Abstract
Effective sampling for severe acute respiratory syndrome 2 (SARS-CoV-2) is a common approach for monitoring disinfection efficacy and effective environmental surveillance. This study evaluated sampling efficiency and limits of detection (LODs) of macrofoam swab and sponge stick sampling methods for recovering infectious SARS-CoV-2 and viral RNA (vRNA) from surfaces. Macrofoam swab and sponge stick methods were evaluated for collection of SARS-CoV-2 suspended in a soil load from 6-in2 coupons composed of four materials: stainless steel (SS), acrylonitrile butadiene styrene (ABS) plastic, bus seat fabric, and Formica. Recovery of infectious SARS-CoV-2 was more efficient than vRNA recovery on all materials except Formica (macrofoam swab sampling) and ABS (sponge stick sampling). Macrofoam swab sampling recovered significantly more vRNA from Formica than ABS and SS, and sponge stick sampling recovered significantly more vRNA from ABS than Formica and SS, suggesting that material and sampling method choice can affect surveillance results. Time since initial contamination significantly affected infectious virus recovery from all materials, with vRNA recovery showing limited to no difference, suggesting that SARS-CoV-2 vRNA can remain detectable after viral infectivity has dissipated. This study showed that a complex relationship exists between sampling method, material, time from contamination to sampling, and recovery of SARS-CoV-2. In conclusion, data show that careful consideration be used when selecting surface types for sampling and interpreting SARS-CoV-2 vRNA recovery with respect to presence of infectious virus.
Keywords: Limit of detection, sampling efficiency, SARS-CoV-2 recovery, surface sampling
Introduction
The ability to reliably detect the presence of an infectious pathogen is a key component of effective infection control strategies to monitor and respond to disease outbreaks. During past outbreaks as seen with severe acute respiratory syndrome–associated coronavirus (SARS-CoV), Middle East respiratory syndrome, and the coronavirus disease 2019 (COVID-19) pandemic (SARS-CoV-2), critical data were collected on the environmental persistence of CoV in aerosols (Liu et al. 2020; Dabisch et al. 2021), in water (Tran et al. 2021), and on contaminated surfaces (Van Doremalen Neeltje et al. 2020; Ardura et al. 2021; Harvey et al. 2021; Marcenac et al. 2021). Though the dominant route of transmission for SARS-CoV-2 is direct contact with respiratory droplets, aerosols, or mucosal secretions from infectious individuals (CDC 2021c; Peng et al. 2022), contaminated environmental surfaces in community and other environments (e.g., hospital rooms, schools, or public transportation) may also play a role in indirect transmission (CDC 2021b).
Studies on the persistence of infectious coronaviruses on surfaces and the role these surfaces play in viral transmission have explored viral stability and survival (Mahl and Sadler 1975; Sizun et al. 2000; Marques and Domingo 2021). Middle East respiratory syndrome and endemic human coronaviruses were shown to be able to persist as long as 9 days (Kampf et al. 2020) and infectious SARS-CoV-2 as long as 3 days (Van Doremalen Neeltje et al. 2020) after deposition onto surfaces. For SARS-CoV-1, fomite transmission was associated with nosocomial spread (Chen et al. 2004). And though the risk of infection via fomite transmission is likely low, infection via contact with contaminated surfaces or fomite transmission of SARS-CoV-2 is still possible (CDC 2021b).
Interpreting respiratory virus sampling studies is challenging because many sampling strategies target either viral nucleic acid (e.g., viral RNA [vRNA]) or infectious virus, with few employing an integrated approach. Strategies using vRNA recovery are most common because vRNA kits are widely available, rapid, and comparatively cheap; however, multiple studies have identified that vRNA presence and concentration do not correlate with an increased presence of infectious virus (Colaneri et al. 2020; Santarpia et al. 2020; Yamagishi et al. 2020; Moore et al. 2021; Zhou et al. 2021). Recently, sampling the exhaled breath of a patient with COVID-19 did not result in detection of infectious virus, despite the confirmation of very high loads of vRNA (Johnson et al. 2022) detected in those samples. This disconnect between vRNA and infectious virus is true for many viral agents, including Ebola; high quantities of Ebola vRNA can be detected but do not correlate with infectious Ebola virus on disinfected surfaces (Cook et al. 2016).
An additional complicating factor in interpreting surveillance studies is the breadth of methods employed. A systematic review of primary studies on SARS-CoV-2 sampling from fomites (in 2020 alone) found significant discrepancies in viral culture and isolation methods, and none of the studies reported successful viral culture despite positive reverse transcription quantitative polymerase chain reaction (RT-qPCR) detection of SARS-CoV-2 vRNA (Onakpoya et al. 2021). Though many of these studies employed swab sampling for vRNA or to detect infectious virus on surfaces (Pena et al. 2021), other methods included sponge (Fernández-de-Mera et al. 2021) or sterile gauze (Santarpia et al. 2020) sampling methods. Despite swab studies sharing similar methodology, variations in moistening agent (e.g., viral transport media, cell culture media, extraction media, saline solutions) and swab type (e.g., Dacron swab, macrofoam swab, flocked swabs) make direct comparisons difficult (Chia et al. 2020; Pasquarella et al. 2020; Razzini et al. 2020; Pena et al. 2021; Zhou et al. 2021).
Developing a unifying method able to detect both vRNA and infectious virus on high-touch surfaces is a key need to enable rapid data collection, comparable across studies, to support the identification of transmission routes and contamination locations and to monitor and identify effective countermeasure strategies. Because no surface sampling method for SARS-CoV-2 had been proven at the onset of this work, methods were adapted from validated norovirus (macrofoam swab; Scherer et al. 2009; Park et al. 2015; Park et al. 2017; Silvestri et al. 2021) and anthrax (cellulose sponge stick; Rose et al. 2011; CDC 2012; Silvestri et al. 2021) sampling methods. Macrofoam swab and sponge stick sampling methods are simple to perform, commercially available, evaluated on multiple surface types, validated for sampling biological agents (albeit not SARS-CoV-2), and have sampling protocols available (CDC 2012; Park et al. 2017). These methods are also commonly used for sampling large surface areas (e.g., walls, desktops, carpet) common to high-touch community environments (Park et al. 2017).
This study evaluated macrofoam swab (hereafter swab) and sponge stick (hereafter sponge) for their ability to sample SARS-CoV-2 contamination (infectious and vRNA) on high-touch surfaces common to community and other environmental surfaces (stainless steel [SS], acrylonitrile butadiene styrene [ABS] plastic, Formica, and bus seat fabric [SF]). Using varied SARS-CoV-2 contamination levels, this study sought to better understand the efficiency of both sampling (swab and sponge) and analysis (vRNA and infectious virus) methods for SARS-CoV-2 detection. Overall, this study sought to inform how sampling method and analysis choice impact how data can be used to monitor transmission risk and countermeasure efficacy.
Methods
Cells and virus
SARS-CoV-2 (isolate USA-WA1/2020) was obtained from the ATCC (American Type Culture Collection), and all work with SARS-CoV-2 was performed within a class II biosafety cabinet within a biosafety level 3 (BSL-3) facility (Battelle Eastern Science and Technology Facility) with appropriate personal protective equipment and appropriate training. Vero cells (ATCC CCL-81, derived from African green monkey kidney) were used for SARS-CoV-2 infectivity assays and were maintained and propagated as described previously (Hardison, Ryan, et al. 2022). Briefly, Vero cells were maintained in complete growth medium (CGM; Dulbecco’s minimal essential medium [DMEM]) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. SARS-CoV-2 for coupon inoculation was prepared with a 5% soil load. SARS-CoV-2 stocks were diluted 60:40 (volume:-volume) in either DMEM (5% FBS) or simulated saliva (5% FBS). Simulated saliva was prepared as described previously (Woo et al. 2010) and modified to include a 10-fold increase in phosphates (final concentration of 15 mM KH2PO4, 24.6 mM K2HPO4). Viral inoculum titer was determined by 50% tissue culture infectious dose (TCID50) assays on Vero cells for each test.
Murine hepatitis virus strain A59 (MHV-A59) and L2 cells were kindly provided by Dr. Julian Leibowitz (Texas A&M College of Medicine, College Station, TX) and 17 Clone 1 (17CL-1) cells were provided by Dr. Susan Baker (Loyola University, Chicago, IL). MHV-A59 was titrated on L2 cells and propagated in 17CL-1 cells as described previously (Hardison, Nelson, et al. 2022). Briefly, cells were maintained and propagated in CGM supplemented with Glutamax at 37 °C, 5% CO2. MHV-A59 was propagated in 17CL-1 cells at a multiplicity of infection of 0.3 in low-FBS media (DMEM with Glutamax, 2% FBS, 1% penicillin-streptomycin). Post-inoculation, virus was adsorbed at room temperature for 1 hr; post-adsorption, low-FBS media was added to each flask and incubated at 37 °C, 5% CO2 until the flask reached 80% to 90% cytopathic effect. Infected flasks were directly frozen (−80 °C), thawed at room temperature, and contents were clarified (2,000 × g, 4 °C, 20 min). Clarified viral lysate was aliquoted and stored at −80 °C in single-use (1 mL) vials.
