Abstract

Antimicrobial resistance (AMR) poses a serious threat to global health. The rapid emergence of resistance contrasts with the slow pace of antimicrobial development, emphasizing the urgent need for innovative drug discovery approaches. This study addresses a critical bottleneck in early drug development by introducing integral solvent-induced protein precipitation (iSPP) to rapidly assess the target–engagement of lead compounds in extracts of pathogenic microorganisms under close-to-physiological conditions. iSPP measures the change in protein stability against solvent-induced precipitation in the presence of ligands. The iSPP method for bacteria builds upon established SPP procedures and features optimized denaturation gradients and minimized sample input amounts. The effectiveness of the iSPP workflow was initially demonstrated through a multidrug target–engagement study. Using quantitative mass spectrometry (LC-MS/MS), we successfully identified known drug targets of seven different antibiotics in cell extracts of four AMR-related pathogens: the three Gram-negative bacteria Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa and the Gram-positive bacterium Staphylococcus aureus. The iSPP method was ultimately applied to demonstrate target–engagement of compounds derived from target-based drug discovery. We employed five small molecules targeting three enzymes in the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway—a promising focus for anti-infective drug development. The study showcases iSPP adaptability and efficiency in identifying anti-infective drug targets, advancing early-stage drug discovery against AMR.
Keywords: mass spectrometry, proteomics, solvent-induced precipitation, antibiotics, MEP pathway, target identification
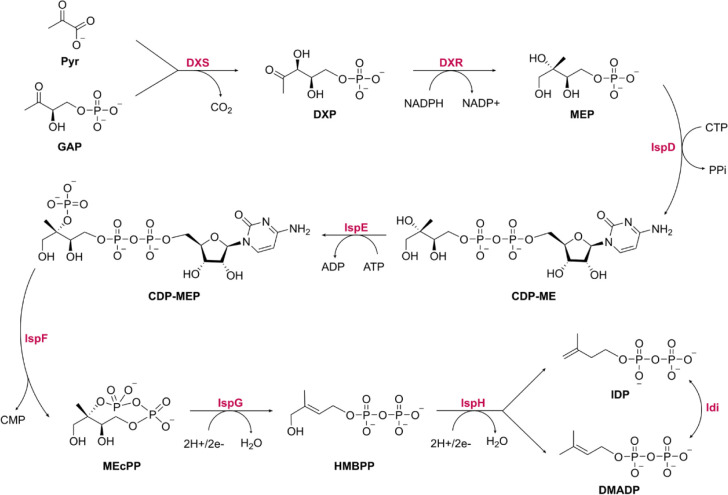
Antibiotic therapy for bacterial infections is among the most significant medical advancements in human history. Although being a cornerstone of contemporary medicine, antibiotic efficacy is increasingly in jeopardy due to the emergence of antimicrobial resistance (AMR). The World Health Organization (WHO) asserts that AMR is a rapidly expanding global issue and it represents one of the primary healthcare concerns.1,2 According to a 2022 study,3 the six primary pathogens responsible for deaths linked to AMR are Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. In 2019, they accounted for 3.57 million deaths globally associated with AMR. The main issue is that resistance emerges rapidly, while the development of antimicrobials requires a substantial amount of time,2 primarily due to insufficient validation of discoveries. One way to tackle AMR is through the identification of new protein target candidates for anti-infective drug discovery. Enzymes involved in the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway represent promising and underexploited targets.4 The MEP pathway is absent in humans but essential in green algae and numerous pathogenic bacteria and apicomplexan protozoa, including important human pathogens. Composed of seven enzymes, it leads to the production of isopentenyl diphosphate (IDP) and its isomer, dimethylallyl diphosphate (DMADP), which are the five-carbon (C5) building units essential for the biosynthesis of all isoprenoids (Scheme 1).
Scheme 1. MEP Pathway and Its Associated Enzyme Idi.
Pyr, pyruvate; GAP, glyceraldehyde 3-phosphate; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXP, 1-deoxy-d-xylulose 5-phosphate; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; MEP, 2-C-methylerythritol 4-phosphate; IspD, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; CTP, cytidine triphosphate; PPi, inorganic diphosphate; CDP-ME, 4-diphosphocytidyl-2-C-methylerythritol; IspE, 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase; CDP-MEP, 4-diphosphocytidyl-2-C-methyl-d-erythritol 2-phosphate; IspF, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; CMP, cytidine monophosphate; MEcPP, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate; IspG, 4-hydroxy-3-methyl-but-2-en-1-yl diphosphate synthase; HMBPP, (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate; IspH, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; IDP, isopentenyl diphosphate; DMADP, dimethylallyl diphosphate; Idi, Isopentenyl-diphosphate delta-isomerase
In addition to the urgent need for new target candidates, a key obstacle throughout the early drug discovery stages to be addressed is the lack of techniques for determining the mode of action (MoA) of antibiotics.5 Proteomics and chemical proteomics approaches are emerging as important tools for overcoming this challenge. By quantitatively measuring protein activity and function in response to, e.g., compound treatment, they provide critical insights into target deconvolution, target–engagement, selectivity, and MoA studies, among others. Ultimately, they can provide a comprehensive picture of protein–ligand interactions and subsequent cellular events.6,7
Numerous mass spectrometry (MS)-based chemical proteomics methods have been developed to screen the proteome for evidence of protein–ligand interactions. One of the most recent additions to that toolbox is the solvent-induced protein precipitation (SPP), which represents a quantitative MS-based proteomics (LC-MS/MS) assay for evaluating proteome-wide target–engagement.8−10 SPP is based on the detection of ligand-induced changes in protein stability upon incubation of a cell lysate with a compound of interest and exposure to increasing concentrations of organic solvents. The denaturation of proteins, and consequently their precipitation, induced by organic solvents is mainly caused by a reduction of the dielectric constant of the solution and destruction of the hydration shell of proteins.11 The compound–protein complex has a lower energy state than the unbound protein and therefore requires more energy to be unfolded, resulting in an increased tolerance to solvent-induced precipitation. The approach, originally demonstrated by Zhang and coworkers8 in human cell extracts, is modification-free, thereby omitting the necessity for any label on the tested compound or target proteins. Akin to other label-free proteome-wide stability assays, such as Thermal Proteome Profiling (TPP),12,13 SPP can be conducted in multiple formats and with different readouts. Most of these formats rely on resolving denaturation curves, which are typically generated by measuring the response of the system at each data point—each representing a single sample across the gradient of the denaturant that contributes to the overall curve, capturing how the system responds at that particular denaturant concentration or condition. Therefore, these techniques are challenging because they typically consume high amounts of sample material, have considerable instrument measurement time, and require intricate data analysis for fitting denaturation curves. Fortunately, these assays can be simplified and streamlined by determining the integral of the denaturation curve. This can be achieved by pooling the soluble fractions of the samples exposed to the selected denaturant gradient after denaturation and precipitation. This compressed approach was initially introduced by Gaetani et al.14 in a TPP study, termed Proteome Integral Solubility Alteration (PISA), and more recently also applied to SPP by Van Vranken et al.9 These studies reported that the approach can result in a compression of the observable effect size, thereby presenting a challenge in the detection of stabilized proteins. To prevent this compression, it is necessary to use an appropriate gradient tailored to the region of the most substantial solubility changes of the known target proteins, as it substantially affects the observed stabilization.9,14
To that end, we developed integral SPP (iSPP) as a procedure that (i) requires only 20 μg of total protein experimental input material per data point, (ii) utilizes a target-specific, empirically selected solvent concentration range for denaturation to maximize the observable effect size of the area-under-the-denaturation-curve readout, and (iii) employs data-independent acquisition (DIA) quantitative MS as sensitive and versatile alternative to antibody-based readouts, especially useful given that the antibodies for bacterial target proteins are frequently not available.
Initially, we explored the iSPP assay to demonstrate target–engagement of seven well-known antibiotics. We established the approach in cell extracts of four key pathogens for deaths associated with AMR: the three Gram-negative bacteria Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa and the Gram-positive bacterium Staphylococcus aureus. The designated protein targets of the model drugs were significantly and selectively stabilized and thus identified as the main interactors (hereafter also referred to as top hits). The entire workflow is highly reproducible with a median coefficient of variation (CV) of 6.1% across all quantified proteins in all performed measurements.
We then confirmed the target–engagement of compounds derived from a target-based drug discovery approach. To this end, we employed two classes targeting enzymes of the MEP pathway: (i) diphosphate derivatives as IspG–IspH inhibitors and (ii) recently discovered reverse β-aza fosmidomycin analogues as DXR inhibitors. The iSPP approach successfully identified the respective MEP enzymes as main targets, together with some potential additional interactors, demonstrating its adaptability and utility in early-stage drug discovery of novel potential anti-infective small molecules.
Results
iSPP Workflow
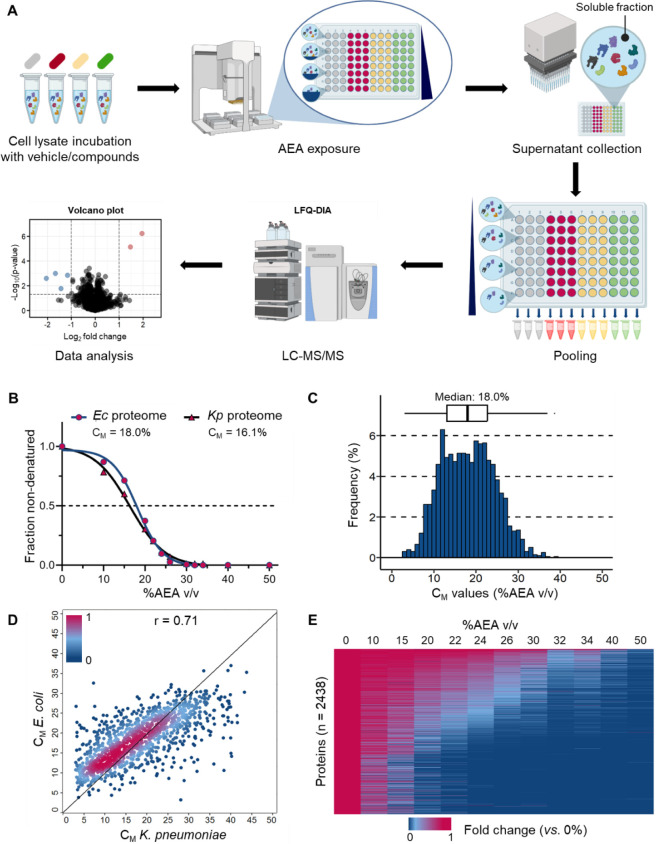
The workflow of the iSPP assay presented in this study is shown in Figure 1A. First, the bacterial proteins are extracted from cell pellets by means of a native lysis buffer containing the non-denaturing detergent IGEPAL CA-630. Aliquots of cell lysates are then incubated with either compound or vehicle, representing distinct conditions. Then, the aliquots are evenly distributed into a 96-well plate, followed by exposure to an increasing acetone/ethanol/acetic acid mixture (50:50:0.1 v/v, abbreviated as AEA), to initiate denaturation and precipitation of proteins. After precipitation, centrifugation is employed to separate soluble proteins from denatured and aggregated ones. Subsequently, equal supernatant volumes are collected. At this stage, the soluble fractions across the AEA gradient are combined (separately for each condition). The pooled samples are then prepared using a standard bottom-up proteomics workflow for quantitative MS readout.15 The integral values of the denaturation curves are determined based on the measured protein intensities in the pooled samples. These values are used to identify stabilized proteins by comparing compound and vehicle-treated groups, serving as an indicator for target–engagement. Stabilized proteins exhibit an increase in soluble protein abundance relative to the vehicle, leading to a positive log2 fold change (log2FC). Conversely, destabilized proteins show a reduction in soluble protein abundance relative to the vehicle, resulting in a negative log2FC. Biological replicates of the same condition allow for reproducibility testing and evaluation of the statistical significance of the observed results.
