Abstract
β-1,3-Glucanase (EC 3.2.1.39) and chitinase (EC 3.2.1.14) mRNAs, proteins, and enzyme activities were expressed specifically in the micropylar tissues of imbibed tomato (Lycopersicon esculentum Mill.) seeds prior to radicle emergence. RNA hybridization and immunoblotting demonstrated that both enzymes were class I basic isoforms. β-1,3-Glucanase was expressed exclusively in the endosperm cap tissue, whereas chitinase localized to both endosperm cap and radicle tip tissues. β-1,3-Glucanase and chitinase appeared in the micropylar tissues of gibberellin-deficient gib-1 tomato seeds only when supplied with gibberellin. Accumulation of β-1,3-glucanase mRNA, protein and enzyme activity was reduced by 100 μM abscisic acid, which delayed or prevented radicle emergence but not endosperm cap weakening. In contrast, expression of chitinase mRNA, protein, and enzyme activity was not affected by abscisic acid. Neither of these enzymes significantly hydrolyzed isolated tomato endosperm cap cell walls. Although both β-1,3-glucanase and chitinase were expressed in tomato endosperm cap tissue prior to radicle emergence, we found no evidence that they were directly involved in cell wall modification or tissue weakening. Possible functions of these hydrolases during tomato seed germination are discussed.
Germination of tomato (Lycopersicon esculentum Mill.) seeds is controlled by interactions between the embryonic radicle tip and the enclosing endosperm cap tissue at the micropylar end of the seed. Considerable evidence indicates that weakening of the endosperm cap tissue is required to allow penetration of the radicle and completion of germination (Groot and Karssen, 1987; Dahal and Bradford, 1990; Toorop et al., 2000). Enzymatic hydrolysis of the endosperm cap cell walls is assumed to be involved in this process, and because the cell walls contain 60% to 70% Man, endo-β-mannanase [(1→4)-β-mannan endohydrolase, E.C. 3.2.1.78] has been studied extensively in relation to tomato seed germination (for review, see Bewley, 1997). A specific endo-β-mannanase gene (LeMAN2) is expressed exclusively in the endosperm cap tissue prior to radicle emergence and endo-β-mannanase activity is consistently associated with germination (Nonogaki et al., 1998, 2000). However, other enzymes may be involved because there are cases where high endo-β-mannanase activity was present yet radicle emergence did not occur (Toorop et al., 1996; Dahal et al., 1997; Still and Bradford, 1997; Toorop et al., 2000). For example, abscisic acid (ABA) inhibited radicle protrusion and completion of tomato seed germination, but did not inhibit LeMAN2 induction (Nonogaki et al., 2000) or the majority of weakening in the endosperm cap (Chen and Bradford, 2000; Toorop et al., 2000). Thus, final cell wall disassembly and tissue weakening in the endosperm cap to allow radicle emergence may require the action of several enzymes.
In tobacco (Nicotiana tabacum) seeds, Meins and colleagues (Vögeli-Lange et al., 1994) demonstrated that a class I β-1,3-glucanase [(1→3)-β-glucan glucanohydrolase, EC 3.2.1.39] gene promoter was specifically active in the endosperm cap tissue of tobacco seeds prior to radicle protrusion. They subsequently confirmed that class I β-1,3-glucanase mRNA accumulation, enzyme activity, and protein content increased just prior to endosperm rupture and demonstrated that both class I β-1,3-glucanase induction and endosperm rupture were inhibited by ABA (Leubner-Metzger et al., 1995). A close correlation between class I β-1,3-glucanase induction and endosperm rupture in response to plant hormones and environmental factors affecting tobacco seed germination constituted strong, but indirect, evidence that class I β-1,3-glucanase may promote radicle protrusion (for review, see Leubner-Metzger and Meins, 1999). More direct evidence of a causal role for class I β-1,3-glucanase during endosperm rupture of tobacco seeds was obtained by sense transformation with a chimeric ABA-inducible class I β-1,3-glucanase transgene, which promoted endosperm rupture upon ABA treatment (Leubner-Metzger and Meins, 2000). Although β-1,3-glucanase expression is often accompanied by expression of chitinase [1, 4-(N-acetyl-β-d-glucosaminide) glycanohydrolase, EC 3.2.1.14] in pathogenesis-related (PR) responses (see below), only very low chitinase activity was detected in germinating tobacco seeds. The absence of significant chitinase accumulation and the close relationship between β-1,3-glucanase expression and completion of germination under diverse conditions led to the hypothesis that β-1,3-glucanase contributed to the hydrolysis of cell wall components resulting in endosperm weakening at the site of radicle protrusion (Vögeli-Lange et al., 1994; Leubner-Metzger et al., 1995; Leubner-Metzger and Meins, 1999).
β-1,3-Glucanase and chitinase are widely distributed in higher plants and have been found during germination of pea (Pisum sativum; Petruzzelli et al., 1999), barley (Hordeum vulgare; Ballance et al., 1976; Høj et al., 1988; Leah et al., 1991), maize (Zea mays; Cordero et al., 1994), and wheat (Triticum aestivum; Caruso et al., 1999). Their expression is regulated by plant hormones that can also influence germination (Rezzonico et al., 1998), and they are implicated in other physiological and developmental processes, including embryogenesis, microsporogenesis, flowering, and abscission (for review, see Leubner-Metzger and Meins, 1999; Neuhaus, 1999). Both β-1,3-glucanase and chitinase are well known as PR proteins, belonging to the PR-2 and PR-3 families, respectively (van Loon, 1999). They are strongly induced when plants respond to wounding or infection by fungal, bacterial, or viral pathogens, and there is compelling evidence that β-1,3-glucanase and chitinase, acting alone and particularly in combination, contribute to plant defenses against fungal infection (for review, see Kombrink and Somssich, 1997; Leubner-Metzger and Meins, 1999; Neuhaus, 1999).
Tomato and tobacco are closely related members of the Solanaceae, and both have been used as model systems to investigate physiological and molecular mechanisms of growth and development. Because tomato has become a preferred model system for studying seed germination physiology (Hilhorst et al., 1998; Bradford et al., 2000), we wished to determine whether the β-1,3-glucanase and chitinase expression patterns characterized in tobacco also occur in association with tomato seed germination and endosperm cap weakening. Based on amino acid sequences of the mature proteins, the β-1,3-glucanases can be grouped into structural classes that differ in sequence homology by at least 40% to 50% (Meins et al., 1992; Simmons, 1994; Høj and Fincher, 1995). At least three structural classes are identified for the highly homologous β-1,3-glucanase isoforms of the Solanaceous species tomato, tobacco, potato (Solanum tuberosum), and pepper (Capsicum annuum). Four cDNAs encoding β-1,3-glucanases have been isolated from tomato. GluB encodes a basic 35-kD class I β-1,3-glucanase that shares approximately 90% sequence identity with tobacco class I β-1,3-glucanases (van Kan et al., 1992). The tomato GluB protein is an intracellular isoform that contains a characteristic carboxy-terminal extension that mediates targeting to the vacuole (van Kan et al., 1992, 1995; Neuhaus, 1999). GluA encodes an extracellular class II β-1,3-glucanase of tomato (van Kan et al., 1992), and two closely related class III β-1,3-glucanase genes (TomPR-Q'a and TomPR-Q'b) were isolated from viroid-infected tomato plants (Domingo et al., 1994). The Solanaceous PR-3 chitinases similarly are classified on the basis of sequence identity (Meins et al., 1992; Neuhaus, 1999) and several cDNAs encoding tomato chitinases are known. Chi9 encodes a class I chitinase and corresponds to a 30-kD intracellular mature protein with approximately 90% sequence identity to tobacco class I chitinases (Danhash et al., 1993). A short carboxy-terminal extension shown to be responsible for the targeting of tobacco class I chitinase to the vacuole is present in the precursor protein of tomato Chi9 (Danhash et al., 1993; Neuhaus, 1999). The cDNAs Chi3, Chi17, and Chi2;1 (identical to Chi14) encode class II tomato chitinases (Danhash et al., 1993; Harikrishna et al., 1996).
Here, we report the identification, expression patterns, tissue localization, and hormonal regulation of β-1,3-glucanase and chitinase during germination of tomato seeds. We also tested the possibility that these enzymes are involved in endosperm cap weakening using puncture force experiments and by studying their ability to hydrolyze isolated cell walls. Although class I isoforms of both β-1,3-glucanase and chitinase are expressed specifically in the endosperm caps of tomato seeds prior to radicle emergence, we conclude that it is unlikely that these enzymes are directly involved in cell wall modification or tissue weakening and discuss other possible roles during germination.