Coupon materials
Coupon materials were obtained and prepared as described previously (Hardison, Nelson, et al. 2022; Hardison, Ryan, et al. 2022). For this study, fatigue-resistant 301 SS (0.01 in. thick; C40 Rockwell hardness; meeting ASTM A666 specifications) and impact-resistant ABS plastic (3/8 in. thick; R101–R109 hardness; meeting UL 94 HB specifications) were purchased from McMaster-Carr, Formica laminate was purchased from Home Depot, and SF was sourced from American Seating (Item # 00333uw1). Materials used as simulated contaminated surfaces included fatigue-resistant 301 SS (0.01 in. thick, C40 hardness; ASTM A666 specifications), impact-resistant ABS plastic (3/8 in. thick, R101–R109 hardness, UL 94 HB specifications), Formica laminate, and SF (# 00333uw1), cut into medium (6-in2) and large (10-in2) coupons. After cutting, coupon materials were prepared as described previously (Hardison, Nelson, et al. 2022; Hardison, Ryan, et al. 2022). Briefly, SS and ABS coupons were cleaned with 70% isopropanol and then wiped with a cloth. SF coupons were not cleaned and were used as received after cutting. After cleaning, coupons were air-dried and packaged in heat-sealed polyethylene and sterilized by electron beam with a dose of 40 kGy. Coupons were used only once for testing.
Coupon size selection
MHV-A59 was used to evaluate whether coupon surface area affected viral recovery via swab and sponge sampling methods. Viral recovery from large (10 × 10 in., 645.2 cm2) and medium (6 × 6 in., 232.2 cm2) SS coupons was performed as described. Three concentrations of MHV-A59 (1.31 × 106, 1.39 × 104, and 1.20 × 102 TCID50), applied to individual medium and large SS coupons, were used to evaluate whether coupon surface area affected viral recovery using swab and stick sampling methods. Inoculation, sampling, and recovery procedures were similar to those described in sections below for SARS-CoV-2. No significant differences were noted in either percentage recovery (Supplemental Table S1, Figure S1) or log reduction in inoculum (Supplemental Table S1) between stick or swab sampling methods; however, the initial inoculum concentration did affect MHV-A59 recovery (p = 0.028, three-way analysis of variance); this effect was independent of method and coupon size and was explored during testing with SARS-CoV-2. These results informed the decision to perform SARS-CoV-2 testing using 6-in. × 6-in. coupons to improve throughput and operations under BSL-3.
Coupon inoculation
Coupons were inoculated in triplicate with viral inoculum (SARS-CoV-2 in simulated saliva) in evenly spaced droplets across each coupon surface (1.0 mL inoculum per coupon). Coupons were inoculated after aseptically laying them flat in secondary containment in a biosafety cabinet. Inoculum concentrations were chosen across a range to identify detection limits and recovery efficacy for each material and sampling method. Initial testing coupons (Figure 1) were inoculated with a target of 1E + 05 TCID50 SARS-CoV-2 per coupon (actual: 4.87E + 04 ± 5.71E + 03 to 2.46E + 05 ± 6.74E + 03 TCID50/coupon; 2.2E + 07 to 8.86E + 07 genome copies [GC]/coupon). Blank coupons and inoculum direct-spike controls were included during each test, to allow detection of cross-contamination and drifts in the titers of viral working stocks.
Figure 1.
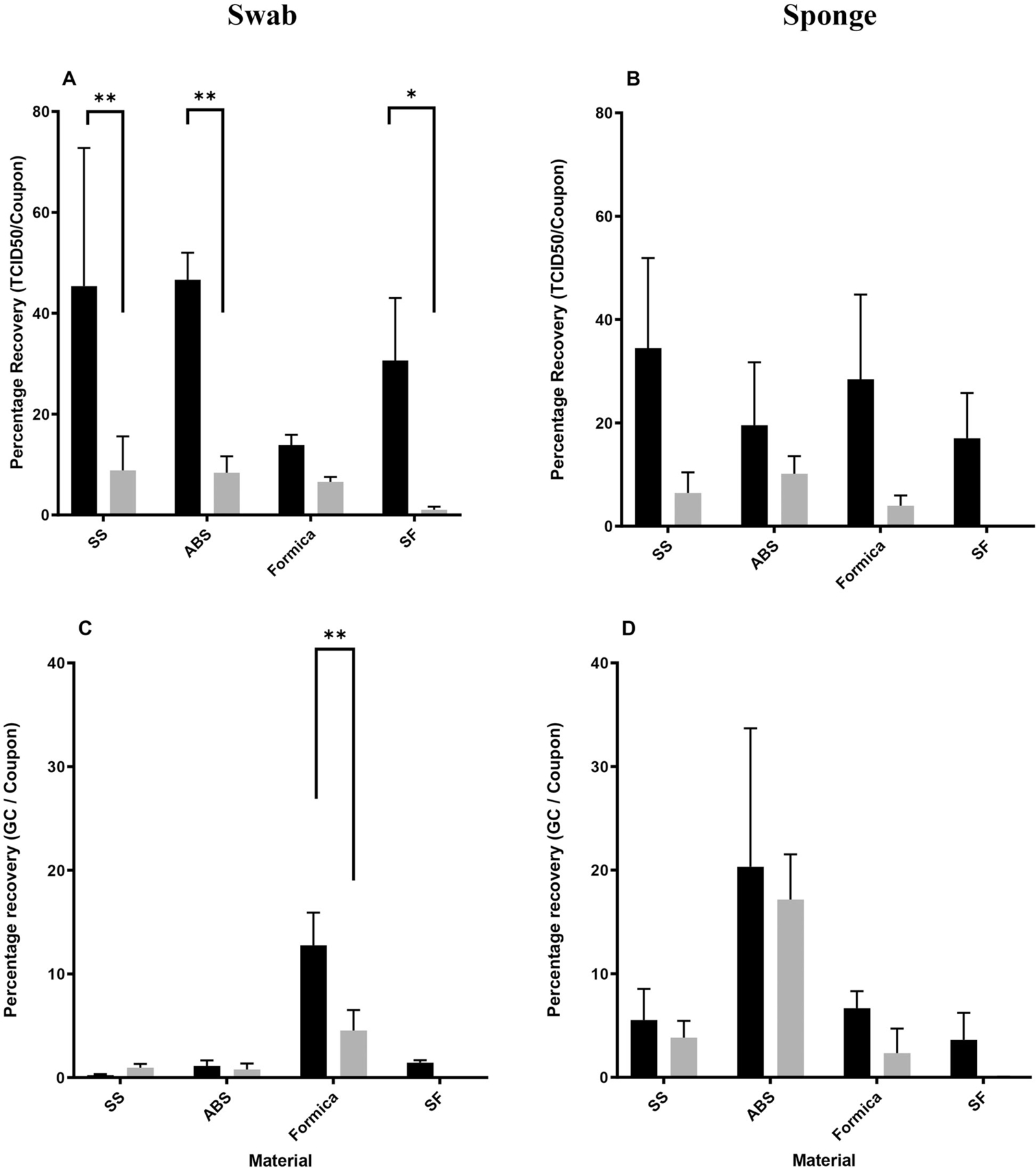
Recovery of (A, B) infectious SARS-CoV-2 and (C, D) vRNA collected from coupons inoculated with SARS-CoV-2 (4.87E + 04 ± 5.71E + 03 TCID50/coupon – 2.46E + 05 ± 6.74E + 03 TCID50/coupon) and sampled via (A, C) macrofoam swab and (B, D) sponge methods. Percentage recovery (mean, standard deviation, n = 3–6) for each material and sampling method was determined using recovered per coupon values over inoculum values. Data represented were recovered at T0 (black bars) and T3 (gray bars). Data are shown as percentage recovery of (A, B) TCID50/coupon or (C, D) GC/coupon (genome copy). *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Sampling methods
Coupons were sampled immediately (T0) or after 3 hr (T3), during which time samples were left undisturbed in the biosafety cabinet at ambient temperature and relative humidity (21.7 °C ± 0.3 °C, 50% ± 2% RH) monitored with Hobo loggers. Swab sampling used dry swabs packed individually. Swabs were soaked overnight at room temperature in 10 mL phosphate-buffered saline, and sampling was performed as described previously (Park et al. 2017). Briefly, the swab was moved across the coupon in horizontal, vertical, and diagonal patterns with a firm uniform force. Polyurethane StickSponges were 1.5 × 3 in. in size and suspended in 10 mL neutralizing buffer. Briefly, the widest part of the wet sponge was wiped on the coupon in an overlapping S pattern, rotating the sponge, in vertical (wide side of the sponge), horizonal (opposite wide side), and diagonal (narrow sides) strokes. Finally, the sponge top was used to wipe the perimeter of the coupon.
