Abstract
Haemophilus ducreyi, the etiologic agent of chancroid, a genital ulcer disease, produces a cell-associated hemolysin whose role in virulence is not well defined. Hemolysin is encoded by two genes, hhdA and hhdB, which, based on their homology to Serratia marcescens shlA and shlB genes, are believed to encode the hemolysin structural protein and a protein required for secretion and modification of this protein, respectively. In this study, we determined the prevalence and expression of the hemolysin genes in 90 H. ducreyi isolates obtained from diverse geographic locations from 1952 to 1996 and found that all strains contained DNA homologous to the hhdB and hhdA genes. In addition, all strains expressed a hemolytic activity. We also determined that hemolysin is expressed in vivo and is immunogenic, as indicated by the induction of antibodies to hemolysin in both the primate and rabbit disease models as well as in human patients with naturally acquired chancroid. Wild-type strain 35000 and isogenic hemolysin-negative mutants showed no difference in lesion development in the temperature-dependent rabbit model. However, immunization of rabbits with the purified hemolysin protein reduced the recovery of wild-type H. ducreyi, but not hemolysin-negative mutants, from lesions. Our study indicates that hemolysin is a possible candidate for vaccine development due to its immunogenicity, expression in vitro and in vivo by most, if not all, strains, and the effect of immunization on reducing the recovery of viable H. ducreyi in experimental disease in rabbits.
Haemophilus ducreyi is the etiologic agent of chancroid, a sexually transmitted disease characterized by genital ulcers and, in more than 50% of cases, inguinal lymphadenopathy (14, 29). This disease is frequently diagnosed in developing countries, where it is often the most common cause of genital ulcers (47). However, outbreaks of chancroid occur in the United States, particularly in inner cities and among those who exchange sex for drugs or money (14, 29). In Africa, chancroid has been shown to increase the risk of acquiring human immunodeficiency virus infection (6, 38, 53), possibly by creating a portal of entry in its host by disrupting the epithelium and/or by increasing the local concentration of CD4+ cells that are targets for infection by the virus (44).
Chancroidal ulcers contain disintegrating epithelial cells, fibroblasts, and inflammatory cells, including macrophages, polymorphonuclear leukocytes, and lymphocytes, as well as viable H. ducreyi (25). The tissue destruction and the ability to survive in the presence of an inflammatory cell infiltrate are consistent with the production of toxins. Several toxins including a cell-associated hemolysin (35, 50) and a secreted cytotoxin, the cytolethal distending toxin (10, 40), have been identified in H. ducreyi 35000. Other virulence factors include lipooligosaccharide (LOS), which may contribute to ulcer formation by enhancing the migration of inflammatory cells to the lesion site and increasing the resistance of H. ducreyi to phagocytosis, and proteins that allow the organism to acquire heme, a nutritional requirement of this organism (7, 15, 45). As with other organisms, the virulence of H. ducreyi is probably multifactorial and dependent on the presence, relative expression, and cell range of several different virulence factors.
The H. ducreyi hemolysin has been cloned and found to be homologous to the pore-forming, calcium-independent hemolysins of Serratia marcescens, Proteus mirabilis, and Edwardsiella tarda (22, 35, 50). The S. marcescens hemolysin requires at least two genes for expression, shlA, which encodes the structural protein for hemolysin, and shlB, which is required for activation and secretion of ShlA (32). Similarly, the H. ducreyi hemolysin is encoded by two genes, termed hhdA and hhdB (35), which presumably have functions analogous to those of the homologous S. marcescens genes. The cell types sensitive to H. ducreyi hemolysin include human epithelial cells, fibroblasts, macrophages, T lymphocytes, and B lymphocytes (1, 34, 54). This target cell range may enable H. ducreyi to cause the tissue destruction characteristic of chancroidal ulcers as well as inhibit the inflammatory and specific immune responses to this organism. H. ducreyi 35000 is able to invade epithelial cells (16, 48), and hemolysin has been shown to enhance invasion by this organism (54), suggesting another role for hemolysin in virulence.
In this study, we surveyed H. ducreyi isolates in our international strain collection for the presence of genes homologous to hhdA and hhdB and for expression of hemolytic activity. We determined that hemolysin is immunogenic in both animal models and chancroid patients. We also evaluated the effectiveness of immunization with purified hemolysin in attenuating ulcer formation and growth of H. ducreyi in rabbits challenged with this organism.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Tables 1 and 2. The construction of strain 35000ΔAPC is described elsewhere (54). This strain is nonhemolytic due to a deletion in the internal PstI site of the hhdA gene, replaced by a cat (chloramphenicol acetyltransferase) cassette as pictured in Fig. 1.
TABLE 1.
H. ducreyi stock cultures and clinical isolates
| Isolate no. | Strain | Geographic site and date of isolation | Reference and/or sourcea |
|---|---|---|---|
| 1 | CIP 542 | Hanoi, Vietnam, 1954b | 24 |
| 2 | 35000 | Winnipeg, Manitoba, Canada, 1976b | 17 |
| 3 | CHIA | VDRL, Atlanta, Ga., 1958, 1953cd | 21, 23 |
| 4 | 2151A | VDRL, Atlanta, Ga., 1962d | 21 |
| 5 | CHIA-A | VDRL, Atlanta, Ga., 1955d | 21 |
| 6 | HD105 | VDRL, Atlanta, Ga., 1962d | 21 |
| 7 | HD167 | VDRL, Atlanta, Ga., 1958d | 21 |
| 8 | HD105 | VDRL, Atlanta, Ga., 1962d | 21 |
| 9 | HD166 | VDRL, Atlanta, Ga., 1958d | 21 |
| 10 | HD167 | VDRL, Atlanta, Ga., 1958d | 21 |
| 11 | HD141 | VDRL, Atlanta, Ga., 1958d | 21 |
| 12 | HD158 | VDRL, Atlanta, Ga., 1958d | 21 |
| 13 | HD109 | VDRL, Atlanta, Ga., 1958d | 21 |
| 14 | HD125 | VDRL, Atlanta, Ga., 1958d | 21 |
| 15 | CH-10-9 | VDRL, Atlanta, Ga., 1954d | 21 |
| 16 | 125Law | VDRL, Atlanta, Ga., 1962d | 21 |
| 17–32 | NOHDf | New Orleans, La., 1989 to 1992 | 21 |
| 33–37 | HMC49–53g | Jackson, Miss., 1994 and 1995 | 21 |
