Significance
Amino acids (AAs) constitute a major component in intracellular biomolecules. More than 25% of the total volume of a mammalian cell was reported to be made of AAs. Here, we study how AAs affect the intracellular liquid–liquid phase separation of proteins, which is relevant to the occurrence of neurodegeneration diseases. We find that AAs play an important role in modulating phase separation in test tubes and live cells. Importantly, we find that three specific AAs, including proline, glutamine, and glycine, significantly depress the formation of stress granules (SGs) in cell lines and primary cells, which can be explained by rendering net homotypic interactions more repulsive. These results deepen our understanding of the underappreciated roles of cellular AAs in modulating intracellular phase separation.
Keywords: liquid–liquid phase separation, protein–protein interactions, biomolecular condensate, stress granule, amino acid
Abstract
Liquid–liquid phase separation (LLPS) is an intracellular process widely used by cells for many key biological functions. It occurs in complex and crowded environments, where amino acids (AAs) are vital components. We have found that AAs render the net interaction between proteins more repulsive. Here, we find that some AAs efficiently suppress LLPS in test tubes (in vitro). We then screen all the proteinogenic AAs and find that three specific AAs, including proline, glutamine, and glycine, significantly suppressed the formation of stress granules (SGs) in U2OS and HeLa cell lines (in vivo) irrespective of stress types. We also observe the effect in primary fibroblast cells, a viable cell model for neurodegenerative disorders. Kinetic studies by live-cell microscopy show that the presence of AAs not only slows down the formation but also decreases the saturating number and prevents the coalescence of SGs. We finally use sedimentation-diffusion equilibrium analytical ultracentrifuge (SE-AUC) to demonstrate that the suppression effects of AAs on LLPS may be due to their modulation in protein–protein and RNA–RNA interactions. Overall, this study reveals an underappreciated role of cellular AAs, which may find biomedical applications, especially in treating SG-associated diseases.
Liquid–liquid phase separation (LLPS) of proteins and nucleic acids (1–4) is a versatile intracellular process, which leads to the formation of biomolecular condensates (BMCs). The occurrence of LLPS was shown to be a widely spread phenomenon in biology (5). The detailed LLPS process and the thus-forming BMCs were first revealed by Brangwynne et al. (6) by studying P granules in germ cells of Caenorhabditis elegans. Subsequently, the fundamental roles of LLPS and BMCs in cellular homeostasis and disease development have been extensively studied in the past decade (7, 8). To study the LLPS process and thus forming BMCs (9), phase-separation-prone proteins are normally purified and incubated in vitro. Jawerth et al. (10) used purified proteins from the FUS family and incubated them in a buffer to study the time-dependent rheological properties of BMCs after the LLPS. Wegmann et al. (11) used purified intrinsically disordered tau protein in vitro to study the LLPS process and the following aggregation process. It is known that in vitro LLPS are quite different from the ones that occur in living cells (in vivo) (4). There are many known intracellular biomolecules, such as phospholipids (12) and ATP (13), that have been shown to affect LLPS in vivo but are typically not present in in vitro experiments of LLPS.
Indeed, there are many small molecules in the cytosol of a cell. Among them, amino acids (AAs) were reported to constitute a major component in cellular biomolecules. More than 25% of the total volume and 6% of the total mass of a mammalian cell were reported to be made of free AAs (14, 15). The total AA concentration in the cytosol of a mammalian cell was also reported to reach around 50 mM in normal conditions (16, 17) and increase significantly to several hundreds of millimoles in response to stress (18, 19). It has been already found that AAs are employed as osmolytes (20) to stabilize protein conformation/folding (21–23) against protein denaturation and aggregation. Yancy et al. (24) reported that AAs such as proline and glutamic acid are employed by water-stressed organisms at an intermediately high concentration (hundreds of millimoles) to reduce the aggregation of proteins. Ignatova and Gierasch (19) also found that the bacteria Escherichia coli regulates the proline concentration to inhibit the initial aggregation process of cellular retinoic acid-binding proteins.
Recently, we have found that proteogenic AAs significantly affect protein–protein interaction by rendering their dispersions more stable (25). In other words, we have found that AAs, at a concentration range similar to the ones found in the cytosol of mammalian cells (tens to hundreds of millimoles), render proteins in vitro more repulsive by using different techniques, such as SE-AUC, Cross-Interaction Chromatography (CIC) and Surface Plasmon Resonance (SPR) (25–28). Here, we studied the effects of AAs in changing protein–protein interaction and modulating the LLPS in test tubes (in vitro) and in cells (in vivo). For the in vitro studies, BSA was chosen as a model folded protein and the low-complexity domain of FUS (FUS267) as a model intrinsically disordered protein (IDP). FUS plays a significant role in various cellular processes (29) as the mutation of FUS is associated with several diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (30, 31). FUS was also reported to be recruited to stress granules (SGs) (32) in response to stress. The protein–protein interaction and LLPS of both BSA and FUS267 were studied in vitro with the presence of proline. We found that the presence of proline stabilized both protein solutions (33), thus suppressing their phase separation in vitro. We further found that proline and various other proteogenic AAs suppressed the formation of SGs in U2OS and HeLa cell lines and the effect is irrespective of stress types (i.e., heat shock or arsenite stress). The effect of AAs on suppressing the formation of SGs was also observed in primary fibroblast cells, a viable cell model for neurodegenerative disorder diseases. The kinetic study by live-cell microscopy experiments showed that the presence of AAs not only slowed down the formation of SGs but also decreased the saturating number of SGs and prevented the coalescence of SGs. These modulation effects of AAs on SGs can be explained by weaker net attractive interaction between the components of SGs (including scaffold protein, IDP, and RNA) after the addition of AAs. We believe that our study reveals the underappreciated role of cellular AAs on intracellular phase separation.
Results
In Vitro BSA and FUS267 System.