Figure 1.
Solvent profiling of the Escherichia coli and Klebsiella pneumoniae proteomes. (A) Schematic representation of the iSPP approach for target–engagement studies (both compound incubation and soluble fractions pooling were omitted during the solvent profiling experiments). Workflow graphic created with BioRender.com. (B) Denaturation curve of the E. coli and K. pneumoniae proteomes. For each data point, the median value of all quantified proteins is shown. (C) Distribution of CM values for E. coli proteins with high-quality denaturation curves (1984 proteins, R2 ≥ 0.8, and plateau ≤ 0.3). (D) Pearson correlation of CM values for 1249 predicted orthologs identified in our experiments between E. coli and K. pneumoniae, showing a strong positive correlation (r = 0.71). Proteins are colored based on the density of the points. (E) Heatmap representation of all E. coli proteins quantified in the performed experiment (2438 proteins). For each protein, its relative abundance (fold change) at the indicated %AEA (v/v) compared to the lowest concentration (0%) is shown. Proteins are sorted by intensity (in descending order).
In this study, we present a protocol that utilizes a low protein input amount, making it applicable to hard-to-culture pathogens. A typical experimental design—such as four distinct conditions, one vehicle control, and three distinct compounds, each in triplicate, distributed across eight solvent concentrations—requires about 2.0 mg of total protein extract, representing 20 μg per data point. Detailed results from the corresponding validation experiments, essential to the minimal input material of the protocol, are provided in Figure S1. We sought to further facilitate the protocol by omitting dimethyl or TMT peptide labeling, deviating from approaches employed in previous SPP publications.8−10 Consequently, we decided to employ DIA16,17 as an LC-MS/MS identification and quantification strategy, which enabled recording a bacterial proteome within a single LC-MS/MS measurement.
Solvent Profiling of E. coli and K. pneumoniae Proteomes
To establish a robust assay for assessing proteome-wide target–engagement in bacterial lysates, we initially focused on examining the effect of increasing concentrations of AEA on bacterial proteome stability. We evaluated the denaturation behavior of the E. coli K12 proteome, the most widely studied model organism in microbiology, as well as that of K. pneumoniae ATCC13883. We adapted the SPP protocols (Zhang et al.,8 2020; Van Vranken et al.,9 2021; Yu et al.,10 2023), which were applied to human cell lysates, by optimizing the AEA range to be compatible with bacterial lysate. This adjustment resulted in a considerably broader solvent gradient compared with that used for human lysates. The physicochemical properties of proteins vary, as proteomes can exhibit considerable diversity in denaturation patterns.18 Therefore, the adjustment of the AEA range is crucial as it ensures compatibility by establishing optimal conditions for different species.
As a result, E. coli and K. pneumoniae lysates were exposed to a wide range of AEA concentrations, from 0% to 50% (v/v) for the purpose of generating complete protein denaturation curves for the entire proteomes. After supernatant collection, soluble fractions were initially resolved and visualized using SDS-PAGE (Figure S2A,C), and in a subsequent experiment, they were analyzed by LC-MS/MS in biological triplicates. The majority of both proteomes appeared to effectively undergo denaturation and precipitation, reaching a bottom plateau at around 35% (v/v) AEA (Figure 1B). In total, we quantified 2438 proteins in E. coli and 2584 in K. pneumoniae by LC-MS/MS readout, achieving proteome coverage of 89% and 81%, respectively, relative to a global proteomic analysis. Subsequently, we used the measured protein intensities across different AEA concentrations (% v/v) to generate protein denaturation curves and calculate the melting concentrations (CM). CM represents the concentration of AEA (% v/v) at which a protein is equally distributed between the folded and unfolded states, based on the assumption that an unfolded protein precipitates. We successfully fitted high-quality denaturation curves (R2 ≥ 0.8 and plateau ≤ 0.3) for 1984 proteins in E. coli and 2033 in K. pneumoniae, corresponding to 81% and 79% of all quantified proteins, respectively, thus generating CM values for each of them. The calculated median CM of the E. coli proteome in the experiment was 18.0% (v/v) AEA, compared to 16.1% (v/v) AEA for K. pneumoniae. As illustrated in Figure 1B, the denaturation curves for these two bacterial proteomes exhibit similar profiles and a nearly complete overlap. Furthermore, we assessed the Pearson correlation of CM values for 1249 predicted orthologs19 identified in our experiments between E. coli and K. pneumoniae, finding a strong positive correlation (r = 0.71, Figure 1D). This result underscores the high degree of similarity in the solvent-induced precipitation profiles of proteins extracted in the same native lysis buffer across these bacterial species. The CM values of the E. coli and K. pneumoniae proteins are provided in Tables S1 and S2, respectively. Subsequently, we further analyzed the data from the model organism E. coli, examining the correlation between CM values and protein abundance. This analysis revealed a weak correlation (r = 0.20), demonstrating that the biophysical stability of a protein is (largely) independent of its abundance (Figure S3A). Furthermore, to evaluate the reproducibility of our findings, we compared the determined CM values across three independent replicates. The results revealed a strong correlation with r ≥ 0.87 in each comparison (Figure S3B–D). Additionally, we also computed the CV of CM values, obtaining a median CV of 5.8%, underscoring the high precision and reliability of the assay (Figure S3E).
We then categorized proteins into three groups based on the distribution of their CM values: (i) the upper quartile (most stable proteins, CM > 22.64% v/v), (ii) the interquartile range (12.99% v/v ≤ CM ≤ 22.64% v/v), and (iii) the lower quartile (least stable proteins, CM < 12.99% v/v). Subsequently, we conducted a functional annotation analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID)20,21 to investigate the potential overrepresentation of specific protein classes within each group. The analysis of group (i) revealed enrichment in hydrolases (n = 79), chaperones (n = 18), isomerases (n = 31), and rotamases (n = 9). This group is representative of E. coli proteins which showed high tolerance to organic solvent-induced precipitation and did not completely denature (up to 40% v/v AEA, Figure 1E). These proteins are mainly involved in the biological processes related to stress response (n = 30), glycolysis (n = 7), translation regulation (n = 7), and lipid metabolism (n = 13). Specifically, among this group, we find the superoxide-radical degradation proteins (SodA, SodB, and SodC), DNA binding proteins involved in bacterial chromosome organization and compaction under extreme environmental conditions (H-NS, HU-alpha, and HU-beta), chaperones (DnaK, Skp, SurA, GroEL, and GroES), Tat translocation system proteins (TatA and TatE), and outer membrane proteins (e.g., BamA, BamC, BamD, and BamE). Notably, as reported by Mateus et al.5 in a TPP study in E. coli, the chaperones, superoxide-radical degradation proteins, Tat translocation system, and outer membrane proteins were also highly resistant against heat denaturation, indicating extraordinarily stable protein structures. In contrast, group (ii) exhibited enrichment in oxidoreductases (n = 124), ligases (n = 39), ribonucleoproteins (n = 32), ribosomal proteins (n = 32), and RNA-binding proteins (n = 65). The proteins in this group are responsible for amino acid (n = 35) and protein biosynthesis (n = 25), tRNA processing (n = 26), the tricarboxylic acid cycle (n = 17), isoprene biosynthesis (n = 7), cell-wall biogenesis/degradation (n = 23), and carbohydrate metabolism (n = 33), among others. Group (iii) displayed an overrepresentation of kinases (n = 40), exonucleases (n = 9), and transferases (n = 131), such as methyltransferases and aminotransferases. This group is primarily involved in rRNA processing (n = 13), folate biosynthesis (n = 5), amino-acid biosynthesis (n = 35), cell cycle, and cell division (n = 19). Mateus et al.5 in their TPP study highlighted low thermal stability in multiple essential proteins, a finding consistent with our observations. Specifically, proteins exhibiting low stability to heat and organic solvents include topoisomerases (GyrB, TopB, and ParC), proteins involved in DNA replication (DnaA, DnaB, and DnaC), and those associated with cell shape (FtsA, FtsZ, FtsI, ZapA, ZapD, and ZapE). Conversely, multiple components of the small ribosomal subunit and the RNA polymerase sigma D factor RpoD, which exhibited low thermal stability in Mateus et al.’s study, clustered within the interquartile range in our solvent profiling.
Application of iSPP to Bacterial Cell Lysates
The iSPP assay was initially validated through a target–engagement study, employing several drugs in proof-of-principle experiments. The anti-infectives rifampicin, ampicillin, piperacillin, imipenem, cefazolin, nafcillin, and fosmidomycin were systematically investigated in pathogen cell lysates (Table 1).
Table 1. Summary of the Tested Drugs (Each at a Concentration of 10 μM) in Bacterial Lysates with the Corresponding Target Proteins/Protein Groups Showna.
| Model drug | AEA range (v/v) | Target protein(s) (CM %AEA) |
|---|---|---|
| Escherichia coli | ||
| Ampicillin | 14–28% | PBPs: PBP1a (13.0), PBP1b (16.9), PBP4 (23.3), PBP5 (15.6), PBP6 (21.0) |
| Rifampicin | 20–34% | RNAP: RpoA (19.9), RpoB (19.3), RpoC (19.3), RpoZ (22.4) |
| Fosmidomycin | 14–28% | DXR: (14.7) |
| Methotrexate | 20–34% | DHFR: (20.5) |
| Klebsiella pneumoniaeandPseudomonas aeruginosa | ||
| Piperacillin, Imipenem | 12–29.5 % | PBPs: PBP1a (6.9), PBP1b (16.5), PBP4 (19.7), PBP5 (14.7), PBP6 (16.6) |
| Fosmidomycin | 12–29.5 % | DXR: NDb |
| Methotrexate | 12–29.5 % | DHFR: (18.3) |
| Staphylococcus aureus | ||
| Cefazolin, Nafcillin | 14–28% | PBPs |
| Rifampicin | 20–34% | RNAP |
Selected AEA range windows for iSPP target–engagement studies in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus are displayed. Determined CM values for known E. coli and Klebsiella pneumoniae target proteins of model drugs are shown in brackets.
ND: Not Determined.
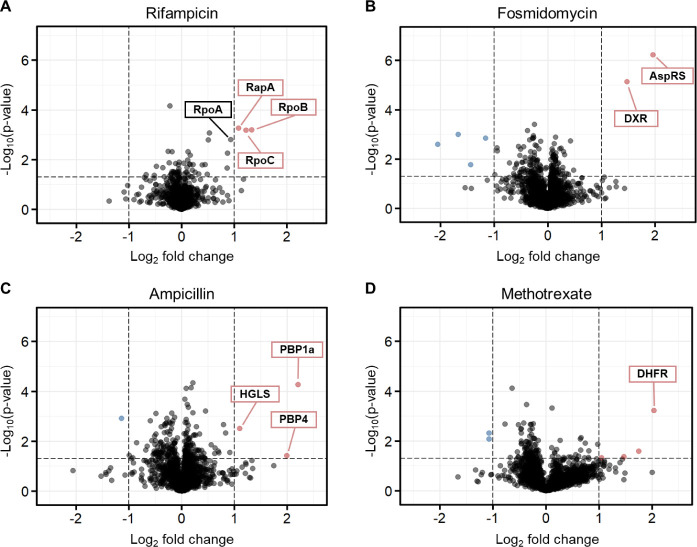
Rifampicin is an ansamycin antibiotic used to treat several types of bacterial infections caused by Gram-negative and Gram-positive bacteria. It interferes with transcription by binding to the β-subunit of the bacterial DNA-dependent RNA polymerase (RNAP), known as RpoB.22 Ampicillin, piperacillin, imipenem, cefazolin, and nafcillin belong to the class of β-lactam antibiotics, exhibiting a spectrum ranging from narrow to broad antibacterial activity.23−26 These antibiotics exert their therapeutic effects by irreversibly binding to membrane-bound penicillin-binding proteins (PBPs). Their inhibition disrupts the integrity of the cell-wall peptidoglycan, leading to bacterial cell lysis and subsequent elimination.27 Fosmidomycin is a natural product having antiparasitic and antibacterial activities.28 Its known target is DXR, which is the most studied enzyme in the MEP pathway. It is responsible for the catalysis of the second and rate-limiting step of the MEP pathway, involving an intramolecular isomerization and reduction to convert the substrate DXP to MEP (Scheme 1). To date, fosmidomycin is the only clinically investigated MEP inhibitor.28 However, its clinical use is limited by unfavorable pharmacokinetic properties, mainly insufficient membrane permeability and rapid clearance.29,30 Additionally, methotrexate (MTX), which is widely used to treat autoimmune and neoplastic diseases, was also included in the study.31 As a chemotherapeutic agent, MTX competitively inhibits human dihydrofolate reductase (DHFR), an enzyme that participates in tetrahydrofolate synthesis. We decided to investigate its target–engagement because of its impact on the growth and diversity of gut bacteria.32,33 Moreover, it has been reported that MTX displays potent activity with low inhibition constant (Ki) values for various bacterial DHFR enzymes.34
We initially evaluated the detectability and abundance of the corresponding known target proteins within the selected bacterial cell lysates through a comprehensive global proteomic characterization using LC-MS/MS. Our analysis led to the identification of 2752 proteins in E. coli (strain K12), 3196 in K. pneumoniae (strain ATCC13883), 4139 in P. aeruginosa (strain PA01), and 1849 in S. aureus (strain Newman). These numbers correspond to UniProt predicted full proteome coverage of 62%, 56%, 74%, and 64%, respectively, for each organism. Notably, this coverage ranks among the highest reported in the literature based on LC-MS/MS analysis, indicating close-to the entirety of the estimated expressed proteome for each bacterium.35−44 All expected target proteins of model drugs were identified and quantified with more than two unique peptides across the bacterial species proving their detectability by LC-MS/MS (Figure S4).
