Summary
Developing microbiome-based markers for pediatric inflammatory bowel disease (PIBD) is challenging. Here, we evaluated the diagnostic and prognostic potential of the gut microbiome in PIBD through a case-control study and cross-cohort analyses. In a Korean PIBD cohort (24 patients with PIBD, 43 controls), we observed that microbial diversity and composition shifted in patients with active PIBD versus controls and recovered at remission. We employed a differential abundance meta-analysis approach to identify microbial markers consistently associated with active inflammation and remission across seven PIBD cohorts from six countries (n = 1,670) including our dataset. Finally, we trained and tested various machine learning models for their ability to predict a patient’s future remission based on baseline bacterial composition. An ensemble model trained with the amplicon sequence variants effectively predicted future remission of PIBD. This research highlights the gut microbiome’s potential to guide precision therapy for PIBD.
Subject areas: Gastroenterology, Microbiome
Graphical abstract
Highlights
-
•
Microbiota dynamics in PIBD patients in active inflammation and remission states
-
•
Prognostic prediction using an ensemble model accurately predicts future remission
-
•
Microbiota variation highlights the need for geographically diverse training data
-
•
Foundation for advancing patient stratification and personalized medicine in PIBD
Gastroenterology; Microbiome
Introduction
The global incidence of pediatric inflammatory bowel disease (PIBD) is on the rise, affecting patients in both Western and newly developed Asian countries.1,2 Patients with PIBD often endure more severe disease trajectories than adults with inflammatory bowel disease (IBD). The complex interplay of genetic predisposition, inappropriate mucosal immunity, and environmental factors underpins the pathogenesis of IBD.3,4,5 Notably, the incidence in Korea has increased that is potentially linked to the adoption of Westernized diets.5 Furthermore, environmental and microbial factors exert greater influence on adolescent-onset IBD than very-early-onset forms of the disease.6 Studies have highlighted that gut microbiome imbalances, characterized by a decrease in commensal bacteria and an increase in potentially harmful microbes, might play a crucial role in the development of IBD.4
The link between the gut microbiome and IBD underscores the potential of microbiome profiling as a pivotal diagnostic tool. This strategy has proven effective for risk assessment, gut dysbiosis identification in newly diagnosed IBD cases, and patient stratification.4,7,8 Evidence suggests that microbiome profiling could be useful for classifying adult patients with IBD.9,10 The prospect of using gut microbiota profiles to predict an individual’s risk of refractory flares and their likelihood of achieving clinical or mucosal remission adds a new dimension to its clinical utility. For instance, studies of treatment-naïve pediatric ulcerative colitis (UC) cohorts revealed associations between microbiota profiles and remission, as well as between the abundance of specific microbial taxa and treatment responses.8,11,12 However, little has been documented regarding the predictive capability of patient responses to treatment or future disease activity.13,14
The baseline healthy microbiome differs between pediatric and adult populations.15 Although increased levels of Proteobacteria, Fusobacterium, and Ruminococcus gnavus have been noted in adults with IBD, pediatric studies often identy elevated Enterococcus.16,17 Geographic variations in microbial markers for adult-onset IBD have been observed that feature both country-specific heterogeneity and some consistent patterns.18,19,20 However, the availability of gut microbiome datasets for patients with PIBD is relatively limited, primarily due to smaller cohort sizes, and unknown global geographic variations. Furthermore, few studies have explored how dysbiosis varies in PIBD with disease activity and medication use.
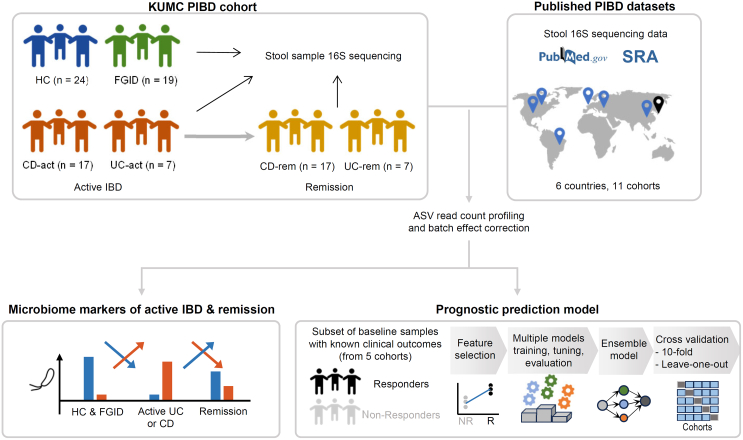
To further investigate the diagnostic and prognostic capabilities of microbiome analyses in PIBD, we comprehensively compared the intestinal microbiomes of children with IBD, those with functional gastrointestinal diseases (FGID), and healthy controls (HC). We then compiled PIBD cohort datasets from published studies worldwide. We developed a prognostic model by integrating our datasets with these global datasets, including microbial profiles and clinical indices (Figure 1). This model predicts a patient’s state of achieving remission based on their microbiome profile and metadata collected during the active IBD phase. Moreover, our study evaluated the geographical specificity of microbial markers by comparing their abundance across nations.
Figure 1.
Overall study design
The microbiota profiles and clinical metadata from a Korean PIBD cohort and 11 published PIBD cohorts were analyzed to identify diagnostic biomarkers. A prognostic model was developed and tested based on the baseline samples with information on subsequent clinical responses. Multiple models were built, performance tested, and an ensemble model was created from the three top scoring models. The prognostic model was evaluated by 10-fold cross-validation and leave-one-study-out analysis. PIBD, pediatric inflammatory bowel disease.
Results
Clinical characteristics
This study’s cohort comprised Korean patients with PIBD (n = 24), those with gut-brain interaction disorders or FGID (n = 19), and HC (n = 24). The IBD category included patients with Crohn's disease (CD; n = 17) and UC (n = 7). The mean ages (for new-onset or exacerbated state) were 14.9 ± 2.6 for the CD-active (CD-act) group and 15.3 ± 2.4 years for the UC-active (UC-act) group. The FGID group, consisting entirely of patients with irritable bowel syndrome (n = 19), had a mean age of 13.4 ± 2.0 years, while the HC group had a mean age of 13.8 ± 2.2 years.
Table 1 details the clinical features of the patients with IBD. Compared to patients with FGID, those with active CD (CD-act) or active UC (UC-act) exhibited significantly higher fecal calprotectin levels (Wilcoxon test; CD-act vs. FGID, p = 3.3E−5; UC-act vs. FGID, p = 9.0E−4). Furthermore, the levels decreased in patients with CD and UC as they transitioned from an active disease state to clinical remission (CR; Figure 2A; Table 1).
Table 1.
Clinical characteristics of patients with CD and UC
| Clinical feature | CD-act | CD-rem | UC-act | UC-rem |
|---|---|---|---|---|
| State | Active | Clinical remission | Active | Clinical remission |
| Number of subjects | 17 | 17 | 7 | 7 |
| Age, years | 14.9 ± 2.6 | – | 15.3 ± 2.4 | – |
| Disease duration between start and endpoint, months | – | 17.0 ± 9.3 | – | 8.07 ± 8.74 |
| Sex, M:F | 9:8 | 9:8 | 6:1 | 6:1 |
| PCDAI | 37.1 ± 15.9 | 3.8 ± 4.5 | – | – |
| PUCAI | – | – | 35.7 ± 23.7 | 1.4 ± 2.4 |
| Calprotectin, mg/kg | 830.8 ± 534.7 | 569.8 ± 764.9 | 655.2 ± 148.3 | 251.5 ± 291.5 |
| SES-CD | 13.53 ± 7.8 | 6.4 ± 5.1 | – | – |
| UCEIS | – | – | 4.3 ± 1.8 | 0.5 ± 0.7 |
| Disease location, n (%) | – | – | – | – |
| Ileal | 1 (6) | – | – | – |
| Colonic | 6 (35) | – | – | – |
| Ileocolonic | 10 (59) | – | – | – |
| Fistula, n (%) | 10 (59) | 2 (12) | – | – |
| Disease extent, n (%) | – | – | – | – |
| Left-sided | – | – | 6 (86) | – |
| Pancolonic | – | – | 1 (14) | – |
| Treatment prior to collection, n (%) | – | – | – | – |
| Anti-tumor necrosis factor alpha | – | 12 (71) | – | 5 (71) |
| Exclusive enteral nutrition | – | 3 (18) | – | – |
| Corticosteroids | 1 | 4 (23) | 2 | 2 (29) |
| Azathioprine | 2 | 9 (53) | 2 | 5 (71) |
| 5-Aminosalicylic acid | – | – | 1 | 3 (43) |
| None | 14 (82) | – | 3 (43) | – |
| Surgery, n (%) | – | 2 (12) | – | 0 |
| Seton operation | – | 1 | – | – |
| Hemicolectomy | – | 1 | – | – |
| Mucosal remission, n (%) | – | 6 (35) | – | 5 (71) |
CD, Crohn disease; PCDAI, Pediatric Crohn’s Disease Activity Index; PUCAI, Pediatric Ulcerative Colitis Activity Index; SES-CD, Simple Endoscopic Score for Crohn’s Disease; UC, ulcerative colitis; UCEIS, Ulcerative Colitis Endoscopic Index of Severity.
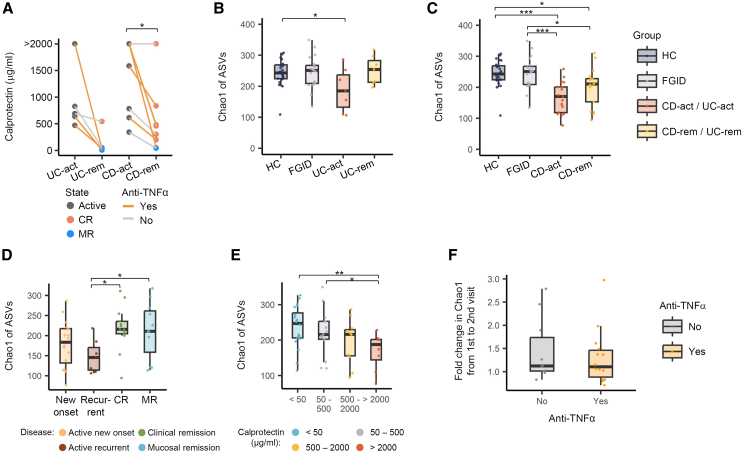
Figure 2.
