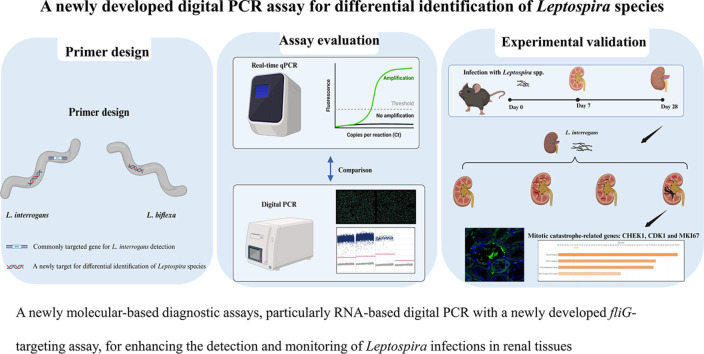
Highlights
-
•
Novel fliG-targeting assay for differential detection with improved sensitivity.
-
•
Improved diagnosis in chronic/latent infection for animal leptospirosis.
-
•
Trace Leptospira detection for better chronic leptospirosis diagnosis.
-
•
Our strategy effectively enables non-invasive Leptospira detection in urine.
Keywords: Leptospirosis, Leptospirosis kidney disease, Leptospira spp., Differential detection, Digital PCR, Latent infection
Abstract
Leptospirosis, a re-emerging zoonotic disease caused by Leptospira spp., poses significant global health and veterinary challenges. Long-term colonization of renal tubules by Leptospira in asymptomatic hosts highlights the need for sensitive detection methods. This study evaluates the chronic or latent Leptospira infections in kidneys using a novel molecular approach to examine individual immune responses differences. Digital PCR strategies employing newly developed primer-probe sets targeting the flagellar fliG gene were used to assess the presence of trace Leptospira in infected murine kidneys and urine samples from laboratory-confirmed leptospirosis patients. RNA-based digital PCR detected leptospires in 58 % (targeting lipl32) and 83 % (targeting fliG) of infected kidneys, demonstrating that the digital PCR strategy targeting the fliG gene offers superior sensitivity. Notably, the newly developed fliG-targeting assay detected as low as 20 fg of Leptospira DNA, offering ten-fold greater sensitivity than traditional qPCR for trace detection. This allows for differential detection of Leptospira species and facilitates monitoring of extremely low bacterial loads with greater sensitivity than conventional methods. We also observed regenerating renal tubules with mitosis and elevated cytokine expression in kidneys with transcriptionally active Leptospira during chronic infection. This approach aids in identifying latent infections and offers insights into individual variations. Our research provides a powerful molecular tool for epidemiological studies and public health surveillance, contributing valuable insights into the prevalence and transmission dynamics of this pervasive zoonotic disease.
Graphical abstract
1. Introduction
Leptospirosis caused by the genus Leptospira infection is the most widespread and neglected zoonotic disease which is also a waterborne disease prevalent in tropical and subtropical regions, accounting for over 70 % of cases occurring (Pappas et al., 2008; Vijayachari et al., 2008; Abela-Ridder et al., 2010; Costa et al., 2015; Karpagam and Ganesh, 2020). Climate change is leading to increased flooding events and a rise in the incidence of leptospirosis. Recent research has discovered that the disease is also associated with semi-arid regions and developed countries, such as the southwestern United States, may be related to the COVID-19 pandemic (Bini Viotti et al., 2020; Sanchez Fernandez et al., 2020; Sykes et al., 2022). Leptospirosis as a neglected burden at the human-animal-environment interface is a typical one health problem posing a public health threat and veterinary concern (Sykes et al., 2022).
This infectious disease affects a broad spectrum of animals, including domestic animals as well as wildlife, such as mice, rats, horses, cows, pigs, dogs, sea lions and cats, and also humans (Hartskeerl and Terpstra, 1996; Cilia et al., 2020). Animal become infected through direct contact with urine contaminated with Leptospira or indirectly through contact with contaminated soil, water, or infected animals (Bradley and Lockaby, 2023). Animal leptospirosis has a broad range of clinical effects, ranging from mild, subclinical infection to multiple-organ failure and death. Subclinical and asymptomatic chronic variants are also the most common forms of animal leptospirosis (Di Azevedo and Lilenbaum, 2021). Leptospira are maintained and transmitted in nature through chronic kidney infections in carrier animals, which excrete the bacteria in their urine for many years (Bradley and Lockaby, 2023). In humans, clinical manifestations range from mild flu-like symptoms to severe multiorgan dysfunction, primarily affecting the kidneys, liver, and lung, while some patients are asymptomatic (Ganoza et al., 2010; Yang et al., 2015; Rajapakse, 2022; Rajaonarivelo et al., 2023). Leptospirosis poses a dual burden as it affects both animals and humans, with the animal burden being greater than human burden (Noguera et al., 2022). Therefore, control and prevention management for leptospirosis should prioritize animal populations. A recent study published in the Journal of Applied Ecology emphasizes mice as a significant yet underestimated source of leptospirosis, highlighting the persistent risk posed by these silent spreaders and the necessity of controlling the spread of the infectious disease (Marie Moinet, 2024). Mice are a common, asymptomatic reservoir of the disease, as a real sanitary threat of both human and animal leptospirosis (Marquez et al., 2019).
The kidney, where conditions in the renal tubules favor Leptospira survival, serves as the preferential site of infection. Pathogenic leptospires undergo colonization within the kidney to initiate immune responses, contributing to the development of leptospirosis kidney disease (Mc and Montgomery, 1952; Matsui et al., 2016; Silva et al., 2020; Chancharoenthana et al., 2022; Chou et al., 2023). These colonization leads to a persistent presence of leptospires in the kidneys (Ratet et al., 2014; Yang et al., 2015; Carrillo-Larco et al., 2019). Research suggests the persistent presence of leptospires in kidneys, potentially due to either latent infection or chronic active infection. The diagnosis of the silent chronic form of animal leptospirosis remains a challenge (Di Azevedo and Lilenbaum, 2021).
Due to the mild and asymptomatic presentation of leptospirosis, this disease is often overlooked. Considering the potential risk of renal disease associated with asymptomatic and chronic leptospiral infections, the utilization of highly sensitive diagnostic methodologies is crucial in addressing leptospirosis. Traditional detection methods such as visualization (direct dark-field microscopy examination), culture and serological techniques (microscopic agglutination test, enzyme-linked immunosorbent assay and Lepto dipstick assay) have been regarded as ineffective because of their low specificity and inadequate sensitivity (Goris and Hartskeerl, 2014; Niloofa et al., 2015; Koizumi, 2020; Martin et al., 2022). While the detection of acute leptospirosis through direct examination, serology and molecular-based techniques is relatively standardized, the diagnosis of chronic leptospirosis, particularly its subclinical form, remains challenging. Current detection methods for Leptospira infections have limited sensitivity and specificity, rendering them unable to detect low bacterial loads in asymptomatic carriers.
Molecular methods, including conventional PCR, real-time quantitative PCR (real-time qPCR) and loop-mediated isothermal amplification (LAMP), are emerging as crucial tools for detecting leptospiral DNA in clinical specimens. Housekeeping genes and pathogen-specific genes serve as typical targets, including 16S rRNA, lipoprotein and flagellar genes such as rrs, lipl32, lfb1, gyrB, and secY, for leptospiral detection (Gokmen et al., 2016; Rupak Nagraik, 2020; Di Azevedo and Lilenbaum, 2021; Bárbara Couto Roloff Padilha, 2022; Pinto et al., 2022; Rethinavelu et al., 2022). The lipl32 gene, which encodes the major outer-membrane lipoprotein LipL32 found only in pathogenic Leptospira species, is currently the most commonly targeted gene for leptospiral detection (Podgorsek et al., 2020; Di Azevedo and Lilenbaum, 2021). Although both pathogenic and nonpathogenic Leptospira species can be detected for the target gene rrs, the gene is highly conserved across the bacterial kingdom, so false-positive results are likely to be detected.
Digital PCR has emerged as a reliable analytical chemistry method, providing greater sensitivity and precision in quantifying low-abundance targets (Salipante and Jerome, 2020; Chen et al., 2021; Lei et al., 2021). There is still lack of research on the application of digital PCR for detecting Leptospira species, especially in cases of latent or chronic infections. This study explores the utility of a digital PCR approach for enhanced sensitivity and precision in detecting low-abundance Leptospira targets, particularly in latent infections. Our findings provide evidence for the effectiveness of a digital PCR approach for the rapid diagnosis of trace amounts of Leptospira infection and its precision in distinguishing between distinct species of Leptospira.
