ABSTRACT
Since the last case of indigenous rubella virus (RuV) was detected in 2009 in the Region of the Americas, sporadic rubella and congenital rubella cases have been confirmed, and subsequently, a low number of associated sequences have been reported. Fifty-one sequences of wild-type RuV, representing four genotypes (1E, 1G, 1J, and 2B), were reported from five countries, with confirmed sources of exposure for 46 cases. Phylogenetic analysis revealed the diversity of these viruses, showing no associations with sustained endemic transmission from previously endemic strains. Notably, 13 sequences were associated with travel from countries where no genetic information of wild-type viruses was available. In addition to sequences from postnatal and congenital infections, 23 sequences were collected from patients with diseases associated with RuV persistent infection. These findings highlight the Region’s success in maintaining rubella elimination, emphasize its valuable contribution to global RuV molecular epidemiology, and address potential challenges in progressing toward the goal of rubella eradication.
Keywords: Rubella virus, molecular epidemiology, genotype, congenital rubella syndrome, disease eradication, Americas
RESUMEN
Desde que en el 2009 se detectara el último caso autóctono de infección por el virus de la rubéola (VRu) en la Región de las Américas, se han confirmado casos esporádicos de rubéola y de rubéola congénita y, posteriormente, se ha notificado un número reducido de secuencias asociadas a este virus. Se notificaron 51 secuencias del VRu de tipo salvaje, que correspondían a cuatro genotipos (1E, 1G, 1J y 2B), procedentes de cinco países, con orígenes de la exposición confirmados en 46 de los casos. El análisis filogenético reveló la diversidad de estos virus y no mostró ninguna asociación con una transmisión endémica sostenida a partir de las cepas que antes habían sido endémicas. Es de destacar que 13 secuencias se asociaron a viajes procedentes de países en los que no se disponía de información genética sobre los virus de tipo salvaje. Además de las secuencias procedentes de infecciones posnatales y congénitas, se recogieron 23 secuencias de pacientes con enfermedades asociadas a la infección persistente por el VRu. Estos resultados ponen de relieve el éxito de la Región a la hora de mantener la eliminación de la rubéola, subrayan su valiosa contribución a la información sobre las características de epidemiología molecular mundial del VRu y abordan los posibles desafíos a los que es preciso hacer frente para avanzar hacia el objetivo de la erradicación de la rubéola.
Palabras clave: Virus de la rubeola, epidemiología molecular, genotipo, síndrome de rubéola congénita, erradicación de la enfermedad, Américas
RESUMO
Desde que o último caso autóctone do vírus da rubéola (RuV) foi detectado em 2009 na Região das Américas, houve confirmação de casos esporádicos de rubéola e rubéola congênita e, posteriormente, registrou-se um pequeno número de sequências associadas. Foram detectadas 51 sequências de RuV tipo selvagem em cinco países, representando quatro genótipos (1E, 1G, 1J e 2B), com fontes confirmadas de exposição em 46 casos. A análise filogenética revelou a diversidade desses vírus, e não se detectou nenhuma associação com a transmissão endêmica sustentada de cepas previamente endêmicas. Vale destacar que 13 sequências estavam associadas a viagens originadas de países onde não havia informações genéticas disponíveis sobre os vírus tipo selvagem. Além das sequências de infecções pós-natais e congênitas, 23 sequências foram coletadas de pacientes com doenças associadas a infecção persistente pelo RuV. Esses achados destacam o sucesso da Região em manter a eliminação da rubéola, enfatizam sua valiosa contribuição para a epidemiologia molecular mundial do RuV e abordam os possíveis desafios no progresso rumo à meta de erradicação da rubéola.
Palavras-chave: Vírus da rubeola, epidemiologia molecular, genótipo, síndrome da rubéola congênita, erradicação de doenças, América
Strong immunization programs and effective surveillance are important for the prevention and control of infectious diseases. One such infectious disease is rubella. Rubella virus (RuV) is an enveloped virus with a single-stranded positive sense RNA genome, approximately 9760 nucleotides in length, in the genus Rubivirus of the Matonaviridae family (1). Postnatal RuV infection is usually associated with mild symptoms such as febrile illness with skin rashes, approximately 50% of which can be subclinical. RuV infection in pregnancy can lead to severe damage to developing fetal organs, causing congenital rubella syndrome (CRS) typically early in gestation. In addition to CRS, persistent RuV infection can also occur postnatally, contributing to a spectrum of clinical manifestations, including encephalitis, Fuchs uveitis, arthritis, and granulomas (2-6).
For rubella, a single dose of vaccine offers long-lasting immunity (1), and a second dose is highly recommended to improve herd immunity and prevent CRS (7). Most Member States in the Region of the Americas adopted the trivalent measles-mumps-rubella (MMR) vaccine in the 2000s, with support from the Pan American Health Organization (PAHO) (https://www.paho.org/en/topics/rubella). Through the implementation of recommended surveillance and immunization strategies, in 2015 the Region of the Americas became the first World Health Organization (WHO) region to achieve the elimination of rubella and CRS (8). The maintenance of rubella elimination was re-verified by the WHO Regional Verification Committee in 2022 and 2023 (9, 10). Currently, the global vision in the 2021 to 2030 Measles and Rubella Strategic Framework is to work toward “a world free from measles and rubella” by achieving and sustaining the regional measles and rubella elimination goals (11).
To support global RuV virologic surveillance, in 2004 the WHO established a standard nomenclature based on 739 nucleotide sequences (nts) from 8731 nts to 9469 nts in the E1 gene of RuV (12, 13). This window provided consistent phylogeny of reference viruses with high-clade credibility values, similar to the full-length sequences of the structural protein coding region (13). Phylogenetic analysis of wild-type and vaccine strains of RuVs using this genotyping window revealed two distinct groups, designated as clade 1 and clade 2. These groups differ by 8% to 10% at the nucleotide level. Clades 1 and 2 can be further subdivided into nine (1B-1J) and three (2A-2C) recognized genotypes, respectively, and each genotype is named alphabetically. Clade 1 also contains a provisional genotype 1a, which comprises RA27/3, the RuV component in the MMR vaccine. Genotype 1a also includes “old-world” strains that were circulating before the 1980s (12, 14). Due to the decreased genetic diversity of wild-type RuV in recent years, a systematic method has been established to subdivide the three dominant circulating genotypes to increase phylogenetic resolution to support the investigation of transmission pathways and identify the source(s) of exposure (15).
Laboratories within the Global Measles and Rubella Laboratory Network are encouraged to submit sequences from rubella and CRS cases to the WHO Rubella Nucleotide Surveillance (RubeNS2) system, initiated in 2005 and maintained by the UK Health Security Agency (16). As of 1 November 2023, there have been 5 934 sequences submitted to RubeNS2 by 50 network laboratories worldwide.
Virologic surveillance data in the Americas played a crucial role in supporting the verification of rubella elimination. Before the confirmation of rubella elimination in 2015, three large-scale outbreaks occurred in the Americas. The first epidemic spanned from 1962 to 1965 in the United States, resulting in approximately 12.5 million cases and 20 000 CRS cases (1). The second outbreak took place in Chile and Peru from 2002 to 2005 with 1 735 cases caused by genotype 1C viruses (17). The third outbreak took place from 2006 to 2008 and was attributed to viruses of genotype 2B. This outbreak led to more than 13 000 rubella cases in Argentina, Brazil, and Chile, and 27 CRS cases reported by Argentina and Brazil (17). The last cases of endemic rubella and CRS in the Americas were reported in 2009 (https://www.paho.org/en/topics/rubella). There has been no evidence of more than 12 months of continuous endemic transmission of rubella since 2010. Therefore, the post-elimination era is defined as starting in 2010 and continuing to the present in this report, even though Regional rubella elimination was officially declared in 2015. With rubella elimination from the Americas nearly 9 years ago, this report documents virologic surveillance in both pre- and post-elimination settings in the Region. Additionally, it addresses the challenges that the global community may face when considering rubella eradication (18).