| 38 | CIP A75 | Lister Institute, London, England, 1952b | Leslie Slaney 31 |
| 39 | CIP A77 | Lister Institute, London, England, 1952b | Leslie Slaney 31 |
| 40 | C148 | Kenya, 1984b | 31 |
| 41 | 409 | Kenya, 1984h | 31 |
| 42 | V-1157 | Seattle, Wash., 1979 | 46 |
| 43 | V-1168 | Seattle, Wash., 1980 | 46 |
| 44 | V-1169 | Seattle, Wash., 1980 | 46 |
| 45 | CF101 | Seattle, Wash., 1981 | 48 |
| 46 | HMC24 | Seattle, Wash., 1981 | HMCi stock collection |
| 47 | HMC25 | Seattle, Wash., 1981 | HMC stock collection |
| 48 | HMC47 | Seattle, Wash., USA, 1995 | 21 |
| 49 | HMC86 | Seattle, Wash., 1992 | HMC stock collection |
| 50 | HMC87 | Seattle, Wash., 1995 | HMC stock collection |
| 51 | HMC88 | Seattle, Wash., 1995 | 21 |
| 52 | HMC89 | Seattle, Wash., 1992 | 21 |
| 53 | LA225 | Los Angeles, Calif., 1982 | 48 |
| 54 | LA228Rj | Los Angeles, Calif., 1982 | 48 |
| 55 | V149/91 | Rwanda, 1991 | 48 |
| 56 | V-180 | Rwanda, 1991 | 48 |
| 57 | HMC68 | Rwanda, 1991 | Peter Piot |
| 58 | HMC37 | England, 1982h | G. R. Kinghorn 28 |
| 59 | HMC38 | England, 1982h | G. R. Kinghorn 28 |
| 60 | HMC39 | England, 1982h | G. R. Kinghorn 28 |
| 61 | HMC48 | Bahamas, 1995 | HMC stock collection |
| 62 | HMC54 | Dominican Republic, 1995 | Margurita Rosada de Quinones |
| 63 | HMC55 | Dominican Republic, 1995 | Margurita Rosada de Quinones |
| 64 | HMC56 | Dominican Republic, 1995 | Margurita Rosada de Quinones |
| 65 | HMC91 | Raleigh, N.C., 1996 | Joan Knapp |
| 66 | HMC 57k | San Francisco, Calif., 1989 | Stephen Morse |
| 67 | HMC59l | San Francisco, Calif., 1989 | Stephen Morse |
| 68 | HMC60 | Florida, 1989 | 21 |
| 69 | HMC61 | Kenya, 1984 | 21 |
| 70 | HMC64 | Kenya, 1984 | 21 |
| 71 | HMC65 | Kenya, 1984 | 21 |
| 72 | HMC62 | Thailand, 1984 | 21 |
| 73 | CH2 | Thailand, 1985 | 48 |
| 74 | CH4 | Thailand, 1985 | Leslie Slaney |
| 75 | CH9 | Thailand, 1985 | Leslie Slaney |
| 76 | CH14 | Thailand, 1985 | Leslie Slaney |
| 77 | CH5 | Thailand, 1985 | Leslie Slaney |
| 78 | CH7 | Thailand, 1985 | Leslie Slaney |
| 79 | ML078 | Kenya, 1985 | Leslie Slaney |
| 80 | ML314 | Kenya, 1985 | Leslie Slaney |
| 81 | ML067 | Kenya, 1985 | Leslie Slaney |
| 82 | ML300 | Kenya, 1985 | Leslie Slaney |
| 83 | 78118 | Winnipeg, Manitoba, Canada, 1975–1977 | Alan Ronald 4 |
| 84 | HMC46 | Kenya, 1995 | Claire Stevens |
| 85 | 54201 | Winnipeg, Manitoba, Canada, 1975–1977 | Alan Ronald 4 |
| 86 | ATCC 27721m | Unknown | ATCC, Manassas, Va. |
| 87 | ATCC 27722 | Unknown | ATCC, Manassas, Va. |
| 88 | 6V | Atlanta, Ga. | Michelle Alfa 3 |
| 89 | 35199 | Winnipeg, Manitoba, Canada | Michelle Alfa 3 |
| 90 | 78226 | Winnipeg, Manitoba, Canada, 1975–1977 | Alan Ronald 4 |
Affiliations: Leslie Slaney, University of Manitoba, Winnipeg, Manitoba, Canada; Peter Piot, Institute of Tropical Medicine, Antwerp, Belgium; G. R. Kinghorn, Sheffield, England; Margurita Rosada de Quinones, Instituto Dermatologico, Santo Domingo, Dominican Republic; Joan Knapp, CDC, Atlanta, Ga.; Stephen Morse, CDC; Alan Ronald, University of Manitoba; Claire Stevens, University of Washington; Michelle Alfa, Boniface General Hospital, Winnipeg, Manitoba, Canada.
Date strains were deposited in strain collection at the Institut Pasteur and geographic site of isolation 4a.
Exact date of isolation confirmed by personal communication with Leslie Slaney.
Date and strain designation as indicated on lyophile stored at the CDC. The strains were lyophilized at the VDRL (Venereal Disease Reference Laboratory), which later became the CDC.
The strain was isolated from bubo pus in 1953, and the lyophile at the VDRL was labeled with the 1958 date.
Sixteen strains: NOHD37, NOHD53, NOHD60, NOHD47, NOHD78, NOHD41, NOHD168, NOHD214, NOHD218, NOHDAZ132, NOHDAZ145, NOHDAZ186, NOHDAZ203, NOHDAZ215, NOHD312, and NOHDSR138.
Five strains: HMC49, HMC50, HMC51, HMC52, and HMC53.
Date strain cited in referenced publication; date of isolation unknown.
HMC, Harborview Medical Center.
Rabbit-passaged derivative of LA228.
Cited as 140142 in CDC stock collection.
Cited as HD192 in CDC stock collection.
Grows only anerobically on S-agar plates.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or referencea |
|---|---|---|
| Strains | ||
| A. pleuropneumoniae | ATCC 27088 | Arnold Smith |
| E. coli DH5α | Host for cloning experiments | GibcoBRL, Gaithersburg, Md. |
| E. coli BL21(DE3) | Host for protein expression | Novagen, Madison, Wis. |
| H. aphrophilus | ATCC 33389 | Arnold Smith |
| H. ducreyi 35000-TcA | Strain 35000 with Tn916 inserted in hhdB, nonhemolytic | 50 |
| H. ducreyi 35000-KmA | Strain 35000 with Tn1545-Δ3 inserted in hhdB, nonhemolytic | 50 |
| H. ducreyi 35000ΔAPC | Strain 35000 with cat cassette in hhdA gene, nonhemolytic | 54 |
| H. haemoglobinophilus | ATCC 19416 | ATCC, Manassas, Va. |
| H. haemolyticus | ATCC 33390 | Arnold Smith |
| H. influenzae | ATCC 33911 | Arnold Smith |
| H. influenzae Rd | Marilyn Roberts | |
| H. parainfluenzae | ATCC 33392 | Arnold Smith |
| H. paraphrophilus | ATCC 29237 | Arnold Smith |
| H. segmis | ATCC 33393 | Arnold Smith |
| T. equigenitalis | ATCC 35865 | Arnold Smith |
| Plasmids | ||
| pUC19 | E. coli cloning vector, Apr | GibcoBRL |
| pCR2.1 | TA cloning vector for cloning PCR products, Ampr Kanr | Invitrogen, San Diego, Calif. |
| pET24+ | Expression plasmid with C-terminal His tag sequence and inducible T7 promoter, no translation initiation signals, Kanr | Novagen |
| pET24a+ | Expression plasmid with C-terminal His tag sequence and inducible T7 promoter, Kanr | Novagen |
| pBCKS | Cmr plasmid derived from pUC19 with KS multiple cloning site | Stratagene, La Jolla, Calif. |
| pPT384-ETa | hhdBA genes cloned into pET24a+ SstI-SalI | This study |
| pLSSK | Shuttle vector with ori, sulA, and strA genes from pLS88 and multiple cloning sites and lacZ gene from pBluescript SK | 54 |
| pPT384 | hhdBA gene region in pTZ18 in same orientation as lac promoter | 50 |
| pPT376BCKS | 5.8-kb BglII fragment containing part of hhdB and all of hhdA cloned into pBCKS SstI-SalI | 50 |
| pETBABN | hhdBA genes cloned into pET24+ BamHI-SalI | This study |
| pLSBAHis+ | hhdBA genes cloned into pLSSK in same orientation as lac promoter. His tag sequence from pET24+ is fused to 3′ end of hhdA | This study |
Affiliations: Arnold Smith, University of Missouri, Columbia; Marilyn Roberts, University of Washington.
