Abstract
Mitochondrial genomes in frogs are crucial in reconstructing phylogenetic relationships and clarifying molecular evolution in these animals. Therefore, we determined and analyzed the complete mitochondrial genome sequence of Zhangixalus omeimontis in this research. The total length of this sequence is 19,782 base pairs, containing a total of 37 genes, which include 22 tRNA genes, 13 protein-coding genes, and 2 rRNA genes, along with two D-loop regions. The mitochondrial genome exhibits a novel rearrangement pattern (tRNASer-ND6-tRNAGlu-Cytb-CR1-ND5-CR2-tRNAThr-tRNALeu-tRNAPro) of genes. The nucleotide base composition of the mitochondrial genome consists of 32.51 % adenine (A), 31.32 % thymine (T), 21.95 % cytosine (C), and 14.21 % guanine (G), exhibiting a bias towards AT content (63.83 %). The phylogenetic tree is constructed using the Bayesian inference (BI) and maximum likelihood (ML) methods. The findings indicated a close relationship between Z. omeimontis and Z. dugritei. The comprehensive mitochondrial genome of Z. omeimontis will be a valuable asset for forthcoming research endeavours focusing on the evolution, taxonomy, and genetic preservation of Zhangixalus.
Keywords: Zhangixalus omeimontis, Rhacophoridae, Genome characterization, Phylogenetic analysis
Specifications Table
| Subject | Biological Sciences |
| Specific subject area | Omics: Genomics |
| Type of data | Table: Gene annotations, base composition Figures: Zhangixalus omeimontis, circular mitogenome map, phylogenetic tree Fasta: Mitogenome sequence Fastq: DNA sequence reads Data Format: Raw and analyzed |
| Data collection | DNA Extraction and Sequencing: Genomic DNA was extracted from the frog toe pad using the TIANamp Animal Genomic DNA Kit (Tiangen Biotech, Beijing). Assembly and annotation: PCR amplification of the 15 pairs of gene primers designed was performed. The PCR amplification fragments were separated by 0.9 % agarose gel electrophoresis and imaged using a gel imager. The purified amplification products were subjected to automated direct sequencing using the Sanger sequencing method and an ABI 3730 sequencer. The sequencing products were assembled into complete mitochondrial genome sequences using DNA Baser software (http://www.DNABaser.com) based on the overlapping regions (150–300 bp overlapping regions) and the annotation process was performed using MitoMaker. Phylogenetic analysis: IQ-tree was used to construct the Maximum Likelihood phylogenetic tree. |
| Data source location | Location: Baicha Village, Bailu Town City: Pengzhou City, Sichuan Province Country: China Latitude and Longitude: 31°13′18.07″N, 103°54′10.28″E Sample Storage Facility: Genomic DNA was deposited in the Ecological Security and Protection Key Laboratory of Sichuan Province with the voucher number JL20200812 collection overseen by Lichun Jiang; contact email: jiang_lichun@126.com). |
| Data accessibility | Repository name: GenBank Data identification number: MZ936366 Direct URL to data: https://www.ncbi.nlm.nih.gov/nuccore/MZ936366/ Repository name: NCBI BioProject Data identification number: PRJNA1001901 Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1001901 Repository name: NCBI BioSample Data identification number: SAMN36836140 Direct URL to data: https://www.ncbi.nlm.nih.gov/biosample/?term= SAMN36836140 Repository name: NCBI SRA Data identification number: SRR25517597 Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/?term= SRR25517597 Repository name: NCBI Genbank Data identification number: MZ936366 Direct URL to data: https://www.ncbi.nlm.nih.gov/nuccore/ MZ936366 Repository name: Zenodo data Data identification number: 10.5281/zenodo.13898565 Direct URL to data: https://zenodo.org/records/13898566 |
| Related research article |
1. Value of the Data
-
•
The complete mitogenome sequence of Zhangixalus omeimontis, native to Emei Mountain in Sichuan, presents a valuable dataset for future endeavors in species identification, molecular taxonomy, conservation efforts, DNA barcoding, and phylogenetic studies.
-
•
These data are useful for analysis of intraspecific divergence, population genomics and phylogeography within the mitochondrial genomes of Z. omeimontis.
-
•
This dataset, encompassing protein-coding sequences, is instrumental in phylogenetic reconstruction, offering heightened molecular clarity and bolstered statistical assurance over sequences derived from partial genes.