This study focuses on developing a novel molecular approach, RNA-based digital PCR targeting fliG gene, to evaluate chronic or latent Leptospira infections in the kidneys, aiming to analyze differences in immune responses and their potential effects on disease progression among affected individuals. Despite the availability of various genetic markers for pathogenic Leptospira detection, such as lipl32, the fliG gene has not yet been explored as a target for molecular approach for Leptospira detection. The fliG gene encodes a protein associated with flagellar structures. Due to its conservation among Leptospira species, it is potentially marker for leptospiral detection. The combination of specific probes for distinguishing Leptospira species with the high sensitivity potential of digital PCR provides new targets for development of improved Leptospira detection methods. This highlights its potential utility in epidemiological surveillance and public health interventions aimed at managing and preventing Leptospira infections.
2. Materials and methods
2.1. Bacterial strains
Leptospira interrogans serovar Copenhageni Fiocruz L1–130 (ATCC number BAA-1198; a pathogenic species) and Leptospira biflexa serovar Patoc (ATCC number 23,582; a nonpathogenic species) were cultured at 28 °C under aerobic conditions in a medium consisting of 10 % Leptospira enrichment Elinghausen-McCullough-Johnson-Harris (EMJH) medium (BD Diagnostics) and 90 % Leptospira medium base EMJM medium. The number of motile leptospires was determined using dark-field microscopy with a Petroff-Hausser counting chamber (Schreier et al., 2009).
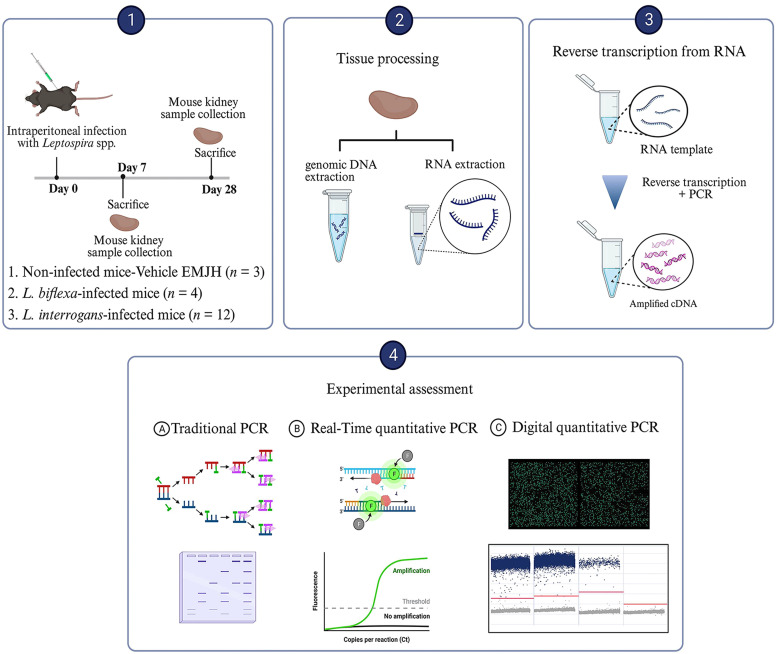
2.2. Chronic leptospiral infection in experimental mice
Seven- to eight-week-old C57BL/6 mice were intraperitoneally injected with 1 × 109 leptospires, while control mice were inoculated with EMJH media. Mice were randomized and allocated to the following groups: I) the control group (mice with base EMJH medium as a negative control; n = 3); II) the nonpathogenic Leptospira group (mice infected with L. biflexa; n = 4); and III) the pathogenic Leptospira group (mice infected with L. interrogans; n = 12). Mice were sacrificed at 7 and 28 days postinfection, after which the kidneys were harvested to extract genomic DNA and total RNA. All animal experiments were conducted under animal biosafety level 2 (ABSL-2) conditions and adhered to relevant guidelines for the use and handling of infected animals. The Institutional Animal Care and Use Committee of the Chang Gung Memorial Hospital in Taiwan approved all animal procedures and experimental protocols (Approval No 2,021,062,203 and No 2,021,121,801).
2.3. Genomic DNA and total RNA extraction
Genomic DNA was extracted from bacterial liquid cultures and whole kidney tissue homogenates using QIAamp DNA extraction kit following the manufacturer's protocol (Cat. No 51,306; Qiagen, Hilden, Germany). Extraction of total RNA from whole kidney tissue homogenates was performed using RNA-BeeTM RNAzol reagent (Tel-Test, Friendswood, TX, USA) with DNaseI digestion according to the manufacturer's protocol. Subsequently, 1 μg of total RNA was reverse transcribed using the Transcriptor cDNA Synthesis kit (Cat. No 04,897,030,001; Roche Diagnostics, Germany). Quantification of both total DNA and RNA was assessed using a NanoDrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE), and the samples were stored at −20 °C until further analysis.
2.4. Molecular detection using PCR and real-time qPCR
PCR was performed with SuperRed PCR Master Mix (BIOTOOLS Co., Ltd., Taiwan) on a PTC-100 programmable thermal controller (M.J. Research Inc., Waltham, Massachusetts, USA). The resulting amplicons were visualized through 2 % gel electrophoresis. Real-time qPCR experiments were performed with TaqMan gene expression assays on an ABI ViiA7™ real-time PCR system (Applied Biosystems).
2.5. Selection of candidate genes and primer design
In order to identify new candidate gene markers for Leptospira, the whole-genome sequences of L. interrogans (NC_005823.1) and L. biflexa (CP000786.1) retrieved from the NCBI database were aligned using Blastn (Zhang et al., 2000) to identify shared and conserved genes across Leptospira species, with a focus on encoding membrane and flagellar proteins. The primers and probes were designed using the online PrimerQuest™ Tool on the Integrated DNA Technologies website (Integrated DNA Technologies Inc., USA) using the default criteria. The primer and probe details are shown in Table 1.
Table 1.
Primer-probe sets sequence designed for detection of Leptospira species.
| Gene target | Primer/probea | Sequence (5′ to 3′)b | Amplicon size (bp)c | Leptospira target | Reference |
|---|---|---|---|---|---|
| lipl32 | lipl32_F | AAGCATTACCGCTTGTGGTG | 242 | L. interrogans | Matsui et al., 2017 |
| lipl32_R | GAACTCCCATTTCAGCGATT | ||||
| lipl32_P | FAM-AAAGCCAGGACAAGCGCCG-BHQ1 | ||||
| fliG | fliG_F | ATACTTCTGCGGGTGGTATTG | 110 | L. interrogans; L. biflexa | This study |
| fliG_R | GCAAGTTCGGGATCTTCTTCT | ||||
| fliG_P1 | FAM-AGTTGAGATTCTAAACTTAGTCG ATCGGGGAACG-BHQ1 | L. interrogans | |||
| fliG_P2 | FAM-ACGGAGAAGACCATCATCGAAGCT-BHQ1 | L. biflexa |
(F), (R) and (P) indicate forward and reverse primer and probe sequences, respectively.
the probe, labelled with a unique fluorescent reporter dye [6-carboxyfluorescein (FAM)] at the 5′-end and a 3′- Black Hole Quencher 1 (BHQ1), was designed to anneal to an internal sequence within the amplified region.
bp, base pairs.
2.6. Digital PCR
Each reaction contained a Probe PCR Master Mix (Qiagen), primers, probes and cDNA template. The digital PCR was performed using the QIAcuity Digital PCR System (Qiagen) and QIAcuity Nanoplate (Qiagen), which are microfluidic dPCR plates that allow for the processing of up to 26,000 partitions/well (Dreo et al., 2014). The QIAcuity Software Suite (Qiagen, version 2.1.7) was used to determine sample thresholds using positive, negative, and no-template control wells with the manual global threshold approach based on the amplitude signal observed in negative control samples.
2.7. Kidney transcript analysis
Relative mRNA expression levels were determined on an ABI ViiA7 real-time PCR system (Applied Biosystems) and calculated using the comparative cycle threshold (Ct) method, with GAPDH serving as an endogenous control.