Coupon extraction
Swabs and sponges were extracted immediately post-sampling. Swab extraction initiated by adding the swab to 5 mL low-FBS DMEM (DMEM supplemented with 2% FBS) followed by vortexing (2 min; 10-sec bursts) at maximum speed (~3,200 rpm). Next, both sides of the swab were expressed along conical tube sides and the swab head was discarded. Sample extracts were inverted three times and passed through Sephadex G-25 via centrifugation (2 min; 1,000 rcf). Stick extraction used a different method. Stick heads were ejected into 45 mL of low-FBS DMEM within a stomacher bag, followed by a 1-min stomach session at ~260 rpm. Extracts were concentrated to 5 mL via 100K MWCO Protein Concentrator followed by Sephadex G-25 filtration as described previously (Hardison, Ryan, et al. 2022). Extracts were immediately tested for viral titer via TCID50, with an aliquot kept at −80 °C for analysis.
Quantification of infectious virus
Virus recovered during testing was quantified by TCID50 assay as previously described (Hardison, Ryan, et al. 2022). Briefly, Vero cells were seeded in 96-well plates in CGM to achieve 80% to 85% confluency the next day. Serially diluted sample extracts (in DMEM supplemented with 2% FBS) were plated (0.1 mL) onto replicate wells of Vero cell 96-well plates (n = 12) and incubated at 37 °C and 5% CO2. Plates were scored for cytopathic effect 48 hr post-infection and viral titer was determined via the Reed-Muench method (Reed and Muench 1938). The TCID50 assay limit of detection (LOD) was 1.8 log10 TCID50 per mL. Percentage recovery of infectious material (Figure 1) was determined by calculating normalized recovery (recovered TCID50/coupon divided by inoculated TCID50/coupon). MHV was titered on L2 cells using TCID50 assays as previous described (Hardison, Nelson, et al. 2022), with results calculated via the Reed-Muench method.
RNA extraction and RT-qPCR
Sample extracts were inactivated prior to removal from the BSL-3 facility via incubation at 75 °C for 45 min (CDC 2021a). Protocol validated as guided by the U.S. Federal Select Agent Program guidance. vRNA was used to extract samples using the QIAamp Viral RNA Extraction kit (140 μL sample) with an 80 μL final eluent suspended in Qiagen Buffer AVE. RT-qPCR was performed using 5 μL of eluent in a final volume of 20 μL per reaction. Reactions were performed using the TaqMan Fast Virus 1-Step Master Mix with a FAM-labeled 60× primer/probe mix specific for the SARS-CoV-2 N1 gene as previously described (Gabitzsch et al. 2021). Standard curves, generated from serial dilutions of synthetic SARS-CoV-2 RNA, were used to calculate GC from test sample Ct values using the standard curve. This value was then converted to GC/coupon by accounting for sample and extraction volume.
Limit of detection calculation
Recovery values (TCID50/coupon or GC/coupon) were plotted against inoculation concentrations, and linear regression was used to generate curves (Supplemental Figure S2). The LOD for each data set was defined as the point where the inoculation concentration (x-value) intersects the assay LOD; the TCID50 assay had an LOD of 64 TCID50/coupon (1.8 log TCID50/mL) and the RT-qPCR assay had a LOD of 4900 GC (3.69 log GC/coupon). LODs were determined for each material and time point (Table 1) using linear regression results (Supplemental Table S2). Assay LODs are represented in Figures 2 to 5 as a dashed line. Assay LOD values are 1.8 log TCID50/coupon for infectious virus recovery and 3.69 log GC/coupon for RT-qPCR.
Table 1.
LOD of infectious and vRNA SARS-CoV-2 for each material and time point tested.
| SARS-CoV-2 surface inoculum |
SARS-CoV-2 recovery |
||||||
|---|---|---|---|---|---|---|---|
| Log10 TCID50 |
Log10 vRNA GC |
Log10 TCID50 |
Log10 vRNA GC |
||||
| Material | Method | Time point | n | Range | Range | LOD | LOD |
| SS | Swab | T 0 | 3 | 2.41–4.73 | 4.91–7.71 | 2.06 | 4.85 |
| T 3 | 6 | 2.41–4.73 | 4.91–7.71 | 2.98 | 4.76 | ||
| Sponge | T 0 | 6 | 2.93–5.29 | 5.63–7.90 | 2.52 | 4.83 | |
| T 3 | 6 | 2.93–5.29 | 5.63–7.90 | 3.33 | 5.24 | ||
| ABS | Swab | T 0 | 6 | 2.67–4.69 | 4.15–7.34 | 2.51 | 5.01 |
| T 3 | 6 | 2.67–4.69 | 5.28–7.34 | 2.99 | 4.39 | ||
| Sponge | T 0 | 6 | 2.63–5.25 | 5.50–7.78 | 2.15 | 4.76 | |
| T 3 | 6 | 2.63–5.25 | 5.50–7.78 | 2.30 | 4.73 | ||
| Formica | Swab | T 0 | 3 | 3.09–5.34 | 5.64–7.73 | 2.30 | 5.12 |
| T 3 | 6 | 3.09–5.34 | 5.64–7.73 | 2.04 | 3.92 | ||
| Sponge | T 0 | 5 | 3.01–5.29 | 5.74–7.94 | 2.39 | 4.97 | |
| T 3 | 5 | 3.01–5.29 | 5.74–7.94 | 2.50 | 5.21 | ||
| SF | Swab | T 0 | 3 | 3.12–5.07 | 5.51–7.50 | 2.50 | 5.41 |
| T 3 | 6 | 3.12–5.58 | 5.51–8.36 | 4.54 | 6.45 | ||
| Sponge | T 0 | 6 | 3.01–5.29 | 5.85–8.13 | 2.77 | 5.19 | |
| T 3 | 6 | 3.01–5.29 | 5.85–8.13 | 5.71a | 6.91 | ||
Infectious SARS-CoV-2 LOD is represented as log TCID50/coupon and vRNA LOD is represented as GC/coupon. LODs listed here are caculated from the regression of each set of data (Figures 2–5, Supplemental Table S2).
LOD is extrapolated from a poor-fitting curve and should be viewed as an estimate.
Figure 2.
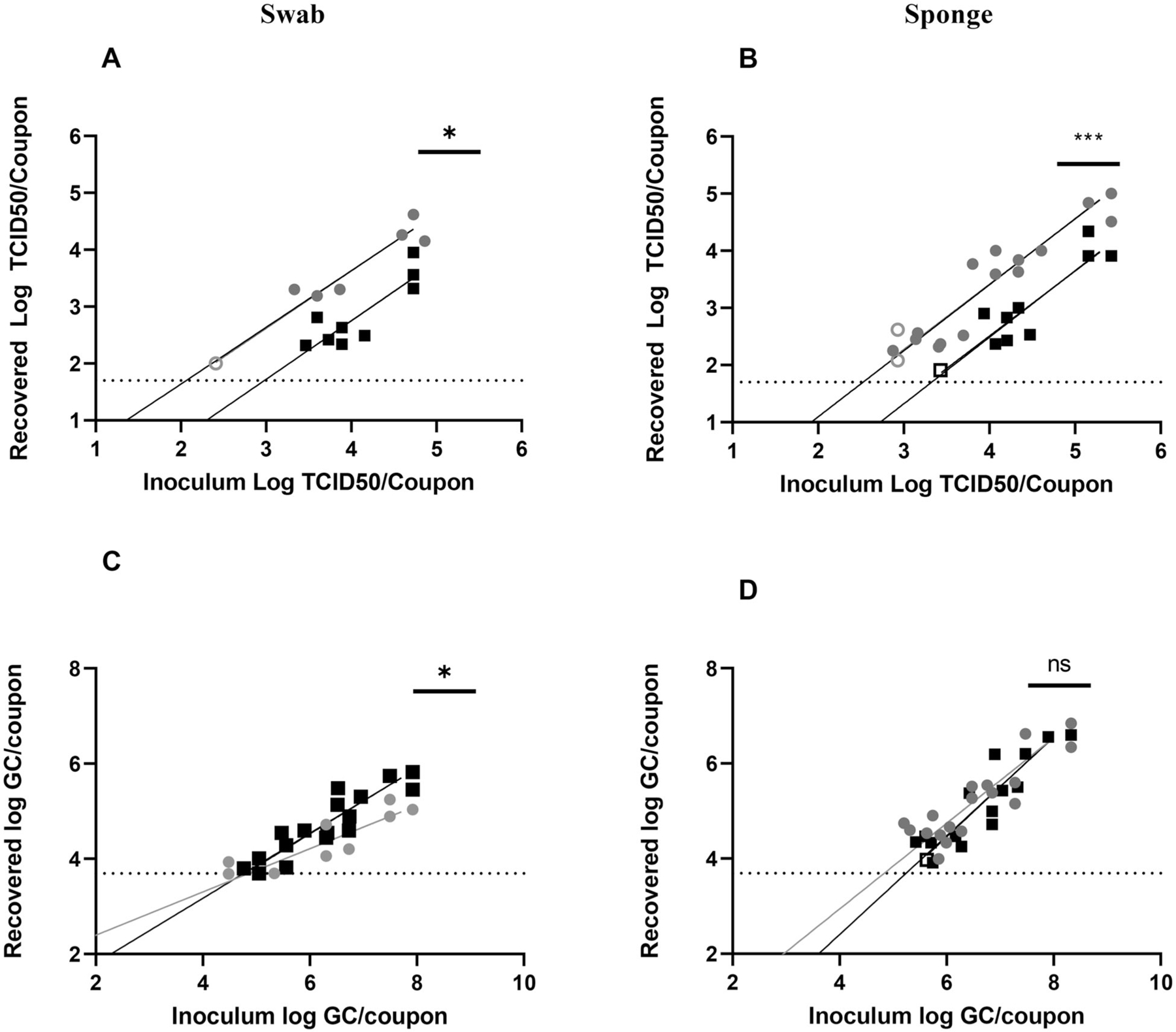
Recovery of SARS-CoV-2 from SS using (A, C) swab and (B, D) sponge methods. Results from T0 recovery (gray circles) or T3 recovery (black squares). Open circles or squares identify samples in which at least one replicate was below the LOD. Data shown on graphs represent individual recovery values. Linear regression lines, shaded in gray (T0) or black (T3), are shown. Details of each linear regression are shown in Supplemental Table S2. Bars on each graph represent significant differences between recovery at T0 and T3, as determined by linear regression comparison (Graphpad Prism v9.3.1). ns: not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Figure 5.
