Abstract
Bacterial endotoxin (lipopolysaccharide [LPS]), a glycolipid found in the outer membranes of gram-negative bacteria, induces the secretion of proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interleukin-1 (IL-1), and IL-6 by monocytes/macrophages. The secretion of these biologically active compounds leads to multiple pathological conditions, such as septic shock. There is substantial evidence that chronic exposure to LPS mediates, at least in part, the tissue destruction associated with gram-negative infection. CD14, a 55-kDa protein, has been identified as an LPS receptor. In conjunction with a serum protein, LPS binding protein (LBP), LPS-CD14 interactions mediate many LPS functions in the inflammatory response. However, CD14 lacks a cytoplasmic domain, or any known signal transduction sequence motif, suggesting the existence of another cell surface domain capable of transducing signals. In this paper, we report a second, CD14-independent LPS binding site, which, based on biological activity, appears to be a functional LPS receptor. Cross-linking experiments were performed to identify LPS binding sites. Two molecules were identified: a 55-kDa protein (CD14) and a second, 78-kDa band. Sequencing of the 78-kDa protein by mass spectroscopic analysis revealed 100% homology with moesin (membrane-organizing extension spike protein). Antibody to CD14 induced partial blocking of the LPS response. However, antimoesin monoclonal antibody completely blocked the LPS-induced TNF-α response in human monocytes, without blocking CD14 binding of LPS. Irrelevant isotype controls had no effect. Additional experiments were performed to evaluate the specificity of the antimoesin blocking. Separate experiments evaluated antimoesin effects on monocyte chemotaxis, IL-1 production in response to IL-1 stimulation, and TNF-α secretion in response to Staphylococcus aureus stimulation. Antimoesin blocked only LPS-mediated events. The data suggest that moesin functions as an independent LPS receptor on human monocytes. The role of moesin in transduction of CD14-mediated signals is discussed.
Septic shock syndrome induced by gram-negative bacteria is a serious problem associated with high morbidity and mortality. In the United States, approximately 500,000 individuals suffer from sepsis annually; of these individuals, 175,000 die due to acute-phase reaction, disseminated intravascular coagulation, multiple organ failure, and shock (8, 31, 35). It is estimated that 50% of the sepsis cases originate from gram-negative bacterial infections (2).
The activity of lipopolysaccharide (LPS), on a cellular level, appears to be mediated by specific receptors (22). The LPS-induced proinflammatory activity of monocyte/macrophages has been shown to be mediated, at least in part, by a surface receptor, CD14, and a serum protein described as LPS binding protein (LBP) (42). However, the amino acid composition of the CD14 receptor does not contain a traditional transmembrane sequence and has been demonstrated to be a glycosylphosphatidylinositol-anchored protein without a cytoplasmic domain. One interpretation of this finding implies the existence of a coreceptor with a cytoplasmic domain to transduce signal across the cell membrane (10).
Experimental evidence has suggested the existence of multiple LPS binding sites and perhaps multiple receptors. For instance, using fluorescein isothiocyanate-labeled LPS (FITC-LPS) to measure LPS binding sites on human monocytes, CD14 blocking monoclonal antibody (in molar excess sufficient to block soluble and cell bound CD14) only partially inhibited FITC-LPS binding, suggesting the presence of other recognition sites (20, 28). Moreover, the addition of anti-CD14 to monocytes stimulated with LPS did not totally inhibit tumor necrosis factor alpha (TNF-α) production, suggesting an independently functioning receptor (7).
LPS-induced protein tyrosine phosphorylation has been shown to be specifically inhibited by anti-CD14 at low concentrations of LPS. However, at higher concentrations of LPS, tyrosine phosphorylation was not impaired, suggesting a lower-affinity CD14-independent pathway (29, 40). Several reports have suggested the existence of other CD14-independent LPS receptors and binding sites, including proteins of 18 (14), 38 to 40 (19), 70 to 80 (17, 18), and 95 to 96 (9, 13) kDa, using a variety of experimental approaches and different cell sources.
More recently, significant work has implicated Toll proteins in LPS-mediated signaling. Toll proteins were originally described in Drosophila as differentiation proteins with high homology to the human interleukin-1 (IL-1) receptor. Yang et al. demonstrated that Toll-like receptor 2 (TLR2) transfected into a human cell line is capable of transducing signals, as measured by translocation of NF-κB (43). The LPS-induced response was also measured in CD14-transfected cells, and the response was enhanced by cotransfection with TLR2, suggesting that TLR2 may act as a coreceptor for CD14. A second study, by Poltorak et al., reported that LPS resistance in C3H/HEJ mice is mediated by a mutation in a gene coding for TLR4 (25). Taken together, these reports suggest that proteins of the Toll family of receptors have mammalian analogues which function alone or in concert with CD14.