RESULTS
Coordinate Accumulation of β-1,3-Glucanase and Chitinase in the Micropylar Tissues of Germinating Tomato Seeds
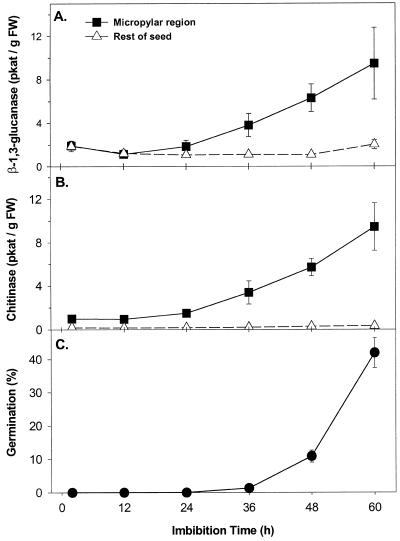
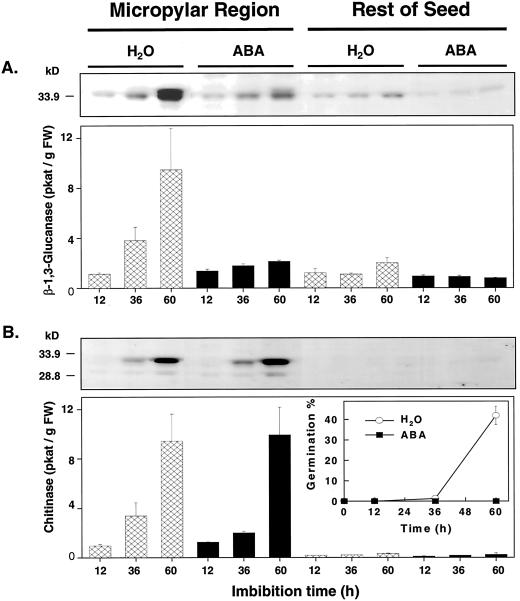
Highly sensitive radiometric assays were used to measure the activities of β-1,3-glucanase and chitinase in imbibed tomato seeds. [3H]Laminarin, an algal β-1,3-glucan known to be digested by all β-1,3-glucanase isoforms (Kauffmann et al., 1987; Beffa et al., 1993), was utilized for the β-1,3-glucanase assays and [3H]chitin as a substrate for chitinase. The reduced forms of these substrates were used, making the activities measured specific for the endo-type isoforms of these enzymes. β-1,3-Glucanase and chitinase activities assayed in protein extracts from tomato cv Moneymaker (MM) seeds began to increase just prior to radicle emergence (Fig. 1). A doubling of β-1,3-glucanase activity (Fig. 1A) and a 4-fold increase in chitinase activity (Fig. 1B) occurred after 36 h of imbibition on water (1% ± 0.1% germinated) and activities continued to increase until at least 60 h (43% ± 5% germinated; Fig. 1C). The increase in both activities occurred only in the micropylar region, not in the rest of the tomato seed (Fig. 1, A and B). Thus, as in the case of germinating tobacco seeds (Leubner-Metzger et al., 1995), β-1,3-glucanase was expressed in the micropylar tissues of tomato seeds just prior to radicle protrusion, but in contrast to the situation in tobacco, coordinate expression of chitinase also occurs in tomato.
Figure 1.
β-1,3-Glucanase and chitinase activity of the micropylar region (endosperm cap and radicle tip) and the rest of tomato seeds (lateral endosperm and embryo) during imbibition and germination. A, Tomato cv Moneymaker seeds were imbibed in water at 25°C and β-1,3-glucanase enzyme activity was assayed radiometrically using reduced [3H]laminarin as substrate. B, Chitinase enzyme activity was assayed in the same seeds using reduced [3H]chitin as substrate. C, The time course of germination (radicle emergence) in water at 25°C. Both germinated and ungerminated seeds were included in enzyme extractions at each time point. Error bars indicate ±se.
A Class I Isoform of β-1,3-Glucanase Is Transcriptionally Induced Exclusively in the Endosperm Caps of Germinating Tomato Seeds
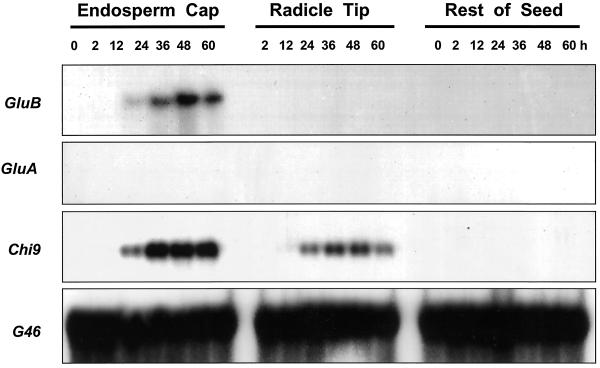
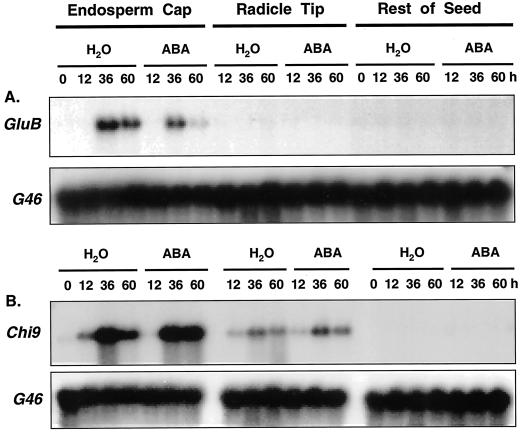
When tomato class I (GluB) and class II (GluA) β-1,3-glucanase cDNAs isolated from tomato leaves (van Kan et al., 1992) were used to screen a germinating tomato seed cDNA library, only plaques hybridizing with the GluB cDNA were identified. A cDNA isolated from these plaques had a sequence identical to that of the GluB from tomato leaves used to screen the library (data not shown). Northern blots containing total RNA isolated from different tissues of MM seeds at 0, 2, 12, 24, 36, 48, and 60 h after imbibition were examined for expression of mRNAs of different tomato β-1,3-glucanase genes. An mRNA band hybridizing to GluB, encoding a class I basic vacuolar β-1,3-glucanase (van Kan et al., 1992), was first detectable at 24 h after imbibition in water and increased until at least 48 h after imbibition (Fig. 2). The GluB mRNA was restricted to the tomato endosperm cap tissue only, not being detected in radicle tips or in the remaining seed tissues. On the other hand, GluA mRNA, which is translated into a 33-kD class II acidic extracellular β-1,3-glucanase (van Kan et al., 1992), was not detected in germinating tomato seeds (Fig. 2). Moreover, mRNAs of Tom PR-Q'a and Tom PR-Q'b, two class III β-1,3-glucanases (Domingo et al., 1994), were not expressed appreciably during tomato seed germination (data not shown).
Figure 2.
Gel-blot hybridization assays of RNA from dissected tissues of tomato cv Moneymaker seeds using riboprobes from cDNAs of different classes of tomato β-1,3-glucanases or chitinase: GluB (class I basic intracellular β-1, 3-glucanase); GluA (class II acidic extracellular β-1, 3-glucanase); and Chi9 (class I basic intracellular chitinase). G46 is a cDNA for a ribosomal protein used to demonstrate equal loading of the lanes (Cooley et al., 1999). Seeds were imbibed for the indicated times in water prior to dissection and extraction of RNA. Total RNA (10 μg) was loaded in each lane. The RNA sample of dry (0 h) micropylar region tissues includes endosperm caps and radicle tips. In addition, no signal was detected using probes for Tom PR-Q'a (class III acidic β-1, 3-glucanase), Tom PR-Q'b (class III basic β-1, 3-glucanase), Chi3 and Chi17 (class II acidic extracellular chitinases), or Chi2;1 (Chi14; class II basic chitinase) (data not shown).
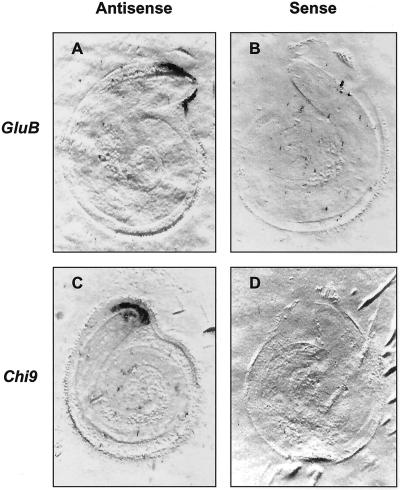
The tissue localization of class I β-1,3-glucanase mRNA within tomato seeds was further confirmed by tissue printing (McClure and Guilfoyle, 1989; Nonogaki et al., 2000). In agreement with the northern blots of RNA extracted from dissected seed parts (Fig. 2), GluB antisense RNA probes hybridized only to the endosperm cap tissue (Fig. 3A).
Figure 3.
Localization of tomato class I β-1,3-glucanase and chitinase mRNAs in individual tomato cv Moneymaker seeds by tissue printing. After 48 h of imbibition in water at 25°C, individual ungerminated tomato seeds were bisected and the cut surface of each half was printed directly on separate nylon membranes. The mirror-image tissue prints were then hybridized with antisense (A and C) or sense (B and D) riboprobes for class I tomato β-1,3-glucanase (GluB; A and B) or class I chitinase (Chi9; C and D).
A rabbit polyclonal antiserum directed against tobacco class I β-1,3-glucanase was used for immunoblot analysis of protein expression during tomato seed germination. The antiserum detected the class I, II, and III tobacco isoforms of the enzyme (Neuhaus et al., 1992; Beffa et al., 1993) and cross-reacted with purified tomato homologs (Fischer et al., 1989). The antiserum identified a major 35-kD protein band in tomato seed protein extracts (Fig. 4A), corresponding in size to tomato class I β-1,3-glucanase (GluB, 35 kD; van Kan et al., 1992). The intensity of this signal increased in endosperm caps from 24 through 60 h in parallel with the increase in β-1,3-glucanase enzyme activity and accumulation of GluB mRNA, whereas the 35-kD protein, GluB mRNA, and enzyme activity were not detected in the rest of the seed (Figs. 1A, 2, 3A, and 4A). When added to protein extracts from tomato micropylar regions, the tobacco antibody blocked all of the induced β-1,3-glucanase activity (data not shown), indicating that it is detecting a tomato β-1,3-glucanase. In addition, when GluB protein was expressed in bacteria as a fusion with maltose-binding protein (42.7 kD), the antiserum detected both the purified fusion protein (77 kD) and the cleaved GluB protein (35 kD; Fig. 4B), demonstrating that the antiserum can detect authentic tomato GluB. Additional fainter bands at 78, 61, and 51 kD are present in extracts from both the endosperm cap and the rest of the seed (Fig. 4A). However, their spatial and temporal presence does not match with the accumulation pattern of β-1,3-glucanase activity.
