Abstract
Reactive oxygen intermediates (ROI) and reactive nitrogen intermediates (RNI) produced by activated macrophages participate in host defense against the facultative intracellular pathogens Mycobacterium tuberculosis and Salmonella typhimurium. To survive within macrophages, such pathogens may have evolved ROI and RNI resistance mechanisms. ROI resistance pathways have been intensively studied. Much less is known about the mechanisms of resistance to RNI. To identify possible RNI resistance genes in M. tuberculosis, a mycobacterial library was expressed in S. typhimurium and subjected to selection by exposure to the NO donor S-nitrosoglutathione (GSNO) in concentrations sufficient to kill the vast majority of nontransformed salmonellae. Among the rare surviving recombinants was a clone expressing noxR3, a novel and previously anonymous M. tuberculosis gene predicted to encode a small, basic protein. Expression of noxR3 protected S. typhimurium not only from GSNO and acidified nitrite but also from H2O2. noxR3 is the third gene cloned from M. tuberculosis that has been shown to protect heterologous cells from both RNI and ROI. This suggests diversity in the repertoire of mechanisms that help pathogens resist the oxidative and nitrosative defenses of the host.
Reactive oxygen intermediates (ROI) and reactive nitrogen intermediates (RNI), produced in large amounts by immunologically activated macrophages, and granulysin, a protein exocytosed by cytotoxic T cells, are among the few effector molecules shown to be required for bacteriostatic or bacteriocidal actions against Mycobacterium tuberculosis as it dwells within host macrophages (1, 2, 13, 20). Of these effectors, the most stringent requirement for control of tuberculosis in mice has been demonstrated for RNI. Murine tuberculosis is markedly exacerbated by the administration of inhibitors of inducible nitric oxide synthase (iNOS; NOS2) or disruption of the gene encoding it (2, 13). Analogous experiments cannot be conducted in human subjects. However, pulmonary alveolar macrophages from people with tuberculosis express iNOS (16, 24), and human pulmonary alveolar macrophages that express iNOS use RNI to kill mycobacteria in vitro (17).
In about 90% of immunocompetent, well-nourished individuals infected with M. tuberculosis, host cells restrict replication of the bacterium well enough that the host remains free of active tuberculosis for his or her lifetime. However, the remaining 10% of infected individuals develop active disease that is life-threatening if untreated. A failure to control the infection with a higher risk of fatality despite treatment occurs in a much higher proportion of infected people who are immunocompromised. This picture suggests a dynamic balance. Host factors, including iNOS, may usually hold the organism in check but fail to sterilize it. Their suboptimal expression may permit the pathogen to accelerate its replication. For its part, the pathogen may variably express mechanisms to resist the bacteriostatic or bacteriocidal actions of host defenses, including RNI. Identification of RNI resistance mechanisms might allow the design of inhibitors that could sensitize the pathogen to RNI at whatever level the host can produce. Theoretically, such inhibitors might permit the immunocompetent host to develop sterilizing immunity instead of chronic, subclinical infection, and might help the immunocompromised host to avoid developing overt disease contagious to others.
To lay the groundwork for testing these ideas, we have been searching for RNI resistance genes in M. tuberculosis by both selection (8) and homology-based strategies (3). To date, one candidate RNI resistance gene has been cloned by each approach. In the present report, we have modified the selection strategy of Ehrt et al. (8) by changing the nitrosative stress and the host bacterium expressing the M. tuberculosis library. This led to the cloning of a novel gene that confers substantial resistance both to RNI and to ROI.
MATERIALS AND METHODS
Reagents.
Reagents were obtained from the indicated sources: GSNO (S-nitrosoglutathione; Alexis Co., San Diego, Calif.); yeast extract, tryptone, and Bacto-Agar (Difco Laboratories, Detroit, Mich.); IPTG (isopropyl-β-d-thiogalactopyranoside) and rapid ligation kits (Boehringer Mannheim Biochemicals, Mannheim, Germany); AlamarBlue (AccuMed International Companies, Westlake, Ohio); TRIzol (Gibco/BRL, Gaithersburg, Md.); zirconium-silica beads (0.1 mm; Biospec Products, Bartlesville, Okla.); restriction endonucleases, T4 DNA ligase, and DNA polymerase large fragment (Klenow) (New England Biolabs, Inc., Beverly, Mass.); PCR and reverse transcription-PCR kits (Perkin-Elmer, Branchburg, N.J.); Ni-nitriloacetic acid (NTA) agarose beads and Miniprep, Maxiprep, and Gel extraction kits for DNA extraction and purification (Qiagen, Inc., Santa Clarita, Calif.); and DNA primers (Oligos, Etc., Inc., Guilford, Conn.). Other chemicals and antibiotics were purchased from Sigma Chemical Co., St. Louis, Mo.
Bacterial strains and growth conditions.
For recombinant DNA manipulations, the following E. coli strains were used as competent cells for transformation: HB101 (Gibco/BRL), XL1-Blue, XL2-Blue (Stratagene, Inc., La Jolla, Calif.), and M15 (Qiagen). Luria-Bertani (LB) broth or LB agar plates containing appropriate antibiotics (ampicillin at 100 μg/ml or kanamycin at 40 μg/ml) were used for their growth. S. typhimurium LT2 (strain SGSC1412; Salmonella Genetic Stock Centre, University of Calgary, Calgary, Alberta, Canada) was used for the functional characterization of M. tuberculosis genes. S. typhimurium 14028 (a kind gift of Ferric Fang) was used for confirmatory experiments where indicated. LB broth or LB agar plates containing appropriate antibiotics (ampicillin at 200 μg/ml) were used for propagation. The following mycobacterial strains were used in this study: M. tuberculosis CB3.3 (10), M. smegmatis mc2155 (a kind gift of William Jacobs), M. bovis ATCC 19210, M. avium ATCC 25291, M. intracellulare ATCC 13950, and M. fortuitum ATCC 6841. Mycobacterial strains were grown in Middlebrook 7H9 broth (Difco) supplemented with 2% glycerol, 0.05% Tween 80, and ADC supplement (Difco) or plated on 7H11 agar (Difco).