Subsequently, following the methodology applied to E. coli and K. pneumoniae, we assessed the denaturation behavior of P. aeruginosa and S. aureus proteomes upon organic solvent exposure. This involved exposing cell lysates to a wide AEA gradient (v/v, 0–50%) and subsequently resolving the soluble fractions using SDS-PAGE (Figure S2B–D). The resulting denaturation profiles guided the selection of the AEA range windows for subsequent iSPP target–engagement experiments. Additionally, we considered the denaturation behavior obtained by LC-MS/MS readout of the E. coli and K. pneumoniae proteins as a proxy for their homologs in the other pathogens (Figures S5–S7). Therefore, based on the determined denaturation curves and CM values for the well-known targets of the model drugs, we narrowed the AEA gradient to the region of the most substantial solubility changes, maximizing the effect size in stability for the known interactors. As a result, for some bacteria, we employed multiple AEA windows given the variations in denaturation behaviors observed for the target proteins of the employed drugs (Table 1). We utilized a more generalized gradient to confirm the engagement of proteins with CM values closely aligned with the median proteome denaturation behavior, thereby representing the optimal AEA concentration range for most proteins. Conversely, we adopted a targeted approach, selecting a higher range of AEA concentrations for confirming the engagement of proteins exhibiting a high tolerance to solvent-induced precipitation. The model drugs tested in the corresponding bacterial lysates and the selected AEA gradients are summarized in Table 1.
All bacterial cell lysates were incubated with 10 μM of the respective drug (n = 3) or vehicle (DMSO or ddH2O, n = 3), to ensure the binding equilibrium. The samples were then exposed to eight different concentrations of AEA, and the resulting soluble fractions were pooled together. The iSPP approach demonstrated high reproducibility of protein intensity measurements, as evidenced by low CV values (Figure S8).
Validation of iSPP in E. coli by Model Drugs
To demonstrate iSPP suitability for the detection of target–engagement in E. coli cell lysate, we employed the assay to confirm targets of four well-characterized drugs (Table 1). The iSPP experiments resulted in an average identification of 2163 proteins under all conditions.
The iSPP experiments confirmed stabilization for all target proteins (DXR, DHFR, and RpoB) and target protein groups (PBPs). Interestingly, for rifampicin, we observed (Figure 2A) the stabilization of other subunits within the RNAP complex in addition to the primary target RpoB, specifically the subunits alpha (RpoA, close-to the predefined threshold) and beta (RpoC). Moreover, the protein RapA, a transcription regulator forming a stable complex with the RNAP core enzyme,45,46 was also identified among the top hits.
Figure 2.
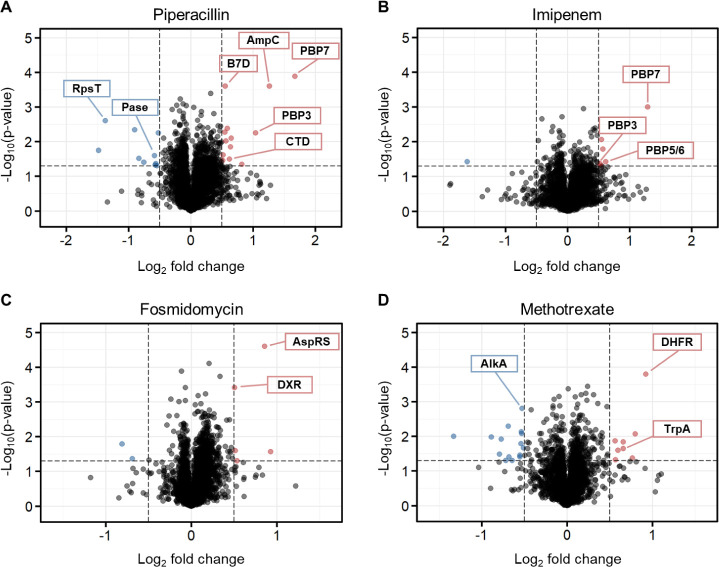
iSPP approach in Escherichia coli cell lysates to identify the protein targets of model drugs. E. coli lysates were incubated with vehicle, rifampicin (A), or methotrexate (D) and then exposed to the AEA gradient 20–34% v/v. Fosmidomycin (B), ampicillin (C), or the corresponding vehicle control-incubated cell lysates were exposed to the AEA gradient 14–28% v/v. All drugs were tested at a concentration of 10 μM. Data are presented as a volcano plot to highlight changes in protein abundance of each drug over vehicle sample vs statistical significance. We implemented criteria to ensure robust identification and selection of proteins exhibiting statistically significant changes in response to the experimental conditions by setting thresholds of a log2 fold change (log2FC) > |1.0| and a p-value < 0.05 (dashed lines). Red, stabilized proteins with log2FC > 1.0 and p-value < 0.05; blue, destabilized proteins with log2FC < −1.0 and p-value < 0.05; gray/black, proteins with −1.0 < log2FC < 1.0 and p-value < 0.05 and proteins with p-value > 0.05.
Our study also unveiled the Aspartate-tRNA Synthetase (AspRS) as a putative target of fosmidomycin. AspRS is an aminoacyl-tRNA synthetase, catalyzing the attachment of aspartate to tRNA(Asp).47 As depicted in Figure 2B, AspRS emerged as one of the two prominently stabilized proteins, alongside the known target DXR. Notably, AspRS was consistently identified as a top hit in both experimental repetitions (n = 2) with the selected gradient (14–28% v/v AEA, Figure S9A). To further assess the stabilization of AspRS by fosmidomycin, we employed an additional AEA range. Specifically, we shifted the gradient to higher solvent concentrations (20–34% v/v AEA). Within this range, only AspRS emerged as the main stabilized protein, while DXR was not identified in the quantitative proteomics data due to its lower CM value (Figure S9B). These findings suggest that EcAspRS could be an additional target of fosmidomycin.
Ampicillin led to a significant stabilization of PBP1a (MrcA) and PBP4 (DacB), the two top hits showing the largest magnitude of change, as well as 2-hydroxyglutarate synthase (HGLS), which is involved in d-lysine metabolism (Figure 2C).48 These results are in accordance with the study performed by Mateus et al.,5 who employed two-dimensional (2D)-TPP to identify targets of ampicillin in E. coli living cells and cell lysates. Their target–engagement study in cell lysates showed significant stabilization of only three proteins: MrcA, DacB, and AmpC, the latter being a β-lactamase.5
Validation of iSPP in K. pneumoniae and P. aeruginosa by Model Drugs
We applied the iSPP workflow to two additional Gram-negative bacteria, K. pneumoniae and P. aeruginosa, using the drugs listed in Table 1. We identified over all conditions an average of 2674 proteins in K. pneumoniae and 3409 in P. aeruginosa, corresponding to 84% and 82% proteome coverage compared to the global proteomic analysis, respectively. Again, the iSPP results clearly identified among the top hits the known target proteins, indicating that the selected drugs bound and stabilized their targets against solvent-induced denaturation (Figures 3,4).
Figure 3.
iSPP approach in Klebsiella pneumoniae cell lysates. K. pneumoniae cell lysates were incubated with vehicle, piperacillin (A), imipenem (B), fosmidomycin (C), or methotrexate (D) and then exposed to the AEA gradient 12–29.5% v/v (10 μM for all drugs). Data are presented as a volcano plot to highlight changes in abundance of each drug over vehicle sample vs statistical significance. The thresholds were set to a log2 fold change (log2FC) > |0.5| and a p-value < 0.05 (dashed lines). Red, stabilized proteins with log2FC > 0.5 and p-value < 0.05; blue, destabilized proteins with log2FC < −0.5 and p-value < 0.05; gray/black, proteins with −0.5 < log2FC < 0.5 and p-value < 0.05 and proteins with p-value > 0.05.
Figure 4.
iSPP approach in Pseudomonas aeruginosa cell lysates. P. aeruginosa cell lysates were incubated with vehicle, piperacillin (A), imipenem (B), fosmidomycin (C), or methotrexate (D) and then exposed to the AEA gradient 12–29.5% v/v (10 μM for all drugs). Data are presented as a volcano plot to highlight changes in abundance of each drug over vehicle vs statistical significance. The thresholds were set to a log2 fold change (log2FC) > |0.5| and a p-value < 0.05 (dashed lines). Red, stabilized proteins with log2FC > 0.5 and p-value < 0.05; blue, destabilized proteins with log2FC < −0.5 and p-value < 0.05; gray/black, proteins with −0.5 < log2FC < 0.5 and p-value < 0.05 and proteins with p-value > 0.05.
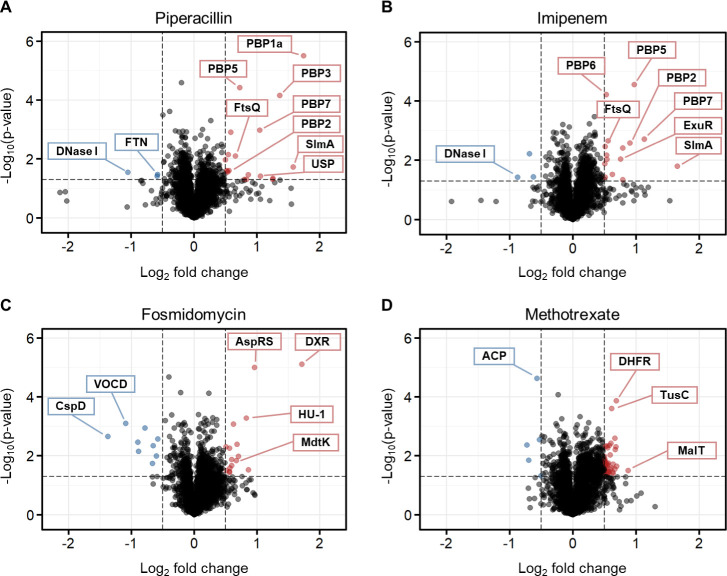
In K. pneumoniae, piperacillin and imipenem showed stabilization of a broad-spectrum of PBPs, with five and four PBPs being stabilized, respectively (Figure 3A,B). The same β-lactams in P. aeruginosa also led to the stabilization of PBPs, with two of them being stabilized by piperacillin and three by imipenem (Figure 4A,B). Notably, PBP7 (PbpG) stood out as the only PBP shared between the two bacterial species and β-lactams. In addition to the expected protein targets, our analysis identified several other proteins exhibiting significant (de)stabilization. In K. pneumoniae, piperacillin was found to stabilize a universal stress protein (USP), necessary for resistance to DNA-damaging agents, and to destabilize ferritin (FTN), an iron-storage protein. Imipenem stabilized the DNA-binding transcriptional repressor ExuR. Additionally, both β-lactams led to the stabilization of the nucleoid occlusion factor SlmA, which prevents Z-ring formation and cell division over the nucleoid, and the cell division protein FtsQ. Conversely, both β-lactams resulted in the destabilization of DNA-specific endonuclease I (DNase I). In P. aeruginosa, as shown in Figure 4A, the β-lactamase AmpC, which confers resistance to penicillins and cephalosporins,49 was stabilized by piperacillin. Moreover, a carboxyltransferase domain-containing protein (CTD) and a band 7 domain-containing protein (B7D) of unknown function were also stabilized. Conversely, phosphoesterase (Pase) and the small ribosomal subunit protein bS20 (RpsT) were destabilized.
