ABSTRACT
Conventional T cell‐directed immunosuppression is the mainstay of standard‐of‐care therapy to prevent graft rejection in clinical organ transplantation. However, it remains ineffective in preventing experimental and clinical organ xenograft rejection. Here, we explored the impact of allogeneic versus xenogeneic antigen stimulation on human T cell responses and gene profile. A comparable proliferative human T cell response was observed in vitro following stimulation with either human or pig cells. Yet, elevated High mobility group box‐1 (HMGB1) levels were following xenogeneic but not allogeneic stimulation, suggesting a pro‐inflammatory response. Next, human peripheral blood mononuclear cells (PBMC) were cultured with allogeneic human, “concordant” xenogeneic monkey, or “discordant” xenogeneic pig, intact cells, or cell lysates. Flow‐sorted CD3+T cells were analyzed for gene expression using NanoString. A distinct pro‐inflammatory gene profile was observed in human CD3+T cells following co‐culture with discordant xenogeneic pig cells, but not concordant xenogeneic monkey cells or allogeneic human cells. Uniquely, stimulation with pig cells induced the expression of the transcription factor NCF4, which promotes inflammasome activation. Pig cell lysate, but not intact pig cells, induced high expression of the DNA‐binding cytokine interleukin‐26 gene. Collectively, these observations highlight the impact of xenogeneic stimulation of human T cells in pig xenograft recipients and concomitant inflammatory responses, which may contribute to immunosuppression‐resistant xenograft rejection. Finally, the impact of genetic engineering of donor pigs on human T cell transcriptomic gene profile is yet to be determined.
Abbreviations
- FDA
Food and Drug Administration
- HMGB1
High mobility group box‐1
- MLR
mixed lymphocyte reaction
- NHP
nonhuman primate
- PBMC
peripheral blood mononuclear cell
1. Introduction
Xenotransplantation remains a viable approach to bridge the gap of organ shortage by providing an alternative and unlimited source of organs [1]. Recent progress in genetic editing, immunosuppressive therapy, organ preservation, and infection control measures have significantly enhanced long‐term life‐supporting pig organ xenograft survival in nonhuman primates (NHPs) [2, 3, 4]. Such remarkable progress laid the foundation for the recent clinical attempts of transplantation (Tx) of life‐supporting pig organs in human patients, where expanded access authorization by the United States Food and Drug Administration (FDA), that is, compassionate use was granted for patients, who were considered ineligible for an allo‐Tx, to receive organ xenografts obtained from genetically engineered (GE) pigs. Two patients received heart xenografts from GE pig donors at the University of Maryland, and two patients received kidney xenografts from GE pig donors at Massachusetts General Hospital and NYU Langone, respectively. In one recipient, pig heart xenograft failure ensued less than 2 months after Tx [5]. There has been a concern that xenografts may have been lost due to rejection, despite the use of anti‐CD154mAb‐based immunosuppression in the second heart xenograft recipient (ATC 2024).
Currently, standard‐of‐care immunosuppression fails to achieve long‐term xenograft protection in preclinical NHP studies [6]. Further approaches have been considered to achieve long‐term xenograft survival, such as, donor pig genetic modifications, selection of recipients with low pre‐Tx levels of anti‐pig antibodies [7], and targeting of the CD40‐CD154 co‐stimulation pathway [7, 8].
Following xenotransplantation, the immune system identifies pig xenograft as foreign and initiates both innate [9, 10] and adaptive [11, 12] immune responses, which leads to a robust inflammatory reaction within the xenograft as well as systemically in the recipient, as shown in NHP studies [13]. Particularly, the magnitude of such inflammatory response is stronger in xenograft compared to allograft recipients [14]. Meanwhile, clinically available immunosuppressive therapy does not prevent systemic inflammatory responses in xenograft recipients, despite effective regulation of xeno‐reactive T cell responses [15].
Here we hypothesize that xenogeneic stimulation of human T cells induces a distinct genetic profile from that induced by allogeneic stimulation. If factual, this may indicate that prevention of pig xenograft rejection requires a distinct therapeutic approach from the conventional immunosuppression standard‐of‐care currently used in clinical allo‐Tx.
2. Methods
2.1. Isolation of Human, Monkey, and Pig Cells
Human peripheral blood mononuclear cells (PBMC) were isolated from buffy coats of blood type O healthy volunteers (Institute for Transfusion Medicine, Pittsburgh, PA, USA). Pig PBMC were isolated from whole blood of farm pigs. Monkey PBMC were isolated from whole blood of naïve juvenile rhesus monkeys. Briefly, human buffy coat, monkey or pig peripheral blood were diluted with phosphate‐buffered saline (PBS) at a 1:1 volume ratio, then overlaid on Ficoll‐Paque Plus (GE Healthcare Life Sciences AB), spun for 25 min at 1500 rpm (room temperature), and buffy coat collected. PBMC were treated with red blood cell lysis buffer (150 mmol/L NH4C; 1 mmol/L KHCO3; 0.1 mmol/L Na2EDTA), and cell viability was evaluated with Trypan blue.