DNA library and plasmids.
A genomic library was constructed from CB3.3, a strain of M. tuberculosis prevalent among clinical isolates in New York City (10), by Sabine Ehrt by using partial digestion of M. tuberculosis chromosomal DNA with Sau3A, followed by ligation of the DNA fragments into the BamHI site of the E. coli vector pBluescript SK(−) (pBS) (8). Truncation constructs of clone 32, termed pNR32dE and pNR32dS, were constructed by digesting pNR32 with EcoRI or SmaI, with self-ligation. To make pNR32dS2, pNR32dS was digested with SacII and self-ligated. To prepare a His-tagged fusion construct, pNR32dS was digested with SmaI and AflIII and blunt ended with Klenow. The 300-bp DNA fragment containing the small open reading frame (ORF) was cloned in frame into the SmaI site of the expression vector pQE30 (Qiagen).
Preparation of chromosomal DNA.
Genomic DNAs were prepared from mycobacteria by using the method described by van Soolingen et al. (23). Briefly, 30 ml of late-log-phase cultures were harvested by centrifugation, washed once in TE buffer (Tris with EDTA, pH 8.0), and resuspended in 1 ml of TE buffer. Samples were divided in half, and lysozyme (50 μl; 10 mg/ml) was added. After incubation at 37°C for 1 h, 70 μl of 10% sodium dodecyl sulfate (SDS) and 10 μl of proteinase K (10 mg/ml) were added, and the samples were incubated at 65°C for 1 h. After the subsequent addition of 100 μl of 5 M NaCl and 80 μl of 10% N-acetyl-N,N,N-trimethylammonium bromide, the samples were incubated at 65°C for 30 min. The mixture was extracted twice with chloroform and precipitated with a 0.6 volume of isopropanol. The resulting high-molecular-weight DNA was treated with RNase, extracted with phenol-chloroform-isoamylalcohol (25:24:1) and chloroform-isoamylalcohol (24:1), precipitated with ethanol, and resuspended in TE buffer.
Isolation of RNA.
RNA was isolated from 30 ml of logarithmically growing mycobacterial cultures. Bacteria were pelleted and resuspended in 1 ml of TRIzol (Gibco/BRL). Zirconium-silica beads (0.5 ml; 0.1 mm diameter) were added, and lysis was performed in a FastPrep FP120 bead beater apparatus (Bio 101) at 6,500 rpm for two cycles of 20 s. The aqueous phase was extracted with chloroform, and the RNA was precipitated with isopropanol. The RNA was treated with 10 U of DNase I (Boehringer Mannheim) for 30 min at 37°C, followed by purification with Qiagen RNEasy columns.
Extraction of proteins.
Mycobacterial cultures (30 ml) were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS), resuspended in 1 ml of PBS containing 1 mM phenylmethylsulfonyl fluoride, and lysed in a FastPrep apparatus as described above. Homogenates were separated from the beads and centrifuged. The insoluble pellet was resuspended in 100 μl of sample buffer (12 mM Tris-HCl, pH 6.8; 5% glycerol; 0.4% SDS; 2.88 mM 2-mercaptoethanol; 0.02% bromphenol blue) and incubated at 65°C for 20 min.
Purification of fusion protein and preparation of antibody.
A pQE30-based vector containing the NR32dS ORF was used to transform Escherichia coli M15 containing pREP4. A transformed colony was grown overnight in 2 ml of LB containing 150 μg of ampicillin and 40 μg of kanamycin per ml and used to inoculate 100 ml of fresh LB containing both antibiotics. IPTG (2 mM) was added when the bacterial culture reached an optical density at 600 nm (OD600) of 0.6 to 0.8. Four hours later, the bacteria were pelleted, resuspended in 10 ml of buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris) at pH 8.0, and lysed by vortexing at room temperature for 1 h. The supernatant was mixed with 2 ml of Ni-NTA resin at room temperature for 30 min and packed into a column. The column was washed three times in 10 ml (each time) of buffer B at pH 6.3, and the His-tagged protein was eluted with buffer B at pH 4.5 in eight 1-ml fractions and concentrated (Microcon 3; Millipore, Inc.). After further purification by SDS-polyacrylamide gel electrophoresis (PAGE), 200 μg of purified protein in complete Freund adjuvant was injected subcutaneously in each of two New Zealand White rabbits, followed by boosts in incomplete Freund adjuvant every 3 weeks. Antisera used here were collected after at least two boosts.
Affinity purification of antibody.
Approximately 300 μg of column-purified fusion protein was further purified by SDS–15% PAGE and transferred to a nitrocellulose membrane. Staining with Ponceau S revealed a single band at about 11.5 kDa, the size predicted for the fusion protein. A strip containing this species was removed and incubated in blocking buffer (3% bovine serum albumin–0.02% NaN3 in PBS) for 1 h at room temperature (0.25 ml of blocking buffer per cm2 of membrane). Antiserum diluted 1:40 in blocking buffer was incubated with the strip for 5 h at room temperature with gentle shaking. The strip was rinsed sequentially in 0.15 M NaCl and PBS for 20 min each, and antibody was eluted with 1 ml of 0.2 M glycine (pH 2.8)–1 mM EGTA for 20 min at room temperature, followed by neutralization with 0.1 volume of 1 M Tris and the addition of 0.1 volume of 10× PBS and 0.02% NaN3.
Immunoblot.
Transformed S. typhimurium were sonicated in 300 mM NaCl–50 mM sodium phosphate (pH 7.6) or boiled in SDS-PAGE sample loading buffer containing 1% SDS and 100 μM β-mercaptoethanol. Mycobacteria were lysed in a FastPrep beater. Lysates (100 to 200 μg of protein) were subjected to 15% SDS-PAGE, transferred to a nitrocellulose membrane, and reacted with affinity-purified antibody in 5% nonfat milk in Tris-buffered saline with 0.1% Tween 20. Bound antibody was detected by enhanced chemiluminescence.
Bacterial survival after exposure to nitrosative, oxidative, and other stresses.