We first studied the effect of proline on bovine serum albumin (BSA) (a folded protein). Sedimentation-diffusion equilibrium analytical ultracentrifuge (SE-AUC) (34) was used to measure the second virial coefficient (B22) (35), which quantitatively characterized protein–protein interaction (33). The measurement principle is based on the equation of state (Eq. 1):
| [1] |
where ∏ is the osmotic pressure, ρ is the number density, and kT is the product of the Boltzmann constant by the temperature. B22 and B23 are both second virial coefficients, where the notation 1 is assigned to the solvent, 2 is assigned to solute 2 (proteins), and 3 to solute 3 (AAs). In this case, B22 indicates protein–protein interaction. A positive change (ΔB22 > 0) indicates that the protein solution becomes more stable or more rigorously that the difference between interprotein repulsion and attraction grows in favor of the former while ΔB22 < 0 indicates that the solution becomes less stable because it implies that the net interaction between proteins are more attractive in solution or more rigorously that the difference between interprotein repulsion and attraction grows in favor of the latter. The procedure to obtain B22 is illustrated in Fig. 1A. Linear fitting of the curve of versus is performed instead of directly fitting the curve Π versus to avoid noisy data due to low protein concentrations (27), SI Appendix, Fig. S1A. As shown in Fig. 1B, we find that the larger the proline concentration, the larger ΔB22. When no proline is present in the solution, B22 for BSA–BSA interaction is , corresponding to a BSA diameter of ~8.3 nm if effective hard-sphere interaction potential is assumed (36, 37). This value is in good agreement with the hydrodynamic diameter of BSA (7.8 nm) in water (38). With the addition of 1 M proline, the value of B22 is increased by up to 28% (from to ) and the effective hard sphere diameter increased by 8% (from 8.3 nm to 9.0 nm), which indicates a more stable BSA solution. The value of B23, which indicates BSA–proline interaction, is however constant, SI Appendix, Fig. S1B. Following literature procedures (39) (experimental details in Materials and Methods), polyethylene glycol (PEG 4,000 Da) was added into the BSA solution as a crowding agent to induce the phase separation of BSA. The system spontaneously separates into two phases, a BSA-rich droplet phase and a BSA-poor dilute phase. The two phases were separated by centrifugation, Fig. 1C. UV-vis spectroscopy was used to measure the BSA concentration in the BSA dilute phase. As shown in Fig. 1D, we found that the presence of proline in the BSA solution increased the BSA concentration in the dilute phase. We also analyzed the BSA droplet phase by widefield optical microscopy (experimental details in Materials and Methods, 60 mins of incubation time before microscopy experiments to allow for sufficient droplet sedimentation on the sample well bottom) as a function of proline concentration. We also found that the BSA concentration, as well as the partition coefficient of BSA in the droplet phase, decreased with higher proline concentration by using fluorescence-labeled BSA proteins (SI Appendix, Fig. S2 A and B). The recruitment of poly-lysine (K10) was also weakened by the addition of proline (SI Appendix, Fig. S2C). The effect of proline can be however counteracted by a higher PEG concentration (SI Appendix, Fig. S2 D and E). The increase in BSA concentration in the dilute phase as well as the decline in BSA concentration in the BSA-rich droplets upon the addition of proline both indicate a more stable BSA solution.
Fig. 1.
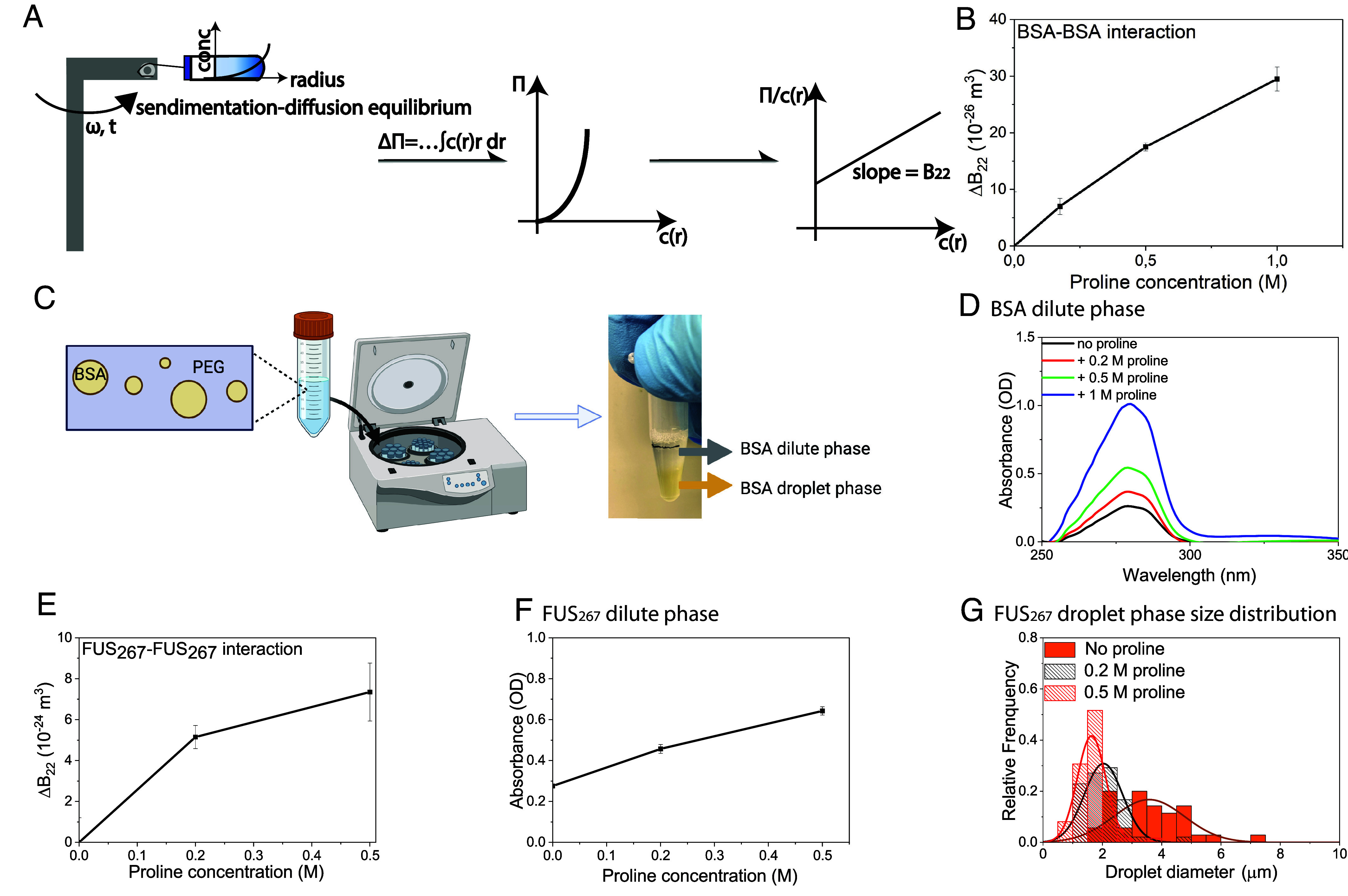
The effect of proline in BSA–BSA interaction and phase separation, and FUS267–FUS267 interaction and phase separation. (A) Schematic representation of the procedures for the calculation of B22 by SE-AUC. (B) The change of B22 (ΔB22) for BSA–BSA interaction with different proline concentrations in BSA solution (33) (C) Schematic representation of the procedures for the separation of the BSA dilute and droplet phases by centrifugation after the spontaneous phase separation of BSA with 4,000 Da polyethylene glycol (PEG) as a crowding agent. (D) UV-vis spectroscopy of the BSA dilute phase with different proline concentrations. (E) Change of B22 (ΔB22) for FUS267–FUS267 interaction with different proline concentrations (0.2 and 0.5 M) in FUS267 solution. (F) Absorbance at 275 nm of the FUS267 dilute phase as a function of proline concentrations (0, 0.2, and 0.5 M). (G) FUS267 droplet size distribution as a function of proline concentrations (0, 0.2, and 0.5 M).
We also studied the effect of proline on LLPS of the low-complexity domain of fused in sarcoma [FUS267, an intrinsically disordered protein (40). As shown in Fig. 1E, ΔB22 for FUS267–FUS267 interaction increased with higher proline concentrations (from 0 to 0.2 and 0.5 M), indicating a more stable FUS267 solution after the addition of proline. The LLPS of FUS267 was induced according to the literature procedure (40) (experimental details in Materials and Methods). As shown in Fig. 1F, the FUS267 concentration in the dilute phase increased with higher proline concentrations, as measured via UV-VIS spectroscopy. By using widefield optical microscopy, we also observed that the smaller the size of the FUS267 droplets is, the higher the proline concentration (Fig. 1G, the related widefield optical microscopy images shown in SI Appendix, Fig. S2F).
