Abstract
The SPINDLY (SPY) protein of Arabidopsis is a negative regulator of gibberellin (GA) response. The SPY protein has 10 copies of the tetratricopeptide repeat (TPR) at the N terminus. TPR motifs function as protein-protein interaction domains. Several spy alleles are affected only in the TPR region suggesting that protein-protein interactions mediated by this domain are important for proper GA signaling. We have used a reverse genetics approach to further investigate the role of the TPR domain. The TPR domain of SPY was overexpressed in wild-type, gai, and spy plants. Expression of the TPR domain alone is not sufficient to rescue spy mutants. Expression of the TPR domain in a wild-type background produces phenotypes similar to those caused by loss-of-function spy mutants including resistance to GA biosynthesis inhibitors, short hypocotyl length, and early flowering. The dwarfing of the floral shoot internodes caused by the gai mutation was suppressed by expression of the TRP domain. Expression of the TPR domain had no effect on the abundance of endogenous SPY mRNA. The TPR domain was found to interact with SPY both in vitro and in yeast two-hybrid assays. These data indicate that the TPR domain of SPY can participate in protein-protein interactions and that these interactions are important for the proper functioning of SPY.
Gibberellins (GAs) are tetracyclic diterpeneoid plant hormones that are required for many aspects of growth and development (Hooley, 1994; Swain et al., 1997). Bioactive GAs are believed to be perceived at the plasma membrane (Hooley et al., 1991; Gilory and Jones, 1994; Lovegrove et al., 1998). Using cell biological, pharmacological, and genetic approaches, a number of potential components of the GA signaling pathway have been identified (Thornton et al., 1999; Lovegrove and Hooley, 2000). The total number of components in the GA pathway and the function of the different components remain to be defined.
Genetic studies in Arabidopsis have shown that the SPINDLY (SPY) protein plays a role in the GA response pathway (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Thornton et al., 1999). All known spy alleles are recessive and, to varying degrees, suppress all of the phenotypes caused by GA deficiency (Jacobsen and Olszewski, 1993; Silverstone et al., 1997; Swain et al., 2001). Therefore, SPY is hypothesized to be a negative regulator of GA signaling. Consistent with this hypothesis, the barley (Hordeum vulgare) ortholog of SPY, when expressed in aleurone cells under the control of a strong promoter, is able to suppress the expression of a β-glucoronidase (GUS) reporter gene driven by an α-amylase promoter (Robertson et al., 1998).
The SPY protein has significant similarity to O-linked GlcNAc transferase (OGT) from animals (Kreppel et al., 1997; Lubas et al., 1997). OGTs are cytosolic and nuclear enzymes that modify proteins by adding a single GlcNAc in an O-linkage to Ser and/or Thr. A large body of evidence indicates that O-GlcNAc protein modification is a regulatory modification (Hart, 1997; Comer and Hart, 2000), suggesting that posttranslational O-GlcNAc modification of protein(s) plays a role in GA signaling (Thornton et al., 1999).
Both SPY and animal OGT have two distinct domains, the tetratricopeptide repeat (TPR) domain and the OGT catalytic domain. SPY and animal OGTs have between nine and 11 TPRs at their N termini (Jacobsen et al., 1996; Kreppel et al., 1997; Lubas et al., 1997). The TPR motif is a degenerate 34 amino acid sequence with amino acids at eight positions that are similar in size and hydrophobicity (Lamb et al., 1995; Blatch and Lässle, 1999). TPR-containing proteins have been identified in organisms from all kingdoms and are believed to act as scaffolds for the assembly of multiprotein complexes (Das et al., 1998; Pratt, 1998). The crystal structures of two TPR domains have been determined (Das et al., 1998; Scheufler et al., 2000). Each TPR is composed of a pair of α-helices, which are packed in an antiparallel arrangement. The TPRs fold into a right-handed super-helical structure that binds the interacting proteins.
TPR protein-containing complexes participate in diverse processes including cell cycle control, repression of transcription, response to stress, and the import of proteins into organelles (Lamb et al., 1995; Blatch and Lässle, 1999). The individual TPRs of a TPR domain can interact with different proteins (Lamb et al., 1994; Tzamarias and Struhl, 1995; Young et al., 1998; Gounalaki et al., 2000). For example, in yeast (Saccharomyces cerevisiae), subunits of the anaphase-promoting complex, CDC16, CDC23, and CDC27, can be co-immunoprecipitated as a protein complex (Lamb et al., 1994). Mutations in the seventh TPR of CDC27 reduce its ability to interact with CDC23, but do not change the interaction with CDC16.
Protein-protein interactions occurring at the TPRs play an important role in the functioning of the protein complex. The expression of a truncated version of human phosphatase 5 (PP5) consisting only of the TPR domain blocks glucocorticoid signaling in a dominant negative manner. The inhibition of glucocorticoid signaling occurs because the truncated PP5 replaces full-length PP5 in the glucocorticoid receptor complex (Chen et al., 1996).