2. Background
Zhangixalus omeimontis (Omei Treefrog, https://amphibiaweb.org/species/4485) is one of the widely distributed species in the family Rhacophoridae, which is endemic to know from Sichuan, Yunnan, Guizhou, Hunan, Hubei and Guangxi Provinces in central and southern China, and inhabits at an altitude of 700–2000 m in wet mountainous with lush forest [1]. Although Z. omeimontis has been the subject of numerous behavioral ecology studies in recent years, limited information is currently available regarding its mitochondrial genome. Mitochondrial DNA has been used as a molecular marker to study population genetic structure and the evolutionary history of species [2]. Previous investigations into the phylogenetic relationships within Rhacophoridae have relied on partial nuclear (recombination activating gene-1; brain-derived neurotrophic factor, BDNF; rhodopsin exon-1, Rhod MHC class I, Microsatellite) [3,4] and mitochondrial gene (Cytb, ND2, COI, 12S rRNA, 16S rRNA, and trnV etc.) [[5], [6], [7]] sequences, yet these have provided insufficient molecular resolution to clarify the lineage among Rhacophorus species and their relatives. A subsequent effort to refine the phylogenetic tree with complete mitochondrial genome data included the majority of Rhacophoridae species, but omitted Zhangixalus representatives. Notably, research conducted by Jiang et al. [8]. introduced a new genus, Zhangixalus, challenging previous classifications that placed this genus within Rhacophorus. Consequently, the mitochondrial genomic relationship between Zhangixalus and its closest relatives remains to be fully elucidated. In this study, we conducted the sequencing and analysis of the entire mitochondrial genome of Z. omeimontis to clarify its taxonomic placement within the Rhacophoridae family. This endeavour aims to provide a significant genetic asset for forthcoming phylogenetic investigations encompassing the genera Zhangixalus, Rhacophorus, and Rhacophoridae.
3. Data Description
3.1. Mitogenome organization
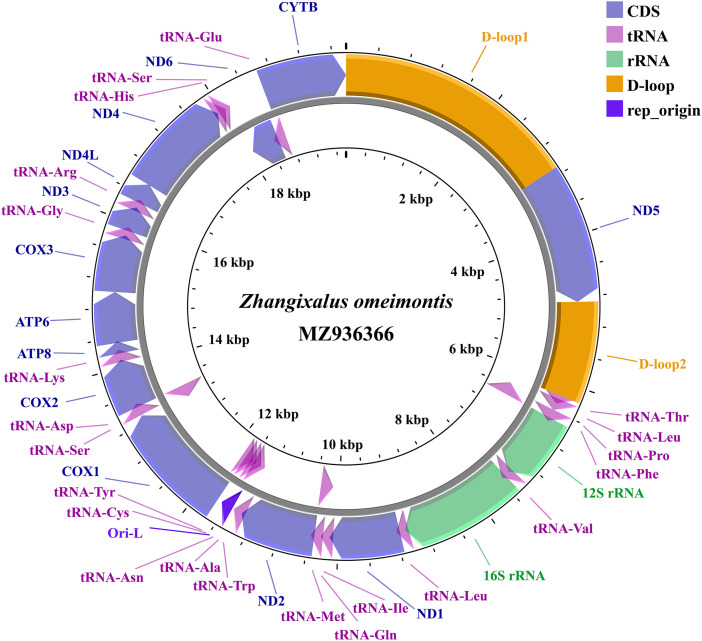
The complete mitochondrial genome sequence of Z. omeimontis is 19,782 base pairs in length (GenBank Accession Number: MZ936366) and comprises 2 rRNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, 13 protein-coding genes (PCGs), and two control regions known as D-loops (Fig. 1 and Table 1). These genetic components exhibit comparable lengths to their homologous genes found in amphibians [9]. The composition and arrangement of the control region varied considerably within this genus. Its mitogenome demonstrates a new gene rearrangement pattern (tRNASer-ND6-tRNAGlu-Cytb-CR1-ND5-CR2-tRNAThr-tRNALeu-tRNAPro). The nucleotide base composition of the mitogenome is characterized by a higher proportion of adenine (A) at 32.51 %, followed by thymine (T) at 31.32 %, cytosine (C) at 21.95 %, and guanine (G) at 14.21 %. The combined adenine and thymine content, known as A + T content, accounts for 63.83 % of the overall nucleotide composition. The sequence displays a low positive AT-skew (0.0186) and negative GC-skew (−0.2140) (Table 2). This pattern is consistent with findings in other vertebrate species.
Fig. 1.
