Signalling by FGFR2 in disease | 0.08176046 |
| ST‐EPN‐RELA | |
| Mitochondrial biogenesis | 0.09483678 |
| Keratan sulfate biosynthesis | 0.09483678 |
| ST‐EPN‐YAP1 | |
| GRB7 events in ERBB2 signalling | 0.00282658 |
| Downregulation of ERBB2:ERBB3 signalling | 0.00463631 |
| Constitutive signalling by aberrant PI3K in cancer | 0.00463631 |
| ERBB2 activates PTK6 signalling | 0.00463631 |
| ERBB2 regulates cell motility | 0.00463631 |
| PI3K events in ERBB2 signalling | 0.00516862 |
| PI3K/AKT signalling in cancer | 0.00573294 |
| PI5, PP2A, and IER3 Regulate PI3K/AKT signalling | 0.00573294 |
| Negative regulation of the PI3K.AKT network | 0.00573294 |
| Signalling by ERBB2 TMD/JMD mutants | 0.00573294 |
| Signalling by ERBB2 KD mutants | 0.00639273 |
| SHC1 events in ERBB2 signalling | 0.00639273 |
| Downregulation of ERBB2 signalling | 0.00639273 |
| Signalling ERBB2 in cancer | 0.00639273 |
| Signalling by ERBB2 | 0.01909767 |
| Signalling PTK6 | 0.02077429 |
| Signalling by nonreceptor tyrosine kinases | 0.02077429 |
| Signalling by ERBB4 | 0.02290672 |
| PIP3 activates AKT signalling | 0.03364767 |
| Intracellular signalling by second messengers | 0.04900415 |
| Formyl peptide receptors bind formyl peptides and many other ligands | 0.05411417 |
| Calcitonin‐like ligand receptors | 0.05411417 |
| PI3K events in ERBB4 signalling | 0.07362005 |
| OAS antiviral response | 0.07362005 |
| Interferon alpha/beta signalling | 0.07703115 |
| Diseases of signal transduction by growth factor receptors and second messengers | 0.07703115 |
| SHC1 events in ERBB4 signalling | 0.07703115 |
| GRB2 events in ERBB2 signalling | 0.07703115 |
| TFAP2 (AP‐2) family regulates the transcription of growth factors and their receptors | 0.07703115 |
| Interleukin‐4 and Interleukin‐13 signalling | 0.07867710 |
| Regulation of TP53 activity through methylation | 0.07867710 |
| Intrinsic pathway of fibrin clot formation | 0.07867710 |
| Interferon‐gamma signalling | 0.07867710 |
| PIWI‐interacting RNA (piRNA) biogenesis | 0.07867710 |
| Long‐term potentiation | 0.07867710 |
| ESR‐mediated signalling | 0.07867710 |
| Cytokine signalling in immune system | 0.08308865 |
| DNA damage recognition in GG‐NER | 0.09674191 |
As there is a complex link between the role of immune cells in tumour development and progression [17], using the methylation data, we estimated immune cell composition between subgroups and between sexes within subgroups. There were statistically significant differences in immune cell composition within the tumour based on a molecular subgroup (Figure S1). There were also significant differences in immune cell type present between sexes within subgroups as well. In PF‐EPN‐B, there were differences in B lymphocyte and eosinophil composition. In PF‐SE, there was a difference in cytotoxic T lymphocytes. In SP‐MPE, there was a difference in eosinophil composition (Figure 3).
FIGURE 3.
