Abstract
The human IRGM gene has been linked to inflammatory diseases including sepsis and Crohn’s disease. Decreased expression of human IRGM, or the mouse orthologues Irgm1 and Irgm2, leads to increased production of a number of inflammatory chemokines and cytokines in vivo and/or in cultured macrophages. Prior work has indicated that increased cytokine production is instigated by metabolic alterations and changes in mitochondrial homeostasis; however, a comprehensive mechanism has not been elucidated. In the studies presented here, RNA deep sequencing and quantitative PCR were used to show that increases in cytokine production, as well as most changes in the transcriptional profile of Irgm1−/− bone marrow-derived macrophages (BMM), are dependent on increased type I IFN production seen in those cells. Metabolic alterations that drive increased cytokines in Irgm1−/− BMM - specifically increases in glycolysis and increased accumulation of acyl-carnitines - were unaffected by quenching type I IFN signaling. Dysregulation of peroxisomal homeostasis was identified as a novel upstream pathway that governs type I IFN production and inflammatory cytokine production. Collectively, these results enhance our understanding of the complex biochemical changes that are triggered by lack of Irgm1 and contribute to inflammatory disease seen with Irgm1-deficiency.
Keywords: macrophage, peroxisome, cytokine induction, GTPase, interferon
The Immunity-Related GTPases (IRGs) are a family of large GTPases that are thought to function similar to dynamins in regulating membrane dynamics in cells. Membrane processes in which they have been implicated include autophagy and mitophagy (1, 2, 3, 4, 5, 6), mitochondrial fission (6, 7), processing of vacuoles that contain pathogens (8, 9, 10, 11), vesicle formation and transport (11, 12, 13, 14, 15, 16), and cell motility (17, 18). However, the specific mechanisms through which they influence these processes are not clear. Regulation of membrane processes by IRGs is important in immune and inflammatory contexts. Transcription of IRG genes is potently induced by interferon (IFN)-γ and/or lipopolysaccharide, and polymorphisms in the human IRGM gene are associated with several inflammatory diseases including Crohn’s disease (CD) (19, 20, 21, 22, 23), Mycobacterium tuberculosis infection (3, 24), sepsis (4), and non-alcoholic fatty liver disease (25).
Decreased expression of IRGM proteins leads to increases in inflammatory cytokine production, which is thought to be pivotal for the involvement of IRGMs in the aforementioned inflammatory diseases. For instance, disease loci for IRGM are linked to increases in serum tumor necrosis factor (TNF), a key cytokine in CD pathogenesis (26). A global deletion of the mouse orthologue Irgm2 leads to increased production of IL-1α, IL-1β, and IFNɣ in lipopolysaccharide (LPS)-injected mice (a model of sepsis), while deletion of Irgm1 leads to increases in TNF, MIP-1α, and GM-CSF following LPS exposure (27). Additionally, in Irgm1−/− mice, intestinal levels of IL-6, S100A, IL-18, and CXCL1 are elevated relative to wild-type levels following infection with Citrobacter rodentium (an inflammatory bowel disease model), while multiple serum cytokines and chemokines are elevated tonically in those same mice in absence of experimental infection (28). Most of these cytokines and chemokines are likewise elevated in macrophages purified from Irgm1−/− or Irgm2−/− mice and then activated in culture (27, 28). Elevated production of inflammatory cytokines in Irgm1−/− macrophages is dependent on metabolic alterations in those cells that include increased glycolysis and impaired fatty acid catabolism, as cytokine levels are dampened when the macrophages are exposed to a glycolytic inhibitor (2-deoxyglucose) or fatty acid synthase inhibitors (C75 or cerulenin). Because Irgm1-deficient macrophages also exhibit altered mitochondrial homeostasis—manifested as impaired mitophagy (29) and altered mitochondrial fusion dynamics (28)—it has been suggested that impaired mitochondrial health may be an important driver of the metabolic impairments that subsequently lead to increases in inflammatory cytokines. The details of this mechanism are, nevertheless, not clear. In the current work, we examined in more depth how Irgm1 deficiency in primary macrophages triggers elevated cytokine production.
Results
A role for type I IFN in mediating proinflammatory cytokine production in Irgm1-deficient macrophages
Recently published work has suggested that impaired mitophagy in Irgm1-deficient cells triggers the production of type I IFN. This occurs either through soiling of the cytoplasm with mitochondrial DNA that either engages cGAS/STING, or through a TLR7-dependent mechanism (29). Elevation of type I IFN production in this manner is a major contributor to the striking susceptibility to M. tuberculosis, Listeria monocytogenes, and Salmonella typhimurium seen in Irgm1−/− mice, as susceptibility is largely rescued in IFNAR−/−/Irgm1−/− mice lacking type I IFN signaling. (In contrast, decreased resistance to Toxoplasma gondii is maintained) (30). Bone marrow-derived macrophages (BMM) from Irgm1−/− mice also manifest a type I IFN gene expression pattern with increased expression of many type I IFN-regulated genes including Mx1, which is often used as a marker of type I IFN signaling (29, 31).
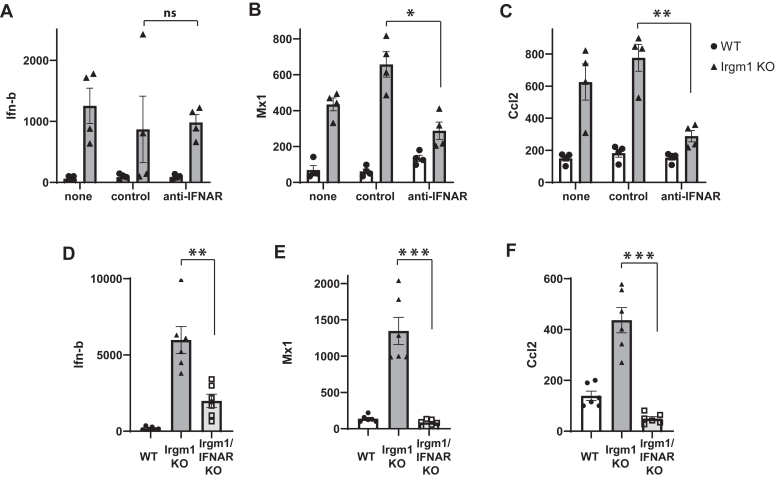
As alluded to above, we previously described a chronic, sterile inflammation in Irgm1-deficient mice and BMM that is manifested as increased production of a broad number of inflammatory cytokines (28). Because IFNs regulate the expression of thousands of genes (32), we reasoned that elevated production of type I IFN in Irgm1-deficient cells may in turn stimulate the production of some of the other cytokines and chemokines that are increased with Irgm1-deficiency. We tested this here by dampening type I IFN signaling in macrophages and examining cytokine gene expression with a focus on Ccl2 (Mcp-1), one of the chemokines that we found previously to be elevated in Irgm1−/− BMM (28). We disrupted type I signaling in two ways: by use of type I IFN receptor neutralizing antibodies (33) or by introducing a genetic deletion of the type I IFN receptor gene (IFNAR) onto the Irgm1−/− background. With the neutralizing antibody approach (Fig. 1, A–C), enhanced expression of both Mx1 and Ccl2 in Irgm1−/− BMM was significantly decreased. Ifnb mRNA expression, which was markedly elevated in Irgm1−/− BMM as compared to WT BMM, was unaffected by the IFNAR neutralizing antibody (Fig. 1A). Genetic deletion of IFNAR (Fig. 1, D–F) led to even more dramatic effects with complete loss of the elevated Mx1 and Ccl2 expression seen in Irgm1−/− cells. There was also an unexpected decrease in Ifnb expression, perhaps suggesting a feedback regulatory mechanism. Together, these results support the hypothesis that enhanced production of Ccl2 and similar cytokines in Irgm1−/− macrophages is mediated by increases in type I IFN.