2.2. Mixed Lymphocyte Reaction (MLR)
Co‐cultures were carried out in round‐bottom, 96‐well plates with serum‐free, AIM‐V medium (Invitrogen, Carlsbad, CA, USA). Human PBMC were used as responders at 0.2 × 106 cells/well. Irradiated human or pig PBMC were used as stimulators at stimulator–responder ratios of 1:1. 3H‐thymidine (1 µCi/well) was added to each well during the last 16 h of incubation. Cells were harvested on glass‐fiber filter mats. Next, they were analyzed by beta‐scintillation counting on a liquid scintillation counter (PerkinElmer, Waltham, MA, USA). The mean of triplicate results was expressed as 3H‐thymidine incorporation values (presented in units of counts per minute [CPM]).
2.3. Luminex Multiplex Immunoassays for Cytokine Levels
Supernatants were collected from MLR cultures on Day 3. Human cytokine and chemokine levels were assessed using custom LEGENDplex Luminex kit (BioLegend, San Diego, CA, USA).
2.4. Western Blotting for HMGB1
Western blot analysis for High mobility group box‐1 (HMGB1) was performed as previously described [16]. Briefly, supernatant in MLR co‐cultures was collected on Day 3 and stored at −80°C for Western blot analysis. Primary polyclonal rabbit antibody to HMGB1 (1:5000; BD Biosciences) was used. Membranes were developed with the SuperSignal West Pico Chemiluminescent Kit (Pierce) and exposed to film.
2.5. Flow Sorting of T Cells for RNA Analysis
Human PBMCs were cultured with either human PBMC, monkey PBMC, or pig PBMC stimulators. In tandem, cell lysates from human, monkey, and pig stimulator cells were also prepared using the freeze–thaw technique. Same simulator cell numbers (human, monkey, or pig) were used for cultures using intact cells or lysate stimulation and used at 1 (responder):1 (stimulator) ratio to co‐cultures. All stimulator cells (including cells used for lysate preparation) were labeled with PKH (Sigma) and irradiated (as for the MLR experiments). For lysate preparation, cell suspensions were exposed to multiple cycles (5–10 cycles) of freezing (dry ice) followed by thawing at room temperature. The absence of intact cells was confirmed using Trypan blue staining. If intact cells were detected, freeze–thaw cycles were repeated. As a control, human PBMC were either cultured alone or stimulated with lipopolysaccharide (LPS) (10 ng/mL) 24 h before harvest. Responder cells were harvested on Day 5 and washed twice with sodium azide‐free medium; CD3+T cells were flow‐sorted for RNA analysis (Figure S1).
2.6. RNA Isolation and Gene Expression Analysis
RNA was isolated and quantified using NanoString nCounter GX Human Immunology V2 assay, which accounts for around 600 genes (NanoString Technologies, Seattle, WA, USA) and normalized using nSolver by following the manufacturer's instructions. Normalized gene expression data were imported to Partek Flow v11.0.23.1105 for filtering and downstream analysis. Genes whose expression did not vary between samples were excluded from downstream analysis. Unsupervised hierarchical clustering was performed on both the gene expression and samples. Principal component analysis (PCA) was performed for dimensionality reduction. Differential gene expression was performed using gene specific analysis (GSA) in Partek Flow. Volcano plots were drawn to visualize differential gene expression between different samples.
2.7. Statistical Analyses
Significance was determined by paired, two‐tailed Student's t‐tests. Analyses were carried out using GraphPad Prism version 4 (GraphPad Software, San Diego, CA, USA). p < 0.05 was considered significant.
3. Results
3.1. Human PBMC Response to Xenogeneic Pig Cells Versus Allogeneic Human Cells
Three individual human responder PBMC were cultured with either irradiated human allogeneic PBMC (n = 6) or irradiated pig xenogeneic PBMC (n = 3). After culture, cells were harvested on Days 3, 4, 5, and 6 to assess the proliferative response of human PBMC to human versus pig stimulators (Figure 1). Proliferation of human responder cells gradually increased to reach a maximum level on Day 5, followed by decreased proliferation on Day 6. Notably, human PBMC proliferation in response to human stimulators was not significantly greater than that to pig stimulators.
FIGURE 1.
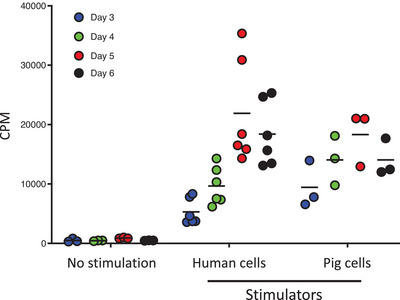
Human PBMC response to allogeneic versus xenogeneic stimulation. In MLR, human PBMC were cultured either alone, with irradiated allogeneic human PBMC, or with irradiated xenogeneic pig PBMC. Responder cells were harvested on Days 3, 4, 5, and 6 to assess for proliferation using 3H incorporation. CPM = counts per minute; MLR = mixed lymphocyte reaction; PBMC = peripheral blood mononuclear cell.