METHODS
Data sources
RuV sequences were obtained from the US National Institutes of Health’s genetic sequence database, GenBank (www.ncbi.nlm.nih.gov/genbank/) and from the WHO Rubella Virus Nucleotide Surveillance database, RubeNS2 (https://who-gmrln.org/rubens2). All of the sequences reported to RubeNS2 are also available in GenBank, except for one: RVs/Cartagena.COL/31.12. Permission to use RVs/Cartagena.COL/31.12 was authorized by the National Institute of Health of Colombia, and permission to analyze and assign lineages to the sequences in the RubeNS2 database was granted by the steering committee. Sequences originating from vaccine viruses or laboratory strains, including recombinant viruses, were excluded from the dataset. The inclusion criterion for this report was limited to sequences that covered the complete genotyping window. In instances where multiple sequences were available from a single case, only a singular sequence was chosen for analysis.
The epidemiologic information, e.g., country of exposure, is included as one of the data elements associated with the case reported to the US Centers for Disease Control and Prevention (19). In Canada, epidemiological information for rubella cases is collected by the Canadian measles/rubella surveillance system (https://www.canada.ca/en/public-health/services/surveillance.html#a5) (20). Some of the cases included in this analysis have been described elsewhere (4, 21, 22). For sequences from other countries, the sources of exposure and diseases associated with RuV infection were collected from GenBank and/or RubeNS2.
Phylogenetic analyses and lineage assignment
Sequence alignment and phylogenetic analysis were performed using Molecular Evolutionary Genetics Analysis software, version 7 (23). The genotypes and/or lineages of sequences were determined using characterized reference virus sequences defined in the previous studies through the neighbor-joining method and bootstrap test with 1 000 replicas (15, 24-26).
RESULTS
Sixteen countries in the Americas contributed a total of 228 RuV sequences with the complete genotyping window to GenBank and RubeNS2 from 1961 to 2023. Of these sequences, 207 entries were sourced from GenBank, while 20 were exclusively found in RubeNS2.
Molecular epidemiology in the pre-elimination era
From 1961 to 2009, a total of 155 RuV sequences from eight genotypes (1a, 1B, 1C, 1D, 1E, 1G, 1J, and 2B) were reported by 15 countries in the Americas (Table 1). Except for one sequence collected from a patient with chronic arthritis, all sequences were obtained from cases associated with acute or congenital infections. Nineteen sequences collected before 1984 belonged to genotype 1a and were exclusively contributed by the United States.
TABLE 1. Genotypes and lineages of rubella virus (RuV) detected in the countries of the Region of the Americas, by era of detection,1961 through 2023.
|
Country |
Genotype (lineage)/yr |
|
|---|---|---|
|
1961-2009 (pre-elimination) |
2010-2023 (post-elimination) |
|
|
Argentina |
1B/98; 2B(L0)/08-09a |
|
|
Bahamas |
1E(L0)/97 |
|
|
Belize |
1C/94 |
|
|
Brazil |
1a/99-09; 1G(L0)/03,04; 1J/05; 2B(L0)/06-09a |
|
|
Canada |
1a/84, 85d; 1D/87; 1E(L0)/97; 1G(L0)/05b; 2B(L0)/07b |
1E(L2)/19; 1G(L1b)/23b; 1J/11,14b; |
|
Chile |
1a/99 1C/05a; 1E(L0)/99; 2B(L0)/07a |
2B(L2c)/11b |
|
Colombia |
2B(L2c)/12b |
|
|
Ecuador |
1C/99 |
|
|
El Salvador |
1C/02 |
|
|
Guyana |
1E(L0)/97 |
|
|
Honduras |
1C/00 |
|
|
Mexico |
1C/97 |
2B(L2c)/12b |
|
Panama |
1C/99 |
|
|
Peru |
1C/04-05a |
|
|
Suriname |
1E(L0)/98 |
|
|
United States |
1a/61-00; 1B/99; 1C/90-00; 1D/98; 1E(L0)/97-00b,c; 1E(L1)/08b; 1E(L2)/05b; 1E(L4)/08; |
1a/16-20d; 1G(L0)/14; 1G(L1b)/12-17b,c; 1G(L2a)/10b; 1J/10b,15b; |
Genotypes related to large-scale outbreaks.
Known or likely importation(s) from other countries.
Source of exposure for some sequences was not identified.
RuV persistent infection.
Unclassified; not closely related to any of the recognized genotypes.
Source: Prepared by the authors using data from GenBank and RubeNS2.
From 1984 to 2009, a total of 13 genotype 1a viruses were reported in Brazil, Canada, Chile, and the United States. The most recent occurrence of wild-type genotype 1a viruses was reported by Brazil in 2009. In addition to genotype 1a, 123 sequences of other genotypes were reported by 15 countries. The most frequently detected genotypes during this period were 1C (n=42) and 2B (n=46), contributing to large-scale outbreaks in the Americas from 2002 to 2008. Other genotypes, such as 1B (n=2), 1D (n=2), and 1G (n=6) were endemic in specific countries during this period (17), while a few cases of 1J (n= 6) were imported (27).
Viruses of genotype 1E with four lineages were found during 1984 to 2009: L0 (n=16), L1 (n=1), L2 (n=1), and L4 (n=1). Cases associated with 1E-L1 and 1E-L2 were known to be imported from China and Malaysia, respectively. Except for one importation from Ukraine, viruses of 1E-L0 were detected in six countries in the Americas, and a few were associated with small-scale outbreaks or sporadic cases in the United States (27).
Notably, a genotype 2B virus (JN635293) was first detected in 2000 in the United States, and was imported from India (27). This virus belonged to lineage 2B-L2a (15) and is distinct from the virus that caused outbreaks in South America, which belonged to 2B-L0 (Table 1; Figure 1).
FIGURE 1. Phylogenetic trees of wild-type rubella virus sequences detected in the Region of the Americas A. Genotype 1E.
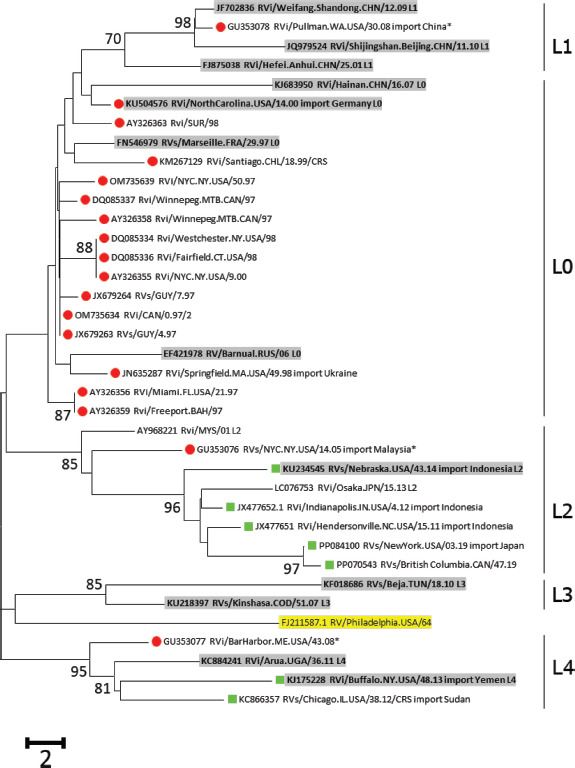
Note: The phylogenetic analysis of sequences from the Americas with the lineage reference sequences (shaded) of Genotype 1E, Genotype 1G, and Genotype 2B. Trees were rooted with RVi/Pennsylvania.USA/64 (JF727653; highlighted). Sequences from 2010-2023 are green squares; sequences before 2010 are red circles; sequences from 2004-2010 in the United States are marked by asterisks. The percentage of replicate trees with values >70%, in which the associated lineages clustered together in the bootstrap test (1 000 replicates), is shown next to the node.