Figure 1.
Increased expression of Ccl-2 in Irgm1-deficient macrophages requires type I IFN signaling.A–C, bone marrow-derived macrophages (BMM) from WT and Irgm1−/− mice were activated with IFN-ɣ for 24 h and were simultaneously exposed to no antisera (none), a control/non-immune antiserum (control), or anti-type I IFN receptor antiserum (anti-IFNAR). Cells were collected and quantitative PCR was used to measure expression of the indicated mRNAs relative to ActA expression. The results were normalized to one of the WT control samples and expressed as a percent of that sample. Four separate experiments were performed using macrophages prepared from distinct mice in each experiment, and the results were combined. D–F, BMM from WT, Irgm1−/−, and IFNAR/Irgm1−/− mice were activated with IFN-ɣ for 24 h. Cells were collected and used to quantify the expression of the indicated mRNAs using quantitative PCR. Shown are the combined results of two experiments performed with a total of 6 BMM cultures per genotype prepared from distinct mice. Error bars indicate SEM. ns, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Possible impact of type I IFN on metabolic and mitochondrial dysfunction in Irgm1-deficient macrophages
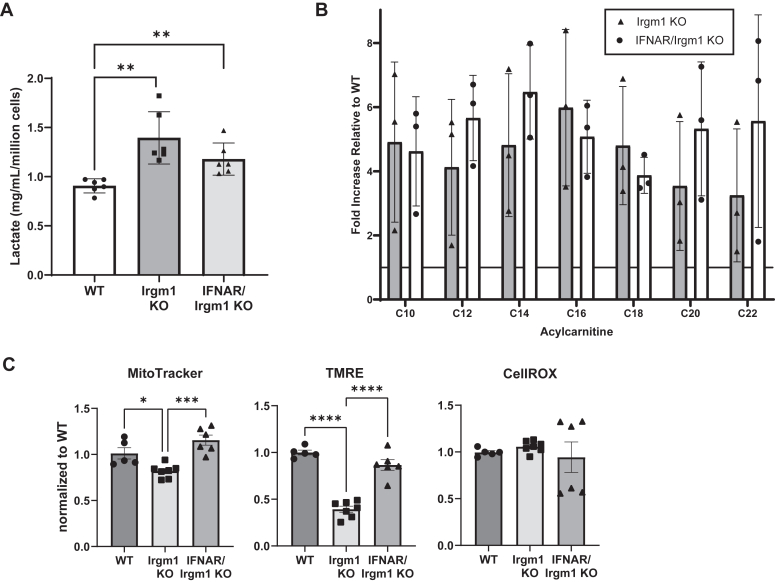
Because the increased glycolysis and acylcarnitine accumulation initiated by Irgm1 deficiency are necessary for the subsequent increase in inflammatory cytokine production (28), we next asked whether type I IFN may instigate those metabolic changes. Using lactate production as a surrogate for glycolysis, we found lactate production to be elevated in both IFN-ɣ-activated Irgm1−/− and IFNAR/Irgm1−/− BMM (relative to levels in activated WT BMM) (Fig. 2A). As a readout for fatty acid oxidation, we measured acyl-carnitines using a mass-spectrometry-based metabolic profiling approach. Multiple medium- and long-chain acyl-carnitines were found to be elevated in both IFN-ɣ-activated Irgm1−/− and IFNAR/Irgm1−/− BMM, ranging from about 3 to 6 times the levels found in WT BMM (Fig. 2B). Thus, by these measures, the metabolic alterations that are triggered by Irgm1 deficiency in macrophages are not dependent on increased type I IFN production, and likely lie upstream of type I IFN production in the signaling cascade triggered by Irgm1 deficiency.
Figure 2.
Metabolic changes in Irgm1-deficient macrophages do not require type I IFN signaling.A, Groups of 3 WT, Irgm1−/−, and IFNAR/Irgm1−/− BMM isolated from distinct mice were activated with IFN-γ for 24 h. Conditioned media were used for lactate measurements that were normalized to cell counts. The results shown are representative of three separate experiments. B, groups of 3 WT, Irgm1−/−, and IFNAR/Irgm1−/− BMM isolated from distinct mice were activated with IFN-γ for 24 h and simultaneously exposed to 0.5 mM L-carnitine and 100 μM 1:1 oleate/palmitate. Lysates were prepared from the cells, and acyl-carnitines were measured using MS/MS. Shown are average acyl-carnitine levels for the indicated genotypes relative to WT levels. Acyl-carnitine levels were not statistically different between Irgm1−/− and IFNAR/Irgm1−/− BMM (p > 0.05 in all cases). Shown is one representative experiment out of two. C, WT and Irgm1−/− BMM were activated with IFN-ɣ for 24 h. The cells were stained with Mitotracker Green, TMRE, or CellROX, which were quantified by flow cytometry. Shown are the combined results of the two experiments. Data points represent BMM isolated from distinct mice. Error bars indicate SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Prior work has suggested that diminished removal of poorly functioning mitochondria in Irgm1−/− BMM due to impaired mitophagy leads to the type I IFN signature (29). Here we tested whether the increase in type I IFN may influence mitochondrial functioning in Irgm1−/− BMM. To do so, we used flow cytometry to capture key aspects of mitochondrial health. Irgm1−/− BMM as compared to WT BMM displayed a striking reduction in mitochondrial membrane potential (TMRE) and a small but statistically significant decrease in mitochondrial mass, though little difference in reactive oxygen species (CellROX) (Fig. 2C). Mitochondria from IFNAR/Irgm1−/− BMM did not display statistical changes in mitochondrial mass and membrane potential relative to WT BMM (Fig. 2C). Thus, increased type I IFN signaling may contribute to altered mitochondrial health in Irgm1-deficient cells.
Transcriptional profile of IFNAR/Irgm1−/− macrophages
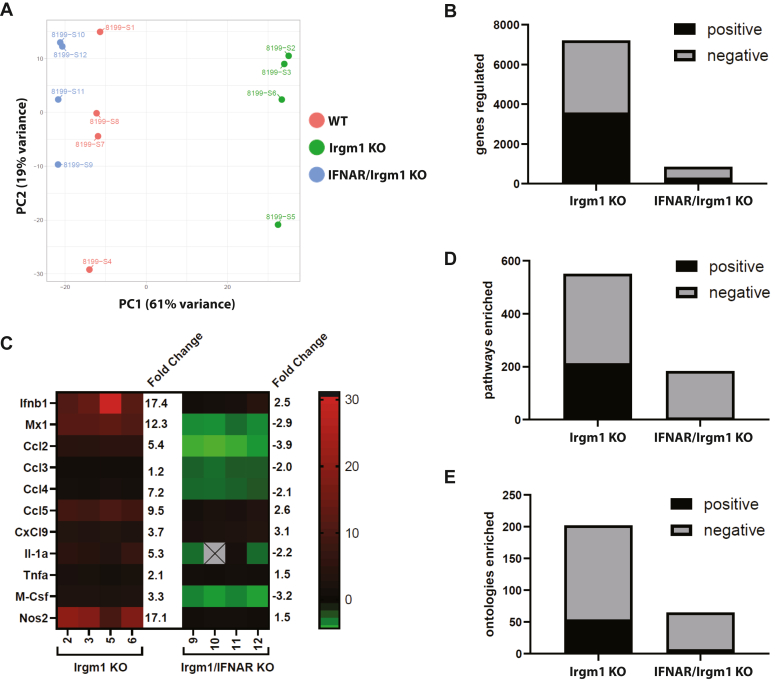
To gain more insight into the transcriptional changes that are initiated by Irgm1 deficiency and their dependence on type I IFN, we performed deep RNA sequencing and Gene Expression Enrichment Analysis (GSEA) comparing BMM cultured from WT, Irgm1−/−, and IFNAR/Irgm1−/− mice (Fig. 3 and Table 1). Principal component analysis showed that the gene expression profiles from Irgm1−/− BMM cultures were markedly different from those of WT BMM (Fig. 3A), while those from IFNAR/Irgm1−/− were closer to those of WT BMM (Fig. 3A). Comparing Irgm1−/− BMM to WT BMM, 7206 genes showed statistically significant differences in gene transcription, yet comparing IFNAR/Irgm1−/− to WT BMM, there were only 846 that were differentially transcribed (Fig. 3B, FDR < 0.05). Among the identified genes, there were multiple inflammatory cytokines that were markedly elevated in Irgm1−/− BMM relative to WT BMM that were not statistically elevated in IFNAR/Irgm1−/− BMM (Fig. 3C). These data are consistent with the quantitative RT-PCR Ccl2 analyses (Fig. 1) and suggest that most transcriptional changes, including the sterile inflammation seen in Irgm1−/− BMM and mice, are mediated by increased Type I IFN production.