We have identified a number of spy alleles affecting only the TPR domain (Jacobsen et al., 1996; T.S. Tseng and N.E. Olszewski, unpublished data), suggesting the involvement of SPY's TPRs in GA signaling. Although genetically SPY has been shown to negatively regulate GA signaling, the role of the TPR and OGT domains is undefined.
This work has reinvestigated the role of the TPR domain in GA signal transduction. It was reported previously that overexpression of the TPR domain of SPY did not cause any obvious phenotype (Jacobsen et al., 1998); however, the construct used in the study did not include the 5′-untranslated leader from SPY. Since publishing the previous work, we have found that inclusion of the 5′-untranslated leader in constructs where the expression of SPY is driven by either its own promoter or the cauliflower mosaic virus (CaMV) 35S promoter enhances the ability of the constructs to rescue spy mutants (Swain et al., 2001). These results suggest that SPY's 5′-untranslated leader is important for its proper expression. This conclusion is further supported by analysis of SPY::GUS reporter constructs differing in this region (S.E. Swain and N.E. Olszewski, unpublished data). Therefore, we examined how GA signaling was affected by ectopic expression of SPY's TPR domain using a construct containing the 5′-untranslated leader of SPY. In addition, we have examined the ability of the TPRs to function as a protein-protein interaction domain.
RESULTS
Expression of the SPY TPR Domain Does Not Rescue spy Mutants
A transgene expressing the TPR domain under the control of the CaMV 35S promoter (Fig. 1A) was introduced into gai, spy-3, and spy-6 mutants, and wild-type Arabidopsis. Twenty-one wild-type Columbia lines, three gai lines, five spy-3 lines, and three spy-6 lines, containing a single transgene locus, were identified. All of the gai and spy lines and eight of the Columbia lines were made homozygous for the transgene locus and used in the studies described below.
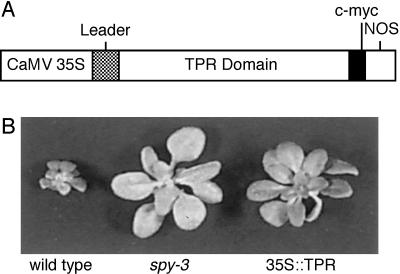
Figure 1.
The effects of GA biosynthesis inhibitors on the vegetative growth of plants ectopically expressing the TPR domain of SPY. A, Map of the gene construct for overexpression the TPR domain of SPY. The components of the gene include the CaMV 35S promoter (CaMV 35S), the 5′-untranslated leader of SPY (Leader), the region of the SPY gene that encodes the TPR region (TPR Domain), a c-myc epitope tag (c-myc), and the nopaline synthase gene polyadenylation sequence (NOS). B, Three-week old plants of wild type, spy-3, and wild-type TPR-overexpressing line 7b8 were continuously grown in the presence of 10−7 m uniconazole.
All of the spy-3 and spy-6 transgenic lines germinated in the presence of a concentration of paclobutrazol (35 mg L−1) that is sufficient to inhibit the germination of wild-type Arabidopsis (not shown) indicating that overexpression of the TPR domain alone is not sufficient to rescue the germination phenotype of spy plants.
Expression of the SPY TPR Domain Confers Resistance to GA Biosynthesis Inhibitors
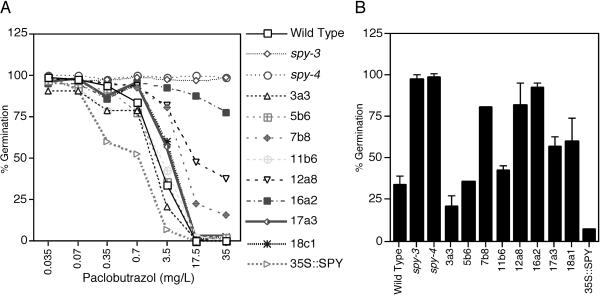
spy mutants are resistant to both the dwarfing and germination-inhibiting effects of GA biosynthesis inhibitors (Jacobsen and Olszewski, 1993). If expression of the SPY TPR domain impairs SPY activity by disrupting protein-protein interactions, expression of it is predicted to phenocopy the effects of spy mutations. Therefore, we examined the sensitivity of germination of the TPR-expressing lines to the GA biosynthesis inhibitor paclobutrazol. Five of the eight transgenic lines expressing the TPR domain in a wild-type background were less sensitive to paclobutrazol (Fig. 2). None of the transgenic lines were as resistant to paclobutrazol as spy mutants and three of the lines were similar to wild type. In contrast, and consistent with our previous observations (Swain et al., 2001), a line overexpressing the full-length SPY protein (Fig. 2B; line 35S::SPY) was more sensitive to paclobutrazol than wild type.
Figure 2.