Gut microbiota alpha diversity of patients with UC and CD compared to healthy controls and patients with functional gastrointestinal diseases
(A) The fecal calprotectin concentration of the patients was measured at diagnosis (UC-act, CD-act) and remission (UC-rem, CD-rem). The remission state was further categorized into CR and MR based on endoscopic examination findings. Data points are shown in different colors, and the subset of patients who received anti-TNF-α treatment can be distinguished by line color. Fecal calprotectin concentration data included five patients with UC and nine patients with CD. Paired Wilcoxon test: CD-rem vs. CD-act, p = 0.022; UC-rem vs. UC-act, p = 0.063. (BE) Chao1 richness index calculated from read counts per 16S ASV.
(B) Comparison among HC and patients with FGID, UC-act, and UC-rem.
(C) Comparison among HC and patients with FGID, CD-act, and CD-rem.
(D) Comparison among patients with IBD based on diagnostic status. Active IBD states were further classified into new-onset and recurrent types. Remission states were classified into clinical and mucosal. Wilcoxon test: CR vs. recurrent IBD, 1.5-fold, p = 0.01; MR vs. recurrent IBD, 1.4-fold, p = 0.04.
(E) Comparison within IBD patients based on fecal calprotectin levels. Wilcoxon test: very high vs. low, p = 0.006; very high vs. low-mid, p = 0.03.
(F) Fold changes in ASV Chao1 index from active to remission state divided by patient usage of the anti-TNF-α agent. Significance of the difference was tested by Wilcoxon’s signed rank test. The pairs of groups with values of p < 0.05 are indicated by horizontal brackets. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Boxplots in (B–F) represent interquartile range, along with the individual data points shown as dots. A full table of statistical values is available as Table S1. Anti-TNF-α, anti-tumor necrosis factor alpha; ASV, amplicon sequence variant; CD, Crohn disease; CD-act, active CD; CD-rem, remission CD; CR, clinical remission; FGID, functional gastrointestinal disease; HC, healthy controls; IBD, inflammatory bowel disease; MR, mucosal remission; UC, ulcerative colitis; UC-act, active UC; UC-rem, remission UC.
Gut microbiota diversity in the PIBD cohort
We assessed the alpha diversity of fecal microbiota in patients with PIBD across active (UC-act and CD-act) and remission (UC-rem and CD-rem) disease states compared to those with FGID and HC. Using the Chao1 richness index based on the composition of 16S amplicon sequence variants (ASVs), we found that diversity was similar between the HC and the patients with FGID (Wilcoxon test; 1.01-fold, p = 0.64). However, significantly lower diversity was observed in the microbiota of patients, 1.34-fold lower in UC-act (p = 0.04) and 1.45-fold lower in CD-act (p = 1.4E-6) versus those in the HC/FGID group (Figures 2B and 2C; Table S1). This indicates that reduced gut microbial diversity is associated with active IBD. Interestingly, although not statistically significant, patients in remission exhibited relatively higher Chao1 indices than those in an active state, suggesting that microbial diversity tends to recover with CR in IBD patients (paired Wilcoxon test; UC-rem vs. UC-act, 1.37-fold, p = 0.097; CD-rem vs. CD-act, 1.24-fold, p = 0.092).
Similarly, the nonparametric Shannon diversity index showed no difference between HC and patients with FGID (1.01-fold; p = 0.84) but it was significantly lower in patients with active IBD versus the HC/FGID group, further supporting the association between reduced diversity and active disease (Wilcoxon test; HC/FGID vs. UC-act, 1.15-fold, p = 0.16; HC/FGID vs. CD-act, 1.17-fold, p = 0.012). However, no significant difference was observed in the Shannon index between the active and remission states, indicating that the recovery of microbial diversity might not be uniform across all diversity metrics (UC-act vs. UC-rem, p = 0.26; CD-act vs. CD-rem, p = 0.84).
To explore the correlation between microbiome alpha diversity and PIBD prognosis, we categorized disease activity into four states: new-onset active, recurrently active, CR, and mucosal remission. Notably, the Chao1 index was significantly higher in the clinical and mucosal remission states compare to the recurrently active state, highlighting a potential association between increased microbial diversity and favorable disease outcomes (Figure 2D).
Furthermore, we categorized the samples by fecal calprotectin levels as follows: low (<50 mg/kg), low-mid (50–500 mg/kg), high (500–2,000 mg/kg), and very high (>2,000 mg/kg). A decreasing trend in the Chao1 index was observed with increasing calprotectin levels, with significant differences between samples with very high calprotectin levels and those with low or low-mid levels (Figure 2E).
Finally, we found no significant difference in the magnitude of microbiota richness recovery (i.e., fold increase from the active to remission state within each patient) between patients receiving anti-tumor necrosis factor alpha (anti-TNF-α) therapy and those who did not. This suggests that the effect of microbial diversity recovery may be independent of treatment modality (Figure 2F).
Microbial taxonomic composition of the PIBD cohort
We investigated fecal microbial beta-diversity in the 16S ASV dataset and observed that the variation in microbial composition was significantly associated with disease states and inflammation levels. The microbial compositions of patients with UC-act and CD-act were significantly different from those of HC and the patients with FGID (Figure 3A). However, the ASV composition did not significantly differentiate between active and remission states within the UC and CD cohorts (Figures 3B and 3C).
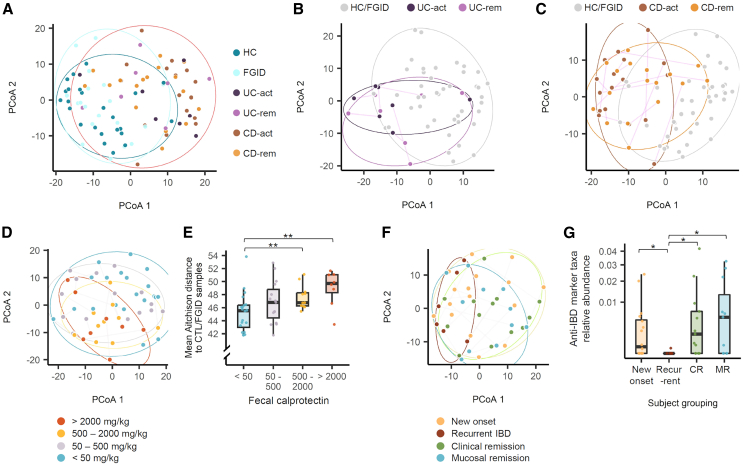
Figure 3.
Gut bacterial community variation associated with patients’ clinical features
(A) PCoA of gut microbiota using read counts per 16S ASV. Aitchison distance was used to quantify inter-sample differences. Adonis 2 test: UC-act vs. HC and FGID, R2 = 0.046, p = 0.001; CD-act vs. HC and FGID, R2 = 0.078, p = 0.001.
(B and C) Individual trajectories within the patients with UC (B) and CD (C) over two sampling time points. Adonis 2 test: UC-act vs. UC-rem, R2 = 0.069, p = 0.63; CD-act vs. CD-rem, R2 = 0.038, p = 0.078.
(D and E) Comparison of fecal samples based on fecal calprotectin levels using PCoA (D) and mean distance to healthy controls (E). Adonis 2 test: low vs. high, R2 = 0.052, p = 0.01; low vs. very high, R2 = 0.076, p = 0.001.
(F and G) Comparison of new-onset patients, those with recurrent active IBD, those in CR, and those in MR. In (A–D) and (F), ellipses are drawn around each sample group using the geom_mark_ellipse function of the ggforce package, and statistical significance of inter-group variation was tested by Adonis2 permutational multivariate analysis of variance implemented in the vegan package based on Aitchison distance. In (A), the patients with IBD (UC-act, UC-rem, CD-act, and CD-rem) are circumscribed together in a single ellipse. In (E) and (G), the boxplots represent interquartile range, along with the individual data points shown as dots. The pairs of groups with values of p < 0.05 as determined by the Wilcoxon test are indicated by horizontal brackets: ∗p < 0.05, ∗∗p < 0.01. ASV, amplicon sequence variant; CD, Crohn disease; CD-act, active CD; CD-rem, remission CD; CR, clinical remission; MR, mucosal remission; PCoA, principal coordinate analysis; UC, ulcerative colitis; UC-act, active UC; UC-rem, remission UC.
Significant differences in ASV composition were observed between samples with low fecal calprotectin levels and those with high or very high levels (Figure 3D). Furthermore, the gut microbial compositions of the four calprotectin-based groups increasingly diverged from those of HC and the patients with FGID as evidenced by the mean Aitchison distance (Figure 3E).
Significant differences in gut microbial composition were also observed between new-onset and recurrent IBD cases (R2 = 0.064, p = 0.02) as well as between recurrent IBD and remission states (both clinical and mucosal; R2 = 0.083, p = 0.007 for recurrent vs. CR; R2 = 0.10, p = 0.004 for recurrent vs. mucosal remission) (Figure 3F). The microbiome shifts among patients with recurrent IBD were not influenced by any specific medications received.