Complete mitochondrial genome organization and gene arrangement of Z. omeimontis. Gene encoded on H- and l- strands with inverse arrow directions were shown outside and inside the circle, respectively. The complete mitogenome of Z. omeimontis is 19,782 bp with the inclusion of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, origin of l-strand replication (Ori-L) and control region (D-loop).
Table 1.
Mitogenomic organization of Z. omeimontis.
| Gene | Position |
Size | IGN (bp) | Codon |
Direction | ||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| D-loop1 | 1 | 3104 | 3104 | 0 | H | ||
| ND5 | 3105 | 4883 | 1779 | 0 | ATG | TAA | H |
| D-loop2 | 4884 | 6187 | 1304 | 0 | H | ||
| tRNA-Thr | 6188 | 6258 | 71 | 26 | H | ||
| tRNA-Leu | 6285 | 6356 | 72 | 25 | H | ||
| tRNA-Pro | 6382 | 6450 | 69 | 0 | L | ||
| tRNA-Phe | 6451 | 6520 | 70 | 0 | H | ||
| 12S rRNA | 6521 | 7445 | 925 | 0 | H | ||
| tRNA-Val | 7446 | 7514 | 69 | 0 | H | ||
| 16S rRNA | 7515 | 9081 | 1567 | 0 | H | ||
| tRNA-Leu | 9082 | 9155 | 74 | 0 | H | ||
| ND1 | 9156 | 10,116 | 961 | 0 | ATG | T– | H |
| tRNA-Ile | 10,117 | 10,187 | 71 | −1 | H | ||
| tRNA-Gln | 10,187 | 10,257 | 71 | −1 | L | ||
| tRNA-Met | 10,257 | 10,325 | 69 | 0 | H | ||
| ND2 | 10,326 | 11,363 | 1038 | 0 | ATT | TAA | H |
| tRNA-Trp | 11,364 | 11,433 | 70 | 0 | H | ||
| tRNA-Ala | 11,434 | 11,504 | 71 | 1 | L | ||
| tRNA-Asn | 11,506 | 11,578 | 73 | 0 | L | ||
| Ori-L | 11,579 | 11,603 | 25 | 0 | H | ||
| tRNA-Cys | 11,604 | 11,668 | 65 | 0 | L | ||
| tRNA-Tyr | 11,669 | 11,735 | 67 | 4 | L | ||
| COI | 11,740 | 13,293 | 1554 | −13 | ATA | AGG | H |
| tRNA-Ser | 13,281 | 13,351 | 71 | 2 | L | ||
| tRNA-Asp | 13,354 | 13,422 | 69 | 0 | H | ||
| COII | 13,423 | 14,106 | 684 | 20 | ATG | TAA | H |
| tRNA-Lys | 14,127 | 14,197 | 71 | 0 | H | ||
| ATP8 | 14,198 | 14,362 | 165 | −22 | ATG | TAA | H |
| ATP6 | 14,341 | 15,034 | 694 | 0 | ATG | T– | H |
| COIII | 15,035 | 15,818 | 784 | 0 | ATG | T– | H |
| tRNA-Gly | 15,819 | 15,886 | 68 | 0 | H | ||
| ND3 | 15,887 | 16,226 | 340 | 0 | ATG | T– | H |
| tRNA-Arg | 16,227 | 16,294 | 68 | 3 | H | ||
| ND4L | 16,298 | 16,582 | 285 | −7 | ATG | TAA | H |
| ND4 | 16,576 | 17,938 | 1363 | 0 | ATG | T– | H |
| tRNA-His | 17,939 | 18,009 | 71 | 0 | H | ||
| tRNA-Ser | 18,010 | 18,076 | 67 | 4 | H | ||
| ND6 | 18,081 | 18,569 | 489 | 0 | ATG | AGA | L |
| tRNA-Glu | 18,570 | 18,637 | 68 | 2 | L | ||
| Cytb | 18,640 | 19,782 | 1143 | 0 | ATG | TAA | H |
Notes: Data are given as Z. omeimontis. IGN, intergenic nucleotides; negative numbers indicate that adjacent genes overlap.
Table 2.