The germination of SPY TPR seeds is less sensitive to inhibition by paclobutrazol. A, Germination of wild-type, spy mutants, and TPR-expressing lines, and an SPY-overexpressing line on different concentrations of paclobutrazol was scored. B, The germination rates of TPR-expressing lines, SPY-overexpressing line, spy-3, spy-4, and wild-type seeds sown on 3.5 mg L−1 of paclobutrazol. Data are means ± se of three independent experiments. In some cases, the se is too small for the se bar to be visible.
The TPR-expressing lines were also more resistant to the inhibition of leaf expansion by uniconazole, a GA biosynthesis inhibitor that acts in the same manner as paclobutrazol. After 2 d of germination, the plants were grown on 10−7 m uniconazole. After 3 weeks, the lines expressing the TPR domain were noticeably larger than non-transgenic control lines (Fig. 1B).
Expression of the SPY TPR Domain Causes a Short Hypocotyl Phenotype
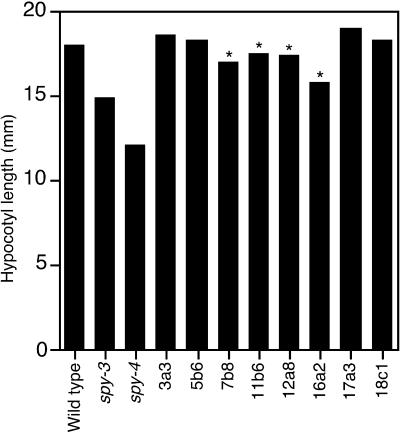
When grown in the dark, spy seedlings have shorter hypocotyls than wild type, and seedlings overexpressing the complete SPY protein have longer hypocotyls than wild type (Swain et al., 2001). After 2 weeks of growth on a defined medium in the dark, the hypocotyls of four of the TPR-expressing lines were significantly shorter than those of wild-type plants but not as short as those of spy-3 or spy-4 plants (Fig. 3). The remaining four transgenic lines had hypocotyl lengths that were indistinguishable from wild type.
Figure 3.
Expression of the TPR domain of SPY inhibits hypocotyl elongation. The length of the hypocotyl of TPR-expressing lines, spy-3, spy-4, and wild-type seedlings was measured after 2 weeks of growth in the dark. spy mutants have shorter hypocotyls than the wild-type plants. The asterisks above the bars indicate the four transgenic lines with hypocotyl lengths that are significantly shorter (t test, P < 0.001) than the wild type. The average values from three different experiments are shown. In all cases, the se is too small for the se bar to be visible.
Expression of the SPY TPR Domain Accelerates Flowering
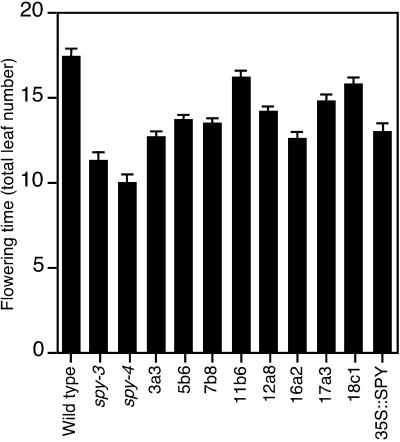
GA treatment and loss of SPY function both accelerate the flowering of plants grown under either long or short days (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996). The TPR-expressing lines together with wild type, spy-3, and spy-4 were grown under long-day conditions and the total number of leaves present when the petals on the first flower were fully expanded was recorded as an indicator of flowering time (Fig. 4). All eight of the TPR-expressing lines flowered earlier than wild type but none of them flowered as early as the spy mutants. Lines 16a2, 12a8, and 7b8, which had the most resistance to paclobutrazol (Fig. 2), flowered earlier than the lines with less resistance to paclobutrazol.
Figure 4.
Expression of the TPR domain of SPY accelerates flowering. The total leaf number of plants at flowering was recorded for TPR-expressing, wild-type, spy-3, spy-4, and SPY-overexpressing plants grown under long-day conditions. Data are means ± se of three independent experiments.
Expression of the SPY TPR Domain Suppresses the gai Mutation
The gai mutation causes semidwarfism due to a reduction in the response to GA (for review, see Sun, 2000). Because the spy mutation suppresses gai (Jacobsen et al., 1996; Table I), the effect of expression of the TPR domain in the gai background on the average length of the internodes below the oldest flower of the fully elongated floral stem was determined (Table I). The three TPR-expressing gai lines examined all had average internode lengths that were longer than untransformed gai. Although the 35S::TPR construct suppressed gai as effectively as spy-3, it was not as effective as the spy-4 mutation.
Table I.