2.8. Histology
Mouse kidneys were formalin-fixed and paraffin-embedded for the hematoxylin and eosin (H&E) staining to evaluate inflammatory changes and kidney structural injury by light microscopy.
3. Results
3.1. Chronically Leptospira-infected mice: renal immune responses induced by leptospira spp. infection
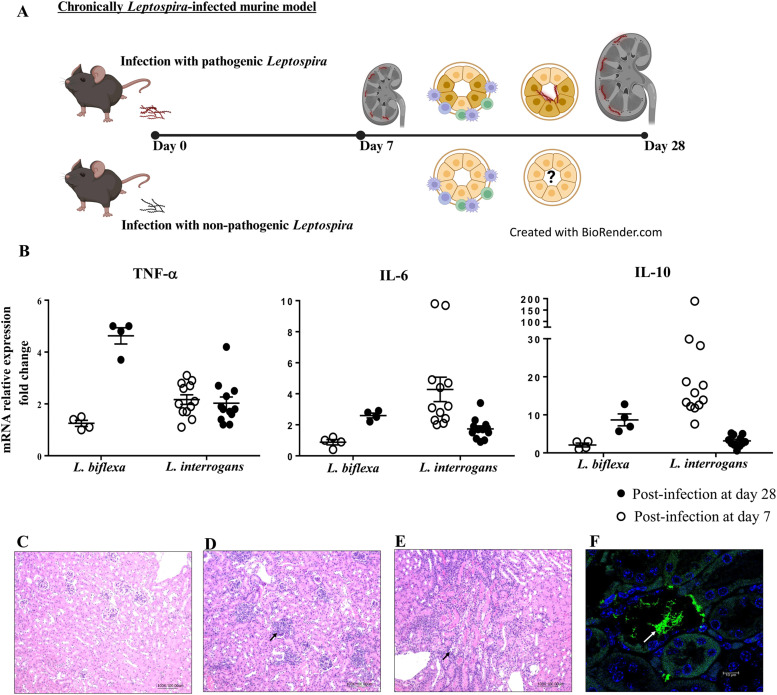
To understand how Leptospira establish persistence or latency in reservoir host species, we utilized mice as an infection model. Mice serve as long-term carriers and transmitters of Leptospira spp., typically without developing disease (Richer et al., 2015; Gomes-Solecki et al., 2017; Hamond et al., 2022). Fig. 1A illustrates the strategic approach of infecting C57BL/6 mice with Leptospira via intraperitoneal injection. It highlights the subsequent changes in immune responses and bacterial colonization within the kidneys, following infection with either pathogenic or non-pathogenic Leptospira species. Fig. 1B shows that pathogenic Leptospira infection triggers an elevated immune response characterized by increased levels of pro-inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6), following decreased anti-inflammatory cytokine (IL-10) levels over the 28-day infection period. This heightened inflammatory response, as evidenced by immune cell infiltration and bacterial colonization in renal tubules (Fig. 1C-F), can lead to chronic infection and tissue damage. In contrast, nonpathogenic Leptospira spp. infection also induces an immune response, as shown by elevated levels of TNF-α, IL-6, and IL-10 in the kidneys (Fig. 1B). Interestingly, the expression of the anti-inflammatory cytokine IL-10 appears to increase over time in nonpathogenic Leptospira infection, suggesting a potentially role in regulating the immune response. In addition, the H&E-stained tissue section also observed immune cell infiltration in kidneys infected with nonpathogenic Leptospira spp. (Fig. 1D).
Fig. 1.
Experimental design for investigating the impact of Leptospira infection on renal immune responses. (A) Schematic view of the study of immune responses and bacterial colonization in the kidneys following infection with either pathogenic or nonpathogenic Leptospira spp. (B) The immune responses to Leptospira infection are characterized by the mRNA expression of pro-inflammatory cytokines (TNF-α and IL-6) and the anti-inflammatory cytokine IL-10. Representative renal histological findings in uninfected (C) and infected (D∼F) mice. These tissues were formalin-fixed and paraffin-embedded for histology and immunofluorescence study. Light microscopy of kidney tissues, stained with the hematoxylin and eosin (Magnification, 100 X). After infection at 28 days, inflammatory infiltration is observed in the nonpathogenic L. biflexa-infected (D) and pathogenic L. interrogans-infected (E) mice. (F) Bacterial adherence to the tubule lumen was found in the kidneys from mice infected with L. interrogans at 28 days post-infection. Confocal images of immunofluorescent staining (magnification of 400X). Anti-leptospiral LipL32 (green; white arrow) and DAPI staining (blue).
Although immune responses are observed in kidneys infected with nonpathogenic Leptospira, it remains challenging to determine whether nonpathogenic Leptospira are present in trace amounts due to the lack of specific detection targets for nonpathogenic Leptospira spp. Current methods for detecting nonpathogenic Leptospira spp., such as 16S rRNA analysis, are prone to cross-reactivity with other bacteria, potentially leading to false-positive results and hindering accurate assessment of their presence in the kidneys. There is a need to develop more specific and sensitive detection methods for Leptospira that can simultaneously detect pathogenic and non-pathogenic Leptospira species for providing a more precise understanding of whether these bacteria establish latent or chronic (active form) infections in tissues.
3.2. Comparative Leptospira detection in infected kidneys: advantages of RNA-based digital PCR
This study compared digital PCR (targeting the lipl32 gene, which is unique to pathogenic Leptospira species) with conventional and real-time qPCR for detecting and quantifying leptospires in infected kidneys (Fig. 2). In our evaluation of digital PCR targeting the lipl32 gene to detect pathogenic Leptospira in infected kidneys, we observed the following results at the DNA and RNA levels. Using digital PCR with RNA-based analysis, leptospires were detected in 58 % (7 out of 12) of infected mouse kidneys on both days 7 and 28. In contrast, DNA-based analysis using digital PCR identified leptospires in 55 % (6 out of 11) of infected kidneys on day 7 and in 42 % (5 out of 12) on day 28. When using real-time qPCR, RNA-based analysis detected leptospires in 0 % (0 out of 12) of infected kidneys on day 7 and in only 8 % (1 out of 12) on day 28 (Table 2). These findings reveal a statistically significant difference (p < 0.05, Wilcoxon Signed Rank Test) between DNA- and RNA-based analysis on day 28 using digital PCR, supporting the conclusion that RNA-based digital PCR provides superior sensitivity for detecting Leptospira in kidney tissues. Additionally, the RNA-based method offers greater sensitivity and reduces false positive results. However, it is notable that digital PCR targeting the lipl32 gene did not detect bacteria in kidneys infected with nonpathogenic Leptospira, despite the presence of immune cell infiltration.
Fig. 2.
The evaluation of digital PCR for the detection of pathogenic Leptospira infection in kidneys. Our study employed a digital PCR approach designed to target the lipl32 gene, which is specific only to pathogenic species of Leptospira. This methodology, combined with conventional and real-time qPCR, was utilized for the detection and quantification of pathogenic leptospires in the kidneys of mice subjected to experimental Leptospira infection. Negative control mice were used as a reference for noninfected status, while mice with nonpathogenic L. biflexa infection served as an infection control group. Figure was created with biorender.com (accessed on Jan 2024).
Table 2.
Proportion of infected mice assessed by examining the presence of Leptospira in the kidneys.
| Target gene | Post-infection (days) | Groups | Genomic DNA extracted from kidneys |
Total RNA extracted from kidneys |
||||
|---|---|---|---|---|---|---|---|---|
| Traditional PCRa | Real-time PCRa | Digital PCRa | Traditional PCRa | Real-time PCRa | Digital PCRa | |||
| lipl32 | 7 | Without infection | 0 (0/3) | 0 (0/3) | 0 (0/3) | 0 (0/3) | 0 (0/3) | 0 (0/3) |
| L. biflexa infection | 0 (0/4) | 0 (0/4) | 0 (0/4) | 0 (0/4) | 0 (0/4) | 0 (0/4) | ||
| L. interrogans infection | 0 (0/12) | 8 (1/12) | 55 (6/11) | 0 (0/12) | 0 (0/12) | 58 (7/12) | ||
| 28 | Without infection | 0 (0/3) | 67 (2/3) | 0 (0/3) | 0 (0/3) | 0 (0/3) | 0 (0/3) | |
| L. biflexa infection | 0 (0/4) | 50 (2/4) | 0 (0/4) | 0 (0/4) | 0 (0/4) | 0 (0/4) | ||
| L. interrogans infection | 0 (0/12) | 33 (4/12) | 42 (5/12) | 50 (6/12) | 8 (1/12) | 58 (7/12) | ||
% of mice: Percentage (no. mice with the presence of Leptospira in the kidneys/no. total mice).