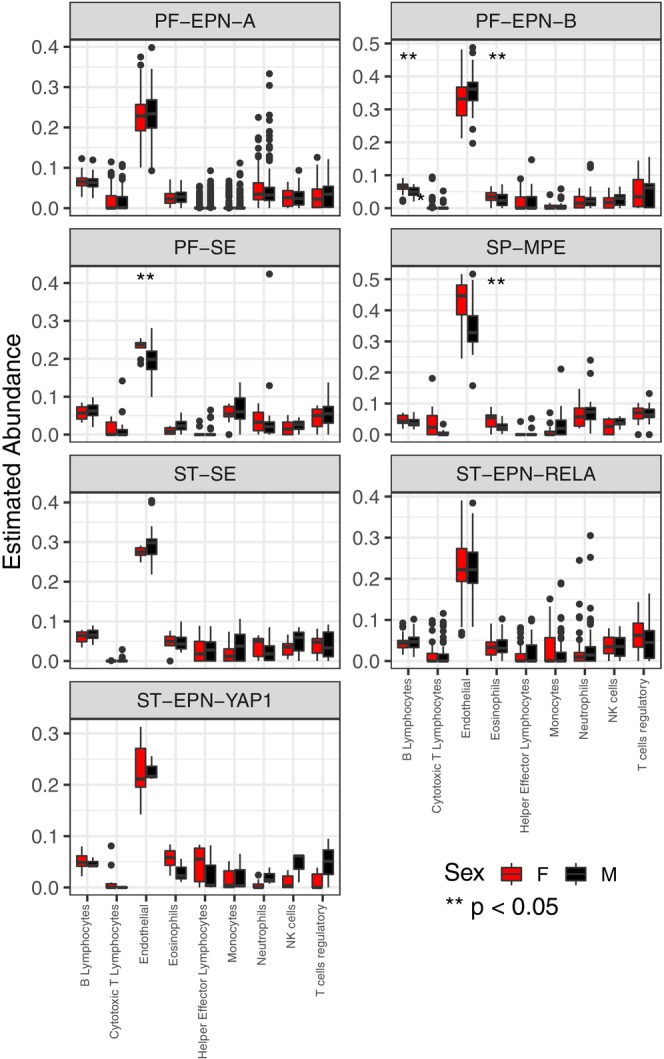
MethylCIBERSORT stromal signature matrix within ependymoma subgroups from the Pajtler data. Boxplots compare ependymoma subgroup means for each cell type. Significance was calculated using pairwise Wilcoxon Rank Sum tests. p values (Boxplots here compare female (red) and male (black) means for each cell type in each ependymoma subgroup as labelled. ** p value used for significance was ≤ 0.05.
In the validation MCI dataset (n = 103), sex differences in DNA methylation were assessed in four molecular subgroups of ependymoma that were the exact same molecular subgroups with statistically significant sex differences in DNA methylation in the Pajtler dataset. The FDR‐adjusted p values indicated that these four molecular subgroups of ependymoma in MCI also had statistically significant differences in DMPs which overlapped with those seen in the Pajtler dataset between males and females for the following subgroups: PF‐EPN‐A (n = 36), PF‐EPN‐B (n = 11), SP‐MPE (n = 5) and ST‐EPN‐RELA (n = 15). This can be seen in Figure 4.
FIGURE 4.
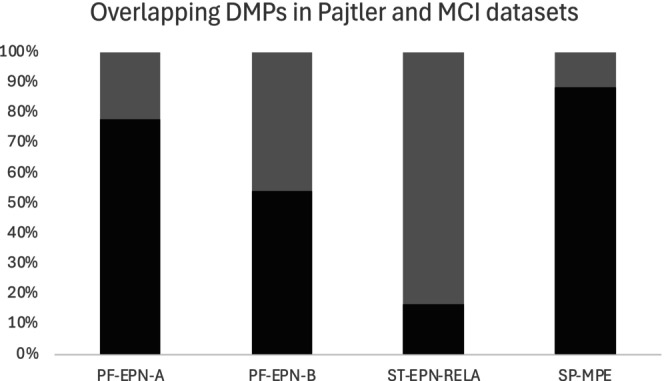
Percent of overlapping DMPs found within Pajtler and MCI datasets. Here, 100% shows the sex DMPs in each subgroup in the Pajtler dataset, and the grey bar indicates the overlapping sex DMPs that were also statistically significantly differentially (FDR < 0.05) methylated in the MCI dataset in each subgroup.
4. Discussion
From 492 primary ependymoma samples, we identified sex differences in 5‐year OS within all cases and in ST‐EPN‐RELA with females having better OS than males. We also identified sex differences in methylation within each subgroup, excluding SP‐EPN. PF‐SE had the higher number of DMPs by sex (n = 283), followed by PF‐EPN‐A (n = 240), SP‐MPE (n = 56), ST‐EPN‐RELA (n = 36), PF‐EPN‐B (n = 25), ST‐EPN‐YAP1 (n = 18) and ST‐SE (n = 10). In the MCI validation cohort, statistically significant sex differences in DNA methylation were identified in four ependymoma subgroups: PF‐EPN‐A (n = 36), PF‐EPN‐B (n = 11), SP‐MPE (n = 5) and ST‐EPN‐RELA (n = 15). These four subgroups were the only subgroups that had greater than 10 samples and thus were the only subgroups included in the analysis in the validation cohort. Unsupervised hierarchical clustering was not driven by other clinical factors, suggesting that there is a true sex difference in DMPs independent of clinical factors.
