Abstract
A 119-kb bacterial artificial chromosome from the JOINTLESS locus on the tomato (Lycopersicon esculentum) chromosome 11 contained 15 putative genes. Repetitive sequences in this region include one copia-like LTR retrotransposon, 13 simple sequence repeats, three copies of a novel type III foldback transposon, and four putative short DNA repeats. Database searches showed that the foldback transposon and the short DNA repeats seemed to be associated preferably with genes. The predicted tomato genes were compared with the complete Arabidopsis genome. Eleven out of 15 tomato open reading frames were found to be colinear with segments on five Arabidopsis bacterial artificial chromosome/P1-derived artificial chromosome clones. The synteny patterns, however, did not reveal duplicated segments in Arabidopsis, where over half of the genome is duplicated. Our analysis indicated that the microsynteny between the tomato and Arabidopsis genomes was still conserved at a very small scale but was complicated by the large number of gene families in the Arabidopsis genome.
Tomato (Lycopersicon esculentum) is one of the most important and intensely studied model dicot plants. Compared with the pioneering research in disease resistance and fruit maturation, little is know about the microsyntenic relationship of tomato and other plant species as revealed by sequencing long contiguous stretches of genomic DNA. To date, the longest tomato sequence reported in GenBank is a 105-kb bacterial artificial chromosome (BAC) sequence from the chromosome 2 ovate region (Ku et al., 2000). In contrast, the Arabidopsis genome has been sequenced (The Arabidopsis Genome Initiative [AGI], 2000; http://www.Arabidopsis.org/; http://www.kazusa.or.jp/) and provides a good opportunity for comparative genomics. It is time to answer the questions such as: To what extent can the knowledge gained from the Arabidopsis genome sequence be applied to understand the tomato genome, a species that separated from Arabidopsis more than 100 million years ago (MYA; Gandolfo et al., 1998; Yang et al., 1999)? The sequence of the 105-kb BAC from tomato chromosome 2, containing the ovate locus, was compared with the Arabidopsis genome, shedding interesting light on the synteny of the genomes of these two model species (Ku et al., 2000). Considering the genome size of tomato (approximately 970Mb), more comparative sequence analysis will be required before we can develop a comprehensive and confident comparison of the genomes of these two dicot plants.
In our effort to map-base clone the tomato JOINTLESS gene, a gene that controls the development of pedicel abscission zones (Mao et al., 2000a), we sequenced a 118,813-bp BAC clone encompassing the tomato JOINTLESS region on chromosome 11. In addition to the 15 putative open reading frames (ORFs) detected, repetitive sequences, especially small DNA elements such as foldback transposons, were also described in detail here. A comparison of the predicted tomato genes with the Arabidopsis genome showed that segmented gene colinearity between the two species was preserved, but located on different Arabidopsis chromosomes or on the same chromosome but physically separated. Unlike the chromosome 2 ovate locus, the JOINTLESS region seemed to be synteneous to the non-duplicated portion of the Arabidopsis genome.
RESULTS AND DISCUSSION
Sequence Characteristics of the Tomato JOINTLESS Locus
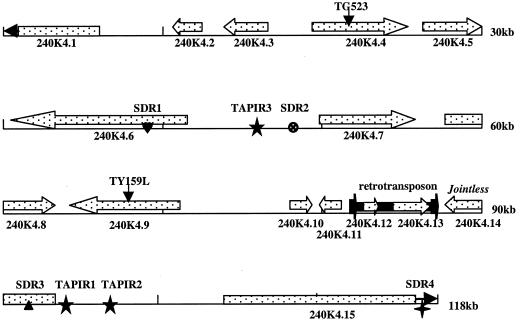
Tomato BAC clone 240K04 (GenBank accession no. AF292003.), containing the JOINTLESS locus, was isolated from an L. esculentum cv Heinz1706 BAC library (Budiman et al., 2000) by screening with jointless-linked RFLP markers TG523 and TY159L. Previous work showed that the genomic region containing the jointless locus has a low genetic/physical ratio of less than 100 kb per centiMorgan (cM) as demonstrated by yeast artificial chromosome physical mapping (Zhang et al., 1994), in contrast to the whole genome average of 750 kb cM−1 (Tanksley et al., 1992). The tomato genomic sequence on BAC 240K04 was found to be 118,813 bp long and contained TG523 and TY159L, genetically 1 cM apart, but separated by only approximately 50 kb (Fig. 1). In total, 15 putative genes (including two that belong to one retrotransposon) were predicted on 240K04, giving a gene density in this region as one every 8 kb. This ratio was close to a recent estimation of one gene per 6.2 kb at the ovate region of tomato chromosome 2 (Ku et al., 2000) and nearly twice that of Arabidopsis chromosome 2 (one gene per 4.4 kb; Lin et al., 1999). BAC 240K04 had an average GC content of approximately 32% with the 15 putative gene coding regions having a higher GC content of 42%. These data were comparable to a previous study where the whole genome GC content was estimated as 37% and coding regions as 46% (Messeguer et al., 1991). Concerning the repetitive sequences on BAC 240K04, the only known transposable element was a copia-like retrotransposon. Small DNA elements, however, were found frequently when the sequence of 240K04 was searched against GenBank, reminding us of small elements like miniature inverted-repeat elements (MITEs) in rice and maize (Bureau and Wessler, 1994; Bureau et al., 1996). These DNA elements (short DNA repeats [SDRs]) were of great interest because of their frequent association with gene sequences. In particular, some SDRs were directly involved in the mutation of tomato genes such as jointless (Mao et al., 2000a) and yellow flesh (Fray and Grierson, 1993).