The incubation of fosmidomycin with the two Gram-negative lysates resulted in the identification of the expected target DXR and the putative target AspRS, which is consistent with our previous observations in E. coli (Figures 3C, 4C). Noteworthy, DXR exhibited a larger stabilization effect in K. pneumoniae (log2FC = 1.7) than in P. aeruginosa (log2FC = 0.5). As a reference, the half-maximal inhibitory concentration (IC50) values of fosmidomycin reported in the literature indicate stronger inhibition for KpDXR, IC50 = 20 nM, compared to PaDXR, IC50 = 150 nM.28 Moreover, in K. pneumoniae, we observed the stabilization of the DNA-binding protein HU-beta (HU-1), responsible for wrapping DNA to prevent its denaturation under extreme environmental conditions. We also identified the stabilization of multidrug resistance protein MdtK, a multidrug efflux pump likely functioning as a Na+/drug antiporter. Major proteins that found to be destabilized include the cold shock-like protein CspD and a VOC domain-containing protein (VOCD). The latter is part of the fosfomycin resistance enzyme family, which confers resistance to the antibiotic.
Finally, MTX led to the stabilization of the expected target, DHFR, in both Gram-negative lysates (Figures 3D, 4D). Additionally, in K. pneumoniae, we observed the stabilization of transcriptional regulator MalT and the tRNA 2-thiouridine synthesizing protein C (TusC), which is part of a complex pathway that catalyzes the conversion of uridine into 2-thiouridine. This modification is important for the structure and function of tRNA molecules. Conversely, we saw the destabilization of the acyl carrier protein (ACP), which functions as a carrier of the growing fatty acid chain in fatty acid biosynthesis. In P. aeruginosa, stabilization of the tryptophan synthase alpha chain (TrpA), involved in amino acid biosynthesis, was observed. As for destabilized proteins, we identified the DNA-3-methyladenine glycosylase II (AlkA), which is involved in the hydrolysis of alkylated DNA.
Validation of iSPP in S. aureus by Model Drugs
After establishing the iSPP approach in Gram-negative bacteria, we assessed its applicability to the Gram-positive bacterium S. aureus. As one of the most pathogenic Gram-positives, S. aureus is responsible for a wide range of infections, from mild skin infections to life-threatening conditions such as septicemia, pneumonia, endocarditis, and osteomyelitis.50
For S. aureus, we identified over all conditions 1752 proteins in the iSPP assay (95% coverage of the total detected proteome in the global analysis) and identified the designated targets of the tested antibiotics (Figure S10).
Cefazolin and nafcillin induced the stabilization of two PBPs. PBP3 (FtsI) emerged as the top hit for both β-lactams, as illustrated in Figure S10A,B. Notably, an efflux transporter of the Bcr/CflA family was found to be destabilized by both β-lactams. Members of this family with known activity include Bcr (bicyclomycin resistance protein) in E. coli,51 Flor (chloramphenicol and florfenicol resistance) in Salmonella typhimurium,52 and CmlA (chloramphenicol resistance) found in the Pseudomonas plasmid R1033.53
Incubation of S. aureus lysate with rifampicin resulted in the highly significant stabilization of the two RNAP subunits RpoB and RpoC (Figure S10C). Additionally, RpoA exhibited stabilization, close-to the predefined threshold.
Target–engagement of MEP Pathway Inhibitors in E. coli
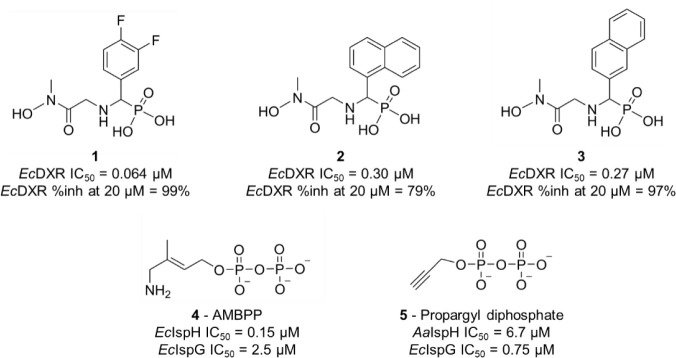
One of the primary objectives of our study was to leverage the iSPP platform for target–engagement studies of small molecules in early drug discovery stages. To that end, we employed compounds targeting enzymes within the MEP pathway, which is a valuable focus for the development of new antimicrobial agents. Due to the unfavorable pharmacokinetic properties of fosmidomycin, intensive efforts for optimizations based on its lead structure have resulted in DXR inhibitors.28 Recently, a series of β-aza-reversed fosmidomycin analogues were discovered by the Van Calenbergh group. In our study, we investigated the target–engagement of compounds 1–3 (Schemes 2, S1) in E. coli cell lysates. The analogues showed potent in vitro enzyme inhibitory activity against EcDXR with nanomolar IC50 values.
Scheme 2. Chemical Structures and Enzymatic Inhibition of Reverse β-Aza Fosmidomycin Analogues 1–3, AMBPP 4, and Propargyl Diphosphate 5.
Ec, Escherichia coli ; Aa, Aquifex aeolicus
Additionally, we included small molecules targeting the final two enzymes in the MEP pathway: (E)-4-amino-3-methylbut-2-en-1-yl diphosphate (AMBPP, 4) and propargyl diphosphate 5 (Schemes 2, S2). The penultimate enzyme in the pathway, IspG, catalyzes the reductive dehydroxylation of MEcPP to form HMBPP (Scheme 1), while IspH, the final enzyme, converts HMBPP into a mixture of the two isoprenoid precursors IDP and DMADP. Both proteins are oxygen-sensitive [4Fe-4S] cluster-containing metalloenzymes, oxidoreductases that catalyze a 2e– reduction and the elimination of a water molecule.54
AMBPP was originally designed as an analogue of HMBPP, the substrate of IspH.55,56 Noteworthy, AMBPP is the most potent IspH inhibitor published to date.57 It is a slow-binding competitive inhibitor displaying a Ki value in the nanomolar range (Ki = 54 nM, E. coli IspH; IC50 = 0.15 μM) that binds to the fourth unique iron of the [4Fe-4S]2+ cluster with its amino functional group.55,56 In contrast, propargyl diphosphate was rationally designed as a mechanism-based fragment. As reported in the literature, its interaction with the reduced [4Fe-4S] cluster was elucidated through electron paramagnetic resonance (EPR), revealing the formation of a π-complex between its alkyne group and the apical iron (Aquifex aeolicus IspH; IC50 = 6.7 μM).58,59 Additionally, these two compounds inhibit EcIspG by interacting with its [4Fe-4S] cluster. AMBPP demonstrates moderate EcIspG inhibition in vitro (IC50 = 2.5 μM),54 while the propargyl diphosphate shows good potency with an IC50 of 0.75 μM from which a Ki value of 0.33 μM was estimated.60
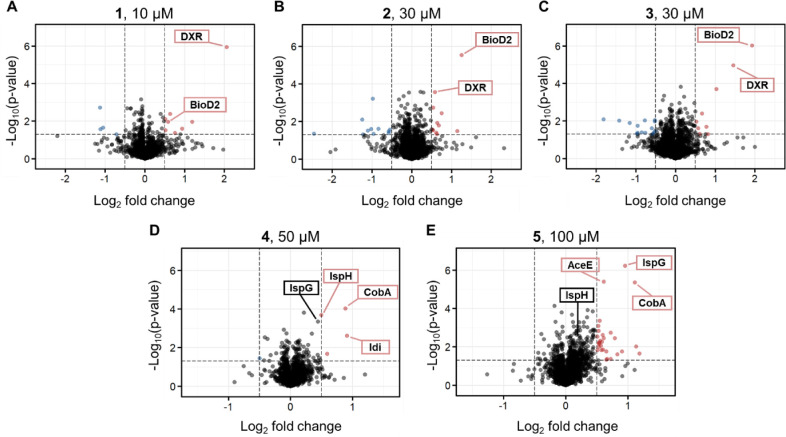
In our study, these five MEP pathway inhibitors were tested in E. coli using the previously chosen generalized gradient (14–28% v/v AEA) because the CM values of the MEP enzymes predominantly clustered within the interquartile range in the functional annotation analysis of E. coli solvent profiling. Our iSPP approach revealed the stabilization of the expected MEP enzymes DXR, IspG, and IspH, aligning with their respective in vitro enzymatic activities, along with the identification of some additional targets (Figure 5). Fosmidomycin analogues 1–3 were incubated with E. coli cell lysate at a concentration approximately 100 times their respective IC50 values. The statistical analysis revealed stabilization of the expected target DXR for all analogues (Figure 5A–C). The most potent inhibitor 1 exhibited selective DXR stabilization, representing the top hit with a log2FC of 2.05 (Figure 5A). Additionally, BioD2 emerged as a second target, particularly for 2 and 3 (Figure 5B,C). BioD2 is an ATP-dependent dethiobiotin synthetase involved in biotin synthesis. The biotin synthesis pathway is crucial for the pathogenesis of several important human pathogens, and it is absent in mammals. Therefore, BioD2 and the other enzymes in this pathway are attractive targets for novel therapeutic agents.61 Interestingly, BioD2 is a metalloenzyme, suggesting a role for the hydroxamate moiety present in 1–3 in its Mg2+ chelation.62
Figure 5.
iSPP approach in Escherichia coli cell lysates to identify (A–C) the putative targets of fosmidomycin analogues (1–3, 10, 30, and 30 μM, respectively) and (D, E) diphosphate derivatives (4 and 5, 50 and 100 μM, respectively). E. coli cell lysates were incubated with vehicle or the above-mentioned compounds and then exposed to the AEA gradient 14–28% v/v. Data are presented as a volcano plot to highlight changes in abundance of each compound over vehicle vs statistical significance. The thresholds were set to a log2 fold change (log2FC) > |0.5| and a p-value < 0.05. Red, stabilized proteins with log2FC > 0.5 and p-value < 0.05; blue, destabilized proteins with log2FC < −0.5 and p-value < 0.05; gray/black, proteins with −0.5 < log2FC < 0.5 and p-value < 0.05 and proteins with p-value > 0.05.