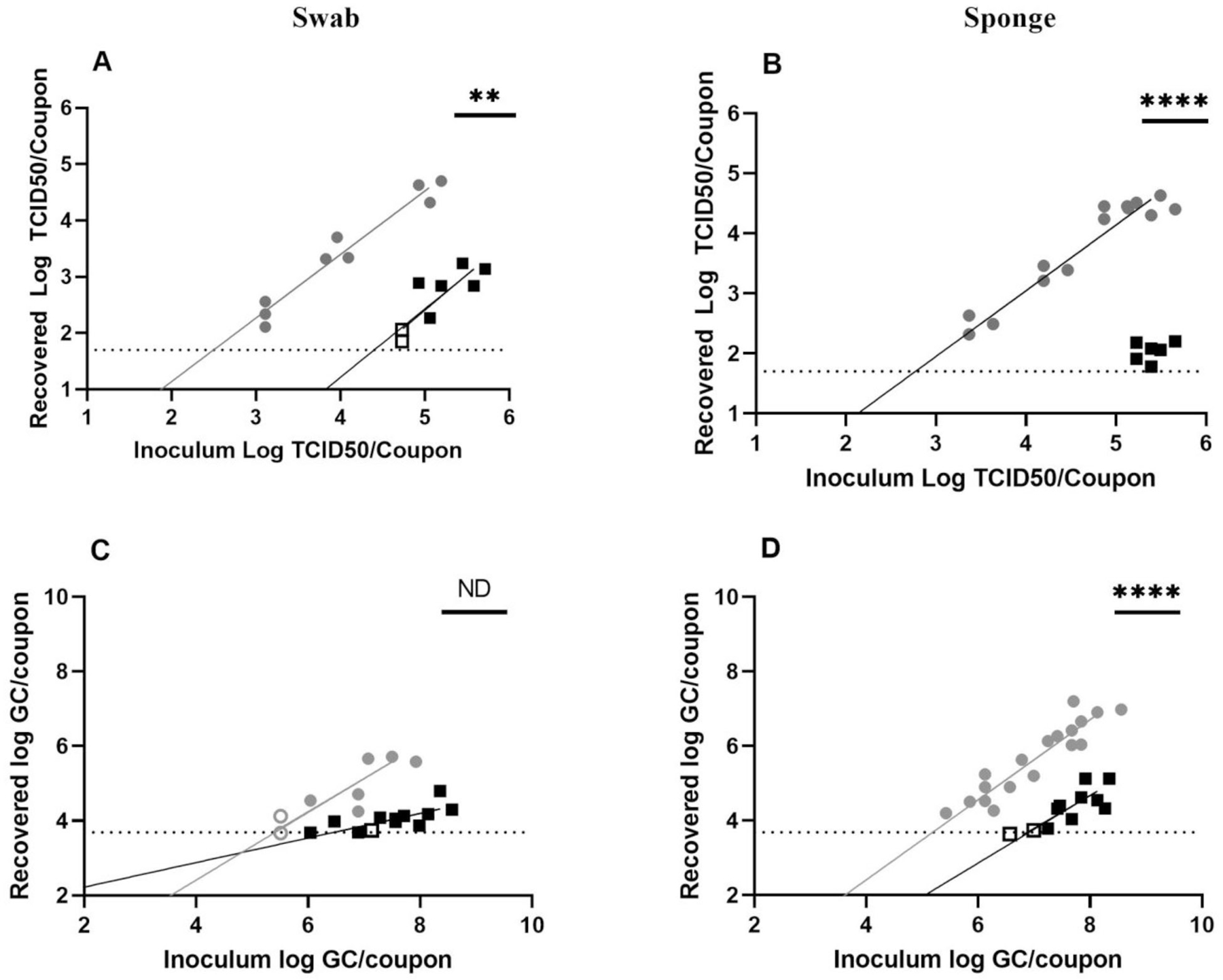
Sampling of SARS-CoV-2 on seat fabric. Recovery from (A, C) swab and (B, D) sponge methods with results from T0 recovery (gray circles) or T3 recovery (black squares). Open circles or squares identify samples in which at least one replicate was below the LOD. Data shown on graphs represent individual recovery values. Linear regression lines, shaded in gray (T0) or black (T3), are shown. Details of each linear regression are shown in Supplemental Table S2. Bars on each graph represent significant differences between recovery at T0 and T3, as determined by linear regression comparison (Graphpad Prism v9.3.1). ns: not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. ND indicates that regression comparison was not possible owing to difference in line slopes.
Analysis and statistics
The study design allowed for several comparisons to be made for each sampling method and surface material type for both infectious virus and vRNA recovery. The matrix shown in Table 1 enabled the recovery, including LOD comparison, from the four different surfaces as a function of sampling method (swab or sponge) and time post-inoculation of the surface (T0 vs. T3). The number of samples and test inoculum ranges for each test are also provided in Table 1. The Results section presents these comparisons divided by material type.
Inoculation of the surfaces with different concentrations of infectious virus allowed for LODs to be determined for each sampling method on each surface type, for both infectious virus and vRNA as a function of time after surface inoculation. Inoculum levels were lowered, in an iterative approach, until nondetects were achieved for each sampling method and material type.
Statistical analysis to determine significance of SARS-CoV-2 recovery (Figure 1) used a combination of ordinary two-way analysis of variance and Holm-Sidak’s t-test (calculated in GraphPad Prism 7.0e or 9.3.1, GraphPad Software, San Diego, CA, www.graphpad.com). Individual and multiple comparison Holm-Sidak t-tests were performed (Prism 7.0e) to compare results varying in one category (e.g., one material across multiple sampling times). Percentage recovery of infectious material and vRNA (Figure 1) was determined by calculating normalized recovery (recovered TCID50/coupon divided by inoculated TCID50/coupon or recovered GC/coupon divided by inoculated GC/coupon). Linear regression was performed on recovery data and regression comparison was performed (Prism 7.0e) to compare slope and y-intercepts between T0 and T3 data sets for each material tested.
Results
Recovery of SARS-CoV-2 from direct inoculation on swab and sponge materials
A similar, albeit not statistically significant, difference was seen for MHV-A59 recovery efficiency (54% ± 29% recovery swab; 38% ± 11% recovery sponge). Recovery efficiency of SARS-CoV-2 vRNA was significantly less than infectious virus for both swab (vRNA recovery 5.9% ± 0.2%; n = 3, p = 0.0091) and sponge (vRNA recovery 4.1% ± 1.8%; n = 8). To determine whether sample processing affected vRNA recovery, samples were collected before and after the Sephadex gel filtration step in the analysis method. Samples collected post-filtration showed a loss of 0.78 to 0.85 log10 vRNA GC/sample; only 16% ± 2% of inoculated vRNA was recovered after filtration. Interestingly, filtration did not affect infectious recovery as severely, with only a loss of 0.1 to 0.21 log TCID50/sample; 70.2% ± 12% of inoculated virus was recovered after filtration. These data suggest that Sephadex filtration may reduce the ratio of viral vRNA to infectious virus (pre-filtration, 640 GC:IU; post-filtration, 140.67 GC:IU).