B. Genotype 1G.
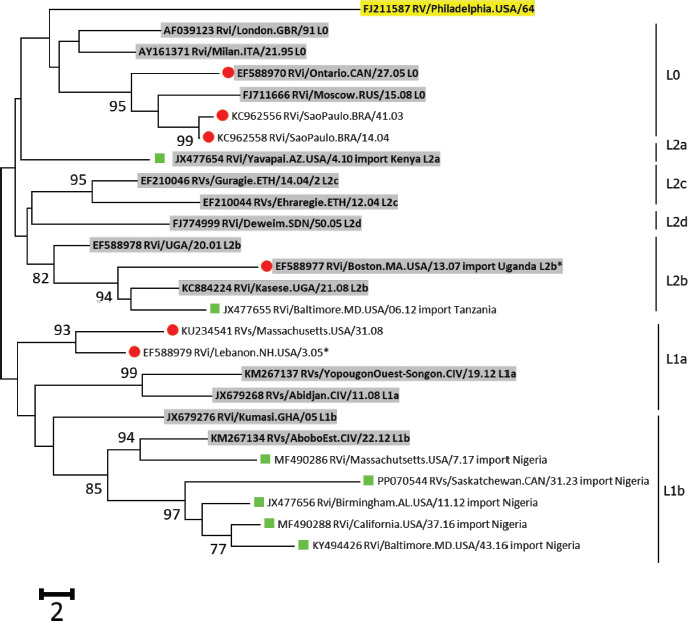
Note: The phylogenetic analysis of sequences from the Americas with the lineage reference sequences (shaded) of Genotype 1E, Genotype 1G, and Genotype 2B. Trees were rooted with RVi/Pennsylvania.USA/64 (JF727653; highlighted). Sequences from 2010-2023 are green squares; sequences before 2010 are red circles; sequences from 2004-2010 in the United States are marked by asterisks. The percentage of replicate trees with values >70%, in which the associated lineages clustered together in the bootstrap test (1 000 replicates), is shown next to the node.
C. Genotype 2B.
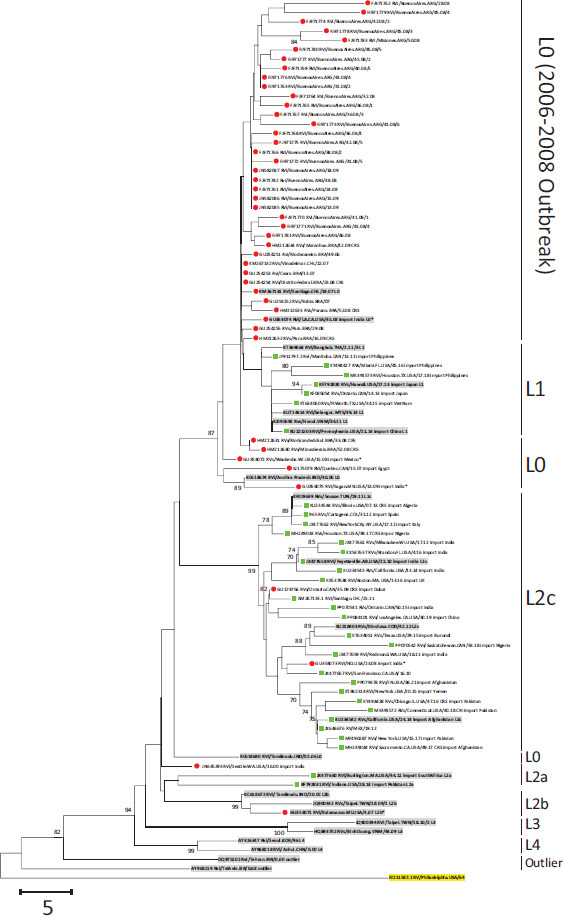
Note: The phylogenetic analysis of sequences from the Americas with the lineage reference sequences (shaded) of Genotype 1E, Genotype 1G, and Genotype 2B. Trees were rooted with RVi/Pennsylvania.USA/64 (JF727653; highlighted). Sequences from 2010-2023 are green squares; sequences before 2010 are red circles; sequences from 2004-2010 in the United States are marked by asterisks. The percentage of replicate trees with values >70%, in which the associated lineages clustered together in the bootstrap test (1 000 replicates), is shown next to the node.
Source: Prepared by the authors using data from GenBank and RubeNS2.
Molecular epidemiology in the post-elimination era
Between 2010 and 2023, five countries in the Americas (Canada, Chile, Colombia, Mexico, and the United States) reported 73 sequences representing six genotypes (1a, 1E, 1G, 1J, 2B, and 2C) (Table 1). Except for the 22 sequences of genotypes 1a and 2C from patients with persistent infections, the remaining 51 sequences of genotypes 1E, 1G, 1J, and 2B were associated with postnatal or congenital infections. Epidemiologic investigations were conducted by epidemiology staff regarding measles and rubella elimination in each country to identify the sources of exposure for 46 of the 51 cases; sources included countries in the five other WHO Regions (Table 2).
TABLE 2. Sequences obtained from rubella and CRS cases during routine surveillance reported by countries in the Region of the Americas, 2010-2023.
|
Sequence IDa |
Virus name |
Genotype (lineage)b |
Reporting country |
Year |
Diseasee |
Global RuV virologic surveillancef |
|
|---|---|---|---|---|---|---|---|
|
RVi/Manitoba.CAN/13.11 |
2B (L1) |
Canada |
Philippines |
2011 |
CRS |
||
|
RVs/BritishColumbia.CAN/25.11 |
1J |
Canada |
Unknown |
2011 |
|||
|
RVs/Ontario.CAN/14.13 |
2B (L1) |
Canada |
Japan |
2013 |
(25) |
||
|
RVs/Ontario.CAN/09.14 |
1J |
Canada |
Philippines |
2014 |
|||
|
RVs/Ontario.CAN/50.15 |
2B (L2c) |
Canada |
India |
2015 |
CRS |
(35) |
|
|
RVs/Saskatchewan.CAN/33.18 |
2B (L2c) |
Canada |
Nigeriag |
2018 |
CRS |
||
|
RVs/British Columbia.CAN/47.19 |
1E (L2) |
Canada |
Unknown |
2019 |
|||
|
RVs/Saskatchewan.CAN/31.23 |
1G (L1b) |
Canada |
Nigeriag |
2023 |
CRS |
||
|
RVi/Santiago.CHL/25.11 |
2B (L2c) |
Chile |
India |
2011 |
(35) |
||
|
965 |
RVs/Cartagena.COL/31.12 |
2B (L2c) |
Colombia |
Spain |
2012 |
(36) |
|
|
RVs/CDMX.MEX/19.12 |
2B (L2c) |
Mexico |
Unknown |
2012 |
|||
|
RVi/Miami.FL.USA/21.10 |
1J |
United States |
Philippines |
2010 |
|||
|
RVi/Yavapai.AZ.USA/4.10 |
1G (L2a) |
United States |
Kenya |
2010 |
(15) |
||
|
RVi/SanFrancisco.CA.USA/16.10 |
2B (L2c) |
United States |
Unknown |
2010 |
|||
|
RVi/Fayetteville.AR.USA/22.10 |
2B (L2c) |
United States |
India |
2010 |
|||
|
RVi/Hendersonville.NC.USA/15.11 |
1E (L2) |
United States |
Indonesia |
2011 |
|||
|
RVi/Redmond.WA.USA/18.11 |
2B (L2c) |
United States |
India |
2011 |
|||
|
RVi/Burlington.MA.USA/44.11 |
2B (L2a) |
United States |
South Africa |
2011 |
(15) |
||
|
RVi/Indianapolis.IN.USA/4.12 |
1E (L2) |
United States |
Indonesia |
2012 |
|||
|
RVi/Baltimore.MD.USA/06.12 |
1G (L2b) |
United States |
Tanzaniag |
2012 |
CRS |
||
|
Rvi/Birmingham.AL.USA/11.12 |
1G (L1b) |
United States |
Nigeriag |
2012 |
CRS |
||
|
RVs/Milwaukee.WI.USA/17.12 |
2B (L2c) |
United States |
India |
2012 |
(15) |
||
|
RVi/NewYorkCity.NY.USA/17.12 |
2B (L2c) |
United States |
Italy |
2012 |
GenBank |
||
|
RVs/Chicago.IL.USA/38.12 |
1E (L4) |
United States |
Sudan |
2012 |
CRS |
GenBank |
|
|
RVs/Hawaii.USA/17.13 |
2B (L1) |
United States |
Japan |
2013 |
(25) |
||
|
RVi/Indiana.USA/28.13 |
2B (L2a) |
United States |
Pakistang |
2013 |
|||
|
RVi/Buffalo.NY.USA/48.13 |
1E (L4) |
United States |
Yemeng |
2013 |
CRS |
||
|
RVs/Illinois.USA/07.13 |
2B (L2c) |
United States |
Algeria |
2013 |
CRSh |
RubeNS |
|
|
RVi/Pennsylvania.USA/22.14 |
2B (L1) |
United States |
China |
2014 |
|||
|
RVs/California.USA/24.14 |
2B (L2c) |
United States |
Unknown |
2014 |
|||
|
RVs/California.USA/13.14 |
2B (L2c) |
United States |
India |
2014 |
|||
|
RVs/Nebraska.USA/43.14 |
1E (L2) |
United States |
Indonesia |
2014 |
(28) |
||
|
Rvi/NewYork.USA/20.15 |
2B (L2c) |
United States |
Yemeng |
2015 |
|||
|
RVs/Florida.USA/09.15 |
1J |
United States |
Philippines |
2015 |
(34) |
||
|
RVs/FtWorth.TX.USA/34.15 |
2B (L1) |
United States |
Vietnam |
2015 |
(34) |
||
|
RVs/Texas.USA/29.15 |
2B (L2c) |
United States |
Burundig |
2015 |
|||
|
RVs/Brandon.FL.USA/4.16 |
2B (L2c) |
United States |
India |
2016 |
(35) |
||
|
RVi/Boston.MA.USA/13.16 |
2B (L2c) |
United States |
United Kingdom |
2016 |
RubeNS |
||
|