Effect of TPR expression on internode length
| Genotypea | 35S∷TPR Line | Internode Length |
|---|---|---|
| mm | ||
| Wild type | – | 15.30 ± 0.72 |
| gai | – | 6.06 ± 0.20 |
| gai spy-4 | – | 16.67 ± 0.72 |
| gai spy-3 | – | 9.54 ± 0.82 |
| gai | 36a/1 | 9.73 ± 0.66 |
| gai | 38a/2 | 13.64 ± 0.50 |
| gai | 39a/4 | 10.36 ± 0.62 |
The average length of the internodes between the youngest rosette leaf and the oldest flower of the fully elongated floral shoot was determined. All data are means ± se; n ≥ 14.
All mutations have been backcrossed into the ecotype Columbia at least three times.
The Endogenous SPY Gene Is Not Silenced in the TPR Lines
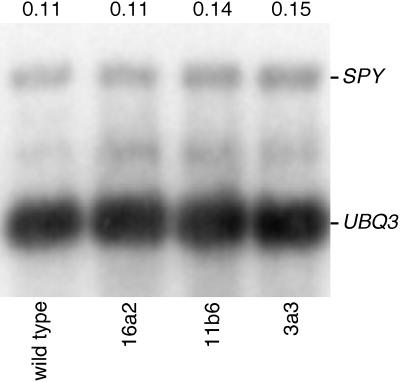
The results from the above studies indicate that plants containing the 35S::TPR construct are weak phenocopies of spy mutants. Although these results are consistent with the hypothesis that expression of the TPR domain interferes with SPY function, they are also consistent with the hypothesis that the presence of the transgene silences the expression of the endogenous SPY gene. Three of the transgenic lines that exhibited paclobutrazol resistance were examined and found to contain SPY TPR RNA (not shown). Due to its low abundance, the endogenous SPY message was not detectable in this experiment. When examined by quantitative reverse transcriptase (RT)-PCR, however, the SPY mRNA level in the lines was indistinguishable from wild type (Fig. 5), indicating that the expression of the endogenous SPY gene was not silenced in the TPR-expressing lines.
Figure 5.
Expression of the TPR domain of SPY does not affect the expression of the endogenous SPY gene. RT-PCR was used to quantitate the amount of SPY mRNA present in wild-type plants and three lines expressing the SPY TPR domain. Each reaction contained primers that amplify both cDNA derived from the endogenous SPY mRNA (SPY) and UBIQUITIN 3 mRNA (UBQ3) and was terminated when the amplification of both products was linear with respect to cycle number (see “Materials and Methods”). The primers to the SPY mRNA do not amplify the mRNA that encodes the TPR domain of SPY. An autoradiogram of a blot containing the PCR products that was probed with 32P-labeled SPY and UBQ3 probes is shown. The signal from each product was quantitated and the number above each lane indicates the ratio of the hybridization to RT-PCR products. Because both sets of primers flank introns, the products from genomic DNA will be larger than the products from cDNA. No product from genomic DNA was detected in any experiment.
The TPR Domain of SPY Interacts with Itself
Animal OGT is a homotrimer (Kreppel et al., 1997). Although the TPR repeats of OGT have been shown to be essential for the assembly or stability of the trimer (Kreppel and Hart, 1999), direct interactions between the TPR domains have not been reported. SPY is also likely to be a homotrimer (T.M. Thornton and N.E. Olszewski, unpublished data). We tested the possibility that SPY's TPR domain interacts with other SPY subunits, using both a yeast two-hybrid assay and an in vitro interaction assay.
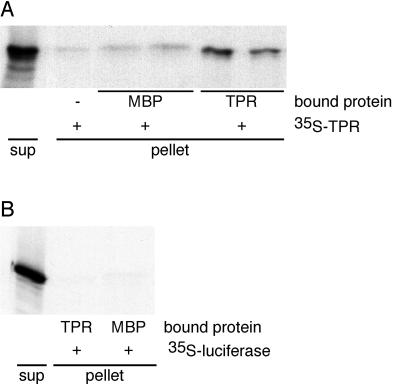
In vitro interaction assays were carried out with maltose-binding protein (MBP) and MBP-TPR fusion protein, and 35S-labeled TPR (Fig. 6A). Although nonspecific interaction of 35S-labeled TPR with both the MBP and the empty resin was detected, over three independent experiments the MBP-TPR protein retained 4.8 ± 0.6 times more 35S-labeled TPR protein than MBP. In vitro co-immunoprecipitation assays detected interaction between the TPR protein and full-length SPY (not shown). We further tested the specificity of the TPR-TPR interaction by measuring the interaction of MBP and MBP-TPR with luciferase (Fig. 6B). In two experiments, no difference in the amount of luciferase retained by either MBP or MBP-TPR was detected, suggesting that luciferase does not interact with the TPR protein.
Figure 6.
In vitro interaction assays of the TPR domain of SPY. A, Equal volumes of empty resin (−), resin bound with either MBP or MBP-TPR (TPR) were incubated with 35S-labeled TPR protein. The bound 35S-labeled TPR protein (pellet), together with 20% of the supernatant (sup), was subjected to SDS-PAGE and the 35S-labeled TPR protein was visualized by fluorography. The experiments were repeated three times with similar results. B, 35S-Labeled luciferase was incubated with resin containing MBP or MBP-TPR.