Base composition and AT/GC skewness of mitogenome of Z. omeimontis.
| Sequence | size (bp) | A% | G% | T% | C% | A + T % | G + C % | AT skew | GC skew |
|---|---|---|---|---|---|---|---|---|---|
| Mitogenome | 19,782 | 32.51 | 14.21 | 31.32 | 21.95 | 63.83 | 36.17 | 0.0186 | −0.2140 |
| Protein-coding protein | 11,276 | 29.23 | 13.82 | 32.86 | 24.10 | 62.09 | 37.91 | −0.0584 | −0.2711 |
| tRNAs | 1535 | 30.49 | 21.30 | 28.60 | 19.61 | 59.09 | 40.91 | 0.0320 | 0.0414 |
| rRNAs | 2492 | 36.08 | 17.82 | 24.52 | 21.59 | 60.59 | 39.41 | 0.1907 | −0.0957 |
| Control region | 4408 | 36.39 | 14.27 | 35.16 | 14.18 | 71.55 | 28.45 | 0.0171 | 0.0032 |
Among the 37 genes, the ND6 gene and eight transfer RNA (tRNA) genes (tRNAPro, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer and tRNAGlu) were located on the light strand, the rest of the genes are encoded on the heavy strand. For 13 PCGs, ND2 uses ATT as the start codon, COI uses ATA as the start codon, and the codon of 11 PCGs (ND1, ND3, ND4, ND4L, ND5, ND6, COII, COIII, ATP6, ATP8 and Cytb) are started with ATG. The stop codon of PCG is generally TAA/TGG or incomplete. In the study, the six PCGs (ATP8, COII, ND2, ND5, ND4L and Cytb) ended with TAA as a stop codon. But the stop codon of COI is AGG, and the stop codon of ND6 is AGA. The rest five genes (ATP6, COIII, ND1, ND3, and ND4) are found to be incomplete T-stop codons (Table 1), which may be presumably completed by posttranscriptional polyadenylation with polyA tail [10,11]. The 22 tRNA genes are interspersed along the whole genome, and range in size from 65 bp (tRNACys) to 73 bp (tRNALeu). Among the 2 rRNA genes, 12S rRNA is located between tRNAPhe and tRNAVal with 925 bp length, and 16S rRNA is located between tRNAVal and tRNALeu with 1567 bp length. The mitochondria sequence contains two D-loop regions (lengths of 3104 bp and 1304 bp), which are located between Cytb and ND5, ND5 and tRNAThr, respectively. Moreover, a putative origin of the Light-strand replication (OL) as the small non-coding region, is located between tRNAAsn and tRNACys within the WANCY tRNA cluster with the length of 25 bp, it is similar to most vertebrate mitogenomes [12].
3.2. Phylogenetic analysis
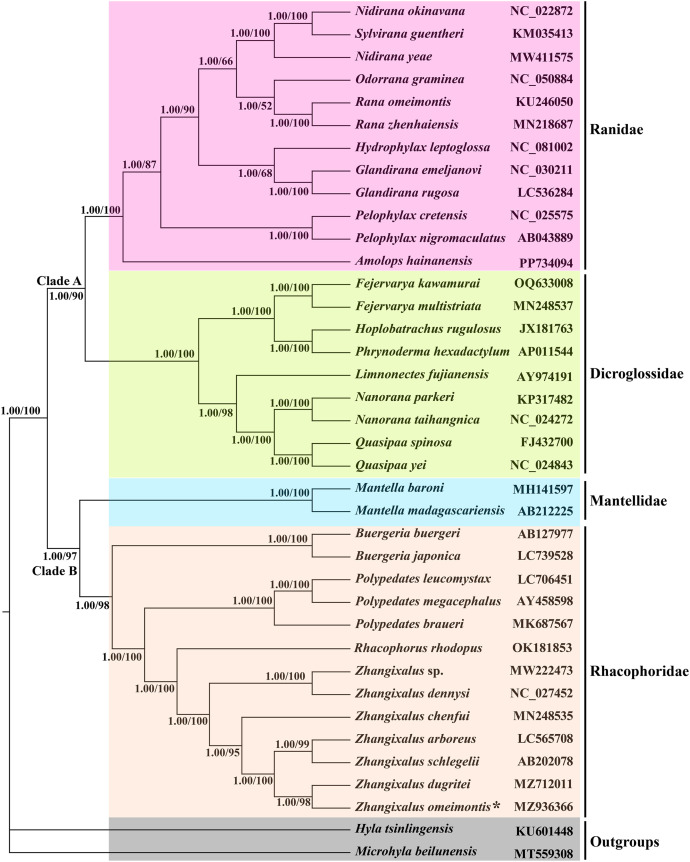
Based on comprehensive mitochondrial DNA genome sequences of 38 species, four families of amphibians (Dicroglossidae, Ranidae, Mantellidae, and Rhacophoridae) have been divided into two distinct clades (groups), as illustrated in Fig. 2. Clade A includes 14 genera: Amolops, Glandirana, Hydrophylax, Sylvirana, Odorrana, Rana, Pelophylax, Nidirana, Fejervarya, Hoplobatrachus, Limnonectes, Nanorana, Quasipaa, and Phrynoderma. Clade B comprises five genera: Zhangixalus, Rhacophorus, Polypedates, Buergeria, and Mantella. Additionally, two species (Microhyla beilunensis and Hyla tsinlingensis) are considered outgroups to this classification. Through Bayesian inference and maximum likelihood analysis, it has been determined that Z. omeimontis and Z. dugritei are closely related and form a sister group with Z. arboreus and Z. schlegelii. This suggests a close evolutionary relationship among these species. Notably, the clustering of Zhangixalus and Rhacophorus in the phylogenetic tree supports the monophyletic (originating from a single common ancestor) nature of the Zhangixalus genus, according to the research findings.