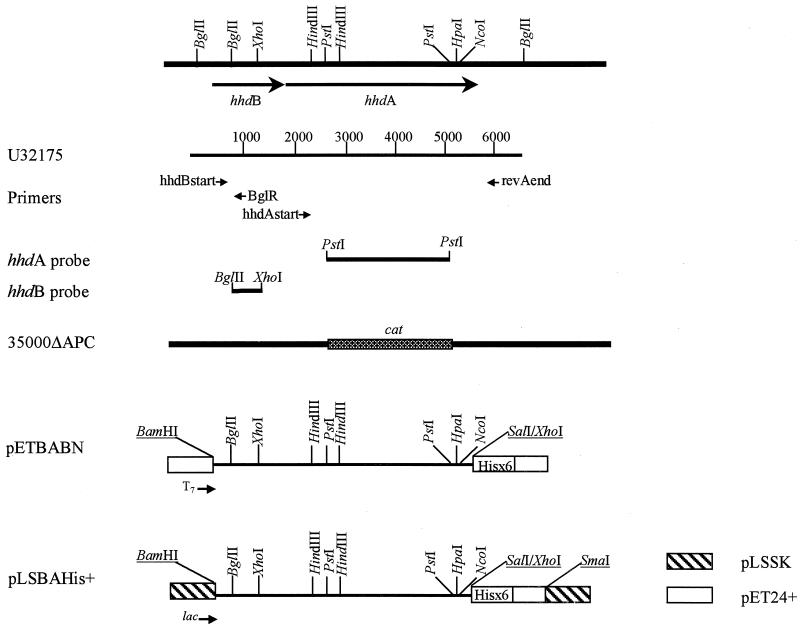
FIG. 1.
Diagram of H. ducreyi hemolysin gene region from strain 35000, the hemolysin-negative derivative of strain 35000, constructs containing the hhdA and hhdB genes, and locations of primers and probes used in this study. Restriction sites used for cloning that were added as a result of PCR amplification or from multiple cloning sites are underlined. The cat cassette, used to replace the PstI fragment internal to the hhdA gene in 35000ΔAPC, is necessarily not drawn to scale. U32175 is the GenBank accession number for hhdB and hhdA (35); numbers on the diagram indicate base pairs.
Ninety stock cultures and clinical isolates were obtained from diverse geographic locations between 1952 and 1996 (Table 1), and all are maintained in the stock culture collection at Harborview Medical Center. All isolates used in this study were identified as H. ducreyi based on the following characteristics: pleomorphic gram-negative rods by Gram stain reaction, characteristic colony morphology, negative reaction in the porphryin test (Innovative Diagnostic Systems, L.P., Norcross, Ga.), and a positive reaction in the taxonomic spot blot test (51). To avoid changes that might appear with repeated subculturing, isolates were always revived from frozen stock solutions and subcultured only once before use in subsequent experiments.
H. ducreyi 35000 was grown on chocolate agar, which consisted of GC agar base (Difco Laboratories, Detroit, Mich.) with 1% hemoglobin (BBL Microbiology Systems, Cockeysville, Md.) and 1% XV factor enrichment (PML Microbiologicals, Tualatin, Oreg.) added after autoclaving. All other H. ducreyi clinical isolates were grown on S-agar plates, which are chocolate agar plates supplemented with 5% fetal bovine serum (21). Interestingly, strain ATCC 27721 (obtained from the American Type Culture Collection [ATCC]) grew only anaerobically after 3 days incubation at 35°C on S-agar plates incubated with a GasPak H2 and CO2 generator envelope (BBL) in a GasPak jar with a catalyst; we were unable to grow this strain on S-agar plates in a candle jar, similar to the other H. ducreyi strains in our collection, or in a microaerophilic environment generated by a GasPak H2 and CO2 generator envelope in GasPak jars with the catalyst removed (42). However, strain ATCC 27721 was also able to grow on S-agar plates incubated for 4 days at 37°C in a Bactron anaerobic/environmental chamber (Sheldon Manufacturing Inc., Cornelius, Oreg.) with an atmosphere of 5% H2, 5% CO2, and 90% N2, confirming its anaerobic growth requirement.
H. ducreyi 35000-TcA, 35000-KmA, and 35000ΔAPC were routinely grown on chocolate agar without antibiotic selection and maintained their nonhemolytic phenotype under these conditions. Actinobacillus pleuropneumoniae, Taylorella equigenitalis, and Haemophilus species other than H. ducreyi (Table 2) were cultivated on chocolate agar plates. Horse blood agar plates (HBAPs), the plate media used for evaluating hemolysis of all isolates, are bilayer plates containing horse blood as described elsewhere (50). Chocolate agar plates containing 100 μg of streptomycin per ml were used to select for strain 35000 containing pLSSK and its derivatives. For quantitation of hemolytic activity, H. ducreyi isolates were grown overnight in Hd broth (51) and then diluted 1:5 and grown for 4 to 6 h to achieve maximal hemolysin expression as described previously (50). For purification of the His-tagged hemolysin, H. ducreyi 35000-KmA with pLSBAHis+ was grown on either chocolate agar or charcoal agar plates (51) with 100 μg of streptomycin per ml for 24 to 48 h.
Escherichia coli strains (Table 2) were grown in L broth and L agar (27). When appropriate, the following antibiotic concentrations (in micrograms per milliliter) were added to the media: ampicillin, 100; kanamycin, 30; chloramphenicol, 30; and streptomycin, 100.
DNA manipulation.