S. typhimurium LT2 transformed with plasmids encoding M. tuberculosis sequences was stored on plates at 4°C for no more than 4 weeks before a fresh round of transformation. Freshly inoculated overnight cultures of transformed bacteria were diluted to an OD600 of 0.02 to 0.04 (∼106 to 107 CFU/ml). Survival assays were carried out in LB at pH 5.0 for GSNO and sodium nitrite and at pH 7.0 for all other test compounds. Bacteria (1 ml) were incubated with GSNO (1 to 10 mM) at 37°C with shaking at 225 rpm. Aliquots were taken at the indicated time points for serial dilution and plating onto LB agar plates containing 200 μg of ampicillin per ml or for fluorescence-based microplate assay (18) as detailed below. For other stresses, triplicate cultures (100 μl) were incubated with test compounds in Corning 96-well polystyrene plates in a 37°C humidified incubator supplemented with 5% CO2; the pH did not vary over the course of the assay. At the indicated time points, 10 μl from each well was transferred to 100 μl of LB–10% AlamarBlue with ampicillin in new 96-well plates. These read-out plates were stored at 4°C overnight and then transferred to 37°C and shaken at 75 rpm while fluorescence was recorded hourly in a microplate reader (Millipore). Progress curves were compared to a standard growth curve of the same strain. Surviving bacterial numbers derived from counting colonies on plates were expressed as “CFU,” while numbers calculated from AlamarBlue assay (18) were expressed as “calculated CFU.”
RESULTS
Cloning of noxR3 from M. tuberculosis.
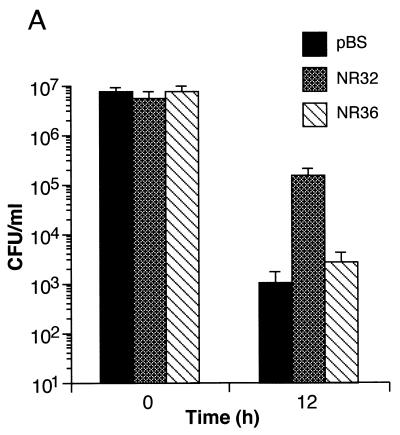
S. typhimurium isolates transformed with an M. tuberculosis genomic library were exposed to GSNO, a physiologic form of RNI produced by activated macrophages and known to be bactericidal when taken up by salmonellae (7). Exposure to GSNO (10 mM) at pH 5.0 for 24 h in LB reduced by 5 to 6 log10 the CFU of bacteria transformed with vector pBS alone compared to growth in the absence of GSNO. Individual plasmids containing M. tuberculosis inserts were recovered from surviving colonies transformed with the genomic library. The selection process was repeated at least once with bacteria freshly transformed with the recovered plasmids. Clone NR32, which contained a 1.35-kb insert, conferred a consistent survival advantage of 2 log10 over the control at 12 h, while the pBS vector-transformed control declined in viability by a factor of 4 log10 (Fig. 1A). Results were similar with another strain (S. typhimurium 14028 [data not shown]). Clone NR36 contained the same M. tuberculosis insert as NR32 except for a 150-bp truncation at the 5′ end. Clone NR36 conferred little resistance at 12 h compared to vector-transformed bacteria (Fig. 1A). Clone NR36 was either a false-positive result in an early screen or suffered this deletion during the process of repeat screening. Both clones NR32 and NR36 were in the same orientation as, and presumably driven by, the lacZ promoter.
FIG. 1.
Characterization of a plasmid insert from M. tuberculosis that protects S. typhimurium from GSNO. (A) Protection of S. typhimurium from GSNO by transformation with M. tuberculosis-derived insert NR32 but not by the closely related, spontaneously truncated insert, NR36. Salmonella isolates transformed with pBS vector alone, NR32, or NR36 were exposed for 12 h to 10 mM GSNO (pH 5.0), and surviving bacteria were enumerated. The means ± the standard deviation (SD) of four independent experiments are shown. (B) Nucleotide sequence of NR32. Arrows denote the start of NR32 (position 1) and NR36 (position 151). The amino acid sequence is shown for a small ORF unique to NR32. (C) Hydrophilicity plot of the ORF in panel B.
The 1,352-bp clone NR32 matched 100% to an anonymous sequence of unknown function in the M. tuberculosis genome database (H37Rv sequence; GenBank accession number Z79701/MTCY277, complementary orientation of sequences 17204 to 18555). NR32 overlaps Rv1498c (complementary to sequences 16901 to 17518), encompasses Rv1499 (corresponding to sequences 17933 to 18403), and overlaps Rv1500 (nucleotides 18448 to 19476). The NR32 sequence contains several hypothetical ORFs in both forward and reverse orientations. The deletion of the first 150 bp from clone 32 that created clone NR36 affected only one hypothetical ORF, whose 270-bp sequence was predicted to encode a novel peptide of 88 amino acids (9,342 Da) (Fig. 1B) arranged in alternating hydrophilic and hydrophobic segments (Fig. 1C). The predicted pI of 11.74 reflects the presence of 10 Arg residues.
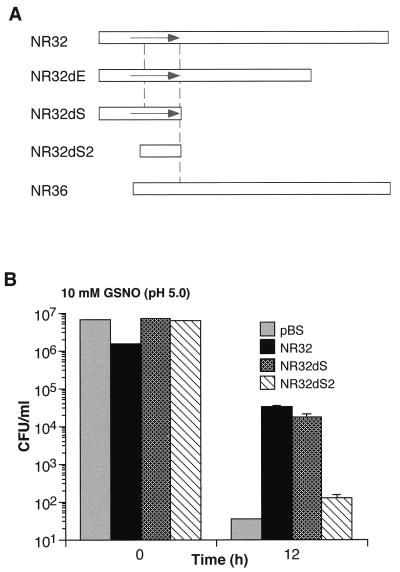
To determine the functional relevance of this ORF, we prepared several truncated versions of clone NR32 and tested the ability of the truncation mutants to confer a survival advantage on S. typhimurium exposed to GSNO. One 3′ truncation generated clone NR32dS, which retained 375 bp of the 5′ sequence of NR32, just enough to include the small ORF. A smaller 3′ truncation generated NR32dE, which retained 992 bp of the 5′ sequence of NR32. Salmonellae transformed with plasmids containing either of these two truncation mutants and exposed to 10 mM GSNO (pH 5.0) for 12 h behaved similarly to salmonellae transformed with NR32. The survival was at least 2 to 3 log10 higher than that of salmonellae transformed with the pBS vector alone or with clone 36 (Fig. 2). A 174-bp 5′ truncation of clone NR32dS eliminated most of the ORF. This double mutant, termed NR32dS2, conferred no survival advantage (Fig. 2). Thus, truncation analysis suggested that the ORF at the 5′ end of clone NR32 was necessary and sufficient for the phenotype of resistance to GSNO. The M. tuberculosis gene encoded by this clone was named noxR3 (nitrogen oxides and oxygen intermediate resistance) based on this and subsequently characterized features.