Figure 1.
A graphical display of the predicted ORFs and the DNA elements on BAC 240K04. Arrows for each ORF indicate the coding orientation. The pointed arrows on 240K04.1 and 240K04.15 indicate that these two ORFs are 3′ incomplete and the blunt arrows flanking the retrotransposon represent the two LTRs.
Genes
Fifteen putative genes were predicted by gene prediction programs such as GenScan and their functions were determined by searching the nonredundant protein database in GenBank (Table I, Fig. 1). ORFs 240K04.1 and 240K04.15 were incomplete because they were located at either end of the BAC. Six predicted genes (240K04.2, 3, 5, 6, 8, and 11) had no significant match in GenBank. However, five had significant expressed sequence tag (EST) matches with The Institute for Genomic Research tomato gene index (Lycopersicon Gene Index, http://www.tigr.org/tdb/lgi/), and thus were annotated as unknown proteins. In total, 11 out of 15 putative genes had significant EST matches in the LGI, including ORFs 240K04.12 and 240K04.13, that apparently belong to the same retrotransposon as delimited by the two LTRs as described below. In the tomato ovate region, however, only six out of the 17 ORFs were found to have EST matches (Ku et al., 2000). ORFs 240K04.6 and 240K04.7 may be from the same gene family because a portion of their sequences were significantly similar to each other. Based on the GenBank search results, we were able to assign putative functions to nine ORFs including JOINTLESS, a member of the MADS-box transcription factor gene family (Table I).
Table I.
Detailed annotation analysis of 15 ORFs in tomato BAC 240K04
| Name | Approximate Coding Region | Best Homology (GenBanka) | Best Blastp E Value | Best The Institute for Genomic Research Expressed Sequence Tag (EST) | Putative Protein Function |
|---|---|---|---|---|---|
| 240K04.01 | <6,761–6,831 | T05632 | 2e–31 | AW929546 | Putative permease |
| 240K04.02 | 12,852–19,435 | No homology | – | AW219175b | Unknown protein |
| 240K04.03 | 17,278–16,871 | No homology | – | TC43589 | Unknown protein |
| 240K04.04 | 22,829–26,608 | NP_009196 | 8e–90 | AW223638b | Putative suppressor of yeast gcr2 |
| 240K04.05 | 27,444–29,478 | AC006340 | 1e–68 | TC41345b | Unknown protein |
| 240K04.06 | 42,332–30,012 | AAD22346.1 | e–160 | TC39148b | Unknown protein |
| 240K04.07 | 50,514–55,503 | T05634 | e–120 | TC40844 | Putative centromere protein |
| 240K04.08 | 57,336–64,386 | P77253 | 8e–10 | AI487713b | Unknown protein |
| 240K04.09 | 70,688–65,478 | T06805 | e–105 | TC40402b | Putative auxin growth promotor |
| 240K04.10 | 77,615–79,017 | CAB80933 | 5e–51 | TC40659 | Putative protein phosphatase |
| 240K04.11 | 81,356–80,189 | AP000381 | e–122 | TC45720b | Unknown protein |
| 240K04.12 | 82,223–83,281 | AP002459 | 3e–64 | BE463242b | Putative polyprotein |
| 240K04.13 | 84,217–86,904 | BAA90383.1 | e–156 | TC41502b | Putative polyprotein |
| 240K04.14 | 93,204–88,295 | AAD22365 | 4e–75 | TC40481b | Putative MADS-box gene |
| 240K04.15 | 104,210–117,994 | AJ001729 | 2e–25 | AW929351b | Similar to TH65 protein |
As performed in Dec. 2000.
Containing amino acid identity > 95% over 50 amino acids.
Repetitive Sequences
Previous work showed that the tomato genome contains mostly low copy number sequences (Zamir and Tanksley, 1988). Ganal et al. (1988) estimated by hybridization with the major tomato repeated sequences that the total amount of highly repeated sequences might lie between 10% and 15%. An analysis of 1,205 random BAC end sequences also showed that the repetitive sequences in the tomato genome is around 12% (Budiman et al., 2000). Although a DNA database search of the 240K04 sequence did not detect large blocks of repetitive sequences, regions of small DNA segments were found with high similarity to more than 20 gene sequences from tomato or potato Solanum spp., indicating the repetitive nature of these DNA sequences.