Figure 4.
β-1,3-Glucanase and chitinase protein accumulation in the micropylar region and the rest of tomato seeds during imbibition and germination. Tomato cv Moneymaker seeds were imbibed in water at 25°C for the times indicated. Western blots of tomato seed proteins are shown using antiserum to tobacco class I β-1,3-glucanase (A) and to tobacco class I chitinase (C), respectively. The left-most lane in A contains authentic tobacco 33-kD class I β-1,3-glucanase (Gln I) and in C contains tobacco class I chitinase A (34 kD) and chitinase B (32 kD; Chn I). The bacterially expressed fusion proteins of maltose-binding protein and tomato β-1,3-glucanase (pMAL-GluB; B) and of maltose-binding protein and chitinase (pMAL-Chi9; D) were immunoblotted with the antisera used in A and C, respectively. The left lanes in B and D were loaded with the purified fusion proteins (approximately 77 kD), whereas the right lanes were loaded with the same samples after protease cleavage of the fusion protein to separate the maltose-binding protein from the tomato proteins.
Together, these results demonstrate convincingly that the endo-type β-1,3-glucanase activity that appears specifically in the tomato endosperm cap prior to radicle emergence is due to the expression of the class I GluB gene and that expression is regulated primarily at the level of transcription.
A Class I Isoform of Chitinase Is Transcriptionally Induced in the Endosperm Caps and Radicle Tips of Germinating Tomato Seeds
Only mRNA hybridizing to the tomato class I basic chitinase cDNA (Chi9) encoding an intracellular chitinase (Danhash et al., 1993) was detected in RNA samples from imbibed tomato seeds (Fig. 2). Chi9 mRNA first appeared at 24 h of imbibition and accumulated from 36 through 60 h. Like GluB, Chi9 mRNA accumulated primarily in the endosperm caps, although it was also present at lower abundance in radicle tips (Fig. 2). This localization of Chi9 mRNA was confirmed by tissue printing, where a strong signal was present in the endosperm cap region and a weaker signal was detected in the radicle tip (Fig. 3C). No hybridization was detected in the rest of the seed. Among the other chitinase genes tested, no accumulation of class II acidic extracellular chitinase mRNAs (Chi3 and Chi17) or of class II basic extracellular chitinase mRNA (Chi2;1 = Chi14; Danhash et al., 1993; Harikrishna et al., 1996) was detected in any seed tissues (data not shown).
Immunoblot analysis using an antiserum to tobacco class I chitinase that detects the class I but not the other chitinase isoforms of tobacco (Shinshi et al., 1987; Beffa et al., 1995) detected a major 33-kD band in tomato micropylar tissue extracts that accumulated in parallel with increasing chitinase activity (Fig. 4C). In addition, a 30-kD band could be seen after longer imbibition times. However, no chitinase protein accumulation was detected in the rest of the seed over the same period (Fig. 4C). Additional bands at molecular mass positions greater than approximately 55 kD were sometimes detected, but their appearance did not correlate with chitinase activity. When Chi9 protein was expressed in bacteria as a fusion with maltose-binding protein, the antiserum detected both the purified fusion protein and the cleaved Chi9 protein (Fig. 4D), demonstrating that the antiserum can detect authentic tomato Chi9.
In parallel with the results for GluB, the chitinase activity that appears in the tomato endosperm cap and radicle tip prior to radicle emergence is due to expression of the class I Chi9 gene, and expression is regulated primarily at the level of transcription.
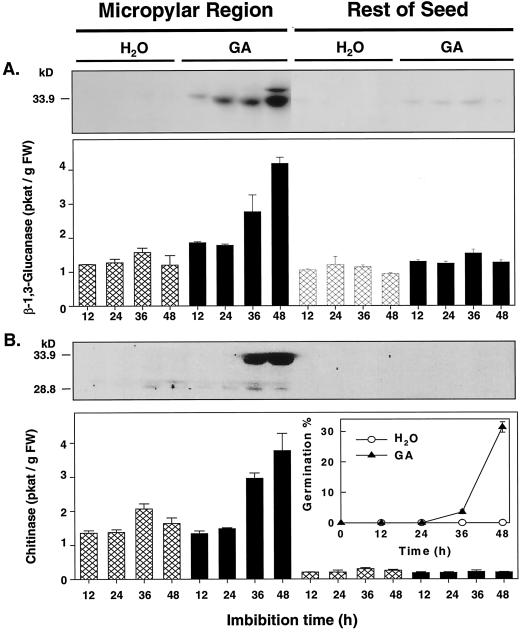
β-1,3-Glucanase and Chitinase Induction in the Micropylar Regions of gib-1 Tomato Seeds Requires Gibberellin (GA)
Germination of the GA-deficient gib-1 tomato mutant requires supplementary GA (Fig. 5B, inset; Groot and Karssen, 1987). Only very low enzyme activities or protein accumulations of both β-1,3-glucanase (Fig. 5A) and chitinase (Fig. 5B) were detected in any part of water-imbibed gib-1 seeds during the first 48 h of imbibition. However, when imbibed in 100 μm GA4+7, which initiated seed germination, β-1,3-glucanase and chitinase activities as well as the corresponding class I antigens increased to levels similar to that in wild-type MM seeds, and in the same tissue-localized manner (Fig. 5). The micropylar region, but not the rest of the seed, showed increases in enzyme activities and protein amounts for both enzymes in the presence of GA. Results were the same when calculated on a protein basis or on a seed part basis (data not shown).
Figure 5.
Enzyme activities and immunoblot assays of β-1,3-glucanase (A) and chitinase (B) of gib-1 tomato seeds after different imbibition times in water or in 100 μm GA4+7 at 25°C. The inset in B shows the germination time courses for seeds in either water or GA. Error bars indicate ±se.
ABA Suppresses Both mRNA and Protein Accumulation of β-1,3-Glucanase, But Not of Chitinase, in Germinating Tomato Seeds
In tobacco, ABA inhibited class I β-1,3-glucanase gene expression in imbibed seeds, but chitinase was not expressed even in the absence of ABA (Leubner-Metzger et al., 1995). Because ABA is effective in inhibiting germination but does not block endo-β-mannanase expression in tomato seeds (Dahal et al., 1997; Nonogaki et al., 2000), it was of interest to determine the effect of ABA on GluB and Chi9 expression in tomato. GluB mRNA abundance in seeds imbibed in 100 μm ABA was 68% and 27% of that in water-imbibed seeds at 36 and 60 h, respectively (Fig. 6A). In addition, both β-1,3-glucanase activity and protein accumulation in micropylar tissues of MM seeds were suppressed by ABA (Fig. 7A). Chi9 mRNA accumulation, however, was essentially unaffected in either endosperm caps or radicle tips by imbibition in the ABA solution (Fig. 6B), and chitinase activity and protein accumulation were not significantly altered (Fig. 7B). These results were confirmed by experiments showing that although increasing concentrations of ABA proportionately inhibited both germination and GluB mRNA accumulation, the abundance of Chi9 mRNA was relatively unaffected by up to 100 μm ABA (data not shown).
Figure 6.
Gel blot analyses of tomato class I β-1,3-glucanase (A) and class I chitinase (B) mRNA expression in response to 100 μm ABA treatment during tomato cv Moneymaker seed imbibition and germination. Seeds were imbibed for the indicated times in water or 100 μm ABA prior to dissection and extraction of RNA. The membranes were hybridized to riboprobes for either tomato class I β-1,3-glucanase (GluB; A) or class I chitinase (Chi9; B). G46 is a cDNA for a ribosomal protein to demonstrate equal loading of lanes with 10 μg of total RNA. The RNA sample of dry micropylar region tissues (0 h) includes both endosperm caps and radicle tips.
Figure 7.
Enzyme assays and protein gel-blot analyses of tomato class I β-1,3-glucanase (A) and class I chitinase (B) in response to 100 μm ABA treatment during tomato cv Moneymaker seed imbibition and germination. The inset in B shows the germination time courses for seeds in either water or ABA. Error bars indicate ±se.
Tomato Endosperm Cap Weakening Is Not Inhibited by ABA
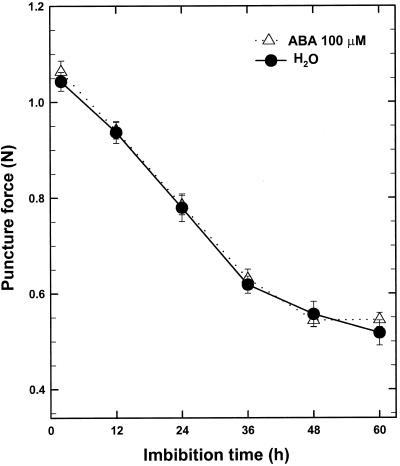
A prediction of the hypothesis of Leubner-Metzger et al. (1995) is that if β-1,3-glucanase contributes to tomato endosperm weakening, the force required to puncture the endosperm cap should be reduced coincident with increasing β-1,3-glucanase activity. Therefore, we compared the change in puncture force of germinating tomato endosperm caps in water with those imbibed in 100 μm ABA. Although ABA inhibited MM tomato seed germination and β-1,3-glucanase accumulation, physical weakening of endosperm caps was neither prevented nor delayed (Fig. 8). Furthermore, the decline in endosperm cap strength began within 12 h of imbibition (Fig. 8), well before the increase in either β-1,3-glucanase or chitinase activity (Fig. 1).