3.3. A newly developed digital PCR method for Leptospira detection: selection of Leptospira targets
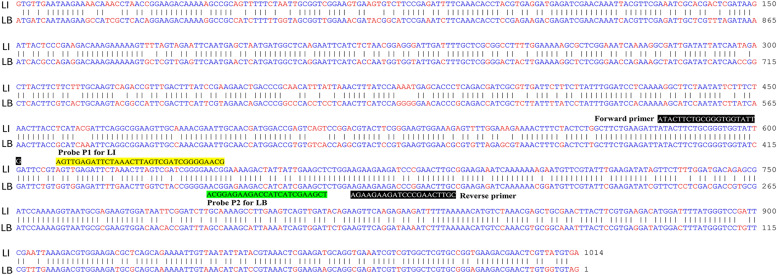
To identify novel Leptospira candidate gene markers, we designed new sets of primers and probes capable of simultaneously detecting and discriminating between pathogenic and nonpathogenic Leptospira species. The whole-genome sequences of L. interrogans and L. biflexa were downloaded from the NCBI database. A BLASTn search and sequence alignment were conducted to identify shared and conserved genes, particularly those encoding outer membrane, surface, and flagellar proteins, between pathogenic and nonpathogenic Leptospira species (Zhang et al., 2000). Comparative sequence analysis of the L. interrogans and L. biflexa genomes using a BLASTn-based alignment method revealed 79 % identity between the nucleotide sequence of fliG (LIC10023), encoding a flagellar motor switch protein from L. interrogans, and that of fliG2 (LEPBI_I3423) from L. biflexa (an alignment obtaining an E-value of zero). Alignment of the DNA sequence using the Martinez/Needlemen-Wunsch alignment method by MegAlign (DNASTAR, USA), with a gap penalty of 1.10 and a gap length penalty of 0.33, revealed that the sequence similarity index between LIC10023 and LEPBI_I3423 was 78.3 % (Fig. 3). Orthologous sequences to the selected nucleotide sequences were identified between L. interrogans and L. biflexa through whole-genome reciprocal best hit BLASTx analysis (Moreno-Hagelsieb and Latimer, 2008), indicating that protein sequences exhibiting >90 % identity and 99 % coverage were regarded as homologs. Hence, the orthologous gene pairs LIC10023 and LEPBI_I3423 between L. interrogans and L. biflexa were used for primer and probe design via the PrimerQuest™ Design Tool. We identified nucleotide sequence variant sites between the retrieved homologous sequences and then designed specific probes accordingly.
Fig. 3.
Sequence alignment for each primer/probe pair using representative Leptospira species. Alignment of the DNA sequence of the fliG gene from L. interrogans serovar Copenhageni str. Fiocruz L1–130 (NCBI accession number: NC_005823.1) and the fliG2 gene from L. biflexa serovar Patoc strain 'Patoc 1 (Paris)' (NCBI accession number: CP000786.1) was performed. Alignment of the DNA sequence using the Martinez/Needlemen-Wunsch alignment method by MegAlign (DNASTAR, USA) revealed that the sequence similarity index was 78.3 %. Identical nucleotides are shown as blue letters, and different nucleotides aligned at the same position are shown as red letters. The positions of the forward and reverse primers are indicated as white letters on a black background. The nucleotides corresponding to the probe for the target gene from L. interrogans (LI) are highlighted in yellow, while those for the target gene from L. biflexa (LB) are highlighted in green.
3.4. Validation of newly designed primer-probe sets for Leptospira species detection
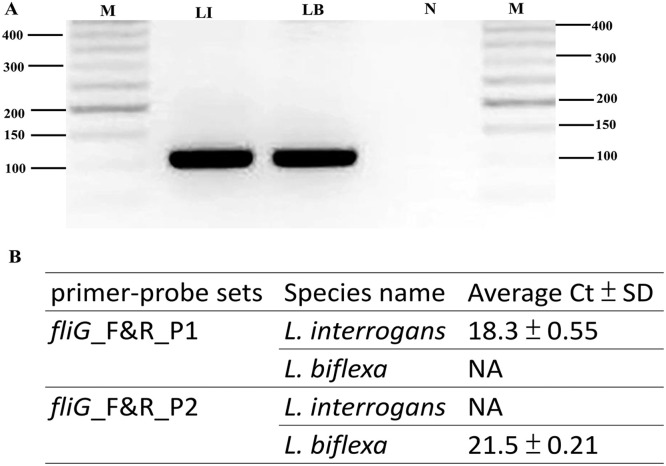
The ability of these primers and probes to identify leptospiral sequences was initially evaluated using the BLASTn program against the NCBI sequence database. To further assess the utility of the novel primer pairs targeting the fliG gene developed in this study, we performed PCR analysis using the primer pairs and amplification procedure with genomic DNA from L. interrogans and L. biflexa (Fig. 4). The novel primer pairs (fliG_F and fliG_R) successfully yielded PCR products of the expected 110 bp size (Fig. 4A), demonstrating their effectiveness in amplifying both pathogenic and nonpathogenic Leptospira species.
Fig. 4.
Validation of novel primer-probe sets for Leptospira species detection. (A) PCR amplification was performed using the primer pair fliG_F and fliG_R. Each reaction contained 25 ng of Leptospira genomic DNA under standard conditions. The amplification assay consisted of 30 cycles of 95 °C for 15 s, 60 °C for 75 s, and 72 °C for 75 s. Subsequently, the samples were maintained at 72 °C for 10 min, and the PCR products were separated via agarose gel electrophoresis and photographed. The genomic DNA from L. interrogans yielded a 110 bp fragment (Lane LI); genomic DNA from L. biflexa amplified a 110 bp fragment (Lane LB); negative control (water; Lane N); M, molecular size determined using a 50-bp ladder molecular marker. (B) RT‒PCR amplification was performed using the primer pairs fliG_F and fliG_R combined with different probes designed in this study. Duplicate Ct values were averaged, and the standard deviation (SD) of the Ct values was calculated. NA: samples with no amplification detected.
To confirm the specificity of novel probes targeting the fliG gene for the differential detection of L. interrogans and L. biflexa, we performed real-time qPCR analysis using novel primer pairs (fliG_F and fliG_R) and designed probes against genomic DNA preparations from Leptospira species. The specificity of the novel probes (fliG_P1 for L. interrogans and fliG_P2 for L. biflexa) was evaluated via real-time qPCR analysis. This analysis revealed proper amplification of the fliG gene from L. interrogans using the fliG_P1 probe, with no amplification observed for L. biflexa. Similarly, proper amplification of L. biflexa was achieved using the fliG_P2 probe, while no amplification was detected for L. interrogans (Fig. 4B).
3.5. Evaluation of the sensitivities of Leptospira quantification and the clinical validation of digital PCR using novel primer-probe sets
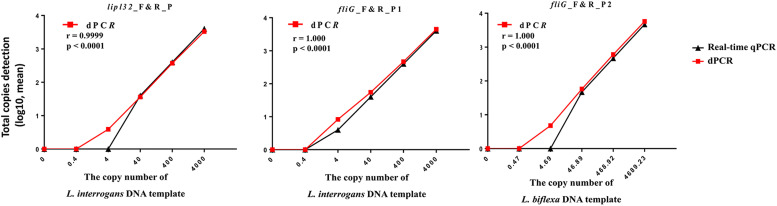
Leptospira genomic DNA was serially diluted and analyzed using both real-time qPCR and digital PCR to measure copy number. The accuracy of Leptospira DNA quantification by digital PCR was assessed by evaluating linearity within the detection range. This involved a tenfold serial dilution at concentrations ranging from 0.02 ng to 2 fg of Leptospira genomic DNA (for L. interrogans, the total genome copy input ranged from 4000 to 0.4; for L. biflexa, the total genome input of 4000 to 0.4 copy number estimates ranged from 4689.23 to 0.47). The copy number of fliG detected by digital PCR perfectly correlated (Pearson's correlation coefficient r = 1.00) with the theoretically calculated number of Leptospira copies, showing that fliG exhibited excellent linearity between the target input and measured values (Fig. 5). Notably, this method displayed greater sensitivity than real-time qPCR for quantitatively measuring leptospiral copy numbers, particularly at low concentrations (20 fg of input DNA; estimated copy number of 4.0) (Fig. 5). Real-time qPCR using the published primer-probe set targeting the lipl32 gene detected as little as 200 fg of purified genomic DNA from L. interrogans. In comparison, our newly designed primers and probes targeting the fliG gene detected as little as 20 fg of purified genomic DNA from L. interrogans. Table 3 summarizes the results obtained from the newly developed primer pair and probe for evaluating the sensitivity of the digital PCR assays. These findings demonstrate that our novel primers and probes targeting the fliG gene offer superior sensitivity for L. interrogans detection compared to those targeting the lipl32 gene. Additionally, digital PCR assays provide greater overall sensitivity for Leptospira detection than real-time qPCR assays.