It has been shown that the pro‐inflammatory HMGB1 protein is released following co‐culture of human T cells with pig endothelial cells [17]. On Day 3, we assessed HMGB1 production in the supernatant of human and pig co‐cultures (Figure S2). High levels of HMGB1 were detected in supernatants of human PBMC cultured with pig cells, but not with human cells. These observations suggest a stronger pro‐inflammatory response following xenogeneic pig stimulation of human PBMC.
3.2. Unique Pro‐Inflammatory Transcriptional Changes in Human T Cells Following Stimulation With Xenogeneic Pig Cells
Next, we aimed to evaluate the impact of allogeneic (human) versus xenogeneic (monkey or pig) stimulation on gene expression in human CD3+T cells. Hierarchical clustering was performed on both genes and samples to assess in an unsupervised fashion whether exposure to allogeneic or xenogeneic, intact cells or cell lysates, leads to disparate gene expression changes in human CD3+T cells (Figure 2A). As controls, gene expression in human CD3+T cells cultured either alone or with LPS were measured.
FIGURE 2.
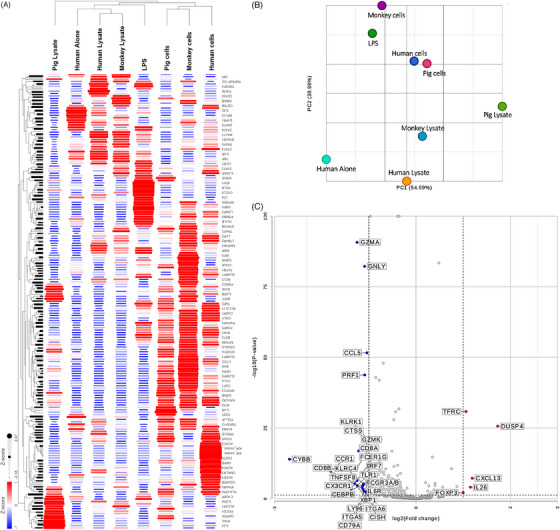
Gene expression in xeno‐ versus allo‐stimulated human T cells. (A) Hierarchical clustering of samples and genes based on detected gene expression using the NanoString nCounter GX Human Immunology V2 assay. Dendrograms represent sample (columns) clusters. (B) Principal component analysis of samples was performed to validate hierarchical clustering results. (C) Volcano plot of differential gene expression in human T cells co‐cultured with pig lysates versus human T cells co‐cultured with pig cells. Thresholds for p < 0.05 and |fold change| >1.5 are represented with dotted lines.
Gene expression profiles of human CD3+T cells cultured with cell lysates were different from those from intact cells, regardless of whether it was allogeneic human, xenogeneic (concordant) monkey, or xenogeneic (discordant) pig cells (Figure 2A). The dendrograms on the rows clearly cluster human CD3+T cells co‐cultured with pig, monkey, or human cells. Another cluster contained samples from human CD3+T cells cultured with monkey lysates or human lysates. Most surprising was the sample of human CD3+T cells cultured with pig lysates as it did not cluster with other samples and most of the genes were expressed less than in the other samples, but few were uniquely upregulated. We performed PCA to project our samples in a 2D space based on gene expression profiles in different samples (Figure 2B). Distinctly, gene expression in human CD3+T cells cultured with pig lysate was quite different from all other samples as the pig lysate sample was solitarily located at the right edge of the PCA space.
Furthermore, differential gene expression analysis visualized by volcano plot in Figure 2C indicates that, as compared to stimulation with pig cells, pig lysate stimulation led to upregulation of interleukin (IL)‐26, CXCL13, DUSP4, and TFRC and lacked the upregulation of genes such as GZMA, PRF1, CD8A, XBP1, and CX3CR1 seen in CD3+T cells stimulated by pig cells.
In an effort to corroborate gene expression changes in human CD3+T cells with functional analysis, we measured cytokine levels on Day 3 in human PBMC co‐cultures with either pig or human cells (shown in Figure S3). By gene expression, IFNγ was upregulated in both human CD3+T cells cultured with allogeneic or pig cells, while TNFα was downregulated in allo‐stimulated CD3+T cells but upregulated in pig‐stimulated CD3+T cells (Figure S3A). Congruently, while IFNγ cytokine levels were not significantly different, TNFα cytokine levels were significantly higher in the pig cell‐stimulated PBMC sample (Figure S3B).