Figure 3.
Role for Type I IFN signaling in transcriptomic changes in Irgm1-deficient macrophages. BMM from four distinct mice each for C57Bl/6 wild-type (WT), Irgm1−/− (Irgm1 KO), and IFNAR/Irgm1−/− (IFNAR/Irgm1 KO) were activated with IFN-γ for 24 h. mRNA was prepared and used for 16S RNA deep sequencing and Gene Set Enrichment Analysis. A, principal component analysis; B, the number of genes with statistically significant differences in expression in the indicated genotype relative to WT; LRT, FDR < 0.05. C, a heat map of select inflammatory chemokines and cytokines that have statistically significant increases in expression in Irgm1−/− BMM (relative to WT) that are reversed in IFNAR/Irgm1−/− BMM. The average fold change in expression is indicated. D, the number of gene pathways that were enriched in the indicated genotype relative to WT; fold change > 1.25; t test, P, FDR < 0.05. E, the number of ontologies that were enriched in the indicated genotype relative to WT; fold change > 1.25; t test, FDR < 0.05.
Table 1.
Normalized Enrichment Scores (NES) for select gene pathways and ontologies enriched by Irgm1 and/or IFNAR deletion in bone marrow macrophages
| Rescued by escued by IFNAR deletion | ||
|---|---|---|
| Gene Pathway/Ontology | Irgm1−/− vs WT BMM | Irgm1−/−/IFNAR−/− vs WT BMM |
| DER IFN Alpha Response Up | 2.00a | −1.61a |
| GOBP Response to Type I IFN | 2.04a | −1.65a |
| GOBP Cytokine Production | 1.63a | ns |
| GOBP Inflammatory Response | 1.54a | ns |
| Unaffected by IFNAR deletion | ||
| Gene Pathway/Ontology | Irgm1−/−vs WT BMM | Irgm1−/−/IFNAR−/−vs WT BMM |
| WP Aerobic Glycolysis | 1.53b | 1.70b |
| Biocarta PPARA Pathway | 1.52b | 1.68b |
| GOBP Protein Refolding | 1.96a | 2.02a |
| GOMF Heat Shock Protein Binding | 1.80a | 1.92a |
| Reactome Hsf1 Activation | 1.91a | 2.66a |
| GOBP Cell Cycle Process | −1.63a | −1.58a |
| GOBP Organelle Fission | −1.77a | −1.66a |
| Reactome Golgi to ER Retrograde Transport | −1.61b | −1.41b |
p < 0.001; ns, not significant.
p < 0.05.
A similar pattern was seen in the gene sets that were identified as differentially enriched with GSEA (Fig. 3, D and E, FDR < 0.05). There were 551 gene sets that were enriched in Irgm1−/− BMM relative to WT BMM, while there were only 183 enriched in IFNAR/Irgm1−/− BMM compared to WT BMM. Similarly, there were 202 gene ontologies enriched in Irgm1−/− BMM and only 65 enriched in IFNAR/Irgm1−/− BMM. These results imply that increased type I IFN drives the majority of the biological processes that are altered with Irgm1 deficiency. However, a closer look at some of the specific gene sets/ontologies (Table 1) revealed that although gene sets related to type I IFN signaling and inflammation were indeed rescued with the IFNAR deletion, there were several gene sets that were not, potentially representing gene pathways upstream of type I IFN production that might drive the IFN production and ultimately the inflammation seen with Irgm1 deficiency. Some of these were expected. For instance, aerobic glycolysis was enriched in both Irgm1−/− BMM and IFNAR/Irgm1−/− BMM, which was consistent with our above analysis of lactate production (Fig. 2A). There were also enriched gene sets related to protein refolding and heat shock responses; these likely stem from the long-held observation that mouse IRGM proteins act to stabilize a number of other IRG proteins (known as GKS IRG proteins) so that in the absence of Irgm1 or other IRGM proteins, those GKS IRG proteins form cellular aggregates (10, 34, 35, 36), ostensibly triggering heat shock and unfolded protein responses. There were also pathways related to organelle fission; these are in line with the presumed function of IRG proteins as dynamins as well as the specific observations from our labs and others that Irgm1 controls mitochondrial fission. An unexpected gene pathway that was enriched was the Peroxisome Proliferator-Activated Receptor (PPAR)-α Pathway, which we explored further.
Peroxisomal dysfunction in Irgm1−/− macrophages
Peroxisomes are single membrane organelles that play essential roles in lipid metabolism including fatty acid catabolism and synthesis of cholesterol and plasmalogens (37). The biochemical reactions that occur in peroxisomes produce significant amounts of reactive oxygen species like hydrogen peroxide that are detoxified by catalase produced in the organelles. Peroxisome synthesis is driven by PPAR pathways. Prior publications have indicated that a fraction of cellular Irgm3 (36) and human IRGMd (6) localizes to peroxisomes, but otherwise, there have been no previous connections of IRGM proteins to peroxisomal health. Perturbation of the PPAR-α pathway suggests that peroxisome synthesis and/or function may be altered with Irgm1-deficiency.