Fig. 2.
The phylogenetic tree inferred by the Bayesian inference (BI), Phylogenetic relationships of Rhacophorus omeimontis and other 37 species based on 13 protein-coding genes (PCGs), 2 rRNA genes and 22 tRNA genes. Microhyla beilunensis (Microhylidae) and Hyla tsinlingensis (Hylidae) are used as an outgroup. GenBank accession numbers and bootstrap values and posterior probabilities of nodes are shown on the tree. The asterisks indicate new sequences generated in this study.
3.3. Nonsynonymous and synonymous substitution
The nonsynonymous (Ka) and synonymous (Ks) substitution rates (Ka/Ks ratio) analysis across 13 protein-coding genes of the mitochondrial genome of Z. omeimontis were found to be less than 1, suggesting that strong purifying selection is acting on most of the mitochondrial protein-coding genes (Table 3). This is consistent with the idea that mitochondrial proteins play critical roles in energy production and other essential cellular processes, and that mutations that disrupt their function are likely to be deleterious and selected against.
Table 3.
The Ka/Ks values among the Zhangixalus species.
| ATP6 | ATP8 | COI | COII | COIII | Cytb | ND1 | ND2 | ND3 | ND4 | ND4L | ND5 | ND6 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AB202078-vs-LC565708 | 0.0940 | 0.3166 | 0.0109 | 0.0411 | 0.0337 | 0.0353 | 0.0560 | 0.0819 | 0.0848 | 0.0676 | 0.1347 | 0.0495 | 0.0731 |
| AB202078-vs-MN248535 | 0.1119 | 0.3678 | 0.0173 | 0.0419 | 0.0253 | 0.0318 | 0.0792 | 0.1219 | 0.1059 | 0.1802 | 0.2479 | 0.0559 | 0.1342 |
| AB202078-vs-MZ712011 | 0.1124 | 0.3177 | 0.0148 | 0.0266 | 0.0167 | 0.0373 | 0.0448 | 0.1194 | 0.0505 | 0.0823 | 0.1496 | 0.0502 | 0.1469 |
| AB202078-vs-MZ936366 | 0.1012 | 0.2218 | 0.0183 | 0.0222 | 0.0134 | 0.0237 | 0.0633 | 0.0918 | 0.0622 | 0.0793 | 0.1201 | 0.0606 | 0.1085 |
| LC565708-vs-MN248535 | 0.0789 | 0.5846 | 0.0166 | 0.0329 | 0.0326 | 0.0371 | 0.0632 | 0.1243 | 0.0855 | 0.1615 | 0.1223 | 0.0576 | 0.1459 |
| LC565708-vs-MZ712011 | 0.0933 | 0.3793 | 0.0126 | 0.0290 | 0.0232 | 0.0347 | 0.0472 | 0.1093 | 0.0819 | 0.0773 | 0.1459 | 0.0470 | 0.1342 |
| LC565708-vs-MZ936366 | 0.0507 | 0.2440 | 0.0178 | 0.0147 | 0.0212 | 0.0233 | 0.0540 | 0.0991 | 0.0800 | 0.0688 | 0.0905 | 0.0564 | 0.0949 |
| MN248535-vs-MZ712011 | 0.0672 | 0.3061 | 0.0091 | 0.0310 | 0.0274 | 0.0311 | 0.0549 | 0.1255 | 0.0679 | 0.1507 | 0.1570 | 0.0489 | 0.1707 |
| MN248535-vs-MZ936366 | 0.0950 | 0.3629 | 0.0104 | 0.0353 | 0.0278 | 0.0244 | 0.0579 | 0.1323 | 0.0782 | 0.1492 | 0.1828 | 0.0887 | 0.1361 |
| MN248535-vs-MZ936366 | 0.0318 | 0.1680 | 0.0204 | 0.0105 | 0.0336 | 0.0351 | 0.0503 | 0.0934 | 0.0622 | 0.0763 | 0.1084 | 0.0570 | 0.0762 |
3.4. Discussion and conclusion
By utilizing Sanger sequencing and assembly techniques, the complete mitochondrial genome of Z. omeimontis has been determined to be 19,782 base pairs in length. The gene arrangements and composition exhibit similarities to the presumed ancestral mitochondrial genome arrangement of Buergeria buergeri within the Rhacophoridae family [9], as well as to those of various other previously analyzed tree frog specie [2,9,13]. A Bayesian inference tree was constructed using the complete mitochondrial genomes of Z. omeimontis and 37 other species indicates that Z. omeimontis, Z. dugritei, Z. arboreus, Z. schlegelii, Z. sp., and Z. dennysi form a sister-group mitochondrial relationship with Rhacophorus rhodopus, P. braueri and P. megacephalus, then gnues Buergeria species, consistent with the findings of Jiang et al. [8], Chen et al. [14], and Dufresnes et al. [15]. Including additional closely related taxa in future comprehensive mitochondrial genome-based phylogenetic analyses may enhance our understanding