Recovery of SARS-CoV-2 from stainless steel
Recovery from nonporous stainless steel was compared between immediate (T0) and 3 hr post-inoculation (T3) for both swab and sponge sampling. SARS-CoV-2 recovery from SS showed generally increased recovery of infectious SARS-CoV-2 at T0 for swab (Figure 1A) and sponge (Figure 1B) sampling versus T3. However, increased recovery was only statistically significant for swabs (Figure 1A, p = 0.0071). Interestingly, recovery of SARS-CoV-2 vRNA, measured in GC, did not show any significant difference at T0 by either swab (Figure 1C) or sponge (Figure 1D). Recovery of vRNA from SS was substantially decreased when compared to infectious recovery for each of the two sampling methods.
To assess whether SARS-CoV-2 recovery from SS varied across inoculation concentrations, recovery of infectious virus (Figures 2A, 2B) and vRNA (Figures 2C, 2D) was evaluated over several orders of inoculum magnitudes. Regardless of inoculation concentration and sampling method (Figures 2A, 2B), infectious SARS-CoV-2 recovery from SS was significantly higher at T0 than at T3. Linear regression showed significant high recovery of infectious SARS-CoV-2 at T0 for both swab (p = 0.0167) and sponge (p = 0.0002) sampling. Interestingly, no difference in recovery of SARS-CoV-2 vRNA was observed between T0 and T3 data sets (Figures 2C, 2D).
Linear regression was also used to determine the LOD for infectious SARS-CoV-2 and vRNA recovery (Table 1). These data (Table 1) reflected the importance of time for recovery of infectious virus, with the more sensitive LOD seen at T0 for both sponge and swab methods. The data (except Formica with swab) showed that infectious SARS-CoV-2 LODs increased with time after inoculation of the surface. However, this trend was not observed with vRNA SARS-CoV-2 recovery except carpet SF.
Recovery of SARS-CoV-2 from ABS
Initial sampling results from ABS at T0 and T3 appeared similar to sampling results from SS. Recovery of infectious SARS-CoV-2 from ABS demonstrated greater recovery at T0 for swab sampling (Figure 1A, p = 0.0047), whereas increased recovery via sponge sampling (Figure 1B, NS) only showed a trending increase (i.e., difference in recovery between time points was not statistically significant). Like SS, vRNA recovery from ABS did not show any difference between T0 and T3 via either swab (Figure 1C) or sponge (Figure 1D) methods. Interestingly, recovery of vRNA from ABS was significantly higher when using the sponge method at both T0 (p < 0.0001) and T3 (p = 0.0003) compared to using the swab method. ABS was the only material in which sampling recoveries were significantly different between sampling methods.
Testing across inoculation concentrations showed that infectious SARS-CoV-2 was recovered at higher efficiency at T0 than at T3 when sampled with the swab method (Figure 3A, p = 0.0409) but not with the sponge method (Figure 3B). No differences in vRNA recovery were observed over inoculation concentrations for either swab (Figure 3C) or sponge (Figure 3D) method.
Figure 3.
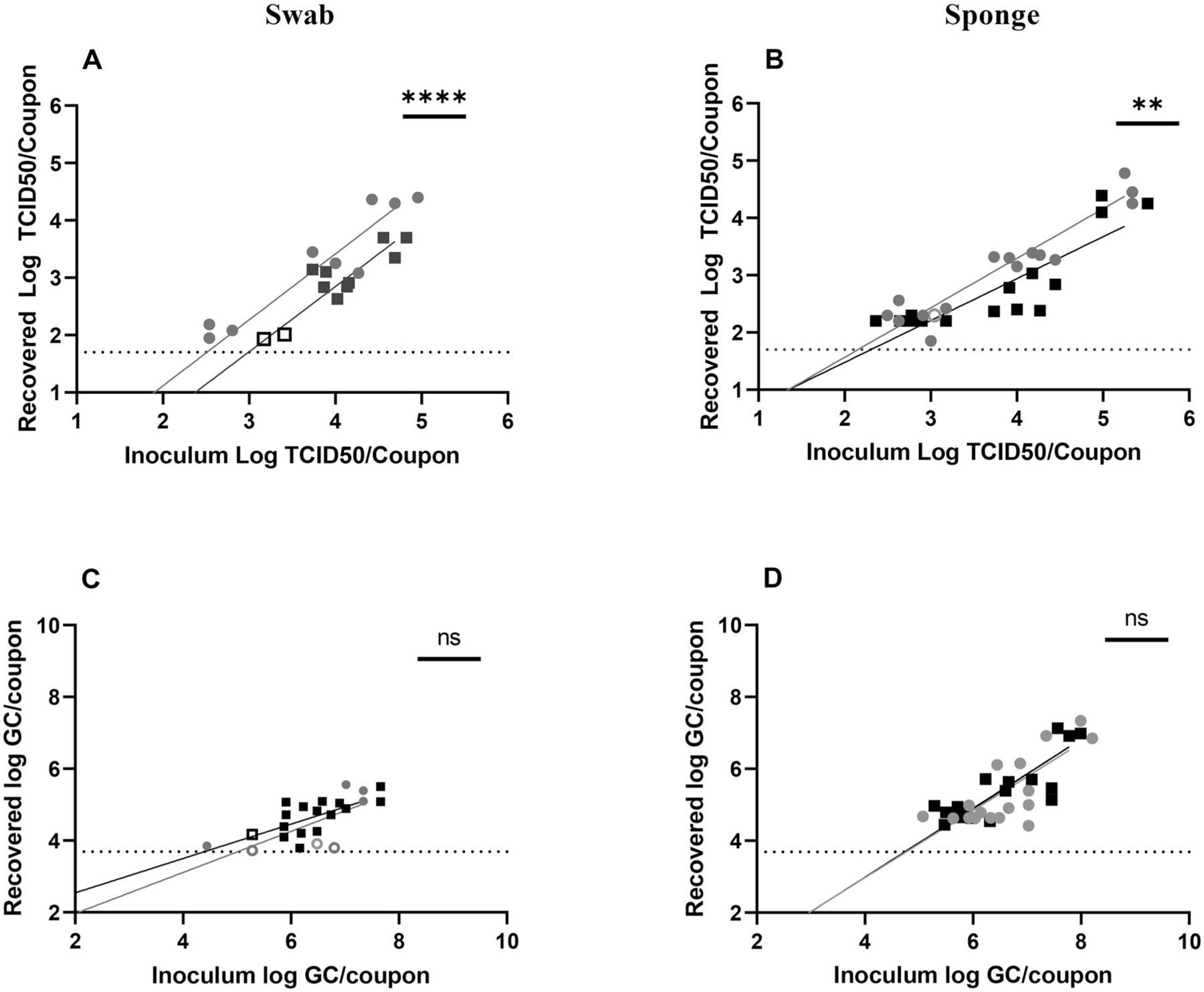
Sampling of SARS-CoV-2 on ABS plastic. Recovery from (A, C) swab and (B, D) sponge methods with results from T0 recovery (gray circles) or T3 recovery (black squares). Open circles or squares identify samples in which at least one replicate was below the LOD. Data shown on graphs represent individual recovery values. Linear regression lines, shaded in gray (T0) or black (T3), are shown. Details of each linear regression are shown in Supplemental Table S2. Bars on each graph represent significant differences between recovery at T0 and T3, as determined by linear regression comparison (Graphpad Prism v9.3.1). ns: not significant, *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Linear regression from the data in Figure 3 was used to calculate the LOD for infectious SARS-CoV-2 and vRNA recovery for each sampling method (Table 1). Though the infectious virus LODs were slightly lower for swab (T0, 2.51 log TCID50/coupon; T3, 2.99 TCID50/coupon) and sponge (T0, 2.15 TCID50/coupon; T3, 2.3 TCID50/coupon) methods when compared with SS LODs, differences between sampling methods were not as stark. Aligning with the data in Figures 1 and 3, calculated LODs for vRNA recovery were similar for T0 and T3 when sampling with the sponge method (T0, 4.76 GC/coupon; T3, 4.73 GC/coupon). Though not statistically significant, swab sampling on average was more sensitive at T3 than at T0 (T0, 5.01 GC/coupon; T3, 4.39 GC/coupon).
Recovery of SARS-CoV-2 from Formica
Sampling from Formica showed a slightly greater recovery at T0 than at T3 (Figures 1A, 1B); however, no differences were statistically significant. Recovery of vRNA via swab did show greater recovery at T0 (Figure 1C, p = 0.0020), although sponge recovery did not exhibit any statistical difference (Figure 1D). Sponge and swab recovery were similar for recovery of infectious virus and vRNA recovery at both T0 and T3.