Based on these previous reports, the mechanism of action of LPS stimulation of monocytes is beginning to unfold. However, it is unclear if Toll-like proteins constitute the only class of protein capable of binding and transducing LPS signals, or if other molecules, some previously reported, have similar properties. In particular, the mechanism by which the signal for LPS binding is transferred across the plasma membrane remains an area of intense interest. In this paper, we report the isolation and characterization of another apparent LPS receptor, using a cross-linking strategy. Measurement of a variety of functions mediated by different agonists suggest specificity of the LPS-mediated interaction. Interestingly, monoclonal antibody to this molecule, identified as moesin (membrane-organizing extension spike protein), completely blocks the monocyte response to LPS, suggesting a role for moesin in the transduction of all LPS-induced signals, including those mediated by CD14. We explore the possibility that moesin may function as a signal transducing coreceptor for CD14.
MATERIALS AND METHODS
Reagents.
All reagents and buffers were purchased from Sigma Co. (St. Louis, Mo.). CD14 antibody MY4 was purchased from Coulter (Hialeah, Fla.), and antibody to moesin was purchased from Transduction Laboratories (Lexington, Ky.). Cross-linking reagents were obtained from Pierce Chemical Co. (Rockford, Ill.).
Monocyte cell culture and TNF-α production.
All patient samples were obtained after approval of the Internal Review Board at Boston University Medical Center. Peripheral venous blood was obtained from healthy volunteers, using heparin (10 U/ml) as the anticoagulant. Mononuclear cells were separated on Mono-Poly resolving medium, washed twice in phosphate-buffered saline (PBS), and resuspended in RPMI 1640 medium supplemented with 5% human type AB serum (culture medium). Cells were seeded into six-well tissue culture plates (Costar, Cambridge, Mass.) at a concentration of 2 × 107 cells per well and incubated in a humidified 5% CO2 atmosphere for 2 h. Nonadherent cells were aspirated, and the wells were washed three times with warm PBS. After the final wash, 2 ml of culture medium was added to each well (28).
TNF-α ELISA.
The release of TNF-α by macrophages was quantified using a commercial enzyme-linked immunosorbent assay (ELISA) kit (BioSource, Camarillo, Calif., or Cistron Inc., Pine Brook, N.J.) or by an ELISA assay developed in our laboratory as previously described (28). Ninety-six-well ELISA plates (Maxisorp; Nunc, Naperville, Ill.) were coated with mouse anti-human TNF-α monoclonal antibody (R&D Systems, Minneapolis, Minn.) in coating buffer (carbonate-bicarbonate buffer [pH 9.6]) by overnight incubation at 4°C. The wells were blocked overnight (4°C) with 2% bovine serum albumin in coating buffer, and samples were added. After overnight incubation (4°C), goat anti-TNF-α polyclonal antibody (R&D Systems) was added, followed by a donkey anti-goat horseradish peroxidase conjugate (Sigma). o-Phenylenezdiamine was used as the substrate. The reaction was stopped by addition of 4 N sulfuric acid, and optical density was measured with a Vmax microplate reader (Molecular Devices) at 490 to 600 nm. Samples with optical density values falling outside the standard range were assayed again at an appropriate dilution.
Cross-linking.
FITC-LPS from Escherichia coli O55:B5 and unlabeled LPS from the same strain were obtained from Sigma and used at a concentration of 3 μg of FITC/mg of LPS. LPS was also extracted from fresh cultures of E. coli O55:B5 by the hot phenol-water method and further purified by cesium chloride isopycnic density gradient centrifugation as previously described (23). Conjugation to FITC was accomplished by the method of Skelly et al. (30). Purified and conjugated LPS was sonicated three times for 10 s. Five hundred microliters of a 0.2% solution (in water) of sulfosuccinimidyl 2-(p-azidosalicylamido) o-1,3-dithioproprionate (SASD; Pierce Chemical Co.) was added according to the protocol of Wollenweber and Morrison (41), followed immediately by 0.1 M borate buffer (pH 8.5). The mixture was kept at room temperature for 30 min and sonicated three more times for 30 s, and an additional incubation with 0.4 mg SASD was performed under the same conditions. The FITC-LPS-ASD complex was separated from excess SASD by centrifugation at 2,000 × g for 2 min and subsequent dialysis of the cleared supernatant against PBS (pH 7.2) overnight at 4°C, stored, and aliquoted (9 × 100 μl) at −80°C. All reactions were carried out under reduced light, using a 25-W red light source. Selected wells of adherent mononuclear cells were incubated with anti-CD14 monoclonal antibody MY4 (Coulter) at a final concentration of 2.5 μg/ml and incubated at 37°C for 1 h with gentle shaking; then FITC-LPS-ASD (1 μg/ml) was added to all wells. Alternating wells also received a 100-fold excess of unlabeled LPS. The incubation was continued for 30 min with gentle shaking and placed on ice for photolysis as suggested by Tobias (34). Samples were subjected to photolysis by irradiation with shortwave UV light (Ultra-products Inc., San Gabriel, Calif.) from a distance of 1 cm for 10 min at 20°C, with occasional shaking. Samples were washed three times with cold PBS, and cells were lysed in a solution of 0.5% Triton X-100, 0.5% Nonidet P-40, 50 mM Tris (pH 8.0), 0.15 M NaCl, and 1 μl of freshly added phenylmethylsulfonyl fluoride for 20 min at 4°C. Nuclei were then pelleted by centrifugation at 12,000 × g for 15 min at 4°C. Equal aliquots of sample were placed in loading buffer (0.3 M sodium dodecyl sulfate, 20% sucrose-bromophenol blue) and applied to a 10% discontinuous mini-sodium dodecyl sulfate-polyacrylamide gel. LPS-cross-linked complexes were detected after Western blot transfer and staining with anti-fluorescein horseradish peroxidase conjugate (ECL system; Amersham).