There is an increased incidence of ependymoma in males overall [9]. When looking at incidence and survival outcomes based on sex, there is often no distinction between subgroups, particularly in epidemiologic analyses as these data are lacking. In our sample set, we observed a male excess in diagnoses in all subgroups except PF‐EPN‐B and ST‐EPN‐YAP1. There are noted worse outcomes in males compared to females in ependymoma historically [10, 18]. We did see worse overall survival in males than in females, although this was marginally nonsignificant. We also saw worse PFS in males than in females overall. Within subgroups, there was also worse OS seen in males compared to females in one subgroup, ST‐EPN‐RELA, which comprises 70% of paediatric supratentorial ependymoma cases [4].
Concerning our investigation into sex differences in methylation, there was one gene, RFTN1, shared between all subgroups that had sex DMPs. The RFTN1 gene has been associated with T‐cell binding and T‐cell activation [19] and other T‐cell signalling pathways through the T‐cell receptor [20]. With increased proportions of CD8+ and CD4+ T cells in samples with higher RFTN1 expression, and subsequently more chemotactic factors, RFTN1 may enhance antitumour immunity [19]. Although not much is known about the expression of RFTN1 in brain tumours, there is information in other cancer types. RFTN1 is known to increase likelihood of progression in gastric cancer with higher expression leading to increased proliferation and decreased apoptosis [21]. RFTN1 expression is increased in skin cancer development as well [22]. It is downregulated in breast cancer [23] and ovarian cancer [24]. Increased RFTN1 expression is noted to be a negative prognostic factor in renal cancers [25]. Overactive T cells from increased RFTN1 expression within the brain could lead to neuroinflammation and may cause altered immune regulation as well as increased proliferation and decreased apoptosis of tumour cells. With continued understanding and further research with this gene, it could become a potential diagnostic biomarker or therapeutic target for certain cancer types. RFTN1 is also associated with double‐stranded RNA binding, B‐cell receptor signalling and toll‐like receptor signalling [25, 26].
We were able to classify reactome pathways from the sex‐DMPs that were mapped to genes in each subgroup of ependymoma (Table 3). Significant pathways in the PF‐EPN‐B subgroup were RNA polymerase transcription and promoter clearance as well as SMAD domain mutants in cancer. SMAD mutations are associated with pancreatic and other GI cancers [27]. Mutations in the MH1 domain of SMAD can affect DNA binding [27]. Alterations to DNA binding proteins affect transcription factors and are associated with the development and progression of certain cancers. These transcription factors can then be targets for therapeutic intervention [28]. Whereas mutations in the MH2 region can interfere with TGF‐beta signalling [29]. Alteration to TGF‐beta signalling can lead to tumour development and progression as well [30]. Overexpression of TGF‐beta in brain tumours like ependymoma could lead to increased cell proliferation as well as increased tumour invasion and angiogenesis. Top pathways in PF‐SE include FGFR2 signalling and activation. FGFR is associated with high‐risk paediatric ependymomas [31]. FGFR‐targeted therapies are another promising option for future studies in paediatric ependymoma [32] and may show sex‐specific effects through an interaction with the underlying DNA methylation status of the gene.
Top pathways identified from sex‐DMP genes in ST‐EPN‐YAP1 include ErbB2 signalling as well as PI3K signalling. ErbB2 pathway is a tyrosine kinase pathway that is involved with cell growth and differentiation [27]. Receptors for this pathway are found in many ependymoma samples in children, with higher expression causing increased proliferation and subsequently worse outcomes [33]. Lapatinib is ErbB1 and ErbB2 inhibitors that can cross the blood–brain barrier; however, it was not specifically tested in ependymoma in vitro [33]. There was a phase 1 trial looking at lapatinib in refractory paediatric brain tumours which showed it was well‐tolerated [34], but subsequent studies found no significant outcome differences between this drug and standard therapy [35, 36]. The PI3K pathway is associated with growth factors and induces cell division and apoptosis. In ependymoma, there is more expression of genes in this pathway in supratentorial tumours as opposed to posterior fossa or spinal ependymomas and higher expression of proteins in this pathway can be used as a marker of worse PFS [37]. There has not been information about PI3K expression by ependymoma subgroups over tumour locations. This could also be a potential therapeutic target in this patient population. The pathways associated with the RFTN1 gene do not appear to be associated with top pathways in each subgroup, and thus, the functional significance of the pathways within each subgroup remains unclear.