Simple Sequence Repeats (SSRs)
There were 14 SSRs along the 118.8-kb sequence. Four of them were simple repeats, three (TA)n, and one (TTA)8. The remaining were mixtures of different repeats derived from nucleotide conversions or substitutions such as (TG)8(TA)8 at 2,243 bp, (TTA)5 TA(TAA)2(TTA)2 at 69,378 bp, and (TA)17TC(TG)7 at 75,565 bp. These SSRs gave a density of one SSR per 8.5 kb. In addition, (AT)n-type SSRs were the main class of SSRs, consistent with a previous report that (AT)n SSRs were more frequently found in coding regions in tomato (Broun and Tanksley, 1996).
Retrotransposons
One copia-like LTR retrotransposon, named Lere1, was present on BAC 240K04. Lere1 was 5,532 bp long with two 276-bp LTRs that were 97% identical. A six-frame translation of the Lere1 sequence revealed numerous stop codons within the polyprotein region indicating that Lere1 was highly degraded, though the major components of a retrotransposon such as the endonuclease (or integrase) domain, the reverse transcriptase domain, and the RNase domain were still recognizable. Two predicted ORFs (240K04.12 and 240K04.13) represented most of the contiguous amino acid regions from all three forward frames. 240K04.13 had strong homology to known copia-like retrotransposons. However, 240K04.12 did not contain any major domains but was simply homologous to annotated Arabidopsis sequences. A DNA-DNA comparison revealed that Lere1 was not identical to any retrotransposons previously cloned in tomato. Nevertheless, there were two tomato genomic sequences, the vacuolar invertase genes from Lycopersicon pimpinelliforlium (Z12028.1) and L. esculentum (Z12027.1; Elliott et al., 1993), which contain sequences corresponding to the polyprotein region of Lere1. It is interesting that a TBlastN search of the LGI revealed that one EST match (TC14149) had 94% protein sequence identity along 251 amino acids, indicating that either a similar retrotransposon was still active in tomato or a retrotransposon (or part of it) was co-expressed in TC14149.
Foldback Transposon Tomato Anionic Peroxidase Inverted Repeat (TAPIR)
Three regions of BAC 240K04 were found to be similar to TAPIR elements (Hong and Tucker, 1998). Four copies of TAPIR have been reported as inverted repeats in the non-coding regions of tomato polygalacturonase (TAPG) 1, 2, and 4, as well as the first intron of tomato anionic peroxidase (X15853). An updated GenBank search using TAPIR1 as a query showed additional tomato sequences bearing TAPIR elements (Table II).
Table II.
Analysis of TAPIR nad SDR repeat elements in tomato BAC 240K04
| Name and Location | Length (bp)/AT content | Feature | Shared by and the Locations |
|---|---|---|---|
| TAPIR1, 93801 | 335, 64% | Foldback | AF001000 L.e. TAPG 1, 5′ |
| AF001001 L.e. TAPG 2, 5′ | |||
| TAPIR2, 95674 | – | – | AF001002 L.e. TAPG 4, 3′ |
| X15853, L.e. TAP1, intron | |||
| TAPIR3, 46351 | – | – | AF179442 L.e. plasma membrane H+-ATPase isoform LHA2, 5′ |
| AJ002236 L. pim. Cf-9 resistance gene cluster, intron | |||
| SDR1, 39050 | 300, 60% | N/A | M88487 L.e. ACC synthase, 5′ |
| X67143 L.e. yellow flesh mutant mRNA for phytoene synthase, 3′ half | |||
| SDR2, 48401 | 400, 72% | gagaaagagagaaa Motifs | AF001001 L.e. TAPG 2, 3′Z12028 L. pim. gene encoding vacuolar invertase, 3′ |
| Z12027 L.e. gene for vacuolar invertase, 3′ | |||
| SDR3, 91001 | 150, 84% | N/A | AJ272307 L. pen. lin 5 gene for beta fructosidase, first intron |
| X13497 S.t. wound-induced genes, intergenic: 3′WIN1 and 5′WIN2 | |||
| AJ006379 L.e. subtilase (sbt2) gene, 5′ | |||
| AJ277064 L.e. PR-5 gene for pathogenesis-relatedprotein, 5′ | |||
| M59427 L. per proteinase inhibitor I gene, 5′ | |||
| U68072 L.e. HMG2 gene, 5′ before retrotransposon ToRTL | |||
| M37304 L.e. polygalacturonase, exons 1–9, fourth intron | |||
| AF220602 L. pim. Rio Grande 76R Pto locus, repeat region | |||
| AF273333 L.e. clone BAC 19, intron | |||
| SDR4, 116711 | 250, 47% | SolSINE | AF034411, L.e. cytosolic Cu, Zn superoxidedismutase, intron |
| (Rebatchouk and Narita, 1997) | U75644 L.e. farnesyl-protein transferase beta subunit, three copies, introns | ||
| Y10603 L.e. ldh2 gene, intron | |||
| Z27233 S.t. (STAC1) gene for ACC acid synthase, 3′ |
L.e., L. esculentum; L. pim., L. pimpinellifolium; L. pen., Lycopersicon pennellii; S.t., Solanum tuberosum; L. per., Lycopersicon peruvianum; ACC, 1-aminocyclopropane-1-carboxylate; TAP: tomato anionic peroxidase; N.A., not applicable.