To determine whether certain microbial markers are associated with the inflammatory state, we identified bacterial genera that were depleted in the active inflammation condition (compared to HC and patients with FGID) and relatively restored in remission or vice versa. In patients with UC, two Firmicutes genera, Oscillibacter species, and an unnamed taxonomic genus from the Ruminococcaceae family, were depleted in the UC-act condition and recovered in the UC-rem condition (Figure S1). In patients with CD, six genera were identified: five exhibited depletion in the CD-act condition and recovery in the CD-rem condition (Collinsella and Gordonibacter from Actinobacteria, a Eubacterium clade including Eubacterium uniforme and Eubacterium ventriosum, a Ruminococcus clade containing Ruminococcus bromii, Sporobacter, and an unnamed taxonomic genus from Ruminococcaceae), while one showed enrichment in the CD-act condition and a decrease in the CD-rem condition (Gemella from Lachnospiraceae) (Figure S2).
Genera that were depleted in IBD and recovered in remission (anti-IBD markers) were significantly reduced in patients with recurrence compared to those in remission states (vs. CR, p = 0.01; vs. mucosal remission, p = 0.03). However, this depletion was not significant in patients with new-onset disease (vs. CR, p = 0.48; vs. mucosal remission, p = 0.32) (Figure 3G).
Taxonomic markers associated with IBD and remission across multiple cohorts
To validate our findings of a relatively small Korean cohort, we assessed publicly available 16S V3–4 or V4 amplicon sequencing data from pediatric CD and UC cohorts described in 11 previous studies worldwide. These studies included data from countries such as Brazil, China, the Czech Republic, the UK, the USA, and Canada, along with a multinational cohort (n. samples = 2,401; n. subjects = 1,199).16,21,22,23,24,25,26,27,28,29 After excluding data generated from non-stool sample types, such as ileal or rectal biopsies, and patients not differentiated into CD or UC, we included six cohorts in the differential abundance marker analysis (n. samples = 1,670; n. subjects = 664; Table S2).
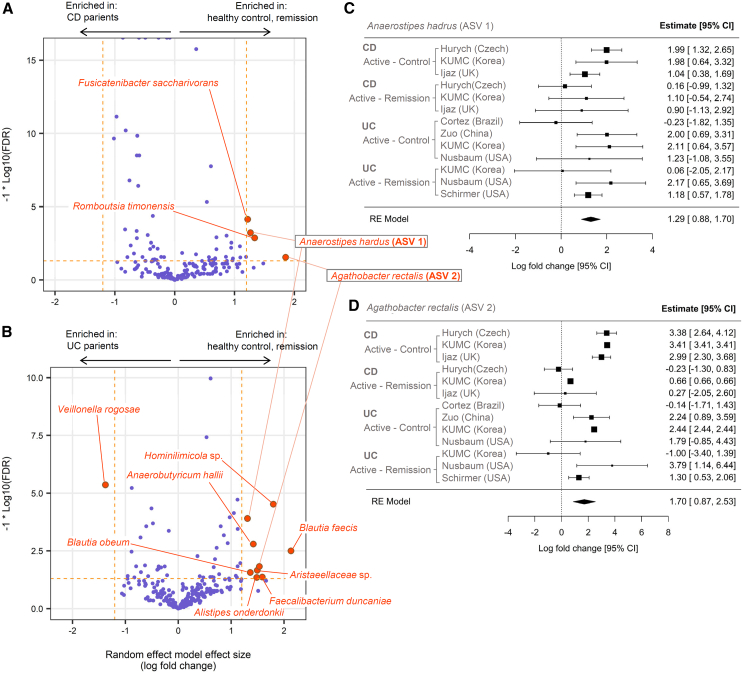
We leveraged these multiple datasets to identify ASVs that could serve as potential diagnostic microbial markers. Our screening process aims to identify the ASVs with robust “anti-IBD” or “pro-IBD” associations. An anti-IBD or pro-IBD association was defined as depletion or enrichment in active IBD compared to both the healthy and the remission groups based on meta-analysis of multiple cohorts. In each individual cohort, we tested the differential abundance of ASVs either between active IBD and HC or between active IBD and remission states using ANCOM-BC and ALDEx2. Results from six individual ANCOM-BC analyses (HC vs. CD, 3 cohorts; CD vs. remission, 3 cohorts), summarized using a random effects model, identified four ASVs consistently displaying an anti-CD pattern, applying a meta-analysis effect size (log fold change) threshold of 1.2 and false discovery rate (FDR) cutoff of 0.05 (Figure 4A). Likewise, nine ASVs with anti-UC pattern were identified from seven individual ANCOM-BC analyses (HC vs. UC, 4 cohorts; UC vs. remission, 3 cohorts) (Figure 4B). Two ASVs, each classified as Anaerostipes hadrus (ASV 1 in Figure 4) and Agathobacter rectalis (ASV 2 in Figure 4), showed both anti-CD and anti-UC patterns (Figures 4C and 4D). There was no ASV discovered to have pro-CD pattern, whereas there was a single pro-UC ASV, which was classified as Veillonella rogosae (Figure 4B; see Table S3 for full list of differential ASVs selected in meta-analysis).
Figure 4.
Microbial markers of active CD and UC derived from meta-analysis of cohorts
(A) Effect size and false discovery rate (FDR) values of the ASVs estimated by random effect model combining ANCOM-BC results from three CD-HC comparisons and three CD-remission comparisons. The ASVs that pass the thresholds for FDR (<0.05) and effect size (absolute value of log fold change >1.2) are emphasized with larger point size and different color. Note that positive effect sizes mean higher abundance in either healthy control or remission state, depending on the analyzed dataset, compared to the active CD. Horizontal dashed line in orange corresponds to FDR value 0.05. Two vertical dashed lines in orange correspond to log fold change −1.2 and 1.2, respectively. Taxonomic names are displayed only for the ASVs that pass the FDR and log fold change thresholds.
(B) Same plot as (A) drawn for UC datasets.
(C and D) Forest plot representing meta-analysis summarization of the two selected ASVs. The square point and horizontal line range represent the estimated fold change and its 95% confidence interval, respectively. The ASV 1 (Anaerostipes hadrus) and ASV 2 (Agathobacter rectalis) were selected from (A) and (B) as these two were commonly detected as markers in both panels. The meta-analysis was performed with the rem function of the R package metafor, using the ANCOM-BC’s effect size and standard error values as inputs. See also Figures S1–S3 and Tables S3 and S4. ASVs, amplicon sequence variants; CD, Crohn disease; HC, healthy control; PIBD, pediatric inflammatory bowel disease; UC, ulcerative colitis.
We also manually searched for the ASVs that repeatedly showed significant differential abundance in active CD or UC from individual differential abundance tests. From the CD cohorts, we identified the four ASVs that most frequently passed the FDR threshold of 0.1 (3 out of 3 HC vs. CD analyses; 2 out of 3 CD vs. remission analyses). Among these, only the Roseburia hominis ASV strictly followed the pattern of depletion in CD and recovery in remission (Figure S3A). From the UC cohorts, an ASV belonging to the Gemmiger genus alone was the single most repeatedly recovered taxon (4 out of 4 HC vs. UC analyses; 2 out of 3 UC vs. remission analyses). This Gemmiger ASV strictly followed the pattern of depletion in UC and recovery in remission (Figure S3B). None of those frequently detected differential abundance markers had “pro-IBD” pattern (Table S4).
Prediction of future remission cases using machine learning approach
From global PIBD datasets, we extracted baseline stool-derived samples from patients with active IBD who had not yet received the treatment of interest in each study. These patients were later categorized based on whether they achieved remission or their disease remained refractory, encompassing both CD and UC cases.23,24,25,26 By integrating these data with our cohort of patients with CD-rem and UC-rem, we compiled a meta-analysis dataset comprising 174 patients who later achieved remission and 111 who did not, including 214 UC and 71 CD patients from the UK, USA, Czech Republic, and Korea (Table S5).23,24,25,30,31 The prognostic training panel consisted of subjects undergoing diverse treatment regimens, including exclusive enteral nutrition (n = 21), 5-aminosalicylic acid (n = 65), corticosteroids (n = 140), anti-TNF-α agents (n = 17), fecal microbiota transplantation (n = 7), and some cases with missing information (n = 7).
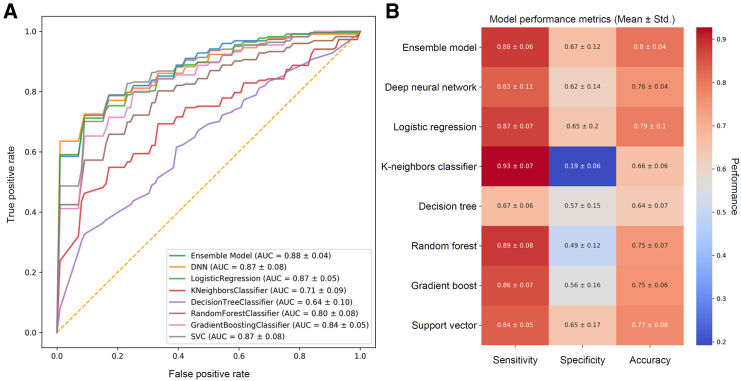
Using this multi-cohort dataset, we developed eight different machine learning (ML) models, namely deep neural networks (DNN), logistic regression (LR), k-nearest-neighbor classifier (KNN), decision tree classifier (DT), random forest classifier (RFC), gradient boosting classifier (GB), support vector machine (SVC), and an ensemble model consisting of the top three models (DNN, LR, SVC) to explore whether the data collected from previous and current studies could predict future remission cases. We initially trained the DNN model using 16S ribosomal RNA (rRNA) ASVs. A filter based on point-biserial correlation analysis (p < 0.05) against the patients’ future remission state in the training dataset was applied, resulting in 705 ASVs. The complete list of ASVs selected by the point-biserial correlation test is presented in Table S6. These filtered feature profiles were then used to train the DNN, which was validated through rigorous 10-fold cross-validation (Figure 5A).
Figure 5.
Comparison of various models trained with patients’ future remission states based on the microbiota profile sampled during the active disease state
(A) Receiver operating characteristic curves of the eight machine learning models trained on the baseline microbiota 16S ASV read counts and clinical features to predict patient remission.