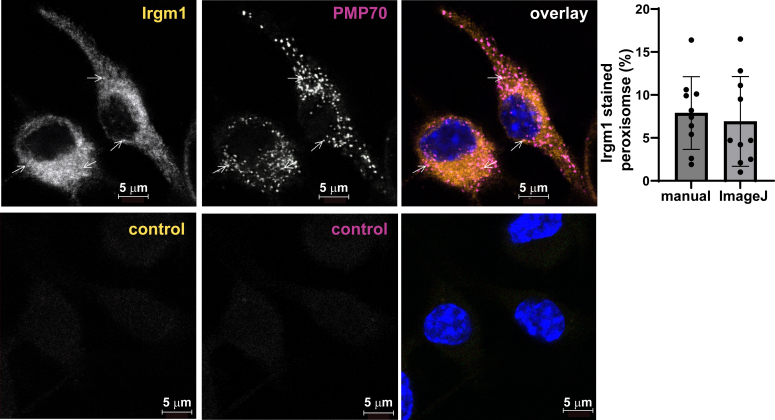
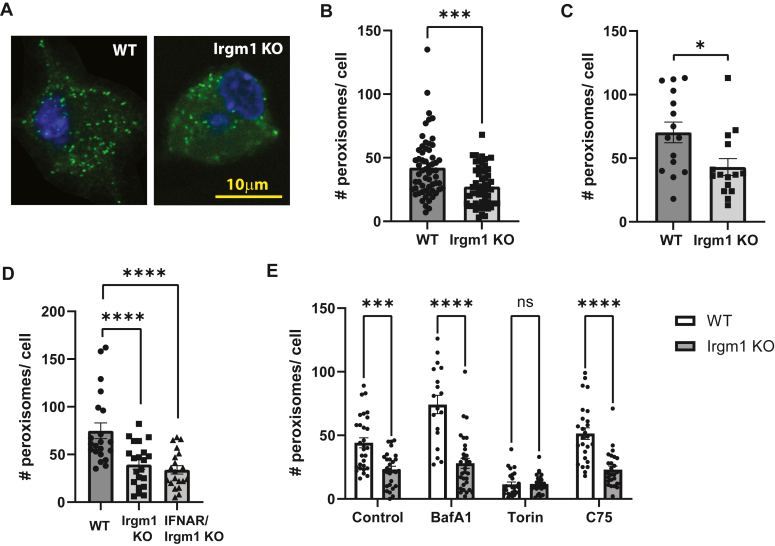
To explore a role for Irgm1 in peroxisomal biology, we first assessed whether Irgm1 localized to peroxisomes in WT BMM (Fig. 4). We and others have previously found the bulk of Irgm1 to localize to the Golgi apparatus (15, 16, 38), with additional localization to other organelles including mitochondria (6, 7) and lysosomes (39). Here we found that Irgm1 decorated 7 to 8% of the peroxisomes, potentially suggesting a direct role in peroxisome biology. Peroxisomes were next compared in WT and Irgm1−/− BMM. There were no apparent differences in the appearance of individual peroxisomes by immunofluorescence microscopy (Fig. 5A); however, there was a striking 50% decrease in peroxisome numbers in Irgm1−/− BMM compared to WT BMM (Fig. 5B). Peroxisomes were also enumerated in live cells by transfection with a fluorescent peroxisomal construct. By this approach, peroxisome numbers were likewise found to be reduced by about 50% in Irgm1−/− BMM (Fig. 5C). Peroxisomes were next enumerated in IFNAR/Irgm1−/− BMM and found to be decreased to about the same extent as those in Irgm1−/− BMM (Fig. 5D), indicating that this effect of Irgm1 deficiency is not dependent on increased type I IFN signaling. A reduction in peroxisome numbers could result from reduced peroxisome biogenesis or increased peroxisome turnover via autophagy (i.e. pexophagy). Because of the established associations of Irgm1 with autophagic flux, we explored the latter by enumerating peroxisomes following treatment with the autophagic inducer, Torin-1, and the inhibitor, bafilomycin A1 (Baf A) (Fig. 5E). As expected, peroxisome numbers in WT BMM were markedly reduced when autophagy was stimulated with Torin-1 (40), but there was only a modest impact in Irgm1−/− BMM. Also as expected, inhibition of autophagy with Baf A led to a marked increase in peroxisome numbers. In contrast, Irgm1−/− BMM were largely insensitive to Baf A, suggesting either that the dominant pathway dictating peroxisome numbers with Irgm1 deficiency may not be autophagic, or that any impact is on very distal autophagic flux. Finally, we also tested how treating Irgm1−/− BMM with the fatty acid synthase inhibitor, C75, affected peroxisome numbers (Fig. 5E). As mentioned earlier, we previously associated Irgm1-deficiency in macrophages with increased acyl-carnitine accumulation, an effect that drives inflammatory cytokine production and is mitigated by treatment with C75 (28). In the current studies, C75 had no effect on reduced peroxisome levels in Irgm1−/− BMM, suggesting that this metabolic change in Irgm1-deficient macrophages occurs downstream of the change in peroxisome homeostasis.
Figure 4.
Irgm1 localizes to a subset of peroxisomes in macrophages. WT BMM activated with IFN-γ for 24 h were fixed and stained with anti-Irgm1 (green) or anti-PMP70 (red) antibodies. Shown are representative confocal images; arrows mark select peroxisomes that showed Irgm1 staining. Irgm1 localization to peroxisomes was quantified in 10 cells using both manual counting and ImageJ/JACoP analysis, as indicated. Error bars indicate SD. The lower control panels show representative images of the cells stained with secondary antibodies only. The nuclei in the overlay images are stained with DAPI.
Figure 5.
Peroxisome numbers are decreased with Irgm1 deficiency. BMM from the indicated mice were cultured on coverslips and either activated with IFN-γ for 24 h and used for immunocytochemistry with anti-PMP70 antibodies (A–B, D–E), or transfected with a fluorescent peroxisome plasmid, activated with IFN-γ, and used for live fluorescent imaging (C). Shown are: A, representative images of WT and Irgm1−/− (Irgm1 KO) BMM. B, Peroxisome counts from WT and Irgm1−/− BMM; data from 4 separate experiments using cells from distinct mice were combined. C, peroxisome counts from WT and Irgm1−/− BMM. D, peroxisome counts from WT, Irgm1−/−, and IFNAR/Irgm1−/− BMM. E, peroxisome counts from WT and Irgm1−/− BMM; the cells were treated with bafilomycin A1 (0.1 μM), Torin-1 (1 μM), or C75 (10μΜ), simultaneously with IFN-γ as indicated. Data points represent individual cells. Error bars indicate SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
While these results establish a role for Irgm1 in peroxisomal homeostasis, Irgm1 may or may not affect the biochemical or cellular functioning of individual peroxisomes. We explored this by examining peroxisome-related gene sets and ontologies in the GSEA detailed above. Comparing WT to Irgm1−/− BMM, there was no statistical difference in gene expression in multiple gene sets related to peroxisome function including GOBP Protein Import into Peroxisome Matrix (FDR = 0.26), GOBP Protein Targeting to Peroxisome (FDR = 0.64), GOBP Peroxisome Organization (FDR = 0.99), KEGG_Peroxisome (FDR = 0.34), and Reactome Synthesis of Bile Acids and Bile Salts (FDR = 0.59). Thus, by these measures, Irgm1 does not seem to affect many basic peroxisomal functions.
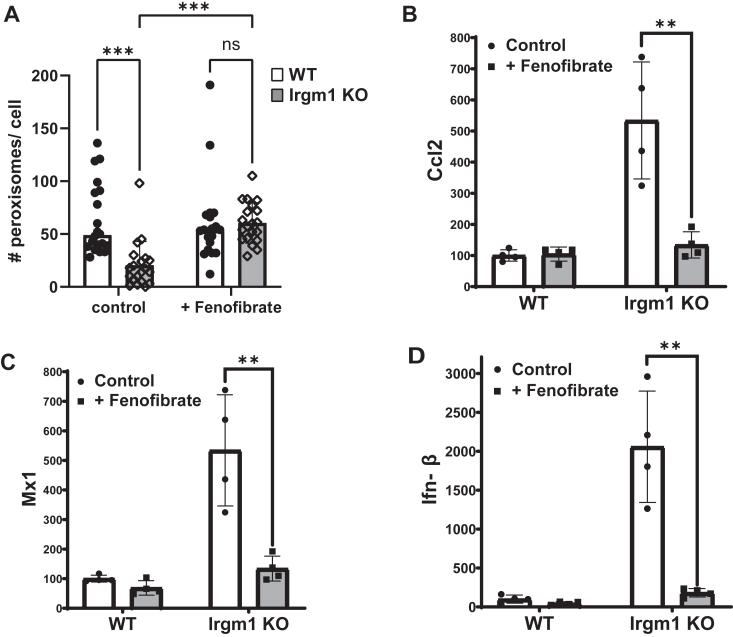
We reasoned that even if individual peroxisomes function normally in Irgm1−/− BMM, a decrease in peroxisome levels will decrease peroxisome function in the aggregate, potentially leading to increased inflammation. The scientific literature indicates that through their roles in fatty acid metabolism and ROS homeostasis, peroxisomes play an important role in inflammation, and a reduction of peroxisomes has been shown to be pro-inflammatory in other contexts (41, 42). To test this possibility, we treated cells with the PPARα agonist, fenofibrate (43) in an effort to increase peroxisome levels. Exposure of Irgm1−/− BMM to fenofibrate normalized peroxisome numbers, bringing them close to WT levels while having little impact on peroxisome numbers in WT BMM (Fig. 6A). When cytokine expression was assessed, the PPARα agonist dramatically lowered Ccl2, Mx1, and Ifnb mRNA levels in Irgm1−/− BMM close to the levels seen in WT BMM (Fig. 6, B–D). These results suggested that the reductions in peroxisome levels in Irgm1−/− BMM contribute to the increased type I IFN responses and subsequent stimulation of inflammatory cytokine production.