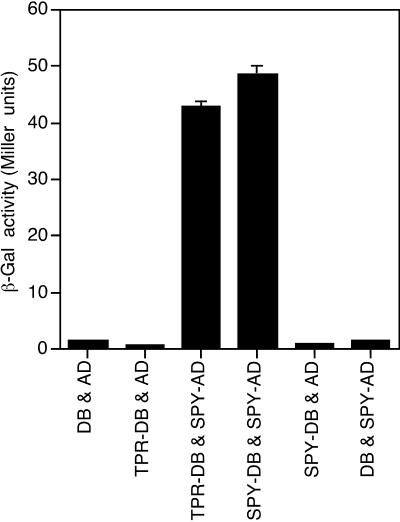
The interaction between the TPR domain and SPY was confirmed using the yeast two-hybrid system (Fields and Song, 1989; Chien et al., 1991). A construct expressing the TPR domain as part of a fusion with the GAL4 DNA-binding domain and a construct expressing full-length SPY protein as part of a fusion with the GAL4 activation domain were introduced into the yeast strain HF7c. The resulting strain had more β-galactosidase activity than control strains (Fig. 7) and, unlike the control strains, was able to grow on medium lacking His (not shown). These results confirm that the TPR domain can interact with the full-length SPY protein. We also found that full-length SPY fused to the GAL4 DNA activation domain interacted with full-length SPY fused to the GAL4 DNA-binding domain.
Figure 7.
The TPR domain of SPY interacts with the SPY protein in a yeast two-hybrid assay. The β-galactosidase activity of yeast strains containing plasmids expressing a GAL4 DNA binding domain: TPR fusion protein (TPR-DB), GAL4 DNA binding domain: SPY fusion protein (SPY-DB), the GAL4 DNA binding domain (DB), GAL4 activation domain:SPY fusion protein (SPY-AD), or the activation domain (AD) was determined. The proteins expressed in each strain are indicated below. In some cases, the se is too small for the se bar to be visible. The experiment was repeated two additional times with similar results.
DISCUSSION
TPR domains are protein-protein interaction motifs (Lamb et al., 1995; Blatch and Lässle, 1999). In Arabidopsis and barley, SPY has been shown to negatively regulate GA signaling (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Robertson et al., 1998). We tested the hypothesis that the TPR domain of SPY is involved in protein-protein interactions that are critical for GA signaling.
Eight spy alleles affecting the TPR domain have been identified (Jacobsen et al., 1996; T.S. Tseng and N.E. Olszewski, unpublished data). All of these alleles affect TPR 6, 8, and/or 9 and leave the other repeats unchanged. Because the mutations are either small in frame deletions or miss-sense mutations, mutants carrying the mutations are expected to produce a SPY protein. Although some spy alleles are affected in the C-terminal OGT domain, it remains a formal possibility that SPY's only role in GA signaling is to facilitate the formation of a GA signaling complex via protein interactions through the TPRs. This model predicts that expression of the TPRs alone will be sufficient to rescue spy alleles. Because expression of the TPR domain did not restore the ability of paclobutrazol to inhibit the germination of spy-3 and spy-6 seeds, this model was not supported.
There are several other mutually nonexclusive models for the functioning of the TPR domain. The TPR domain may participate in the formation of a functional SPY enzyme, it may interact with SPY's substrates, or it may interact with proteins that regulate the cellular localization or substrate specificity of SPY. If the TPR domain participates in these interactions, expression of the TPR domain alone will impair SPY's activity and interfere with GA signaling.
Eight lines expressing the TPR domain in a wild-type background have been characterized. These lines, to various extents, exhibit phenotypes consistent with a loss of SPY function. Although not all of the lines exhibited every one of these phenotypes, collectively the lines were resistant to inhibition of germination (Fig. 2) and vegetative dwarfing (Fig. 1B) by GA biosynthesis inhibitors, had short hypocotyls (Fig. 3), and flowered early (Fig. 4). All of these defects are also exhibited by spy mutants. Analysis of three of the TPR-expressing lines that exhibit defects in several GA responses indicates that they are expressing the TPR RNA. Moreover, these lines have wild-type levels of SPY mRNA (Fig. 5), indicating that the defects in GA signaling are not due to silencing of the endogenous SPY gene. These results support the hypothesis that the TPR domain plays a critical role in GA signaling.
Additional support for the role of the TPR domain in GA signaling comes from the analysis of three gai lines expressing the TPR domain. The gai mutation, which causes semidwarfism by reducing the response to GA (Sun, 2000), is suppressed by spy (Jacobsen et al., 1996). The average length of internodes of the floral shoot was longer in the three independent 35S::TPR gai lines analyzed than in untransformed gai (Table I), indicating that expression of the TPR domain suppresses gai.