(B) Comparison of classification performance of the eight machine learning models tested based on sensitivity, specificity, and accuracy metrics. See also Figure S4 and Tables S5 and S6. ASVs, amplicon sequence variants.
Consequently, the ensemble model demonstrated well-rounded performance across various metrics, including sensitivity, specificity, and accuracy, compared to the other seven ML models (Figure 5B). Interestingly, the selected ASVs rarely overlapped with those identified through differential abundance analysis (Table S6), suggesting that these bacteria are associated exclusively with treatment response rather than in the disease state itself.
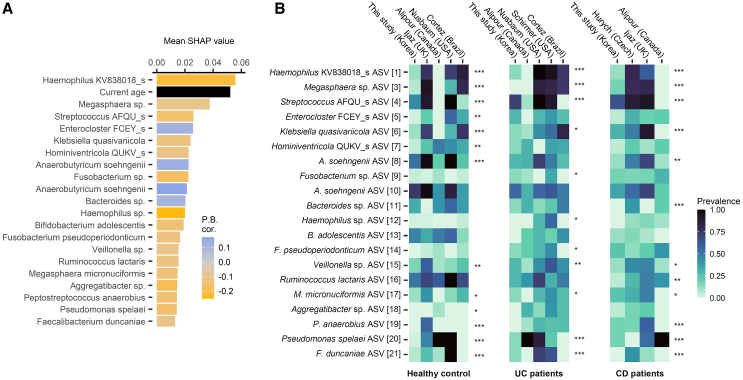
Given the ensemble model’s effectiveness, we assessed the contribution scores of each amplicon within the ASV data using Shapley additive explanation (SHAP) values. The top 20 ASVs with the highest impact on our ensemble model prediction are listed in Figure 6A. Incorporating both microbial and clinical features, our model revealed significant impacts of age on the likelihood of treatment response, with age being the second most important feature. This aligns well with studies where IBD can develop at any age but is most common at ages 11–16 years and that males are prone to more severe IBD complications.32,33 Several of the top 20 bacteria were previously identified as potentially pathogenic or beneficial, with a comprehensive literature review provided in Table S7. Moreover, these ASVs varied significantly in abundance across the analyzed UC and CD cohorts. Specifically, cohort-specific abundance variations were observed in 11 ASVs among patients with UC and 12 ASVs among patients with CD as determined by the Kruskal-Wallis test (p < 0.05) (Figure 6B). The identified predictive ASV markers were both disease- and cohort-specific, highlighting the potential for targeted therapeutic interventions. Although 10-fold cross-validation confirmed the model’s robustness, we further tested our ensemble model with leave-one-study-out cross-validation. In leave-one-study-out evaluation, our prognostic model predicted CR with an AUC of 0.89, accuracy of 0.82, specificity of 0.52, and sensitivity of 0.9 (Figure S4). This external validation offered a robust estimate of how well the model generalizes to new, unseen studies. It evaluates the model’s performance in a more realistic scenario where the training and test data come from different distributions, thus assessing the future applicability of the model to new cohort data. This will ensure that the model is not overfitted onto a specific dataset but makes accurate predictions across diverse populations and conditions. At the same time, this may indicate the prospects of the model for wider application in different research contexts, increasing the utility value or reliability in practical applications.
Figure 6.
Key microbial markers of the prognostic model and their prevalence in different cohorts
(A) The features with the highest SHAP score of feature importance in the ensemble model. The top 21 features are shown, including one clinical variable and 20 microbial ASVs. Point-biserial correlation between the feature and the future response is indicated as a color gradient; the features with positive values had higher values in patients who achieve remission (i.e., responders).
(B) Heat maps showing the prevalence of each ASV in the CD, UC, and HC populations of different cohorts. The ASVs with the highest SHAP scores are included in the figure. The taxonomy label was given at the lowest resolved rank. Kruskal-Wallis test significance is marked at the top of each heatmap column: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Table S7 for the literature review of the taxa represented by these ASVs. ASVs, amplicon sequence variants; CD, Crohn disease; HC, healthy control; SHAP, Shapley additive explanation; UC, ulcerative colitis.
Discussion
Dysbiosis in the microbiota is one of the key factors contributing to the development of IBD, alongside genetic and environmental influences.3,4,5 The highly interconnected nature and converging effects of these three factors make treating IBD particularly challenging. Nevertheless, studies have shown that the microbiota is deeply associated with IBD prognosis.13,14 In this study, we aimed to elucidate the microbiota dynamics associated with active inflammation and remission by investigating microbiota sequencing data alongside clinical indices from patients with PIBD. The study can be largely divided into two parts. First, using the data from our Korean pediatric cohort and global pediatric cohorts, we investigated differences between patients with PIBD and HC, as well as between patients in the active disease and remission states, to identify potential diagnostic markers. Second, we tested various ML models and developed an ensemble model using baseline microbiota and clinical features of the patients with known clinical outcomes collected from four different cohorts from three countries as training data. Highly contributing features in the ensemble model were then selected to identify any overlaps with diagnostic markers with the aim of determining how many diagnostic markers could also serve as prognostic markers and identifying any nondiagnostic markers involved in IBD prognosis. Furthermore, by teasing apart the regional abundances of the top-contributing features, we elucidated prognostic markers that can be used universally on a global or regional scale.
In the Korean pediatric cohort, microbial diversity was reduced during active IBD and restored upon remission (Figures 2B and 2C). The overall community composition also shifted during the active IBD phases (Figures 3B and 3C). This reduction in microbial diversity correlated with inflammatory severity, as indicated by the levels of calprotectin, a marker that is widely used for IBD screenings and mucosal inflammation predictions (Figures 2E and 3E). Reduced gut microbial diversity may both result from and contribute to the disease, as inflammation-induced microenvironments favor dysbiosis-related microbes, whereas lower microbial diversity is associated with IBD relapse.34,35 Patients with recurrent or exacerbated IBD exhibited less microbial diversity and an abundance of anti-IBD marker taxa compared to those with new-onset IBD, although the statistical significance was marginal (p = 0.20 for diversity, Figure 2D; p = 0.05 for anti-IBD taxa abundance; Figure 3G). This could reflect the longer disease duration and increased risk of poor outcomes in terms of exacerbations. Furthermore, the recurrent CD group demonstrated a higher Simple Endoscopic Score for Crohn’s Disease than the new-onset group (20.3 ± 7.64 vs. 12.10 ± 7.31, respectively), although the Pediatric Crohn’s Disease Activity Index (PCDAI) values were similar (37.14 ± 17.62 vs. 36.67 ± 3.82, respectively), suggesting that more severe mucosal inflammation may be correlated with less diversity and a lower abundance of anti-IBD marker taxa. This finding aligns with previously reported associations between less diversity and higher calprotectin levels.11
Next, we compared HC, patients with active disease, and patients with disease in remission. Using the statistical framework for meta-analysis, we identified Anaerostipes hadrus and an Agathobacter rectalis as the shared markers of CD and UC that are depleted in active IBD and recovered in remission states in most analyzed cohorts. Both species are butyrate-producing bacteria that are abundant in human gut,36,37 and in the case of A. rectalis (formerly called “Eubacterium rectale”), previous studies have demonstrated its immunomodulatory functions.36,38 Our differential abundance analysis approach assumed that pro-inflammatory microbes would be enriched during active disease and decreased in remission, whereas anti-inflammatory microbes would deplete during active disease and get restored in remission. However, we discovered that taxa that were depleted in active IBD were not always restored in remission. For example, Waltera intestinalis and Eubacterium ventriosum that were consistently depleted in CD compared to healthy controls tend to be further depleted in the remission conditions (Figure S3A). It is important to note that the discovered taxonomic markers varied based on the analysis approach. For instance, the ASVs selected based on the summarized effect size and FDR from meta-analysis statistics using random effects model did not overlap with the ASVs that most frequently passed the FDR cutoff in individual cohort analyses.39
Using the machine learning framework, we developed a prognostic prediction model that classifies patients into remission and non-remission groups based on ASV profiles sampled during the active disease state. While a few studies have employed various machine learning techniques, such as personalized prediction of patient responses to specific therapy or a general prognosis of future clinical outcomes in patients with PIBD (Table 2), conflicting results have been reported due to small sample sizes. Inconsistencies in microbial markers across patient populations may stem from various factors, such as geographic and age-related microbiota variations, methodological inconsistencies across datasets, or inherent disease heterogeneity.40,41,42,43 Strategies for cross-cohort meta-analyses using multiple heterogeneous cohorts have been proposed to address these issues, yet limitations remain, including confounding factors from diverse environments and batch effects caused by different technologies.40 To minimize methodological and age-related impacts, we focused exclusively on pediatric datasets and included only Illumina-platform-derived amplicon data targeting the V4 region of the 16S rRNA gene. We selectively included only informative taxa to streamline the microbial feature set as described by previous studies.44,45 Interestingly, our ensemble model identified age as a significant contributing factor, affirming the model’s robustness, as childhood-onset IBD tends to be more aggressive and rapidly progressive compared to adult-onset IBD.46 Despite a marginal decrease in performance upon leave-one-study-out cross-validation, the model demonstrated reasonable performance (82% accuracy). We hypothesize that this slight underperformance may be attributable to region-specific taxa that significantly influenced the model.
Table 2.