The reason that TAPIRs were of interest was that one TAPIR element was found to be associated with a 939-bp deletion in the promoter and first exon of the jointless mutant allele (Mao et al., 2000a), indicating that TAPIR could be a transposable element. With its strong secondary structure and a middle loop, TAPIR was very similar to a type III foldback transposon (Fig. 2a; Rebatchouk and Narita, 1997). To prove that TAPIR did transpose in the tomato genome, we designed primers flanking the three TAPIR loci on BAC 240K04 (TAPIR1, TAPIR 2, and TAPIR3) and PCR amplified these regions from both cultivated and wild tomatoes (Fig. 2b). At the TAPIR1 locus, the wild tomato species Lycopersicon hirsutum had a smaller band compared with the other tomato species or cultivars. This DNA fragment was cloned and sequenced. The results showed that in L. hirsutum, the TAPIR element was absent at the site corresponding to the TAPIR1 locus of the control cultivated tomato, indicating that the TAPIR may have transposed. Like other transposition events that often cause the deletion of neighboring sequences, the deletion in the jointless mutant may well have been caused by the transposition of TAPIR1 at the 5′ region of the JOINTLESS gene (Mao et al., 2000a).
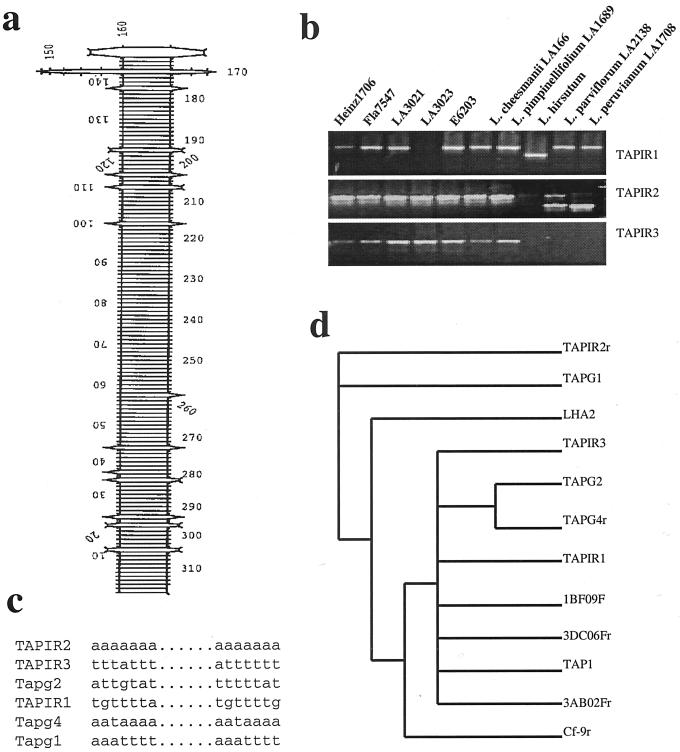
Figure 2.
Characterization of foldback transposon TAPIR. a, Strong secondary structure of the TAPIR element. b, A PCR survey of the three TAPIR loci in various tomato lines/species. Primers used are: TAPIR1f, 5′GAT AGT TAA AGA TGC GCC TAA C3′; TAPIR1r, 5′CGT GTG GGT GTA TAT CTA TTC3′; TAPIR2f, 5′GGT AGA TAG GCA AAA GTT TC3′; TAPIR2r, 5′GAA TAG ATA TAC ACC CAC ACG3′; TAPIR3f, 5′CAC TTG TTA TAC ACT TGT GAT GG3′; and TAPIR3r, 5′CCT TGT TGG GTA TTT GCA TGT G3′. c, The sequence of the direct repeats (DRs) flanking the tomato TAPIRs. d, A phylogenetic tree developed using the middle loop sequences of 12 TAPIRs. 1BF09F, 3DC06F, and 3AB02F were three BAC ends. The lowercase r indicates the reverse strand.
The transposition events of TAPIRs can be indicators of the evolutionary relationship among the tomato species. In Figure 2b, the investigation among various tomato species of the three loci corresponding to TAPIR1, 2, and 3 on 240K04 showed that polymorphism was more likely to be found in the wild species L. hirsutum, Lycopersicon parviflorum, and L. peruvianum. The other two wild species, Lycopersicon cheesemanii and L. pimpinefollium, displayed no polymorphism at all. This demonstrated that the genomes of the latter two wild species were more similar to the modern cultivars than the former three, concordant with the results of a previous phylogenetic study using RFLPs (Miller and Tanksley, 1990). Insertions of SDRs at certain loci of the cultivated tomato genomes could be very recent events, after the cultivated tomatoes were further separated from the wild species.