In Vivo U2OS and HeLa Cell Lines.
After the in vitro experiments with BSA and FUS267, we studied the effect of proline on the intracellular LLPS in vivo by employing SGs (41, 42), a well-known example of BMCs that form via the phase separation of a diverse set of proteins and RNAs. In this study, the SGs in U2OS cells were induced in response to oxidative stress by the addition of sodium arsenite (NaAs). It is noteworthy that SGs contain a variety of client proteins, including FUS (43). In the positive control row of Fig. 2A, we show fluorescence microscopy images of fixed U2OS cells that developed SGs after the oxidative stress for 45 mins (indicated by the arrowheads in the alexa fluor channel). We pretreated the cells with 200 mM proline for 2 h before the stress. After the pretreatment, the intracellular proline concentration increased to ~130 mM, as estimated by the LC-MS, SI Appendix, Fig. S3 (the details of intracellular amino acid extraction protocol and LC-MS quantification analysis are described in SI Appendix, SI1). We believe that the significant proline concentration increase in the cytosol is due to facilitated diffusion (44) of AAs, a process of spontaneous passive transport of AAs across the cell membrane via specific transmembrane integral protein transporters (45). Cell viability after the pretreatment with proline was also measured by the cell proliferation assay (MTT assay), as described in SI Appendix, SI2. It was found that the viability of the cells did not change after the uptake of proline (SI Appendix, Fig. S4). NaAs was then added to the cells and incubated for ~45 mins at room temperature to induce the formation of SGs (the details of cell culture, cell stress, and fixing procedures described in Materials and Methods). As shown in Fig. 2A, we observed that the number of SGs in U2OS cells decreased with the pretreatment of 200 mM proline compared to the positive control condition (the treatment of only 200 mM proline without the later addition of NaAs did not change the distribution of G3BP in the cytosol, SI Appendix, Fig. S5). We further quantified the number change of SGs per cell after the proline pretreatment (detailed analysis method and script described in Materials and Methods). As shown in Fig. 2B, we found that the pretreatment of proline at concentrations larger than 200 mM leads to a statistically significant decrease in the average number of SGs per cell (P < 0.0001). We then screened all the other neutral proteinogenic AAs (with a solubility higher than 100 mM) for their effects on SGs and we found that the pretreatment with glutamine (200 mM) and glycine (400 mM) also decreased the SG number significantly (P < 0.0001), Fig. 2C. The pretreatment with serine, valine, and alanine at 400 mM did not lead to any significant change in the average number of SGs per cell but led to a significant decrease in the average size of SGs, SI Appendix, Fig. S6. Charged proteinogenic AAs were also screened. As shown in SI Appendix, Fig. S7, negatively charged aspartic acids (20 mM) decreased both the number of SGs (P < 0.01) and the size of SGs (P < 0.1) while histidine (100 mM) decreased the size of SGs (P < 0.01). We also observed that the presence of AAs prevents the formation of SGs in response to heat shock stress, Fig. 2A. As shown in Fig. 2D, proline, glutamine, and glycine all significantly decreased the number of SGs per cell after the heat shock treatment at 43 °C, 5% CO2 for 1 h. The effect of proline on the average number of SGs per cell was also tested in HeLa cells and the same phenomenon was observed (Fig. 2E). Proline, glutamine, and glycine all significantly decreased the average number of SGs per cell in HeLa cells (P < 0.0001), Fig. 2 F and G. Apart from proline (Fig. 2B), the effect of glutamine and glycine on the average number of SGs per cell was also found to be concentration dependent (SI Appendix, Fig. S8).
Fig. 2.
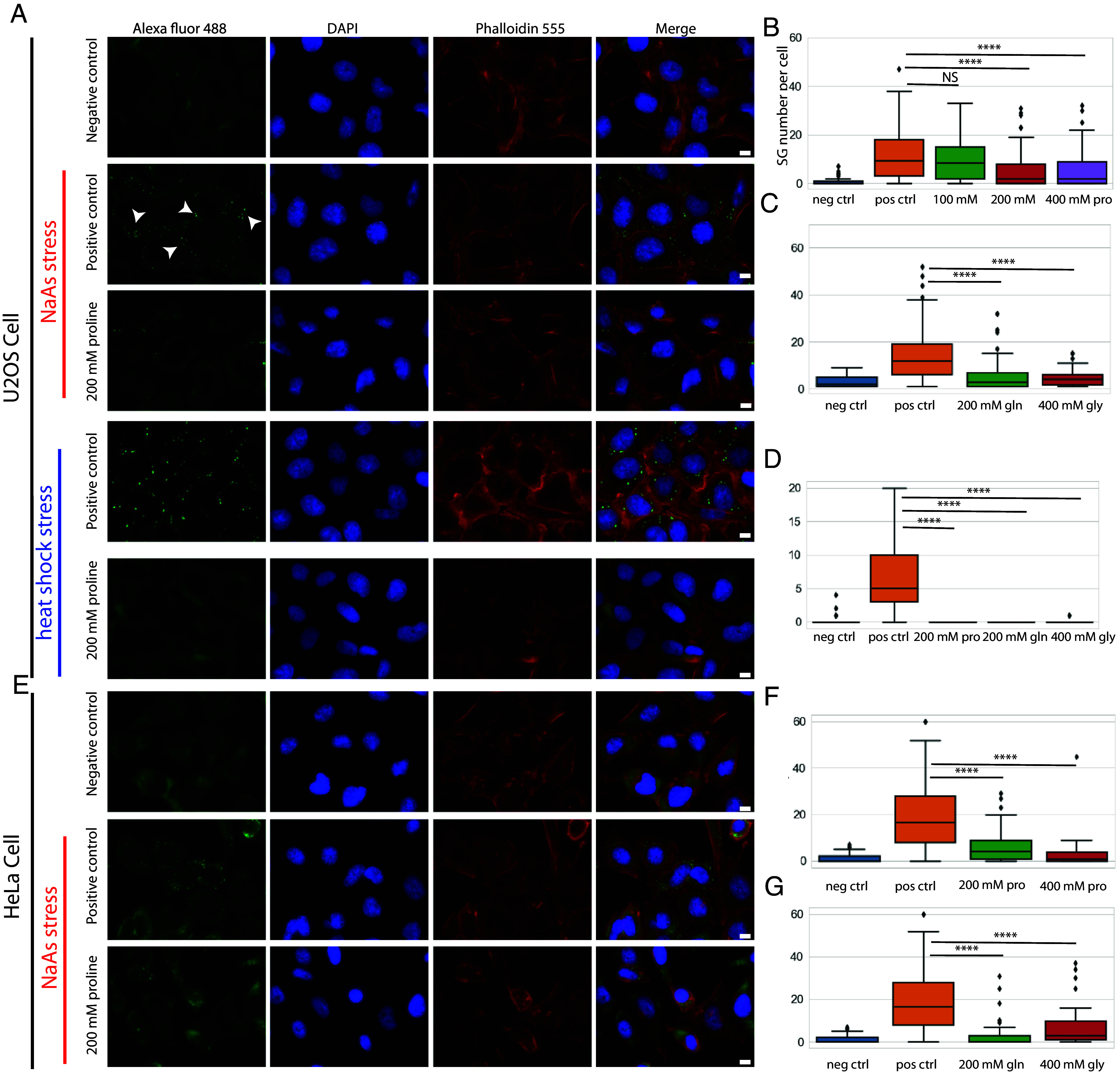
Immunofluorescence microscopy showing the effect of proline and other neutral amino acids (AAs) on stress granules (SGs) in U2OS and HeLa cells. (A) U2OS cells exhibited a significantly reduced number of SGs following the pretreatment with 200 mM proline for 2 h compared to positive control where no exogenous AAs were added in response to NaAs and heat shock stress. (B) The number of SGs per cell after the pretreatment with 100, 200, and 400 mM proline in response to NaAs stress. (C) The number of SGs per cell after the pretreatment with other neutral proteinogenic AAs including glutamine (200 mM) and glycine (400 mM) in response to NaAs stress. (D) The number of SGs per cell after the pretreatment with 200 mM proline, 200 mM glutamine, and 400 mM glycine in response to heat shock stress. (E) HeLa cells exhibited a significantly reduced number of SGs following the pretreatment with 200 mM proline for 2 h compared to positive control where no exogenous AAs were added in response to NaAs stress. (F) The number of SGs per cell after the pretreatment with 200 and 400 mM proline in response to NaAs stress. (G) The number of SGs per cell after the pretreatment with glutamine (200 mM) and glycine (400 mM) in response to NaAs stress. (****P value < 0.0001; NS: no significance, P value > 0.05). Arrowheads: SGs. (Scale bar, 10 μm.) Positive control means that stressing the cells without the pretreatment with AAs and negative control means that pretreatment with AAs without stressing the cells.