Microbiome-based prediction models for PIBD in previous studies
| Author and year | Task | Model type | Countries | Cohort description | Model input | Model output | Model performance |
|---|---|---|---|---|---|---|---|
| Kolho et al. 2015 | Prediction of fecal calprotectin level | Linear mixed effect model | Finland |
N = 94 - Control, n = 26 - CD, n = 36 - UC, n = 26 - IBD-U, n = 6 |
Fecal microbiome | Predicted fecal calprotectin level | It is possible to predict calprotectin levels using selected bacterial taxa (AUC = 0.85). |
| Kolho et al. 2015 | Prediction of anti-TNF-α response | Linear mixed effect model | Finland | Subset of the cohort: anti-TNF-α receivers, n = 11 - Responders, n = 6 - Non-responders, n = 5 |
Fecal microbiome: -Two taxa abundance (Clostridium sphenoides and Haemophilus) |
Predicted fecal calprotectin level | It is possible to predict the patient response to anti-TNF-α using the two selected bacterial taxa (AUC = 0.88). |
| Douglas et al. 2018 | Diagnosis | Random forest | UK |
N = 40 - Control, n = 20 - CD, n = 20 |
Intestinal tissue microbiome | Diagnosis of CD | Prediction accuracy was highest with genus-level 16S profiles (84.2%).a |
| Douglas et al. 2018 | Prediction of response to induction treatment | Random forest | UK | CD, n = 20 | Intestinal tissue microbiome | The probability the sample is from a responder | Prediction accuracy was highest with the model combining different feature types (94.4%).a |
| Hyams et al. 2019 | Prediction of response to anti-TNF-α | Multiple imputation multivariate logistic regression with LASSO variable selection | USA and Canada | UC, N = 386 - With biologics, n = 177 - Responders, n = 150 |
Fecal and rectal tissue microbiome: Host side: - Clinical indices - Gene expressions in rectal tissue |
The probability the sample is from those who achieve CS-free remission at week 52 | Clinical data alone (e.g., week 4 remission, PUCAI, baseline hemoglobin) predicted week 52 remission with an AUC of 0.68. Adding host gene expression and microbial features improved accuracy to an AUC of 0.75. |
| Jones et al. 2020 | Prediction of response to EEN | Random forest | Canada | CD, N = 19 - All received EEN - Responders, n = 13 |
Fecal microbiome | The probability the sample is from those who achieve sustained remission until week 24 | Sustained remission can be predicted based on ASVs (AUC = 0.74) but not with other taxonomic levels or shotgun-based profiles. The predictions were improved by the addition of species richness (AUC = 0.83) and further improved by the addition of disease location and behavior (AUC = 0.9). |
| Wang et al. 2021 | Diagnosis | Random forest | China |
N = 93 - IBD, n = 66 - Controls, n = 27 |
Fecal microbiome | Diagnosis of IBD | In the training set, predictions based on 11 OTUs achieved an AUC of 0.88. The validation dataset including IBD (n = 14) and IBS (n = 48) achieved an AUC of 0.84. |
| Zuo et al. 2022 | Diagnosis | Random forest | USA |
N = 42 - UC, n = 19 - Control, n = 23 |
Fecal microbiome | Diagnosis of UC | The best prediction was made with pathway abundance (AUC = 0.95). - Genus composition (AUC = 0.91) - Species composition (AUC = 0.91) The addition of sex- and age-related variables did not improve the model’s performance. |
| Dhaliwal et al. 2023 | Prediction of escalation to anti-TNF-α | Cox proportional hazards regression | Canada | UC, N = 96 - Anti-TNF-α due to non-response to CS, n = 54 - Anti-TNF-α among CS responders, n = 24 - Clinical remission, n = 62 |
Clinical variables, Fecal microbiome |
Hazard ratio and significance value per input clinical variable | Hypoalbuminemia, greater PUCAI, older age, and male sex were significant predictors of escalation to anti-TNF-α. The baseline microbiome was not predictive of escalation to anti-TNF-α. |
| Ventin-Holmberg et al. 2022 | Prediction of response to anti-TNF-α | Regression (PathModel function in R package mare) | Finland | IBD, n = 30 - CD, n = 25 - UC, n = 2 - IBD-U, n = 3 Final cohort used in the model - Anti-TNF-α responders, n = 5 - Anti-TNF-α non-responders, n = 13 |
Fecal microbiome | Probability the sample is from a responder | The Week 6 response to anti-TNF-α can be predicted by the baseline fecal calprotectin level and Ruminococcus count (AUC = 0.89) - The baseline Ruminococcus count alone gives a slightly less accurate prediction (AUC = 0.79). |
| Our study | Prediction of response to induction treatment | Deep neural network, logistic regression, support vector machine | Multinational cohorts: Korea, USA, Canada, and UK; external validation: Czech | IBD, N = 248 - Responders, n = 147 - Non-responders, n = 101 |
Fecal microbiome Host side: - Age, sex, calprotectin level, and disease severity - Usage of anti-TNF-α, 5-ASA, AZA, EEN, and steroids |
Probability the sample is from a responder | The ensemble model performed well (AUC = 0.9), demonstrating that the presence or absence of commensal and pathogenic bacteria can influence future PIBD remission or relapse. |
5-ASA, 5-aminosalicylic acid; ASV, amplicon sequence variant; AUC, area under the curve; AZA, azathioprine; CD, Crohn disease; CS, corticosteroids; DNN, deep neural network; EEN, exclusive enteral nutrition; IBD-U, inflammatory bowel disease–unclassified; IBS, irritable bowel syndrome; N/A, not available; OTUs, operational taxonomic units; PIBD, pediatric inflammatory bowel disease; PUCAI, Pediatric Ulcerative Colitis Activity Index; TNF-α, tumor necrosis factor alpha; UC, ulcerative colitis.
Note that this study did not include the ASV profiles in the performance comparison.
Investigation of such a prognostic prediction model also raised intriguing questions about the microbiome features contributing to the predictability of IBD remission. Our ensemble model suggests that several microbial species and genera may contribute to future PIBD prognosis predictions. The results encompass a wide array of organisms, including both well-established and under-appreciated taxa in the context of PIBD. Among the top features whose abundance was indicative of future clinical remission, Bifidobacterium adolescentis and Faecalibacterium duncaniae were previously reported to be associated with PIBD. These species are known for their anti-inflammatory properties and ability to produce short-chain fatty acids (SCFAs), particularly butyrate, which plays a crucial role in maintaining intestinal homeostasis and barrier function.22 The inclusion of Anaerobutyricum soehngenii, a producer of both butyrate and propionate, among the top features further highlights the potential importance of SCFA production in PIBD prognosis.47
Conversely, our model also identified several potentially harmful bacteria. For instance, Fusobacteria species, including Fusobacterium pseudoperiodonticum, were previously implicated in IBD pathogenesis, potentially through enhanced mucosal invasiveness and pro-inflammatory effects.48 Similarly, Haemophilus and Streptococcus species were highly associated with PIBD and exacerbated intestinal inflammation.49,50 It is also intriguing to see Klebsiella quasivariicola in our model as a high contributing factor. While a direct connection between Klebsiella species and PIBD has never been reported, they have been associated with intestinal inflammation as well.51
We combined several cohorts to create the training dataset for our prognostic model, resulting in a dataset with varied treatment regimens and remission definitions. For example, remission in the UK cohort52 was defined as PCDAI <10 after 8 weeks of exclusive enteral nutrition, whereas the US cohort25 defined it as Pediatric Ulcerative Colitis Activity Index <10 with no corticosteroid therapy in the preceding 28 days at week 52, with patients receiving either 5-aminosalicylic acid monotherapy or a combined corticosteroid-5-aminosalicylic acid regimen. Due to this variability, our model’s predictions do not directly correspond to a specific treatment-endpoint combination. Instead, this model might predict a patient’s general responsiveness to certain treatment strategies. As the model considers specific treatment options as input variables, we could predict the likelihood of achieving remission based on a patient’s age and microbiota profile by simulating the prediction with different treatment scenarios. This capability potentially expands the model’s utility beyond traditional predictions.
The complex microbial signature captured by our model reflects the multifaceted nature of PIBD pathogenesis. It suggests that the disease progression is influenced not only by the reduced population of beneficial, SCFA-producing, bacteria but also by the proliferation of potentially harmful, pro-inflammatory species. Overall, this intricate balance may provide key insights for personalized treatment and prognosis prediction in PIBD.
In conclusion, this study provides insights into the microbiota dynamics associated with active inflammation and remission states of patients with PIBD. Additionally, we developed a prognostic prediction model using an ensemble framework that accurately identifies patients in whom remission may occur in the future based on the given ASV profile sampled during the active disease state. Our study highlights variations in the gut microbiome landscapes in PIBD cohorts from different countries, suggesting the need for further research into the role of microbiome variation in the geographic stratification of PIBD rates. Despite this, our microbiome-based prognostic model yields promising results. The exacerbation rate in PIBD has remained high at 50%–70% within a span of 2 years.2 Our findings offer the potential for stratifying high-risk groups and predicting outcomes based on biomarkers, thus enabling the establishment of proactive treatment strategies in advance. This approach might contribute to personalized precision medicine for patients with PIBD. Further investigations into these PIBD-related bacteria will deepen our knowledge of disease progression and microbiome-guided therapy.
Limitations of study
Our study has several limitations. (1) Despite the strong performance in most metrics, the ensemble model presented in this study exhibits low specificity. Given the limited number of samples available in PIBD cohorts, we believe there is still room for improvement. (2) Although the machine learning approach identified important features that significantly impacted prognosis prediction, this does not imply that the features with high scores are causal factors. More rigorous and controlled experiments are necessary to establish true causality. (3) Related to the previous point, this research is entirely data-driven. Although we have discussed potential biological mechanisms underlying the high-scoring features, their true biological relevance must be validated through carefully controlled experiments.
Resource availability
Lead contact
Requests for further information and resources should be directed to and will be fulfilled by the lead contact, Jung Ok Shim (shimjo@korea.ac.kr).
Materials availability
This study did not generate new reagents or cultured cell lines.
Data and code availability
-
•
The 16S amplicon sequences from stool samples collected in our Korean cohort are publicly available, and the accession numbers are listed in the key resources table. Sample information related to the Korean cohort data is available in Table S8. The accession numbers of the stool sequencing data from published projects used in our study are listed in the key resources table, with the associated sample metadata provided in Table S2.