The incubation of E. coli lysate with AMBPP 4 and propargyl diphosphate 5 was conducted at high concentrations (50 and 100 μM, respectively), considering the dual inhibition of IspG and IspH and their distinct potencies. Incubation with AMBPP 4 resulted in the stabilization of IspH, with this enzyme identified among the top hits (Figure 5D). IspG did not surpass the set fold change threshold for being identified as a hit candidate. Conversely, propargyl diphosphate 5 induced a marked stabilization of IspG, while IspH did not exhibit stabilization (log2FC = 0.16, Figure 5E). Furthermore, this analysis revealed significantly stabilized additional targets. Specifically, both inhibitors exhibited stabilization of the corrinoid adenosyltransferase (CobA), the enzyme responsible for transferring an adenosyl moiety from MgATP to a broad range of corrinoid substrates.63 The observed stabilization suggests a possible binding of the diphosphate groups in 4 and 5 to the ATP binding site of CobA. Additionally, 4 stabilized the isopentenyl-diphosphate delta-isomerase (Idi), an enzyme associated with the MEP pathway, responsible for the isomerization of IDP to its electrophilic isomer DMADP (Scheme 1).64 IspH and Idi share a high degree of structural similarity in substrates, which provides a plausible explanation for association.
Discussion
In the current era marked by soaring AMR and an imperative need for novel anti-infectives, MS-based proteomics can play a decisive role in identifying and validating protein–ligand interactions. The SPP approach was established by Zhang et al.8 in human cell extracts as an LC-MS/MS-based method to assess target–engagement by measuring a protein’s resistance to solvent-induced denaturation and aggregation. Subsequently, Van Vranken and coworkers9 applied the compressed format to SPP, which relies on pooling multiple aliquots of the lysate upon exposure to an increasing concentration of AEA (in our study referred to as iSPP). As a result, the SPP assay has emerged as a valuable alternative to TPP.9,65 Both approaches are modification-free and analogous in their ability to induce protein unfolding and aggregation, yet they differ in their denaturation principles.8,65
In our study, we have established a comprehensive workflow for iSPP target–engagement in the three Gram-negative bacteria E. coli, K. pneumoniae, and P. aeruginosa and the Gram-positive bacterium S. aureus. We utilized DIA quantitative mass spectrometry, which offers advantages over other acquisition methods, including accurate label-free proteome quantification, a low number of missing values, and cost-effectiveness.16,66,67 A key aspect of this study involved adapting the protocol to utilize minimal protein input amounts, specifically 20 μg of total protein per data point. This adaptation makes the workflow particularly well-suited for hard-to-culture pathogens and, more broadly, for biological systems characterized by slow growth rates and limited protein yields.
Our ultimate goal was to exploit the assay for the target–engagement study of small molecules derived from target-based drug discovery. To that end, we first assessed the E. coli and K. pneumoniae proteomes’ response to organic solvents by performing a solvent proteome profiling. We identified 2438 E. coli and 2584 K. pneumoniae proteins and generated high-quality denaturation curves and CM values for 1984 and 2033 of them, respectively. We then conducted a functional annotation analysis on E. coli proteins categorized into three groups based on their CM values: the upper quartile (most stable proteins), the interquartile range, and the lower quartile (least stable proteins). We identified overrepresented protein classes and related biological processes within each group. Notably, the MEP enzymes responsible for isoprene biosynthesis fell into the interquartile range.
Subsequently, we utilized the model drugs rifampicin, ampicillin, piperacillin, imipenem, cefazolin, nafcillin, fosmidomycin, and methotrexate to demonstrate the iSPP target–engagement approach employing a rational selection of target-specific AEA gradient ranges based on the denaturation behavior of the corresponding target proteins and their respective CM values (Table 1). All of the established targets were stabilized and identified as top hits. Our findings, following E. coli incubation with ampicillin, exhibited an overlap in the stabilization of PBP1a (MrcA) and PBP4 (DacB) with the results obtained by Mateus et al.5 in a 2D-TPP study. The consistency between 2D-TPP and iSPP results underscores the validity of iSPP as target–engagement assay for bacteria. Furthermore, certain proteins that may not exhibit drug-binding stabilization during thermal denaturation could be responsive to organic solvent-induced denaturation and vice versa.65 We also observed a phenomenon reminiscent of thermal proximity coaggregation (TPCA), initially reported by Tan and colleagues68 in Cellular Thermal Shift Assay (CETSA) experiments. TPCA involves the comelting of proteins within the same complex, where the denaturation of one component can destabilize others, resulting in similar melting curves. In our solvent proteome profiling study of E. coli, we observed multiple subunits (RpoA, RpoB, and RpoC) of the RNAP core enzyme displaying nearly identical denaturation curves and comparable CM values (Figure S5). Furthermore, these subunits, along with the RNAP-associated protein RapA,45,46 exhibited similar stabilization by rifampicin in our iSPP experiments (Figure 2A). This result shows the potential for detecting the costabilization of protein–protein interactors as a consequence of ligand binding.
Additionally, we demonstrated the importance of AEA gradient selection for successful target–engagement studies in an iSPP format. To that end, we explored suboptimal AEA range windows beyond those listed in Table 1. We applied the targeted AEA range of 20–34% v/v in E. coli cell lysate for fosmidomycin and the generalized AEA range of 14–28% v/v in both E. coli and S. aureus cell lysates for rifampicin. These windows were not tailored to the regions exhibiting the most substantial solubility changes for the corresponding target proteins, thereby diminishing the observed effect size in stability and precluding their identification among the top hits (Figure S9).
Finally, we assessed the target–engagement for inhibitors of the MEP enzymes. Our experiments led to the identification of the putative targets EcDXR, EcIspG, and EcIspH as main stabilized proteins, along with some potential additional targets, such as BioD2, CobA, and Idi. Some of these additional targets can confer a positive attribute to anti-infective agents, potentially resulting in a decreased development of resistance. Consequently, the β-aza-reversed fosmidomycin analogues 1–3 represent promising EcDXR inhibitors, providing an interesting step forward for this class. The HMBPP analogue 4 and the propargyl diphosphate 5 are potent in vitro inhibitors of the enzymes IspH and IspG, respectively, and represent a starting point for the development of novel inhibitors with improved drug-like properties.
The results of our iSPP experiments also highlighted some limitations related to the compressed format. Specifically, achieving robust stabilization of target protein(s) upon certain compound–protein interactions can be difficult, as observed specifically for DXR in P. aeruginosa lysate following incubation with fosmidomycin. Denaturation curve experiments using the standard SPP assay have the potential to overcome this limitation. However, they would require additional time for MS measurements and data analysis. In our study, we also observed the presence of multiple (de)stabilized proteins alongside the designated targets for some conditions, e.g., imipenem and fosmidomycin in K. pneumoniae. For proteins such as the multidrug efflux pump MdtK and the fosfomycin resistance enzyme VOCD, the observed (de)stabilization may be linked to binding events. However, in other cases, the (de)stabilization does not necessarily indicate engagement but rather arises from an artifact associated with the low signal-to-noise ratio, potentially leading to false-positive hits. Destabilization may also occur when ligands bind to a (partially) unfolded state of a protein, thereby reducing its stability—a phenomenon previously observed in thermal shift assays (TSAs), as reported, for instance, by Cimmperman and colleagues.69 Alternatively, destabilization might result from downstream effects on proteins caused by upstream target inhibition.10,70 However, since the experiments performed in our study were conducted in cell lysates, the downstream effects are less relevant due to lower metabolic activity compared to live cells, indicating that the observed destabilized proteins need to be interpreted with great care. Whether conducting iSPP analysis in living bacterial cells is feasible remains unexplored, and this will be tested in the future. Such an analysis holds great potential and would provide valuable insights into the effects of compounds in living bacteria as well as the concentration dependency of changes in solvent stability.
During our target–engagement studies of model drugs, we also encountered potential additional targets, such as the aminoacyl-tRNA synthetase AspRS, which exhibited stabilization by fosmidomycin in various experimental iterations in E. coli (across multiple gradients), K. pneumoniae, and P. aeruginosa extracts. Table S3 contains a list of (de)stabilized proteins of all the model anti-infectives used in our iSPP experiments that could be useful for potential exploration of off-targets or cytotoxicity studies. Although not the primary focus of our investigation, the iSPP assay serves as a valuable tool for target deconvolution, e.g., following phenotypic screenings. Nonetheless, conducting these studies and obtaining a comprehensive target profile via iSPP may require the use of multiple solvent range windows or various compound concentrations.
Conclusions
In this study, we demonstrated the potential of the chemical proteomics iSPP assay for supporting drug discovery and development of novel anti-infectives. The solvent profiling of E. coli and K. pneumoniae proteomes revealed a strong positive correlation, indicating a high degree of similarity in the solvent-induced precipitation profiles of proteins across these bacterial species. Our iSPP method builds upon established SPP procedures and implements optimized denaturation gradients and minimized sample input amounts required for target–engagement studies. The solvent concentration range was specifically tailored to the region where the known target proteins exhibited the most substantial solubility changes, ensuring that the observable effect size was maximized. Through DIA-based quantitative mass spectrometry, we successfully identified established drug targets for seven antibiotics in cell extracts of four AMR-related pathogens, underscoring the assay’s adaptability and applicability across multiple infectious disease models. Additionally, the iSPP approach confirmed target–engagement of compounds developed through target-based drug discovery for MEP pathway enzyme inhibitors. We hope the assay will serve as a valuable tool to tackle the global AMR emergence and will enable faster and more affordable discovery and development of lead compounds against primary human pathogens.
Methods
EcDXR Activity Assay
The EcDXR activity was measured through a DXS–DXR coupled assay previously described, using M. tuberculosis DXS (MtDXS) as the coupling enzyme.71EcDXR and MtDXS were expressed and purified as previously described.72,73 First, a MtDXS reaction was prepared with a final concentration of 250 μM d-l-glyceraldehyde 3-phosphate, 500 μM pyruvate, 300 μM thiamine diphosphate, and 1.5 μM of MtDXS, in an assay buffer composed of 200 mM HEPES pH 8, 2 mM MgCl2. The MtDXS reaction mixture was incubated at 25 °C for 90 min. The DXR reaction was then started with the addition of EcDXR and NADPH to final concentrations of 75 nM and 150 μM, respectively. The activity was monitored through the decrease in fluorescence of NADPH oxidation using a plate reader SYNERGY H1 (BioTEK) with wavelengths 340/450 nm (excitation/emission) for 15 min at 30 °C. Initial velocity was calculated through the slope of the linear region in the first 10 min of reaction using OriginPro8. For dose–response assays, EcDXR was previously incubated for 15 min with a serial dilution of the desired compound starting at 50 μM. 1% DMSO was used as a negative control whereas 50 μM of fosmidomycin was used as a positive control. The inhibition percentage was measured using the following equation:
The percentage of inhibition was calculated for each compound concentration, and the dose–response curve was fitted using the Hill1 model with OriginPro8. Half-maximal inhibitory concentration (IC50) values were then calculated using the interpolation method. All experiments were conducted in triplicates.
Bacterial Culture and Cell Extract Preparation
Escherichia coli (strain K12), Klebsiella pneumoniae (strain ATCC13883), Pseudomonas aeruginosa (strain PA01), and Staphylococcus aureus (strain Newman) cell lysates were obtained from the Helmholtz Institute for Pharmaceutical Research Saarland (HIPS). All bacteria were grown aerobically overnight (ON) with agitation at 200 rpm and 37 °C in lysogeny broth. Cells were washed and resuspended in 3× cell pellet volume with ice-cold lysis buffer containing 50 mM Tris/HCl pH 7.5, 5% glycerol, 150 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.8% IGEPAL CA-630, and 1× Halt Protease and Phosphatase Inhibitor Cocktails (Thermo Fisher Scientific). The resuspended cell pellet was vortexed for 30 s, incubated for 30 min on ice, and further lysed via homogenization (BANDELIN SONOPULS mini20) with 5 cycles at 70% amplitude for 30 s. Each cycle was followed by 90 s pause where the suspension was kept at 4 °C. The lysate was then centrifuged twice for 30 min at 4 °C 17 000× g, and the supernatant was collected (centrifuge MIKRO 220R, Hettich). The protein lysates were diluted to 0.8 mg/mL (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific) using lysis buffer without IGEPAL CA-630, resulting in a reduced percentage of 0.4% IGEPAL CA-630, and then stored at −80 °C until further use.