Figure 8.
Time course of the decrease in endosperm cap puncture force of tomato cv Moneymaker seeds incubated in water or 100 μm ABA at 25°C. The endosperm caps were dissected from ungerminated seeds imbibed for the indicated times.
β-1,3-Glucanase and Chitinase Do Not Hydrolyze Isolated Tomato Endosperm Cell Walls
Another question relevant to the roles of β-1,3-glucanase and chitinase in germination is whether there are substrates in the endosperm cell walls on which they can act, potentially contributing to tissue weakening. To test this, cell walls were isolated from tomato endosperm caps directly after the start of imbibition and were incubated with the enzymes. Table I shows the amounts of reducing sugars released from the cell walls incubated with class I β-1,3-glucanases and/or chitinases of tomato and tobacco. Defined substrates (carboxymethyl [CM]-pachyman and CM-chitin-Remazol Brilliant Violet 5R [RBV]) were used to confirm enzyme activity, and bacterially expressed tomato endo-β-mannanase fusion protein (pMAL-LeMAN2) was a positive control enzyme known to be active on isolated endosperm cap cell walls (Nonogaki et al., 2000). Both bacterially expressed pMAL-GluB or purified tobacco β-1,3-glucanase released large amounts of reducing sugars into reaction buffer when compared with their boiled counterparts using defined substrate. Because there was no statistical difference between boiled and blank samples, the boiling treatment was effective in eliminating the activities of these β-1,3-glucanases. Similar data were obtained from defined substrate incubated with either bacterially expressed tomato chitinase or purified tobacco chitinase, although the activity remaining after boiling was somewhat higher (Table I). However, there were no significant differences between active and boiled enzyme samples when endosperm cap cell walls were used as a substrate for β-1,3-glucanase, chitinase, or both enzymes together (Table I). The isolated cell walls, which were composed of approximately 60% Man, 20% Glc, 12% Ara, and 5% Gal (Dahal et al., 1997), were hydrolyzed by endo-β-mannanase (pMAL-LeMAN2) (Table I). Because LeMAN2 is induced earlier following imbibition than is GluB (Nonogaki et al., 2000; Figs. 1, 2), it is possible that endo-β-mannanase action on the cell walls is required to expose substrates for GluB. However, pMAL-GluB did not hydrolyze the cell wall material remaining after extensive incubation with pMAL-LeMAN2 (Table I).
Table I.
Activity of tomato and tobacco class I β-1,3-glucanase and class I chitinase on defined substrates and on isolated tomato endosperm cap cell walls
| Enzyme | Boiled | Substrate
|
||
|---|---|---|---|---|
| CM-pachyman | CM-chitin-RBV | Endosperm cap cell walls | ||
| μg Glc-equivalent reducing sugars | ||||
| None | – | 3.8 ± 0.3c | 21.2 ± 4.3b | 3.3 ± 0.4e |
| pMAL-GluB | No | 240 ± 12b | – | 3.6 ± 0.2e |
| Yes | 6.6 ± 0.6c | – | 4.1 ± 0.3de | |
| Tobacco Gln | No | 491 ± 12a | – | 3.8 ± 0.5e |
| Yes | 7.0 ± 0.3c | – | 3.6 ± 0.4e | |
| pMAL-Chi9 | No | – | 36.2 ± 0.9a | 3.5 ± 0.4e |
| Yes | – | 23.7 ± 0.4b | 4.5 ± 0.6de | |
| Tobacco Chn | No | – | 44.0 ± 1.6a | 7.9 ± 0.6bed |
| Yes | – | 23.6 ± 1.7b | 5.4 ± 0.9cde | |
| GluB + Chi9 | No | – | – | 7.0 ± 1.1bcde |
| Yes | – | – | 6.8 ± 0.7bcde | |
| Gln + Chn | No | – | – | 10.4 ± 0.9b |
| Yes | – | – | 8.7 ± 0.4bc | |
| pMAL-LeMAN2 | No | – | – | 74.2 ± 2.1a |
| Yes | – | – | 5.1 ± 0.5cde | |
| LeMAN2 → GluB | No | – | – | 3.4 ± 0.8e |
| Yes | – | – | 3.3 ± 0.2e | |
Tomato β-1,3-glucanase expressed in bacteria as a fusion protein with maltose-binding protein (pMAL-GluB), β-1,3-glucanase purified from tobacco leaves (Gln), bacterially expressed tomato chitinase fusion protein (pMAL-Chi9), chitinase purified from tobacco leaves (Chn), combinations of the expressed (GluB + Chi9) and purified (Gln + Chn) enzymes, and bacterially expressed tomato endo-β-mannanase fusion protein (pMAL-LeMAN2) were incubated for 36 h at 30°C with the indicated substrates and reducing sugars released were quantified after trifluoroacetic acid hydrolysis. Tomato Moneymaker seeds directly after the start of imbibition were used to isolate endosperm cap cell walls, which were incubated with the enzymes as indicated. LeMAN2 → GluB: cell walls first incubated with endo-β-mannanase and the remaining insoluble material incubated with pMAL-G1uB. Means ± se (n = 3) are shown. Means within each substrate category (columns) were compared first by ANOVA, and if significant, mean differences were identified by Duncan's Multiple Range Test. Means within each substrate group followed by the same letter are not significantly different (P < 0.05).
DISCUSSION
In agreement with previous results with tobacco (Vögeli-Lange et al., 1994; Leubner-Metzger et al., 1995, 1996), β-1,3-glucanase mRNA, protein, and enzyme activity were expressed specifically in the micropylar endosperm at an early stage of tomato seed germination. However, in contrast to the case with tobacco seeds, where little chitinase activity was detected in imbibed seeds (Leubner-Metzger et al., 1995), chitinase mRNA, protein, and enzyme activity accumulated in the micropylar region of imbibed and germinating tomato seeds essentially in parallel with β-1,3-glucanase. Although the majority of chitinase mRNA was present in the endosperm cap tissue, a smaller accumulation of mRNA occurred also in the radicle tips. Thus, both β-1,3-glucanase and chitinase were expressed in tomato seeds prior to radicle emergence specifically in the endosperm cap tissue undergoing weakening or in the adjacent radicle tip.
Several lines of evidence support the conclusion that increases in β-1,3-glucanase and chitinase activities during tomato seed germination result from transcriptional induction of the class I isoforms of these enzymes. Only cDNAs hybridizing to the class I β-1,3-glucanase GluB gene were identified by screening a germinating seed cDNA library. Northern analyses using specific cDNAs representing the known classes of tomato β-1,3-glucanase and chitinase genes (van Kan et al., 1992; Danhash et al., 1993; Harikrishna et al., 1996) revealed mRNA accumulation for the GluB gene and the class I chitinase Chi9 gene, but no accumulation of the known class II (GluA, Chi3, Chi17, and Chi2;1) and class III (TomPR-Q'a and TomPRQ'b) mRNAs was detected. GluB mRNA expression was localized exclusively in the micropylar endosperm and correlated temporally and spatially with the accumulation of endo-type β-1,3-glucanase activity and of a major 35-kD immunoreactive protein in this tissue, which strongly suggests that it is the class I β-1,3-glucanase (GluB, 35 kD) reported by van Kan et al. (1992). Enzyme activity inhibition experiments using this antiserum further support the view that the class I-type GluB gene accounts for most, if not all, of the tomato seed endo- β-1,3-glucanase activity. Similar mRNA and immunological evidence indicates that the accumulation of endo-type chitinase activity and of a 33-kD protein is due to the induction of the class I Chi9 chitinase gene. Only Chi9 mRNA was detected in tomato seeds, and because tomato and tobacco class I chitinases share approximately 90% amino acid sequence identity, it is likely that the anti-tobacco class I chitinase antiserum, which does not detect the other tobacco isoforms (Shinshi et al., 1987; Beffa et al., 1995), cross-reacted with the tomato class I chitinase Chi9. A 30-kD antigen also detected during late imbibition was likely due to posttranslational processing, as already described for Chi9 (Sticher et al., 1992; Danhash et al., 1993). Finally, the differential effects of ABA on GluB and Chi9 expression (see below) are consistent with the regulation of the corresponding class I genes in tobacco cells (Rezzonico et al., 1998).
Our results do not agree with those of Morohashi and Matsushima (2000), who detected β-1,3-glucanase activity in micropylar tissues of tomato cv First Up seeds only after radicle protrusion had occurred, and chitinase activity was low and did not increase during germination. Our time course experiments using specific cDNAs for the known structural classes of tomato β-1,3-glucanases and chitinases and antisera for Solanaceous β-1,3-glucanases and chitinases revealed that the mRNAs, proteins and enzyme activities of the class I isoforms of β-1,3-glucanase and chitinase began to increase in the micropylar endosperm cap prior to radicle protrusion. Tissue prints clearly illustrated the presence of GluB and Chi9 mRNAs in the micropylar tissues prior to radicle emergence (Fig. 3). We found that the reducing sugar and colorimetric activity assays used by Morohashi and Matsushima (2000) were less sensitive than the radiometric assays employed here (data not shown). This, or differences between the tomato cultivars used, may explain why the initial increases in both β-1,3-glucanase and chitinase activities that we detected prior to radicle emergence were not seen in their study.