In Vivo Primary Fibroblast Cells.
To address the relevance of our findings of the effects of AAs on SGs to disease treatment, we performed experiments using primary human dermal fibroblast cells, a viable cell model for studying neurodegenerative disorders associated with SGs (46, 47). Many SGs formed in the cytosol of the primary fibroblast cells (indicated by arrowheads) after the incubation with NaAs (1 mM) for 2 h at RT (positive control, Fig. 3B), compared to no SG formation in the negative control (Fig. 3A). With the pretreatment of proline (200 mM), glutamine (200 mM), and glycine (400 mM) before the NaAs stress, the formation of SGs was significantly suppressed (Fig. 3 C, D, and E). The average number of SGs per cell in fibroblast cells was significantly decreased after the pretreatment with these AAs (P < 0.0001), Fig. 3F.
Fig. 3.
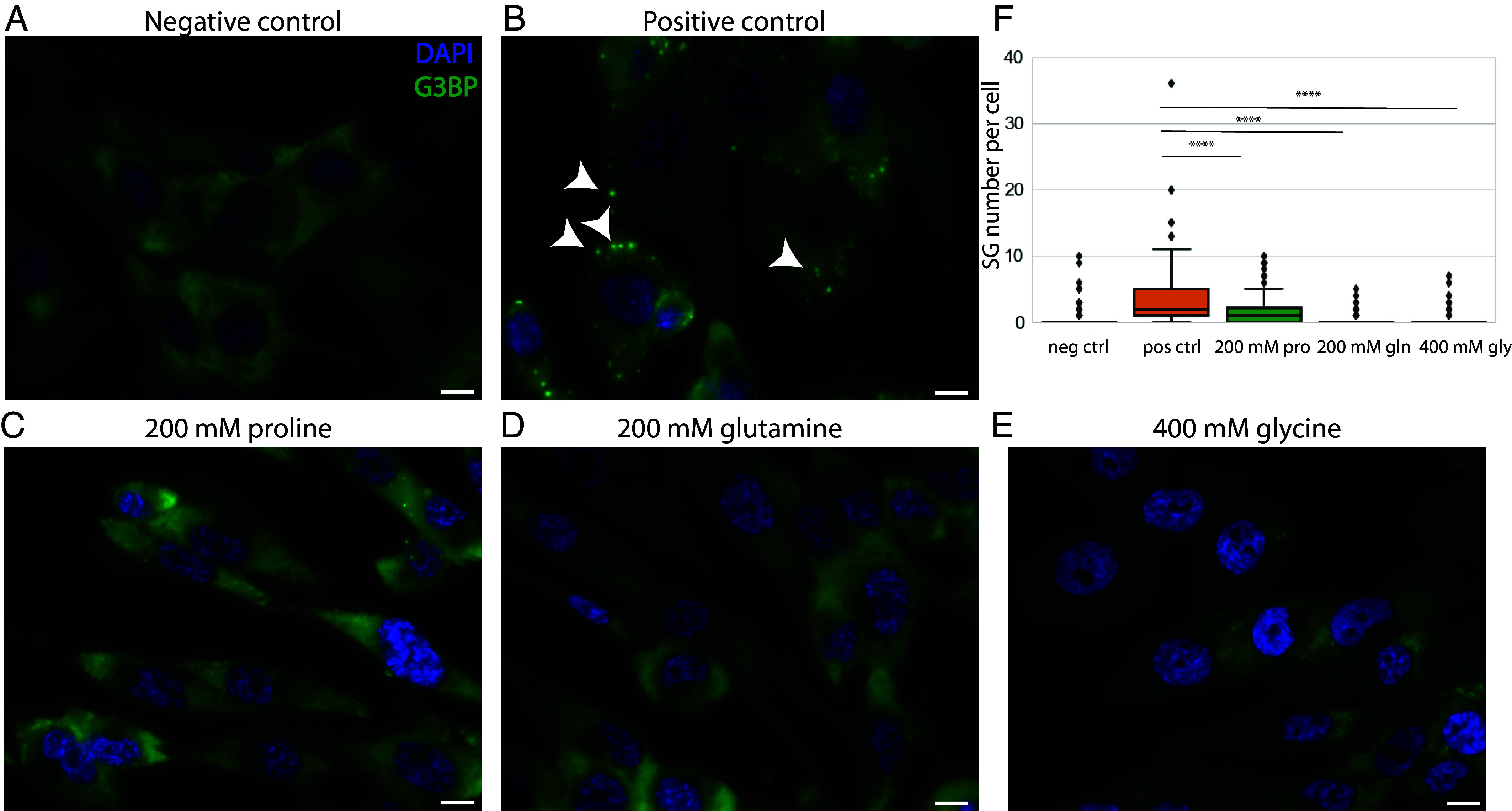
Immunofluorescence microscopy showing the effect of proline and other neutral AAs on SGs in primary fibroblast cells. (A and B) fibroblast cells exhibited a significant number of SGs following the incubation with NaAs (1 mM) for 2 h at RT; (C–E) primary fibroblast cells exhibited a significantly reduced number of SGs following the pretreatment with 200 mM proline (C), 200 mM glutamine (D), 400 mM glycine (E) for 2 h (F). The number of SGs per cell after the pretreatment with proline (200 mM), glutamine (200 mM), and glycine (400 mM) (****P value < 0.0001). (Scale bar, 10 μm.) Arrowheads: SGs.