of the evolutionary relationships among Zhangixalus, Rhacophorus, and Polyedates [[16], [17], [18]]. It is crucial to acknowledge that alterations in taxonomic sampling could potentially influence the species relationships depicted in the phylogenetic tree. Therefore, further investigations incorporating extensive taxon sampling are imperative to accurately validate the phylogenetic connections within the genera Zhangixalus and Rhacophorus. Our findings significantly contribute to understanding the genetic diversity and evolution of Zhangixalus, offering valuable insights for future studies in this field.
4. Experimental Design, Materials and Methods
4.1. Biological sample
The Z. omeimontis used in this study was collected in Baicha village, Bailu Town, Pengzhou City, Sichuan Province, China (31°13′18.07″N, 103°54′10.28″E) in August 2020, and it was identified according to morphological keys (Fig. 3) [19]. The distribution of this species is shown in Figure S1. Upon capturing Z. omeimontis in their natural habitat, the interdigital webbing was sterilized with alcohol, approximately 30 mg of webbing was excised, disinfected once more, and then returned to the wild. Subsequently, the sample was preserved in 95 % ethanol and deposited in the Ecological Security and Protection Key Laboratory of Sichuan Province with the voucher number JL2020081208 (http://zdsys.mnu.cn/; collection overseen by Lichun Jiang; contact email: jiang_lichun@126.com).
Fig. 3.
The specimen of Zhangixalus omeimontis from Baicha Village, Bailu Township, Pengzhou City, Sichuan Province, China (Photo by Lichun Jiang).
4.2. DNA extraction and sequencing
Genomic DNA was extracted from the webs of the frog toe using the TIANamp Animal Genomic DNA Kit (TIANGEN, Beijing) following the operation instructions. The mitogenome sequences of relative species of Zhangixalus and Rhacophorus were referenced and aligned using ClustalW software. Partial PCR primers were designed based on the alignments of the relatively conserved regions. Another part of the primers derived from the literature [20]. Then, PCR amplification was performed using the designed 15 pairs of gene primers. The PCR amplification fragments were separated by 0.9 % agarose gel electrophoresis and the fragments were scanned with a gel imager. Each purified amplification products were subjected to automated direct sequencing using an ABI 3730 sequencer based on the Sanger sequencing method. DNA Baser software (http://www.DNABaser.com) was used to assemble the whole mitogenome sequence based on the overlapping portions (150–300 bp in the overlapping region) of the sequencing products. The annotation process was executed using MITOS online tool. The mitogenome sequence and gene annotations were submitted to the National Center for Biotechnology Information GenBank database under the accession number MZ936366. The CGView online server (https://proksee.ca/) was chosen to draw the mitogenome map.
4.3. Phylogenetic analysis
To study the phylogenetic relationship of Z. omeimontis, we used the BI method to structure a phylogenetic tree and analysis based on 37 genes (2 rRNA genes, 22 tRNA genes, and 13 PCGs) of 38 species (13 Rhacophoridae, 2 Mantellidae, 9 Dicroglossinae, 12 Ranidae, and 2 outgroups). The BI analysis was performed using MrBayes v3.2. The SequenceMatrix was used to splice sequences of the same species. BI of nucleotide acid datasets was performed using the model of GTR + G + I (nst = mixed; rates = invgamma). The program commences by executing four Monte-Carlo Markov Chains for 500,000 generations in a random manner. Output trees were sample freq every 1000 generations and the first 25 % of samples were discarded as burn-in.