Plasmid DNA was extracted by using a Wizard Miniprep kit (Promega, Madison, Wis.). Standard techniques were used for (27) restriction digests, ligations, agarose gels, and Southern blots. PCR amplification was accomplished in a Perkin-Elmer 240 thermocycler (Perkin-Elmer, Branchburg, N.J.), using primers manufactured by GibcoBRL (Gaithersburg, Md.) and listed in Table 3. E. coli was made competent for the uptake of DNA by the rubidium chloride method (18). Electroporation of H. ducreyi was accomplished by a modification of the method of Hansen et al. (19) as described elsewhere (50), using selection with 100 μg of streptomycin per ml for pLSSK derivatives.
TABLE 3.
Oligonucleotide primers used in this study
| Name | Target | Positiona | Strand | Sequence 5′-3′b | Comment |
|---|---|---|---|---|---|
| hhdBstart | hhdBA | 209–237 (U32175) | + | gga tccaag gag GAT AAT ATG AGA AGA TGT GAA ATT ATA AC | BamHI site and RBS added |
| revAend | hhdBA | 5421–5398 (U32175) | − | gtc gac TCG AAT GGC CAT CTT AGC ATC GAC | SalI added |
| hhdAstart | hhdA | 1889–1909 (U32175) | + | gga tccaag gag ATA CAT ATA TGA AAA AAT GGA AGC C | BamHI site and RBS added |
| BglR | hhdBA | 785–764 (U32175) | − | CCA AAT CTA AAT GAA TAC CGC C | |
| 1545-1 | Tn1545-Δ3 | Left end of Tn1545-Δ3 (5) | − | GAT AAA GTG TGA TAA GTC CAG | |
| 1545-2 | Tn1545-Δ3 | Right end of Tn1545-Δ3 (5) | − | CAC ATA GAA TAA GGC TTT ACG | |
| Tn916 | Tn916 | Right end of Tn916 (9) | − | GAG TGG TTT TGA CCT TGA |
Position on target sequences. Numbers are based on given reference or GenBank citation.
Homologous sequences are in uppercase, restriction sites are underlined, and ribosome binding sites (RBS) are double underlined.
Plasmid constructions.
Plasmids used in this study are listed in Table 2 and pictured in Fig. 1. Plasmids that are intermediate constructions are described in the text only. Plasmid pPT384-Eta is derived from pPT384 (50) and consists of the 7.7-kb insert encoding both hhdB and hhdA cloned into pET24a+ in the same orientation as the T7 promoter, using SstI and SalI. Plasmid pPT376BCKS is derived from pPT376 (50) and consists of the 5.8-kb insert from this plasmid cloned into pBCKS by using SstI and SalI. This 5.8-kb fragment encodes all of hhdA but only a part of hhdB and is in the orientation opposite the lac promoter of the vector.
Plasmid pLSBAHis+, containing hhdB and hhdA fused to a 3′ His tag sequence, was constructed in several steps (Table 2 and Fig. 1). First, a 5.2-kb PCR product containing the hhdA and hhdB genes was amplified from chromosomal DNA isolated from strain 35000 with primers hhdBstart and revAend (Table 3) under the following reaction conditions: 50 mM Tris, (pH 9.2), 16 mM (NH4)2SO4, 5 mM MgCl2, 500 μM deoxynucleoside triphosphates, 0.3 μM primers, and 0.5 μl Long Extend polymerase (Boehringer Mannheim, Indianapolis, Ind.). The cycling conditions were an initial denaturation at 92°C for 2 min; 30 cycles of 92°C for 45 s, 63°C for 1 min, and 68°C for 5 min; and a final 7-min extension at 68°C. This 5.2-kb PCR product was cloned into pCR2.1 by using a TA cloning kit (Invitrogen), generating pCRBA, and transformed into E. coli DH5α. To confirm that no errors were introduced by PCR, the insert of pCRBA was sequenced from the vector into the hemolysin sequence to the BglII site at 686 bp from the 5′ end and to the NcoI site at 5,185 bp from the 3′ end (numbers based on hemolysin sequence assigned GenBank accession no. U32175). The insert in pCRBA was then cloned into pUC19 in the orientation opposite the lac promoter, using BamHI and SalI, generating pBA. To ensure that the middle segment of pBA contained no errors generated by PCR, the 4,499-bp sequence between the BglII and NcoI sites in pBA was replaced with the comparable sequence in pPT384, after cutting both plasmids with BglII and NcoI and isolating the appropriate fragments. The resulting plasmid, pBABN, was digested with BamHI and SalI, and the fragment containing the hemolysin genes was cloned into the BamHI and XhoI sites of pET24+, producing pETBABN, which contained an in-frame fusion of the 3′ end of hhdA with the vector’s His tag sequence (Fig. 1).
For expression in H. ducreyi, hemolysin genes with the His tag fusion from pETBABN were cloned into the H. ducreyi shuttle vector, pLSSK. To accomplish this, the BamHI-SmaI fragment of pETBABN containing the hhdB and hhdA genes along with vector sequence 3′ of the His tag, was cloned into pLSSK similarly cut with BamHI and SmaI. This plasmid, pLSBAHis+, was isolated by Wizard minipreps (Promega) and transformed into H. ducreyi 35000-KmA by electroporation.
Dot blot hybridization.
For detection of hemolysin genes in different isolates of H. ducreyi bacteria were incubated for 24 h on S-agar plates and then suspended to the approximate turbidity of a 1.0 McFarland standard (33). Ten microliters of this suspension was mixed with 200 μl of a 1.5 M NaCl–0.5 M NaOH solution and incubated for 30 min at room temperature to lyse the cells and denature the DNA; then the entire sample was applied to Maximum Strength Nytran+ filters (Schleicher & Schuell, Keene, N.H.) with a vacuum dot blot apparatus. After drying for 2 h at 80°C, the blots were hybridized to DNA probes radiolabeled with 32P by using a random primer labeling kit (GibcoBRL). Hybridization was accomplished as described previously (30), using high-stringency conditions (50% formamide, wash at 65°C).
The three probes used for these assays were whole-cell DNA from CIP 542, the H. ducreyi type strain, a probe internal to hhdA, and a probe internal to hhdB. Whole-cell DNA from CIP 542 was prepared by standard techniques (41). The hhdA probe was the 2,574-bp PstI fragment internal to the hhdA gene (Fig. 1) purified from pPT376BCKS. The hhdB probe was the 482-bp BglII-XhoI (in the hhdB gene [Fig. 1]) fragment isolated from pPT376BCKS by SstI (in the polylinker of pBCKS)-XhoI digestion and gel purification. Radiolabeled gel-purified fragments were tested with dot blots of E. coli DH5α(pBCKS) and showed minimal hybridization, indicating that the probe DNA contained minimal cross-reacting vector DNA.
Diagnostic PCR.