RVi/Baltimore.MD.USA/43.16 |
1G (L1b) |
United States |
Nigeriag |
2016 |
CRS |
||
|
RVs/Miami.FL.USA/45.16 |
2B (L1) |
United States |
Philippines |
2016 |
GenBank |
||
|
RVs/Chicago.IL.USA/47.16 |
2B (L2c) |
United States |
Pakistan |
2016 |
CRS |
RubeNS |
|
|
RVi/California.USA/37.16 |
1G (L1b) |
United States |
Nigeriag |
2016 |
CRS |
||
|
RVi/Massachutsetts.USA/7.17 |
1G (L1b) |
United States |
Nigeriag |
2017 |
CRS |
||
|
RVs/New York.USA/15.17 |
2B (L2c) |
United States |
Pakistan |
2017 |
RubeNS |
||
|
RVs/Houston.TX.USA/39.17 |
2B (L2c) |
United States |
Nigeriag |
2017 |
CRS |
||
|
RVs/Sacramento.CA.USA/49.17 |
2B (L2c) |
United States |
Afghanistang |
2017 |
CRS |
||
|
RVs/Connecticut.USA/40.18 |
2B (L2c) |
United States |
Pakistan |
2018 |
CRI |
RubeNS |
|
|
RVi/Houston.TX.USA/17.18 |
2B (L1) |
United States |
Philippines |
2018 |
(34) |
||
|
RVs/NYC.New York.USA/03.19 |
1E (L2) |
United States |
Japan |
2019 |
(34) |
||
|
RVs/LosAngeles.CA.USA/30.19 |
2B (L2c) |
United States |
China |
2019 |
(24) |
||
|
RVs/IN.USA/38.21 |
2B (L2c) |
United States |
Afghanistang |
2021 |
Abbreviations: CRI, congenital rubella infection; CRS, congenital rubella syndrome; RubeNS2, Rubella Nucleotide Surveillance system; RV, rubella virus; RuV, rubella virus.
Permission to use RVs/Cartagena.COL/31.12, exclusively in RubeNS2, was granted by the National Reference Laboratory, Colombia. All other sequences were available in GenBank.
Epidemiological investigation identified international travel history that would be consistent when the case was likely to have been exposed to rubella virus.
Unknown: case is known to be imported but the source could not be identified.
Only sequences clearly associated with CRS/CRI are denoted; otherwise, the sequences are associated with acquired rubella.
Reports of the genotype (or lineage, as applicable) from the country of exposure or by other countries that reported exposures in the same country.
Genetic data of wild-type RuV was not available.
Sequence was collected from a cataract extracted from a CRS case born outside of the country.
Source: Prepared by the authors using data from GenBank and RubeNS2.
Phylogenetic analyses revealed that the viruses detected in the Americas in the post-elimination era exhibited diverse genetic clusters within each genotype, distinct from the indigenous strains circulating in the pre-elimination era (Figure 1). The genotype 2B viruses from this period included 2B-L1 (n=7) from countries in the West Pacific Region; 2B-L2a (n=2) from South Africa and Pakistan; and 2B-L2c (n=23) from countries in the five other Regions (Table 2). Viruses of genotype 1G from this period were all imported from Africa and comprised 1G-L2a (n=1; from Kenya), 1G-L2b (n=1; from Tanzania), and 1G-L1b (n=5; from Nigeria). Except for one sequence, the sources of exposure for all genotype 1E viruses from the post-elimination era are well-documented, and the viruses belong to 1E-L2 (n=5; from countries in the Southeast Asian and West Pacific WHO Regions and 1E-L4 (n=2 from countries in the Eastern Mediterranean Region).
The phylogenetic analysis of viruses detected in the Americas during this period aligns with global RuV molecular epidemiology, as reported by the countries of exposure and/or by other countries where viruses from the same origin were detected (Table 2). In the United States, two viruses imported from China were identified, and their lineages were consistent with the documented RuV molecular epidemiology in China during the same periods (24). Similarly, viruses imported from Japan were found in the same phylogenetic lineages as those documented in the country during the same period (25). On the other hand, although genetic data on circulating RuV were not available from Indonesia, three importations from Indonesia were detected in the United States from 2011 to 2014. These viruses are in the same phylogenetic lineage as those described among travelers from Indonesia to Japan in 2017 (28). Virologic surveillance in the Americas also documented wild-type RuV that had not been reported from the countries of exposure. For example, 13 sequences were imported from countries (Afghanistan, Burundi, Nigeria, Tanzania, and Yemen) where no wild-type RuV genotypes are documented (15, 17, 29).
RuV sequences from diseases associated with persistent infection
In addition to sequences from routine rubella and CRS surveillance, 23 RuV sequences from persistent infections were obtained from patients presenting with chronic arthritis (n=1), Fuchs uveitis (n=3), and granulomas (n=19), and have been reported in Canada and the United States between 1985 and 2020 (Table 3). The phylogeny of these sequences reveals diverse genetic groups. The viruses from patients with Fuchs uveitis were genotypes 2C, 1G (1G-L0), and 1E (1E-L4). Among the sequences from patients with granulomas, in addition to the 17 vaccine-derived sequences (genotype 1a), one sequence was found to be genotype 2C and was closely related to the sequence from a virus found in a patient with Fuchs uveitis. Notably, one of the sequences from a patient with granulomas (MT249313) was not closely related to any of the recognized genotypes (Figure 2) (5). No rubella cases or persistent infection from contacts of these patients were reported.