Amino acid sequencing and mass spectroscopic analysis.
LPS was cross-linked to monocytes from 15 healthy donors, using the SASD method and a preparative gel run to obtain sufficient material for the sequence analysis. Protein was extracted directly from the gel by an in-gel digestion method (15). The protein was reduced by submerging the gel in 100 μl of 0.01 M dithiothreitol in 0.1 M Tris buffer (pH 8.5), with gentle shaking for 1 h at 50°C. The buffer was then replaced by 100 μl of 0.015 M N-isopropyliodoacetamide (5) in 0.1 M Tris (pH 8.5) and left in the dark for 30 min. The alkylating solution was then discarded, and the gel washed four times with 500 μl of 0.5 M Tris (pH 8.5) containing 50% acetonitrile for 30 min with shaking and dried completely in a Speed-Vac concentrator. Forty microliters of digestion buffer (0.2 μg of endoproteinase Lys-C in 0.05 M Tris [pH 8.5]) was added to the dried gel and incubated for 20 h at 37°C. The supernatant and three washes with 50% acetonitrile in 0.1% trifluoroacetic acid were combined for analysis. The preparation was fractionated by high-pressure liquid chromatography (HPLC) after drying and reconstitution in 0.1% trifluoroacetic acid. Seven peaks were individually collected and subjected to amino acid sequencing in a Perkin-Elmer sequencer (product no. 4949). To confirm the sequence data, matrix-assisted laser desorption ionization mass spectroscopic analysis was performed on the digest and on 10% of selected fractions, using a Perceptive Voyager PE-RP mass spectrometer.
Flow cytometry.
Flow cytometry was performed on a FACScan fluorescence-activated cell sorter, using FITC-labeled MY4 (anti-CD14 antibody; Coulter) or antimoesin antibody (Transduction Laboratories). FITC-labeled mouse immunoglobulin G2b (IgG2b) was used as the irrelevant antibody control for MY4, and IgG1 was used as the irrelevant antibody control for antimoesin. Human monocytes were isolated (2.5 × 106 cells/ml in PBS [pH 7.2] containing 0.02% azide) and incubated with FITC-MY4 at a concentration of 2.5 μg/ml. This concentration yielded optimal staining in preliminary experiments. Cells were preincubated with 250 μg of antimoesin per ml for 10 min, or the antimoesin was added at the same time as the CD14 ligand. One million cells (0.4 ml) were analyzed by fluorescence-activated cell sorting for each condition, in triplicate.
Chemotaxis.
Peripheral venous blood was separated by Ficoll-Hypaque centrifugation. Cells were suspended in an assay medium consisting of Gey’s balanced salt solution supplemented with 2% bovine serum albumin at a concentration of 2.5 × 106 cells per ml. The cell suspension was preincubated for 30 min with either antimoesin (50 μg/ml) or irrelevant antibody (IgG1; 50 μg/ml) and then placed in the upper compartment of a modified Boyden chamber separated by a 5-μm-pore-size Micropore filter. The lower compartment contained the synthetic chemotactic peptide formylmethionylleucylphenylalanine (FMLP; 2 × 10−8 M). Chemotaxis was evaluated by counting the number of cells that accumulated on the distal surface of the filter after a 60-min incubation. This method of quantification was found to correlate well with other methods of cell enumeration such as leading-front or through-the-filter counting (33). Ten high-power fields (magnification of ×400) were counted for each of triplicate filters. Statistical differences between conditions were determined by analysis of variance (ANOVA). Random migration was determined under the same conditions without chemotactic stimulus.
IL-1β autocrine stimulation of IL-1β release.