-
•
Codes used for all analysis and a standalone command-line tool for the prognostic prediction introduced in this study are available at https://github.com/smha118/IBD_remission_study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This work was supported and funded by the Kun-hee Lee Child Cancer & Rare Disease Project, Republic of Korea (no. 22C-011-0100) and supported by a National Research Foundation of Korea grant funded by the Korean government (Ministry of Science and ICT) (no. NRF-2018R1C1B5047245). S.M.H. was supported by the QCB Collaboratory Fellowship at University of California, Los Angeles. J.H. and O.C. were supported by the National Institute of Virology and Bacteriology project (Program EXCELES, ID Project No. LX22NPO5103) funded by the European Union Next Generation EU.
Author contributions
K.L.: conceptualization, data curation, formal analysis, methodology, and writing–original draft. S.M.H.: formal analysis, methodology, and writing–original draft. G.K.: formal analysis and writing–review. J.H. and O.C.: data curation and writing–review. J.O.S.: conceptualization, data curation, methodology, formal analysis, writing–original draft, and supervision. J.O.S. takes responsibility for the integrity of the work as a whole. All authors approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| DNeasy PowerSoil Pro Kit | QIAGEN | 47014 |
| Oligonucleotides | ||
| 16S rRNA forward primer | CCTACGGGNGGCWGCAG | 341F |
| 16S rRNA reverse primer | GACTACHVGGGTATCTAATCC | 805R |
| Deposited data | ||
| 16S amplicon data (Korea) | This study | BioProject: PRJNA917086 |
| 16S amplicon data (Brazil) | Cortez et al.27 | BioProject: PRJNA610934 |
| 16S amplicon data (Canada) | Alipour et al.24 | BioProject: PRJNA298762 |
| 16S amplicon data (China) | Wang et al.22 | CRA005251 |
| 16S amplicon data (Czech) | Hurych et al.31 | BioProject: PRJNA958468 |
| 16S amplicon data (Israel) | Turner et al.53 | BioProject: PRJNA532645 |
| 16S amplicon data (UK) | Ijaz et al.52 | BioProject: PRJEB18780 |
| 16S amplicon data (UK) | Douglas et al.21 | BioProject: PRJEB21933 |
| 16S amplicon data (USA) | Gevers et al.28 | BioProject: PRJNA237362 BioProject: PRJNA205152 |
| 16S amplicon data (USA) | Nusbaum et al.16 | BioProject: PRJNA438164 |
| 16S amplicon data (USA) | Zuo et al.23 | BioProject: PRJNA759642 |
| 16S amplicon data (USA/Canada) | Schirmer et al.25 | BioProject: PRJNA436359 |
| Software and algorithms | ||
| fastp | https://github.com/OpenGene/fastp | 0.21.0 |
| Usearch | http://www.drive5.com/usearch/ | 11.0.667 |
| Vsearch | https://github.com/torognes/vsearch | 2.21.1 |
| MMseqs2 | https://github.com/soedinglab/MMseqs2 | 15-6f452 |
| Phyloseq | https://joey711.github.io/phyloseq/ | 1.22.3 |
| MMUPHin | https://github.com/biobakery/MMUPHin | 1.19.1 |
| Scikit-learn | https://scikit-learn.org/ | 1.2.0 |
| Keras | https://keras.io/ | 2.11.0 |
| Python | https://www.python.org/ | 3.10.4 |
| scikeras | https://github.com/adriangb/scikeras | 0.10.0 |
| SHAP | https://github.com/shap/shap | 0.41.0 |
| Vegan | https://github.com/vegandevs/vegan | 2.6.6 |
| metafor | https://wviechtb.github.io/metafor/ | 4.6–0 |
| ALDEx2 | https://github.com/ggloor/ALDEx_bioc | 1.38.0 |
| ANCOM-BC | https://github.com/FrederickHuangLin/ANCOMBC | 2.8.0 |
Experimental model and study participant details
Study design
This is a prospective observational study of pediatric-onset inflammatory bowel disease. The inclusion criteria included patients with new-onset or recurrent (exacerbated) PIBD (CD or UC), diagnosed before the age of 18 years, without a history of biologics treatment. Exclusion criteria were patients with indeterminate colitis and those who did not achieve clinical remission.
Sample size and group allocation
The final sample size included 24 PIBD patients: 17 patients with CD and 7 with UC. They received their standard treatment in real-world practice without any study-specific allocation. Additionally, age-matched patients with FGID (n = 19) and HC (n = 24), all ethnically Korean and including both males and females, were enrolled as control groups. Data from the FGID and HC data were also used in another study.39
Participants demographics and data collection
All participants were of Korean ethnicity, including both male and female patients. Stool samples and clinical data were collected at two time points for each PIBD patient. We defined initial samples collected at enrollment, which were either at the time of diagnosis or during an exacerbated state, as representing the active disease state (referred to as CD-act and UC-act samples throughout the manuscript). Follow-up samples collected during clinical remission, characterized by achieving PCDAI or PUCAI score of less than 10 and taken at an interval of at least 2 months, were defined as the remission state and referred to as CD-rem and UC-rem samples.
We collected detailed clinical data at both enrollment and follow-up, including age, sex, ethnicity, weight, height, clinical indices (PCDAI or PUCAI), fecal calprotectin levels, endoscopic scores (Simple Endoscopic Score for Crohn’s Disease or Ulcerative Colitis Endoscopic Index of Severity), and treatment details. Demographic and clinical characteristics of the patients, as well as management details, are provided in Tables 1 and S8.
For the control groups, stool samples were collected once from each participant.
Disease severity classification
Disease severity was categorized as inactive/remission, mild, moderate, or severe based on the following thresholds: a PCDAI score of <10, 10–27.5, 30–37.5, and ≥40, and a PUCAI score of <10, 10–34, 35–64, and ≥65, respectively.
Sex and gender reporting
Our study included both male and female participants to enhance generalizability, and we did not observe any sex-specific effects in the results.
Ethics statement
Informed consent was obtained from the children and their parents, and the Institutional Review Board of Korea University Guro Hospital approved this study (no. 2020GR0509).
Method details
Stool sample collection
Participants were asked to collect a teaspoon of their stool using the dedicated spoon provided in the stool container. Samples from inpatients were collected and immediately frozen on site at −20°C. Outpatients were asked to store their samples in the refrigerator before shipping them to our center. The samples were individually delivered via courier service in a dry ice box within 1–2 h, and then frozen at −20°C. The entire process must be completed within 12 h.
Microbiota sequencing
Deoxyribonucleic acid (DNA) was extracted for 16S rRNA gene sequencing. After being diluted in 10 mL of phosphate-buffered saline, samples were filtered and vibrated for 24 h. Microbial genomic DNA was extracted from stool samples using a PowerSoil DNA Isolation Kit (MO BIO Laboratory, San Diego, CA, USA) following the manufacturer’s instructions. Bacterial 16S rRNA genes were amplified with the primers targeting V3–V4 hypervariable regions.54 Amplicon libraries were sequenced on a MiSeq platform (Illumina, San Diego, CA, USA).
Public datasets
In addition to the sequencing data from the stool samples in this study, we searched a worldwide database and obtained gut microbiota 16S amplicon sequencing datasets that were publicly released from previous studies of PIBD cohorts. Specifically, we queried the NCBI PubMed (www.ncbi.nlm.nih.gov/pubmed) database on March 12, 2022, using the following terms: (“Inflammatory bowel disease”[Title/Abstract] OR “Crohn’s disease”[Title/Abstract] OR “ulcerative colitis”[Title/Abstract]) AND (microbiota OR microbiome OR “bacterial community” OR “bacterial communities” OR “microbial community” OR “microbial communities”) AND (pediatric OR pediatric OR adolescent OR adolescence OR children) AND 16S. After manually reviewing the retrieved articles, we recruited the datasets that met the following criteria: (a) 16S V3–4 or V4 region was targeted and (b) sequenced in Illumina, (c) run accessions were matched with the subject-level metadata; and (d) study subjects were not adults. Once the NCBI SRA accession numbers and the associated sample metadata were collected, we downloaded the raw sequencing reads in fastq format using Kingfisher v0.0.1 (wwood.github.io/kingfisher-download). The final list of sequencing runs and associated metadata analyzed in this study is provided in Table S2.
Sequence data analyses
All analyzed 16S amplicon datasets, including our own, had paired-end sequencing layouts. First, a pair of fastq files from each sample was preprocessed with Fastp 0.21.0 to trim adapter sequences, remove low-quality reads, and merge the paired reads into one full amplicon sequence.54 Next, we used Usearch version 11.0.667 to sequentially perform read orientation, truncate 20 bp from each end to remove the primer regions, and filter low-quality reads based on quality scores.55 The preprocessed reads from all samples from the same study (i.e., each cohort) were pooled together and dereplicated using the “--derep_fulllength” command of Vsearch version 2.21.1.56 We generated ASVs from the pooled-dereplicated reads of each study by applying the “--cluster_unoise –minsize 5” and “--uchime3_denovo” commands sequentially using Vsearch version 2.21.1.56 We assigned taxonomy to the ASVs using the “usearch11 -sintax -strand plus -sintax_cutoff 0.6” command with the EzBioCloud database50 as taxonomic reference and created read count tables for each study using the “--usearch_global --otutabout” command of Vsearch version 2.21.1. We performed cross-cohort unification of the ASVs by sequence clustering using the MMseqs2 command “easy-cluster --cluster-mode 2 --cov-mode 1 -c 0.9 --min-seq-id 1”.57 We defined Operational Taxonomic Units at a 97% cutoff from the cross-cohort ASVs using the MMseqs2 “--cluster-mode 2 --cov-mode 1 -c 0.9 --min-seq-id 0.97” command. Finally, aggregated read count tables at species and higher taxonomic ranks were calculated from the ASV-level read count table using the “aggregate_taxa” operator provided in the phyloseq R package.58 In the analysis incorporating multiple public datasets, we performed batch effect correction on the ASV read count matrix before launching the downstream analysis to eliminate possible study-level effects. We used adjust_batch function of MMUPHin R package, with cohort name (i.e., study name) as the batch variable and disease state as the covariate.59 The adjusted read counts were used in downstream analyses of differential abundance and prognostic model training.