Formation Kinetics of SGs.
To reveal the effect of AAs on the formation process kinetics of SGs, we performed live-cell microscopy of U2OS cells constitutively expressing GFP-tagged G3BP1, a marker protein of SGs (48, 49) with the presence of proline and glutamine. As shown in the control set of Fig. 4A (qualitatively) and 4B (quantitatively), SGs started to form at 15 mins and saturate at 25 mins, after which they started to coalesce, leading to a decrease in the number of SGs (indicated as lag, growth, and coalescence phases in Fig. 4B). The coalescence events are indicated inside the red squares in Fig. 4A. In contrast, the proline pretreatment for 2 h led to a decrease in the SG formation rate. In addition, the saturating number of SGs per cell (maximum number of SGs before the coalescence dominates) decreased by more than half (from 20 to 8 SGs per cell) and the coalescence event became trivial as no significant decrease in the number of SGs was observed. When the proline was added at the same time as NaAs, we also observed a lower rate of SG formation, decreased saturating number of SGs, and diminished coalescence of SGs. However, the effect was less pronounced when compared to the 2 h pretreatment with proline. This may be because the diffusion process of proline into the cells took time to reach equilibrium. When we posttreated the cells with proline 15 mins after the addition of NaAs, we observed an immediate suppression of the SG formation rate. The SG number saturation was reached at 40 mins, which is 15 mins later than the control set. Then, 200 mM glutamine also showed similar effects, as shown in Fig. 4C (related live cell images in SI Appendix, Fig. S9). It is noteworthy that the pretreatment with 200 mM glutamine also delayed the formation of SGs. This delay effect however was not found in the cotreatment and posttreatment experiments.
Fig. 4.
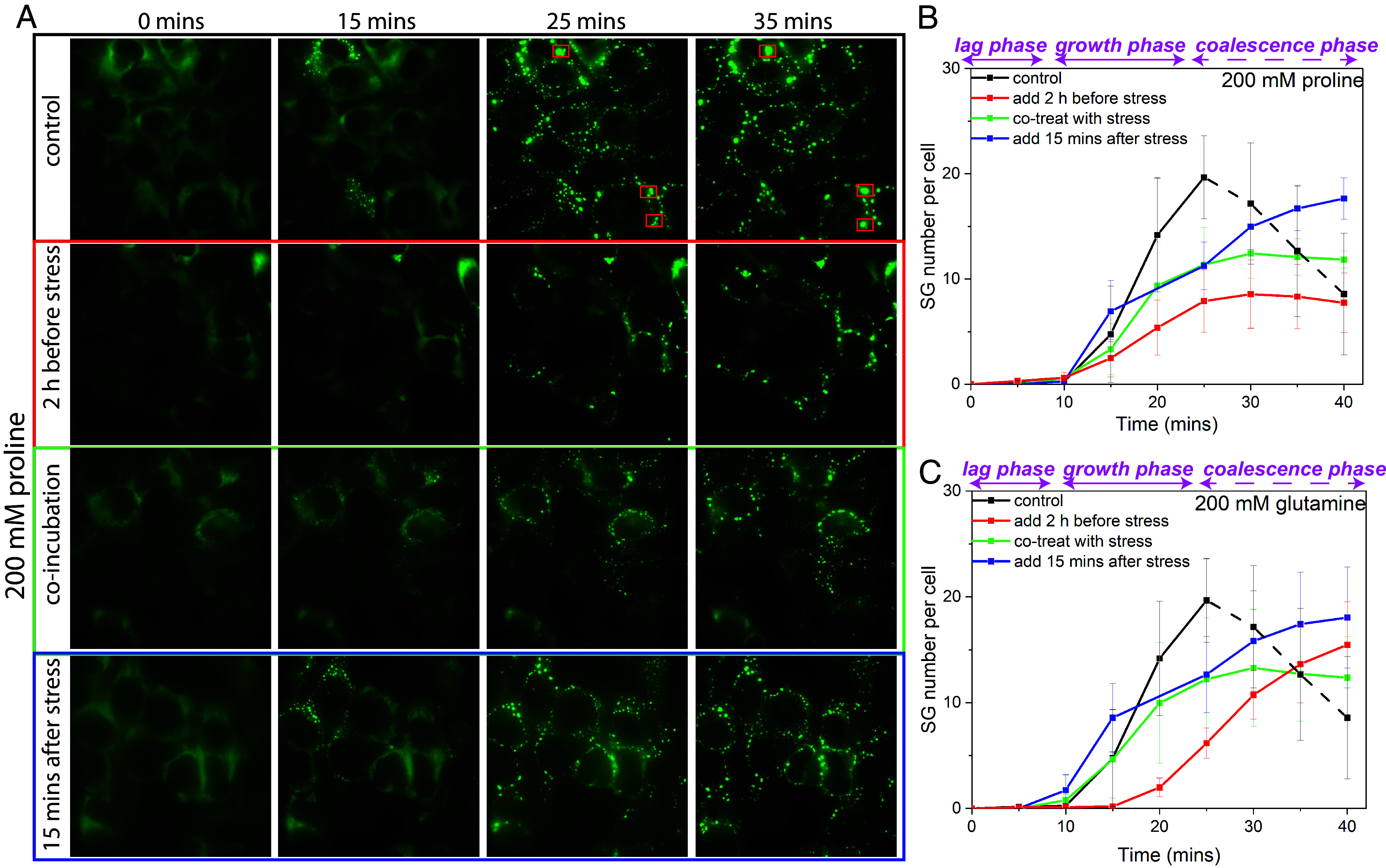
Live cell microscopy of GFP-tagged G3BP1 showing the effect of proline and glutamine (200 mM) on the formation process of SGs in U2OS cells. (A) The pretreatment for 2 h, the cotreatment, and the posttreatment (15 mins after the addition of NaAs) with 200 mM proline all suppressed the formation of SGs compared to positive control (no exogenous AAs were added) by live cell microscopy of U2OS cells with GFP tagged G3BP1 (the coalescence events indicated in the red squares). (B) Time series of the number of SGs per cell after the addition of NaAs including positive control (no proline treatment) and 200 mM proline pre-/co-/posttreatment (indicated as lag, growth and coalescence phases). (C) Time series of the number of SGs per cell after the addition of NaAs including positive control (no glutamine treatment) and 200 mM glutamine pre-/co-/posttreatment (related live cell images in SI Appendix, Fig. S9). (Scale bar, 10 μm.)
Interactions for the Formation of SGs.