FIG. 2.
Confirmation of the functional ORF in insert NR32. (A) Schematic of NR32 (top bar), the spontaneously arising 5′ truncation mutant NR36 (bottom bar), and engineered 3′ and 5′ truncations (three middle bars). Arrow denotes the position and orientation of the putative functional ORF. (B) Protection of S. typhimurium from GSNO by transformation with NR32 and NR32dS but not by NR32dS2 or pBS vector alone. Experiment was conducted as described for Fig. 1. The means ± the SD of triplicates in a representative experiment are shown.
Expression of noxR3.
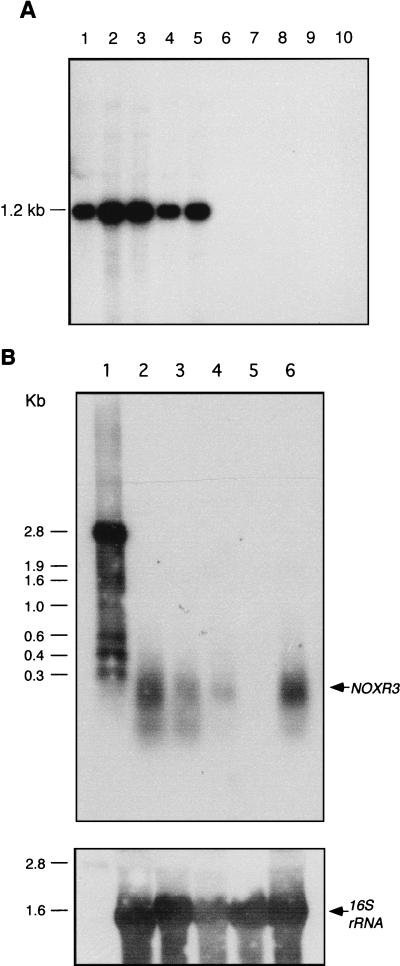
The distribution of noxR3 was studied in M. tuberculosis strains H37Ra and CB3.3; the other members of the M. tuberculosis complex, M. bovis bacillus Calmette-Guérin (BCG), M. africanum, and M. microti; the M. avium complex species M. avium and M. intracellulare; and the environmental mycobacterial species M. kansasii, M. chelonae, and M. smegmatis. A 267-bp PCR product encompassing the noxR3 ORF was used as the DNA probe in Southern hybridization analysis of genomic DNAs digested with SmaI. A hybridization signal at ∼1.2 kb was present only in the species belonging to the M. tuberculosis complex (Fig. 3A). A noxR3 DNA probe hybridized with a transcript of ∼0.3 kb in RNA from all members of the M. tuberculosis complex tested, including M. tuberculosis CB3.3, M. africanum, M. bovis BCG, and M. microti but not with RNA from M. smegmatis (Fig. 3B). A loading control for 16S rRNA showed that the negative result with M. smegmatis was not due to insufficient RNA (Fig. 3B).
FIG. 3.
Mycobacterial expression of noxR3 at the genomic and RNA levels. (A) Southern blot. Genomic DNAs (1 μg) digested with SmaI were loaded onto a 0.9% agarose gel, electrophoresed, transferred to a filter, and hybridized with a 267-bp DNA fragment containing the noxR3 sequence. Lanes: 1, M. tuberculosis CB3.3; 2, M. tuberculosis H37Ra; 3, M. bovis BCG; 4, M. africanum; 5, M. microti; 6, M. avium; 7, M. intracellulare; 8, M. kansasii; 9, M. chelonae; 10, M. smegmatis. (B) Northern blot. In the upper panel, RNAs (10 μg) were hybridized to a 250-bp digoxigenin-labeled DNA fragment containing the noxR3 sequence. Lanes: 1, digoxigenin-labeled RNA size markers; 2, M. africanum; 3, M. bovis BCG; 4, M. microti; 5, M. smegmatis; 6, M. tuberculosis CB3.3. The lower panel shows a loading control. The same membrane was stripped and reprobed with a 16S rRNA gene-specific digoxigenin-labeled DNA fragment.
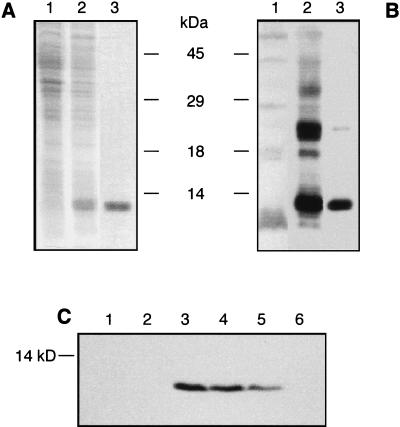
To analyze the expression of noxR3 at the protein level, a hexahistidine-tagged fusion protein (predicted and observed mass of ∼11.5 kDa) was overexpressed in E. coli and purified by nickel column chromatography (Fig. 4A). NH2-terminal amino acid sequencing confirmed that 10 amino acids following the 6 His and 13 linker residues conformed to the predicted ORF encoded by the M. tuberculosis gene. The column-purified protein was subjected to SDS-PAGE, and the molecular mass region containing the sole stainable band (11.5 kDa) was used to raise a rabbit antiserum. The antiserum reacted with the fusion protein in the recombinant E. coli both before and after its purification (Fig. 4B).