Gibberellin and ABA differentially affected the expression of β-1,3-glucanase and chitinase during tomato seed germination. Gibberellin-deficient gib-1 mutant seeds cannot complete germination unless exogenous GA is provided (Groot and Karssen, 1987). β-1,3-glucanase activity and protein amounts also increased prior to the initiation of radicle emergence only in GA-treated seeds (Fig. 5). GA-induced germination of tobacco seeds in the dark similarly was associated with class I β-1,3-glucanase accumulation (Leubner-Metzger et al., 1996). However, in contrast to tobacco seeds, GA also resulted in an increase in chitinase protein and activity in gib-1 tomato seeds coinciding with the initiation of radicle emergence (Fig. 5). Although these results are consistent with the induction of a number of hydrolytic enzymes in the tomato endosperm cap in response to GA (Groot et al., 1988; Bradford et al., 2000), they are not proof that GA directly induces expression of class I β-1,3-glucanase and chitinase. Because the response of these genes to GA is 12 to 24 h slower than that of endo-β-mannanase (LeMAN2; Nonogaki et al., 2000) or expansin (LeEXP4; Chen and Bradford, 2000) genes, it is possible that expression of GluB and Chi9 is a consequence of the stimulation of germination by GA, rather than being a direct response to GA.
In contrast to the effect of GA, ABA inhibited seed germination and the expression of class I β-1,3-glucanase of tomato (Figs. 6 and 7) and tobacco (Leubner-Metzger et al., 1995). Furthermore, sense transformation of tobacco with a chimeric ABA-inducible class I β-1,3-glucanase transgene resulted in the promotion of endosperm rupture upon ABA treatment, providing evidence that class I β-1,3-glucanase contributes to endosperm rupture of tobacco seeds (Leubner-Metzger and Meins, 2000). However, in tomato seeds endosperm weakening was unaffected by ABA (Fig. 8), whereas β-1,3-glucanase activity was largely inhibited, and weakening was initiated well before any increase in β-1,3-glucanase activity was evident. Therefore, it is unlikely that the enzyme is directly involved in the initial weakening of tomato endosperm caps. Chitinase, on the other hand, was not expressed during tobacco seed germination (Leubner-Metzger et al., 1995), and its expression in tomato seeds was unaffected by ABA (Figs. 6 and 7). Because tobacco seeds germinate without appreciable chitinase activity, and tomato seeds were inhibited from germinating by ABA without any effect on chitinase expression, it is unlikely that chitinase plays a mechanistic role in the germination process.
If β-1,3-glucanase and chitinase were to be involved in cell wall degradation associated with endosperm rupture, substrates for these enzymes should be present in cell walls of the endosperm cap. β-1,3-Glucan (callose) is present in large quantities in the outer cell walls of the endosperm of muskmelon seeds (Yim and Bradford, 1998) and in seed coats of other species (e.g. Bhalla and Slattery, 1984; Bevilacqua et al., 1987). It is also present as small callosic deposits scattered through the starchy endosperm of cereal caryopses (Høj and Fincher, 1995). However, β-1,3-glucan was not detected in tomato seeds by histochemical staining with aniline blue (Beresniewicz et al., 1995; K.O. Yim and K.J. Bradford, unpublished data). Although chitin is not thought to be a component of plant cell walls, a possible target for chitinase includes arabinogalactan proteins that are proposed to have multiple roles in plant development, including embryogenesis (e.g. Domon et al., 2000; Majewska-Sawka and Nothnagel, 2000). To test whether substrates for these enzymes might be present, crude cell walls isolated from tomato endosperm caps dissected from seeds soon after the start of imbibition were treated with β-1,3-glucanase and chitinase from tomato or tobacco. We did not detect significant release of reducing sugars from isolated cell walls with these β-1,3-glucanases or chitinases used either alone or in combination (Table I). Thus, we found no evidence of substrates for β-1,3-glucanase or chitinase in tomato endosperm cap cell walls.
It has been proposed that tomato endosperm cap weakening is a biphasic process (Karssen et al., 1989; Toorop et al., 2000). The first phase is characterized by major ABA-independent weakening and is associated with endosperm cap-specific expression of endo-β-mannanase (Nonogaki et al., 2000), expansin (Chen and Bradford, 2000), and xyloglucan endotransglycosylase (Bradford et al., 2000) genes. A second phase just prior to radicle emergence is proposed to be under the control of ABA and could involve further weakening of the endosperm cap by other enzymes. If there is a second ABA-sensitive phase of tomato endosperm cap weakening, inhibition of β-1,3-glucanase expression by ABA would be consistent with the hypothesis that this enzyme is involved in the final processes that control radicle protrusion. However, while the tomato pregerminative endo-β-mannanase fusion protein (pMAL-LeMAN2; Nonogaki et al., 2000) released reducing sugars into the incubation buffer from isolated endosperm cap cell walls, the remaining wall material was not susceptible to hydrolysis by β-1,3-glucanase (Table I). Whether β-1,3-glucanase contributes to a second phase of endosperm weakening in tomato seed germination remains to be demonstrated.
On the other hand, the simultaneous and tissue-specific expression of both β-1,3-glucanase and chitinase in germinating tomato seeds suggests that they may represent a prophylactic mechanism for protection against microbial infection, as has been proposed previously (Høj et al., 1988; Fincher, 1989; Leah et al., 1991; Cordero et al., 1994; Høj and Fincher, 1995; Caruso et al., 1999; Petruzzelli et al., 1999; Morohashi and Matsushima, 2000). In addition to the physical barriers, such as lignified testae or pericarp tissues, that are effective during seed dispersal, germinating seeds may utilize physiological strategies to counter microbial invasion, as a germinating seed containing carbohydrate, protein, and lipid reserves is an attractive target for microorganisms in the surrounding soil. Because both β-1,3-glucanase and chitinase are induced specifically in the micropylar endosperm of germinating tomato seeds just prior to radicle emergence, it is feasible that they are expressed in anticipation of radicle penetration through this tissue, which will expose the inner tissues of the seed and create an avenue for entry of microorganisms into the storage reserves of the lateral endosperm. The localized expression of β-1,3-glucanase and chitinase in the endosperm tissues that will be in direct contact with the biotic environment might constitute an effective barrier to microbial entry into the seed. There is compelling evidence that β-1,3-glucanase and chitinase, acting directly by degrading pathogen cell walls or indirectly by releasing oligosaccharide elicitors of defense reactions, can help defend plants against fungal infection (e.g. Leah et al., 1991; Jach et al., 1995; Jongedijk et al., 1995; Lawrence et al., 1996). Concentrating such a defensive mechanism in the “front-line” tissues of the micropylar endosperm would obviate the need to mount a similar defense throughout the remaining endosperm tissue, particularly because the defense would only be required until the latter had been mobilized to the growing seedling.
The functions of β-1,3-glucanase and chitinase during tomato seed germination remain unresolved. Does their presence actually deter microbial infection during seed germination? Are they a wound response to cell wall hydrolysis during endosperm cap weakening? Do β-1,3-glucanase and/or chitinase release elicitor oligosaccharides that serve as signaling molecules in the regulation of the final step of radicle protrusion? What signals regulate expression of these genes to so precisely localize their appearance in space and time? To what extent do the enzymes and their localization vary from species to species in seeds with endosperm-limited germination? Further studies are in progress to address these questions.
MATERIALS AND METHODS
Plant Material and Seed Germination
Seeds of both tomato (Lycopersicon esculentum Mill. cv Moneymaker) and its isogenic GA-deficient gib-1 mutant were produced in the field in Davis, CA. After extraction and drying, the seeds were stored at −20°C until used. As described previously (Ni and Bradford, 1993), tomato seeds were sown on two layers of germination blotters in 9-cm-diameter petri dishes moistened with 12 mL of distilled water, GA4+7 (Abbott Laboratories, North Chicago), or (±) ABA (Sigma, St. Louis) solution and incubated at 25°C in the dark.
Tomato seeds were dissected into either two parts (micropylar region and the rest of seed containing the lateral endosperm and the remainder of the embryo) for protein extraction or into three parts (the endosperm cap, radicle tip, and the rest of seed) for RNA isolation (Nonogaki et al., 1992; Cooley et al., 1999) after different imbibition times at 25°C. Tissues were frozen immediately in liquid nitrogen upon dissection and stored at −80°C until used.
Preparation of Protein Extracts
Each protein sample replicate was prepared from a batch of 500 tomato seeds with three replications for each time point. Tissue extracts were prepared and clarified as described by Leubner-Metzger et al. (1995). In brief, tissues were first homogenized with a prechilled mortar and pestle and transferred to a tube containing 1 mL (for micropylar tissues) or 4 mL (for the rest of seed tissues) of extraction buffer (200 mm Tris/HCl, 0.25 mm EDTA, 5 mm 1, 4-dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride [pH 8.0]). The homogenate was centrifuged at 12,000g at 4°C for 15 min. The supernatant was retained and represented the soluble protein fraction. Protein concentration was determined by the Bradford reagent (Bio-Rad, Hercules, CA) using bovine gamma globulin as standard. All samples were stored at −80°C until used.
β-1,3-Glucanase and Chitinase Activity Assays
β-1,3-Glucanase and chitinase activities were assayed radiometrically using [3H]laminarin and [3H]chitin reduced with NaBH3 as the substrates, respectively, as described in Keefe et al. (1990). Purified tobacco (Nicotiana tabacum) class I β-1,3-glucanase and chitinase were used to calibrate the activity assays.