TAPIR elements have another feature common to known transposons, i.e. they are flanked by short DRs. Comparison of the flanking sequences of intact TAPIRs showed that TAPIR DRs were usually 7 bp long and mainly composed of A/T bases (Fig. 2c), indicating that A/T-rich regions could be the preferable insertion targets for TAPIRs. In addition to the three copies present in BAC 240K04 (Fig. 1), another nine tomato genomic sequences, all of which were sequences of identified genes, were found to bare TAPIR elements in their 5′/3′ or intron regions (Table II). A phylogenetic tree was generated using the sequences corresponding to the loop region of each TAPIR because this part of the sequence appeared to be the most heterogeneous. Eight out of 12 TAPIRs tested clustered into a major group including TAPIR1 and TAPIR3 (Fig. 2d). Therefore, TAPIRs may be derived from the same ancestor. The remaining four seemed to be more differentiated. It is unclear whether the sequence differences occurred after their insertion, or TAPIRs had different origins in their origination by “trapping” a different piece of genomic DNA in between the two inverted repeats.
Type III foldback transposons such as TAPIR are similar to MITEs such as maize Stowaway that also have strong secondary structure and are thought to be associated with genes in both monocot and dicot plants (Bureau and Wessler, 1994). Though multiple cis elements such as ethylene and auxin response elements have been found in TAPIR sequences near tomato TAPG genes (Hong and Tucker, 1998), no essential function was found for TAPIRs in regulating adjacent down stream genes (Hong et al., 2000). This may imply that the association of TAPIRs with tomato genes does not mean that they have particular functions in gene regulation but is simply due to the preferable sequence composition for TAPIR insertion sites at these positions. A recent study of Arabidopsis transposons also demonstrated that A/T-rich sequences, which in many cases were intergenic and intron sequences, were the preferred locations for transposons (Le et al., 2000).
SDRs
Four SDRs were identified in the sequence of BAC 240K04 that had no similarity in sequence and secondary structure to any known transposable elements (Table II). The lengths of SDRs ranged from 150 to 400 bp. A BlastN search of GenBank showed that four kinds of SDRs were present in 18 tomato genomic DNAs. Like TAPIR elements, SDRs were found mainly located at a gene's 5′, 3′, or introns. The prevalence of SDRs indicated that these elements could be unidentified transposons. We tested the SDR1 locus in various tomato species using a similar PCR experiment as described in Figure 2b and found that wild tomato species L. hirsutum, L. parviflorum, and L. peruvianum had shorter products (data not shown), indicating that SDR1, like TAPIR, could be a transposable element too. This hypothesis was further supported by the discovery that half of the mRNA sequence of the phytoene synthase from the yellow flesh mutant (X67143.1) was a nearly full-length SDR1. Fray and Grierson (1993) reported that the yellow flesh mutant allele contains a highly repeated genomic DNA sequence. Aberrant transcripts containing part of the TAPIR1 sequence were also observed from the jointless allele (Mao et al., 2000a).
SDR3 was A/T rich and only 150 bp long. There were eight tomato genomic sequences in GenBank containing this element, including the 5′ region of the tomato HMG2 (3-hydroxy-3-methylglutaryl CoA reductase 2) gene that also contains a retrotransposon (ToLTR) at the 5′ end. SDR3 had four significant matches in the LGI (AI776920, AI487358, AW036511, and AW220080), indicating that this small element was often positioned close enough to be included in the transcript of a gene. Among the wild relatives of cultivated tomato species such as L. esculentum, L. pimpinefollium is considered the most closely related (Miller and Tanksley, 1990). The coding sequences of the vacuolar invertase in these two species have been shown to be identical (Elliott et al., 1993). The conservation of repetitive sequences that were associated with the gene in the two species, including the segments of retrotransposon Lere1 sequence at their 5′ region and the small element SDR2 at their 3′ region, may further indicate that they are very similar at the genomic sequence level.
It appeared that SDRs were specific to only members of the family Solanaceae. SDR3 and SDR4 each had a copy in a potato gene (Table II). Other than that, SDRs were absent from the non-tomato genomic sequences. The constraint of SDRs in the Solanaceae family may indicate that the occurrence of these elements was a recent event, after the separation of the Solanaceae from the remaining plant families.
Gene Colinearity of Tomato BAC 240K04 and the Arabidopsis Genome
Four criteria were used for synteny analysis: homology, physical distance, colinearity, and gene orientation. This means that all the genes considered had significant homology with each other, were physically close, and were in the same order and transcriptional orientation. First, the protein sequences from the fifteen putative tomato genes from BAC 240K4 were searched against the Arabidopsis sequences in GenBank using TBlastN. All but one (240K4.03) of the putative genes had significant similarity with the Arabidopsis genome with an expectation value of E < 10−10 (Table III). Besides 240K04.12 and 240K04.13 that belong to the retrotransposon polyprotein, six more ORFs had more than five Arabidopsis matches with an E value less than 10−10, indicating that they were members of various gene families. An example was 240K04.14, the MADS-box gene whose MADS-box domain is present in numerous MADS-box transcription factors in Arabidopsis. ORF 240K04.9, a putative auxin-independent growth promoter, had more than 25 Arabidopsis matches with E<10−10. The multiple Arabidopsis matches of tomato ORFs were apparently due to the high number of gene families in the Arabidopsis genome—more than 37% of Arabidopsis genes have >five members (AGI, 2000). Gene families may cause problems when to determine the Arabidopsis orthologue for the correspondent tomato gene based on sequence homology. Therefore, in our microsynteny study, homology is only one of the four criteria evaluated.