To determine the presence and size of the hemolysin genes as well as to confirm the identity of strains 6V, 78226, 35199, and ATCC 27721 as H. ducreyi, diagnostic PCRs were performed. For each strain, bacteria were removed from one chocolate plate with a sterile swab and suspended in sample transport medium (Roche Diagnostic Systems, Branchburg, N.J.). An equal volume of sample diluent (Roche Diagnostic Systems) was added; the suspension vortexed and then incubated at room temperature for 10 min. Fifty microliters of this sample was added to the PCR mixture of a final volume of 100 μl. H. ducreyi PCR, which amplifies a product unique to H. ducreyi, was performed as described elsewhere (47).
Amplification of hhdA, which resulted in a 3,532-bp product, was accomplished with primers hhdAstart and revAend (Table 3) with the conditions used for plasmid constructions (see above). In addition, hhdB was amplified with 0.3 μM primers hhdBstart and BglR (Table 3), 200 μM each deoxynucleoside triphosphate, 3.1 mM MgCl2, 1× Taq buffer (Promega), and 1 μl of Taq polymerase (Promega) with the same cycling conditions as for hhdA. A negative control included for all of these amplifications was a sample processed from an uninoculated chocolate plate incubated in the same candle jar (or anaerobe jar in the case of strain ATCC 27721) as the strains studied and treated with sample transport medium and sample diluent in parallel with the bacterial samples.
Purification of hemolysin.
His-tagged hemolysin was purified from strain 35000-KmA(pLSBAHis+) in parallel with a mock purification of 35000-KmA(pLSSK). Bacteria were removed from the plate with a cotton swab, suspended in cell lysate buffer (6 M urea, 10 mM Tris, 100 mM NaH2PO4 [pH 8.0]), and then mixed for 1 to 2 h at room temperature on a gyratory platform shaker (New Brunswick Scientific, Edison, N.J.). Cell debris was removed by centrifugation at 5,000 × g, and the supernatant, (cell lysate) was applied to Ni-nitrilotriacetic acid (NTA) resin (Qiagen, Santa Clarita, Calif.). The resin was washed with 6 M urea–10 mM Tris–100 mM NaH2PO4 (pH 6.3) until the optical density at 280 nm (OD280) of the eluate was ≤0.01. The His-tagged hemolysin was eluted with 5 ml of 6 M urea–10 mM Tris–100 mM NaH2PO4 (pH 5.8) and 5 ml of 6 M urea–10 mM Tris–100 mM NaH2PO4 (pH 4.5), and 1.0-ml fractions were collected and monitored by OD280. Fractions containing protein were combined, and protein concentrations determined with the Bradford assay (Bio-Rad, Hercules, Calif.). Bacteria from 5 to 10 plates yielded approximately 0.1 to 1 mg of purified protein. Purified hemolysin was used as antigen on Western blots for testing human, primate, and rabbit sera, to immunize rabbits for generation of polyclonal antibodies, and in rabbit challenge experiments.
Immunization with hemolysin and production of polyclonal rabbit serum to His-tagged hemolysin.
His-tagged hemolysin purified by Ni-NTA chromatography as described above was concentrated and desalted with 10-kDa Centriprep or 50-kDa Centricon concentrators (Amicon, Beverly, Mass.) as instructed by the manufacturer. The buffer was changed to 10 mM HEPES (pH 7.4)–0.85% NaCl. The protein concentration was determined by the Bio-Rad assay, approximately 100 to 150 μg of protein was combined with MPL (monophosphoryl lipid A from Streptomyces minnesota)-TDM (synthetic trehalose dicorynomycolate)-CWS (cell wall of mycobacterium) adjuvant (Sigma, St. Louis, Mo.) and injected into New Zealand White rabbits at intradermal, intramuscular, intraperitoneal, and subcutaneous sites according to the manufacturer’s protocol. Three rabbits were immunized with identical injections and were boosted with identical injections at 4 and 8 weeks. Five milliliters of blood was collected before initial immunization and at 4, 8, and 10 weeks postinjection. Serum was then collected and frozen in aliquots.
Animal and human sera.
Sera from primates previously infected with H. ducreyi has been described elsewhere (49). Pooled rabbit sera, a gift from Eric Hansen (University of Texas, Dallas), was obtained from two animals infected intradermally with H. ducreyi 35000 three times over a 3-month period (18a). Normal rabbit serum from uninfected rabbits was also obtained from Eric Hansen. Human sera, a gift from Chen-Yen Chen (Centers for Disease Control and Prevention [CDC], Atlanta, Ga.), was obtained from patients enrolled in a study of a chancroid outbreak in Jackson, Miss. (8, 29). Among these patients, two groups of sera were selected for testing based on their reaction in the absorption enzyme immune assay (EIA) and multiplex PCR as previously determined (8). One group (five cases) was positive for H. ducreyi infection by both assays. The second group (five controls) was negative for H. ducreyi infection by both assays. Of the controls, two were positive for Treponema pallidum and three were positive for herpes simplex virus.
SDS-PAGE and Western analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8% polyacrylamide gels as described previously (26) and visualized with Coomassie Blue GelCode reagent (Pierce, Rockport, Ill.). For Western blot analysis, proteins were transferred to nitrocellulose (Schleicher & Schuell) in 0.5× Towbin buffer with 20% methanol and 0.1% SDS (52) with a transblot apparatus (Novex, San Diego, Calif.) for 1.5 h at 22 V. The membranes were stained with 0.05% Ponceau S in 5% acetic acid to mark lanes and standards (Mark 12; Novex) and then destained with 1× TTBS (0.1% Tween 20, 20 mM Tris [pH 7.5], 137 mM NaCl). The nitrocellulose was then incubated with 5% nonfat dry milk in 1× TTBS overnight at 4°C, incubated with primary serum diluted 1:1,000 in 1× TTBS for 2 h at room temperature, washed in 1× TTBS, and then incubated with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Sigma) or goat anti-human immunoglobulin G (used with both monkey and human sera; Sigma) diluted 1:30,000 in 1× TTBS. Immunoreactive proteins were detected after incubation with the colorimetric substrates nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as instructed by the manufacturer (Boehringer Mannheim). The dried nitrocellulose filters were scanned with a Microtek (Redondo Beach, Calif.) ScanWizard, and the images were analyzed with NIH Image software (National Institutes of Health Bethesda, Md.) to determine the intensity of the immunoreactive bands. Because the intensity value is a ratio of the pixel density based on a 256-pixel gray scale, no units are given.
Hemolysin assays.
Hemolysin activity was assayed by detection of released hemoglobin from horse erythrocytes at OD540. Erythrocytes were washed in 0.85% NaCl–10 mM CaCl2 until the supernatant was clear. Hemolysin eluted from the Ni-NTA column was adjusted to neutral pH with 1 M Tris (pH 8.0) and serially diluted twofold in assay buffer (0.85% NaCl, 10 mM Tris [pH 7.5]) in microtiter plates, followed by addition of a 1% solution of erythrocytes and incubation at 37°C for 1 h. After incubation, unlysed erythrocytes were removed by centrifugation and the OD540 of the resulting solution was measured on a microtiter plate reader (Titertek Multiskan MC; Flow Laboratories, Inc., McLean, Va). Controls of 100 and 0% lysis were determined in similar wells containing erythrocytes in distilled water and assay buffer, respectively. Included as a control was a blank purification from H. ducreyi 35000-KmA(pLSSK).