There are known sex differences in brain tumours and genomic differences discussed in the literature [38, 39]. Through methylation differences and hormonal changes, there is a difference in the tumour environment and tumorigenesis among different sexes [38]. In our study, we saw the highest number of sex‐DMPs in PF‐SE which are often seen in adults with overall good outcomes [40]. The next highest number of sex‐DMPs was seen in PF‐EPN‐A which is most commonly seen in children with a male predominance and overall poor survival [6]. It remains unclear whether epigenetic changes associated with sex differences are driving outcomes found in the epidemiologic literature.
While we have a large discovery dataset and a validation dataset for ependymoma, which is a rare paediatric brain tumour, there are limitations to this study. We were not able to obtain survival data for the validation dataset from MCI [13]. We also were unable to obtain enough cases in the additional cohort to validate all the different subgroups of ependymoma. This study is not a population‐based study, but rather a clinical cohort, and thus, it may not be representative of the distribution of ependymoma subgroups in the general population as the genomic profiling of these tumours requires tumour tissue availability, which may be differently distributed in a population‐based setting where surgery is not an option for all individuals with ependymoma depending on tumour location and size. There may be other differences between sex including other clinical criteria, risk factors, or response to therapy which may contribute to potential differences seen in outcomes between males and females, but this information was not available for evaluation within our study.
In conclusion, we were able to identify differences in DMPs within subgroups of ependymoma with PF‐SE having the highest number of DMPs, closely followed by PF‐EPN‐A. We did find overall better outcomes in females overall and within ST‐EPN‐RELA. There was one sex differentially methylated gene (RFTN1) common to all subgroups, and there were pathway differences among four of the subgroups. Many of the identified pathways are known to have associations with ependymoma and are potential options for future therapeutic targets in the most high‐risk cases.
Author Contributions
Shelby Mestnik: formal analysis (supporting), writing – original draft (lead). Natali Sorajja: conceptualization (supporting), data curation (lead), investigation (lead), writing – original draft (supporting), writing – review and editing (equal). Zhanni Lu: formal analysis (equal), writing – review and editing (equal). Lauren J. Mills: formal analysis (equal), writing – review and editing (equal). Lindsay Williams: conceptualization (lead), data curation (supporting), investigation (supporting), writing – review and editing (equal).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Figure S1.
Shelby Mestnik and Natali Sorajja contributed equally to this work.
Data Availability Statement
Data is publicly available through Pajtler et al. GEO Series accession number GSE65362 as well as through the molecular characterization initiative (MCI).
References
- 1. Home–GEO–NCBI . accessed August 2, 2024, https://www.ncbi.nlm.nih.gov/geo/.
- 2. Vitanza N. A. and Partap S., “Pediatric Ependymoma,” Journal of Child Neurology 31, no. 12 (2016): 1354–1366, 10.1177/0883073815610428. [DOI] [PubMed] [Google Scholar]
- 3. Khatua S., Ramaswamy V., and Bouffet E., “Current Therapy and the Evolving Molecular Landscape of Paediatric Ependymoma,” European Journal of Cancer 2017, no. 70 (1990): 34–41, 10.1016/j.ejca.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 4. Zschernack V., Jünger S. T., Mynarek M., et al., “Supratentorial Ependymoma in Childhood: More Than Just RELA or YAP,” Acta Neuropathologica 141, no. 3 (2021): 455–466, 10.1007/s00401-020-02260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. “Childhood Ependymoma–NCI,” (2024), https://www.cancer.gov/types/brain/patient/childhood‐ependymoma.