FIG. 4.
Expression of NOXR3 protein in transformed E. coli and S. typhimurium. (A) Expression and purification of NOXR3 fusion protein monitored on Coomassie blue-stained SDS–15% PAGE. Lane 1, lysate (50 μg of protein) of uninduced E. coli transformed with pQE30-NR32ORF1; lane 2, lysate (50 μg of protein) of E. coli as in lane 1 but after 4 h of induction with 2 mM IPTG; lane 3, NOXR3 fusion protein (2 μg) after chromatographic purification. (B) Immunoblot with rabbit anti-NOXR3 antiserum raised against the antigen illustrated in panel A, lane 3. Lane 1, lysate (100 μg of protein) of uninduced E. coli transformed with pQE30-NR32ORF1; lane 2, lysate (100 μg of protein) of E. coli as in lane 1 but after 4 h of induction with 2 mM IPTG; lane 3, NOXR3 fusion protein (0.5 μg) after chromatographic purification. (C) Immunoblot of lysates (200 μg of protein per lane) of transformed S. typhimurium with affinity-purified antibody from the antiserum characterized in panel B. Transformation was by pBS vector containing the following: lane 1, no insert; lane 2, NR36; lane 3, NR32; lane 4, NR32dE; lane 5, NR32dS; and lane 6, NR32dS2.
The anti-NOXR3 antiserum was affinity purified on antigen and used to immunoblot lysates of transformed S. typhimurium. A band migrating at ∼10 kDa (the size predicted for NOXR3 without fusion residues) was recognized in salmonellae transformed with plasmids containing clones NR32, NR32dE, or NR32dS but not with the vector alone or plasmids containing clones NR36 or NR32dS2. The amount of NOXR3 protein appeared to be greater in salmonellae transformed with NR32 and NR32dE than those transformed with NR32dS when the same amount of lysate protein was compared under denaturing and reducing conditions (Fig. 4C). When lysates were prepared by sonication without SDS, NOXR3 protein was detected only in the insoluble portion, suggesting either that the NOXR3 had aggregated or that it was associated with bacterial membranes or cell walls (not shown). Thus, the same ORF that was inferred to be functional from truncation analysis was expressed as a protein with the predicted N-terminal amino acid sequence and was expressed selectively in those Salmonella organisms that displayed enhanced resistance to GSNO.
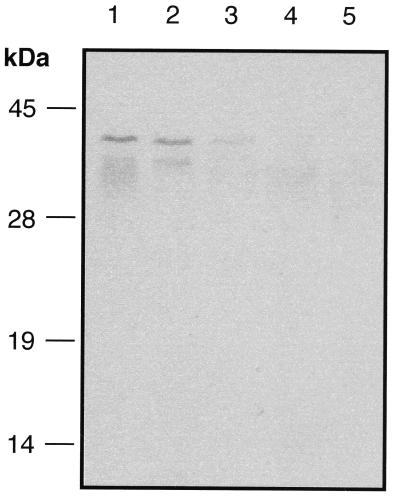
Finally, we immunoblotted lysates of mycobacteria with affinity-purified anti-NOXR3 immunoglobulin G. A single discrete polypeptide was detected that migrated with an apparent molecular mass of about 40 kDa in lysates from M. tuberculosis CB3.3, M. bovis BCG, and M. microti (Fig. 5). In contrast, no reactivity was detected in lysates from M. avium, M. intracellulare (Fig. 5), or M. fortuitum (not shown). Because the ∼40-kDa species was only seen in species that contained the noxR3 gene, the immunoreaction might be authentic, even though the apparent mass was unexplained.
FIG. 5.
Possible expression of NOXR3 protein in mycobacteria. Mycobacteria were cultured in vitro under standard conditions. Lysates (100 μg) were separated by SDS–15% PAGE and transferred to nitrocellulose membranes. Affinity-purified anti-NOXR3 antibody was used for immunodetection. Lanes: 1, M. tuberculosis CB3.3; 2, M. bovis BCG; 3, M. microti; 4, M. avium; 5, M. intracellulare.
noxR3 confers resistance not only to GSNO but also to NO2− and H2O2.
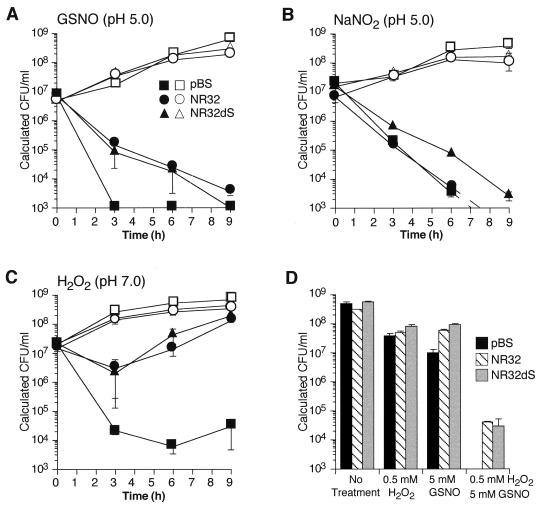
Next, we wanted to see whether the resistant phenotype conferred by noxR3 might extend beyond the stress used for selection. Salmonellae transformed with NR32, NR32dS, or vector alone all grew at the same rate when no stress was applied (Fig. 6A). As before, transformation of Salmonella with either NR32 or NR32dS conferred relative resistance to GSNO, such that >1 log10-fold more bacteria survived than when the Salmonella was transformed with pBS vector alone (Fig. 6A). Another physiologic form of RNI produced by activated macrophages is NO2−. In mildly acidic conditions, resembling the pH of the phagosome of activated macrophages, a proportion of NO2− is protonated to nitrous acid. The latter dismutates to give back NO, the additional radical NO2, and higher oxides of nitrogen (8, 22). That NO2− can be bactericidal at a mildly acidic pH has been appreciated for decades (14). The ability of NO2− at pH 5.0 to kill Salmonella was confirmed in the present studies (Fig. 6B). Remarkably, however, transformation with clone NR32 conferred no protection against NO2− in the same experiments in which the same transformants were protected from GSNO at the same pH (compare Fig. 6A and B). This suggests that GSNO and NO2− do not constitute interchangeable forms of RNI with regard to Salmonella. However, some protection against NO2− at pH 5.0 was afforded by transformation with NR32dS (Fig. 6B). This may signify that expression of a smaller amount of recombinant NOXR3 (see Fig. 4C) was optimal in Salmonella confronted with NO2− or that products of the downstream sequence exerted a negative effect on survival in NO2−.