Fig. 5.
Dynamic ranges and sensitivity for digital PCR assays targeting lipl32 and fliG genes. The total copy number of each sample was determined using a digital PCR (absolute total copies detection; log values) and estimated from the Ct values obtained from real-time qPCR (data are presented as means; detailed values are provided in Table 3). The X-axis represents the 10-fold dilution series of Leptospira genomic DNA, and the numbers of copies were theoretically calculated using the copy number calculator tool on the Technology Networks server (https://www.technologynetworks.com/tn/tools/copynumbercalculator). Correlation analysis between the theoretically calculated Leptospira copies (X-axis) and the copy number measured in the dilution series using both real-time qPCR and digital PCR (Y-axis, displayed as the mean values; n ≥ 3). Pearson's correlation coefficient (r) and p value are indicated. Statistical analyses were performed using Graph-Pad Prism 6.0 software (GraphPad Software, Inc.).
Table 3.
The sensitivity evaluation of newly designed primers and probes using real-time qPCR and digital PCR.
| Leptospira species |
L. interrogans |
L. interrogans |
L. biflexa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| primer-probe sets |
lipl32_F&R_P |
fliG_F&R_P1b |
fliG_F&R_P2 |
|||||||||
| Assays | real-time qPCR | dPCR | real-time qPCR | dPCR | real-time qPCR | dPCR | ||||||
| Quantitya | Ct mean ± SDc | RSD (%)d | Total copies mean ± SDe | RSD (%)d | Ct mean ± SDc | RSD (%)d | Total copies mean ± SDe | RSD (%)d | Ct mean ± SDc | RSD (%)d | Total copies mean ± SDe | RSD (%)d |
| 20,000 | 29.53 ± 0.17 | 0.56 | 3,334.47 ± 110.85 | 3.32 | 28.99 ± 0.05 | 0.16 | 4,484.67 ± 163.12 | 3.64 | 31.45 ± 0.06 | 0.20 | 5,800 ± 448.54 | 7.73 |
| 2000 | 33.34 ± 0.08 | 0.24 | 372.50 ± 47.22 | 12.68 | 32.42 ± 0.15 | 0.46 | 464.47 ± 24.17 | 5.20 | 34.45 ± 0.78 | 2.27 | 602.20 ± 38.56 | 6.40 |
| 200 | 36.71 ± 0.45 | 1.21 | 35.92 ± 2.98 | 8.29 | 35.38 ± 0.23 | 0.66 | 55.07 ± 10.25 | 18.61 | 38.31 ± 0.30 | 0.79 | 57.88 ± 8.05 | 13. 90 |
| 20 | NA | NA | 3.93 ± 0.85 | 19.68 | 39.37 ± 0.10 | 0.24 | 8.25 ± 0.37 | 4.47 | NA | NA | 4.74 ± 0.95 | 20. 12 |
| 2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
10-fold serial dilution (from 0.02 ng to 2 fg) of genomic DNA extracted from Leptospira species.
Developed in this study.
the cycle threshold (Ct) values (Ct < 40, positive; otherwise, negative); Mean ± SD, all the Ct values were the average of the three experiments ± standard deviation; NA: samples with no amplification detected.
RSD (%), relative standard deviation; used to analyze the precision of detection work, RSD (%) < 25 %, indicated this system has good repeatability.
Total number of copies detected by digital PCR (dPCR); Mean ± SD, all the average of the six experiments ± standard deviation; NA: samples with no amplification detected.
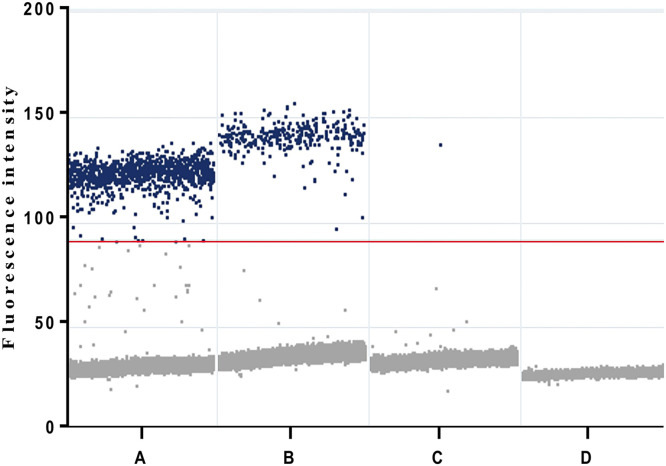
To demonstrate the clinical application of the assays, we performed a confirmed patient diagnosed using the Leptospira IgG/IgM test kit. Urine samples collected before and after the initiation of antibiotic therapy were tested using digital PCR with our novel primer-probe sets (Fig. 6). The results showed that 71.8 copies per ml of Leptospira detected in the urine before antibiotic therapy and 0.1 copies per ml on day 10 after therapy, demonstrating that our strategy indeed effectively be used for non-invasive detection of Leptospira in urine specimens.
Fig. 6.
Raw droplet scatter plot for the clinical validity of digital PCR assays targeting fliG genes in urine specimens from a laboratory-confirmed leptospirosis patient. Digital PCR for detection of Leptospira from A: positive control (cells infected with L. interrogans), B: urine specimens (before the initiation of antibiotic therapy), C: urine specimens (after the initiation of antibiotic therapy) and D: no template control. Positive droplets are obviously observed in the urine sample from before the initiation of antibiotic therapy. Blue dots represent the positive droplets, above the red horizontal threshold. Gray dots represent the negative droplets. The threshold for the detection of positive results was set at an amplitude of 90. Estimated copy numbers: Urine samples collected before the initiation of antibiotic therapy: 71.8 copy numbers per ml; Urine samples collected after the initiation of antibiotic therapy at day 10: 0.1 copy numbers per ml.
3.6. Leptospira latent infection detection and the impact of long-term Leptospira spp. presence on renal tissue
We performed an RNA-based assessment of Leptospira species identified and quantified in the kidneys of mice with chronic leptospiral infections using digital PCR coupled with newly designed primer-probe sets. In a digital PCR assay, leptospires targeting the lipl32 and fliG genes were detected in 58 % (7 out of 12) and 83 % (10 out of 12), respectively, of the kidneys from infected mice on day 7 (Table 4). The lipl32 transcript copy numbers detected in the kidneys of infected mice on day 7 ranged from 4.2 to 34.4 copies per μg of tissue, while the range of the fliG transcript copy numbers was 2.2 to 10.8 copies. In 41 % (5 out of 12) of the infected mice, the presence of the lipl32 gene was not detected; however, very low levels of pathogenic Leptospira spp. were detected using a primer-probe set targeting the fliG gene (Table 4).
Table 4.