TABLE 3. Rubella sequences from cases associated with persistent infections.
|
Sequence ID |
Virus name |
Genotype (lineage) |
Disease |
Source |
|---|---|---|---|---|
|
RVi/Vancouver.CAN/85 |
1a |
Chronic arthritis |
||
|
RVs/Scranton.PA.USA/19.13 |
2C |
Fuchs uveitis |
||
|
RVs/SanFrancisco.CA.USA/49.14 |
1G (L0) |
Fuchs uveitis |
Germany |
|
|
RVs/California.USA/43.16 |
1a |
Granuloma |
||
|
RVi/California.USA/43.16 |
1a |
Granuloma |
||
|
RVs/Louisiana.USA/27.17 |
1a |
Granuloma |
||
|
RVs/Louisiana.USA/27.17/2 |
1a |
Granuloma |
||
|
RVs/RhodeIsland.USA/9.17 |
1a |
Granuloma |
||
|
RVi/RhodeIsland.USA/9.17 |
1a |
Granuloma |
||
|
RVi/Louisiana.USA/27.17 |
1a |
Granuloma |
||
|
RVs/Alberta.CAN/4.17/GR |
1a |
Granuloma |
||
|
RVs/Maryland.USA/6.17 |
1E (L4) |
Fuchs uveitis |
Ethiopia |
|
|
RVi/RhodeIsland.USA/32.17 |
1a |
Granuloma |
||
|
RVs/Oregon.USA/05.18 |
1a |
Granuloma |
||
|
RVi/Oregon.USA/05.18 |
1a |
Granuloma |
||
|
RVs/Marlton.NJ.USA/33.18 |
1a |
Granuloma |
||
|
RVi/RhodeIsland.USA/7.18 |
1a |
Granuloma |
||
|
RVi/California.USA/12.18 |
1a |
Granuloma |
||
|
RVs/GreenBay.WI.USA/44.19 |
1a |
Granuloma |
||
|
RVs/Kenosha.WI.USA/41.19 |
2C |
Granuloma |
||
|
RVs/Greenfield.WI.USA/45.19 |
1a |
Granuloma |
||
|
RVs/Philadelphia.PA.USA/46.19 |
Unclassifieda |
Granuloma |
||
|
RVi/Oregon.USA/49.20 |
1a |
Granuloma |
The sequence presents 10% to 12% difference from any of the sequence in Clade 2 (5).
Source: Prepared by the authors using data from GenBank.
FIGURE 2. Phylogenetic tree of sequences associated with diseases by RuV persistent infection.
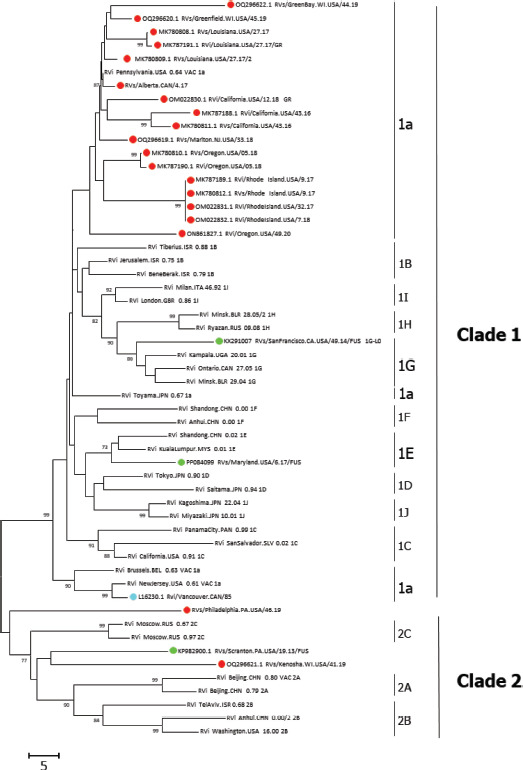
Note: The genetic relationship analysis between the sequences from persistent infection and 32 reference RuV sequences was based on the standard genotyping window and the bootstrap test (1 000 replicates) (23). Bootstrap support values >70% are shown at the nodes. Sequences are labeled according to associated diseases, including chronic arthritis (cyan), granuloma (red), or Fuchs uveitis syndrome (green). Distances were computed using the number of differences method.
Source: Prepared by the authors using data from GenBank.
DISCUSSION
Molecular epidemiology has proven invaluable in the rubella elimination program, offering essential evidence when combined with epidemiological data. It aids in identifying endemic strains, tracking importations, and documenting the interruption of endemic virus transmission before elimination. In the United States, virologic surveillance played a crucial role in supporting rubella elimination, complementing epidemiological evidence (27, 30). Analyzing viruses in the Americas by assigning phylogenetic lineages demonstrated the absence of sustained transmission of any indigenous strain in the post-elimination era, confirming the maintenance of elimination status (17, 30). Nevertheless, Regional MMR vaccination coverage has not achieved the expected goal of 95% in recent years (https://www.paho.org/en/immunization/immunization-data-and-statistics), and universal administration of the rubella-containing vaccine worldwide remains incomplete. Wild-type virus continues to circulate in many parts of the world, leaving susceptible children at risk (31).
Before elimination, eight of the 13 RuV genotypes were detected in the Americas. Shifts in circulating genotypes have been observed in the Region since the 1960s (27). Genotype 1a was known to be the predominant genotype in the United States before 1980 (27). Due to limited genetic information during this period, it is conceivable that genotype 1a was prevalent in the other parts of the Americas as well. From 2002 to 2009, large outbreaks by genotypes 1C (2002-2005) and 2B (2006-2008) occurred in multiple countries in the Region of the Americas (17). Besides these three genotypes, endemic transmission of viruses of genotypes 1B, 1E, and 1G, likely from importations, was also documented in Brazil, the United States, and Canada, respectively (17, 27).
From 2010 to 2023, only four genotypes were detected. Indigenous strains identified before 2009 were no longer found, and none of the importations led to large-scale outbreaks. Challenges persist, including asymptomatic infection, timely specimen collection, low viral load, and insufficient sequence information in some countries, complicating the tracking of sources of infections by genetic identity. Additionally, as elimination programs progress, the anticipated inactivity or extinction of recognized genotypes may reduce genetic diversity, leading to a loss of useful geographic associations (16). A method to subdivide genotypes globally has proven to be an invaluable tool for discerning genetic differences among viruses, particularly in monitoring RuV molecular epidemiology in China, Japan, and Uganda (24-26). Using this approach, viruses in the Americas appeared in diverse genetic groups, consistent with virologic surveillance reported by other countries. Additionally, the Region documented sequences from cases with recent travel (during the likely exposure period) to countries where genotype data have either not been available or have not been updated recently.
In addition to sequences from rubella or CRS cases in the Americas, a few sequences from cases of persistent RuV infection were reported in Canada and the United States. Seventeen of 23 sequences were closely related to RA27/3, the RuV vaccine strain, and were isolated from patients with primary immune deficiencies (4, 6). The phylogeny of one of the sequences from a foreign-born patient with persistent infection was consistent with the genetic characteristics of the virus documented in the patient’s home country (3). Interestingly, while there is no documented RuV virologic surveillance before 1961, the three sequences from older patients, each associated with different diseases related to rubella persistent infection, revealed that Clade 2 virus may have circulated in the Americas during the 1950s (2). Notably, genotype 2C, exclusively detected in Russia (13) was found in two of the patients, and one sequence was not closely associated with any recognized genotype (5). Although there is no evidence of transmission among close contacts of these cases, likely due to high immunization coverage in North America, the identification of these cases and the broad spectrum of diseases associated with rubella persistence highlight challenges in the consideration of global RuV eradication (18). The development of effective therapeutic antivirals for persistent RuV infections (32) and the creation of subunit vaccines using modern vaccinology are crucial steps to prevent the spread of the virus among unvaccinated/non-immune individuals.