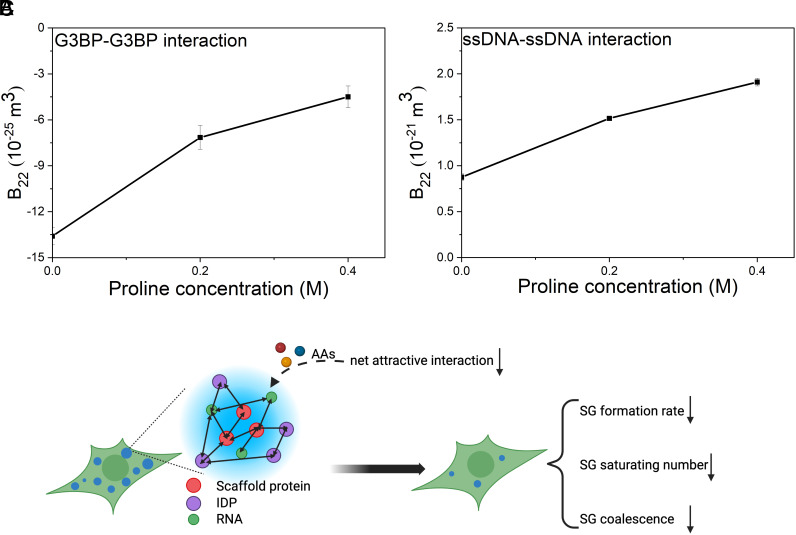
In light of the finding that proline stabilizes the protein–protein interactions in solution for the low-complexity domain of FUS which is commonly recruited in SGs (50) (Fig. 1E), we hypothesize that AAs suppress the formation of SGs because the presence of AAs renders the net interaction for SG components more repulsive. To prove that, we studied the effects of proline on G3BP–G3BP interaction [key scaffold protein for the formation of SGs (51)] as well as ssDNA–ssDNA interaction (nucleic acid to model a large variety of RNA recruited in SGs). As shown in Fig. 5A, B22 for G3BP–G3BP interaction in solution (25 mM Tris-HCl buffer at pH 7.6) increased with higher proline concentrations (from 0 to 0.2 and 0.4 M), indicating a less net G3BP attractive interaction after the addition of proline. As shown in Fig. 5B, B22 for ssDNA–ssDNA interaction in solution (1× TE buffer at pH 7.0) also increased with higher proline concentrations (from 0 to 0.2 and 0.4 M), indicating a more net ssDNA repulsive interaction after the addition of proline.
Fig. 5.
The effect of proline in G3BP–G3BP interaction (A) and ssDNA–ssDNA interaction (B) in solution and the proposed mechanism for the modulation of AAs on the formation of SGs (C). The SGs are formed in the cytosol as a result of complex interactions among an assembly of proteins (including scaffold proteins which are crucial for the formation of SGs and a large variety of IDP) as well as many RNA in response to stress. The presence of AAs is proposed to render net attractive forces between these SG components weaker and thus modulate the formation of SGs. The modulation effect is composed of three aspects: i) lower SG formation rate; ii) smaller SG saturating number; and iii) less SG coalescence.
Discussion and Conclusions.
We find that proline stabilizes the interaction for both BSA (a folded protein) and FUS267 (an intrinsically disordered protein) in solution. This is established by our findings that B22 in both cases becomes larger with a higher proline concentration. This is consistent with our recent finding that AAs have a general role in stabilizing protein dispersions (25) (the only exception that we found so far is α-synuclein, SI Appendix, Fig. S10). The increase in B22 directly implies an increase in the osmotic pressure in the solution, which in turn implies a decrease in the protein chemical potential. An obvious consequence of this latter fact is the proteins’ tendency to phase separate changes. Indeed, we have shown here that the addition of proline diminished the LLPS of both BSA and FUS267 in vitro. The data presented here show a clear effect at a concentration of 200 mM, which is approximately four times higher than the typical intracellular amino acid concentration in normal conditions (~50 mM) but at the same concentration range (several hundreds of mM) that is found when osmotic stress is applied to cells (24).
Because of these findings, we ventured into investigating the effect of intracellular AAs on the formation of SGs in living cells (in vivo). We found that a variety of neutral AAs at concentrations of hundreds of millimolar can either reduce the average number (proline, glutamine, and glycine) or average size (serine, valine, and alanine) of SGs in two cell lines (i.e., U2OS and HeLa cells). We also found the modulation effect of AAs on SGs of primary fibroblast cells. The cell type represents a viable cell model for several neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and ALS. Furthermore, live cell microscopy study of proline and glutamine revealed the multiple effects of these AAs on the SG formation process: They i) reduce the SG formation rate; ii) decrease the SG saturating number; and iii) prevent SG coalescence. These modulation effects of AAs on SGs can be explained by the lower net attractive force between SG components upon the addition of AAs. We corroborated this hypothesis by studying the effects of proline on G3BP–G3BP interaction (key scaffold protein for the formation of SGs) as well as ssDNA–ssDNA interaction (nucleic acid to model a large variety of RNA recruited in SGs) (Fig. 5C). A further proteomics study by using proximity labelling (52) will provide a full composition change of SGs after the treatment of AAs. Given that the cell is also rich in peptides, we will further study whether they can also modulate the LLPS in vivo in the next step of our research, which could be connected with the metabolic cycle of cells. We will also study why specific AAs reduce the number of SGs while other AAs only reduce the size of SGs. The problem is complex as many effects could lead to this difference. We currently employ the combination of NMR and LC-MS to study the binding position and binding affinity of different AAs as well as the partitioning of different AAs in protein condensates. Overall, we believe that this improved understanding of AAs’ effects in tuning intracellular protein phase separation may provide new knowledge of intracellular complex biophysics (4, 7) and provide a new strategy for the treatment of neurodegenerative disorders associated with SGs.
Materials and Methods
Materials.
Bovine serum albumin (BSA) (A7638), polyethylene glycol (PEG) 4 kDa (A16151), and all the AAs that were used in this study were purchased from Sigma-Aldrich. BSA, Rhodamine labeled was purchased from Nanocs. Recombinant Human G3BP protein was purchased from Abcam (ab103304). FUS low complexity domain (LCD) (residues 1 to 267) was kindly provided by Dufresne Lab (40). U2OS human osteosarcoma cells (wild type) were kindly provided by Dufresne Lab (48). U2OS cells stably expressing GFP-tagged G3BP1 were kindly provided by Pelkmans Lab(49). HeLa cells (CCL-2™) and Primary Dermal Fibroblast Normal; Human, Neonatal (PCS-201-010™) were purchased from ATCC. ssDNA (p4844) was purchased from Tilibit Nanosystems GmbH. Both mouse anti-G3BP (ab56574) and Alexa fluor 488 anti-mouse IgG (ab150113) were purchased from Abcam.
SE-AUC Experiments.
Using SE-AUC experiments for BSA–BSA interaction as an example, a BSA solution (20 mg/ml) in phosphate buffer (pH 7, 50 mM) was mixed with proline at a series of concentrations (0, 0.2, 0.5, and 1 M). The final solutions (60 µl) were added into the AUC cells (3 mm pathlength). The sedimentation–diffusion equilibria at a proper angular velocity (44,000 rpm), 20 °C were reached for these samples after 24 h. The protein concentration gradient was obtained and converted into the equation of state (EOS) curve (Eq. 1) using the well-known relation (35, 53) (Eq. 2), where c is the mass concentration of the solute species (kg/m3), µ is the chemical potential (J/kg), ω is the angular velocity (rad/s), d is the density (kg/m3), ri is the arbitrary radial position (m), rm is the meniscus radial position (m), and r is the radial position (m), respectively.
| [2] |
The final step is to calculate , as shown in Eq. 3 (rearranged from Eq. 1). The slope of the curve ( vs ) equals the value of B22.
| [3] |
The change of B22 (ΔB22) with the proline concentration series is thus calculated.