Table III.
TBlastN sequence comparison of 15 tomato ORFs with the Arabidopsis genome
| Name | No. of Matches with E < 10−10 | Syntenic Homologue | Chromosome | TBlastN E Value |
|---|---|---|---|---|
| 240K4.01 | 11 | F15K20 | II | e–48 |
| F20D10 | IV | e–92 | ||
| 240K4.02 | 1 | K19O4 | V | e–22 |
| 240K4.03 | 0 | |||
| 240K4.04 | 1 | K19O4 | V | e–235 |
| 240K4.05 | 1 | F14M13 | II | e–100 |
| 240K4.06 | 9 | F14M13 | II | e–211 |
| K19O4 | V | e–5 | ||
| F20D10 | IV | e–10 | ||
| 240K4.07 | 2 | F20D10 | IV | e–140 |
| 240K4.08 | 1 | F12B17 | V | e–56 |
| 240K4.09 | >25 | MNA5F22K18 | VIV | e–113e–112 |
| 240K4.10 | 0 | F12B17 | V | e–7 |
| 240K4.11 | 5 | K17E12MHK7 | IIIV | e–152e–146 |
| 240K4.12 | 15 | F3L12 | II | e–78 |
| 240K4.13 | >25 | F5A13 | I | 0.0 |
| 240K4.14 | >25 | F14M13 | II | e–72 |
| F22K18 | IV | e–23 | ||
| 240K4.15 | 23 | MNA5 | V | e–117 |
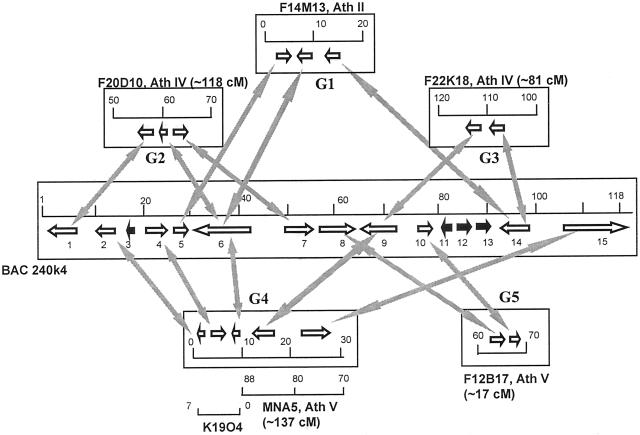
Figure 3 shows the network of microsynteny of the tomato ORFs on BAC 240K04 with the Arabidopsis genome. Regions in Arabidopsis were not labeled with the names of the ORFs due to the fact that in both tomato and Arabidopsis most of the ORFs were derived from the gene prediction programs and may not be correspondent ORF by ORF. Because the borders of ORFs on tomato BAC 240K04 were distinguishable we believe that the correspondent regions in Arabidopsis genome should represent separate ORFs. Five Arabidopsis DNA segments were found to contain clusters of genes that were physically close to each other and were in the same order as their homologs in tomato (G1–G5). The coding orientations of the corresponding genes were exactly the same in both genomes, meeting the criteria set above. The number of genes in each segment were from two to five. G3 and G5 contained two genes in less than 10 kb that were significantly similar to the tomato genes. The largest segment G4 contained five genes that were colinear with those on the tomato BAC 240K04. The ORF 240K04.6 had nine matches in the Arabidopsis genome. Three of them were located in the three syntenic segments from the Arabidopsis chromosomes II, IV, and V, respectively. Another ORF 240K04.14, the MADS-box gene, was associated with two Arabidopsis syntenic segments, G1 and G3. The presence of the MADS-box genes in the vicinity of other tomato homologs in Arabidopsis may be coincidental due to a large number of such genes and their highly conserved DNA binding domain. However, the identical coding orientation in both Arabidopsis and tomato when compared with the other gene(s) in the same syntenic segment supported that these MADS-box genes did fall within syntenic clusters in Arabidopsis.
Figure 3.
A schematic display of the Arabidopsis syntenic segments to tomato BAC 240K04. The number of centiMorgans (cM) indicate the approximate positions of the corresponding Arabidopsis BAC/P1-derived artificial chromosome clones on the Arabidopsis genome. Black arrows indicate ORFs that do not fall in any colinear segments. Arrows on the tomato BAC indicate approximate positions of the predicted ORFs, whereas arrows on Arabidopsis BAC/P1-derived artificial chromosome clones indicate major homologous regions.
Overall, the five Arabidopsis syntenic segments were from three chromosomes. G2 and G3 were from chromosome IV and more than 37 cM apart. G4 and G5 were from chromosome V and more than 120 cM apart. However, no duplicated Arabidopsis segments were observed for these syntenic clusters, compared with the tomato chromosome 2 ovate region where the Arabidopsis syntenic segments were duplicated (Ku et al., 2000). Thus, the Arabidopsis segments syntenic to the tomato JOINTLESS region may fall in the portion of the genome that was not duplicated because the duplicated part in the Arabidopsis genome comprises only 60% (AGI, 2000). It is interesting that none of the genes in the Arabidopsis syntenic segments contained a gene that had a different coding orientation to its tomato counterpart. In the tomato chromosome 2 ovate region, however, three out of 12 syntenic tomato genes were located on the opposite strand in Arabidopsis (Ku et al., 2000). This may indicate that the duplicated portion of the Arabidopsis genome could be subject to less selection pressure because of gene copy redundancy and therefore may accommodate more genetic rearrangements.