Quantification and statistical analysis
Development of ensemble model to predict future IBD remission cases
A total of 347 samples were used to generate the models. Here we included only samples taken from patients in an active IBD state. The remission status of each sample was obtained from patient data from the current study (n = 23) and four additional studies.16,24,25,26 For metadata, we gathered the following information: i) current disease status (UC or CD); ii) calprotectin range; iii) disease severity; iv) patient age; v) patient sex; and vi) four interventions (antibiotics, anti-TNF-α, 5-ASA, AZA, and steroids). The calprotectin range scheme included four categories: <250, 250–500, 500–2000, and >2000 (mg/kg). Categorical values in the metadata were converted to numeric values using the LabelEncoder function of the scikit-learn library (v1.2.0). Subsequently, the 16S rRNA data were log-normalized with a pseudo-count of 1 to circumvent the sparse nature of the amplicon data.
Next, a point-biserial correlation analysis was performed to filter the elements in the abundance data that were highly associated with remission/non-remission status. We used cutoff values of absolute correlation coefficient >0.1 and p < 0.05. Metadata was added after the point-biserial selection of features. The DNN was constructed with the Keras library (v2.11.0) in Python (v3.10.4) and converted into a scikit-learn readable object using scikeras (v0.10.0). Since the DNN model predicts a binary output (remission or not), we used the rectified linear unit for the activation function except for the output layer, where sigmoid was used. We used binary_crossentropy as a loss function and adaptive moment estimation for model optimization. Six additional ML models, namely: Logistic Regression, GaussianNB, KNeighborsClassifier, DecisionTreeClassifier, RandomForestClassifier, and support vector machine. Hyperparameter tuning was performed on each model using RandomizedSearchCV from the scikit-learn library (v1.2.0). The range of parameters tested on each model is listed in Table S9. Lastly, an ensemble model was generated using the best parameters selected for each model using VotingClassifier with the AUC of each model as weights where only the models above 0.85 of AUC were added and weighted based on their performance rank.
Four performance metrics (receiver operating characteristic/AUC, sensitivity, specificity, and accuracy) were measured with 10-fold and “LeaveOneGroupOut” cross-validation using a prebuilt function in the scikit-learn library (v1.2.0). Based on the performance metrics, the ensemble model was chosen as the best model. After selecting the best model, we used SHAP (v0.41.0) to measure feature importance within the model to assess the contribution score of each feature using SHAP (v0.41.0).30
Statistical analysis
Alpha diversity was calculated from ASV read count tables using the Chao1 index with the “estimate_richness” function of the phyloseq package. To account for variable sequencing depth, we rarefied the ASV read count tables to the smallest number of reads per single sample 100 times using the “rarefy_even_depth” function of the phyloseq package and used the median Chao1 index as each sample’s diversity score. Intersample dissimilarities of composition were measured with Aitchison distance calculated from unrarefied read count tables using the “vegdist” function in the vegan package.60 The overall relationship was visualized with PCoA coordinates determined using the “pcoa” function in the ape package, while testing of the correlation with disease metadata was performed using the “adonis2” function of the vegan package. Intergroup differences in the above metrics were tested using the “wilcox.test” and “kruskal.test” functions. We used the ANCOM-BC and the ALDEx2 methods to discover the taxa that were differentially abundant between groups. To summarize differntial abundance test results from multiple cohorts to discover robustly differential markers, we used random effects model developed for meta-analysis. In this meta-analysis we used ANCOM-BC results as ALDEx2 tended to give conservative lists (less markers) and ANCOM-BC always included ALDEx2 markers. For each ASV, we derived meta-analysis effect size and p values using rma function (method = “REML”) implemented in the metafor R pacakge, with the effect size (i.e., log fold change) and the standard error values written by ANCOM-BC input. In the visualization of the resulting differentially abundant taxa, we used the Wilcoxon rank-sum test of the proportion of reads to mark the significance.53,61
Additional resources
This study was registered with the Clinical Research Information Service of the Korea Center for Disease Control and Prevention and the World Health Organization International Clinical Trials Registry Platform (no. KCT0008372 https://cris.nih.go.kr/cris/search/detailSearch.do?seq=24512&search_page=L).
Published: November 22, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111442.
Supplemental information
References
- 1.Sýkora J., Pomahačová R., Kreslová M., Cvalínová D., Štych P., Schwarz J. Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J. Gastroenterol. 2018;24:2741–2763. doi: 10.3748/wjg.v24.i25.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choe Y.J., Han K., Shim J.O. Treatment patterns of anti-tumour necrosis factor-alpha and prognosis of paediatric and adult-onset inflammatory bowel disease in Korea: a nationwide population-based study. Aliment. Pharmacol. Ther. 2022;56:980–988. doi: 10.1111/apt.17125. [DOI] [PubMed] [Google Scholar]
- 3.Shim J.O., Seo J.K. Very early-onset inflammatory bowel disease (IBD) in infancy is a different disease entity from adult-onset IBD; one form of interleukin-10 receptor mutations. J. Hum. Genet. 2014;59:337–341. doi: 10.1038/jhg.2014.32. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narula N., Wong E.C.L., Dehghan M., Mente A., Rangarajan S., Lanas F., Lopez-Jaramillo P., Rohatgi P., Lakshmi P.V.M., Varma R.P., et al. Association of ultra-processed food intake with risk of inflammatory bowel disease: prospective cohort study. BMJ. 2021;374 doi: 10.1136/bmj.n1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutta A.K., Chacko A. Influence of environmental factors on the onset and course of inflammatory bowel disease. World J. Gastroenterol. 2016;22:1088–1100. doi: 10.3748/wjg.v22.i3.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vatn S., Carstens A., Kristoffersen A.B., Bergemalm D., Casén C., Moen A.E.F., Tannaes T.M., Lindstrøm J., Detlie T.E., Olbjørn C., et al. Faecal microbiota signatures of IBD and their relation to diagnosis, disease phenotype, inflammation, treatment escalation and anti-TNF response in a European Multicentre Study (IBD-Character) Scand. J. Gastroenterol. 2020;55:1146–1156. doi: 10.1080/00365521.2020.1803396. [DOI] [PubMed] [Google Scholar]
- 8.Pascal V., Pozuelo M., Borruel N., Casellas F., Campos D., Santiago A., Martinez X., Varela E., Sarrabayrouse G., Machiels K., et al. A microbial signature for Crohn’s disease. Gut. 2017;66:813–822. doi: 10.1136/gutjnl-2016-313235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manandhar I., Alimadadi A., Aryal S., Munroe P.B., Joe B., Cheng X. Gut microbiome-based supervised machine learning for clinical diagnosis of inflammatory bowel diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2021;320:G328–G337. doi: 10.1152/ajpgi.00360.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyams J.S., Davis Thomas S., Gotman N., Haberman Y., Karns R., Schirmer M., Mo A., Mack D.R., Boyle B., Griffiths A.M., et al. Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet. 2019;393:1708–1720. doi: 10.1016/S0140-6736(18)32592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Höyhtyä M., Korpela K., Saqib S., Junkkari S., Nissilä E., Nikkonen A., Dikareva E., Salonen A., de Vos W.M., Kolho K.-L. Quantitative Fecal Microbiota Profiles Relate to Therapy Response During Induction With Tumor Necrosis Factor α Antagonist Infliximab in Pediatric Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2023;29:116–124. doi: 10.1093/ibd/izac182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart L., Farbod Y., Szamosi J.C., Yamamoto M., Britz-McKibbin P., Halgren C., Zachos M., Pai N. Effect of Exclusive Enteral Nutrition and Corticosteroid Induction Therapy on the Gut Microbiota of Pediatric Patients with Inflammatory Bowel Disease. Nutrients. 2020;12 doi: 10.3390/nu12061691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ananthakrishnan A.N., Luo C., Yajnik V., Khalili H., Garber J.J., Stevens B.W., Cleland T., Xavier R.J. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe. 2017;21:603–610.e3. doi: 10.1016/j.chom.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah R., Cope J.L., Nagy-Szakal D., Dowd S., Versalovic J., Hollister E.B., Kellermayer R. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microb. 2016;7:384–396. doi: 10.1080/19490976.2016.1190073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis E.C., Dinsmoor A.M., Wang M., Donovan S.M. Microbiome Composition in Pediatric Populations from Birth to Adolescence: Impact of Diet and Prebiotic and Probiotic Interventions. Dig. Dis. Sci. 2020;65:706–722. doi: 10.1007/s10620-020-06092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nusbaum D.J., Sun F., Ren J., Zhu Z., Ramsy N., Pervolarakis N., Kunde S., England W., Gao B., Fiehn O., et al. Gut microbial and metabolomic profiles after fecal microbiota transplantation in pediatric ulcerative colitis patients. FEMS Microbiol. Ecol. 2018;94 doi: 10.1093/femsec/fiy133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sartor R.B., Wu G.D. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayorga L., Serrano-Gómez G., Xie Z., Borruel N., Manichanh C. Intercontinental Gut Microbiome Variances in IBD. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms231810868. https://www.mdpi.com/1422-0067/23/18/10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pesoa S.A., Portela N., Fernández E., Elbarcha O., Gotteland M., Magne F. Comparison of Argentinean microbiota with other geographical populations reveals different taxonomic and functional signatures associated with obesity. Sci. Rep. 2021;11:7762. doi: 10.1038/s41598-021-87365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Xu Z.Z., He Y., Yang Y., Liu L., Lin Q., Nie Y., Li M., Zhi F., Liu S., et al. Gut Microbiota Offers Universal Biomarkers across Ethnicity in Inflammatory Bowel Disease Diagnosis and Infliximab Response Prediction. mSystems. 