Human monocytes were obtained from healthy donors, isolated, purified, and placed in six-well plates as described above. Cells were preincubated with 100 μg each of either antimoesin or irrelevant antibody per ml, then stimulated with 10 ng of human recombinant IL-1β (R&D Systems) per ml for 4 h at 37°C in a humidified 5% CO2 atmosphere, washed three times with warm Dulbecco’s phosphate-buffered saline and placed in fresh medium for 8 h. As a positive control, cells were incubated with 1 μg of E. coli LPS per ml under similar conditions. The secreted IL-1β protein was quantified with a commercial ELISA kit.
TNF-α production in response to stimulation with Staphylococcus aureus.
Human monocytes were prepared as described above. The cells were then incubated with heat-killed S. aureus at a concentration of 108 bacteria/ml for 16 h, and the secreted TNF-α protein was quantified with a commercial ELISA kit. Stimulation with LPS served as a positive control, as described above.
Statistical analysis.
Statistical significance in the ELISA was analyzed by one-way ANOVA. A comparison between dose and kinetic responses of the different LPS preparations was evaluated by repeated-measures ANOVA.
RESULTS
Cross-linking.
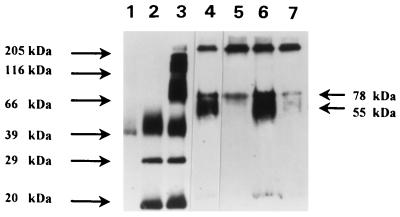
We used a cross-linking strategy similar to that used to isolate and characterize LBP to identify LPS binding proteins on the surface of human monocytes (34). After cross-linking and Western blot analysis, two bands were identified as binding LPS (Fig. 1). The controls used in these experiments included MY4 (anti-CD14), irrelevant isotype antibody, and excess unlabeled LPS, to determine which band corresponded to CD14 and to control for nonspecific binding of LPS. The 55-kDa band corresponding to CD14 was inhibited by anti-CD14 antibody. The 78-kDa band was not blocked by MY4; however, the intensity of staining was slightly reduced, suggesting that optimal binding of the 78-kDa protein may be dependent on CD14 binding. The 78-kDa band was cut from the gel for sequence analysis.
FIG. 1.
Western blot analysis of cross-linking of LPS to the monocyte surface. Lanes: 1, FITC-LPS; 2, FITC-labeled low-molecular-weight markers; 3, FITC-labeled high-molecular-weight markers; 4, monocytes incubated with FITC-LPS-ASD plus irrelevant antibody IgG2b; 5, monocytes incubated with FITC-LPS-ASD plus MY4; 6, monocytes incubated with FITC-LPS-ASD; 7, monocytes incubated with FITC-LPS-ASD plus excess unlabeled LPS. Molecular masses are indicated at the left and right. Two strongly staining bands are apparent when monocytes are labeled with FITC-LPS-ASD (lane 6). The monoclonal antibody to CD14 (MY4, 2.5 mg/ml) inhibits binding of FITC-LPS-ASD at 55 kDa (lane 5); excess unlabeled LPS displaces binding of the labeled LPS at both 55 and 78 kDa (lane 7), demonstrating competition of binding by unlabeled LPS.
Purification and sequencing.
To obtain sufficient material for sequencing, monocytes from 15 healthy donors were purified and FITC-LPS-ASD cross-linked as described above. The membrane fraction was run in a sulfo-link column (Pierce), using anti-FITC to remove the receptor complexes. The proteins (CD14 and 78-kDa protein) were separated by gel filtration and isolated by cleavage of the ASD linkage with 2-mercaptoethanol. The purified LPS binding proteins were digested with endoproteinase Lys-C, and the molecular weight and sequence were determined by HPLC and mass spectroscopy. The 55-kDa band was confirmed to be CD14. The primary sequence of the 78-kDa fragments analyzed revealed 100% homology with moesin (Fig. 2).
FIG. 2.
Primary sequence of moesin. The boxed areas are the sequences obtained for the various 78-kDa protein peptides derived from endopeptidase digestion and HPLC fractionation. The sequences of the 78-kDa peptides demonstrate 100% homology to the published sequence of moesin.
Flow cytometry.
In a separate set of flow cytometry experiments, inhibition of CD14 binding by antimoesin was evaluated in order to rule out cross-reactivity of antimoesin with CD14. MY4 was used as the CD14 ligand and was labeled with FITC. Results revealed no inhibition of binding to CD14 by antimoesin antibody at 100-fold excess concentrations (data not shown).
Role of moesin in LPS signaling.