Although the duplicated Arabidopsis genome may fit the model of polyploidization and subsequent gene loss (AGI, 2000; Ku et al., 2000), we observed that ORF 240K04.6 was associated with three syntenic segments from three Arabidopsis chromosomes. In each segment, 240K04.6 was in the middle of the syntenic cluster and therefore the possibility of a false correlation was low, although ORF 240K04.6 had nine significant matches with the Arabidopsis sequences. The presence of one ORF in more than two synteneous segments supported the hypothesis that other alternatives such as independent segmental duplication may also be possible in shaping the Arabidopsis genome (AGI, 2000). We also noticed that three Arabidopsis segments (G1, G3, and G4) were spanning the tomato region that contained the retrotransposon (represented by 240K04.12 and 240K04.13). However, none of these segments contained a retrotransposon in their syntenic context, indicating that the transposition event in tomato occurred after the divergence of these two species about 100 MYA (Gandolfo et al., 1998; Yang et al., 1999).
CONCLUSIONS
The analysis of the 118,813-bp region from the tomato JOINTLESS region revealed that the tomato genome may contain an abundance of SDRs or small transposons. The presence of some of these elements may be very recent events as demonstrated by the PCR experiment to investigate the related loci in the wild tomatoes. Although SDRs were frequently associated with tomato genes, they did not appear to have functions in regulating adjacent genes. Unlike in rice and maize where small DNA elements, such as MITEs, have been found in large numbers and associated with genes too (Bureau and Wessler, 1994; Wessler et al., 1995; Mao et al., 2000b), the Arabidopsis genome has fewer numbers of such elements (Casacuberta et al., 1998; Ade and Belzile, 1999; Le et al., 2000). Therefore, the large number of such elements may contribute significantly in the expansion of the genomes of rice, maize, and tomato.
Comparison of the tomato JOINTELSS locus with the Arabidopsis genome demonstrated that small syntenic segments could be easily found in the genomes of these two dicot plants that have been diverged more than 100 MYA. In particular, the strict conservation of the gene-encoding orientations in these colinear segments indicates that the region may represent an ancestral block of genes. It is interesting that the comparison of this region did not reveal duplicated segments in Arabidopsis in which more than 60% of its genome is duplicated. It is tempting to put forward a hypothesis about the synteny between the tomato and Arabidopsis genomes, but a clearer picture will be visible only when we have additional long stretches of tomato sequence available for investigation.
MATERIALS AND METHODS
Tomato (Lycopersicon esculentum) BAC Library Screening
Hybridization of high-density tomato BAC filters using TG523 and TY159L insert is as described elsewhere (Mao et al., 2000a). DNAs of positive clones for insert check and Southern analysis were isolated using the standard alkali lysis method (Sambrook et al., 1989).
Construction of a Shotgun Library of Clone 240K4 and Sequencing
A maxiprep of tomato BAC clones was performed using alkali lysis and further purified by CsCl gradient centrifugation using a standard protocol (Sambrook et al., 1989). BAC DNA was nebulized under 6.5 pounds per square inch for 4 min and end repaired. Fragments of 2 to 5 kb were cut out from an agarose gel, purified, and ligated to pBluescipt. The ligation products were electroporated into DH10B competent cells (Gibco BRL, Rockville, MD). Clones were picked randomly and stored in 96-well microtiter plates.
DNA sequencing was performed on ABI 377XL automatic sequencers. Templates of shotgun clones were prepared by the AutoGen (Integrated Separation Systems, Portsmouth, NH) from 3-mL overnight cultures. DNA from each preparation was finally dissolved in 80 μL of water or Tris EDTA and 4 μL was used for sequencing reactions using the BigDye Cycle Sequencing Kit following manufacturer's instructions (ABI, Columbia, MD).
Sequence Analysis
Production sequencing of approximately 2,700 shotgun clones was performed with an additional 200 reactions for finishing. Phred (Ewing and Green, 1998; Ewing et al., 1998) was used for base calling from the trace files, and Consed (Gordon et al., 1998) was used to assemble the sequences into contigs. The final consensus sequence was searched against GenBank using BlastN and BlastX algorithms (Altschul et al., 1997) through the Web site of the National Center for Biotechnology Information (www.ncbi.nlm.gov). Gene structures were predicted with Genscan, Genscan+ (Chris Burge, Massachusetts Institute of Technology, Cambridge, http://CCR-81.mit.edu/GENSCAN.html), and GeneMarkHMM (Mark Borodovsky, Georgia Tech, Atlanta, http://genemark.biology.gatech. edu/GeneMark/). Arabidopsis databases was searched through The Arabidopsis Information Resource Web site (http://www.Arabidopsis.org/) and The Kazusa Arabidopsis data opening site (http://www.kazusa.or.jp/kaos/).