- 6. Pajtler K. W., Witt H., Sill M., et al., “Molecular Classification of Ependymal Tumors Across All CNS Compartments, Histopathological Grades, and Age Groups,” Cancer Cell 27, no. 5 (2015): 728–743, 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neumann J. E., Spohn M., Obrecht D., et al., “Molecular Characterization of Histopathological Ependymoma Variants,” Acta Neuropathologica 139, no. 2 (2020): 305–318, 10.1007/s00401-019-02090-0. [DOI] [PubMed] [Google Scholar]
- 8. Chen R., Smith‐Cohn M., Cohen A. L., and Colman H., “Glioma Subclassifications and Their Clinical Significance,” Neurotherapeutics 14, no. 2 (2017): 284–297, 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williams L. A., Richardson M., Marcotte E. L., Poynter J. N., and Spector L. G., “Sex‐Ratio Among Childhood Cancers by Single‐Year of Age,” Pediatric Blood & Cancer 66, no. 6 (2019): e27620, 10.1002/pbc.27620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams L. A. and Spector L. G., “Survival Differences Between Males and Females Diagnosed With Childhood Cancer,” JNCI Cancer Spectrum 3, no. 2 (2019): pkz032, 10.1093/jncics/pkz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kulis M. and Esteller M., “DNA Methylation and Cancer,” in Advances in Genetics, vol. 70 (Elsevier, 2010), 27–56, 0065‐2660, 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 12. Moss R. M., Sorajja N., Mills L. J., et al., “Sex Differences in Methylation Profiles Are Apparent in Medulloblastoma, Particularly Among SHH Tumors,” Frontiers in Oncology 13 (2023): 1113121, 10.3389/fonc.2023.1113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. “Childhood Cancer Data Initiative (CCDI): Molecular Characterization Initiative,” dbGap Study Accession: phs002790.v1.p1.
- 14. Chakravarthy A., Furness A., Joshi K., et al., “Pan‐Cancer Deconvolution of Tumour Composition Using DNA Methylation,” Nature Communications 9, no. 1 (2018): 3220, 10.1038/s41467-018-05570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newman A. M., Steen C. B., Liu C. L., et al., “Determining Cell Type Abundance and Expression From Bulk Tissues With Digital Cytometry,” Nature Biotechnology 37, no. 7 (2019): 773–782, 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Rourke K., “NCI Launches the Molecular Characterization Initiative for Pediatric Tumors,” Cancer 128, no. 16 (2022): 3012, 10.1002/cncr.34381. [DOI] [PubMed] [Google Scholar]
- 17. Lu C., Liu Y., Ali N. M., Zhang B., and Cui X., “The Role of Innate Immune Cells in the Tumor Microenvironment and Research Progress in Anti‐Tumor Therapy,” Frontiers in Immunology 13 (2023): 260, 10.3389/fimmu.2022.1039260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soon W. C., Goacher E., Solanki S., et al., “The Role of Sex Genotype in Paediatric CNS Tumour Incidence and Survival,” Child's Nervous System 37, no. 7 (2021): 2177–2186, 10.1007/s00381-021-05165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xin H., Zhang M., Miu L., Zhou L., Li Z., and Tang L., “RFTN1: An Potential Inhibitor of the Immunity System in TME of Breast Cancer Improving the Tumor Immune Escape,” (2024), 10.21203/rs.3.rs-4243522/v1. [DOI]
- 20. von Haller P. D., Donohoe S., Goodlett D. R., Aebersold R., and Watts J. D., “Mass Spectrometric Characterization of Proteins Extracted From Jurkat T Cell Detergent‐Resistant Membrane Domains,” Proteomics 1, no. 8 (2001): 1010–1021, 10.1002/1615-9861(200108)1. [DOI] [PubMed] [Google Scholar]
- 21. “RFTN1 Facilitates Gastric Cancer Progression by Modulating AKT/p38 Signaling Pathways–PubMed,” accessed August 15, 2024, https://pubmed.ncbi.nlm.nih.gov/35490655/.
- 22. Alameda J. P., Navarro M., Ramírez Á., et al., “IKKα Regulates the Stratification and Differentiation of the Epidermis: Implications for Skin Cancer Development,” Oncotarget 7, no. 47 (2016): 76779–76792, 10.18632/oncotarget.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gkretsi V., Papanikolaou V., Zacharia L. C., Athanassiou E., Wu C., and Tsezou A., “Mitogen‐Inducible Gene‐2 (MIG2) and Migfilin Expression Is Reduced in Samples of Human Breast Cancer,” Anticancer Research 33, no. 5 (2013): 1977–1981. [PubMed] [Google Scholar]
- 24. Sherman‐Baust C. A., Becker K. G., W. H. Wood, III , Zhang Y., and Morin P. J., “Gene Expression and Pathway Analysis of Ovarian Cancer Cells Selected for Resistance to Cisplatin, Paclitaxel, or Doxorubicin,” Journal of Ovarian Research 4 (2011): 21, 10.1186/1757-2215-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. “Expression of RFTN1 in Cancer–Summary–The Human Protein Atlas,” accessed August 15, 2024, https://www.proteinatlas.org/ENSG00000131378‐RFTN1/pathology.