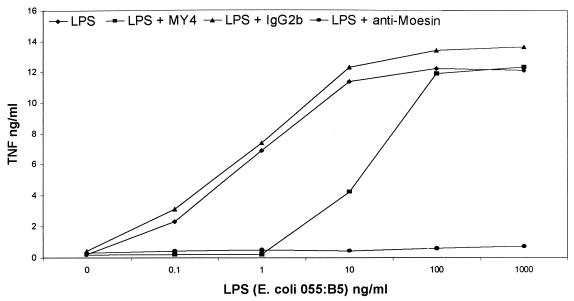
Antibody inhibition experiments were performed to determine the effect of antimoesin antibody on the biological response to LPS binding to monocytes. In the first experiment, TNF-α secretion was measured after stimulation with LPS. Freshly isolated monocytes from healthy donors were pretreated with antimoesin, MY4, and irrelevant antibody controls. MY4 inhibited the response to LPS only at low concentrations (Fig. 3), although, on a molar basis, the concentration of antibody was sufficient to bind any free, soluble CD14 that might be available from the serum in the reaction mixture. Conversely, antimoesin inhibited the LPS response at all concentrations, suggesting a requirement for moesin for the transduction of the CD14-mediated signal.
FIG. 3.
Blocking of LPS stimulation (1 μg/ml) of TNF-α secretion by antibody to CD14 and antibody to moesin. Anti-CD14 blocks the LPS-induced response at low concentrations, whereas antimoesin completely blocks LPS stimulation of TNF secretion. The irrelevant antibody controls had no effect.
Antimoesin blocking of cell functions unrelated to LPS binding.
We reasoned that since moesin is a structural protein in fairly large quantity on the cell surface, antimoesin may have been paralyzing the cell, inhibiting all receptor-mediated functions. To determine the specificity of blocking by antimoesin, we used three different agonists with separate functional outcomes and tested the effects of antimoesin on chemotaxis to formulated peptide, IL-1 secretion in response to IL-1 stimulation, and TNF-α secretion in response to S. aureus stimulation.
Chemotaxis.
Chemotaxis was analyzed by a modified Boyden chamber assay using FMLP as the chemoattractant. There was no significant inhibition of chemotaxis to FMLP by either antimoesin or irrelevant antibody (IgG1) (each at 50 μg/ml); values for FMLP, antimoesin, IgG1, and random migration (PBS) were 26.1 ± 5.4, 21.4 ± 4.3, 21.5 ± 4.2, and 1.5 ± 0.7 cells per high-power field, respectively.
IL-1β autocrine stimulation of IL-1β release.
Inhibition by antimoesin of autocrine stimulation of IL-1 release was evaluated by ELISA. The results revealed that antimoesin had no effect on IL-1-stimulated monocyte function. Values for IL-1, IL-1 plus antimoesin, IL-1 plus IgG1, LPS, and LPS plus antimoesin were 16.32 ± 0.78, 16.42 ± 1.1, 16.54 ± 1.2, 18.7 ± 1.9, and 0.85 ± 0.7 ng/ml, respectively.
TNF-α production in response to stimulation with S. aureus.
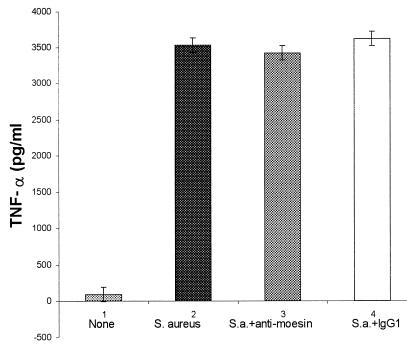
S. aureus is a potent, LPS-independent stimulator of monocyte secretion of TNF-α. Evaluation of TNF-α secretion by ELISA revealed that antimoesin inhibited LPS-mediated stimulation but not S. aureus stimulation (Fig. 4).
FIG. 4.
Stimulation of TNF-α secretion of monocytes by S. aureus. Antimoesin is used to inhibit the monocyte response. Antimoesin markedly inhibits the LPS-induced response but has no effect on S. aureus-induced secretion.
DISCUSSION
In this study, we report the identification and characterization of a new LPS binding site on human monocytes. Furthermore, we provide evidence to suggest that this new binding site, identified as moesin, has functional properties consistent with that of a receptor and may play a role as a signal transducing coreceptor for CD14.
The suggestion of a second LPS receptor independent of CD14 has come from experiments in which it was demonstrated that anti-CD14 blocking antibody was effective only at low concentrations of LPS (28). The lack of blocking by anti-CD14 antibody at higher LPS concentrations cannot be explained by excess soluble CD14 in the assay from serum sources, since the antibody is in molar excess to the possible contribution of soluble CD14 from serum (7, 20, 40). More recently, exciting new work implicating human analogues of Drosophila Toll proteins as receptors, and possibly signaling coreceptors for CD14, has provided support for the suggestion that other LPS binding sites may play an important role in the biologic response to bacterial endotoxin.