Figure 6.
PPARα agonist, fenofibrate, normalizes type I IFN signaling in Irgm1-deficient macrophages.A, WT and Irgm1−/− BMM plated on coverslips were activated with IFN-γ for 24 h in the presence or absence of fenofibrate (50 μM). The cells were then stained with anti-PMP70 antibodies and peroxisomes were enumerated. Data points represent individual cells. Shown is representative of two studies. B–D, WT and Irgm1−/− BMM isolated from 3 to 4 distinct mice per genotype were activated with IFN-ɣ for 24 h in the presence or absence of fenofibrate. Cells were collected and used to quantify Ccl-2, Mx-1, and Ifnb mRNA using quantitative PCR. Shown is representative of two experiments. Error bars indicate SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
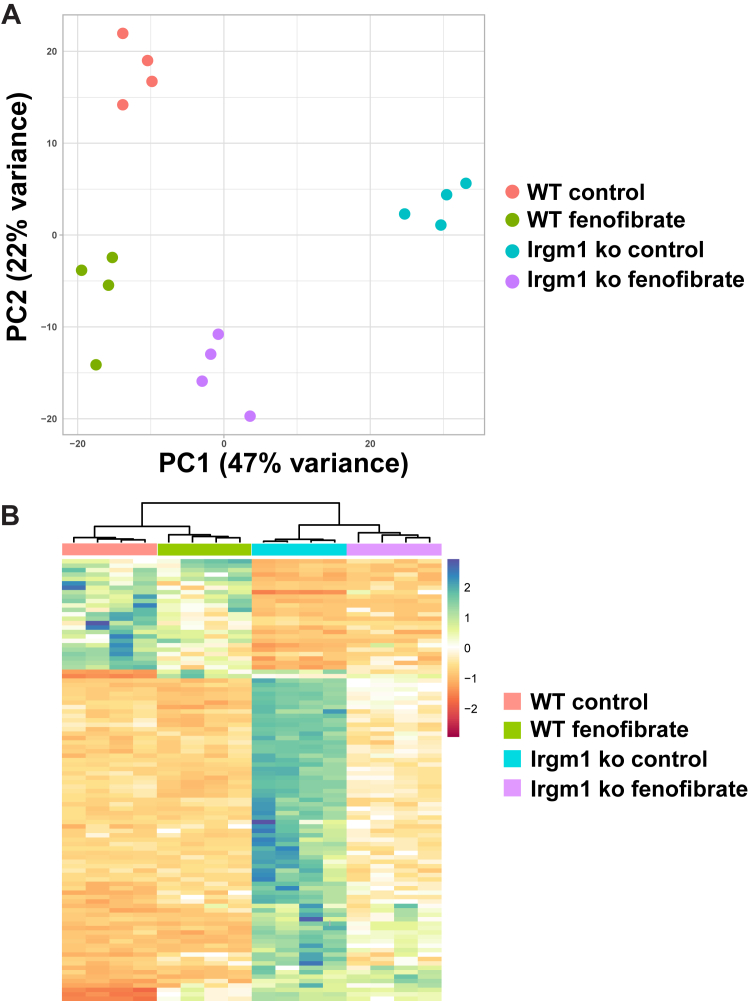
We examined the impact of fenofibrate more broadly by assessing gene expression through RNA-Seq and GSEA. Both the principal component analysis (PCA) (Fig. 7A) and the heat map of gene expression (Fig. 7B) illustrated that in WT and Irgm1−/− BMM following fenofibrate treatment, there was a substantial shift of gene expression in Irgm1−/− BMM so that it more closely aligned with expression in WT BMM. The RNA-Seq data validated the effects on type I IFN production and signaling, as the increases in Ifnb, Mx1, and Mx2 mRNAs in Irgm1−/− BMM compared to WT BMM were substantially muted, changing from 36.0-, 13.6-, and 11.2-fold increases in control cells (LRT, FDR < 0.0001) to 4.08-, 2.04-, and 1.88-fold increases. Further, most of the gene pathway/ontology enrichment seen in control Irgm1−/− BMM compared to WT BMM (Table 1) was obviated following fenofibrate treatment including GOBP cytokine production, GOBP Inflammatory Response, Biocarta PPARA Pathway, GOBP Protein Refolding, GOMF Heat Shock Protein Binding, GOBP Cell Cycle Process, GOBP Organelle Fission, and Reactome Golgi to ER Retrograde Transport (all displaying non-significant differences, t test, FDR > 0.05). The only exceptions that maintained enrichment following fenofibrate treatment were WP Aerobic Glycolysis (NES 2.09, t test, FDR < 0.0001) and Reactome Hsf1 Activation (NES 1.80, , t test, FDR < 0.0001).
Figure 7.
Fenofibrate normalizes gene expression patterns in Irgm1-deficient macrophages. Cultures of WT and Irgm1−/− BMM isolated from distinct mice were activated with IFN-γ for 24 h in the absence (Control) or presence of fenofibrate (50 μM). RNA was isolated from the cells and used for deep sequencing. Shown is (A) principal component analysis and (B) heat map of gene expression. Other details are described in the text.
Discussion
Changes in the expression of IRGM proteins have been associated with various inflammatory diseases in humans and mouse models of disease (44). While the underlying mechanisms are unclear, our previous work implicated that metabolic changes in Irgm1−/− cells, specifically increases in glycolysis and decreases in fatty acid turnover, are pivotal events that drive increased inflammatory cytokine production (28). Because work from several labs has identified changes in mitochondrial homeostasis with Irgm1 deficiency (6, 7, 28, 29, 31, 45), a working hypothesis has been that lack of removal of dysfunctional mitochondria with Irgm1 deficiency leads to decreased fatty acid catabolism and decreased oxidative phosphorylation, the latter prompting increases in glycolysis. Our current work adds key details to this mechanism. Leveraging the recent finding of increased type I IFN signaling in Irgm1-deficient cells, we found that the production of an array of inflammatory cytokines and chemokines in Irgm1-deficient macrophages is a consequence of increased type I IFN signaling, rather than occurring independently. Type I IFN signaling, in fact, appears to be the major driver of transcriptional changes for the majority of pathways that are altered in Irgm1−/− macrophages. Nevertheless, there are several gene pathways/ontologies that are enriched in both Irgm1−/− BMM and IFNAR/Irgm1−/− BMM that represent upstream mechanisms that likely drive type I IFN production – these include increased glycolysis, increased unfolded protein responses, impaired organelle fission, and altered peroxisome homeostasis.
Peroxisomes and mitochondria are distinct organelles that share some important cellular functions, including fatty acid metabolism and ROS scavenging (46). They are both dynamic organelles subject to fission and fusion events mediated by dynamin-like proteins, possibly including Irgm1. When the organelles become dysfunctional, they are both removed by autophagic processes. In the current work, we have identified that peroxisome homeostasis is dramatically altered in activated macrophages when Irgm1 is absent. Our data suggest that the decrease in peroxisome numbers and ostensibly reduced levels of peroxisome functioning in the cells contribute to type I IFN production and the subsequent inflammatory cytokine production.