Protein Gel-Blot Analysis
SDS-PAGE was performed using 5% (w/v) acrylamide stacking gels and 10% (w/v) separation gels [1.5% (w/w) N,N′-methylene-bis(acrylamide)] according to Laemmli (1970). Equal volumes (5 μL) of the soluble protein extracts were applied to each lane. Purified tobacco class I β-1,3-glucanase (33 kD) or class I chitinase A and B proteins (32 or 34 kD; 10 μg per lane) were used as positive controls. Proteins were electrotransferred from SDS-PAGE gels to polyvinylidene fluoride membranes (Immobilon-P, Millipore, Bedford, MA) in 20% (v/v) methanol, 48 mm Tris, 39 mm Gly, and 0.0375% (w/v) SDS via a semidry system (Trans-Blot SD, Bio-Rad). The membranes were saturated with 5% (w/v) skim milk in phosphate buffered saline (12.5 mm sodium phosphate and 150 mm NaCl, pH 7.4) overnight, then were incubated with either rabbit anti-tobacco β-1,3-glucanase antiserum or rabbit anti-tobacco chitinase antiserum at 1:1,000 dilution in blocker at 25°C for 1 h. The tobacco β-1,3-glucanase antibody used for immunoblot analyses detects the class I, II, and III isoforms of tobacco β-1,3-glucanase (Neuhaus et al., 1992; Beffa et al., 1993); however, the tobacco chitinase antibody was specific for tobacco class I chitinase only (Shinshi et al., 1987; Beffa et al., 1995). Antigen-antibody complexes were detected by incubating with a 1:2,000 dilution of goat anti-rabbit IgG-horseradish peroxidase secondary antibody (Sigma) in blocker for 1 h at 25°C. Detection of the complexes was by chemiluminescence using the Renaissance western-blot chemiluminescence reagents (DuPont-NEN, Boston). After detection, the membranes were stained with 0.5% (w/v) amido black and destained in 40% (v/v) methanol and 10% (v/v) glacial acetic acid solution to show protein profiles.
RNA Gel-Blot Analysis
Total RNA was isolated from batches of 1,000 to 1,500 seed parts by the phenol/SDS method (Ausubel et al., 1987). The RNA concentration was determined by UV absorbance. The denatured RNA samples were fractionated by electrophoresis through 1.3% (w/v) formaldehyde-agarose gels and transferred to a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Piscataway, NJ). The hybridization probes used were: tomato class I basic intracellular β-1,3-glucanase (GluB, accession no. M80608), tomato class II acidic extracellular β-1,3-glucanase (GluA, accession no. M80604; van Kan et al., 1992), tomato class III acidic β-1,3-glucanase (Tom PR-Q'a, accession no. X7405), tomato class III basic β-1,3-glucanase (Tom PR-Q'b, accession no. X74906), tomato class I basic intracellular chitinase (Chi9, accession no. Z15140), tomato class II acidic extracellular chitinase (Chi3, accession no. Z15141), tomato class II acidic extracellular chitinase (Chi17, accession no. Z15139; Danhash et al., 1993), and tomato class II basic chitinase (Chi2;1 = Chi14, accession no. U30465; Harikrishna et al., 1996). A tomato cDNA (G46) detecting a constitutively expressed mRNA coding for a ribosomal protein (Cooley et al., 1999), was used as RNA equal-loading control. After prehybridization in 6× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate), 0.5% (w/v) SDS, 1× Denhardt's reagent, and 0.1 mg mL−1 denatured salmon sperm DNA at 68°C, the blots were hybridized with 32P-labeled riboprobes prepared from EcoRI- (GluA, GluB, Tom PR-Q'b, Chi3, and Chi17), BamHI- (Chi9 and G46), SmaI- (Tom PR-Q'a), or XhoI- (Chi2;1) linearized plasmids by in vitro transcription using MAXIscript kits (Ambion, Inc., Austin, TX). Hybridized membranes were washed with 2× SSC and 0.1% (w/v) SDS at room temperature for 30 min, and then with 1× SSC and 0.1% (w/v) SDS at 68°C for 30 min, followed by three washes with 0.2× SSC and 0.1% SDS at 68°C for 30 min, and analyzed by autoradiography (Sambrook et al., 1989). The intensity of labeling was measured with a Storm PhosphoImager (Molecular Dynamics, Sunnyvale, CA) and is expressed in arbitrary units.
Tissue Printing
Tissue printing was conducted as described previously (Nonogaki et al., 2000). Imbibed tomato seeds were longitudinally bisected and the cut surface of each half-seed was pressed onto separate positively charged nylon membranes (Hybond N+; Amersham Pharmacia Biotech) for 60 s. The digoxigenin-labeled riboprobes were synthesized separately from both sense and antisense strands of tomato class I β-1,3-glucanase (GluB; van Kan et al., 1992) or chitinase (Chi9; Danhash et al., 1993) cDNAs through in vitro transcription (Boehringer Mannheim, Indianapolis). After UV cross linking, prehybridization, hybridization, washing, and detection were performed following the manufacturer's guidelines (Boehringer Mannheim) except that the signal was colorimetrically detected using 0.025 mg mL−1 5-bromo-4-chloro-3-indolyl-phosphate, 0.1 mg mL−1 nitroblue tetrazolium, and 2 mm MgCl2 in 0.18 m Tris-HCl buffer, pH 8.8. One membrane was hybridized with the antisense probe and its counterpart print was hybridized with sense probe as a control for nonspecific binding.
cDNA Library Screening
A cDNA library constructed from mRNA from gib-1 tomato seeds imbibed in 100 μm GA4+7 for 24 h (Nonogaki et al., 2000) was screened with α-(32P)-dATP-labeled GluA or GluB probes made from a random priming reaction. Phagemids (pBK-CMV) containing cloned DNA inserts were isolated from positive λZAP plaques by in vivo excision following infection of Escherichia coli cells with R408 helper phage, according to the manufacturer's instructions (Stratagene, La Jolla, CA). The cDNAs were sequenced at the Advanced Plant Genetics Facility (University of California, Davis).
Puncture Force Determination
The strength, i.e. puncture force, of tomato endosperm caps was measured using a TA-XT2 Texture Analyzer (Stable Micro Systems, Godalming, UK) fitted with a custom-made steel probe (0.5 mm diameter) according to Groot and Karssen (1987). After different imbibition times, seeds were bisected and the embryonic axes were removed from the micropylar halves. The micropylar tip was then placed on the probe and the force required for probe penetration through the cap was measured. Each time point represents the mean of 50 measurements.
Fusion Protein Preparation
The open reading frames (except the 5′ signal peptides) encoded by tomato class I 1,3-glucanase (GluB) and chitinase (Chi9) genes were amplified by PCR using a BamHI site-linked forward primer (5′-CGGGATCCAATAGGTGTTGT-3′ and 5′-CGGGATCCGAGCAATGTGGTTCA-3′, respectively) and an XbaI site-linked reverse primer (5′-CFFFATCCGAGCAATGTGGTTCA-3′ and 5′-CGTCTAGATTACATAATATCCAAC-3′, respectively). After BamHI and XbaI double digestion, the PCR products were cloned into maltose-binding protein expression vector pMAL-c2 (New England Biolabs, Beverly, MA) and transformed into BL 21 E. coli, a proteinase-deficient strain. Transformed colonies were screening by ampicillin-selecting plates and were confirmed by restriction mapping.
The procedures of fusion protein induction and purification were carried out according to Nonogaki et al. (2000) modified from the manufacturer's manual (New England Biolabs). Overnight cultures (1% [v/v]) of the transformed bacteria were inoculated into rich broth plus ampicillin and incubated on a shaker at 300 rpm for 4 h at 37°C. Then isopropyl-β-d-thiogalactoside (Boehringer Mannheim) was added to 2 mm final concentration to induce overexpression of the fusion proteins. The cultures were grown in the same condition for 2 h. The E. coli cells were harvested by 6,000g centrifugation at 4°C and the bacterial pellets resuspended in sonication buffer (50 mm sodium phosphate buffer, pH 7.2, containing 0.3 m NaCl and 1 mg mL−1 lysozyme; Boehringer Mannheim). After −20°C overnight freezing and thawing in cold water, the lysates were clarified by centrifugation at 10,000g for 10 min. The supernatants were collected and purified through a maltose-binding protein affinity column (amylose resin, New England Biolabs). The targeted proteins were then eluted by washing the resin with column buffer containing 10 mm maltose. The fusion proteins were dialyzed against 10 mm Tris-HCl buffer (pH 7.5) overnight at 4°C to remove maltose before use. To cleave the tomato proteins from maltose-binding protein, Factor Xa (final concentration 200 μg mL−1, New England Biolabs) was added to the purified fusion protein fraction and incubated at room temperature for 4 d.