We cross-linked FITC-LPS to the putative new receptor by using a photoactivatable cross-linking agent, SASD, which resulted in identification of a second protein (in addition to CD14) with an apparent molecular mass of 78 kDa. The protein was purified by elution from the gel and sequenced after internal digestion of the single protein. Sequencing of the resultant peptide fragments revealed 100% homology with a known protein, moesin, a member of the ERM, or band 4.1, gene superfamily, which includes talin, ezrin, radixin, protein 4.1, and merlin. All of these proteins are associated with the submembranous cytoskeleton, although moesin is known to have a cell surface domain and to be capable of signal transduction (12, 21, 24, 32). Members of this family of proteins are widely expressed in different tissues and cells; they have been found to be localized to filopodia and other membranous protrusions that are important for ligand recognition, signal transduction, and motility. Most previous reports have focused on moesin as a linking protein of the submembranous cytoskeleton (27, 36, 37). It is found primarily in the core of microextensions such as filopodia, microvilli, microspikes, and retraction fibers. The cellular distribution is variable, but it has been found in macrophages, lymphocytes, fibroblasts, endothelial cells, epithelial cells, and neuronal cell lines (1). However, it is found primarily in leukocytes and endothelial cells (1). The subcellular distribution of moesin follows closely the dynamic changes in cell shape that take place during attachment, spreading, and cell movement.
Interestingly, although moesin was first identified as a structural protein, it was later found to be associated with the receptor for measles virus (4). Subsequently, a mouse monoclonal antibody was produced by immunization with bovine moesin. Treatment of cells with the monoclonal antibody effectively prevented measles virus infection, although there is some question about whether there may have been cross-reactivity with CD46 (3). Other studies suggest that although CD46 is the primary measles receptor, association of CD46 with moesin is necessary for transduction of the signal (6, 26). In another system, the transduction of signal after binding of ligand to CD44 appears to be mediated by association of CD44 with moesin (38, 44). The gene for moesin has been cloned and sequenced (16). Moesin also has been implicated in binding of human immunodeficiency virus type 1 envelope protein gp120 (11), in rheumatoid arthritis (39), and in Rho kinase phosphorylation (12).
Antibody inhibition experiments were performed to determine the relationship of CD14 binding and moesin binding of LPS to the biologic response. Anti-CD14 (MY4) is known to block the biologic response at low concentrations of antibody. One explanation is the binding of LPS to an independent, lower-affinity receptor (possibly moesin) at higher concentrations of LPS. It was expected that antimoesin would not block the biologic response at low LPS concentrations due to LPS binding to CD14. However, antimoesin blocked the biologic response of monocytes at all concentrations of LPS tested, suggesting that moesin is involved in the transduction of the CD14 signal. While studies to elucidate the interaction of CD14 and moesin in the cell membrane remain to be done, the finding that inhibition of CD14 binding of LPS by MY4 also partially reduces binding of LPS to moesin suggests that a physical interaction of CD14 and moesin is necessary for optimal binding of LPS. Competitive inhibition experiments revealed that anti-CD14 and antimoesin do not cross-react with moesin and CD14, respectively.
Experiments were carried out to determine the specificity of the antimoesin inhibition of monocyte function. Since moesin is a structural membrane protein, it is possible that antimoesin has a more global effect, inhibiting not only LPS-mediated events but also a wide variety of receptor-mediated events. We examined two monocyte functions unrelated to TNF-α secretion stimulated by two unrelated receptors, chemotaxis stimulated by FMLP and IL-1 secretion stimulated by IL-1, and found that neither was inhibited by antimoesin. We also investigated TNF-α secretion stimulated independently of LPS, using heat-killed S. aureus; antimoesin had no effect on TNF-α secretion mediated by a molecule, other than LPS.
In conclusion, the data suggest that moesin is a second, independent receptor for LPS on the surface of human monocytes capable of stimulating a biologic response (TNF-α secretion). There is also indirect evidence, obtained through antibody inhibition experiments, that moesin is a necessary coreceptor for the transduction of the CD14 signal. Further studies are required to determine the exact nature of the interaction between LPS, LBP, CD14, Toll, and moesin.
ACKNOWLEDGMENTS
We acknowledge the help and advice of Douglas Golenbock and the assistance of Mary Ann Gawinowicz with performance of the gas chromatography mass spectroscopy sequence analysis.
This work was supported in part by PHS grants DE06436, DE10709, and DE07206.