Global RuV molecular epidemiology faces numerous challenges. Obtaining genetic information is hindered by the low shedding and high guanine and cytosine nucleotides content (approximately 70%) in the RNA genome of the virus. Despite this, many surveillance efforts overlook collecting specimens for genetic characterization, especially during outbreaks. PAHO recommends collecting serum specimens for every suspected measles or rubella case, along with at least one specimen for viral detection (33). Between 2010 and 2023, RuV sequences were obtained from 51 of 137 confirmed cases, with 45 sequences from North America in the past decade (https://www.paho.org/en/topics/rubella). Thus, there is an urgent need to enhance specimen collection and reporting of sequences regionally and globally. This is particularly vital in WHO Regions where the RuV vaccine has not been introduced in all countries and/or routine immunization has not yet reached 95% across all Member States (WHO Immunization Data Portal; https://immunizationdata.who.int/), leading to a high number of reported rubella cases without genetic information or with genetic data not captured in RubeNS2 and/or GenBank (16).
Conclusions
Virologic surveillance plays a crucial role in verifying the sustained maintenance of rubella elimination status in the Region of the Americas. Countries in the Americas have made a substantial contribution to global RuV molecular epidemiology by integrating genetic data with surveillance for imported cases, providing crucial information for countries with incomplete genetic data of wild-type RuV. The gaps in global RuV molecular epidemiology present challenges in tracing infection sources during suspected importations and/or chain of transmissions. As more countries commit to the goal of rubella and CRS elimination, improving RuV virologic surveillance will be essential to providing robust support for global rubella control and elimination efforts.
Disclaimer.
Authors hold sole responsibility for the views expressed in the manuscript, which may not necessarily reflect the opinion or policy of the RPSP/PAJPH and/or the Pan American Health Organization. In addition, the findings and conclusions in this report are those of the authors and do not necessarily represent the views of the United States Department of Health and Human Services.
Acknowledgements.
The authors are grateful for the support of the contributing laboratories in GenBank and the RubeNS2, as well as the members of the Global Measles and Rubella Laboratory Network in Argentina, Brazil, Chile, Colombia, Ecuador, El Salvador, Honduras, Mexico, Panama, and Peru. The authors specifically acknowledge Dr. Sergio Yebrail Gomez Rangel at the National Institute of Health in Colombia and the steering committee members and chairpersons of the Rubella Nucleotide Surveillance system for granting permissions to use the sequences.
Funding Statement
This article was supported by a grant and cooperative agreement (NU66GH002171) from the US Centers for Disease Control and Prevention.
Footnotes
Funding.
This article was supported by a grant and cooperative agreement (NU66GH002171) from the US Centers for Disease Control and Prevention.
REFERENCES
- 1.Winter AK, Moss WJ. Rubella. Lancet. 2022;399(10332):1336–46. doi: 10.1016/S0140-6736(21)02691-X. [DOI] [PubMed] [Google Scholar]; Winter AK, Moss WJ. Rubella. Lancet. 2022;399(10332):1336-46. [DOI] [PubMed]
- 2.Abernathy E, Peairs RR, Chen M-h, Icenogle J, Namdari H. Genomic characterization of a persistent rubella virus from a case of Fuch’ uveitis syndrome in a 73 year old man. J Clin Virol. 2015;69:104–9. doi: 10.1016/j.jcv.2015.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abernathy E, Peairs RR, Chen M-h, Icenogle J, Namdari H. Genomic characterization of a persistent rubella virus from a case of Fuch’ uveitis syndrome in a 73 year old man. J Clin Virol. 2015;69:104-9. [DOI] [PMC free article] [PubMed]
- 3.Doan T, Wilson MR, Crawford ED, Chow ED, Khan LM, Knopp KA, et al. Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens. Genome Med. 2016;8(1):90. doi: 10.1186/s13073-016-0344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Doan T, Wilson MR, Crawford ED, Chow ED, Khan LM, Knopp KA, et al. Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens. Genome Med. 2016;8(1):90. [DOI] [PMC free article] [PubMed]
- 4.Murguia-Favela L, Hiebert J, Haber RM. “Noninfectious” cutaneous granulomas in primary immunodeficiency patients and association with rubella virus vaccine strain. J Cutan Med Surg. 2019;23(3):341–2. doi: 10.1177/1203475419825780. [DOI] [PubMed] [Google Scholar]; Murguia-Favela L, Hiebert J, Haber RM. “Noninfectious” cutaneous granulomas in primary immunodeficiency patients and association with rubella virus vaccine strain. J Cutan Med Surg. 2019;23(3):341-2. [DOI] [PubMed]
- 5.Shields BE, Perelygina L, Samimi S, Haun P, Leung T, Abernathy E, et al. Granulomatous dermatitis associated with rubella virus infection in an adult with immunodeficiency. JAMA Dermatol. 2021;157(7):842–7. doi: 10.1001/jamadermatol.2021.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shields BE, Perelygina L, Samimi S, Haun P, Leung T, Abernathy E, et al. Granulomatous dermatitis associated with rubella virus infection in an adult with immunodeficiency. JAMA Dermatol. 2021;157(7):842-7. [DOI] [PMC free article] [PubMed]
- 6.Perelygina L, Chen MH, Suppiah S, Adebayo A, Abernathy E, Dorsey M, et al. Infectious vaccine-derived rubella viruses emerge, persist, and evolve in cutaneous granulomas of children with primary immunodeficiencies. PLoS Pathog. 2019;15(10):e1008080. doi: 10.1371/journal.ppat.1008080. [DOI] [PMC free article] [PubMed] [Google Scholar]; Perelygina L, Chen MH, Suppiah S, Adebayo A, Abernathy E, Dorsey M, et al. Infectious vaccine-derived rubella viruses emerge, persist, and evolve in cutaneous granulomas of children with primary immunodeficiencies. PLoS Pathog. 2019;15(10):e1008080. [DOI] [PMC free article] [PubMed]
- 7.Kanai M, Kamiya H, Okuno H, Sunagawa T, Tanaka-Taya K, Matsui T, et al. epidemiology of congenital rubella syndrome related to the 2012-2013 rubella epidemic in Japan. J Pediatric Infect Dis Soc. 2022;11(9):400–3. doi: 10.1093/jpids/piac043. [DOI] [PubMed] [Google Scholar]; Kanai M, Kamiya H, Okuno H, Sunagawa T, Tanaka-Taya K, Matsui T, et al. epidemiology of congenital rubella syndrome related to the 2012-2013 rubella epidemic in Japan. J Pediatric Infect Dis Soc. 2022;11(9):400-3. [DOI] [PubMed]
- 8.Pan American Health Organization. Americas Region is declared the world’s first to eliminate rubella 2015. [[Accessed 20 March 2024]]. Available from: https://www.paho.org/en/news/29-4-2015-americas-region-declared-worlds-first-eliminate-rubella.; Pan American Health Organization. Americas Region is declared the world’s first to eliminate rubella 2015. [Accessed 20 March 2024]. Available from: https://www.paho.org/en/news/29-4-2015-americas-region-declared-worlds-first-eliminate-rubella
- 9.Pan American Health Organization Regional Framework for the Monitoring and Re-Verification of Measles, Rubella, and Congenital Rubella Syndrome Elimination in the Americas. 2022. [[Accessed 20 March 2024]]. Available from: https://iris.paho.org/handle/10665.2/55074.; Pan American Health Organization. Regional Framework for the Monitoring and Re-Verification of Measles, Rubella, and Congenital Rubella Syndrome Elimination in the Americas. 2022. [Accessed 20 March 2024]. Available from: https://iris.paho.org/handle/10665.2/55074.