Solvent Proteome Profiling for LC-MS/MS Readout
E. coli and K. pneumoniae cell lysates were thawed on ice. The lysates were distributed into a 96-well plate, 20 μg total protein per each data point, and then exposed to 12 increasing concentrations of acetone/ethanol/acetic acid mixture (AEA, v/v) from 0% to 50% (0, 10, 15, 20, 22, 24, 26, 30, 32, 34, 40, and 50%, performed on the Bravo Automated Liquid Handling Platform, Agilent) in a final reaction volume of 50 μL. Samples were incubated at 37 °C at 750 rpm for 20 min (ThermoMixer C, Eppendorf). Precipitated proteins were removed by centrifugation at 4402× g for 35 min (centrifuge 5920R, Eppendorf). An equal volume of each soluble fraction was collected (Bravo Automated Liquid Handling Platform) and prepared for LC-MS/MS analysis. All experimental conditions were conducted in triplicates.
Solvent Proteome Profiling for SDS-PAGE Readout
E. coli, K. pneumoniae, P. aeruginosa, and S. aureus cell lysates were thawed on ice and then exposed to an increasing concentration of AEA from 0% to 50% (v/v). For E. coli cell lysate, 14 aliquots were prepared (0, 7, 10, 13, 16, 19, 22, 25, 28, 31, 34, 37, 40, and 50%). For K. pneumoniae, P. aeruginosa, and S. aureus cell lysates, 13 aliquots were prepared (0, 8, 11, 14, 17, 20, 23, 26, 29, 32, 35, 40, and 50%). The samples were then processed as described above. Upon supernatant collection, the soluble fractions were dried down in a concentrator plus (Eppendorf) and then resuspended to a final protein concentration of 1.0 mg/mL with 2× NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) containing 25 mM DTT. Proteins were resolved on NuPAGE 4–12% Bis-Tris Protein Gels (Thermo Fisher Scientific). Gels were stained ON with Coomassie-staining solution (ROTI Blue, Carl Roth). Images were acquired on a ChemiDoc MP Imaging System (BIO-RAD).
iSPP Assay
All bacterial cell lysates were thawed on ice and then distributed in aliquots. Each aliquot was then incubated with the corresponding compound (solubilized in DMSO or ddH2O) or vehicle control (DMSO or ddH2O, respectively), at the desired concentration (Table 1). The samples were incubated at room temperature (RT) for 30 min on an end-over-end shaker. Rifampin, ampicillin (trihydrate), and methotrexate were purchased from Selleckchem. Piperacillin and imipenem were purchased from Sigma-Aldrich. Fosmidomycin (sodium), nafcillin (sodium monohydrate), and cefazolin (sodium) were purchased from MedChemExpress. Each aliquot was then distributed into eight wells, treated with an increasing concentration of AEA (v/v) from 14% to 28% (14, 16, 18, 20, 22, 24, 26, and 28%), 20% to 34% (20, 22, 24, 26, 28, 30, 32, and 34%), or 12% to 29.5% (12, 14.5, 17, 19.5, 22, 24.5, 27, and 29.5%). Samples were incubated at 37 °C and 750 rpm for 20 min. Precipitated proteins were removed by centrifugation at 4400× g for 35 min. An equal volume of each resulting soluble fraction was pooled into a single sample (Bravo Automated Liquid Handling Platform) and prepared for LC-MS/MS analysis. All experimental conditions were conducted in triplicates.
Sample Preparation for LC-MS/MS Analysis
The soluble fractions were dried and then resuspended to a final protein concentration of 1.0 mg/mL with 5% SDS buffer containing 50 mM TEAB at pH 7.5. The bacterial cell lysates for global proteomic analysis were diluted 1:1 to a final protein concentration of 0.4 mg/mL with 10% SDS buffer. To reduce disulfide bonds, 10 mM DTT was added to all samples, followed by incubation for 30 min at 35 °C and 700 rpm on the ThermoMixer C. Protein alkylation was performed with 55 mM chloroacetamide and 30 min incubation at RT. Samples were acidified by adding phosphoric acid to a final concentration of 2.5% and subsequently diluted 7-fold with 90% methanol in 100 mM TEAB pH 7.5. The samples were transferred to an S-trap column (Protifi) and washed 4× with the same buffer. Sequencing Grade Modified Trypsin (Promega) in TEAB pH 8.5 was added to the S-trap column at a ratio of 1:10 (trypsin/protein), and the digestion reaction was carried out ON at 37 °C. Peptides were eluted with 50 mM TEAB pH 8.5, followed by 0.1% formic acid and then 50/50/0.1 v/v acetonitrile (ACN)/water/FA. Samples were dried down, and peptides were resuspended with 0.5% FA. Peptides were desalted on the Bravo Automated Liquid Handling Platform using C18 cartridges (5 μL bed volume, Agilent) and the standard AssayMAP peptide cleanup v2.0 protocol. Briefly, C18 cartridges were primed with 100 μL of 50/50/0.1 (v/v) acetonitrile (ACN)/water/FA and equilibrated with 50 μL of 0.1% FA at a flow rate of 10 μL/min. The samples were loaded at 5 μL/min, followed by an internal cartridge wash with 0.1% FA at a flow rate of 10 μL/min. Peptides were eluted with 50 μL of 60/40/0.1 (v/v) acetonitrile (ACN)/water/FA at a flow rate of 5 μL/min. Samples were dried and stored at −80 °C until further use.
Liquid Chromatography and Mass Spectrometry Data Acquisition
All samples were solubilized in 0.1% FA before being injected in volumes equivalent to 1 μg on a Dionex UltiMate 3000 nano System (Thermo Fisher Scientific) coupled online to a Q Exactive Plus (Thermo Fisher Scientific) equipped with an Orbitrap mass analyzer. Peptides were delivered to a trap column (75 μm × 2 cm, packed in-house with ReproSil-Pur 120 ODS-3 resin, Dr. Maisch). Subsequently, they were separated on an analytical column (75 μm × 55 cm, packed in-house with Reprosil-Gold 120 C18, 3 μm resin, Dr. Maisch) at a flow rate of 300 nL/min using a 100 min gradient, ranging from 2% to 32% solvent B (0.1% FA, 5% DMSO in HPLC-MS-grade acetonitrile) in solvent A (0.1% FA, 5% DMSO in HPLC-MS-grade water). The column oven temperature was set at 50 °C. The QE plus instrument was operated in data-independent acquisition (DIA), in positive ionization mode. Full scan spectra (m/z 400–1000) were acquired in centroid mode at an Orbitrap resolution of 70 000, an AGC target set to 3e6, and a maximum injection time of 20 ms. Subsequently, DIA scans were collected by utilizing 30 windows, with a 1 Da window overlap. HCD collision was set to 27%, loop count to 30, Orbitrap resolution to 35 000, AGC target to 3e6, and a maximum injection time set to automatic.
Peptide and Protein Identification and Quantification
Raw LFQ-DIA files were processed with DIA-NN (v. 1.8.1). They were analyzed in library-free mode using the UniProt FASTA files for each organism: Escherichia coli K12, taxon identifier: 83333; Pseudomonas aeruginosa PAO1, taxon identifier: 208964; Klebsiella pneumoniae ATCC13883, taxon identifier: 1125630 (ATCC13883 proteome is redundant to the reference HS11286); Staphylococcus aureus Newman, taxon identifier: 93061 (Newman proteome is redundant to the reference NCTC 8325/PS 47); and canonical versions, not older than 5 months prior to MS measurements. The raw files were digested selecting Trypsin/P as enzyme specificity with two maximal missed cleavages. Peptide length was restricted from 7 to 30 peptides, and the precursor m/z range was set from 300 to 1800. Cysteine carbamidomethylation was selected as a fixed modification, and methionine oxidation and N-terminal acetylation were selected as variable modifications. The maximum number of variable modifications was set to three, and a match between runs (MBR) was enabled. All other parameters were set to default, including the 1% precursor FDR. Cross-run normalization (RT-dependent) was enabled for raw files of the iSPP experiments.
Solvent Profiling Data Analysis
Protein intensity values of each replicate were normalized to their median abundance and expressed as a ratio to the lowest AEA concentration sample using Excel software. Statistical analyses of data and plot generation were then performed in GraphPad Prism (v. 8.3.0) and RStudio (v. 4.3.2) using dplyr package (v. 1.1.4) and ggplot2 package (v. 3.5.0).74,75 The sigmoidal denaturation curves were generated using a nonlinear regression model and then filtered according to the following criteria: denaturation curves must reach a plateau of ≤0.3, the coefficient of determination (R2) must be ≥0.8, and a valid slope must be obtained. The functional annotation analysis was performed on the Database for Annotation, Visualization and Integrated Discovery (DAVID).20,21
iSPP Data Analysis
Protein intensity values of biological replicates across all conditions were normalized to their median abundance and log2-transformed. Missing values were imputed from a normal distribution (width of 0.3, downshift of 1.5) for vehicle controls only. Moreover, p-values were obtained by a two-sample t test over replicates with a permutation-based false discovery rate correction (FDR 0.05) on Perseus (v. 2.0.10.0). The volcano plots were generated in RStudio by EnhancedVolcano package (v. 1.20.0), plotting proteins by statistical significance (vertical axis, −log10p-value) and magnitude of change (horizontal axis, log2 fold change) of the quantified LFQ-DIA protein intensities for each compound condition over vehicle control.76
Acknowledgments
This work was funded by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 860816. We thank Victor Gawriljuk and Prof. Dr. Matthew Groves at the University of Groningen for the activity data on the enzyme EcDXR. We also thank Simone Amann and Dr. Jörg Haupenthal at the Helmholtz Institute for Pharmaceutical Research (HIPS) for providing bacterial cell lysates. TOC graphic and Figure 1A were created with BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.4c00417.
Additional information on the iSPP protocol adaptation and related data (Figures S1, S8–S10); solvent proteome profiling (Figures S2 and S3); denaturation curves for the expected target proteins of the model drugs and compounds tested (Figures S4–S7); synthetic procedure for compounds 1–5 (Schemes S1 and S2); and purity assessments (Figures S11–S14) (PDF)
Melting concentration (CM) values for Escherichia coli (Table S1) and Klebsiella pneumoniae (Table S2) proteomes; proteins destabilized/stabilized in the iSPP confirmation experiments by model drugs (Table S3) (XLSX)
The authors declare the following competing financial interest(s): Hannes Hahne is co-founder and shareholder of OmicScouts GmbH, a proteomics and chemical proteomics-focused contract research organization.