Quantification of leptospires and cytokine expression in the kidneys of infected mice. We employed a digital PCR approach specifically targeting the lipl32 gene (for detecting pathogenic Leptospira spp.) and the fliG gene (for differential detection of pathogenic and nonpathogenic Leptospira spp.) to assess the number of leptospires in kidneys. To evaluate the effect of the Leptospira presence on cytokine expression in infected mice, we quantified the expression levels of cytokines, IL-10, IL-6 and TNF-α, using a real-time qPCR assay.
| After infection at day 7 |
Without infection |
Nonpathogenic L. biflexa infection |
Pathogenic L. interrogans infection |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | #2 | #3 | #1 | #2 | #3 | #4 | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 | #11 | #12 | ||
| lipl32 | Concentration (copies/ μg RNA) |
0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.3 | 15.2 | 0.0 | 0.0 | 0.0 | 4.2 | 6.6 | 34.4 | 0.0 | 6.7 | 9.8 | 0.0 |
| fliG | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.1 | 8.4 | 2.2 | 8.8 | 4.4 | 4.4 | 6.6 | 8.7 | 0.0 | 12.9 | 0.0 | 6.5 | 4.4 | |
| IL-10a | Expression level (Fold change) |
1.0 | 1.0 | 1.0 | 1.4 | 1.0 | 2.9 | 3.0 | 13.9 | 17.8 | 7.6 | 12.2 | 12.6 | 28.2 | 18.7 | 15.8 | 189.0 | 29.9 | 13.3 | 11.8 |
| IL-6a | 1.0 | 1.0 | 1.0 | 0.4 | 0.9 | 1.2 | 1.0 | 4.4 | 3.2 | 2.4 | 2.0 | 2.1 | 4.7 | 9.8 | 4.9 | 2.3 | 9.7 | 3.1 | 2.8 | |
| TNF-αa | 1.0 | 1.0 | 1.0 | 1.0 | 1.4 | 1.5 | 1.1 | 2.6 | 1.7 | 1.4 | 1.7 | 2.1 | 3.1 | 2.7 | 2.0 | 2.8 | 2.9 | 1.9 | 1.1 | |
| After infection at day 28 |
Without infection |
Nonpathogenic L. biflexa infection |
Pathogenic L. interrogans infection |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | #2 | #3 | #1 | #2 | #3 | #4 | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 | #11 | #12 | ||
| lipl32 | Concentration (copies/ μg RNA) |
0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1680.4 | 77.8 | 135.1 | 597.2 | 49.3 | 13.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 285.8 |
| fliG | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 2.2 | 2.2 | 2.2 | 4.4 | 4.4 | 2.2 | 2.1 | 0.0 | 0.0 | 2.2 | 6.5 | |
| IL-10a | Expression level (Fold change) |
1.0 | 1.0 | 1.0 | 6.9 | 5.7 | 12.8 | 9.3 | 2.9 | 0.6 | 3.4 | 2.5 | 5.0 | 5.2 | 3.8 | 2.5 | 2.1 | 0.6 | 4.8 | 1.8 |
| IL-6a | 1.0 | 1.0 | 1.0 | 2.5 | 2.2 | 2.8 | 2.9 | 1.7 | 1.1 | 2.3 | 1.9 | 3.4 | 1.8 | 1.5 | 1.5 | 1.0 | 0.9 | 1.7 | 2.0 | |
| TNF-αa | 1.0 | 1.0 | 1.0 | 5.0 | 3.7 | 5.0 | 4.8 | 1.8 | 1.2 | 2.3 | 1.8 | 4.2 | 2.5 | 1.9 | 1.7 | 1.2 | 1.6 | 2.7 | 1.4 | |
TaqMan probes were used for the indicated genes: Mm00439614_m1 (interleukin (IL)−10), Mm01210733_m1 (IL-6), Mm99999068_m1 (TNF-α) and Mm99999915_g1 (GAPDH).
We examined the association between Leptospira quantity and cytokine expression levels in the kidneys of infected mice. Specifically, we investigated whether the bacterial load within the kidneys correlates with the expression levels of renal cytokines, such as IL-10, IL-6, and TNF-α, induced by infection. After Leptospira infection on day 7, we observed increased expression of IL-10, along with elevated IL-6 levels, in the kidneys of mice infected with pathogenic Leptospira spp. However, this pattern was not found in the kidneys of mice infected with nonpathogenic Leptospira. After nonpathogenic Leptospira infection on day 28, an increase in IL-10 expression was observed in the infected kidneys, along with elevated levels of IL-6 and TNF-α, whereas the presence of nonpathogenic Leptospira spp. was not detected in the infected kidneys (Table 4). After Leptospira infection on day 7, histopathological analysis revealed inflammatory cell infiltration in the kidneys of all mice infected with pathogenic Leprospira species, where leptospires were present. Furthermore, 50 % (6 out of 12) of the mice infected with pathogenic Leptospira species exhibited tubular degeneration and necrotic lesions. After Leptospira infection on day 28, the regeneration of renal tubules was observed in the infected kidneys, except for one mouse (#10 in Table 4), in which leptospires were not detected and cytokine expression remained normal.
As shown in Supplementary Fig. 1, histopathological examination of infected kidney sections revealed regenerating renal tubules accompanied by mitosis in kidneys where leptospiral presence (as detected by the fliG gene) and transcriptionally active leptospires (as detected by the lipl32 gene) were detected, along with increased expression of cytokines (see #3 from mice infected with pathogenic Leptospira species in Table 4). This observation prompted a comprehensive analysis of cell cycle-related genes using the RNA-seq data previously generated for our investigation (GSE111249) (Chou et al., 2018). Pathway analysis revealed that cell cycle pathway pathways, including “Cell cycle checkpoints”, “Mitotic Prometaphase”, “Mitotic metaphase and anaphase” and “Mitotic G1 phase and G1/S transition” are involved in the effects of long-term Leptospira presence on renal tissue. Further studies are needed to understand how these genes contribute to leptospirosis kidney diseases.
4. Discussion
This study introduces a novel digital PCR method for detecting active and latent Leptospira infections with enhanced sensitivity and specificity. The comparative analysis of Leptospira detection in infected kidneys demonstrates that digital PCR, both DNA- and RNA-based, is more effective than traditional and real-time qPCR methods, with the RNA-based approach offering superior efficacy over the DNA-based methods. This study provided insights into the dynamics of leptospiral presence in infected kidneys over time. Notably, our results revealed that leptospires remained detectable by detecting the lipl32 gene in the kidneys of more than half of the infected mice even at 28 days after infection, highlighting the chronic infection of the pathogen in renal tissues. This finding emphasizes the importance of continued monitoring and timely intervention in chronic leptospirosis kidney disease.
We further developed novel primer-probe sets targeting the fliG gene for Leptospira detection, which exhibited heightened sensitivity in comparison to the conventional lipl32 gene. Through PCR and real-time qPCR analyses, we confirmed the ability of these primer-probe sets to detect and discriminate between L. interrogans and L. biflexa. By employing tenfold dilutions of Leptospira genomic DNA to test novel primer-probe sets targeting the fliG gene, the results demonstrated high sensitivity, highlighting its superior performance in detecting L. interrogans. The integration of a digital PCR approach with our newly developed fliG-targeting assay enables simultaneous detection and differentiation of pathogenic and nonpathogenic Leptospira species, representing a valuable tool for Leptospira detection.
The quantification of transcript copy numbers revealed the dynamic range of Leptospira abundance in infected kidneys. Notably, in a subset of infected mice in which the lipl32 gene was not detected, the fliG gene-targeted primer-probe set successfully identified very low levels of Leptospira spp. Our findings challenge the previously notion of rapid clearance of nonpathogenic Leptospira in living host (Surdel et al., 2022). As identified by the novel primers and probes targeting the fliG gene, only one out of four mice had detectible levels of nonpathogenic Leptospira at 7 days postinfection by the novel primers and probes targeting the fliG gene. This finding suggests that nonpathogenic Leptospira may persistent in kidney tissues for 7 days than previously thought, highlighting the importance of sensitive detection methods. Moreover, the differential detection patterns observed between the lipl32 and fliG genes highlight the importance of utilizing multiple target genes for comprehensive pathogenic Leptospira detection. Interestingly, at 28 days postinfection, trace amounts of fliG gene expression were detected in the kidneys of some mice infected with pathogenic Leptospira, while lipl32 gene expression was undetectable. This suggests the potential presence of latent infection in leptospirosis kidney disease. These findings emphasize the importance of longitudinal monitoring of leptospiral infections and the dynamic interplay between pathogen clearance and persistence in host tissues. The precise quantification of leptospiral abundance in infected tissues provides important information about disease progression, host-pathogen dynamics, and the efficacy of therapeutic interventions.
Our previous studies revealed global alterations in renal gene expression in response to leptospiral infection, revealing that leptospires induced immune-related pathways within damaged kidneys (Chou et al., 2018, 2021, 2023). Histopathological examination of kidneys with detectable leptospiral presence and active transcriptional activity revealed regenerating renal tubules accompanied by mitosis, indicative of ongoing tissue repair processes. Elevated cytokine expression suggests interplay between the renal milieu, tissue injury, and leptospiral infection.