- 10.Pan American Health Organization [[Accessed 20 March 2024]];Immunization Newsletter. 2003 45(1) Available from: https://iris.paho.org/handle/10665.2/57594. [Google Scholar]; Pan American Health Organization. Immunization Newsletter. 2003;45(1). [Accessed 20 March 2024]. Available from: https://iris.paho.org/handle/10665.2/57594
- 11.World Health Organization Global vaccine action plan 2011-2020. [[Accessed 20 March 2024]]. Available from: https://www.who.int/teams/immunization-vaccines-and-biologicals/strategies/global-vaccine-action-plan.; World Health Organization. Global vaccine action plan 2011-2020 [Accessed 20 March 2024]. Available from: https://www.who.int/teams/immunization-vaccines-and-biologicals/strategies/global-vaccine-action-plan
- 12.World Health Organization Rubella virus nomenclature update: 2013. Wkly Epidemiol Rec. 2013;88(32):337–43. [PubMed] [Google Scholar]; World Health Organization. Rubella virus nomenclature update: 2013. Wkly Epidemiol Rec. 2013;88(32):337-43. [PubMed]
- 13.World Health Organization Wkly Epidemiol Rec. Standardization of the nomenclature for genetic characteristics of wild-type rubella viruses. 2005;80(14):126–32. [PubMed] [Google Scholar]; World Health Organization. Standardization of the nomenclature for genetic characteristics of wild-type rubella viruses. Wkly Epidemiol Rec. 2005;80(14):126-32. [PubMed]
- 14.Frey TK, Abernathy ES, Bosma TJ, Starkey WG, Corbett KM, Best JM, et al. molecular analysis of rubella virus epidemiology across three continents, North America, Europe, and Asia, 1961–1997. J Infect Dis. 1998;178(3):642–50. doi: 10.1086/515370. [DOI] [PubMed] [Google Scholar]; Frey TK, Abernathy ES, Bosma TJ, Starkey WG, Corbett KM, Best JM, et al. molecular analysis of rubella virus epidemiology across three continents, North America, Europe, and Asia, 1961–1997. J Infect Dis. 1998;178(3):642-50. [DOI] [PubMed]
- 15.Rivailler P, Abernathy E, Icenogle J. Genetic diversity of currently circulating rubella viruses: a need to define more precise viral groups. J Gen Virol. 2017;98(3):396–404. doi: 10.1099/jgv.0.000680. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rivailler P, Abernathy E, Icenogle J. Genetic diversity of currently circulating rubella viruses: a need to define more precise viral groups. J Gen Virol. 2017;98(3):396-404. [DOI] [PMC free article] [PubMed]
- 16.Brown KE, Rota PA, Goodson JL, Williams D, Abernathy E, Takeda M, Mulders MN. Genetic characterization of measles and rubella viruses detected through global measles and rubella elimination surveillance, 2016-2018. MMWR Morb Mortal Wkly Rep. 2019;68(26):587–91. doi: 10.15585/mmwr.mm6826a3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brown KE, Rota PA, Goodson JL, Williams D, Abernathy E, Takeda M, Mulders MN. Genetic characterization of measles and rubella viruses detected through global measles and rubella elimination surveillance, 2016-2018. MMWR Morb Mortal Wkly Rep. 2019;68(26):587-91. [DOI] [PMC free article] [PubMed]
- 17.Icenogle JP, Siqueira MM, Abernathy ES, Lemos XR, Fasce RA, Torres G, Reef SE. Virologic surveillance for wild-type rubella viruses in the Americas. J Infect Dis. 2011;204(Suppl 2):S647–51. doi: 10.1093/infdis/jir431. [DOI] [PubMed] [Google Scholar]; Icenogle JP, Siqueira MM, Abernathy ES, Lemos XR, Fasce RA, Torres G, Reef SE. Virologic surveillance for wild-type rubella viruses in the Americas. J Infect Dis. 2011;204(Suppl 2):S647-51. [DOI] [PubMed]
- 18.Reef SE, Icenogle JP, Plotkin SA. The path to eradication of rubella. Vaccine. 2023;41(50):7525–31. doi: 10.1016/j.vaccine.2023.11.014. [DOI] [PubMed] [Google Scholar]; Reef SE, Icenogle JP, Plotkin SA. The path to eradication of rubella. Vaccine. 2023;41(50):7525-31. [DOI] [PubMed]
- 19.US National Center for Immunization and Respiratory Diseases Updated 6 March 6 2020. Manual for the Surveillance of Vaccine-Preventable Diseases: Centers for Disease Control and Prevention, Department of Health and Human Services. [[Accessed 20 March 2024]]. Available from: https://www.cdc.gov/vaccines/pubs/surv-manual/chpt14-rubella.html.; US National Center for Immunization and Respiratory Diseases. Manual for the Surveillance of Vaccine-Preventable Diseases: Centers for Disease Control and Prevention, Department of Health and Human Services. Updated 6 March 6 2020. [Accessed 20 March 2024]. Available from: https://www.cdc.gov/vaccines/pubs/surv-manual/chpt14-rubella.html
- 20.Macey JF, Tam T, Lipskie T, Tipples G, Eisbrenner T. Rubella elimination, the Canadian experience. J Infect Dis. 2011;204(Suppl 2):S585–92. doi: 10.1093/infdis/jir406. [DOI] [PubMed] [Google Scholar]; Macey JF, Tam T, Lipskie T, Tipples G, Eisbrenner T. Rubella elimination, the Canadian experience. J Infect Dis. 2011;204(Suppl 2):S585-92. [DOI] [PubMed]
- 21.Saboui M, Hiebert J, Squires SG, Guay M, Barcellos P, Thom A, Li YA. Re-verifying the elimination of measles, rubella and congenital rubella syndrome in Canada, 2016-2020. Can Commun Dis Rep. 2021;47(11):476–8. doi: 10.14745/ccdr.v47i11a06. [DOI] [PMC free article] [PubMed] [Google Scholar]; Saboui M, Hiebert J, Squires SG, Guay M, Barcellos P, Thom A, Li YA. Re-verifying the elimination of measles, rubella and congenital rubella syndrome in Canada, 2016-2020. Can Commun Dis Rep. 2021;47(11):476-8. [DOI] [PMC free article] [PubMed]
- 22.Government of Canada . Elimination of Measles Rubella and Congenital Rubella Syndrome in Canada Documentation and Verification Report: Executive Summary. Ottawa: Government of Canada; [[Accessed 20 March 2024]]. Available from: https://www.canada.ca/en/public-health/services/immunization/vaccine-preventable-diseases/elimination-measles-rubella-congenital-rubella-syndrome-canada-documentation-verification-report.html. [Google Scholar]; Government of Canada. Elimination of Measles Rubella and Congenital Rubella Syndrome in Canada Documentation and Verification Report: Executive Summary. Ottawa: Government of Canada. [Accessed 20 March 2024]. Available from: https://www.canada.ca/en/public-health/services/immunization/vaccine-preventable-diseases/elimination-measles-rubella-congenital-rubella-syndrome-canada-documentation-verification-report.html
- 23.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870-4. [DOI] [PMC free article] [PubMed]
- 24.Zhu Z, Cui A, Zhang Y, Mao N, Liu Y, Liu L, et al. Transmission dynamics of the rubella virus circulating in china during 2010–2019: 2 lineage switches between genotypes 1e and 2b. Clin Infect Dis. 2021;73(7):1157–64. doi: 10.1093/cid/ciab339. [DOI] [PubMed] [Google Scholar]; Zhu Z, Cui A, Zhang Y, Mao N, Liu Y, Liu L, et al. Transmission dynamics of the rubella virus circulating in china during 2010–2019: 2 lineage switches between genotypes 1e and 2b. Clin Infect Dis. 2021;73(7):1157-64. [DOI] [PubMed]