- 26. “RFTN1 Raftlin, Lipid Raft Linker 1 [ Homo sapiens (Human)]—Gene–NCBI,” accessed November 25, 2024, https://www.ncbi.nlm.nih.gov/gene/23180.
- 27. Milacic M., Beavers D., Conley P., et al., “The Reactome Pathway Knowledgebase 2024,” Nucleic Acids Research 52, no. D1 (2024): D672–D678, 10.1093/nar/gkad1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shiroma Y., Takahashi R., Yamamoto Y., and Tahara H., “Targeting DNA Binding Proteins for Cancer Therapy,” Cancer Science 111, no. 4 (2020): 1058–1064, 10.1111/cas.14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fleming N. I., Jorissen R. N., Mouradov D., et al., “SMAD2, SMAD3 and SMAD4 Mutations in Colorectal Cancer,” Cancer Research 73, no. 2 (2013): 725–735, 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- 30. Massagué J., “TGFβ in Cancer,” Cell 134, no. 2 (2008): 215–230, 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown L. M., Ekert P. G., and Fleuren E. D. G., “Biological and Clinical Implications of FGFR Aberrations in Paediatric and Young Adult Cancers,” Oncogene 42, no. 23 (2023): 1875–1888, 10.1038/s41388-023-02705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lötsch D., Kirchhofer D., Englinger B., et al., “Targeting Fibroblast Growth Factor Receptors to Combat Aggressive Ependymoma,” Acta Neuropathologica 142, no. 2 (2021): 339–360, 10.1007/s00401-021-02327-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shonka N. A., “Targets for Therapy in Ependymoma,” Targeted Oncology 6, no. 3 (2011): 163–169, 10.1007/s11523-011-0170-0. [DOI] [PubMed] [Google Scholar]
- 34. Fouladi M., ClintonF S., Blaney S. M., et al., “Phase I Trial of Lapatinib in Children With Refractory CNS Malignancies: A Pediatric Brain Tumor Consortium Study,” Journal of Clinical Oncology 28, no. 27 (2010): 4221–4227, 10.1200/JCO.2010.28.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DeWire M., Fouladi M., Turner D. C., et al., “An Open‐Label, Two‐Stage, Phase II Study of Bevacizumab and Lapatinib in Children With Recurrent or Refractory Ependymoma: A Collaborative Ependymoma Research Network Study (CERN),” Journal of Neuro‐Oncology 123, no. 1 (2015): 85–91, 10.1007/s11060-015-1764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fouladi M., Stewart C. F., Blaney S. M., et al., “A Molecular Biology and Phase II Trial of Lapatinib in Children With Refractory CNS Malignancies: A Pediatric Brain Tumor Consortium Study,” Journal of Neuro‐Oncology 114, no. 2 (2013): 173–179, 10.1007/s11060-013-1166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rogers H. A., Mayne C., Chapman R. J., Kilday J. P., Coyle B., and Grundy R. G., “PI3K Pathway Activation Provides a Novel Therapeutic Target for Pediatric Ependymoma and Is an Independent Marker of Progression‐Free Survival,” Clinical Cancer Research 19, no. 23 (2013): 6450–6460, 10.1158/1078-0432.CCR-13-0222. [DOI] [PubMed] [Google Scholar]
- 38. Sun T., Warrington N. M., and Rubin J. B., “Why Does Jack, and Not Jill, Break His Crown? Sex Disparity in Brain Tumors,” Biology of Sex Differences 3 (2012): 3, 10.1186/2042-6410-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun T., Plutynski A., Ward S., and Rubin J. B., “An Integrative View on Sex Differences in Brain Tumors,” Cellular and Molecular Life Sciences 72, no. 17 (2015): 3323–3342, 10.1007/s00018-015-1930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lombardi G., Della Puppa A., Pizzi M., et al., “An Overview of Intracranial Ependymomas in Adults,” Cancers 13, no. 23 (2021): 6128, 10.3390/cancers13236128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Data Availability Statement
Data is publicly available through Pajtler et al. GEO Series accession number GSE65362 as well as through the molecular characterization initiative (MCI).