4.4. Ka and Ks analysis
The analysis of Ka/Ks was conducted as follows: 1) Coding DNA sequences (CDS) and protein sequences of 13 protein-coding genes (PCGs) from five species were collected from GenBank; 2) Multiple sequence alignment (MSA) was performed at the protein level using MAFFT software to ensure that the alignment reflects functional conservation; 3) The aligned protein sequences were used to guide the nucleotide (CDS) alignment, ensuring that the DNA alignment remains in-frame and avoids gaps that could lead to inaccurate Ka/Ks estimations; 4) Using the aligned DNA sequences, Ka and Ks substitution rates were calculated with the Ka/Ks Calculator tool.
Limitations
Not applicable.
Ethics Statement
This study was carried out in accordance with the animal care and use committee at the Mianyang Normal University. Efforts were taken to minimize suffering and included administering anesthesia. The study did not involve endangered or protected species. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Credit Author Statement
Qinggang Mei: Investigation, Collect samples, Analyze the data, Writing-Original draft; Yi Qing: Investigation, Collect samples, Contribute analysis tools; Yiming Deng: Investigation; Collect samples; Contribute analysis tools; Organize tables and beautify pictures, Administration project; Dongmei Zhao: Investigation, Analyze the data, Prepare ffgures and tables; Lichun Jiang: Conceive and designed the experiments, Write the paper, Visualization, Supervision, Administration project, Funding acquisition. All authors were involved in drafting the paper and final version approval. The contributions are ranked in order.
Acknowledgments
This research was supported by the Natural Science Foundation Project of Science and Technology Department of Sichuan Province (No. 2023NSFSC0206), and the Research Project of Ecological Security and Protection Key Laboratory of Sichuan Province (No. ESP2003).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2024.111154.
Appendix. Supplementary materials
Figure S1. Species distribution map of the Omei Treefrog, Zhangixalus omeimontis.
Data Availability
Zenodo dataZenodo data (Original data).
References
- 1.Chaabane A., Verneau O., Preez L.D. Indopolystoma n. gen. (monogenea, polystomatidae) with the description of three new species and reassignment of eight known polystoma species from asian frogs (anura, rhacophoridae) Parasite. 2019;26:67. doi: 10.1051/parasite/2019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang M., Lv T., Duan R., Zhang S., Li H. The complete mitochondrial genome of rhacophorus dennysi (anura: rhacophoridae) and phylogenetic analysis, Mitochondrial DNA. DNA Mapp. Seq. Anal. 2016;27:3719–3720. doi: 10.3109/19401736.2015.1079873. [DOI] [PubMed] [Google Scholar]
- 3.Meegaskumbura M., Meegaskumbura S., Bowatte G., Manamendra-Arachchi K., Pethiyagoda R., Hanken J., Schneider C.J. Taruga (anura: rhacophoridae), a new genus of foam-nesting tree frogs endemic to sri lanka. Ceylon J. Sci. (Biol. Sci.) 2011;39:75–94. doi: 10.4038/cjsbs.v39i2.2995. [DOI] [Google Scholar]
- 4.Zhao M., Wang Y., Shen H., Li C., Chen C., Luo Z., Wu H. Evolution by selection, recombination, and gene duplication in MHC class I genes of two rhacophoridae species. BMC Evol. Biol. 2013;13:113. doi: 10.1186/1471-2148-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J.-T., Li Y., Murphy R.W., Rao D.-Q., Zhang Y.-P. Phylogenetic resolution and systematics of the asian tree frogs, rhacophorus (rhacophoridae, amphibia) Zool. Scr. 2012;41:557–570. doi: 10.1111/j.1463-6409.2012.00557.x. [DOI] [Google Scholar]
- 6.Liu Y., Yang M., Jiang Y., Han F., Li Y., Ni Q., Yao Y., Xu H., Zhang M. The near complete mitochondrial genome of white-lipped treefrog, polypedates braueri (anura, rhacophoridae), Mitochondrial DNA. DNA Mapp. Seq. Anal. 2017;28:271–272. doi: 10.3109/19401736.2015.1118075. [DOI] [PubMed] [Google Scholar]