None of the defects in GA response of the TPR-expressing lines was as strong as the defects of spy mutants. This could occur if the level of TPR expression is not sufficient to completely block the interaction of full-length SPY and its partners, or if the full-length SPY protein has a higher affinity for its partners than does the TPR protein. It is also possible that the TPR domain is less stable than the full-length protein. Although the TPR RNA is relatively easy to detect on northern blots, for reasons that are not apparent, we have not been able to detect the TPR protein when western blots are probed with antibodies against the c-myc epitope tag. However, this does not indicate that the TPR protein is unstable relative to SPY because we are only able to detect SPY from Arabidopsis if it is first enriched by partial purification.
In contrast to the other phenotypes of TPR-expressing lines, the short hypocotyl phenotype of these lines is not consistent with the hypothesis that SPY represses GA response. This phenotype is also observed in spy mutants and lines overexpressing full-length SPY have longer hypocotyls than wild type (Swain et al., 2001). It is interesting that GA-deficient ga1 spy double mutants have a longer hypocotyl than ga1 seedlings, indicating that spy suppresses the effects of GA deficiency in this organ (Swain et al., 2001). The simplest explanation for these results is that, in addition to its role in GA response, SPY performs additional function(s) in the hypocotyl. The hypothesis that SPY is involved in more than just GA signaling is also supported by the observations that spy mutants and plants overexpressing SPY exhibit additional phenotypes that cannot be explained based on our current understanding of GA action (Swain et al., 2001).
The TPR motifs of OGT are important for the assembly or stability of the OGT trimers (Kreppel and Hart, 1999) and also play a role in the substrate specificity of the enzyme (Lubas and Hanover, 2000). TPRs 3 to 6 are responsible for trimerization of rat OGT to form the holoenzyme. Deletion of the first six TPRs results in the formation of monomeric OGT, although the monomer still has the OGT activity (Kreppel and Hart, 1999). Similar deletion of the TPRs of human OGT changes the substrate specificity (Lubas and Hanover, 2000).
Like the animal OGT, the SPY holoenzyme is likely to be a homotrimer (T.M. Thornton and N.E. Olszewski, unpublished data). We have found that the TPR domain is able to interact with SPY both in vitro (Fig. 6) and in yeast (Fig. 7), supporting the participation of the TPR domain in the assembly of the holoenzyme. We have also found that SPY from plants is present in an 850-kD complex (T.M. Thornton and N.E. Olszewski, unpublished data), suggesting that it may be associated with other plant proteins. In the future, it will be necessary to examine the effects of TPR expression on the assembly of holoenzyme and the 850-kD complex to determine the precise mechanism(s) by which TPR expression interferes with GA responses.
MATERIALS AND METHODS
Plant Material
All the seeds used for phenotypic characterization were harvested from plants grown in a growth chamber under cool-white fluorescent lights (18 h of light, 6 h of darkness [18L/6D]) at 22°C. Unless indicated otherwise, experimental plants were also grown under these conditions.
Constructs Overexpressing the TPR Domain of SPY and Plant Transformation
The 5′-untranslated leader and the protein coding region extending to the MspAI site located downstream of the TPR domain and a HincII to SacI restriction fragment encoding a 6× c-myc tag (stock no. CD3-128; Arabidopsis Biological Resource Center, Columbus, OH) were cloned into pOCA121 to produce pOCA121-TPR (Fig. 1A).
Wild-type plants of Arabidopsis ecotype Columbia, spy-3 and spy-6 in the same background, and homozygous gai plants produced after backcrossing the gai mutation into the Columbia ecotype three times were vacuum infiltrated with Agrobacterium tumefaciens strain C58C1 containing the helper plasmid pMP90 and pOCA121-TPR, following a procedure similar to the one described by Ye et al. (1999). Transformed T1 plants were selected on medium containing 1× Murashige and Skoog salts (Sigma, St. Louis) and 50 μg mL−1 of kanamycin (Sigma). Homozygous T3 seeds were isolated from T2 lines that based on segregation of kanamycin resistance contained a single transgene locus and seedlings derived from these seeds were used in all of the following studies.
Analysis of Resistance to GA Biosynthesis Inhibitors
Paclobutrazol was diluted in distill water to make a stock solution of 35 mg L−1. Serial dilutions were made from the stock solution. Seeds were sown on glass fiber disc (GF/C, Whatman, Clifton, NJ) and placed upon a single sheet of filter paper in 150-mm plastic petri dishes with 8 mL of paclobutrazol solution. After stratification at 4°C for 3 d, seeds were allowed to germinate at room temperature (22°C) for 1 week under continuous light. Seedlings that had expanded cotyledons were scored as having germinated.
When vegetative resistance to uniconazole was determined, seeds were surface-sterilized and sown on 1× Murashige and Skoog medium. After stratification at 4°C for 3 d, seeds were allowed to germinate at 22°C under long- day conditions (18L/6D) for 2 d, and uniconazole was then added to a final concentration of 10−7 m. Both wild-type and spy-3 plants were included on the same plates as controls.