The means through which Irgm1 affects peroxisomes was not identified in the current studies. Peroxisome homeostasis is regulated by a series of PEX proteins that control various aspects of organelle formation. However, Irgm1 does not seem to regulate their expression as RNA-Seq data for multiple PEX proteins showed either no significant difference in expression or very small <1.3-fold differences in Irgm1−/− BMM compared to WT BMM, including PEX1, PEX2, PEX3, PEX5, PEX6, PEX7, PEX10, PEX11a, PEX12, PEX13, PEX14, and PEX19. Only PEX 16 showed a 2.1-fold higher expression in Irgm1−/− BMM compared to WT BMM (FDR < 0.0001). Therefore, it seems unlikely that Irgm1 influences homeostasis by PEX-regulated mechanisms of organelle formation. Published reports have indicated that Irgm1 promotes autophagic turnover and fission of mitochondria, likely through its presumed dynamin-like function. Thus, it is quite possible that Irgm1 may affect peroxisomes in a similar manner, particularly given our evidence of direct association (Fig. 4). We note that at the level of fluorescence microscopy, we did not see a shift in the peroxisomal fission/fusion balance—the organelles appeared punctate in both WT and Irgm1−/− BMM—however, it is possible that the kinetics of our experiments were not optimized to capture subtle differences. Irgm1 may alternatively influence peroxisomes through its autophagic function, yet the levels of peroxisomes in Irgm1−/− BMM appeared relatively insensitive to the autophagic inducer and inhibitor that we tested. It is interesting in this context that mice lacking the autophagy protein, Atg16L1, share enhanced colitis and perturbed Paneth cell functioning with Irgm1−/− mice, as well as also displaying increased PPAR signaling (47).
While mitochondrial dysfunction likely plays a significant role in inflammation triggered by Irgm1 deficiency as we and others have posited, a few observations suggest that it is not the sole driver. First, as discussed, peroxisomes appear to play a significant role, and their dysfunction lies upstream of increased production of type I IFN and other inflammatory cytokines. Second, as we also demonstrated in the current studies, decreased mitochondrial mass and membrane potential in Irgm1−/− BMM are dependent on Type I IFN production, suggesting these phenotypes are caused by an increase in type I IFN signaling. In addition, we previously found changes in the mitochondrial fission/fusion balance in Irgm1−/− BMM to be dependent on increased glycolysis (28). These findings place at least a few facets of mitochondrial dysfunction in Irgm1−/− macrophages downstream of other functional changes in those cells. There are certainly complex signaling and feedback responses that occur among peroxisomes, mitochondria, and metabolic pathways in inflammatory contexts. Our observations emphasize the need for further work to determine how Irgm1 influences peroxisomal and mitochondrial function, and the relevance to IRGM protein-regulated inflammation.
Experimental procedures
Mice and cell culture
Irgm1−/− (48) and IFNAR/Irgm1−/− (29, 30) have been described previously and were backcrossed to C57BL/6 mice (wild-type). All mice were housed and maintained under procedures approved by the Institutional Animal Care and Use Committees at the Duke University and Durham VA Medical Centers.
Primary murine BMM were isolated from the tibia and femurs of 2- to 4-month-old male and female mice and cultured according to standard procedures described previously (17). The bone marrow was flushed from the bones using a 27G needle fitted to a syringe filled with DMEM (Life Technologies); the marrow was dispersed by drawing through the needle three to four times; and red cells were lysed with ACK lysing buffer (Life Technologies). Adherent cells were cultured for 6 days in BMM medium [DMEM supplemented with 10% (v/v) FBS (Hyclone) and 20% (v/v) L929 cell-conditioned medium]. The cells were cultured on Petri dishes that were not cell culture-treated, resulting in cultures that were loosely adherent and easily removed from the plates with cell dissociation buffer (13150–016, Gibco/Thermo Fisher). 24 h prior to all experiments, the cells were placed in a medium lacking L929-conditioned media [DMEM (11995, Gibco/ThermoFisher), supplemented with 10% (v/v) FBS and 100 units/ml penicillin/100 μg/ml streptomycin (15140, GIBCO/ThermoFisher)] on coverslips or cell culture plates. All primary cells and cell lines were grown and maintained in a humidified atmosphere of 5% CO2 at 37 °C. Where appropriate, the following were added to the growth media as indicated in the text: 100 U/ml interferon (IFN)-γ (IF005, EMD Millipore), IFNAR neutralizing antibodies, 50 μM fenofibrate (F6020, Sigma), 1 μM Torin-1 (inh-tor1, InvivoGen), 0.1 μM bafilomycin A1 (SC201550, Santa Cruz Biotechnology), and/or 10 μΜ C75 (C5490, Sigma).
Quantitative RT-PCR
Total RNA was extracted from cultured BMM using an RNeasy Mini Kit (Qiagen) per the manufacturer’s instructions. Complementary DNA was generated using SuperScript II reverse transcriptase (Invitrogen) or an iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCR was performed for each sample in duplicate, using TaqMan Gene Expression Master Mix (Applied Biosystems) or SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). The following primer sets were used:
Ccl2 forward 5′-CTGTTCACAGTTGCCGGCTG-3′, reverse 5′- TGAATGAGTAGCAGCAGGTGAG-3′
Mx1 forward 5′-GAGCTGATTTCTTCATCCAGTCAC-3′, reverse 5′- GTGCCTGCACAGATTATTCACAGAA-3′
IfnB forward 5′-AGCTCCAAGAAAGGACGAACA-3′, reverse 5′-GCCCTGTAGGTGAGGTTGAT-3′
Actb forward 5′-CGGTTCCGATGCCCTGAGGCTCTT-3′ reverse 5′-CGTCACACTTCATGATGGAATTGA-3′ (49)
Relative gene expression was determined using the comparative threshold cycle (Ct) method (50). Briefly, fold change was calculated using the equation ΔΔCt = [(Ct gene of interest − Ct actb)Irgm1 KO − mean(Ct gene of interest − Ct actb)Control WT]. Actb was used as an internal control. The control WT sample(s) was set at a baseline of “1”.
Metabolite measurements
BMM were cultured in triplicate on 100 mm cell culture plates to near confluence. To measure acyl-carnitines, the culture medium was supplemented with 0.5 mM L-carnitine and 100 μΜ of a 1:1 oleate/palmitate stock complexed to 0.14% (w/v) BSA for 24 h. The cells were lifted with rubber scrapers and collected in 0.3 ml of 0.6% (v/v) formic acid. An equal volume of acetonitrile was then added to the lysates, which were then stored at −80 °C. Targeted mass spectrometry-based metabolic profiling was performed at the Duke Sarah W. Stedman Nutrition and Metabolism Center Mass Spectrometry Laboratory as previously described (51, 52, 53). Acyl-carnitines were measured by direct-injection electrospray tandem mass spectrometry (MS/MS) using a Xevo TQDMS system (Waters) equipped with an Acquity UPLC system, and a data system running MassLynx 4.1 software (Waters) (51, 52). Metabolite data were normalized to the total protein content in each sample as determined by a Pierce BCA Protein Assay Kit (23225, Pierce Thermo Fisher Scientific).- Lactate was measured as we previously described (54). In brief, conditioned media from BMM cultures were diluted 200 times in lactate assay buffer (100 mM Tris HCl, 20 mM KCl pH 8.5), and 50 μl of the diluted sample was added to a 96 well plate. To each sample or lactate standard (0–50 μg/ml), 50 μl of detection reagent were added [2.25 U/ml lactate dehydrogenase (Sigma-Aldrich), 2.7 U/ml Diaphorase (Innovative research, Novi, MI), 2 mM NAD (Sigma-Aldrich), and 28 μM Resazurin (Sigma-Aldrich)] in lactate assay buffer. After incubation for 30 min at 37 °C, fluorescence was measured at EX: 544/EM: 590 nm using a FLUOstar Omega plate reader (BMG Labtech).