Droplet Formation In Vitro.
BSA droplet.
The method for BSA droplet formation is adjusted from the work by Testa et al. (39). Briefly, 100 µL MilliQ water (with different concentrations of proline), 125 µL of 4× potassium phosphate buffer (400 mM potassium phosphate and 800 mM KCl, pH 7), and 75 µL of 20% (w/v) BSA were mixed in a 1.5 mL Eppendorf tube. Then, 200 µL of 60% (w/v) PEG 4 kDa was added to make BSA droplet suspension. The droplet suspension was further stabilized for 60 mins (to allow for sufficient droplet sedimentation to the sample well bottom) and the imaging was done quickly on the bottom of the sample well by Olympus Cell xCellence microscope, using ×60 oil objective with a numerical aperture (NA) of 1.35 and a Hamamatsu ORCA 03 g camera. The BSA dilute phase was further separated for UV-vis spectroscopy after centrifugation at 10,000 g for 10 min.
FUS Droplet.
The method for FUS droplet formation is adjusted from the work by Ijavi et al. (40). Briefly, the 2 mM FUS stock (more specifically, FUS low complexity domain (LCD) (residues 1 to 267) with 6 M urea in the buffer (50 mM HEPES and 150 mM NaCl, pH 7) was diluted 200 times with the buffer (with different concentrations of proline) to induce the phase separation. The droplet suspension was further stabilized for 60 mins and the imaging was done quickly on the bottom of the sample well by Olympus Cell xCellence microscope, using ×60 oil objective with a numerical aperture (NA) of 1.35 and a Hamamatsu ORCA 03 g camera. The FUS concentration in the dilute phase was measured by measuring the absorbance at 275 nm in an analytical ultracentrifuge (at 9,000 rpm). The droplet size distribution was measured by a simple MATLAB script. In the script, the diameter of a droplet is measured by clicking 2 points on the droplet sphere.
Cell Culture.
U2OS human osteosarcoma cells, HeLa cells (CCL-2™), and U2OS cells stably expressing GFP-tagged G3BP1 were grown in Dulbecco’s modified eagle medium (high glucose, GlutaMAX™ supplement), supplemented with 5% or 10% fetal bovine serum at 37 °C in 5% CO2. Primary Dermal Fibroblast cells were grown in Fibroblast Growth Medium (C-23110 from PromoCell) at 37 °C in 5% CO2.
Immunofluorescence Microscopy.
After the treatment with 1 mM sodium arsenite (Sigma-Aldrich) for ~45 mins at RT (arsenite stress) or at 43 °C, 5% CO2 for 1 h (heat shock stress) to induce stress granule formation, wild-type U2OS cells cultured in µ-Slide 18 Well Glass Bottom (ibid, 81817) were fixed with 4% formaldehyde for 15 min, permeabilized with 0.25% Triton X-100 in phosphate-buffered saline (PBS) for 15 mins and blocked with 20 mg/ml bovine serum albumin (Sigma-Aldrich) in 0.25% Triton X-100. The fixed cells were first incubated overnight at 4 °C with primary antibodies of mouse anti-G3BP (1:300, Abcam) and then incubated for 1 h at RT with the secondary antibodies of Alexa fluor 488 anti-mouse IgG (1:300, Abcam). Then, DNA was stained by the incubation in DAPI (1:1000, Sigma) for 15 min at RT and F-actin was stained by the incubation in Alexa Fluor™ 555 Phalloidin (1:500, Fischer Scientific). The fixed cells were imaged by Olympus Cell xCellence microscope, using ×60 oil objective with a numerical aperture (NA) of 1.35 and a Hamamatsu ORCA 03 g camera.
Live Cell Imaging.
Single U2OS cells stably expressing GFP-tagged G3BP1 were cultured in µ-Slide 18 Well Glass Bottom (ibid, 81817) and allowed for growth overnight. The cells were then switched into live-cell imaging media [FluoroBrite™ DMEM with 10% FBS, 10 mM DL-Lactic acid sodium, and ProLong™ Live Antifade Reagent (1:100)] and stabilized at 37 °C in 5% CO2 for 2 h. The cells were imaged at 37 °C and 5% CO2 after 0.5 mM sodium arsenite was added. Imaging was done on an Olympus Cell xCellence microscope with a cage incubator (37 °C and 5% CO2), using ×60 oil objective with a numerical aperture (NA) of 1.35 and an Andor Ixon 3 camera.
Stress Granule and Cell Detection.
Image analysis of the fixed and live images was performed using Fiji (54) and custom scripts (https://dx.doi.org/10.5281/zenodo.8081112) and has been automated to allow for the efficient and reproducible processing of a large number of cells (>100 cells). Briefly, cells were segmented using cellpose (55, 56) and the SGs were detected using a fixed threshold or local maxima, respectively, for the fixed and live images. The optimal values for the fixed threshold or local maxima were obtained when 1) a minimal change was detected in the granule number on varying the threshold value and 2) most of the granules were captured without a minimal number of artificial granules.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
X.X. and F.S. acknowledge the support of the European Union’s Horizon 2020 Research and Innovation program under grant agreement no.101017821 (LIGHT-CAP). A.A.R. acknowledges the support of the Ambizione Grant 202214 from the Swiss NSF. We would like to thank the EPFL BioImaging & Optics Core Facility and in particular José Artacho for his assistance in imaging and Dr. Romain Guiet for his assistance in image processing.
Author contributions
X.X. and F.S. designed research; X.X., A.A.R., and L.R.J. performed research; A.A.R. and K.A.R. contributed new reagents/analytic tools; X.X., E.R.D., and F.S. analyzed data; and X.X., A.A.R., E.R.D., and F.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Datasets data have been deposited in Zenodo (https://doi.org/10.5281/zenodo.10825310) (57). Scripts and datasets are available at https://dx.doi.org/10.5281/zenodo.8081112) (58).