Whole H. ducreyi cells assayed for hemolytic activity were washed and resuspended in 0.85% NaCl–10 mM CaCl2 and assayed in this same buffer. Hemolytic values for whole cells were calculated from the average of duplicate samples as described by Wood et al. (54).
Rabbit experiments.
For immunization experiments and testing the virulence of H. ducreyi 35000 compared to that of strain 35000ΔAPC, the temperature-dependent rabbit model was used as described elsewhere (39), with slight modifications. Briefly, the backs of rabbits were shaved and then inoculated with 104 to 107 CFU of H. ducreyi 35000 and 35000ΔAPC, with two inoculations per dilution. Bacteria used for inoculation were scraped from chocolate plates and suspended in inoculation buffer (10 mM Tris [pH 7.5], 0.85% NaCl) and then adjusted with inoculation buffer to obtain a similar OD600. The inoculum size was further confirmed by plating serial 10-fold dilutions on chocolate agar and did not differ significantly between strains 35000 and 35000ΔAPC in each experiment. Each rabbit was inoculated with both strains 35000 and 35000ΔAPC, housed at 15 to 17°C, and scored by an observer blinded to the inoculum 4 days postinoculation by the criteria of Purcell et al. (39): 0 = no change; 1 = erythema; 2 = induration; 3 = nodule; 4 = necrosis. In these experiments, three rabbits immunized with hemolysin (see above) and three naive rabbits were injected intradermally with serial 10-fold dilutions of 104 to 107 CFU of H. ducreyi 35000 and 35000ΔAPC. At day 4, the lesions were scored by an observer blinded to the inoculum and cultured by aspiration or biopsy. If aspirated, 100 μl of inoculation buffer was injected into the ulcer, aspirated back into the same syringe, and then plated in entirety on chocolate agar. Each syringe was then rinsed with 100 μl of inoculation buffer, which was used to inoculate the same plate as the original specimen. If biopsied, 3-mm (experiment 3) or 4-mm (experiment 2) punch biopsies were collected into 200 μl of inoculation buffer and vortexed vigorously, and then the entire 200 μl volume was plated.
Statistics.
The numbers of lesions with viable bacteria were analyzed by the chi-square test with Excel (Microsoft, Redmond, Wash.) to determine P values for differences in expected versus actual values. Sample standard deviations were calculated for all means.
RESULTS
Hemolysin genes and hemolytic activity are present in all H. ducreyi strains tested.
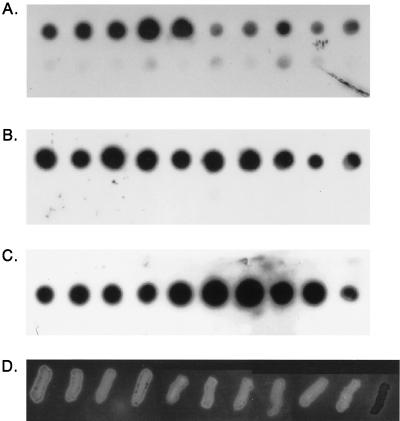
Eighty-seven H. ducreyi isolates (Table 1, isolates 1 to 87) were selected from diverse geographic locations, including the United States, Bahamas, Thailand, Kenya, England, Rwanda, the Dominican Republic, and Canada, as well as diverse dates of isolation, from 1952 to 1996, and were confirmed as H. ducreyi, both by phenotypic tests and by taxonomic spot blots (Fig. 2A and data not shown). The 87 isolates tested hybridized with whole-cell DNA from the H. ducreyi type strain CIP 542, while other Haemophilus and related species (listed in Table 2) did not hybridize, indicating that all isolates were indeed H. ducreyi. All 87 H. ducreyi isolates tested also hybridized with both the hhdB and hhdA probes, indicating that they contained DNA homologous to both genes necessary for expression of active hemolysin. Representative spot blots of DNA from 10 H. ducreyi strains as well as DNA from other species probed with whole-cell DNA (Fig. 2A), the hhdB gene (Fig. 2B), and the hhdA gene (Fig. 2C) are presented in Fig. 2.
FIG. 2.
Identification of H. ducreyi isolates, detection of homology to hemolysin genes, and phenotypic expression of hemolysin in geographically and temporally diverse isolates of H. ducreyi. (A to C) Spot blots of H. ducreyi and other Haemophilus species probed with whole-cell DNA from H. ducreyi type strain CIP 542 (A), the hhdB gene (B), and the hhdA gene (C). Strains and species analyzed: top row, CIP542, 35000, LA228R, CF101, V-1168, V149/91, V-180, HMC56, CHIA, and V-1157; bottom row, H. influenzae Rd, H. parainfluenzae, H. aphrophilus, H. paraphrophilus, H. segmis, A. pleuropneumoniae, H. haemoglobinophiles, T. equigenitalis, H. haemolyticus, and H. influenzae ATCC 33911. (D) Hemolysis of H. ducreyi isolates on HBAPs. Isolates on HBAPs correspond to those in the top rows of panels A to C except strain 35000-KmA, the right-most strain in panel D, which was added as a nonhemolytic control. The photographs of the Southern blots and the HBAP were scanned with a Microtek Scanner III and captured with a Micrografx Picture Publisher (Micrografx, Richardson, Tex.).
All 87 strains of H. ducreyi that could be tested were hemolytic on HBAPs, suggesting that all of these H. ducreyi isolates express the hhdA and hhdB genes. This zone of hemolysis was striking in comparison to the hemolysin-negative H. ducreyi transposon mutant, 35000-KmA (Fig. 2D). One strain of H. ducreyi, ATCC 27721, grew very poorly and only on plates incubated anaerobically; therefore, its hemolytic phenotype could not be tested on HBAPs. However, this strain was positive in the liquid hemolysis assay, its DNA hybridized with the hhdA and hhdB probes, and we were able to amplify both hhdA and hhdB with primers hhdBstart and BglR (hhdB) and hhdAstart and revAend (hhdA). These results confirm that even this obligate anaerobic strain contains both hemolysin genes and expresses hemolysin (data not shown). We also further confirmed its identity as H. ducreyi with diagnostic PCR, and the appropriate-size product was amplified.
Three other strains, 6V, 35199, and 78226 (Table 1, isolates 88 to 90), were also analyzed for the presence of hemolysin genes and activity. These strains were selected for further study because they were previously reported as negative for a fibroblast toxin (3) later identified as the cell-associated hemolysin (1, 34). The identity of these three strains as H. ducreyi was further confirmed by H. ducreyi-specific PCR, and the presence of hemolysin gene sequences was indicated by amplification with primers specific for hhdA and hhdB. Strains 35199, 78226, and 6V were weakly hemolytic on HBAPs and produced 4.2, 13.4, and 15.2%, respectively, of the hemolytic activity of H. ducreyi 35000. This activity was clearly greater than that of 35000ΔAPC, a hemolysin-negative mutant, which was <0.5% in simultaneous assays.