5. Conclusions
The fliG-targeting digital PCR assay significantly improves the detection sensitivity of latent Leptospira infections in kidney tissues, demonstrating superior performance compared to conventional qPCR methods. Our research emphasizes a novel molecular approach for the differential detection and tracking of small quantities of Leptospira spp., aiding in the exploration of low bacterial burdens in asymptomatic carriers. This approach provides a deeper understanding of latent and chronic Leptospia infections, addressing a critical public health issue and ultimately aiding in the control of this pervasive zoonotic disease. While this study demonstrates the development of a potentially useful digital PCR assay for detecting Leptospira, a limitation is that the assay has not yet been tested on a large number of clinical specimens or isolates from epidemiological studies. Future research should focus on evaluating this method across a broader range of clinical isolates to confirm its broader applicability, providing valuable insights into the prevalence and transmission dynamics of leptospirosis.
Supplementary material
Figure S1. Histopathologic examination of kidney tissues with the presence of transcriptionally active leptospires. (A) Inflammatory cell infiltration observed in the interstitium. (B) Renal tubule regeneration observed in several adjacent renal tubules (indicated by white arrows) with mitosis observed (highlighted by yellow arrowhead). (Representative section from mouse #3 infected with pathogenic Leptospira species, as indicated in Table 4) (hematoxylin and eosin staining, original magnification ×400).
Declaration of generative AI and AI-assisted technologies in the writing process
Quillbot was used to check grammar and spelling in order to improve readability. After using this tool, the authors reviewed and edited the content and take full responsibility for the content of the publication. Part of the figures were created by BioRender program (www.biorender.com).
Ethical approval statement
This study was approved by the Medical Ethics Committee of Chang Gung Memorial Hospital (approval number: 202301580B0). Patients who tested serologically positive for leptospirosis and provided informed written consent to participate in the study were included for evaluation.
Funding
This work was funded by grants from the Ministry of Science and Technology, Taiwan (MOST 113–2327-B-182–002 and 111–2314-B-182A-072) and Chang Gung Memorial Hospital (CMRPG3L1071 and CMRPG3L1072).
CRediT authorship contribution statement
Li-Fang Chou: Conceptualization, Funding acquisition, Project administration, Writing – original draft. Yi-Chun Liu: Formal analysis, Supervision, Writing – review & editing. Huang-Yu Yang: Investigation, Resources. Ya-Chung Tian: Resources, Supervision. Chih-Ho Lai: Formal analysis, Supervision. Ming-Yang Chang: Resources, Formal analysis. Cheng-Chieh Hung: Software, Validation. Tong-Hong Wang: Methodology, Software. Shen-Hsing Hsu: Methodology, Data curation. Chung-Ying Tsai: Data curation, Visualization. Pei-Yu Hung: Methodology, Validation. Chih-Wei Yang: Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of competing interest
L.-F. C., P.-Y. H. and C.-W. Y. are inventors (Kidney Research Center, Chang Gung Memorial Hospital) on a patent for “Innovative method for nucleic acid detection tracking minute quantities of Leptospira spp. in biological tissues and environmental samples” (US Provisional Patent Application 63/605,750, filed 4 December 2023). The remaining authors declare no competing interests.
Acknowledgements
We sincerely thank the Research Center for Biomedical Science and Engineering at National Tsing Hua University, Taiwan, for providing the digital PCR facilities. We gratefully acknowledge Microscope Core Laboratory and Laboratory Animal Center at the Chang Gung Memorial Hospital, Linkou for technical assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2024.100327.
Contributor Information
Li-Fang Chou, Email: d928209@gmail.com.
Chih-Wei Yang, Email: cwyang@ms1.hinet.net.
Appendix. Supplementary materials
Data availability
I have shared our data at the attached file step
References
- Abela-Ridder B., Sikkema R., Hartskeerl R.A. Estimating the burden of human leptospirosis. Int. J. Antimicrob. Agents. 2010;36(Suppl 1):S5–S7. doi: 10.1016/j.ijantimicag.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Bárbara Couto Roloff Padilha M.M.S., Daiane Drawanz Hartwig Molecular and serological diagnostic of leptospirosis: a review (2014–2020) Res, Soc Dev. 2022;11 doi: 10.33448/rsd-v11i2.25471. [DOI] [Google Scholar]
- Bini Viotti J., Chan J.C., Rivera C., Tuda C. Sporadic leptospirosis case in Florida presenting as Weil;s disease. IDCases. 2020;19:e00686. doi: 10.1016/j.idcr.2019.e00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley E.A., Lockaby G. Leptospirosis and the environment: a review and future directions. Pathogens. 2023;12 doi: 10.3390/pathogens12091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Larco R.M., Altez-Fernandez C., Acevedo-Rodriguez J.G., Ortiz-Acha K., Ugarte-Gil C. Leptospirosis as a risk factor for chronic kidney disease: a systematic review of observational studies. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancharoenthana W., Leelahavanichkul A., Schultz M.J., Dondorp A.M. Going micro in Leptospirosis kidney disease. Cells. 2022;11 doi: 10.3390/cells11040698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Jiang Y., Cao X., Liu C., Zhang N., Shi D. Droplet digital PCR as an emerging tool in detecting pathogens nucleic acids in infectious diseases. Clin. Chim. Acta. 2021;517:156–161. doi: 10.1016/j.cca.2021.02.008. [DOI] [PubMed] [Google Scholar]
- Chou L.F., Yang H.Y., Hung C.C., Tian Y.C., Hsu S.H., Yang C.W. Leptospirosis kidney disease: evolution from acute to chronic kidney disease. Biomed. J. 2023;46 doi: 10.1016/j.bj.2023.100595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou L.F., Chen T.W., Yang H.Y., Tian Y.C., Chang M.Y., Hung C.C., Hsu S.H., Tsai C.Y., Ko Y.C., Yang C.W. Transcriptomic signatures of exacerbated progression in leptospirosis subclinical chronic kidney disease with secondary nephrotoxic injury. Am. J. Physiol. Renal. Physiol. 2021;320:F1001–F1018. doi: 10.1152/ajprenal.00640.2020. [DOI] [PubMed] [Google Scholar]
- Chou L.F., Chen T.W., Yang H.Y., Chang M.Y., Hsu S.H., Tsai C.Y., Ko Y.C., Huang C.T., Tian Y.C., Hung C.C., Yang C.W. Murine renal Transcriptome profiles upon leptospiral infection: implications for chronic kidney diseases. J Infect Dis. 2018;218:1411–1423. doi: 10.1093/infdis/jiy339. [DOI] [PubMed] [Google Scholar]
- Cilia G., Bertelloni F., Fratini F. Leptospira infections in domestic and wild animals. Pathogens. 2020:9. doi: 10.3390/pathogens9070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F., Hagan J.E., Calcagno J., Kane M., Torgerson P., Martinez-Silveira M.S., Stein C., Abela-Ridder B., Ko A.I. Global morbidity and mortality of Leptospirosis: a systematic review. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Azevedo M.I.N., Lilenbaum W. An overview on the molecular diagnosis of animal leptospirosis. Lett. Appl. Microbiol. 2021;72:496–508. doi: 10.1111/lam.13442. [DOI] [PubMed] [Google Scholar]
- Dreo T., Pirc M., Ramsak Z., Pavsic J., Milavec M., Zel J., Gruden K. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 2014;406:6513–6528. doi: 10.1007/s00216-014-8084-1. [DOI] [PubMed] [Google Scholar]