Immunocytochemistry
Cells were fixed on coverslips with 4% paraformaldehyde (w/v) in PBS for 15 min and permeabilized with 0.2% (w/v) saponin in PBS for 10 min. The cells were then stained for 60 min with anti-Irgm1 mouse monoclonal antibody 1B2, (EMD Millipore, MABF2264) and anti-PMP70 rabbit polyclonal antibody (Abcam, AB3421), followed by Alexa Fluor conjugated secondary antibodies [Molecular Probes/Invitrogen, Chicken anti-rabbit IgG AF594 (A-21442) and chicken anti-mouse IgG (A21200)] for an additional 60 min. Cells were imaged either on a Leica SP8 DM6000CS confocal microscope with PMT and GaAsP HyD detectors, an HC PL APO 63x/1.4 Oil CS2 objective, and Leica LAS AF three software (Fig. 4), or Olympus IX70 inverted fluorescence microscope equipped with a Hamamatsu C8484-03G01 digital camera and ASI MS2000 XY Piezo Z stage (Fig. 5). Confocal images were analyzed using Image J (version 1.554f) with the plugin JACoP (version 2.1.4) and Mander's coefficient analysis.
RNA sequencing and gene set enrichment analysis
RNA sequencing was performed by the Duke University Sequencing and Genome Technologies Core Facility. RNA-seq data were processed using the fastp toolkit (55) to trim low-quality bases and sequencing adapters from the 3’ ends of reads, then mapped to GRCm39 (downloaded from Ensembl, version 106) (56) using the STAR RNA-seq alignment tool (57), and reads aligning to a single genomic location were summarized across genes. For genes having an overlap of at least 10 reads, gene counts were normalized, and differential expression was carried out using the DESeq2 (58) Bioconductor (59) package implemented for the R programming environment. Consistent with the recommendation of the DESeq authors, independent filtering (60) was utilized prior to calculating adjusted p-values and moderated log2 fold-changes were derived using the ashr package (61). Gene set enrichment analysis (62) was performed to identify gene ontology terms and pathways associated with altered gene expression for each of the comparisons performed.
Mitochondrial analyses
BMM were washed with complete RPMI media containing HEPES, β-mercacptoethanol (βME), 10% FBS, penicillin/streptomycin, and glutamine (Sigma-Aldrich), and stained in complete RPMI containing 400 nM Mitotracker Green (Thermo-Fisher), 100 nM of TMRE (Sigma-Aldrich), and 4uM of CellROX (Thermo-Fisher) for 30 min at 37 °C. BMM were then washed in 1x PBS and stained with Zombie Violet viability dye (BioLegend) at a 1:750 dilution in 1x PBS. Cells were analyzed by flow cytometry using an Attune NxT cytometer (Thermo-Fisher), quantifying Mitotracker Green, TMRE, and CellROX geometric mean fluorescent intensity in Zombie Violet negative cells (live cells).
Statistical analysis
Student’s t test, one-way ANOVA with Tukey’s Test for multiple comparisons, and two-way ANOVA were performed, as appropriate, using embedded software in Graphpad Prism. DESeq2’s Wald test with a Benjamini-Hochberg adjustment was performed for differential gene expression analysis; GSEA with a Benjamini-Hochberg adjustment was performed for gene set enrichment analysis.
Data availability
The RNA-Seq data are available from NCBI (GEO Accession: GSE276820). All other data supporting the findings of this study are available from the corresponding author upon request.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Yasheng Gao and Ben Carlson and the Duke Light Microscopy Core Facility for the use of the confocal microscope and helpful conversations. We thank the Duke University School of Medicine for the use of the Sequencing and Genomic Technologies Core Facility. We also thank the UNC Flow Cytometry Core Facility for granting access to their flow cytometers.
Author contributions
B. E F., L. R. F., M. M., B. A., A. S., A. B., Y. A., C. T. M., O. R. I., J. A. P., D. S. L., and P. R. investigation, B. E F., L. R. F., M. M., Y. A., C. T. M., J. A. P., and G. A. T. formal analysis. Y. A., C. T. M., O. R. I., D. S. L., N. J. M., M. B. F., and J. C. writing – review & editing; D. S. L. data curation; N. J. M., M. B. F., J. C., and G. A. T. conceptualization. G. A. T. supervision; G. A. T. writing – original draft.
Funding and additional information
Research reported in this publication was supported by the North Carolina Biotech Center Institutional Support Grant 2017-IDG-1025, by the National Institutes of Health grants 1UM2AI30836-01, AI145929, and AI148243, and by the Intramural Research Program of the National Institute of Environmental Health Sciences of the National Institutes of Health through Z01 ES102005. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Clare E. Bryant
References
- 1.Singh S.B., Davis A.S., Taylor G.A., Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 2.Chauhan S., Mandell M.A., Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol. Cell. 2015;58:507–521. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Intemann C.D., Thye T., Niemann S., Browne E.N., Amanua Chinbuah M., Enimil A., et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura T., Watanabe E., Sakamoto T., Takasu O., Ikeda T., Ikeda K., et al. Autophagy-related IRGM polymorphism is associated with mortality of patients with severe sepsis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petkova D.S., Viret C., Faure M. IRGM in autophagy and viral infections. Front. Immunol. 2012;3:426. doi: 10.3389/fimmu.2012.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh S.B., Ornatowski W., Vergne I., Naylor J., Delgado M., Roberts E., et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat. Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry S.C., Schmidt E.A., Fessler M.B., Taylor G.A. Palmitoylation of the immunity related GTPase, Irgm1: impact on membrane localization and ability to promote mitochondrial fission. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiwari S., Choi H.P., Matsuzawa T., Pypaert M., MacMicking J.D. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat. Immunol. 2009;10:907–917. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randow F., MacMicking J.D., James L.C. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunn J.P., Koenen-Waisman S., Papic N., Schroeder N., Pawlowski N., Lange R., et al. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 2008;27:2495–2509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y.O., Khaminets A., Hunn J.P., Howard J.C. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim B.H., Shenoy A.R., Kumar P., Bradfield C.J., MacMicking J.D. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory A., Taylor R.S., Rulong Shen, Hudson Eric, Pei Veronica, Pavlakis George N., et al. The inducibly expressed GTPase localizes to the endoplasmic reticulum, independently of GTP binding. J. Biol. Chem. 1997;272:10639–10645. doi: 10.1074/jbc.272.16.10639. [DOI] [PubMed] [Google Scholar]
- 14.Praefcke G.J., McMahon H.T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 15.Butcher B.A., Greene R.I., Henry S.C., Annecharico K.L., Weinberg J.B., Denkers E.Y., et al. p47 GTPases regulate Toxoplasma gondii survival in activated macrophages. Infect. Immun. 2005;73:3278–3286. doi: 10.1128/IAI.73.6.3278-3286.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens S., Sabel K., Lange R., Uthaiah R., Wolf E., Howard J.C. Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: retargeting to plasma membrane induced by phagocytosis. J. Immunol. 2004;173:2594–2606. doi: 10.4049/jimmunol.173.4.2594. [DOI] [PubMed] [Google Scholar]
- 17.Henry S.C., Daniell X., Indaram M., Whitesides J.F., Sempowski G.D., Howell D., et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) J. Immunol. 2007;179:6963–6972. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 18.Henry S.C., Traver M., Daniell X., Indaram M., Oliver T., Taylor G.A. Regulation of macrophage motility by Irgm1. J. Leukoc. Biol. 2010;87:333–343. doi: 10.1189/jlb.0509299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellcome Trust Case Control, C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkes M., Barrett J.C., Prescott N.J., Tremelling M., Anderson C.A., Fisher S.A., et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts R.L., Hollis-Moffatt J.E., Gearry R.B., Kennedy M.A., Barclay M.L., Merriman T.R. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561–565. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 22.Latiano A., Palmieri O., Cucchiara S., Castro M., D'Inca R., Guariso G., et al. Polymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn's disease. Am. J. Gastroenterol. 2009;104:110–116. doi: 10.1038/ajg.2008.3. [DOI] [PubMed] [Google Scholar]