- 7.Dang N.-X., Sun F.-H., Lv Y.-Y., Zhao B.-H., Wang J.-C., Murphy R.W., Wang W.-Z., Li J.-T. DNA barcoding and the identification of tree frogs (amphibia: anura: rhacophoridae), Mitochondrial DNA. DNA Mapp. Seq. Anal. 2016;27:2574–2584. doi: 10.3109/19401736.2015.1041113. [DOI] [PubMed] [Google Scholar]
- 8.Jiang D., Jiang K., Ren J., Wu J., Li J. Resurrection of the genus leptomantis, with description of a new genus to the family rhacophoridae (amphibia: anura) Asian Herpetol. Res. 2019;10:1–12. doi: 10.16373/j.cnki.ahr.180058. [DOI] [Google Scholar]
- 9.Sano N., Kurabayashi A., Fujii T., Yonekawa H., Sumida M. Complete nucleotide sequence of the mitochondrial genome of schlegel's tree frog rhacophorus schlegelii (family rhacophoridae): duplicated control regions and gene rearrangements. Genes Genet. Syst. 2005;80:213–224. doi: 10.1266/ggs.80.213. [DOI] [PubMed] [Google Scholar]
- 10.Boore J.L. Complete mitochondrial genome sequence of the polychaete annelid platynereis dumerilii. Mol. Biol. Evol. 2001;18:1413–1416. doi: 10.1093/oxfordjournals.molbev.a003925. [DOI] [PubMed] [Google Scholar]
- 11.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 12.Shan X., Xia Y., Kakehashi R., Kurabayashi A., Zou F.-D., Zeng X.-M. Complete mitochondrial genome of amolops mantzorum (anura: ranidae) Mitochondrial DNA Part A, DNA Mapp., Sequenc. Anal. 2016;27:705–707. doi: 10.3109/19401736.2014.913152. [DOI] [PubMed] [Google Scholar]
- 13.Haridy M., Tachikawa Y., Yoshida S., Tsuyuguchi K., Tomita M., Maeda S., Wada T., Ibi K., Sakai H., Yanai T. Mycobacterium marinum infection in japanese forest green tree frogs (rhacophorus arboreus) J. Comp. Pathol. 2014;151:277–289. doi: 10.1016/j.jcpa.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Chen J.-M., Prendini E., Wu Y.-H., Zhang B.-L., Suwannapoom C., Chen H.-M., Jin J.-Q., Lemmon E.M., Lemmon A.R., Stuart B.L., Raxworthy C.J., Murphy R.W., Yuan Z.-Y., Che J. An integrative phylogenomic approach illuminates the evolutionary history of old world tree frogs (anura: rhacophoridae) Mol. Phylogenet. Evol. 2020;145 doi: 10.1016/j.ympev.2019.106724. [DOI] [PubMed] [Google Scholar]
- 15.Dufresnes C., Ambu J., Prasad V.K., Borzée A., Litvinchuk S.N. A phylogeographical framework for zhangixalus gliding frogs, with insight on their plasticity of nesting behaviour. Biol. J. Linn. Soc. 2022;135:40–51. doi: 10.1093/biolinnean/blab143. [DOI] [Google Scholar]
- 16.Li J., Che J., Bain R.H., Zhao E., Zhang Y. Molecular phylogeny of rhacophoridae (anura): a framework of taxonomic reassignment of species within the genera aquixalus, chiromantis, rhacophorus, and philautus. Mol. Phylogenet. Evol. 2008;48:302–312. doi: 10.1016/j.ympev.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z., Li H., Zhu Y., Feng Q., He Y., Chen X. Molecular phylogeny of the family dicroglossidae (amphibia: anura) inferred from complete mitochondrial genomes. Biochem. Syst. Ecol. 2017;71:1–9. doi: 10.1016/j.bse.2017.01.006. [DOI] [Google Scholar]
- 18.Pyron R.A., Wiens J.J. A large-scale phylogeny of amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 2011;61:543–583. doi: 10.1016/j.ympev.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Che J., Wang K. AmphibiaChina: an online database of Chinese amphibians. Zool. Res. 2016;37:57. doi: 10.13918/j.issn.2095-8137.2016.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurabayashi A., Sumida M. PCR primers for the neobatrachian mitochondrial genome. Curr. Herpetol. 2009;28:1–11. doi: 10.3105/018.028.0101. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Species distribution map of the Omei Treefrog, Zhangixalus omeimontis.
Data Availability Statement
Zenodo dataZenodo data (Original data).