FIG. 6.
Survival of S. typhimurium in LB with RNI, ROI, or a combination of RNI and ROI after transformation with or without noxR3. In panels A through C, open symbols depict bacteria not exposed to the experimental stress, while closed symbols depict bacteria stressed as described. S. typhimurium was transformed with pBS containing no insert (squares), with NR32 (circles), or with NR32dS (triangles). (A) Survival with or without exposure to GSNO (5 mM, pH 5.0). (B) Survival with or without exposure to sodium nitrite (7 mM, pH 5.0). (C) Survival with or without exposure to H2O2 (1 mM, pH 7.0). (D) Survival with or without exposure to H2O2 (0.5 mM, pH 7.0), GSNO (5 mM, pH 7.0), or a combination of the two. S. typhimurium was transformed with pBS containing no insert (solid bars), NR32 (striped bars), or NR32dS (gray bars). The means ± the standard error of three independent experiments for panels A, B, and C and the means ± the SD of a representative experiment in triplicate in panel D are shown. Values falling below the limit of detection (103 calculated CFU) were assigned the value of the limit.
A robust phenotype also emerged when salmonellae were exposed to 1 mM H2O2 at neutral pH. Although all transformants declined in viability at 3 h, transformation with NR32 and NR32dS allowed >2 log10-fold more survival than did the vector alone. By 6 h, the former transformants resumed growth, and by 9 h they displayed a net 4 log10-fold survival advantage over the control (Fig. 6C).
At pH 7.0, GSNO (up to 5 mM) had little bactericidal effect on Salmonella transformed with the vector alone, nor did 0.5 mM H2O2. However, their combination was synergistically bactericidal, reducing viability by a factor of >5.4 log10 by 6 h. Expression of noxR3 in either NR32- or NR32dS-transformed bacteria conferred >1 log10-fold protection (Fig. 6D).
Resistance against other stresses.
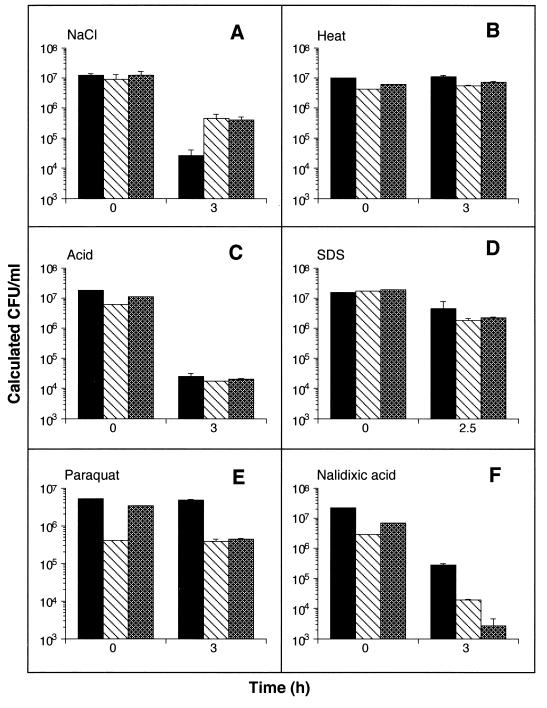
To distinguish whether noxR3 protected Salmonella relatively specifically against RNI and ROI or instead controlled a general survival or repair function, we subjected transformed S. typhimurium to excess sodium chloride, heat, acid, detergent, paraquat, or nalidixic acid. Transformation with NR32 or NR32dS consistently protected against hypertonicity (Fig. 7A). Under the conditions used, heating the bacteria did not reduce viability below that at time zero, but it did prevent the growth seen under nonstressed conditions (compare the difference between 0 and 3 h in Fig. 7B with that in Fig. 6A, B, and C). noxR3-encoding plasmids conferred no resistance against heat stress (Fig. 7B). Heating for longer times or at higher temperatures killed all of the bacteria, while milder conditions had no effect on their growth (not shown). At pH 4.0, bacterial viability was reduced substantially, although not as severely as in many experiments with RNI or ROI. Nonetheless, noxR3 afforded no protection (Fig. 7C). Likewise, the mild toxicity inflicted by SDS was not resisted by noxR3 (Fig. 7D). Finally, noxR3 expression was associated with decreased, rather than increased, survival in bacteria exposed to paraquat (Fig. 7E) or nalidixic acid (Fig. 7F). Thus, expression of noxR3 conferred either a survival advantage or disadvantage or had no effect, depending on the stress. Among the stresses tested, a survival advantage was seen only in nitrosative, oxidative, and hypertonic conditions.
FIG. 7.
Survival of S. typhimurium during exposure to various stresses after transformation with or without noxR3. S. typhimurium were transformed with pBS containing no insert (solid bars), containing NR32 (striped bars), or containing NR32dS (stippled bars) and cultured in LB with the following stresses for the duration of the experiment: 1 M NaCl (A), incubation at 52°C (B), pH 4.0 (achieved by addition of HCl) (C), SDS (5%) (D), paraquat (10 mM) (E), or nalidixic acid (15 μg/ml) (F). The means ± the SD of a representative experiment in triplicate are shown.