- 25.Mori Y, Miyoshi M, Kikuchi M, Sekine M, Umezawa M, Saikusa M, et al. Molecular epidemiology of rubella virus strains detected around the time of the 2012-2013 epidemic in Japan. Front Microbiol. 2017;8:8. doi: 10.3389/fmicb.2017.01513. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mori Y, Miyoshi M, Kikuchi M, Sekine M, Umezawa M, Saikusa M, et al. Molecular epidemiology of rubella virus strains detected around the time of the 2012-2013 epidemic in Japan. Front Microbiol. 2017;8:1513. [DOI] [PMC free article] [PubMed]
- 26.Tushabe P, Bwogi J, Abernathy E, Birungi M, Eliku JP, Seguya R, et al. Descriptive epidemiology of rubella disease and associated virus strains in Uganda. J Med Virol. 2020;92(3):279–87. doi: 10.1002/jmv.25604. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tushabe P, Bwogi J, Abernathy E, Birungi M, Eliku JP, Seguya R, et al. Descriptive epidemiology of rubella disease and associated virus strains in Uganda. J Med Virol. 2020;92(3):279-87. [DOI] [PMC free article] [PubMed]
- 27.Icenogle JP, Frey TK, Abernathy E, Reef SE, Schnurr D, Stewart JA. Genetic analysis of rubella viruses found in the United States between 1966 and 2004: evidence that indigenous rubella viruses have been eliminated. Clin Infect Dis. 2006;43(suppl 3):S133–40. doi: 10.1086/505945. [DOI] [PubMed] [Google Scholar]; Icenogle JP, Frey TK, Abernathy E, Reef SE, Schnurr D, Stewart JA. Genetic analysis of rubella viruses found in the United States between 1966 and 2004: evidence that indigenous rubella viruses have been eliminated. Clin Infect Dis. 2006;43(suppl 3):S133-40. [DOI] [PubMed]
- 28.Kanbayashi D, Kurata T, Nishino Y, Orii F, Takii Y, Kinoshita M, et al. Rubella virus genotype 1e in travelers returning to Japan from Indonesia, 2017. Emerg Infect Dis. 2018;24(9):1763–5. doi: 10.3201/eid2409.180621. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kanbayashi D, Kurata T, Nishino Y, Orii F, Takii Y, Kinoshita M, et al. Rubella virus genotype 1e in travelers returning to Japan from Indonesia, 2017. Emerg Infect Dis. 2018;24(9):1763-5. [DOI] [PMC free article] [PubMed]
- 29.Abernathy ES, Hübschen JM, Muller CP, Jin L, Brown D, Komase K, et al. Status of global virologic surveillance for rubella viruses. J Infect Dis. 2011;204(suppl 1):S524–32. doi: 10.1093/infdis/jir099. [DOI] [PubMed] [Google Scholar]; Abernathy ES, Hübschen JM, Muller CP, Jin L, Brown D, Komase K, et al. Status of global virologic surveillance for rubella viruses. J Infect Dis. 2011;204(suppl 1):S524-32. [DOI] [PubMed]
- 30.Reef SE, Redd SB, Abernathy E, Kutty P, Icenogle JP. Evidence used to support the achievement and maintenance of elimination of rubella and congenital rubella syndrome in the United States. J Infect Dis. 2011;204(suppl 2):S593–7. doi: 10.1093/infdis/jir420. [DOI] [PubMed] [Google Scholar]; Reef SE, Redd SB, Abernathy E, Kutty P, Icenogle JP. Evidence used to support the achievement and maintenance of elimination of rubella and congenital rubella syndrome in the United States. J Infect Dis. 2011;204(suppl 2):S593-7. [DOI] [PubMed]
- 31.Zimmerman LA, Knapp JK, Antoni S, Grant GB, Reef SE. Progress toward rubella and congenital rubella syndrome control and elimination—worldwide, 2012-2020. MMWR Morb Mortal Wkly Rep. 2022;71(6):196–201. doi: 10.15585/mmwr.mm7106a2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zimmerman LA, Knapp JK, Antoni S, Grant GB, Reef SE. Progress toward rubella and congenital rubella syndrome control and elimination—worldwide, 2012-2020. MMWR Morb Mortal Wkly Rep. 2022;71(6):196-201. [DOI] [PMC free article] [PubMed]
- 32.Perelygina L, Buchbinder D, Dorsey MJ, Eloit M, Hauck F, Hautala T, et al. Outcomes for nitazoxanide treatment in a case series of patients with primary immunodeficiencies and rubella virus-associated granuloma. J Clin Immunol. 2019;39(1):112–7. doi: 10.1007/s10875-019-0589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Perelygina L, Buchbinder D, Dorsey MJ, Eloit M, Hauck F, Hautala T, et al. Outcomes for nitazoxanide treatment in a case series of patients with primary immunodeficiencies and rubella virus-associated granuloma. J Clin Immunol. 2019;39(1):112-7. [DOI] [PMC free article] [PubMed]
- 33.Pan American Health Organization . Guidance for testing of measles and rubella in the laboratory network of the Region of Americas. Washington DC: PAHO; 2019. [[Accessed 20 March 2024]]. Available from: https://iris.paho.org/bitstream/handle/10665.2/34932/9789275119976_eng.pdf. [Google Scholar]; Pan American Health Organization. Guidance for testing of measles and rubella in the laboratory network of the Region of Americas. Washington DC: PAHO; 2019. [Accessed 20 March 2024]. Available from: https://iris.paho.org/bitstream/handle/10665.2/34932/9789275119976_eng.pdf
- 34.Knapp JK, Mariano KM, Pastore R, Grabovac V, Takashima Y, Alexander JP Jr., et al. Progress toward rubella elimination–Western Pacific Region, 2000-2019. MMWR Morb Mortal Wkly Rep. 2020;69(24):744–50. doi: 10.15585/mmwr.mm6924a4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Knapp JK, Mariano KM, Pastore R, Grabovac V, Takashima Y, Alexander JP, Jr., et al. Progress toward rubella elimination–Western Pacific Region, 2000-2019. MMWR Morb Mortal Wkly Rep. 2020;69(24):744-50. [DOI] [PMC free article] [PubMed]
- 35.Vaidya SR, Kasibhatla SM, Kamble MB, Munivenkatappa A, Kumbhar NS, Jayaswamy MM, et al. Genetic and antigenic characterization of wild type rubella viruses isolated from India. Vaccine. 2021;39(6):876–81. doi: 10.1016/j.vaccine.2020.12.063. [DOI] [PubMed] [Google Scholar]; Vaidya SR, Kasibhatla SM, Kamble MB, Munivenkatappa A, Kumbhar NS, Jayaswamy MM, et al. Genetic and antigenic characterization of wild type rubella viruses isolated from India. Vaccine. 2021;39(6):876-81. [DOI] [PubMed]
- 36.Seppälä EM, López-Perea N, Torres de Mier MV, Echevarría JE, Fernández-García A, Masa-Calles J. Last cases of rubella and congenital rubella syndrome in Spain, 1997-2016: the success of a vaccination program. Vaccine. 2019;37(1):169–75. doi: 10.1016/j.vaccine.2018.11.017. [DOI] [PubMed] [Google Scholar]; Seppälä EM, López-Perea N, Torres de Mier MV, Echevarría JE, Fernández-García A, Masa-Calles J. Last cases of rubella and congenital rubella syndrome in Spain, 1997-2016: the success of a vaccination program. Vaccine. 2019;37(1):169-75. [DOI] [PubMed]