- 23.Sehgal R., Berg A., Polinski J.I., Hegarty J.P., Lin Z., McKenna K.J., et al. Mutations in IRGM are associated with more frequent need for surgery in patients with ileocolonic Crohn's disease. Dis. colon rectum. 2012;55:115–121. doi: 10.1097/DCR.0b013e31823ccea8. [DOI] [PubMed] [Google Scholar]
- 24.King K.Y., Lew J.D., Ha N.P., Lin J.S., Ma X., Graviss E.A., et al. Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y.C., Chang P.F., Lin H.F., Liu K., Chang M.H., Ni Y.H. Variants in the autophagy-related gene IRGM confer susceptibility to non-alcoholic fatty liver disease by modulating lipophagy. J. Hepatol. 2016;65:1209–1216. doi: 10.1016/j.jhep.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 26.Ajayi T.A., Innes C.L., Grimm S.A., Rai P., Finethy R., Coers J., et al. Crohn's disease IRGM risk alleles are associated with altered gene expression in human tissues. Am. J. Physiol. Gastrointest. Liver Physiol. 2019;316:G95–G105. doi: 10.1152/ajpgi.00196.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finethy R., Dockterman J., Kutsch M., Orench-Rivera N., Wallace G.D., Piro A.S., et al. Dynamin-related Irgm proteins modulate LPS-induced caspase-11 activation and septic shock. EMBO Rep. 2020;21 doi: 10.15252/embr.202050830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt E.A., Fee B.E., Henry S.C., Nichols A.G., Shinohara M.L., Rathmell J.C., et al. Metabolic alterations contribute to enhanced inflammatory cytokine production in Irgm1-deficient macrophages. J. Biol. Chem. 2017;292:4651–4662. doi: 10.1074/jbc.M116.770735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai P., Janardhan K.S., Meacham J., Madenspacher J.H., Lin W.C., Karmaus P.W.F., et al. IRGM1 links mitochondrial quality control to autoimmunity. Nat. Immunol. 2021;22:312–321. doi: 10.1038/s41590-020-00859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rai P., Sharpe M., Ganta C.K., Baker P.J., Mayer-Barber K.D., Fee B.E., et al. IRGM1 supports host defense against intracellular bacteria through suppression of type I interferon in mice. J. Clin. Invest. 2023;133 doi: 10.1172/JCI171982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jena K.K., Mehto S., Nath P., Chauhan N.R., Sahu R., Dhar K., et al. Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS signaling to control interferon response. EMBO Rep. 2020;21 doi: 10.15252/embr.202050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheehan K.C., Lai K.S., Dunn G.P., Bruce A.T., Diamond M.S., Heutel J.D., et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 34.Traver M.K., Henry S.C., Cantillana V., Oliver T., Hunn J.P., Howard J.C., et al. Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J. Biol. Chem. 2011;286:30471–30480. doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papic N., Hunn J.P., Pawlowski N., Zerrahn J., Howard J.C. Inactive and active states of the interferon-inducible resistance GTPase, Irga6, in vivo. J. Biol. Chem. 2008;283:32143–32151. doi: 10.1074/jbc.M804846200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haldar A.K., Saka H.A., Piro A.S., Dunn J.D., Henry S.C., Taylor G.A., et al. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahalingam S.S., Shukla N., Farre J.C., Zientara-Rytter K., Subramani S. Balancing the opposing principles that govern peroxisome homeostasis. Trends Biochem. Sci. 2021;46:200–212. doi: 10.1016/j.tibs.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Springer H.M., Schramm M., Taylor G.A., Howard J.C. Irgm1 (LRG-47), a regulator of cell-autonomous immunity, does not localize to mycobacterial or listerial phagosomes in IFN-gamma-Induced mouse cells. J. Immunol. 2013;191:1765–1774. doi: 10.4049/jimmunol.1300641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maric-Biresev J., Hunn J.P., Krut O., Helms J.B., Martens S., Howard J.C. Loss of the interferon-gamma-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC Biol. 2016;14:33. doi: 10.1186/s12915-016-0255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng J., Chen X., Liu Q., Zhong G., Zhuang M. Ubiquitin ligase MARCH5 localizes to peroxisomes to regulate pexophagy. J. Cell Biol. 2022;221 doi: 10.1083/jcb.202103156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijayan V., Srinu T., Karnati S., Garikapati V., Linke M., Kamalyan L., et al. A new immunomodulatory role for peroxisomes in macrophages activated by the TLR4 ligand lipopolysaccharide. J. Immunol. 2017;198:2414–2425. doi: 10.4049/jimmunol.1601596. [DOI] [PubMed] [Google Scholar]
- 42.Di Cara F., Andreoletti P., Trompier D., Vejux A., Bulow M.H., Sellin J., et al. Peroxisomes in immune response and inflammation. Int. J. Mol. Sci. 2019;20:3877. doi: 10.3390/ijms20163877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lalloyer F., Staels B. Fibrates, glitazones, and peroxisome proliferator-activated receptors. Arteriosclerosis, Thromb. Vasc. Biol. 2010;30:894–899. doi: 10.1161/ATVBAHA.108.179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilla-Moffett D., Barber M.F., Taylor G.A., Coers J. Interferon-Inducible GTPases in host resistance, inflammation and disease. J. Mol. Biol. Mol. Imag. 2016;428:3495–3513. doi: 10.1016/j.jmb.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B., Gulati A.S., Cantillana V., Henry S.C., Schmidt E.A., Daniell X., et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G573–G584. doi: 10.1152/ajpgi.00071.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demarquoy J., Le Borgne F. Crosstalk between mitochondria and peroxisomes. World J. Biol. Chem. 2015;6:301–309. doi: 10.4331/wjbc.v6.i4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cadwell K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collazo C.M., Yap G.S., Sempowski G.D., Lusby K.C., Tessarollo L., Vande Woude G.F., et al. Inactivation of Lrg-47 and Irg-47 reveals a family of interferon γ–inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 2001;194:181–188. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang T.C., Zeitels L.R., Hwang H.W., Chivukula R.R., Wentzel E.A., Dews M., et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 51.An J., Muoio D.M., Shiota M., Fujimoto Y., Cline G.W., Shulman G.I., et al. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat. Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- 52.Wu J.Y., Kao H.J., Li S.C., Stevens R., Hillman S., Millington D., et al. ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J. Clin. Invest. 2004;113:434–440. doi: 10.1172/JCI19574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen M.V., Joseph J.W., Ilkayeva O., Burgess S., Lu D., Ronnebaum S.M., et al. Compensatory responses to pyruvate carboxylase suppression in islet beta-cells. Preservation of glucose-stimulated insulin secretion. J. Biol. Chem. 2006;281:22342–22351. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
- 54.Alwarawrah Y., Danzaki K., Nichols A.G., Fee B.E., Bock C., Kucera G., et al. Irgm1 regulates metabolism and function in T cell subsets. Sci. Rep. 2022;12:850. doi: 10.1038/s41598-021-04442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kersey P.J., Staines D.M., Lawson D., Kulesha E., Derwent P., Humphrey J.C., et al. Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–D97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ignatiadis N., Klaus B., Zaugg J.B., Huber W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods. 2016;13:577–580. doi: 10.1038/nmeth.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens M. False discovery rates: a new deal. Biostatistics. 2017;18:275–294. doi: 10.1093/biostatistics/kxw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-Seq data are available from NCBI (GEO Accession: GSE276820). All other data supporting the findings of this study are available from the corresponding author upon request.