3.3. Pig Cells and Pig Lysate Induce a Distinct Gene Expression in Human CD3+T Cells
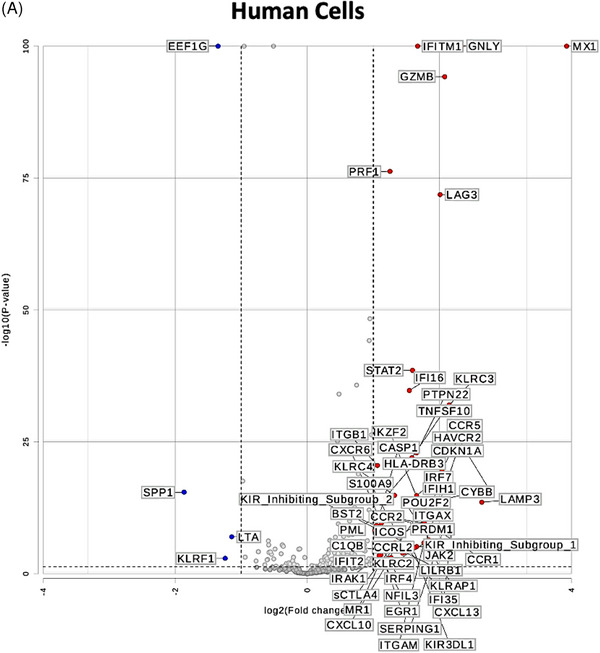
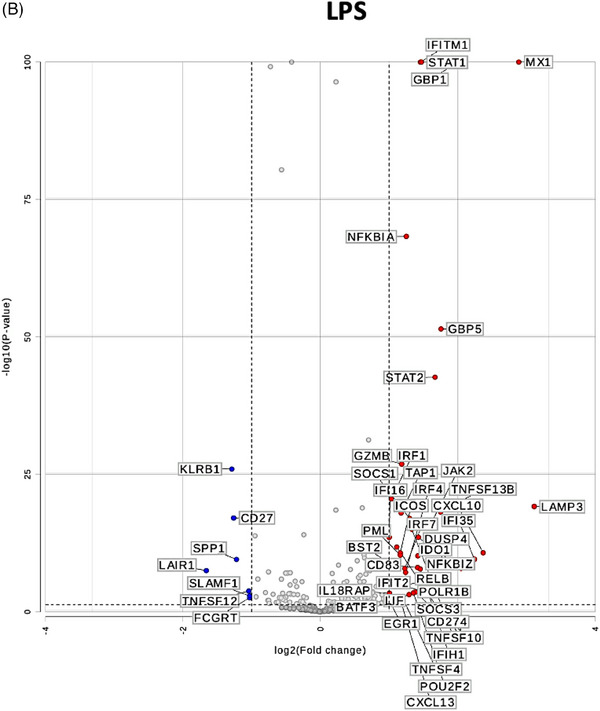
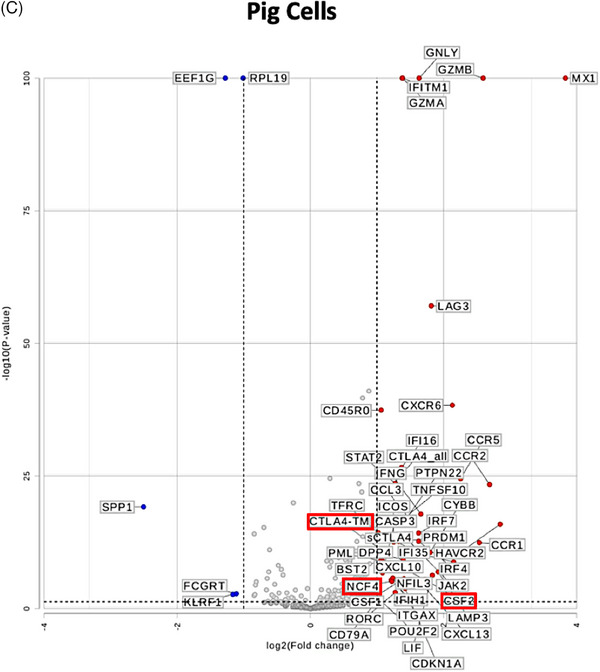
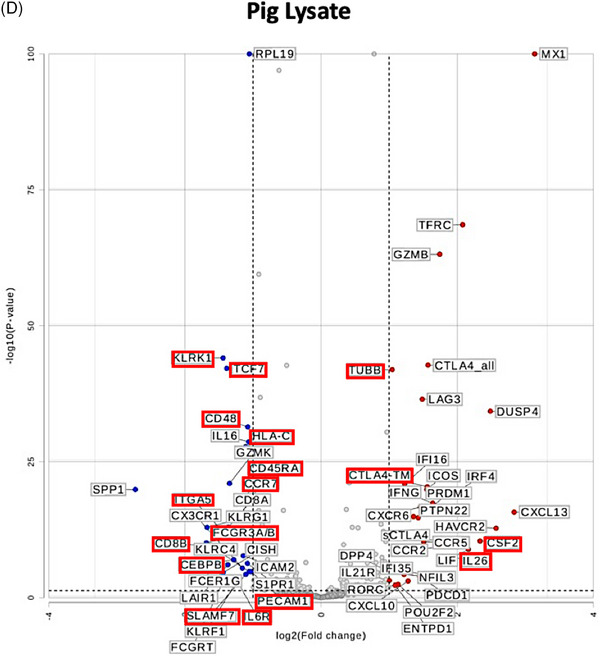
To test whether the gene expression changes in human CD3+T cells are unique to stimulation with pig cells or lysates and not shared with allogeneic or LPS stimulation, we performed differential gene expression analysis as represented in the volcano plots in Figure 3. While using gene expression in human CD3+T cells cultured alone as a universal control, genes that are differentially expressed in CD3+T cells cultured with pig cells (Figure 3C) or lysate (Figure 3D) and stable (|fold change| <1.5) in allogeneic stimulation (Figure 3A) and LPS stimulation (Figure 3B) were pinpointed (outlined in red in Figures 3C and 3D). Both types of stimulation led to upregulation of CTLA4‐TM and CSF2. However, pig cell stimulation uniquely led to upregulation of NCF4, while pig lysate stimulation led to unique upregulation of IL‐26 and TUBB. As seen in Figure 3D, downregulation of many genes is witnessed in human T cells co‐cultured with pig lysate. Those that are uniquely downregulated due to pig lysate stimulation include KLRK1, TCF7, CD48, HLA‐C, CD45RA, CCR7, ITGA5, FCGR2A/B, CD8B, CEBPB, PECAM1, SLAMF7, and IL6R.