Hypocotyl Length Measurements
Seeds were surface sterilized, and transferred to a plastic petri dish containing 1× Murashige and Skoog salts (Sigma) supplemented with 1% (w/v) Suc. After stratification at 4°C for 3 d under dim light, the seeds were then germinated at room temperature (22°C) in the dark. The final hypocotyl length was measured after 2 weeks.
Flowering Time Measurement
Plants grown under long-day conditions (18L/6D) at 22°C were scored as flowering, when the petals of the first flower were visible and fully expanded. Total leaf number was counted as the measurement of flowering time.
Measurement of Internodes of the Floral Shoot
Wild-type, gai, and three independent 35S::TPR gai lines were grown on Rockwool (Growool Horticultural Systems, New South Wales, Australia) containing 1× Hoagland solution until elongation of the main floral shoot was complete. The length of each internode below the oldest (lowest) flower was measured and the mean length of the internodes was calculated.
Quantitation of SPY RNA by RT-PCR
RNA was made from 3-d-old seedlings with TRI reagent (MRC, Cincinnati) according to the manufacturer's instructions. Approximately 3 μg of total RNA was used for the first-strand cDNA synthesis with SuperScript preamplification system (GIBCO-BRL, Rockville, MD), following the manufacturer's instructions. PCR was carried out according to Klimyuk et al. (1993). In the same reaction tube, a fragment of UBQ-3 cDNA (Norris et al., 1993) was amplified with the primer set of 5′-CTCTCCCAAAGCCTAAAGCGA-3′ and 5′-GTCGACTCCTTTTGAATGTTGTA-3′, and a fragment of SPY cDNA was amplified with primers 5′-GCGACCTATCACCATTGGA-3′ and 5′-GAGATCCAGCCATTAGAT-3′. Between cycles 20 and 35, the production of both SPY and UBQ-3 product was linear with respect to cycle number (data not shown); therefore, the PCR products were quantitated after 25 cycles. PCR products were separated in 0.8% (w/v) agarose gel, then transferred to nylon membranes (Osmonics, Westborough, MA). The membranes were probed with 32P-labeled UBQ-3 and SPY probes. Signal intensities were determined with phosphor imager, and the values from the exponential reactions were compared. Both the SPY and UBQ-3 primers flanked introns and, therefore, produced different-sized products from genomic DNA and cDNA. In no case, even when RNA that had not been reverse transcribed was used as the template, was product from SPY or UBQ-3 genomic DNA detected.
In Vitro Interactions of 35S-Labeled TPR Protein with Escherichia coli-Expressed TPR
The plasmid pMAL-TPR was constructed by sub-cloning the SspI to HindIII fragment, encoding the TPR domain from pOCA121-TPR construct into pMAL-c2 (New England Biolabs, Beverly, MA). E. coli (XL1-blue) strains expressing MBP and MBP-TPR fusion protein were induced according to the manufacturer's instructions for 1 h. Cultures expressing MBP-TPR protein were grown at 30°C and cultures expressing MBP protein were grown at 37°C.
MBP-TPR and MBP proteins were affinity purified according to Solinas and Motto (1999) and the manufacturer's instructions (New England Biolabs, Beverly, MA). SPY's TPR domain was in vitro transcribed and translated with 35S-Met, according to Bai and Elledge (1997), in the TNT system (Promega, Madison, WI). The in vitro interaction assays were performed according to Zhao and Sancar (1997). In brief, 35S-TPR protein was incubated with MBP or MBP-TPR that was bound to amylose resin (New England Biolabs) or resin with no bound protein for 1 h at 4°C. Following the incubation, the resin was washed extensively, protein bound to the resin was subjected to SDS-PAGE and 35S-TPR protein was detected by fluorography. The amount of bound 35S-TPR protein was determined by scanning the autoradiogram. The amount of 35S-TPR protein specifically bound to MBP or MBP-TPR was the amount in excess of that bound to empty resin.
Yeast Two-Hybrid Assay
Plasmids, pAS1-CYH2 (Arabidopsis Biological Resource Center), and pACTII (gift from Dr. Tai-ping Sun, Duke University, Durham, NC) containing the DNA binding domain of yeast GAL4 transcription factor (pAS1-CYH2) and the activation domain (ACTII), were used as bait and prey, respectively, in the yeast two-hybrid system. The SPY open reading frame was PCR amplified with a primer (5′-ACAAAACCATGGTGGGACTG-3′) that introduces an NcoI site and a primer (5′-CTGGGTTGACAGCTAGTGGAGTC-3′) that introduces a HincII site and changes the stop codon to a codon encoding a Cys.