Supporting Information
References
- 1.Yewdall N. A., André A. A. M., Lu T., Spruijt E., Coacervates as models of membraneless organelles. Curr. Opin. Colloid Interface Sci. 52, 101416 (2021). [Google Scholar]
- 2.Gao Z., et al. , Liquid-liquid phase separation: Unraveling the enigma of biomolecular condensates in microbial cells. Front. Microbiol. 12, 751880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.André A. A. M., Spruijt E., Liquid-liquid phase separation in crowded environments. IJMS 21, 5908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astoricchio E., Alfano C., Rajendran L., Temussi P. A., Pastore A., The wide world of coacervates: From the sea to neurodegeneration. Trends in Biochem. Sci. 45, 706–717 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Brangwynne C. P., et al. , Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Alberti S., Hyman A. A., Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ijavi M., Physical Characterization of Phase Separated Biomolecular Condensates (ETH Zurich, 2021). [Google Scholar]
- 10.Jawerth L., et al. , Protein condensates as aging Maxwell fluids. Science 370, 1317–1323 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Wegmann S., et al. , Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumelie J. G., et al. , Biomolecular condensates create phospholipid-enriched microenvironments. Nat. Chem. Biol. 20, 302–313 (2023). 10.1038/s41589-023-01474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel A., et al. , ATP as a biological hydrotrope. Science 356, 753–756 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Rollin R., Joanny J.-F., Sens P., Physical basis of the cell size scaling laws. eLife 12, e82490 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberts B., Molecular Biology of the Cell (Garland Science, 2017). [Google Scholar]
- 16.Gauthier-Coles G., et al. , Quantitative modelling of amino acid transport and homeostasis in mammalian cells. Nat. Commun. 12, 5282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergström J., Fürst P., Norée L. O., Vinnars E., Intracellular free amino acid concentration in human muscle tissue. J. Appl. Physiol. 36, 693–697 (1974). [DOI] [PubMed] [Google Scholar]
- 18.Fisher M. T., Proline to the rescue. Proc. Natl. Acad. Sci. U.S A. 103, 13265–13266 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ignatova Z., Gierasch L. M., Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc. Natl. Acad. Sci. U.S A. 103, 13357–13361 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S. H., Ahmad N., Ahmad F., Kumar R., Naturally occurring organic osmolytes: From cell physiology to disease prevention. IUBMB Life 62, 891–895 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Rydeen A. E., Brustad E. M., Pielak G. J., Osmolytes and protein-protein interactions. J. Am. Chem. Soc. 140, 7441–7444 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Street T. O., Bolen D. W., Rose G. D., A molecular mechanism for osmolyte-induced protein stability. Proc. Natl. Acad. Sci. U.S A. 103, 13997–14002 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadmiller S. S., Gorensek-Benitez A. H., Guseman A. J., Pielak G. J., Osmotic shock induced protein destabilization in living cells and its reversal by glycine betaine. J. Mol. Biol. 429, 1155–1161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N., Living with water stress: Evolution of osmolyte systems. Science 217, 1214–1222 (1982). [DOI] [PubMed] [Google Scholar]
- 25.Mao T., et al. , Amino acids stabilizing effect on protein and colloidal dispersions. [arXiv Preprint] (2024). Available at: http://arxiv.org/abs/2404.11574 (Accessed 18 April 2024).
- 26.Winkler P. M., et al. , Modulating weak protein-protein cross-interactions by the addition of free amino acids at millimolar concentrations. J. Phys. Chem. B, 128, 7199–7207 (2024). 10.1021/acs.jpcb.4c01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X., Stellacci F., Amino acids and their biological derivatives modulate protein-protein interactions in an additive way. J. Phys. Chem. Lett. 15, 7154–7160 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong Q., Xufeng X., Stellacci F., Versatile capillary cells for handling concentrated samples in analytical ultracentrifugation. Anal. Chem. 96, 2567–2573 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi A., Takanashi K., FUS interacts with nuclear matrix-associated protein SAFB1 as well as Matrin3 to regulate splicing and ligand-mediated transcription. Sci. Rep. 6, 35195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broustal O., et al. , FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J. Alzheimer’s Dis. 22, 765–769 (2010). [PubMed] [Google Scholar]
- 31.Kwiatkowski T. J., et al. , Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Aulas A., Vande Velde C., Alterations in stress granule dynamics driven by TDP-43 and FUS: A link to pathological inclusions in ALS? Front. Cell. Neurosci. 9, 423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X., et al. , Experimental method to distinguish between a solution and a suspension. Adv. Mater. Interfaces 9, 2200600 (2022). [Google Scholar]
- 34.Xu X., Cölfen H., Ultracentrifugation techniques for the ordering of nanoparticles. Nanomater. 11, 333 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X., de With G., Cölfen H., Self-association and gel formation during sedimentation of like-charged colloids. Mater. Horizons 9, 1216–1221 (2022). [DOI] [PubMed] [Google Scholar]
- 36.van Rijssel J., et al. , Size-dependent second virial coefficients of quantum dots from quantitative cryogenic electron microscopy. J. Phys. Chem. B 118, 11000–11005 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Xu X., Wu B., Cölfen H., de With G., Assembly control at a low Péclet number in ultracentrifugation for uniformly sized nanoparticles. J. Phys. Chem. C 125, 8752–8758 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., et al. , Tracking structural transitions of bovine serum albumin in surfactant solutions by fluorescence correlation spectroscopy and fluorescence lifetime analysis. Soft Matter 11, 2512–2518 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Testa A., et al. , Sustained enzymatic activity and flow in crowded protein droplets. Nat. Commun. 12, 6293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ijavi M., et al. , Surface tensiometry of phase separated protein and polymer droplets by the sessile drop method. Soft Matter 17, 1655–1662 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Glauninger H., Wong Hickernell C. J., Bard J. A. M., Drummond D. A., Stressful steps: Progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell 82, 2544–2556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann S., Kedersha N., Anderson P., Ivanov P., Molecular mechanisms of stress granule assembly and disassembly. Biochimica et Biophysica Acta (BBA)–Mol. Cell Res., 1868, 118876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jain S., et al. , ATPase-Modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooper G. M., “Transport of small molecules” in The Cell: A Molecular Approach (Sinauer Associates, ed. 2, 2000). [Google Scholar]
- 45.Bröer S., Bröer A., Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem. J. 474, 1935–1963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olesen M. A., Villavicencio-Tejo F., Quintanilla R. A., The use of fibroblasts as a valuable strategy for studying mitochondrial impairment in neurological disorders. Transl. Neurodegener. 11, 36 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratti A., et al. , Chronic stress induces formation of stress granules and pathological TDP-43 aggregates in human ALS fibroblasts and iPSC-motoneurons. Neurobiol. Dis. 145, 105051 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Böddeker T. J., et al. , Non-specific adhesive forces between filaments and membraneless organelles. Nat. Phys. 18, 571–578 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P., A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baron D. M., et al. , Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol. Neurodegener. 8, 30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gwon Y., et al. , Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science 372, eabf6548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan C., Knutson S. D., Huth S. W., MacMillan D. W. C., µMap proximity labeling in living cells reveals stress granule disassembly mechanisms. Nat. Chem. Biol. 1–11 (2024). 10.1038/s41589-024-01721-2. [DOI] [PubMed] [Google Scholar]
- 53.Page M. G., Zemb T., Dubois M., Cölfen H., Osmotic pressure and phase boundary determination of multiphase systems by analytical ultracentrifugation. ChemPhysChem 9, 882–890 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stringer C., Wang T., Michaelos M., Pachitariu M., Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Pachitariu M., Stringer C., Cellpose 2.0: How to train your own model. Nat. Methods 19, 1634–1641 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X., Data for “Amino acids modulate liquid-liquid phase separation in vitro and in vivo by regulating protein-protein interactions.” Zenodo. 10.5281/zenodo.10825310. Deposited 16 March 2024. [DOI] [PMC free article] [PubMed]
- 58.Guiet R., Xu X., Stellacci F., Stress granules quantification on fixed and live cells. Zenodo. 10.5281/zenodo.8081112. Deposited 31 January 2024. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Datasets data have been deposited in Zenodo (https://doi.org/10.5281/zenodo.10825310) (57). Scripts and datasets are available at https://dx.doi.org/10.5281/zenodo.8081112) (58).