FIGURE 3.
Differential gene expression analysis uncovers uniquely expressed genes by pig‐stimulated human T cells. Volcano plots of differential gene expression in human T cells co‐cultured with (A) human allogeneic cells, (B) LPS, (C) pig cells, and (D) pig lysates as compared to human T cells cultured alone. Thresholds for p < 0.05 and |fold change| >1.5 are represented with dotted lines. Genes that are differentially expressed in (C) or (D) and similarly expressed (|fold change| <1.5) in (A) and (B) are outlined in red.
4. Discussion
Conventional immunosuppression remains the mainstay of standard‐of‐care therapy in clinical Tx, where T cell‐directed immunosuppression has been successful in the prevention of allograft rejection. However, comparable immunosuppression fails to prevent xenograft rejection in experimental and clinical models. This observation suggests that xenogeneic immune responses may either be stronger or inherently distinct from allogeneic responses. Early studies have demonstrated that in vivo xenogeneic T cell responses are stronger than allogeneic T cell responses [18]. This discrepancy between in vitro and in vivo cellular responses to xenogeneic antigens remains unexplained.
In this study, human PBMC were cultured with human cells or pig cells. The proliferative response of human PBMC to pig cells was not stronger than that to human cells. However, HMGB1 release was observed following human PBMC culture with pig cells, but not human cells, suggesting a pro‐inflammatory response. HMGB1 is a critical mediator of inflammatory, immune and metabolic responses [19]. In vitro, human T cells induce significant HMGB1 release after co‐culture with pig endothelial cells [17]. In vivo, neutralization of extracellular HMGB1 suppresses B cell activation and delays xenograft rejection [20].
Next, human PBMC were cultured with allogeneic human, “concordant” xenogeneic monkey, or “discordant” xenogeneic pig, intact cells or cell lysates. Flow‐sorted CD3+T cells were analyzed for gene expression using NanoString, where utilizing direct detection avoids reverse transcription and amplification biases and is less prone for gene drop‐out as compared to single cell methods, which ensured that our observations are unlikely to be due to technical artifacts.
Following culture with intact pig cells or pig cell lysate, distinct expression of certain genes was observed in human CD3+T cells. Uniquely, intact pig cell stimulation led to upregulation of NCF4 (Neutrophil Cytosolic Factor 4), which plays a key role in inflammasome assembly and activation and further promotes effector T cell function [21]. Notably, inflammasome activation drives macrophage expression of the procoagulant protein tissue factor and systemic activation of coagulation [22]. In NHP xenograft recipients, macrophages infiltrating pig xenografts express tissue factor [9, 23]. Also, the expression of CSF2 (granulocyte‐macrophage colony stimulating factor) which stimulates survival and differentiation of myeloid cells, and CTLA4‐TM (membrane‐bound cytotoxic T‐lymphocyte protein 4) a key regulator of T cell activation, was upregulated in CD3+T cells, compared to stable expression (|fold change| <1.5) following allogeneic stimulation (Figure 3A). In parallel, TNFα cytokine levels were significantly higher following culture with the pig cells, than following human cells (Figure S3B). On the other hand, IL‐26 gene expression was upregulated in response to pig cell lysate stimulation. Of note, IL‐26 is a recently identified member of the IL‐10 cytokine family [24] with pro‐inflammatory [25, 26] and DNA‐binding properties [27, 28]. This was associated with downregulation of SLAMF7 (lymphocyte activation molecular family 7) a key suppressor of inflammation during sepsis [29]. Additionally, TCF7 (T cell factor 1; TCF‐1) expression was downregulated, where inflammation‐induced TCF‐1 downregulation is known to facilitate effector CD8+T cell differentiation [30]. Furthermore, concomitant downregulation of CD45RA and CCR7 expression suggests T cell differentiation towards terminally‐differentiated effector memory T cell phenotype. Interestingly, gene expression of CEBPB, PECAM1, and IL6R, which play key roles in the inflammatory responses, was also downregulated after stimulation with pig cell lysate.