REFERENCES
- 1.Amieva M, Furthmayr H. Subcellular localization of moesin in dynamic filopodia, retraction fibers, and other structures involved in substrate exploration, attachment, and cell-cell contacts. Exp Cell Res. 1995;219:180–196. doi: 10.1006/excr.1995.1218. [DOI] [PubMed] [Google Scholar]
- 2.Danner R L, Elin R J, Hosseini J M. Endotoxemia in human septic shock. Chest. 1991;99:169–176. doi: 10.1378/chest.99.1.169. [DOI] [PubMed] [Google Scholar]
- 3.Devaux P, Gerlier D. Antibody cross-reactivity with CD-46 and lack of cell surface expression suggest that moesin might not mediate measles virus binding. J Virol. 1997;71:1679–1682. doi: 10.1128/jvi.71.2.1679-1682.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunster L, Schneider-Schaulies J, Dehof M, Holers V, Schwartz-Albiez R, ter Meulen V. Moesin, and not the murine functional homologue (Crry/p65) of human membrane cofactor protein (CD46), is involved in the entry of measles virus (strain Edmonston) into susceptible murine cell lines. J Gen Virol. 1995;76:2085–2089. doi: 10.1099/0022-1317-76-8-2085. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara P, Rosenfield J, Guillemot J C, Capdeville J. Internal peptide sequencing. In: Angeletti R H, editor. Techniques in protein chemistry IV. San Diego, Calif: Academic Press Inc.; 1993. pp. 379–387. [Google Scholar]
- 6.Gerlier D, Varior-Krishnan G, Devaux P. CD-46 mediated measles virus entry: a first key to host range specificity. Trends Microbiol. 1995;3:338–345. doi: 10.1016/s0966-842x(00)88972-6. [DOI] [PubMed] [Google Scholar]
- 7.Gessani S, Testa U, Varano B, DiMarzio P, Borghi P, Conti L, Barberi T, Tritavelli E, Martucci R, Seripa D. Enhanced production of LPS-induced cytokines during differentiation of human monocytes to macrophages. J Immunol. 1993;151:3758–3766. [PubMed] [Google Scholar]
- 8.Glauser M P, Zanetti G, Baumgartner J D, Cohen J. Septic shock pathogenesis. Lancet. 1990;338:732–736. doi: 10.1016/0140-6736(91)91452-z. [DOI] [PubMed] [Google Scholar]
- 9.Hampton R Y, Golenbock D T, Raetz R H C. Lipid A binding sites in membranes of macrophage tumor cells. J Biol Chem. 1988;263:14802–14807. [PubMed] [Google Scholar]
- 10.Haziot A, Chen S, Ferrero E, Low M G, Sibert R, Goyert S M. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol. 1988;141:547–552. [PubMed] [Google Scholar]
- 11.Hecker C, Weise C, Schneider-Schaulies J, Holmes H C, ter Meulen V. Specific binding of HIV-1 envelope protein gp120 to the structural membrane proteins ezrin and moesin. Virus Res. 1997;49:215–223. doi: 10.1016/S0168-1702(97)00039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Sukita S T, Tsukita S. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirikae T, Inada K, Hirata M, Yoshida M, Kondo H, Hisatsune K. Identification of RE lipopolysaccharide-binding protein on murine erythrocyte membrane. Microbiol Immunol. 1988;32:33–44. doi: 10.1111/j.1348-0421.1988.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 14.Kirkland T N, Virca G D, Kuus-Reichel T, Tobias P. Identification of lipopolysaccharide-binding proteins in 70Z/3 cells by photoaffinity crosslinking. J Biol Chem. 1990;265:9520–9525. [PubMed] [Google Scholar]
- 15.Krutzsch H C, Inman J K. N-isopropyliodoacetamide in the reduction and alkylation of proteins: use in microsequence analysis. Anal Biochem. 1993;209:109–116. doi: 10.1006/abio.1993.1089. [DOI] [PubMed] [Google Scholar]
- 16.Lankes W, Schwartz-Albiez R, Furthmayer H. Cloning and sequencing of porcine moesin and radixin cDNA and identification of highly conserved domains. Biochim Biophys Acta. 1993;1216:479–482. doi: 10.1016/0167-4781(93)90018-9. [DOI] [PubMed] [Google Scholar]
- 17.Lei M G, Morrison D C. Identification of lipopolysaccharide-binding protein with specificity for an inner core region (kDaO) determinant. FASEB J. 1991;5:A1363. [Google Scholar]
- 18.Lei M G, Morrison D C. Specific endotoxin lipopolysaccharide-binding protein on murine splenocytes. J Immunol. 1988;141:996–1005. [PubMed] [Google Scholar]
- 19.Lei M G, Qureshi N, Morrison D C. Lipopolysaccharide (LPS) binding to 37 kDa and 38 kDa surface proteins on lymphoreticular cells. Immunol Lett. 1988;36:245–250. doi: 10.1016/0165-2478(93)90096-k. [DOI] [PubMed] [Google Scholar]