Data collected across inoculation concentrations showed that more infectious SARS-CoV-2 was collected at T0 via sponge (Figure 4B, p = 0.0088) but not by swab. Though no significant differences were identified initially, a review of the data suggests that increased recovery may be more pronounced at higher inoculation concentrations. A similar pattern was identified for vRNA recovery, with higher recovery at T0 for sponge (Figure 4D, p = 0.0053) but not swab (Figure 4C) sampling. The data in Figure 4D also appear to suggest that recovery at T0 may be more pronounced at higher inoculation concentrations, similar to that observed in Figure 4B. Though these results suggest that inoculation concentration may influence sponge recovery, additional data, particularly at higher concentrations, are needed to confirm.
Figure 4.
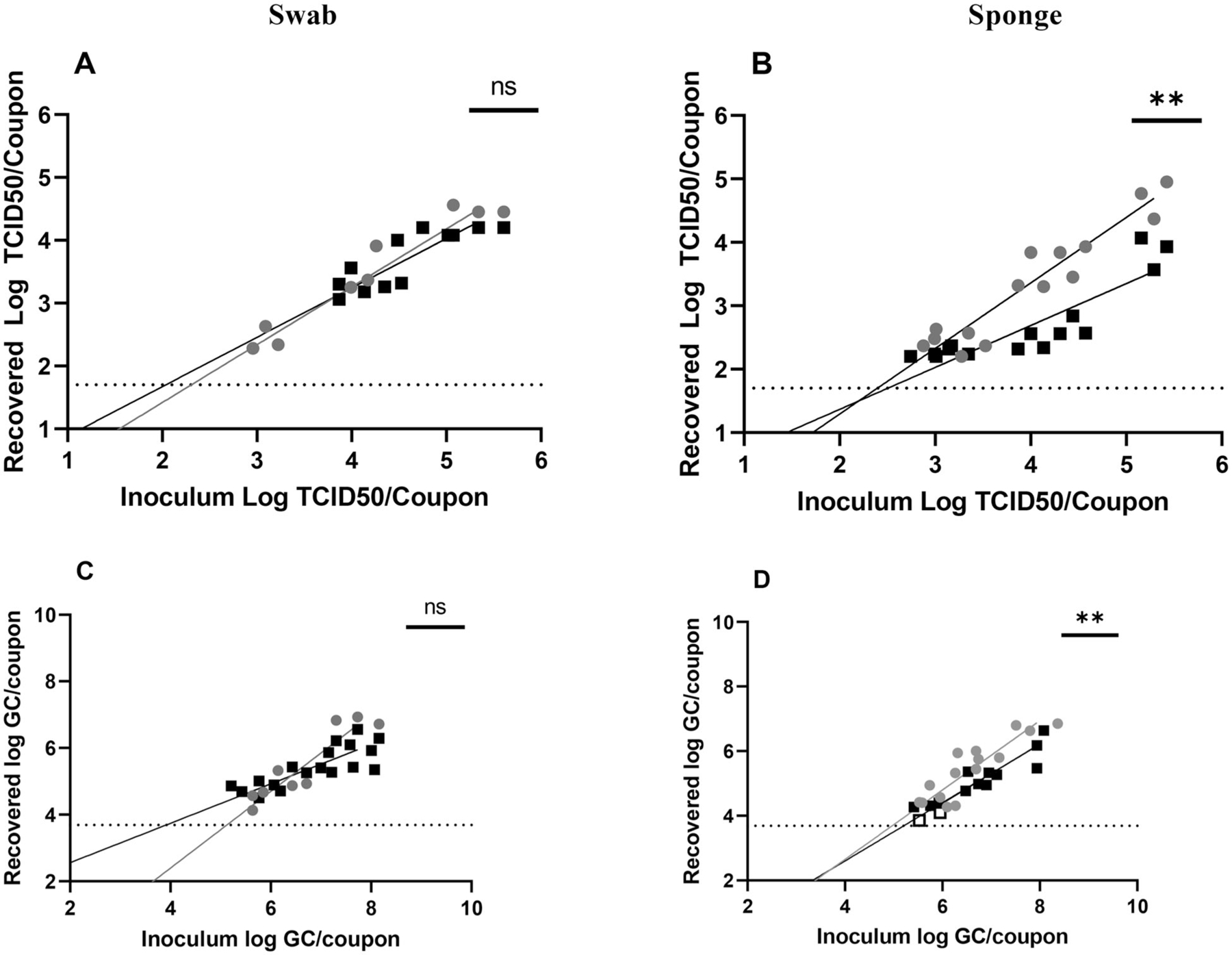
Sampling of SARS-CoV-2 on Formica. Recovery from (A, C) swab and (B, D) sponge methods with results from T0 recovery (gray circles) or T3 recovery (black squares). Open circles or squares identify samples in which at least one replicate was below the LOD. Data shown on graphs represent individual recovery values. Linear regression lines, shaded in gray (T0) or black (T3), are shown. Details of each linear regression are shown in Supplemental Table S2. Bars on each graph represent significant differences between recovery at T0 and T3, as determined by linear regression comparison (Graphpad Prism v9.3.1). ns: not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
LODs (Table 1) were calculated using linear regression (Supplemental Table S2) and demonstrated increased sensitivity (lower LOD values) at T0 for sponge sampling; this was similar to the significant differences seen in the data (Figures 4B, 4D).
Recovery of SARS-CoV-2 from SF
Sampling from SF showed a similar pattern of recovery as SS; that is, increased recovery of infectious SARS-CoV-2 at T0 via swab (Figure 1A, p = 0.0358) but not via sponge (Figure 1B) sampling. Like SS, no significant differences were observed for vRNA recovery (Figures 1C, 1D) or between sponge and swab sampling at either T0 or T3.
Sampling across inoculation concentrations showed greater recovery of infectious SARS-CoV-2 virus at T0 for swab (Figure 5A, p = 0.0011) and sponge (Figure 5B, p < 0.0001) sampling. Recovery of SARS-CoV-2 vRNA was also higher at T0 than at T3 for swab (Figure 5C, p = 0.0307) and sponge (Figure 5D, p < 0.0001) sampling. Notably, recovery of infectious SARS-CoV-2 at T3 was very low, requiring higher inoculation concentrations to be detectable (particularly for sponge sampling; Figure 5B). Recovery from SF was consistently lower than from SS, ABS, and Formica, particularly at the T3 time point.
LODs for SF recovery demonstrated that swab sampling exhibited increased sensitivity at T0 (Table 1) for infectious SARS-CoV-2 and vRNA recoveries, aligning with differences observed in the data (Figures 5A, 5C). Sponge sampling showed increased sensitivity at T0 for vRNA but not infectious for SARS-CoV-2 (Table 1). Note that viral recovery data for T3 (Figure 5B) were limited owing to difficulties recovering infectious virus from the matrix; thus, only the T0 data were able to fit well to a curve (Supplemental Table S2). However, recovery data from T3 were well clustered (Figure 5B) and suggested that infectious virus recovery at T3 is much lower than at T0 and likely had an increased LOD as well (Table 1).
Discussion
The choice of sampling and extraction methods, surface type to sample, quantitative or qualitative survey, and analysis type are key factors to consider when developing a viral surveillance strategy. The choice of assay target (infectious virus or viral RNA) is a critical factor, because surveillance strategies seek to define the presence of an infectious agent. Amid the recent SARS-CoV-2 pandemic, environmental sampling data from high-touch and household surfaces (Marcenac et al. 2021), nonporous surfaces in community spaces (e.g., store door handles; Ardura et al. 2021; Harvey et al. 2021), and hospital settings (Pasquarella et al. 2020) have been heavily leveraged to inform the presence of the SARS-CoV-2 virus, as well as support risk assessment for selecting strategies for opening public and community spaces and to monitor the effectiveness of mitigation strategies (e.g., disinfection; Faezeh et al. 2021). The testing performed in this study utilized a range of virus surface concentrations (from ~1.7 log to ~5.7 log) to determine the effects of virus surface concentration on sampling recovery. The tested range, however, may not fully represent the possible range of virus concentrations in the real world.
The results of this study suggest that infectious SARS-CoV-2 is recovered at a higher percentage than vRNA for both swab and sponge methods (Figure 1), with only some cases where similar percentages of each are recovered (Figure 1C, swab on Formica; Figure 1D, sponge on ABS). Though recovery from most materials was straightforward, recovery from SF, particularly at T3, was difficult and resulted in lower infectious virus (Figures 5A, 5B) and vRNA (Figures 5C, 5D) recovery than other materials. This difficulty mirrors that observed previously (Hardison, Nelson, et al. 2022) and suggests that porous materials may show lower viral recovery than nonporous materials. This study noted that vRNA recovery levels were not consistent with infectious virus recovery levels, with lower percentages of vRNA recovered from SS, Formica, and SF (Figure 1). Material differences were also observed, with swab sampling recovering significantly more vRNA from Formica than ABS or SS (Figure 1C) and sponge sampling recovering more vRNA from ABS than Formica and SS (Figure 1D). These data suggest that both material and method can affect recovery of vRNA.