In vitro “proliferative” human T cell anti‐pig responses are no stronger than anti‐human responses [31]. Lack of GAL (Galactose α1,3 Galactose) expression is associated with reduced human T cell proliferation in vitro [32]. However, these in vitro observations have not been mirrored with improved survival of GE GAL‐knockout organs using standard‐of‐care immunosuppression [15]. While GAL‐knockout pigs provide the backbone for donor pigs used in preclinical NHP studies and in the recent clinical xeno‐Tx cases, combined with various genetic manipulations to prevent complement activation and dysregulation of coagulation [33], additional factors may promote immunosuppression‐resistant xenograft rejection, such as, innate inflammatory responses [13].
Concomitant innate immune responses may play a role in augmenting xenogeneic T cell responses beyond their allogeneic counterparts [34, 35], which may preclude T cell regulation, as shown in allo‐Tx models [36]. In the recent decedent human xenograft recipients, kidney xenografts demonstrated strong evidence of innate immune cell activation [37]. These observations were further supported in vitro [38] and in vivo [13]. In response to inflammation, innate immune cells upregulate tissue factor expression and promote activation of coagulation [39, 40]. Mutually, coagulation factors augment inflammatory responses [41, 42, 43]. In response to xenografts, in vivo reciprocal amplification of inflammation and activation of coagulation may further impede immune cell regulation, requiring higher levels of immunosuppression [44]. Hence, application of conventional immunosuppressive regimen may not be ideal for human xenograft recipients.
Certainly, the current study has some limitations. First, endothelial cells, the likely cell target after Tx were not used as stimulators in these experiments. We aimed to avoid any variability related to antigen expression due to different passages of endothelial cell obtained from wild type pigs versus commercially available human or monkey endothelial cells. Second, cells derived from GE pigs could provide further insight into the impact of various genetic modifications, particularly, transgenic expression of anti‐inflammatory human genes. Third, these observations may not reflect the in vivo dynamics of T cell activation in human allograft versus xenograft recipients. Nonetheless, they demonstrate the nonconformity of T cell responses following allogeneic versus xenogeneic stimulation. Finally, xenogeneic cellular responses observed in this system may not be exclusively T cell‐dependent, as other autologous immune cells, such as, monocytes, may have indirectly influenced human CD3+T cell genetic profile in response to xenogeneic stimulation.
Collectively, data presented herein underscores the inflammatory responses by human immune cells following exposure to discordant xenogeneic pig antigens, but not allogeneic human antigens. These observations justify the monitoring of inflammatory markers and further therapeutic targeting of innate inflammatory responses following clinical xenotransplantation.
Author Contributions
K.I.A.‐D.: data generation and analysis, and writing of the manuscript. M.A.M.: data generation and analysis. M.K.: conducting experiments and data generation. L.L.: conducting experiments. A.P.‐G.: conducting experiments and data generation. M.B.E.: study design, data generation and analysis, and writing of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Supporting Information
Acknowledgments
This work was supported by NIH grant U19 AI131453 and 1R01AI177308‐01. K.I.A.‐D. was supported by the 2023 American Society of Transplantation Career Transition Grant (Grant #998676).
Funding: This study was funded by NIH grant U19 AI131453 and 1R01AI177308‐01A1, 2023 American Society of Transplantation Career Transition Grant (Grant #998676).
Data Availability Statement
The data that support the findings of this study are openly available by the authors upon request.