Assays for Hydrolysis of Tomato Endosperm Cap Cell Walls
Tomato endosperm cap cell walls were prepared by isolating micropylar endosperms (without testae) from MM seeds imbibed in water for no more than 3 h. After dissection, the tissue was pooled in a tube floating on liquid nitrogen and kept at −80°C until use. Endosperm cap tissues were first ground in a mortar, then washed five times each with water, 1 m NaCl, 70% (v/v) ethanol, and chloroform:methanol (3: 1, v/v) in a 1.5-mL microcentrifuge tube and dried at room temperature (Groot et al., 1988; Nonogaki et al., 2000). The wall material was resuspended in 0.1 m citrate-0.2 m phosphate buffer (pH 5.0) containing 0.2% (w/v) bovine serum albumin (Sigma) that had been passed through a 0.22-μm filter (Minipore) to eliminate microbial contamination to make a 1% (w/v) cell wall suspension solution. CM-Pachyman (carboxymethyl-substituted 1,3-glucan; Megazyme, Wicklow, UK; 1% [w/v]) and aqueous CM-chitin-RBV (carboxymethyl-substituted chitin labeled covalently with Remazol Brilliant Violet 5R; Loewe Biochemica GmbH, Munich; final concentration 1 mg mL−1) were used as defined substrates for β-1,3-glucanase and chitinase, respectively. Each assay tube contained 50 μL of the cell wall suspension or defined substrate. Enzyme amounts added to each reaction were 50 μL of pMAL-GluB (0.65 μg μL−1), pMAL-Chi9 (2.22 μg μL−1), or pMAL-LeMAN2 (0.21 μg μL−1) fusion protein (Nonogaki et al., 2000), or 20 μL of purified tobacco class I β-1,3-glucanase (3.78 μg μL−1) or class I chitinase (9.93 μg μL−1). The 33-kD class I β-1,3-glucanase and the 32-kD class I chitinase B were purified from wild-type tobacco cv Havana 425 leaves (Felix and Meins, 1985; Shinshi et al., 1987). The tobacco 34-kD class I chitinase A was purified from leaves of transgenic Nicotiana sylvestris that overproduces the tobacco chitinase (Neuhaus et al., 1991). For boiled enzyme treatments, the enzymes were placed in boiling water for 15 min before addition to assay tubes. After incubating at 30°C for 36 h, an equal volume (100 μL) of 100% cold ethanol was added to terminate the reaction, except tubes containing CM-chitin-RBV, which were stopped by adding 100 μL 1 n HCl. The cell wall samples treated with pMAL-LeMAN2 and pMAL-GluB sequential digestion were incubated with pMAL-LeMAN2 first in the same condition as above. Then, the remaining cell wall substrate was washed five times each with water and 80% (v/v) ethanol and rinsed briefly with reaction buffer. The cell wall material was resuspended in 50 μL reaction buffer and incubated with pMAL-GluB for another 36 h at 30°C. Incubation solutions were separated from insoluble residual substrate by 10,000g centrifugation for 15 min at 4°C, and 100 μL of supernatant solution was dried at 60°C in screw-cap glass tubes. The samples were hydrolyzed at 121°C for 1 h in 1 mL of 2 m trifluoroacetic acid, and the trifluoroacetate was evaporated in an air stream at 40°C (Albersheim et al., 1967; Dahal et al., 1997). The residue was dissolved in 50 μL water and reducing sugars were analyzed by the neocuproine assay using Glc as a standard (Dygert et al., 1965).
ACKNOWLEDGMENTS
The authors are grateful to Dr. Pierre J.G.M. de Wit (Wageningen Agricultural University, The Netherlands), Dr. Pablo Vera (Universidad Politécnica de Valencia, Spain), and Dr. Charles. S. Gasser (University of California, Davis) for kindly providing tomato β-1,3-glucanase and chitinase cDNAs. We also appreciate the assistance Dr. Hiroyuki Nonogaki (Oregon State University, Corvallis) in constructing the fusion protein vectors.
Footnotes
This research was supported in part by the National Science Foundation (grants IBN–9407264 and IBN–9722978 to K.J.B.).
LITERATURE CITED
- Albersheim P, Nevins DJ, English PD, Karr A. A method for analysis of sugars in plant cell wall polysaccharides by GLC. Carbohydr Res. 1967;5:340–345. [Google Scholar]
- Ausubel FM, Brent R, Kinston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 1987. [Google Scholar]
- Ballance GM, Meredith WOS, Laberge DE. Distribution and development of endo-β-glucanase activities in barley tissues during germination. Can J Plant Sci. 1976;56:459–466. [Google Scholar]
- Beffa R, Szell M, Meuwly P, Pay A, Vögeli-Lange R, Metraux J-P, Neuhaus G, Meins F, Jr, Nagy F. Cholera toxin elevates pathogen resistance and induces pathogenesis-related gene expression in tobacco. EMBO J. 1995;14:5753–5761. doi: 10.1002/j.1460-2075.1995.tb00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffa RS, Neuhaus J-M, Meins F., Jr Physiological compensation in antisense transformants: specific induction of an ersatz glucan endo-1,3-β-glucanase in plants infected with necrotizing viruses. Proc Natl Acad Sci USA. 1993;90:8792–8796. doi: 10.1073/pnas.90.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beresniewicz MM, Taylor AG, Goffinet MC, Koeller WD. Chemical nature of a semipermeable layer in seed coats of leek, onion (Liliaceae), tomato and pepper (Solanaceae) Seed Sci Technol. 1995;23:135–145. [Google Scholar]
- Bevilacqua LR, Fossati F, Dondero G. “Callose” in the impermeable seed coat of Sesbania punicea. Ann Bot. 1987;59:335–341. [Google Scholar]
- Bewley JD. Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci. 1997;2:464–469. [Google Scholar]
- Bhalla PL, Slattery HD. Callose deposits make clover seeds impermeable to water. Ann Bot. 1984;53:125–128. [Google Scholar]
- Bradford KJ, Chen F, Cooley MB, Dahal P, Downie B, Fukunaga KK, Gee OH, Gurusinghe S, Mella RA, Nonogaki H. Gene expression prior to radicle emergence in imbibed tomato seeds. In: Black M, Bradford KJ, Vazquez-Ramos J, editors. Seed Biology: Advances and Applications. Wallingford, UK: CAB International; 2000. pp. 231–251. [Google Scholar]
- Caruso C, Chilosi G, Caporale C, Leonardi L, Bertini L, Magro P, Buonocore V. Induction of pathogenesis-related proteins in germinating wheat seeds infected with Fusarium culmorum. Plant Sci. 1999;140:87–97. [Google Scholar]
- Chen F, Bradford KJ. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 2000;124:1265–1274. doi: 10.1104/pp.124.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley MB, Yang H, Dahal P, Mella RA, Downie B, Haigh AM, Bradford KJ. Expression of vacuolar H+-ATPase in response to gibberellin during tomato seed germination. Plant Physiol. 1999;121:1339–1347. doi: 10.1104/pp.121.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero MJ, Raventos D, San Segundo B. Differential expression and induction of chitinases and β-1,3-glucanases in response to fungal infection during germination of maize seeds. Mol Plant-Microbe Interact. 1994;7:23–31. [Google Scholar]
- Dahal P, Bradford KJ. Effects of priming and endosperm integrity on seed germination rates of tomato genotypes: II. Germination at reduced water potential. J Exp Bot. 1990;41:1441–1453. [Google Scholar]
- Dahal P, Nevins DJ, Bradford KJ. Relationship of endo-β-D-mannanase activity and cell wall hydrolysis in tomato endosperm to germination rates. Plant Physiol. 1997;113:1243–1252. doi: 10.1104/pp.113.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhash N, Wagemakers CAM, van Kan JAL, de Wit PJGM. Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Mol Biol. 1993;22:1017–1029. doi: 10.1007/BF00028974. [DOI] [PubMed] [Google Scholar]
- Domingo C, Conejero V, Vera P. Genes encoding acidic and basic class III β-1,3-glucanases are expressed in tomato plants upon viroid infection. Plant Mol Biol. 1994;24:725–732. doi: 10.1007/BF00029854. [DOI] [PubMed] [Google Scholar]
- Domon J-M, Neutelings G, Roger D, David A, David H. A basic chitinase-like protein secreted by embryogenic tissues of Pinus caribaea acts on arabinogalactan proteins extracted from the same cell lines. J Plant Physiol. 2000;156:33–39. [Google Scholar]
- Dygert S, Li LH, Florida D, Thoma JA. Determination of reducing sugars with improved precision. Anal Biochem. 1965;13:367–374. doi: 10.1016/0003-2697(65)90327-1. [DOI] [PubMed] [Google Scholar]
- Felix G, Meins F., Jr Purification, immunoassay and characterization of an abundant cytokinin-regulated polypeptide in cultured tobacco tissues: evidence the protein is a β-1,3-glucanase. Planta. 1985;164:423–428. doi: 10.1007/BF00402956. [DOI] [PubMed] [Google Scholar]
- Fincher GB. Molecular and cellular biology associated with endosperm mobilization in germinating cereal grains. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:305–346. [Google Scholar]
- Fischer W, Christ U, Baumgartner M, Erismann KH, Mosinger E. Pathogenesis-related proteins of tomato: II. Biochemical and immunological characterization. Physiol Mol Plant Pathol. 1989;35:67–83. [Google Scholar]
- Groot SPC, Karssen CM. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta. 1987;171:525–531. doi: 10.1007/BF00392302. [DOI] [PubMed] [Google Scholar]
- Groot SPC, Kieliszewska-Rokicka B, Vermeer E, Karssen CM. Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seeds prior to radicle protrusion. Planta. 1988;174:500–504. doi: 10.1007/BF00634479. [DOI] [PubMed] [Google Scholar]