H. ducreyi hemolysin is expressed poorly in E. coli.
The H. ducreyi hemolysin genes confer a hemolytic phenotype on E. coli cultured on HBAPs as reported previously (50), but very little hemolytic activity was detected in the liquid hemolysis assay, under control of the lac promoter in E. coli DH5α(pPT384-18) or the T7 promoter in E. coli BL21(DE3)(pPT384-ETa) induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). These results are consistent with our observation that expression of hemolysin in E. coli is toxic and causes lysis in its E. coli host. Thus, the detection of hemolysis on plates may be due to release of cell-bound hemolysin by lysis of the E. coli colonies on the HBAPs. In contrast, the liquid hemolysin assay, which uses washed intact cells and shorter incubation times, measures only surface-exposed hemolytic activity.
Expression of the hemolysin genes induced with 0.4 mM IPTG in E. coli BL21(DE3)(pETBABN) showed the accumulation of a 65-kDa protein, not the full-size 125-kDa protein, which could be eluted from nickel resin (data not shown). We concluded from these observations that the H. ducreyi hemolysin is degraded in E. coli, possibly because it is not exported, and that the intact 125-kDa protein could not be easily purified from this organism. Thus, we expressed and purified the His-tagged hemolysin in H. ducreyi in subsequent experiments.
His-tagged hemolysin can be purified from H. ducreyi and is active.
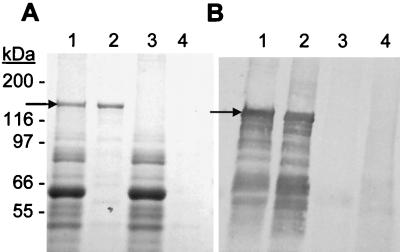
To obtain purified H. ducreyi hemolysin, the His-tagged hemolysin was expressed in H. ducreyi 35000-KmA(pLSBAHis+) and purified with nickel resin. Figure 3A shows the SDS-PAGE profile of purified hemolysin (lane 2, indicated by the arrow) relative to protein profiles of total cell lysates of strain 35000-KmA(pLSBAHis+) (lane 1) and strain 35000-KmA(pLSSK) (lanes 3) and the eluate from a blank purification with 35000-KmA(pLSSK) (lane 4). A 125-kDa protein was purified from strain 35000-KmA(pLSBAHis+) but not from strain 35000-KmA(pLSSK), and the size of this protein is consistent with the predicted molecular weight of HhdA. Figure 3B is a Western blot performed with serum obtained from a rabbit immunized with purified hemolysin and a 125-kDa protein is detected by this polyclonal serum. Numerous smaller protein bands are also detected by the rabbit serum on the immunoblot (Fig. 3B) in extracts of strain 35000-KmA(pLSBAHis+) (lane 1) and in the eluate from the nickel resin (lane 2). However, these immunoreactive bands are not present in the similar samples from strain 35000-KmA(pLSSK) (lanes 3 and 4) and are presumed to be breakdown products of hemolysin.
FIG. 3.
Purification of His-tagged hemolysin from strain 35000-KmA(pLSBAHis+) demonstrated by SDS-PAGE analysis (A) and Western blot analysis with rabbit polyclonal serum against His-tagged hemolysin (B). Lanes: 1, strain 35000-KmA(pLSBAHis+) cell lysate; 2, purified hemolysin from strain 35000-KmA(pLSBAHis+) eluted from Ni-NTA resin; 3, strain 35000-KmA(pLSSK) cell lysate; lane 4, strain 35000-KmA(pLSSK) cell lysate eluted from Ni-NTA resin. Twenty-five micrograms of each cell lysate and equal volumes of Ni column eluates were analyzed on the gel and Western blot. Positions of the H. ducreyi hemolysin protein (arrows) and of the molecular weight protein standards are indicated. Both images were photographed with a Bio-Rad GelDoc 2000 digital camera system equipped with Quantity One 4.0 software (Bio-Rad).
To assess the activity of purified hemolysin, the fraction eluted from the nickel column with the highest A280 was analyzed in a liquid hemolysis assay. The protein concentration of this fraction ranged from 0.1 to 1.0 μg/μl. In these assays, 50% hemolysis was achieved with 0.3 μg of purified hemolysin protein after 1 h of incubation with 1% erythrocytes. No activity was detected in the same fraction from a mock purification with lysate from strain 35000-KmA(pLSSK). This finding further established the identity of the 125-kDa His-tagged protein seen on SDS-PAGE as hemolysin.
Antibodies to hemolysin are present in serum of animals infected with H. ducreyi.
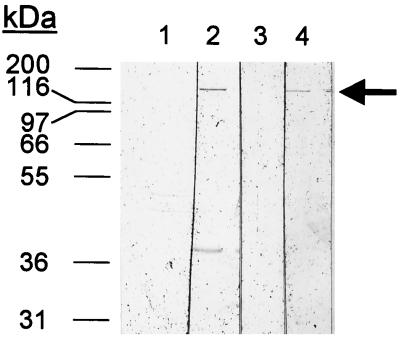
To assess the antibody response to the H. ducreyi hemolysin in animal models of infection, purified hemolysin was reacted with normal and immune sera from rabbits and primates infected with H. ducreyi on Western blots (Fig. 4). In these experiments, rabbits infected repeatedly with H. ducreyi 35000 showed an immune response to hemolysin (lane 2) which was not apparent in the normal (uninfected) sera (Fig. 4, lane 1). This immune response was more pronounced than in rabbits that had been infected with H. ducreyi for only 2 weeks (data not shown). Similarly, primates infected genitally with H. ducreyi CH2 produced antibodies to hemolysin which were detectable in serum obtained at 5 weeks postinfection but not in preimmune serum (Fig. 4, lanes 3 and 4). Similar results were obtained with a second primate infected with strain LA228R (data not shown). A primate injected with heat-killed H. ducreyi produced no antibodies to hemolysin detectable in serum collected 5 weeks postinfection (data not shown).
FIG. 4.
Production of antibodies to hemolysin among rabbits and monkeys infected with H. ducreyi. Each lane contains 200 ng of purified His-tagged hemolysin separated on an 8% polyacrylamide gel, blotted to nitrocellulose, and probed with pooled preimmune rabbit serum (lane 1), serum from rabbits infected three times over 3 months with H. ducreyi 35000 (lane 2), preimmune monkey serum (lane 3), and serum obtained from the same monkey 6 weeks after inoculation of strain CH2 (lane 4).
Chancroid patients have antibodies to hemolysin.