References
- 1. Ekser B., Cooper D. K. C., and Tector A. J., “The Need for Xenotransplantation as a Source of Organs and Cells for Clinical Transplantation,” International Journal of Surgery 23 Pt B (2015): 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iwase H., Liu H., Wijkstrom M., et al., “Pig Kidney Graft Survival in a Baboon for 136 Days: Longest Life‐Supporting Organ Graft Survival to Date,” Xenotransplantation 22, no. 4 (2015): 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anand R. P., Layer J. V., Heja D., et al., “Design and Testing of a Humanized Porcine Donor for Xenotransplantation,” Nature 622, no. 7982 (2023): 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Langin M., Mayr T., Reichart B., et al., “Consistent Success in Life‐supporting Porcine Cardiac Xenotransplantation,” Nature 564, no. 7736 (2018): 430–433. [DOI] [PubMed] [Google Scholar]
- 5. Griffith B. P., Goerlich C. E., Singh A. K., et al., “Genetically Modified Porcine‐to‐Human Cardiac Xenotransplantation,” New England Journal of Medicine 387, no. 1 (2022): 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamamoto T., Hara H., Foote J., et al., “Life‐supporting Kidney Xenotransplantation from Genetically Engineered Pigs in Baboons: A Comparison of Two Immunosuppressive Regimens,” Transplantation 103, no. 10 (2019): 2090–2104. [DOI] [PubMed] [Google Scholar]
- 7. Higginbotham L., Mathews D., Breeden C. A., et al., “Pre‐transplant Antibody Screening and Anti‐CD154 Costimulation Blockade Promote Long‐term Xenograft Survival in a Pig‐to‐Primate Kidney Transplant Model,” Xenotransplantation 22, no. 3 (2015): 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mohiuddin M. M., Singh A. K., Corcoran P. C., et al., “Chimeric 2C10R4 Anti‐CD40 Antibody Therapy Is Critical for Long‐Term Survival of GTKO.HCD46.HTBM Pig‐to‐Primate Cardiac Xenograft,” Nature Communications 7 (2016): 11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ezzelarab M., Garcia B., Azimzadeh A., et al., “The Innate Immune Response and Activation of Coagulation in Alpha1,3‐Galactosyltransferase Gene‐knockout Xenograft Recipients,” Transplantation 87, no. 6 (2009): 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schneider M. K. and Seebach J. D., “Current Cellular Innate Immune Hurdles in Pig‐to‐Primate Xenotransplantation,” Current Opinion in Organ Transplantation 13, no. 2 (2008): 171–177. [DOI] [PubMed] [Google Scholar]
- 11. Kalscheuer H., Onoe T., Dahmani A., et al., “Xenograft Tolerance and Immune Function of Human T Cells Developing in Pig thymus Xenografts,” Journal of Immunology 192, no. 7 (2014): 3442–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scalea J., Hanecamp I., Robson S. C., and Yamada K., “T‐cell‐mediated Immunological Barriers to Xenotransplantation,” Xenotransplantation 19, no. 1 (2012): 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ezzelarab M. B., Ekser B., Azimzadeh A., et al., “Systemic Inflammation in Xenograft Recipients Precedes Activation of Coagulation,” Xenotransplantation 22, no. 1 (2015): 32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwase H., Ekser B., Zhou H., et al., “Further Evidence for Sustained Systemic Inflammation in Xenograft Recipients (SIXR),” Xenotransplantation 22, no. 5 (2015): 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graham M. L., Ramachandran S., Singh A., et al., “Clinically Available Immunosuppression Averts Rejection but Not Systemic Inflammation After Porcine Islet Xenotransplant in Cynomolgus Macaques,” American Journal of Transplantation 22, no. 3 (2022): 745–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsung A., Sahai R., Tanaka H., et al., “The Nuclear Factor HMGB1 Mediates Hepatic Injury After Murine Liver Ischemia‐reperfusion,” Journal of Experimental Medicine 201, no. 7 (2005): 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawahara K., Setoyama K., Kikuchi K., et al., “HMGB1 Release in Co‐Cultures of Porcine Endothelial and Human T Cells,” Xenotransplantation 14, no. 6 (2007): 636–641. [DOI] [PubMed] [Google Scholar]
- 18. Pierson R. N. 3rd, Winn H. J., and Russell P. S., “Xenogeneic Skin Graft Rejection Is Especially Dependent on CD4+ T Cells,” Journal of Experimental Medicine 170, no. 3 (1989): 991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen R., Kang R., and Tang D., “The Mechanism of HMGB1 Secretion and Release,” Experimental & Molecular Medicine 54, no. 2 (2022): 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J. H., Zhao B., Zhu X. H., et al., “Blockade of Extracellular HMGB1 Suppresses Xenoreactive B Cell Responses and Delays Acute Vascular Xenogeneic Rejection,” American Journal of Transplantation 15, no. 8 (2015): 2062–2074. [DOI] [PubMed] [Google Scholar]