Supplementary Material
References
- Gauba A.; Rahman K. M. Evaluation of Antibiotic Resistance Mechanisms in Gram-Negative Bacteria. Antibiotics 2023, 12 (11), 1590. 10.3390/antibiotics12111590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook M. A.; Wright G. D. The Past, Present, and Future of Antibiotics. Sci. Transl. Med. 2022, 14 (657), eabo7793 10.1126/scitranslmed.abo7793. [DOI] [PubMed] [Google Scholar]
- Murray C. J.; Ikuta K. S.; Sharara F.; Swetschinski L.; Robles Aguilar G.; Gray A.; Han C.; Bisignano C.; Rao P.; Wool E.; et al. Global Burden Of Bacterial Antimicrobial Resistance In 2019: a Systematic Analysis. The Lancet 2022, 399 (10325), 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masini T.; Hirsch A. K. H. Development of Inhibitors of the 2C-Methyl-D-Erythritol 4-Phosphate (MEP) Pathway Enzymes as Potential Anti-Infective Agents. J. Med. Chem. 2014, 57, 9740–9763. 10.1021/jm5010978. [DOI] [PubMed] [Google Scholar]
- Mateus A.; Bobonis J.; Kurzawa N.; Stein F.; Helm D.; Hevler J.; Typas A.; Savitski M. M. Thermal Proteome Profiling in Bacteria: Probing Protein State in Vivo. Mol. Syst. Biol. 2018, 14 (7), e8242 10.15252/msb.20188242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner F.; Geddes-McAlister J.; Mann M.; Bantscheff M. The Emerging Role of Mass Spectrometry-Based Proteomics in Drug Discovery. Nat. Rev. Drug Discovery 2022, 21, 637–654. 10.1038/s41573-022-00409-3. [DOI] [PubMed] [Google Scholar]
- Schirle M.; Bantscheff M.; Kuster B. Mass Spectrometry-Based Proteomics in Preclinical Drug Discovery. Chem. Biol. 2012, 19, 72–84. 10.1016/j.chembiol.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Wang Q.; Li Y.; Ruan C.; Wang S.; Hu L.; Ye M. Solvent-Induced Protein Precipitation for Drug Target Discovery on the Proteomic Scale. Anal. Chem. 2020, 92 (1), 1363–1371. 10.1021/acs.analchem.9b04531. [DOI] [PubMed] [Google Scholar]
- Van Vranken J. G.; Li J.; Mitchell D. C.; Navarrete-Perea J.; Gygi S. P. Assessing Target Engagement Using Proteome-Wide Solvent Shift Assays. Elife 2021, 10, 1–21. 10.7554/eLife.70784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.; Chen X.; Xu W.; Li S.; Chai Q.; Zhang Y. Solvent-Induced Proteome Profiling for Proteomic Quantitation and Target Discovery of Small Molecular Drugs. Proteomics 2023, 23, 1–10. 10.1002/pmic.202200281. [DOI] [PubMed] [Google Scholar]
- Cowan D. A. Thermophilic Proteins: Stability and Function in Aqueous and Organic Solvents. Comp. Biochem. Physiol., Part A: mol. Integr. Physiol. 1997, 118 (3), 429–438. 10.1016/S0300-9629(97)00004-2. [DOI] [PubMed] [Google Scholar]
- Becher I.; Werner T.; Doce C.; Zaal E. A.; Tögel I.; Khan C. A.; Rueger A.; Muelbaier M.; Salzer E.; Berkers C. R.; Fitzpatrick P. F.; Bantscheff M.; Savitski M. M. Thermal Profiling Reveals Phenylalanine Hydroxylase As An Off-Target Of Panobinostat. Nat. Chem. Biol. 2016, 12 (11), 908–910. 10.1038/nchembio.2185. [DOI] [PubMed] [Google Scholar]
- Mateus A.; Kurzawa N.; Becher I.; Sridharan S.; Helm D.; Stein F.; Typas A.; Savitski M. M. Thermal Proteome Profiling for Interrogating Protein Interactions. Mol. Syst. Biol. 2020, 16 (3), e9232 10.15252/msb.20199232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetani M.; Sabatier P.; Saei A. A.; Beusch C. M.; Yang Z.; Lundström S. L.; Zubarev R. A. Proteome Integral Solubility Alteration: A High-Throughput Proteomics Assay for Target Deconvolution. J. Proteome Res. 2019, 18 (11), 4027–4037. 10.1021/acs.jproteome.9b00500. [DOI] [PubMed] [Google Scholar]
- Hailemariam M.; Eguez R. V.; Singh H.; Bekele S.; Ameni G.; Pieper R.; Yu Y. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J. Proteome Res. 2018, 17 (9), 2917–2924. 10.1021/acs.jproteome.8b00505. [DOI] [PubMed] [Google Scholar]
- Chapman J. D.; Goodlett D. R.; Masselon C. D. Multiplexed and Data-Independent Tandem Mass Spectrometry for Global Proteome Profiling. Mass Spectrom. Rev. 2014, 33 (6), 452–470. 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- George A. L.; Sidgwick F. R.; Watt J. E.; Martin M. P.; Trost M.; Marín-Rubio J. L.; Dueñas M. E. Comparison of Quantitative Mass Spectrometric Methods for Drug Target Identification by Thermal Proteome Profiling. J. Proteome Res. 2023, 22 (8), 2629–2640. 10.1021/acs.jproteome.3c00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzab A.; Kurzawa N.; Hopf T.; Moerch M.; Zecha J.; Leijten N.; Bian Y.; Musiol E.; Maschberger M.; Stoehr G.; Becher I.; Daly C.; Samaras P.; Mergner J.; Spanier B.; Angelov A.; Werner T.; Bantscheff M.; Wilhelm M.; Klingenspor M.; Lemeer S.; Liebl W.; Hahne H.; Savitski M. M.; Kuster B. Meltome Atlas—Thermal Proteome Stability across the Tree of Life. Nat. Methods 2020, 17 (5), 495–503. 10.1038/s41592-020-0801-4. [DOI] [PubMed] [Google Scholar]
- Altenhoff A. M.; Vesztrocy A. W.; Bernard C.; Train C. M.; Nicheperovich A.; Baños S. P.; Julca I.; Moi D.; Nevers Y.; Majidian S.; Dessimoz C.; Glover N. M. OMA Orthology in 2024: Improved Prokaryote Coverage, Ancestral and Extant GO Enrichment, a Revamped Synteny Viewer and More in the OMA Ecosystem. Nucleic Acids Res. 2024, 52 (D1), D513–D521. 10.1093/NAR/GKAD1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4 (1), 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Sherman B. T.; Hao M.; Qiu J.; Jiao X.; Baseler M. W.; Lane H. C.; Imamichi T.; Chang W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50 (W1), W216–W221. 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli W. Mechanisms of Action and Resistance. Rev. Infect. Dis. 1983, 5 (Supplement_3), S407–S411. 10.1093/CLINIDS/5.SUPPLEMENT_3.S407. [DOI] [PubMed] [Google Scholar]
- Halawa E. M.; Fadel M.; Al-Rabia M. W.; Behairy A.; Nouh N. A.; Abdo M.; Olga R.; Fericean L.; Atwa A. M.; El-Nablaway M.; Abdeen A. Antibiotic Action and Resistance: Updated Review of Mechanisms, Spread, Influencing Factors, and Alternative Approaches for Combating Resistance. Front. Pharmacol. 2024, 14, 14. 10.3389/fphar.2023.1305294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade K. C.; Benjamin D. K. Clinical pharmacology of anti-infective drugs, infectious diseases of the fetus and newborn. Clin. Pharmacol. Anti-Infect. Drugs 2011, 37, 1160–1211. 10.1016/B978-1-4160-6400-8.00037-7. [DOI] [Google Scholar]
- Chen P.; Seth A. K.; Abercrombie J. J.; Mustoe T. A.; Leung K. P. Activity of Imipenem against Klebsiella Pneumoniae Biofilms in Vitro and in Vivo. Antimicrob. Agents Chemother. 2014, 58 (2), 1208–1213. 10.1128/AAC.01353-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karruli A.; Catalini C.; D’Amore C.; Foglia F.; Mari F.; Harxhi A.; Galdiero M.; Durante-Mangoni E. Evidence-Based Treatment of Pseudomonas Aeruginosa Infections: A Critical Reappraisal. Antibiotics 2023, 12, 399. 10.3390/antibiotics12020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L. M.; Silva B. N. M. D.; Barbosa G.; Barreiro E. J. β-Lactam Antibiotics: An Overview from a Medicinal Chemistry Perspective. Eur. J. Med. Chem. 2020, 208, 112829. 10.1016/j.ejmech.2020.112829. [DOI] [PubMed] [Google Scholar]
- Knak T.; Abdullaziz M. A.; Höfmann S.; Alves Avelar L. A.; Klein S.; Martin M.; Fischer M.; Tanaka N.; Kurz T. Over 40 Years of Fosmidomycin Drug Research: A Comprehensive Review and Future Opportunities. Pharmaceuticals MDPI 2022, 15, 1553. 10.3390/ph15121553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chofor R.; Risseeuw M. D. P.; Pouyez J.; Johny C.; Wouters J.; Dowd C. S.; Couch R. D.; Van Calenbergh S. Synthetic Fosmidomycin Analogues with Altered Chelating Moieties Do Not Inhibit 1-Deoxy-d-Xylulose 5-Phosphate Reductoisomerase or Plasmodium Falciparum Growth in Vitro. Molecules 2014, 19 (2), 2571–2587. 10.3390/molecules19022571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister S.; Wiesner J.; Reichenberg A.; Hintz M.; Bietz S.; Harb O. S.; Roos D. S.; Kordes M.; Friesen J.; Matuschewski K.; Lingelbach K.; Jomaa H.; Seeber F. Fosmidomycin Uptake into Plasmodium and Babesia-Infected Erythrocytes Is Facilitated by Parasite-Induced New Permeability Pathways. PLoS One 2011, 6, e19334 10.1371/journal.pone.0019334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronstein B. N. The Mechanism of Action of Methotrexate. Rheum. Dis. Clin. North. Am. 1997, 23 (4), 739–755. 10.1016/S0889-857X(05)70358-6. [DOI] [PubMed] [Google Scholar]
- Letertre M. P. M.; Munjoma N.; Wolfer K.; Pechlivanis A.; McDonald J. A. K.; Hardwick R. N.; Cherrington N. J.; Coen M.; Nicholson J. K.; Hoyles L.; Swann J. R.; Wilson I. D. A Two-Way Interaction between Methotrexate and the Gut Microbiota of Male Sprague-Dawley Rats. J. Proteome Res. 2020, 19 (8), 3326–3339. 10.1021/acs.jproteome.0c00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak R. R.; Alexander M.; Deshpande I.; Stapleton-Gray K.; Rimal B.; Patterson A. D.; Ubeda C.; Scher J. U.; Turnbaugh P. J. Methotrexate Impacts Conserved Pathways in Diverse Human Gut Bacteria Leading to Decreased Host Immune Activation. Cell Host Microbe 2021, 29 (3), 362–377.e11. 10.1016/j.chom.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopytek S. J.; Dyer J. C. D.; Knapp G. S.; Hu J. C. Resistance to Methotrexate Due to AcrAB-Dependent Export from Escherichia Coli. Antimicrob. Agents Chemother. 2000, 44 (11), 3210–3212. 10.1128/AAC.44.11.3210-3212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus A.; Hevler J.; Bobonis J.; Kurzawa N.; Shah M.; Mitosch K.; Goemans C. V.; Helm D.; Stein F.; Typas A.; Savitski M. M. The Functional Proteome Landscape of Escherichia Coli. Nature 2020, 588 (7838), 473–478. 10.1038/s41586-020-3002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee B.; Thakur S. S. Single-Run Mass Spectrometry Analysis Provides Deep Insight into E. Coli Proteome. J. Am. Soc. Mass. Spectrom. 2018, 29 (12), 2394–2401. 10.1007/s13361-018-2066-z. [DOI] [PubMed] [Google Scholar]