The GCG package (University of Wisconsin, Madison, Version 10) command FOLDRNA was used for analyzing secondary structures. The phylogenetic tree was made using PUAPSEARCH and PAUPDISPLAY from multiple sequence alignment generated by PILEUP.
ACKNOWLEDGMENTS
We are grateful to Mareen Milnamow and Ann Hu of the former Novartis Plant Protection, Inc. (Raleigh, NC) for technical help in shotgun library construction. Dr. S.D. Tanksley of Cornell University (Ithaca, NY) kindly provided the genomic DNA of some tomato species. We also thank Drs. Yeisoo Yu and Chunhuan Wan for their help during the course of sequencing and Rob Kingsberry, III (Clemson University, SC) for bioinformatics assistance. The sequence of 240K04 has been deposited in GenBank under the accession number AF292003. Supplemental data are viewable at http://www.genome.clemson.edu/∼lmao/pp_supplement.html.
Footnotes
This work was supported by the National Science Foundation (grant to R.A.W.) and by the Coker Chair in Plant Molecular Genetics (to R.A.W.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
LITERATURE CITED
- Ade J, Belzile FJ. Hairpin elements, the first family of foldback transposons (FTs) in Arabidopsis thaliana. Plant J. 1999;19:591–597. doi: 10.1046/j.1365-313x.1999.00567.x. [DOI] [PubMed] [Google Scholar]
- AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schafer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Tanksley SD. Characterization and genetic mapping of simple repeat sequences in the tomato genome. Mol Gen Genet. 1996;250:39–49. doi: 10.1007/BF02191823. [DOI] [PubMed] [Google Scholar]
- Budiman MA, Mao L, Wood TC, Wing RA. A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res. 2000;10:129–136. [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Ronald PC, Wessler SR. A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc Natl Acad Sci USA. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Casacuberta JM, Puigdomenech P, Monfort A. Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: characterization of the Emigrant family of elements. Plant J. 1998;16:79–85. doi: 10.1046/j.1365-313x.1998.00267.x. [DOI] [PubMed] [Google Scholar]
- Elliott KJ, Butler WO, Dickinson CD, Konno Y, Vedvick TS, Fitzmaurice L, Mirkov TE. Isolation and characterization of fruit vacuolar invertase genes from two tomato species and temporal differences in mRNA levels during fruit ripening. Plant Mol Biol. 1993;21:515–524. doi: 10.1007/BF00028808. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred: I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fray RG, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Lapitan NLV, Tanksley SD. A molecular and cytogenetic survey of major repeated DNA sequences in tomato (Lycopersicon esculentum) Mol Gen Genet. 1988;213:262–268. [Google Scholar]
- Gandolfo MA, Nixon KC, Crepet WL. A new fossil flower from the Turonian of New Jersey: Dressiantha bicarpellatagen. et sp. nov. (Capparales) Am J Bot. 1998;85:964–974. [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hong SB, Sexton R, Tucker ML. Analysis of gene promoters for two tomato poly-galacturonases expressed in abscission zones and the stigma. Plant Physiol. 2000;123:869–881. doi: 10.1104/pp.123.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Tucker ML. Genomic organization of six tomato polygalacturonases and 5′ upstream sequence identity with tap1 and win2 genes. Mol Gen Genet. 1998;258:479–487. doi: 10.1007/s004380050758. [DOI] [PubMed] [Google Scholar]
- Ku HM, Vision T, Liu J, Tanksley SD. Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le QH, Wright S, Yu Z, Bureau T. Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature. 2000a;406:910–913. doi: 10.1038/35022611. [DOI] [PubMed] [Google Scholar]
- Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Woo SS, Sasinowski M, Presting G, Frisch D, Goff S. Rice transposable elements: a survey of 73,000 sequence-tagged-connectors (STCs) Genome Res. 2000b;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer R, Ganal MW, Steffens JC, Tanksley SD. Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol Biol. 1991;16:753–770. doi: 10.1007/BF00015069. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tanksley SD. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor Appl Genet. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- Rebatchouk D, Narita JO. Foldback transposable elements in plants. Plant Mol Biol. 1997;34:831–835. doi: 10.1023/a:1005855008823. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- Yang YW, Lai KN, Tai PY, Li WH. Rates of nucleotide substitution in angiosperm mitochondrialDNA sequences and dates of divergence between Brassicaand other angiosperm lineages. J Mol Evol. 1999;48:597–604. doi: 10.1007/pl00006502. [DOI] [PubMed] [Google Scholar]
- Zamir D, Tanksley SD. Tomato genome is comprised largely of fast-evolving, low copy-number sequences. Mol Gen Genet. 1988;213:254–261. [Google Scholar]
- Zhang H, Martin GB, Tanksley SD, Wing RA. Map-based cloning in crop plants: tomato as a model system: II. Isolation and characterization of a set of overlapping yeast artificial chromosomes encompassing the jointlesslocus. Mol Gen Genet. 1994;244:613–621. doi: 10.1007/BF00282751. [DOI] [PubMed] [Google Scholar]