DISCUSSION
Although the complete sequence of the M. tuberculosis genome is now available, the sequence gives no clue to the function of 16% of the genes (4). Selection strategies are one potentially powerful way to begin to analyze the functions of anonymous genes. In about one-third of the human population, M. tuberculosis resides for decades within macrophages, and thus it is reasonable to anticipate that some M. tuberculosis genes may be devoted to defense against macrophage microbicidal mechanisms. Because of the experimental difficulties, long incubations, and biological hazards associated with mutating, selecting, and transforming M. tuberculosis, we embarked on a two-part strategy to look for candidate RNI resistance genes in this pathogen. The first step was to select genes from a relatively RNI-resistant isolate of M. tuberculosis (10) that impart relative RNI resistance to S. typhimurium, a fast-growing, genetically tractable, relatively RNI-sensitive, facultative intracellular pathogen that serves as a surrogate intermediary. The second step is to knock out and restore candidate RNI resistance genes by allelic replacement in M. tuberculosis itself and to test the phenotype of the resulting mycobacterial strains in vitro and in mice. Here we describe the first of these two steps.
The strategy used in this study was patterned on that used by Ehrt et al. (8), but it yielded a different gene. The use of different nitrosative stresses in the two studies—nitrite (8) versus GSNO—may explain why different genes were recovered, and this in turn may shed light on the emerging diversity of resistance mechanisms to RNI.
Glutathione is the most abundant nonprotein thiol (1 to 5 mM) in macrophages and is actively secreted. Glutathione readily conjugates NO. The product, GSNO, serves to store NO and shield it from reaction with O2 or O2−, to transport NO equivalents across bacterial cell walls (7), and to return NO equivalents to the chemically reactive pool of RNI by transnitrosation or release of NO+ or NO (19). Thus, GSNO is a physiologic form of RNI that is likely to be encountered in the pericellular, intraphagosomal, and cytosolic compartments of macrophages. The same can be said of nitrite. noxR3 encoded by a plasmid with a short insert, NR32dS, afforded protection against both GSNO and nitrite. In contrast, the longer plasmid NR32, which includes additional 3′ sequences, afforded relative resistance to GSNO but not to nitrite. Although it is unclear how the 3′ sequences affected the phenotype, the latter result suggests that GSNO and nitrite are not identical in their impact on the pathogen. As for superoxide (6), products of nitrite may affect both the cell wall and internal targets. GSNO, in contrast, must be taken up before it is bactericidal to Salmonella and thus may preferentially affect internal targets (7).
The production by macrophages of several forms of RNI, coupled with the ability of various forms of RNI to affect the same microorganism in different ways, suggests that pathogens requiring an ability to survive within activated macrophages may need more than one mechanism of RNI resistance. Two RNI resistance gene products indigenous to cultured S. typhimurium have recently been characterized: AhpC (3) and flavohemoglobin (5, 11). In this context, it is all the more interesting that expression of either noxR1 (8) or noxR3 rendered S. typhimurium substantially more resistant to RNI than they already were. In other words, for each transformant, at least two RNI resistance mechanisms were already likely to be operating, yet the addition of a third was consequential.
The biologic significance of the apparent multiplicity of RNI resistance genes will be clearer when we understand how each gene is regulated and when each has been knocked out and reconstituted. We need to learn the subcellular localization of the gene products and by what biochemical mechanisms they impart resistance. Recently, flavohemoglobin has been shown to act as an NO dioxygenase (11). AhpC was first identified as part of an enzyme complex (12). Whether NOXR3 is an enzyme remains to be determined.
After noxR1 (8) and ahpC (3), noxR3 is the third candidate RNI resistance gene cloned from M. tuberculosis. noxR1 has been knocked out and reconstituted in BCG, with a corresponding phenotype of sensitivity and resistance to acidified nitrite (21). Thus, the type of selection strategy employed here for mycobacterial genes expressed in a surrogate enteric bacterial host has the potential to identify genes with the corresponding functions in mycobacteria themselves. ahpC has been knocked down in M. bovis, rendering this pathogen nonvirulent for guinea pigs (25). It remains to be tested whether this reflects loss of resistance to RNI. Knockout of noxR3 in M. tuberculosis, while arduous, now appears warranted. Until this is accomplished, it will remain an open question whether the phenotype associated with the expression of noxR3 in a heterologous host is relevant to its phenotype in the native context.
noxR3 appears to be present only in the genomes of members of the M. tuberculosis complex and not in other nonpathogenic or opportunistic mycobacteria. Although affinity-purified anti-NOXR3 antibody detected NOXR3 protein in transformed Salmonella, no protein was detected in mycobacteria at the expected molecular mass of ∼10 kDa. Instead, a single band was immunoblotted at ∼40 kDa. This polypeptide was only detected in those mycobacteria whose genomes contain a species hybridizing with noxR3. Thus, the possibility must be entertained that the ∼40-kDa species may have been authentic. Perhaps glycosylation accounted for the increase in mass above that of the recombinant protein expressed in E. coli and S. typhimurium. Glycosylation is more common in mycobacteria than in other eubacteria and contributes to the increased mass of another product from M. tuberculosis above that predicted from its nucleotide sequence (15). Alternatively, perhaps SDS-resistant complex formation burdened NOXR3 with exogenous material. Transcriptional read-through can be dismissed by the fact that the NOXR3 transcript was big enough only to account for NOXR3 transcribed from its native promoter. Finally, NOXR3 may have bound to insoluble components of mycobacteria or may have been expressed at levels below the limit of detection after growth without imposed nitrosative or oxidative stress, and the ∼40-kDa antibody-binding reactivity may have been an artifact. In the latter case, it would not be apparent why the ∼40-kDa reactivity was detected only in species of mycobacteria whose genomes contain noxR3. In sum, mycobacterial expression of NOXR3 at the protein level requires further study, even though NOXR3 transcripts were detected in M. tuberculosis.
noxR3 conferred partial resistance to GSNO, H2O2, and salt but not to heat, acid, detergent, paraquat, or nalidixic acid. In comparison, noxR1, another novel gene that also appears to be confined to members of the M. tuberculosis complex (8), conferred resistance to GSNO, nitrite, H2O2, and acid but not to salt, heat, detergent, paraquat, nalidixic acid, or ethanol. The restriction of each gene to a subset of mycobacteria demonstrates that neither of these genes is required for a function common to all mycobacteria. The restriction of the phenotype conferred by each gene demonstrates that neither is a general stress resistance gene. As yet, we have no understanding of the mechanistic basis for the overlapping but distinct resistance patterns conferred by noxR1 and noxR3.