- Harikrishna K, Jampates-Beale R, Milligan SB, Gasser C. An endochitinase gene expressed at high levels in the stylar transmitting tissue of tomatoes. Plant Mol Biol. 1996;30:899–911. doi: 10.1007/BF00020802. [DOI] [PubMed] [Google Scholar]
- Hilhorst HWM, Groot SPC, Bino RJ. The tomato seed as a model system to study seed development and germination. Acta Bot Neerl. 1998;47:169–183. [Google Scholar]
- Høj PB, Fincher GB. Molecular evolution of plant β-glucan endohydrolases. Plant J. 1995;7:367–379. doi: 10.1046/j.1365-313x.1995.7030367.x. [DOI] [PubMed] [Google Scholar]
- Høj PB, Slade AM, Wettenhall REH, Fincher GB. Isolation and characterization of a (1→3)-β-glucan endohydrolase from germinating barley (Hordeum vulgare): amino acid sequence similarity with barley (1→3, 1→4)-β-glucanases. FEBS Lett. 1988;230:67–71. [Google Scholar]
- Jach G, Gornhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Maas C. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 1995;8:97–109. doi: 10.1046/j.1365-313x.1995.08010097.x. [DOI] [PubMed] [Google Scholar]
- Jongedijk E, Tigelaar H, van Roekel JSC, Bres-Vloemans SA, Dekker I, van den Elzen PJM, Cornelissen BJC, Melchers LS. Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica. 1995;85:173–180. [Google Scholar]
- Karssen CM, Haigh A, van der Toorn P, Weges R. Physiological mechanisms involved in seed priming. In: Taylorson RB, editor. Recent Advances in the Development and Germination of Seeds. New York: Plenum Press; 1989. pp. 269–280. [Google Scholar]
- Kauffmann S, Legrand M, Geoffroy P, Fritig B. Biological function of pathogenesis-related proteins: four PR proteins of tobacco have 1,3-β glucanase activity. EMBO. 1987;6:3209–3212. doi: 10.1002/j.1460-2075.1987.tb02637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe D, Hinz U, Meins F., Jr The effect of ethylene on the cell-type-specific and intracellular localization of β-1,3-glucanases and chitinase in tobacco leaves. Planta. 1990;182:43–51. doi: 10.1007/BF00239982. [DOI] [PubMed] [Google Scholar]
- Kombrink E, Somssich IE. Pathogenesis-related proteins and plant defense. In: Carroll GC, Tudzynski P, editors. The Mycota V. Plant Relationships Part A. Berlin: Springer-Verlag; 1997. pp. 107–128. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawrence CB, Joosten MHAJ, Tuzun S. Differential induction of pathogenesis-related proteins in tomato by Alternaria solani and the association of a basic chitinase isozyme with resistance. Physiol Mol Plant Pathol. 1996;48:361–377. [Google Scholar]
- Leah R, Tommerup H, Svendsen I, Mundy J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem. 1991;266:1564–1573. [PubMed] [Google Scholar]
- Leubner-Metzger G, Fründt C, Meins F., Jr Effects of gibberellins, darkness and osmotica on endosperm rupture and class I β-1,3-glucanase induction in tobacco seed germination. Planta. 1996;199:282–288. [Google Scholar]
- Leubner-Metzger G, Fründt C, Vögeli-Lange R, Meins F., Jr Class I β-1,3-glucanases in the endosperm of tobacco during germination. Plant Physiol. 1995;109:751–759. doi: 10.1104/pp.109.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G, Meins F., Jr . Functions and regulation of plant β-1,3-glucanases (PR-2) In: Datta SK, Muthukrishnan S, editors. Pathogenesis-Related Proteins in Plants. Boca Raton, FL: CRC Press; 1999. pp. 49–76. [Google Scholar]
- Leubner-Metzger G, Meins F., Jr Sense transformation reveals a novel role for class I β-1,3-glucanases in tobacco seed germination. Plant J. 2000;23:215–221. doi: 10.1046/j.1365-313x.2000.00773.x. [DOI] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA. The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 2000;122:3–9. doi: 10.1104/pp.122.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle TJ. Tissue print hybridization: a simple technique for detecting organ- and tissue-specific gene expression. Plant Mol Biol. 1989;12:517–524. doi: 10.1007/BF00036966. [DOI] [PubMed] [Google Scholar]
- Meins F, Jr, Neuhaus J-M, Sperisen C, Ryals J. The primary structure of plant pathogenesis-related glucanohydrolases and their genes. In: Boller T, Meins Jr F, editors. Genes Involved in Plant Defense. Berlin: Springer-Verlag; 1992. pp. 245–282. [Google Scholar]
- Morohashi Y, Matsushima H. Development of β-1,3-glucanase activity in germinated tomato seeds. J Exp Bot. 2000;51:1381–1387. [PubMed] [Google Scholar]
- Neuhaus J-M. Plant chitinases. In: Datta SK, Muthukrishnan S, editors. Pathogenesis-Related Proteins in Plants. Boca Raton, FL: CRC Press; 1999. pp. 77–105. [Google Scholar]
- Neuhaus J-M, Flores S, Keefe D, Ahl-Goy P, Meins F., Jr The function of vacuolar β-1,3-glucanase investigated by antisense transformation: susceptibility of transgenic Nicotiana sylvestris plants to Cercospora nicotianae infection. Plant Mol Biol. 1992;19:803–813. doi: 10.1007/BF00027076. [DOI] [PubMed] [Google Scholar]
- Neuhaus J-M, Sticher L, Meins F, Jr, Boller T. A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci USA. 1991;88:10362–10366. doi: 10.1073/pnas.88.22.10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B-R, Bradford KJ. Germination and dormancy of abscisic acid- and gibberellin-deficient mutant tomato (Lycopersicon esculentum) seeds: sensitivity of germination to abscisic acid, gibberellin, and water potential. Plant Physiol. 1993;101:607–617. doi: 10.1104/pp.101.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Gee OH, Bradford KJ. A germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol. 2000;123:1235–1245. doi: 10.1104/pp.123.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Matsushima H, Morohashi Y. Galactomannan hydrolyzing activity develops during priming in the micropylar endosperm tip of tomato seeds. Physiol Plant. 1992;85:167–172. [Google Scholar]
- Nonogaki H, Nomaguchi M, Okumoto N, Kaneko Y, Matsushima H, Morohashi Y. Temporal and spatial pattern of the biochemical activation of the endosperm during and following imbibition of tomato seeds. Physiol Plant. 1998;102:236–242. [Google Scholar]
- Petruzzelli L, Kunz C, Waldvogel R, Meins F, Jr, Leubner-Metzger G. Distinct ethylene- and tissue-specific regulation of β-1,3-glucanases and chitinases during pea seed germination. Planta. 1999;209:195–201. doi: 10.1007/s004250050622. [DOI] [PubMed] [Google Scholar]
- Rezzonico E, Flury N, Meins F, Jr, Beffa R. Transcriptional down-regulation by abscisic acid of pathogenesis-related β-1,3-glucanase genes in tobacco cell cultures. Plant Physiol. 1998;117:585–592. doi: 10.1104/pp.117.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shinshi H, Mohnen D, Meins F., Jr Regulation of a plant pathogenesis-related enzyme: inhibition of chitinase and chitinase mRNA accumulation in cultured tobacco tissues by auxin and cytokinin. Proc Natl Acad Sci USA. 1987;84:89–93. doi: 10.1073/pnas.84.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CR. The physiology and molecular biology of plant 1,3-β-D-glucanases and 1,3–1,4-β-D-glucanases. Crit Rev Plant Sci. 1994;13:325–387. [Google Scholar]
- Sticher L, Hofsteenge J, Milani A, Neuhaus J-M, Meins F., Jr Vacuolar chitinases of tobacco: a new class of hydroxyproline-containing proteins. Science. 1992;257:655–657. doi: 10.1126/science.1496378. [DOI] [PubMed] [Google Scholar]
- Still DW, Bradford KJ. Endo-β-mannanase activity from individual tomato endosperm caps and radicle tips in relation to germination rates. Plant Physiol. 1997;113:21–29. doi: 10.1104/pp.113.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorop PE, Bewley JD, Hilhorst HWM. Endo-β-mannanase isoforms are present in the endosperm and embryo of tomato seeds, but are not essentially linked to the completion of germination. Planta. 1996;200:153–158. [Google Scholar]
- Toorop PE, van Aelst AC, Hilhorst HWM. The second step of the biphasic endosperm cap weakening that mediates tomato (Lycopersicon esculentum) seed germination is under control of ABA. J Exp Bot. 2000;51:1371–1379. [PubMed] [Google Scholar]
- van Kan JAL, Cozijnsen T, Danhash N, de Wit PJGM. Induction of tomato stress protein mRNAs by ethephon, 2,6-dichloroisonicotinic acid and salicylate. Plant Mol Biol. 1995;27:1205–1213. doi: 10.1007/BF00020894. [DOI] [PubMed] [Google Scholar]
- van Kan JAL, Joosten MHAJ, Wagemakers CAM, van den Berg-Velthuis GCM, de Wit PJGM. Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol Biol. 1992;20:513–527. doi: 10.1007/BF00040610. [DOI] [PubMed] [Google Scholar]
- van Loon LC. Occurrence and properties of plant pathogenesis-related proteins. In: Datta SK, Muthukrishnan S, editors. Pathogenesis-Related Proteins in Plants. Boca Raton, FL: CRC Press; 1999. pp. 1–19. [Google Scholar]
- Vögeli-Lange R, Fründt C, Hart CM, Beffa R, Nagy F, Meins F., Jr Evidence for a role of β-1,3-glucanase in dicot seed germination. Plant J. 1994;5:273–278. [Google Scholar]
- Yim KO, Bradford KJ. Callose deposition is responsible for apoplastic semipermeability of the endosperm envelope of muskmelon seeds. Plant Physiol. 1998;118:83–90. doi: 10.1104/pp.118.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]