Additionally, a stark reduction in infectious recovery at the T3 time point was observed for SS (Figures 2A, 2B), ABS (Figures 3A, 3B), Formica (Figure 4B), and SF (Figures 5A, 5B). However, time did not affect vRNA recovery for SS (Figures 2C, 2D), ABS (Figures 3C, 3D), Formica (Figures 4C, 4D), and SF (Figure 5C). Though vRNA recovery was increased at T0 from SF via sponge sampling (Figure 5D), the data were limited owing to the difficulties in working with SF. These results suggest that time has a significant effect on infectious virus recovery and has a reduced, or more limited, effect on vRNA recovery.
These data show that vRNA can remain detectable after viral infectivity has dissipated. One potential explanation for this result is the presence of noninfectious virus particles (e.g., defective particles, partially inactivated virus) above the LOD, whereas infectious virus particles are absent or present below the LOD. These noninfectious particles, long identified in coronaviruses (Makino et al. 1984; Baudoux et al. 1998), could allow a positive RT-qPCR signal despite a lack of infectivity (Cook et al. 2016). The lower vRNA recovery (relative to infectious virus) may be due to increased RNA degradation on fomites (Julian et al. 2011), aggregation of infectious virions, or varied levels of RNA:infectious particle ratios (commonly referred to as the particle to pfu ratio to measure the efficiency by which a virus infects cultured cells). The reported ratios for SARS-CoV-2 vary widely, (Julian et al. 2011; Cook et al. 2016; Colaneri et al. 2020; Ratnesar-Shumate et al. 2021) and have been reported as several hundred RNA genome copies per infective particle (Cotman et al. 2021) to tens of thousands of RNA copies per infective particle (Johnson et al. 2022). In this study, the ratio was affected by Sephadex gel filtration (640 GC:IU pre-gel filtration and 140.67 GC:IU post-gel filtration).
The methods utilized in this study were able to recover infectious virus at T0 and T3, with a decreasing trend over time. These sampling and analysis methods were also able to recover vRNA at T0 and T3, with minimal impact of time on recovery for all materials except seat fabric. Surveillance methods that rely on vRNA detection alone have limited ability to assess virus infectivity on surfaces or time since when the surface was contaminated. Although coronaviruses, including SARS-CoV-2, have been detected on some surfaces for days (Chan et al. 2011; van Doremalen et al. 2013; Warnes et al. 2015; Van Doremalen Neeltje et al. 2020) to weeks (Sizun et al. 2000; Rabenau et al. 2005; Casanova et al. 2010), results without infectivity information can only suggest whether contaminants have been on surfaces at some point in time.
Conclusion
There is a complex relationship between sampling method, material sampled, time from contamination to sampling, sample readout (vRNA vs. infectious virus), and recovery of SARS-CoV-2. Careful choices are key to avoiding data bias and, potentially, missing infectious material on one surface while catching it on another. Because there is no correlation between detecting vRNA and infectious virus, the authors suggest that careful thought be given to sampling strategies relying on vRNA monitoring, with respect to interpretating risk of infection. Additionally, though vRNA can remain detectable over time, infectious SARS-CoV-2 degrades quickly; this differential effect of time complicates interpretation of sampling results when using vRNA analytical methods. A larger evaluation of how presence of infectious virus correlates with vRNA presence over realistic sampling time periods (8–12 hr or 2–5 days), considering surface material, recovery of vRNA, and recovery of infectious virus is warranted.
Supplementary Material
Acknowledgements
We thank Lindsay A. Catlin for assistance with RT-qPCR and K. T. Vandergrif and Dr. Christopher Dibble at Battelle Memorial Institute for helpful technical discussions. The EPA, through its Office of Research and Development, directed the research described herein conducted through contract EP-C-16-014, task order 68HERC20F0241 with Battelle Memorial Institute. It has been subjected to review by the EPA’s Office of Research and Development and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Footnotes
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15459624.2023.2231516. AIHA and ACGIH members may also access supplementary material at http://oeh.tandfonline.com.
Data availability statement
All data are publicly available at data.gov.
References
- Ardura A, Dopico E, Fernandez S, Garcia-Vazquez E. 2021. Citizen volunteers detect SARS-CoV-2 RNA from outdoor urban fomites. Sci Total Environ 787:147719. doi: 10.1016/j.scitotenv.2021.147719. [DOI] [Google Scholar]
- Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol 72(11):8636–8643. doi: 10.1128/JVI.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova LM, Jeon S, Rutala WA, Weber DJ, Sobsey MD. 2010. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl Environ Microbiol 76(9):2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). 2012. Surface sampling procedures for Bacillus anthracis spores from smooth, non-porous surfaces. Centers for Disease Control and Prevention; [updated 2012 Jan 30 2012; accessed 2022 Sept 1]. https://www.cdc.gov/niosh/topics/emres/surface-sampling-bacillus-anthracis.html. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). 2021a. Guidance on the inactivation or removal of select agents and toxins for future use. Centers for Disease Control and Prevention; [updated 2021 Jan 28; accessed 2022 Sept 1]. https://www.selectagents.gov/compliance/guidance/inactivation/index.htm. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). 2021b. SARS-CoV-2 and surface (fomite) transmission for indoor community environments. Centers for Disease Control and Prevention; [updated 2021 Apr 5 accessed 2022 Sept 1]. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/surface-transmission.html. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). 2021c. SARS-CoV-2 transmission. Centers for Disease Control and Prevention; [updated 2021 May 7; accessed 2022 Sept 1]. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/sars-cov-2-transmission.html. [Google Scholar]
- Chan K-H, Peiris JM, Lam S, Poon L, Yuen K, Seto WH. 2011. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv Virol 2011(2011):734690. doi: 10.1155/2011/734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Huang L-M, Chan C-C, Su C-P, Chang S-C, Chang Y-Y, Chen M-L, Hung C-C, Chen W-J, Lin F-Y, et al. 2004. SARS in hospital emergency room. Emerg Infect Dis 10(5):782–788. doi: 10.3201/eid1005.030579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, Lim XF, Lim AS, Sutjipto S, Lee PH, et al. 2020. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun 11(1):2800. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaneri M, Seminari E, Novati S, Asperges E, Biscarini S, Piralla A, Percivalle E, Cassaniti I, Baldanti F, Bruno R, et al. 2020. Severe acute respiratory syndrome coronavirus 2 RNA contamination of inanimate surfaces and virus viability in a health care emergency unit. Clin Microbiol Infect 26(8):1094.e1091–1094.e1095. doi: 10.1016/j.cmi.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BW, Cutts TA, Nikiforuk AM, Leung A, Kobasa D, Theriault SS. 2016. The disinfection characteristics of ebola virus outbreak variants. Sci Rep 6:38293. doi: 10.1038/srep38293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman ZJ, Bowden MJ, Richter BP, Phelps JH, Dibble CJ. 2021. Factors affecting aerosol SARS-CoV-2 transmission via HVAC systems; a modeling study. PLoS Comput Biol 17(10):e1009474. doi: 10.1371/journal.pcbi.1009474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabisch P, Schuit M, Herzog A, Beck K, Wood S, Krause M, Miller D, Weaver W, Freeburger D, Hooper I, et al. 2021. The influence of temperature, humidity, and simulated sunlight on the infectivity of SARS-CoV-2 in aerosols. Aerosol Sci Technol 55(2):142–153. doi: 10.1080/02786826.2020.1829536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faezeh S, Noorimotlagh Z, Mirzaee SA, Kalantar M, Barati B, Fard ME, Fard NK. 2021. The SARS-CoV-2 (COVID-19) pandemic in hospital: an insight into environmental surfaces contamination, disinfectants’ efficiency, and estimation of plastic waste production. Environ Res202: 111809. doi: 10.1016/j.envres.2021.111809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-de-Mera IG, Rodríguez del-Río FJ, de la Fuente J, Pérez-Sancho M, Hervás D, Moreno I, Domínguez M, Domínguez L, Gortázar C. 2021. Detection of environmental SARS-CoV-2 RNA in a high prevalence setting in Spain. Transbound Emerg Dis 68(3):1487–1492. doi: 10.1111/tbed.13817. [DOI] [PubMed] [Google Scholar]