- 20.Lynn W A, Kiu Y, Golenbock D T. Neither CD14 nor serum is absolutely necessary for activation of mononuclear phagocytes by bacterial lipopolysaccharide. Infect Immun. 1993;61:4452–4461. doi: 10.1128/iai.61.10.4452-4461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison D. Endotoxins and disease mechanisms. Annu Rev Med. 1987;38:417–432. doi: 10.1146/annurev.me.38.020187.002221. [DOI] [PubMed] [Google Scholar]
- 23.Morrison D, Lieve L. Fractions of lipopolysaccharide from Escherichia coli O111:B4 prepared by two extraction procedures. J Biol Chem. 1975;250:2911–2919. [PubMed] [Google Scholar]
- 24.Nakamura F, Amieva M, Hirota C, Mizuno Y, Furthmayr H. Phosphorylation of 558T of moesin detected by site-specific antibodies in RAW264.7 macrophages. Biochem Biophys Res Commun. 1996;226:650–656. doi: 10.1006/bbrc.1996.1410. [DOI] [PubMed] [Google Scholar]
- 25.Poltorak A, He X, Smirnova I, Liu M, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 26.Schneider-Schaulies J, Dunster L, Schwartz-Albiez R, Krohne G, ter Meulen V. Physical association of moesin and CD46 as a receptor complex for measles virus. J Virol. 1995;69:2248–2256. doi: 10.1128/jvi.69.4.2248-2256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz-Albiez R, Merling A, Spring H, Moller P, Koretz K. Differential expression of the microspike-associated protein moesin in human tissues. Eur J Cell Biol. 1995;67:189–198. [PubMed] [Google Scholar]
- 28.Shapira L, Takashiba S, Amar S, Van Dyke T E. Porphyromonas gingivalis lipopolysaccharide stimulation of human monocytes: dependence on serum and CD14 receptor. Oral Microbiol Immunol. 1994;9:112–117. doi: 10.1111/j.1399-302x.1994.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 29.Shapira L, Takashiba S, Champagne C, Amar S, Van Dyke T E. Involvement of protein kinase C and protein tyrosine kinase in lipopolysaccharide-induced TNF-alpha and IL-1 beta production by human monocytes. J Immunol. 1994;153:1818–1824. [PubMed] [Google Scholar]
- 30.Skelly R R, Munkenbach P, Morrison D C. Stimulation of T-independent antibody responses by hapten-lipopolysaccharides without repeating polymeric structure. Infect Immun. 1979;23:287–293. doi: 10.1128/iai.23.2.287-293.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone R. Search for sepsis drugs goes on despite past failures. Science. 1994;264:365–367. doi: 10.1126/science.8153620. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K, Sasaki T, Mammoto A, Takaishi K, Kameyama T, Tsukita S, Takai Y. Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J Biol Chem. 1997;272:23371–23375. doi: 10.1074/jbc.272.37.23371. [DOI] [PubMed] [Google Scholar]
- 33.Ternowitz T. Human monocytes in neutrophil chemotaxis in vitro employing 51Cr labeled leukocytes. Scand Microbiol Immunol Pathol Acta. 1985;93:189–193. doi: 10.1111/j.1699-0463.1985.tb02943.x. [DOI] [PubMed] [Google Scholar]
- 34.Tobias P S. Cross-linking of LPS to CD14 on THP-1 cells mediated by LPS-binding protein. J Immunol. 1993;150:3011–3021. [PubMed] [Google Scholar]
- 35.Tracey K J, Lowry S F. The role of cytokine mediators in septic shock. Adv Surg. 1990;23:21. [PubMed] [Google Scholar]
- 36.Tsukita S, Yonemura S, Tsukita S. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci. 1997;22:53–58. doi: 10.1016/s0968-0004(96)10071-2. [DOI] [PubMed] [Google Scholar]
- 37.Tsukita S, Yonemura S. ERM (ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- 38.Tsukita S, Oishi K, Sato N, Kawai A, Tsukita S. ERM family members a molecular linkers between the cell surface glycoprotein CD-44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagatsuma M, Kimura M, Suzuki R, Takeuchi F, Matsuta K, Watanabe H. Ezrin, radixin and moesin are possible auto-immune antigens in rheumatoid arthritis. Mol Immunol. 1996;33:1171–1176. doi: 10.1016/s0161-5890(96)00083-1. [DOI] [PubMed] [Google Scholar]
- 40.Weinstein S, Gold M R, De Franco A L. Bacterial lipopolysaccharide stimulates protein tyrosine phosphorylation in macrophages. Proc Natl Acad Sci USA. 1991;88:4148–4152. doi: 10.1073/pnas.88.10.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wollenweber H, Morrison D. Synthesis and biochemical characterization of photoactivatable, iodinatable, cleavable lipopolysaccharide derivative. J Biol Chem. 1985;260:15068–15074. [PubMed] [Google Scholar]
- 42.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 43.Yang R B, Mark M R, Gray A, Huang A, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature. 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 44.Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol. 1998;140:885–895. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]