- 21. Li L., Mao R., Yuan S., et al., “NCF4 Attenuates Colorectal Cancer Progression by Modulating Inflammasome Activation and Immune Surveillance,” Nature Communications 15, no. 1 (2024): 5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu C., Lu W., Zhang Y., et al., “Inflammasome Activation Triggers Blood Clotting and Host Death Through Pyroptosis,” Immunity 50, no. 6 (2019): 1401–1411. e1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin C. C., Ezzelarab M., Shapiro R., et al., “Recipient Tissue Factor Expression Is Associated With Consumptive Coagulopathy in Pig‐to‐Primate Kidney Xenotransplantation,” American Journal of Transplantation 10, no. 7 (2010): 1556–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knappe A., Hor S., Wittmann S., and Fickenscher H., “Induction of a Novel Cellular Homolog of Interleukin‐10, AK155, by Transformation of T Lymphocytes With Herpesvirus Saimiri,” Journal of Virology 74, no. 8 (2000): 3881–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Donnelly R. P., Sheikh F., Dickensheets H., Savan R., Young H. A., and Walter M. R., “Interleukin‐26: An IL‐10‐Related Cytokine Produced by Th17 Cells,” Cytokine & Growth Factor Reviews 21, no. 5 (2010): 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stephen‐Victor E., Fickenscher H., and Bayry J., “IL‐26: An Emerging Proinflammatory Member of the IL‐10 Cytokine Family With Multifaceted Actions in Antiviral, Antimicrobial, and Autoimmune Responses,” Plos Pathogens 12, no. 6 (2016): e1005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meller S., Di Domizio J., Voo K. S., et al., “T(H)17 Cells Promote Microbial Killing and Innate Immune Sensing of DNA via Interleukin 26,” Nature Immunology 16, no. 9 (2015): 970–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poli C., Augusto J. F., Dauve J., et al., “IL‐26 Confers Proinflammatory Properties to Extracellular DNA,” Journal of Immunology 198, no. 9 (2017): 3650–3661. [DOI] [PubMed] [Google Scholar]
- 29. Wu Y., Wang Q., Li M., et al., “SLAMF7 Regulates the Inflammatory Response in Macrophages During Polymicrobial Sepsis,” Journal of Clinical Investigation 133, no. 6 (2023): e150224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Danilo M., Chennupati V., Silva J. G., Siegert S., and Held W., “Suppression of Tcf1 by Inflammatory Cytokines Facilitates Effector CD8 T Cell Differentiation,” Cell reports 22, no. 8 (2018): 2107–2117. [DOI] [PubMed] [Google Scholar]
- 31. Yamada K., Sachs D. H., and DerSimonian H., “Human Anti‐Porcine Xenogeneic T Cell Response. Evidence for Allelic Specificity of Mixed Leukocyte Reaction and for Both Direct and Indirect Pathways of Recognition,” Journal of Immunology 155, no. 11 (1995): 5249–5256. [PubMed] [Google Scholar]
- 32. Wilhite T., Ezzelarab C., Hara H., et al., “The Effect of Gal Expression on Pig Cells on the Human T‐Cell Xenoresponse,” Xenotransplantation 19, no. 1 (2012): 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cooper D. K. C. and Pierson R. N., “Milestones on the Path to Clinical Pig Organ Xenotransplantation,” American Journal of Transplantation 23, no. 3 (2023): 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yi S., Hawthorne W. J., Lehnert A. M., et al., “T Cell‐activated Macrophages Are Capable of Both Recognition and Rejection of Pancreatic Islet Xenografts,” Journal of Immunology 170, no. 5 (2003): 2750–2758. [DOI] [PubMed] [Google Scholar]
- 35. Fox A., Mountford J., Braakhuis A., and Harrison L. C., “Innate and Adaptive Immune Responses to Nonvascular Xenografts: Evidence That Macrophages Are Direct Effectors of Xenograft Rejection,” Journal of Immunology 166, no. 3 (2001): 2133–2140. [DOI] [PubMed] [Google Scholar]
- 36. Chong A. S. and Alegre M. L., “Transplantation Tolerance and Its Outcome During Infections and Inflammation,” Immunological Reviews 258, no. 1 (2014): 80–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loupy A., Goutaudier V., Giarraputo A., et al., “Immune Response After Pig‐to‐Human Kidney Xenotransplantation: A Multimodal Phenotyping Study,” Lancet 402, no. 10408 (2023): 1158–1169. [DOI] [PubMed] [Google Scholar]
- 38. Ezzelarab M. B., Liu Y. W., Lin C. C., et al., “Role of P‐Selectin and P‐Selectin Glycoprotein Ligand‐1 Interaction in the Induction of Tissue Factor Expression on human Platelets After Incubation With Porcine Aortic Endothelial Cells,” Xenotransplantation 21, no. 1 (2014): 16–24. [DOI] [PubMed] [Google Scholar]
- 39. Hiller E., Saal J. G., and Riethmuller G., “Procoagulant Activity of Activated Monocytes,” Haemostasis 6, no. 6 (1977): 347–350. [DOI] [PubMed] [Google Scholar]
- 40. Niessen F., Schaffner F., Furlan‐Freguia C., et al., “Dendritic Cell PAR1‐S1P3 Signalling Couples Coagulation and Inflammation,” Nature 452, no. 7187 (2008): 654–658. [DOI] [PubMed] [Google Scholar]
- 41. Bokarewa M. I., Morrissey J., and Tarkowski A., “Intra‐articular Tissue Factor/Factor VII Complex Induces Chronic Arthritis,” Inflammation Research 51, no. 9 (2002): 471–477. [DOI] [PubMed] [Google Scholar]
- 42. Bokarewa M. I., Morrissey J. H., and Tarkowski A., “Tissue Factor as a Proinflammatory Agent,” Arthritis Research 4, no. 3 (2002): 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davalos D. and Akassoglou K., “Fibrinogen as a Key Regulator of Inflammation in Disease,” Seminars in Immunopathology 34, no. 1 (2012): 43–62. [DOI] [PubMed] [Google Scholar]
- 44. Byrne G. W., Davies W. R., Oi K., et al., “Increased Immunosuppression, Not Anticoagulation, Extends Cardiac Xenograft Survival,” Transplantation 82, no. 12 (2006): 1787–1791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are openly available by the authors upon request.