For making full-length SPY as a bait and/or a prey construct, the SPY PCR products were digested with NcoI and HincII, then ligated with an HincII to SmaI restriction fragment encoding a 6× c-myc tag isolated from a plasmid (stock no. CD3-128, Arabidopsis Biological Resource Center) into NcoI and SmaI sites of the pAS1-CYH2 or pACTII plasmid.
The PCR product was digested with NcoI and PstI and the fragment containing the TPR domain was sub-cloned to pASI-CYH2 plasmid to produce another bait construct. The bait and prey construct were introduced into the yeast strain HF7c (CLONTECH, Palo Alto, CA) using standard procedures (Bai and Elledge, 1997) and the resulting strains were tested for growth on synthetic media without Trp, Leu, and His (BIO 101, Vista, CA). In addition, β-galactosidase assays were performed according to Guarente (1983), using O-nitrophenyl β-d-galactopyranoside as a substrate.
ACKNOWLEDGMENTS
We thank Tina Thornton and Lynn Hartweck for helpful discussions and critical comments on this manuscript and Angelica Jermakow for technical assistance.
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB–9604126 and MCB–9983583 to N.E.O.).
LITERATURE CITED
- Bai C, Elledge SJ. Searching for interacting proteins with the two-hybrid system I. In: Bartel PL, Fields S, editors. The Yeast Two-Hybrid System. New York: Oxford University Press; 1997. pp. 11–28. [Google Scholar]
- Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Chen M-S, Silverstein AM, Pratt WB, Chinkers M. The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J Biol Chem. 1996;271:32315–32320. doi: 10.1074/jbc.271.50.32315. [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer FI, Hart GW. O-Glycosylation of nuclear and cytosolic proteins. J Biol Chem. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implication for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gilory S, Jones RL. Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol. 1994;104:1185–1192. doi: 10.1104/pp.104.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounalaki N, Tzamarias D, Vlassi M. Identification of residues in the TPR domain of Ssn6 responsible for interaction with the Tup1 protein. FEBS Lett. 2000;473:37–41. doi: 10.1016/s0014-5793(00)01480-0. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hart GW. Dynamic O-linked glycosylation of nuclear and cytosolic proteins. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- Hooley R. Gibberellins: perception, transduction and responses. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- Hooley R, Beale MH, Smith SJ. Gibberellin perception at the plasma membrane of Avena fatua aleurone protoplasts. Planta. 1991;183:274–280. doi: 10.1007/BF00197799. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutation at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE, Meyerowitz EM. SPINDLY's role in the gibberellin response pathway. Symp Soc Exp Biol. 1998;51:73–78. [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JDG. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase: role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Michaud WA, Sikorski RS, Hieter PA. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Lovegrove A, Barratt DH, Beale MH, Hooley R. Gibberellin-photoaffinity labeling of two polypeptides in plant plasma membranes. Plant J. 1998;15:311–320. doi: 10.1046/j.1365-313x.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R. Gibberellin and abscisic acid signaling in aleurone. Trends Plant Sci. 2000;5:102–110. doi: 10.1016/s1360-1385(00)01571-5. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Frank DW, Krause M, Hanover JA. O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase: domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J. The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol. 1993;21:895–906. doi: 10.1007/BF00027120. [DOI] [PubMed] [Google Scholar]
- Pratt WB. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc Soc Exp Biol Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE. Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell. 1998;10:995–1007. doi: 10.1105/tpc.10.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Casamitjana Martínez E, Sun T-p. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997;146:1087–1099. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas G, Motto M. Nonradioactive multi-sample protein-protein interaction assay using an epitope tagging technique. BioTechniques. 1999;26:246–249. doi: 10.2144/99262bm14. [DOI] [PubMed] [Google Scholar]
- Sun T-p. Gibberellin signal transduction. Curr Opin Plant Biol. 2000;3:374–380. doi: 10.1016/s1369-5266(00)00099-6. [DOI] [PubMed] [Google Scholar]
- Swain SM, Reid JB, Kamiya Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997;12:1329–1338. [Google Scholar]
- Swain SM, Tseng T-S, Olszewski NE (2001) Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Thornton TM, Swain SM, Olszewski NE. Gibberellin signal transduction presents… SPY who O-GlcNAc'd me. Trends Plant Sci. 1999;4:424–428. doi: 10.1016/s1360-1385(99)01485-5. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- Ye GN, Stone D, Pang SZ, Creely W, Gonzalez K, Hinchee M. Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J. 1999;19:249–257. doi: 10.1046/j.1365-313x.1999.00520.x. [DOI] [PubMed] [Google Scholar]
- Young JC, Oberman MJ, Hart JU. Specific binding of tetratricopeptide repeat proteins to C-terminal 12-kDa domain of hsp90. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- Zhao S, Sancar A. Human blue–light photoreceptor hCRY2 specifically interacts with protein serine/threonine phosphatase 5 and modulates its activity. Photochem Photobiol. 1997;66:727–731. doi: 10.1111/j.1751-1097.1997.tb03214.x. [DOI] [PubMed] [Google Scholar]