2018;3 doi: 10.1128/mSystems.00188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas G.M., Hansen R., Jones C.M.A., Dunn K.A., Comeau A.M., Bielawski J.P., Tayler R., El-Omar E.M., Russell R.K., Hold G.L., et al. Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn’s disease. Microbiome. 2018;6:13. doi: 10.1186/s40168-018-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Xiao Y., Xu X., Guo L., Yu Y., Li N., Xu C. Characteristics of Fecal Microbiota and Machine Learning Strategy for Fecal Invasive Biomarkers in Pediatric Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.711884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo W., Wang B., Bai X., Luan Y., Fan Y., Michail S., Sun F. 16S rRNA and metagenomic shotgun sequencing data revealed consistent patterns of gut microbiome signature in pediatric ulcerative colitis. Sci. Rep. 2022;12:6421. doi: 10.1038/s41598-022-07995-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alipour M., Zaidi D., Valcheva R., Jovel J., Martínez I., Sergi C., Walter J., Mason A.L., Wong G.K.-S., Dieleman L.A., et al. Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. J. Crohns Colitis. 2016;10:462–471. doi: 10.1093/ecco-jcc/jjv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schirmer M., Denson L., Vlamakis H., Franzosa E.A., Thomas S., Gotman N.M., Rufo P., Baker S.S., Sauer C., Markowitz J., et al. Compositional and Temporal Changes in the Gut Microbiome of Pediatric Ulcerative Colitis Patients Are Linked to Disease Course. Cell Host Microbe. 2018;24:600–610.e4. doi: 10.1016/j.chom.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin H., Peddada S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortez R.V., Moreira L.N., Padilha M., Bibas M.D., Toma R.K., Porta G., Taddei C.R. Gut Microbiome of Children and Adolescents With Primary Sclerosing Cholangitis in Association With Ulcerative Colitis. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.598152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gevers D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M., et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng M.Y., Inohara N., Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundberg S.M., Lee S.-I. In: Advances in Neural Information Processing Systems 30. Guyon I., Luxburg U.V., Bengio S., Wallach H., Fergus R., Vishwanathan S., Garnett R., editors. Curran Associates, Inc.; 2017. A Unified Approach to Interpreting Model Predictions [Internet] pp. 4765–4774.http://papers.nips.cc/paper/7062-a-unified-approach-to-interpreting-model-predictions.pdf [Google Scholar]
- 31.Hurych J., Mascellani Bergo A., Lerchova T., Hlinakova L., Kubat M., Malcova H., Cebecauerova D., Schwarz J., Karaskova E., Hecht T., et al. Faecal Bacteriome and Metabolome Profiles Associated with Decreased Mucosal Inflammatory Activity Upon Anti-TNF Therapy in Paediatric Crohn’s Disease. J. Crohns Colitis. 2024;18:106–120. doi: 10.1093/ecco-jcc/jjad126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasparetto M., Guariso G., Pozza L.V.D., Ross A., Heuschkel R., Zilbauer M. Clinical course and outcomes of diagnosing Inflammatory Bowel Disease in children 10 years and under: retrospective cohort study from two tertiary centres in the United Kingdom and in Italy. BMC Gastroenterol. 2016;16:35. doi: 10.1186/s12876-016-0455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazor Y., Maza I., Kaufman E., Ben-Horin S., Karban A., Chowers Y., Eliakim R. Prediction of disease complication occurrence in Crohn’s disease using phenotype and genotype parameters at diagnosis. J. Crohns Colitis. 2011;5:592–597. doi: 10.1016/j.crohns.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Gong D., Gong X., Wang L., Yu X., Dong Q. Involvement of Reduced Microbial Diversity in Inflammatory Bowel Disease. Gastroenterol. Res. Pract. 2016;2016 doi: 10.1155/2016/6951091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbas-Egbariya H., Haberman Y., Braun T., Hadar R., Denson L., Gal-Mor O., Amir A. Meta-analysis defines predominant shared microbial responses in various diseases and a specific inflammatory bowel disease signal. Genome Biol. 2022;23:61. doi: 10.1186/s13059-022-02637-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu H., Xu X., Fu D., Gu Y., Fan R., Yi H., He X., Wang C., Ouyang B., Zhao P., et al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-κB axis. Cell Host Microbe. 2022;30:1139–1150.e7. doi: 10.1016/j.chom.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Liu D., Xie L.-S., Lian S., Li K., Yang Y., Wang W.-Z., Hu S., Liu S.-J., Liu C., He Z. Anaerostipes hadrus, a butyrate-producing bacterium capable of metabolizing 5-fluorouracil. mSphere. 2024;9 doi: 10.1128/msphere.00816-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Islam S.M.S., Ryu H.-M., Sayeed H.M., Byun H.-O., Jung J.-Y., Kim H.-A., Suh C.-H., Sohn S. Eubacterium rectale attenuates HSV-1 induced systemic inflammation in mice by inhibiting CD83. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.712312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim G.-H., Lee K., Shim J.O. Gut Bacterial Dysbiosis in Irritable Bowel Syndrome: a Case-Control Study and a Cross-Cohort Analysis Using Publicly Available Data Sets. Microbiol. Spectr. 2023;11 doi: 10.1128/spectrum.02125-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herlemann D.P., Labrenz M., Jürgens K., Bertilsson S., Waniek J.J., Andersson A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun S., Wang H., Tsilimigras M.C., Howard A.G., Sha W., Zhang J., Su C., Wang Z., Du S., Sioda M., et al. Does geographical variation confound the relationship between host factors and the human gut microbiota: a population-based study in China. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-038163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Chierico F., Abbatini F., Russo A., Quagliariello A., Reddel S., Capoccia D., Caccamo R., Ginanni Corradini S., Nobili V., De Peppo F., et al. Gut Microbiota Markers in Obese Adolescent and Adult Patients: Age-Dependent Differential Patterns. Front. Microbiol. 2018;9:1210. doi: 10.3389/fmicb.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clooney A.G., Fouhy F., Sleator R.D., O’ Driscoll A., Stanton C., Cotter P.D., Claesson M.J. Comparing Apples and Oranges?: Next Generation Sequencing and Its Impact on Microbiome Analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liñares-Blanco J., Fernandez-Lozano C., Seoane J.A., López-Campos G. Machine Learning Based Microbiome Signature to Predict Inflammatory Bowel Disease Subtypes. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.872671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dadkhah E., Sikaroodi M., Korman L., Hardi R., Baybick J., Hanzel D., Kuehn G., Kuehn T., Gillevet P.M. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol. 2019;6 doi: 10.1136/bmjgast-2019-000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon J.S. Clinical Aspects and Treatments for Pediatric Inflammatory Bowel Diseases. Pediatr. Gastroenterol. Hepatol. Nutr. 2019;22:50–56. doi: 10.5223/pghn.2019.22.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seegers J.F.M.L., Gül I.S., Hofkens S., Brosel S., Schreib G., Brenke J., Donath C., de Vos W.M. Toxicological safety evaluation of live Anaerobutyricum soehngenii strain CH106. J. Appl. Toxicol. 2022;42:244–257. doi: 10.1002/jat.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzgerald R.S., Sanderson I.R., Claesson M.J. Paediatric Inflammatory Bowel Disease and its Relationship with the Microbiome. Microb. Ecol. 2021;82:833–844. doi: 10.1007/s00248-021-01697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teitelbaum J.E., Triantafyllopoulou M. Inflammatory bowel disease and Streptococcus bovis. Dig. Dis. Sci. 2006;51:1439–1442. doi: 10.1007/s10620-005-9053-5. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Q., Su X., Zhang C., Chen W., Wang Y., Yang X., Liu D., Zhang Y., Yang R. Klebsiella pneumoniae Induces Inflammatory Bowel Disease Through Caspase-11-Mediated IL18 in the Gut Epithelial Cells. Cell. Mol. Gastroenterol. Hepatol. 2023;15:613–632. doi: 10.1016/j.jcmgh.2022.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ijaz U.Z., Quince C., Hanske L., Loman N., Calus S.T., Bertz M., Edwards C.A., Gaya D.R., Hansen R., McGrogan P., et al. The distinct features of microbial “dysbiosis” of Crohn’s disease do not occur to the same extent in their unaffected, genetically-linked kindred. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner D., Bishai J., Reshef L., Abitbol G., Focht G., Marcus D., Ledder O., Lev-Tzion R., Orlanski-Meyer E., Yerushalmi B., et al. Antibiotic Cocktail for Pediatric Acute Severe Colitis and the Microbiome: The PRASCO Randomized Controlled Trial. Inflamm. Bowel Dis. 2020;26:1733–1742. doi: 10.1093/ibd/izz298. [DOI] [PubMed] [Google Scholar]
- 54.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 56.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4 doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steinegger M., Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 58.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma S., Shungin D., Mallick H., Schirmer M., Nguyen L.H., Kolde R., Franzosa E., Vlamakis H., Xavier R., Huttenhower C. Population structure discovery in meta-analyzed microbial communities and inflammatory bowel disease using MMUPHin. Genome Biol. 2022;23:208. doi: 10.1186/s13059-022-02753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’hara R., Simpson G., Solymos P., et al. The Comprehensive R Archive Network; 2019. Vegan: Community Ecology Package. [Google Scholar]
- 61.Fernandes A.D., Reid J.N., Macklaim J.M., McMurrough T.A., Edgell D.R., Gloor G.B. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:15. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The 16S amplicon sequences from stool samples collected in our Korean cohort are publicly available, and the accession numbers are listed in the key resources table. Sample information related to the Korean cohort data is available in Table S8. The accession numbers of the stool sequencing data from published projects used in our study are listed in the key resources table, with the associated sample metadata provided in Table S2.
-
•
Codes used for all analysis and a standalone command-line tool for the prognostic prediction introduced in this study are available at https://github.com/smha118/IBD_remission_study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.