- Ganoza C.A., Matthias M.A., Saito M., Cespedes M., Gotuzzo E., Vinetz J.M. Asymptomatic renal colonization of humans in the peruvian Amazon by Leptospira. PLoS Negl. Trop. Dis. 2010;4:e612. doi: 10.1371/journal.pntd.0000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokmen T.G., Soyal A., Kalayci Y., Onlen C., Koksal F. Comparison Of 16S rRNA-PCR-RFLP, LipL32-PCR AND OmpL1-PCR Methods in the diagnosis of Leptospirosis. Rev Inst Med Trop Sao Paulo. 2016;58:64. doi: 10.1590/S1678-9946201658064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Solecki M., Santecchia I., Werts C. Animal models of Leptospirosis: of mice and hamsters. Front Immunol. 2017;8:58. doi: 10.3389/fimmu.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris M.G., Hartskeerl R.A. Leptospirosis serodiagnosis by the microscopic agglutination test. Curr. Protoc. Microbiol. 2014;32 doi: 10.1002/9780471729259.mc12e05s32. Unit 12E 5. [DOI] [PubMed] [Google Scholar]
- Hamond C., Browne A.S., de Wilde L.H., Hornsby R.L., LeCount K., Anderson T., Stuber T., Cranford H.M., Browne S.K., Blanchard G., Horner D., Taylor M.L., Evans M., Angeli N.F., Roth J., Bisgard K.M., Salzer J.S., Schafer I.J., Ellis B.R., Alt D.P., Schlater L., Nally J.E., Ellis E.M. Assessing rodents as carriers of pathogenic Leptospira species in the U.S. Virgin Islands and their risk to animal and public health. Sci. Rep. 2022;12:1132. doi: 10.1038/s41598-022-04846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartskeerl R.A., Terpstra W.J. Leptospirosis in wild animals. Vet. Q. 1996;18(Suppl 3):S149–S150. [PubMed] [Google Scholar]
- Karpagam K.B., Ganesh B. Leptospirosis: a neglected tropical zoonotic infection of public health importance-an updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:835–846. doi: 10.1007/s10096-019-03797-4. [DOI] [PubMed] [Google Scholar]
- Koizumi N. Laboratory diagnosis of Leptospirosis. Methods Mol. Biol. 2020;2134:277–287. doi: 10.1007/978-1-0716-0459-5_25. [DOI] [PubMed] [Google Scholar]
- Lei S., Chen S., Zhong Q. Digital PCR for accurate quantification of pathogens: principles, applications, challenges and future prospects. Int. J. Biol. Macromol. 2021;184:750–759. doi: 10.1016/j.ijbiomac.2021.06.132. [DOI] [PubMed] [Google Scholar]
- Marie Moinet C.R.A., Gasparotto Vinícius P.O., Wilkinson David A., Vallée Emilie, Benschop Jackie, Russell James C. Density matters: how population dynamics of house mice (Mus musculus) inform the epidemiology of Leptospira. J. Appl. Ecol. 2024 doi: 10.1111/1365-2664.14714. [DOI] [Google Scholar]
- Marquez A., Ulivieri T., Benoit E., Kodjo A., Lattard V. House mice as a real sanitary threat of human and animal Leptospirosis: proposal for integrated management. Biomed. Res. Int. 2019;2019 doi: 10.1155/2019/3794876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E.A., Heseltine J.C., Creevy K.E. The evaluation of the diagnostic value of a PCR assay when compared to a serologic micro-agglutination test for Canine Leptospirosis. Front Vet Sci. 2022;9 doi: 10.3389/fvets.2022.815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M., Roche L., Geroult S., Soupe-Gilbert M.E., Monchy D., Huerre M., Goarant C. Cytokine and chemokine expression in kidneys during chronic Leptospirosis in reservoir and susceptible animal models. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc I.W., Montgomery G.L. Renal lesions in Leptospira canicola infection in dogs. J. Pathol. Bacteriol. 1952;64:145–160. doi: 10.1002/path.1700640115. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- Niloofa R., Fernando N., de Silva N.L., Karunanayake L., Wickramasinghe H., Dikmadugoda N., Premawansa G., Wickramasinghe R., de Silva H.J., Premawansa S., Rajapakse S., Handunnetti S. Diagnosis of Leptospirosis: comparison between microscopic agglutination test, IgM-ELISA and IgM rapid immunochromatography test. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera Z.L., Charypkhan D., Hartnack S., Torgerson P.R., Ruegg S.R. The dual burden of animal and human zoonoses: a systematic review. PLoS Negl. Trop. Dis. 2022;16 doi: 10.1371/journal.pntd.0010540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas G., Papadimitriou P., Siozopoulou V., Christou L., Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int. J. Infect. Dis. 2008;12:351–357. doi: 10.1016/j.ijid.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Pinto G.V., Senthilkumar K., Rai P., Kabekkodu S.P., Karunasagar I., Kumar B.K. Current methods for the diagnosis of leptospirosis: issues and challenges. J. Microbiol. Methods. 2022;195 doi: 10.1016/j.mimet.2022.106438. [DOI] [PubMed] [Google Scholar]
- Podgorsek D., Ruzic-Sabljic E., Logar M., Pavlovic A., Remec T., Baklan Z., Pal E., Cerar T. Evaluation of real-time PCR targeting the lipL32 gene for diagnosis of Leptospira infection. BMC Microbiol. 2020;20:59. doi: 10.1186/s12866-020-01744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaonarivelo J.A., Desmoulin A., Maillard O., Collet L., Baudino F., Jaffar-Bandjee M.C., Blonde R., Raffray L., Tortosa P. Clinical manifestations of human leptospirosis: bacteria matter. Front Cell Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse S. Leptospirosis: clinical aspects. Clin. Med. (Lond) 2022;22:14–17. doi: 10.7861/clinmed.2021-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratet G., Veyrier F.J., Fanton d'Andon M., Kammerscheit X., Nicola M.A., Picardeau M., Boneca I.G., Werts C. Live imaging of bioluminescent leptospira interrogans in mice reveals renal colonization as a stealth escape from the blood defenses and antibiotics. PLoS Negl. Trop. Dis. 2014;8:e3359. doi: 10.1371/journal.pntd.0003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethinavelu Gayathri, Vishwakarma A., Mohandass Ramya. Molecular diagnostic methods for the detection of Leptospirosis. J. Pure Appl. Microbiol. 2022;16:782–795. doi: 10.22207/JPAM.16.2.24. [DOI] [Google Scholar]
- Richer L., Potula H.H., Melo R., Vieira A., Gomes-Solecki M. Mouse model for sublethal Leptospira interrogans infection. Infect. Immun. 2015;83:4693–4700. doi: 10.1128/IAI.01115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupak Nagraik A.K., Gupta Shagun, Kumar Dinesh. Pcr based genetic marker for the detection of Leptospira interrogans causing leptospirosis. Vegetos. 2020;33:21–25. doi: 10.1007/s42535-019-00078-5. [DOI] [Google Scholar]
- Salipante S.J., Jerome K.R. Digital PCR-an emerging technology with broad applications in microbiology. Clin. Chem. 2020;66:117–123. doi: 10.1373/clinchem.2019.304048. [DOI] [PubMed] [Google Scholar]
- Sanchez Fernandez P., Kodjo A., Medkour H., Laidoudi Y., Dubourg G., Eldin C., Parola P., Davoust B., Lagier J.C. Vol. 21. IDCases; Marseille, France: 2020. p. e00899. (Autochthonous Human and Animal Leptospirosis). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier S., Triampo W., Doungchawee G., Triampo D., Chadsuthi S. Leptospirosis research: fast, easy and reliable enumeration of mobile leptospires. Biol. Res. 2009;42:5–12. [PubMed] [Google Scholar]
- Silva P.L., Nakajima E., Costa R.M.D., Lee Ho P., Martins E.A., Carvalho E., da Silva J.B. Chemokine expression profiles in liver and kidney of mice with different susceptibilities to leptospirosis. Microb. Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104580. [DOI] [PubMed] [Google Scholar]
- Surdel M.C., Anderson P.N., Hahn B.L., Coburn J. Hematogenous dissemination of pathogenic and non-pathogenic Leptospira in a short-term murine model of infection. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.917962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes J.E., Haake D.A., Gamage C.D., Mills W.Z., Nally J.E. A global one health perspective on leptospirosis in humans and animals. J. Am. Vet. Med. Assoc. 2022;260:1589–1596. doi: 10.2460/javma.22.06.0258. [DOI] [PubMed] [Google Scholar]
- Vijayachari P., Sugunan A.P., Shriram A.N. Leptospirosis: an emerging global public health problem. J. Biosci. 2008;33:557–569. doi: 10.1007/s12038-008-0074-z. [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Hung C.C., Liu S.H., Guo Y.G., Chen Y.C., Ko Y.C., Huang C.T., Chou L.F., Tian Y.C., Chang M.Y., Hsu H.H., Lin M.Y., Hwang S.J., Yang C.W. Overlooked risk for chronic kidney disease after leptospiral infection: a population-based survey and epidemiological cohort evidence. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
I have shared our data at the attached file step