- Gabitzsch E, Safrit JT, Verma M, Rice A, Sieling P, Zakin L, Shin A, Morimoto B, Adisetiyo H, Wong R, et al. 2021. Dual-antigen COVID-19 vaccine subcutaneous prime delivery with oral boosts protects NHP against SARS-CoV-2 challenge. Front Immunol 12:729837–729837. eng. doi: 10.3389/fimmu.2021.729837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RL, Nelson SW, Barriga D, Ghere JM, Fenton GA, James RR, Stewart MJ, Lee S, Calfee MW, Ryan SP, et al. 2022. Efficacy of detergent-based cleaning methods against coronavirus MHV-A59 on porous and non-porous surfaces. J Occup Environ Hyg 19(2):91–101. eng. doi: 10.1080/15459624.2021.2015075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RL, Ryan SP, Limmer RA, Crouse M, Nelson SW, Barriga D, Ghere JM, Stewart MJ, Lee SD, Taylor BM, et al. 2022. Residual antimicrobial coating efficacy against SARS-CoV-2. J Appl Microbiol 132(4):3375–3386. doi: 10.1111/jam.15437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey AP, Fuhrmeister ER, Cantrell ME, Pitol AK, Swarthout JM, Powers JE, Nadimpalli ML, Julian TR, Pickering AJ. 2021. Longitudinal monitoring of SARS-CoV-2 RNA on high-touch surfaces in a community setting. Environ Sci Technol Lett 8(2):168–175. doi: 10.1021/acs.estlett.0c00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TJ, Nishida RT, Sonpar AP, Lin Y-CJ, Watson KA, Smith SW, Conly JM, Evans DH, Olfert JS. 2022. Viral load of SARS-CoV-2 in droplets and bioaerosols directly captured during breathing, speaking and coughing. Sci Rep 12(1):3484. doi: 10.1038/s41598-022-07301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian TR, Tamayo FJ, Leckie JO, Boehm AB. 2011. Comparison of surface sampling methods for virus recovery from fomites. Appl Environ Microbiol 77(19):6918–6925. doi: 10.1128/AEM.05709-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G, Todt D, Pfaender S, Steinmann E. 2020. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect 104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK, Sun L, Duan Y, Cai J, Westerdahl D, et al. 2020. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 582(7813):557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Mahl MC, Sadler C. 1975. Virus survival on inanimate surfaces. Can J Microbiol 21(6):819–823. doi: 10.1139/m75-121. [DOI] [PubMed] [Google Scholar]
- Makino S, Taguchi F, Fujiwara K. 1984. Defective interfering particles of mouse hepatitis virus. Virology. 133(1):9–17. doi: 10.1016/0042-6822(84)90420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcenac P, Park GW, Duca LM, Lewis NM, Dietrich EA, Barclay L, Tamin A, Harcourt JL, Thornburg NJ, Rispens J, et al. 2021. Detection of SARS-CoV-2 on surfaces in households of persons with COVID-19. IJERPH. 18(15): 8184. doi: 10.3390/ijerph18158184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques M, Domingo JL. 2021. Contamination of inert surfaces by SARS-CoV-2: persistence, stability and infectivity. A review. Environ Res 193:110559. doi: 10.1016/j.envres.2020.110559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G, Rickard H, Stevenson D, Aranega-Bou P, Pitman J, Crook A, Davies K, Spencer A, Burton C, Easterbrook L, et al. 2021. Detection of SARS-CoV-2 within the healthcare environment: a multi-centre study conducted during the first wave of the COVID-19 outbreak in England. J Hosp Infect 108:189–196. doi: 10.1016/j.jhin.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onakpoya IJ, Heneghan CJ, Spencer EA, Brassey J, Plüddemann A, Evans DH, Conly JM, Jefferson T. 2021. SARS-CoV-2 and the role of fomite transmission: a systematic review. F1000Res. 10:233–233. eng. doi: 10.12688/f1000research.51590.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GW, Chhabra P, Vinjé J. 2017. Swab sampling method for the detection of human norovirus on surfaces. J Vis Exp (120):e55205. doi: 10.3791/55205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GW, Lee D, Treffiletti A, Hrsak M, Shugart J, Vinjé J. 2015. Evaluation of a new environmental sampling protocol for detection of human norovirus on inanimate surfaces. Appl Environ Microbiol 81(17):5987–5992. doi: 10.1128/AEM.01657-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquarella C, Colucci ME, Bizzarro A, Veronesi L, Affanni P, Meschi T, Brianti E, Vitali P, Albertini R. 2020. Detection of SARS-CoV-2 on hospital surfaces. Acta Biomed 91(9-S):76–78. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena P, Morais J, Quintal Gomes A, Viegas C. 2021. Sampling methods and assays applied in SARS-CoV-2 exposure assessment. Sci Total Environ 775:145903. doi: 10.1016/j.scitotenv.2021.145903. [DOI] [Google Scholar]
- Peng Z, Rojas ALP, Kropff E, Bahnfleth W, Buonanno G, Dancer SJ, Kurnitski J, Li Y, Loomans M, Marr LC, et al. 2022. Practical indicators for risk of airborne transmission in shared indoor environments and their application to COVID-19 outbreaks. Environ Sci Technol 56(2): 1125–1137. doi: 10.1021/acs.est.1c06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenau HF, Cinatl J, Morgenstern B, Bauer G, Preiser W, Doerr HW. 2005. Stability and inactivation of SARS coronavirus. Med Microbiol Immunol 194(1–2):1–6. doi: 10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnesar-Shumate S, Bohannon K, Williams G, Holland B, Krause M, Green B, Freeburger D, Dabisch P. 2021. Comparison of the performance of aerosol sampling devices for measuring infectious SARS-CoV-2 aerosols. Aerosol Sci Technol 55(8):975–986. doi: 10.1080/02786826.2021.1910137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzini K, Castrica M, Menchetti L, Maggi L, Negroni L, Orfeo NV, Pizzoccheri A, Stocco M, Muttini S, Balzaretti CM. 2020. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan, Italy. Sci Total Environ 742:140540. doi: 10.1016/j.scitotenv.2020.140540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27(3):493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Rose LJ, Hodges L, O’Connell H, Noble-Wang J. 2011. National validation study of a cellulose sponge wipe-processing method for use after sampling Bacillus anthracis spores from surfaces. Appl Environ Microbiol 77(23): 8355–8359. doi: 10.1128/AEM.05377-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia JL, Rivera DN, Herrera VL, Morwitzer MJ, Creager HM, Santarpia GW, Crown KK, Brett-Major DM, Schnaubelt ER, Broadhurst MJ, et al. 2020. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci Rep 10(1):12732. doi: 10.1038/s41598-020-69286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer K, Mäde D, Ellerbroek L, Schulenburg J, Johne R, Klein G. 2009. Application of a swab sampling method for the detection of norovirus and rotavirus on artificially contaminated food and environmental surfaces. Food Environ Virol 1(1):42–49. doi: 10.1007/s12560-008-9007-0. [DOI] [Google Scholar]
- Silvestri E, Chambers-Velarde Y, Chandler J, Cuddeback J, Calfee W, Archer J, Shah S. 2021. Collection of surface samples potentially contaminated with microbiological agents using swabs, sponge sticks and wipes (Report No: EPA/600/R-21/051). Washington (DC): U.S. Environmental Protection Agency Office of Research and Development. [Google Scholar]
- Sizun J, Yu MW, Talbot PJ. 2000. Survival of human coronaviruses 229E and OC43 in suspension and after drying on surfaces: a possible source of hospital-acquired infections. J Hosp Infect 46(1):55–60. doi: 10.1053/jhin.2000.0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HN, Le GT, Nguyen DT, Juang R-S, Rinklebe J, Bhatnagar A, Lima EC, Iqbal HM, Sarmah AK, Chao H-P. 2021. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ Res 193:110265. doi: 10.1016/j.envres.2020.110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bockel D, Munier C, Turville S, Badman S, Walker G, Stella A, Aggarwal A, Yeang M, Condylios A, Kelleher A, et al. 2020. Evaluation of commercially available viral transport medium (VTM) for SARS-CoV-2 inactivation and use in point-of-care (POC) testing. Viruses. 12(11): 1208. doi: 10.3390/v12111208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, et al. 2020. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med 382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N, Bushmaker T, Munster VJ. 2013. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Eurosurveillance. 18(38):20590. doi: 10.2807/1560-7917.ES2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- Warnes SL, Little ZR, Keevil CW, Colwell R. 2015. Human coronavirus 229E remains infectious on common touch surface materials. mBio. 6(6):e01697–01615–e01615. doi: 10.1128/mBio.01697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo MH, Hsu YM, Wu CY, Heimbuch B, Wander J. 2010. Method for contamination of filtering facepiece respirators by deposition of MS2 viral aerosols. J Aerosol Sci 41(10):944–952. doi: 10.1016/j.jaerosci.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi T, Ohnishi M, Matsunaga N, Kakimoto K, Kamiya H, Okamoto K, Suzuki M, Gu Y, Sakaguchi M, Tajima T, et al. 2020. Environmental sampling for severe acute respiratory syndrome coronavirus 2 during a COVID-19 outbreak on the Diamond Princess cruise ship. J Infect Dis 222(7):1098–1102. doi: 10.1093/infdis/jiaa437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Otter JA, Price JR, Cimpeanu C, Meno Garcia D, Kinross J, Boshier PR, Mason S, Bolt F, Holmes AH, et al. 2021. Investigating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surface and air contamination in an acute healthcare setting during the peak of the coronavirus disease 2019 (COVID-19) pandemic in London. Clin Infect Dis 73(7):e1870–e1877. doi: 10.1093/cid/ciaa905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available at data.gov.