Both noxR1 and noxR3 conferred cross-protection against oxidative stress as well as against nitrosative stress. This has been observed with RNI resistance mechanisms in other bacteria (3, 9) and may reflect that molecular targets for RNI overlap with those for ROI, including thiols, transition metal centers, lipids, and DNA.
It will be important to determine which RNI and ROI resistance genes are expressed during infection of the human host by M. tuberculosis and at what stage in the course of the disease.
ACKNOWLEDGMENTS
We thank Lei Chen, Ferric Fang, and Qiao-wen Xie for helpful advice; F. Fang for S. typhimurium 14028; W. R. Jacobs, Jr., for M. smegmatis mc2155; Lei Chen for review of the manuscript; and Aihao Ding and Jenny Zhang for expert assistance during final stages of the experiments.
This work was supported by NIH grant HL51967 to C.N., a fellowship to J.R. via Medical Scientist Training Program grant GM07739, and a fellowship to G.S.J. via the Molecular Medicine Training Program supported by the Lucille Markey Charitable Trust.
REFERENCES
- 1.Adams L B, Dinauer M C, Morgenstern D E, Krahenbuhl J L. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tubercle Lung Dis. 1997;78:237–246. doi: 10.1016/s0962-8479(97)90004-6. [DOI] [PubMed] [Google Scholar]
- 2.Chan J, Ying Y, Magliozzo R S, Bloom B R. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Xie Q, Nathan C. Alkylhydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol Cell. 1998;1:795–805. doi: 10.1016/s1097-2765(00)80079-9. [DOI] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin H, Holryod S, Horsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J H, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Crawford J J, Goldberg D E. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem. 1998;273:12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- 6.De Groote M A, Ochsner U A, Shiloh M, Nathan C, McCord J M, Dinauer M D, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Groote M A, Testerman T, Xu Y, Stauffer G, Fang F C. Homocysteine antagonism of nitric oxide-related cytostasis in Salmonella typhimurium. Science. 1996;272:414–417. doi: 10.1126/science.272.5260.414. [DOI] [PubMed] [Google Scholar]
- 8.Ehrt S, Shiloh M S, Ruan J, Choi M, Gunzburg S, Nathan C, Xie Q-W, Riley L. An antioxidant gene from Mycobacterium tuberculosis. J Exp Med. 1997;186:1885–1896. doi: 10.1084/jem.186.11.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang F C. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest. 1997;99:2812–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman C R, Quinn G C, Kreiswirth B N, Perlman D C, Salomon N, Schluger N, Lutfey M, Berger J, Poltoratskaia N, Riley L W. Widespread dissemination of a drug-susceptible strain of Mycobacterium tuberculosis. J Infect Dis. 1997;176:478–484. doi: 10.1086/514067. [DOI] [PubMed] [Google Scholar]
- 11.Gardner P R, Gardner A M, Martin L A, Salzman A L. Nitric oxide dioxygenase: an enzymatic function for flavohemoglobin. Proc Natl Acad Sci USA. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobson F S, Morgan R W, Christman M F, Ames B N. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]
- 13.MacMicking J D, North R J, LaCourse R, Mudgett J S, Shah S K, Nathan C F. Identification of NOS2 as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacMicking J, Xie Q-W, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 15.Menozzi F D, Bischoff R, Fort E, Brennan M J, Locht C. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc Natl Acad Sci USA. 1998;95:12625–12630. doi: 10.1073/pnas.95.21.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson S, da G. Bonecini-Almeida M, Lapa e Silva J R, Nathan C, Xie Q-W, Mumford R, Weidner J R, Calaycay J, Geng J, Boechat N, Linhares C, Rom W, Ho J L. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med. 1996;183:2293–302. doi: 10.1084/jem.183.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nozaki Y, Hasegawa Y, Ichiyama S, Nakashima I, Shimokata K. Mechanism of nitric oxide-dependent killing of Mycobacterium BCG in human alveolar macrophages. Infect Immun. 1997;65:3644–3647. doi: 10.1128/iai.65.9.3644-3647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiloh M S, Ruan J, Nathan C. Evaluation of phagocyte function and bacterial numbers by a fluorescence-based microplate assay. Infect Immun. 1997;65:3193–3198. doi: 10.1128/iai.65.8.3193-3198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamler J S. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 20.Stenger S, Hanson D A, Teitelbaum R, Dewan P, Niazi K R, Froelich C J, Ganz T, Thoma-Uszynski S, Melian A, Bogdan C, Porcelli S A, Bloom B R, Krensky A R, Modlin R L. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282:121–125. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 21.Stewart G R, Erht S, Riley L W, Dale J W, McFadden J. Program and abstracts of the 38th Interscience Congress on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Allelic exchange of the antioxidant gene noxR1 in Mycobacterium bovis BCG generates a macrophage-sensitive mutant, abstr. B-52; p. 59. [Google Scholar]
- 22.Stuehr D J, Nathan C F. Nitric oxide: a macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C-H, Liu C-Y, Lin H-C, Yu C-T, Chung K F, Kuo H P. Increased exhaled nitric oxide in active pulmonary tuberculosis due to inducible NO synthase upregulation in alveolar macrophages. Eur Respir J. 1988;11:809–815. doi: 10.1183/09031936.98.11040809. [DOI] [PubMed] [Google Scholar]
- 25.Wilson T, de Lisle G W, Marcinkeviciene J A, Blanchard J S, Collins D M. Antisense RNA to ahpC, an oxidative stress defence gene involved in isoniazid resistance, indicates AhpC of Mycobacterium bovis has virulence properties. Microbiology. 1998;144:2687–2695. doi: 10.1099/00221287-144-10-2687. [DOI] [PubMed] [Google Scholar]