- Soufi B.; Krug K.; Harst A.; Macek B. Characterization of the E. Coli Proteome and Its Modifications during Growth and Ethanol Stress. Front. Microbiol. 2015, 6, 128127. 10.3389/fmicb.2015.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reales-Calderón J. A.; Sun Z.; Mascaraque V.; Pérez-Navarro E.; Vialás V.; Deutsch E. W.; Moritz R. L.; Gil C.; Martínez J. L.; Molero G. A Wide-Ranging Pseudomonas Aeruginosa PeptideAtlas Build: A Useful Proteomic Resource for a Versatile Pathogen. J. Proteomics 2021, 239, 104192. 10.1016/j.jprot.2021.104192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear M. C.; Seidel L.; Krieger J. R.; Geddes-McAlister J.; Levesque R. C.; Khursigara C. M. Quantitative Proteomics Reveals Unique Responses to Antimicrobial Treatments in Clinical Pseudomonas Aeruginosa Isolates. mSystems 2023, 8 (5), e00491–23 10.1128/msystems.00491-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright B. W.; Kamath K. S.; Krisp C.; Molloy M. P. Proteome Profiling of Pseudomonas Aeruginosa PAO1 Identifies Novel Responders to Copper Stress. BMC Microbiol. 2019, 19 (1), 1–13. 10.1186/s12866-019-1441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher D.; Hempel K.; Sievers S.; Zühlke D.; Pané-Farré J.; Otto A.; Fuchs S.; Albrecht D.; Bernhardt J.; Engelmann S.; Völker U.; van Dijl J. M.; Hecker M. A Proteomic View of an Important Human Pathogen-Towards the Quantification of the Entire Staphylococcus Aureus Proteome. PLoS One 2009, 4 (12), e8176 10.1371/journal.pone.0008176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z.; Fan L.; Zhong Y.; Guo J.; Dong X.; Xu X.; Wang C.; Su Y. Quantitative Proteomics Reveals Reduction in Central Carbon and Energy Metabolisms Contributes to Gentamicin Resistance in Staphylococcus Aureus. J. Proteomics 2023, 277, 104849. 10.1016/j.jprot.2023.104849. [DOI] [PubMed] [Google Scholar]
- Sukumaran A.; Pladwig S.; Geddes-McAlister J. Zinc Limitation in Klebsiella Pneumoniae Profiled by Quantitative Proteomics Influences Transcriptional Regulation and Cation Transporter-Associated Capsule Production. BMC Microbiol. 2021, 21 (1), 1–15. 10.1186/s12866-021-02091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L.; Yang X.; Chen H.; Mo Z.; Li Y.; Wei S.; Zhao Z. Molecular Characteristics and Quantitative Proteomic Analysis of Klebsiella Pneumoniae Strains with Carbapenem and Colistin Resistance. Antibiotics 2022, 11 (10), 1341. 10.3390/antibiotics11101341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhodolets M. V.; Jin D. J. Interaction between RNA Polymerase and RapA, a Bacterial Homolog of the SWI/SNF Protein Family. J. Biol. Chem. 2000, 275 (29), 22090–22097. 10.1074/jbc.M000056200. [DOI] [PubMed] [Google Scholar]
- Liu B.; Zuo Y.; Steitz T. A.; Yang W. Structural Basis for Transcription Reactivation by RapA. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (7), 2006–2010. 10.1073/pnas.1417152112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J.; Plateau P.; Blanquet S. Metabolism of D-Aminoacyl-TRNAs in Escherichia Coli and Saccharomyces Cerevisiae Cells. J. Biol. Chem. 2000, 275 (42), 32535–32542. 10.1074/jbc.M005166200. [DOI] [PubMed] [Google Scholar]
- Thompson M. G.; Blake-Hedges J. M.; Pereira J. H.; Hangasky J. A.; Belcher M. S.; Moore W. M.; Barajas J. F.; Cruz-Morales P.; Washington L. J.; Haushalter R. W.; Eiben C. B.; Liu Y.; Skyrud W.; Benites V. T.; Barnum T. P.; Baidoo E. E. K.; Scheller H. V.; Marletta M. A.; Shih P. M.; Adams P. D.; Keasling J. D. An Iron (II) Dependent Oxygenase Performs the Last Missing Step of Plant Lysine Catabolism. Nat. Commun. 2020, 11 (1), 2931. 10.1038/s41467-020-16815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A. AmpC β-Lactamases. Clin. Microbiol Rev. 2009, 22 (1), 161. 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y. C.; Bae J. S.; Otto M. Pathogenicity and Virulencef Staphylococcus Aureus. Virulence 2021, 12, 547–569. 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley J.; Hyatt L. S.; Ainley K.; Parish J. H.; Herbert R. B.; White G. R. Cloning and Sequence Analysis of an Escherichia Coli Gene Conferring Bicyclomycin Resistance. Gene 1993, 127 (1), 117–120. 10.1016/0378-1119(93)90625-D. [DOI] [PubMed] [Google Scholar]
- Braibant M.; Chevalier J.; Chaslus-Dancla E.; Pagès J. M.; Cloeckaert A. Structural and Functional Study of the Phenicol-Specific Efflux Pump FloR Belonging to the Major Facilitator Superfamily. Antimicrob. Agents Chemother. 2005, 49 (7), 2965–2971. 10.1128/AAC.49.7.2965-2971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette L.; Champetier S.; Buisson J. P.; Roy P. H. Characterization of the Nonenzymatic Chloramphenicol Resistance (CmlA) Gene of the In4 Integron of Tn1696: Similarity of the Product to Transmembrane Transport Proteins. J. Bacteriol. 1991, 173 (14), 4493–4502. 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F.; Wang K.; Li J.; Wang W.; Liu Y. L.; Amin S.; Oldfield E. Inhibition of the 4Fe-4S Proteins IspG and IspH: An EPR, ENDOR and HYSCORE Investigation. Chem. Sci. 2014, 5 (4), 1642–1649. 10.1039/C3SC53301H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens-Botzong A.; Janthawornpong K.; Wolny J. A.; Tambou E. N.; Rohmer M.; Krasutsky S.; Poulter C. D.; Schünemann V.; Seemann M. Biosynthesis of Isoprene Units: Mössbauer Spectroscopy of Substrate and Inhibitor Binding to the [4Fe-4S] Cluster of the LytB/IspH Enzyme. Angew. Chem. 2011, 123 (50), 12182–12185. 10.1002/ange.201104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janthawornpong K.; Krasutsky S.; Chaignon P.; Rohmer M.; Poulter C. D.; Seemann M. Inhibition of IspH, a [4Fe-4S]2+ Enzyme Involved in the Biosynthesis of Isoprenoids via the Methylerythritol Phosphate Pathway. J. Am. Chem. Soc. 2013, 135 (5), 1816–1822. 10.1021/ja309557s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobelius H.; Bianchino G. I.; Borel F.; Chaignon P.; Seemann M. The Reductive Dehydroxylation Catalyzed by IspH, a Source of Inspiration for the Development of Novel Anti-Infectives. Molecules 2022, 27, 708. 10.3390/molecules27030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Wang W.; No J. H.; Zhang Y.; Zhang Y.; Oldfield E. Inhibition of the Fe4S4-Cluster-Containing Protein IspH (LytB): Electron Paramagnetic Resonance, Metallacycles, and Mechanisms. J. Am. Chem. Soc. 2010, 132 (19), 6719–6727. 10.1021/ja909664j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Wang K.; Liu Y. L.; No J. H.; Li J.; Nilges M. J.; Oldfield E. Bioorganometallic Mechanism of Action, and Inhibition, of IspH. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (10), 4522–4527. 10.1073/pnas.0911087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Li J.; Wang K.; Huang C.; Zhang Y.; Oldfield E. Organometallic Mechanism of Action and Inhibition of the 4Fe-4S Isoprenoid Biosynthesis Protein GcpE (IspG). Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (25), 11189–11193. 10.1073/pnas.1000264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirithanakorn C.; Cronan J. E. Biotin, a Universal and Essential Cofactor: Synthesis, Ligation and Regulation. FEMS Microbiol. Rev. 2021, 45 (4), 1–18. 10.1093/femsre/fuab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S.; Hélesbeux J.-J.; Larcher G.; Duval O. Hydroxamate, a Key Pharmacophore Exhibiting a Wide Range of Biological Activities. Mini-Rev. Med. Chem. 2013, 13 (9), 1311–1326. 10.2174/13895575113139990007. [DOI] [PubMed] [Google Scholar]
- Bauer C. B.; Fonseca M. V.; Holden H. M.; Thoden J. B.; Thompson T. B.; Escalante-Semerena J. C.; Rayment I. Three-Dimensional Structure of ATP: Corrinoid Adenosyltransferase from Salmonella Typhimurium in Its Free State, Complexed with MgATP, or Complexed with Hydroxycobalamin and MgATP. Biochemistry 2001, 40 (2), 361–374. 10.1021/bi002145o. [DOI] [PubMed] [Google Scholar]
- Hahn F. M.; Hurlburt A. P.; Poulter C. D. Escherichia Coli Open Reading Frame 696 Is Idi, a Nonessential Gene Encoding Isopentenyl Diphosphate Isomerase. J. Bacteriol. 1999, 181 (15), 4499–4504. 10.1128/JB.181.15.4499-4504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski M. M.; Reinhard F. B. M.; Franken H.; Werner T.; Savitski M. F.; Eberhard D.; Molina D. M.; Jafari R.; Dovega R. B.; Klaeger S.; Kuster B.; Nordlund P.; Bantscheff M.; Drewes G. Tracking Cancer Drugs in Living Cells by Thermal Profiling of the Proteome. Science 2014, 346 (6205), 1255784. 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- Figueroa-Navedo A. M.; Ivanov A. R. Experimental and Data Analysis Advances in Thermal Proteome Profiling. Cells Rep. Methods 2024, 4 (2), 100717. 10.1016/j.crmeth.2024.100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Smith L. S.; Zhu H. J. Data-Independent Acquisition (DIA): An Emerging Proteomics Technology for Analysis of Drug-Metabolizing Enzymes and Transporters. Drug Discovery Today: Technol. 2021, 39, 49–56. 10.1016/j.ddtec.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. S. H.; Go K. D.; Bisteau X.; Dai L.; Yong C. H.; Prabhu N.; Ozturk M. B.; Lim Y. T.; Sreekumar L.; Lengqvist J.; Tergaonkar V.; Kaldis P.; Sobota R. M.; Nordlund P. Thermal Proximity Coaggregation for System-Wide Profiling of Protein Complex Dynamics in Cells. Science 2018, 359 (6380), 1170–1177. 10.1126/science.aan0346. [DOI] [PubMed] [Google Scholar]
- Cimmperman P.; Baranauskiene L.; Jachimovičiute S.; Jachno J.; Torresan J.; Michailoviene V.; Matuliene J.; Sereikaite J.; Bumelis V.; Matulis D. A Quantitative Model of Thermal Stabilization and Destabilization of Proteins by Ligands. Biophys. J. 2008, 95 (7), 3222–3231. 10.1529/biophysj.108.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vranken J. G.; Li J.; Mintseris J.; Gadzuk-Shea M.; Gygi S. P.; Schweppe D. K. Large-Scale Characterization of Drug Mechanism of Action Using Proteome-Wide Thermal Shift Assays. eLife 2024, 13, RP95595. 10.7554/ELIFE.95595.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humnabadkar V.; Jha R. K.; Ghatnekar N.; De Sousa S. M. A High-Throughput Screening Assay for Simultaneous Selection of Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Synthase (Dxs) or 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr). J. Biomol. Screen 2011, 16 (3), 303–312. 10.1177/1087057110394845. [DOI] [PubMed] [Google Scholar]
- Brammer L. A.; Smith J. M.; Wades H.; Meyers C. F. 1-Deoxy-D-Xylulose 5-Phosphate Synthase Catalyzes a Novel Random Sequential Mechanism. J. Biol. Chem. 2011, 286 (42), 36522–36531. 10.1074/jbc.M111.259747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierse R. M.; Oerlemans R.; Reddem E. R.; Gawriljuk V. O.; Alhayek A.; Baitinger D.; Jakobi H.; Laber B.; Lange G.; Hirsch A. K. H.; Groves M. R. First Crystal Structures of 1-Deoxy-D-Xylulose 5-Phosphate Synthase (DXPS) from Mycobacterium Tuberculosis Indicate a Distinct Mechanism of Intermediate Stabilization. Sci. Rep. 2022, 12 (1), 7221. 10.1038/s41598-022-11205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H.ggplot2: Elegant Graphics for Data Analysis; Springer Cham, 2016. 10.1007/978-3-319-24277-4. [DOI] [Google Scholar]
- Wickham H.; François R.; Henry L.; Müller K.; Vaughan D.. Dplyr: A Grammar of Data Manipulation. R package version 1.1.4. 2023, https://github.com/tidyverse/dplyr, https://dplyr.tidyverse.org.
- Blighe K.; Rana S.; Lewis M.. EnhancedVolcano Version 1.10.0: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling; R-Package, 2021. 10.18129/B9.bioc.EnhancedVolcano. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.