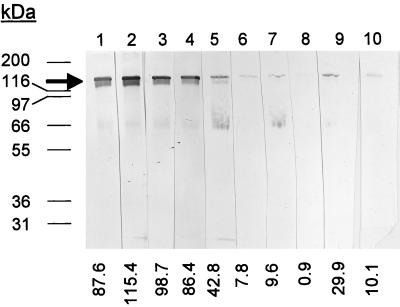
An analysis of the immune response to hemolysin generated during infection was performed with sera from humans with naturally acquired chancroid. In these experiments, sera from human chancroid patients showed a clear reaction to hemolysin compared to sera from patients with genital ulcer disease due to T. pallidum or HSV, who showed a much weaker response (Fig. 5). The intensities of the bands, indicated below the lanes in Fig. 5, clearly differed between the chancroid-positive patients and the controls. For chancroid-positive patients, the range of intensity was 42.8 to 115.4, with a mean of 86.2 ± 26.9. For chancroid-negative patients, the range was 0.9 to 29.9, with a mean of 11.7 ± 10.8. The low level of reactivity on the chancroid-negative patients in this assay may be due either to past chancroidal infection or to infection with a bacterium (such as Proteus or Serratia) that produces related hemolysin proteins that may share cross-reacting epitopes.
FIG. 5.
Production of antibodies to H. ducreyi hemolysin by patients diagnosed with chancroid. Each lane contains 200 ng of purified His-tagged hemolysin separated on an 8% polyacrylamide gel, blotted to nitrocellulose, and probed with human serum from five different chancroid-positive patients (lanes 1 to 5) and from five different chancroid-negative controls (lane 6 to 9). The blots were scanned with a Microtek scanner, and intensities of the bands are shown at the bottom.
Ulcers induced by H. ducreyi 35000 and a hemolysin-deficient hhdA deletion mutant in the temperature-dependent rabbit model are similar.
Lesions induced by strains 35000 and 35000ΔAPC, a nonhemolytic mutant with a deletion in hhdA (54), were compared as part of separate experiments (see below) to assess the protective effect of immunization with hemolysin protein in the rabbit model. We found no significant differences in the severity of lesions induced by strains 35000 and the hemolysin-negative mutant, 35000ΔAPC, in three separate experiments (Table 4, control rabbits). Thus, the mean lesion scores of strain 35000 were 4.0, 3.7, and 3.5 for inocula of 107, 106, and 105 CFU, respectively. In comparison, lesion scores of 3.8, 3.7, and 3.7 were obtained for inocula of 107, 106, and 105 CFU, respectively, for strain 35000ΔAPC in the same unimmunized rabbits.
TABLE 4.
Lesion scores for control and immunized rabbits infected with H. ducreyi 35000 and 35000ΔAPC
| Expt | Strain | Lesion scorea
|
|||||
|---|---|---|---|---|---|---|---|
| Control rabbits
|
Immunized rabbits
|
||||||
| 107b | 106 | 105 | 107 | 106 | 105 | ||
| 1 | 35000 | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 | 3.0 |
| 2 | 35000 | 4.0 | 3.5 | 3.5 | 4.0 | 4.0 | 3.0 |
| 3 | 35000 | 4.0 | 3.5 | 3.0 | 4.0 | 3.5 | 2.5 |
| Mean ± SD | 4.0 ± 0 | 3.7 ± 0.3 | 3.5 ± 0.5 | 4.0 ± 0.0 | 3.8 ± 0.3 | 3.0 ± 0.3 | |
| 1 | 35000ΔAPC | 4.0 | 3.0 | 3.5 | 4.0 | 4.0 | 3.0 |
| 2 | 35000ΔAPC | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 | 3.5 |
| 3 | 35000ΔAPC | 3.5 | 4.0 | 3.5 | 4.0 | 4.0 | 2.5 |
| Mean ± SD | 3.8 ± 0.3 | 3.7 ± 0.6 | 3.7 ± 0.3 | 4.0 ± 0 | 4.0 ± 0.0 | 3.0 ± 0.5 | |
Average score from two lesions per animal. One animal was used in each experiment, for a total of three control and three experimental animals.
Inoculum (CFU).
Similar results were obtained with strains 35000-TcA and 35000-KmA, hemolysin-negative isogenic mutants with transposon insertions in hhdB (data not shown). In these experiments, only naive rabbits were injected, and no significant differences were detected between ulcer scores of wild-type and mutant strains.
Sampling method affects recovery of organisms in lesions induced by H. ducreyi 35000 and 35000ΔAPC in the temperature-dependent rabbit model.
To assess survival of H. ducreyi in the rabbit model, bacteria were recovered from lesions by two methods, aspiration and biopsy. Aspirates were withdrawn from the pustular material in the center of the lesion (experiments 1, 2, and 3), while biopsies were taken from the periphery of the lesions (experiment 2 and 3). In these experiments, strain 35000 was isolated from 64% (9 of 14) and strain 35000ΔAPC was recovered from 15% (2 of 13) of aspirates analyzed from control (unimmunized) rabbits (Table 5). When 10 biopsies were analyzed, the differences between strains 35000 and 35000ΔAPC were less striking: strain 35000 was isolated from 90% of biopsies, while strain 35000ΔAPC was isolated from 70%. When the total number of cultures were analyzed, strain 35000 was recovered from 75% (18 of 24) of lesions induced by 35000 and strain 35000ΔAPC was recovered from 39% (9 of 23) of lesions induced by 35000ΔAPC (P = 0.03).
TABLE 5.
Immunization with hemolysin decreases recovery of H. ducreyi 35000 but not 35000ΔAPC in the temperature-dependent rabbit model
| Expt | Recovery of H. ducreyia
|
|||||
|---|---|---|---|---|---|---|
| 35000
|
35000ΔAPC
|
|||||
| Control rabbits | Immunized rabbits | P | Control rabbits | Immunized rabbits | P | |
| Aspirates | ||||||
| Expt 1 | 4/6b | 0/6b | 0/6b | 0/6b | ||
| Expt 2 | 3/4c | 0/3b | 0/3b | 2/3b | ||
| Expt 3 | 2/4c | 0/4c | 2/4c | 1/4c | ||
| Total | 9/14 | 0/13 | 0.0026 | 2/13 | 3/13 | 0.4795 |
| Biopsies | ||||||
| Expt 2 | 4/4c | 2/3b | 2/4c | 2/3b | ||
| Expt 3 | 5/6b | 1/6b | 5/6c | 5/6b | ||
| Total | 9/10 | 3/9 | 0.0429 | 7/10 | 7/9 | 0.7518 |
| Totald | 18/24 | 3/22 | 0.0004 | 9/23 | 10/22 | 0.6942 |
Number of lesions with viable organisms/number of lesions sampled.
Lesions sampled were from 105-, 106-, and 107-CFU inocula.
Lesions sampled were from 104-, 105-, 106-, and 107-CFU inocula.
Aspirates and biopsies combined.
Immunization with hemolysin protein affects recovery of organisms but not lesion development in the temperature-dependent rabbit model.
To assess the effect of immunization with hemolysin on lesions induced by H. ducreyi in the temperature-dependent rabbit model, we challenged rabbits with strains 35000 and 35000ΔAPC and monitored lesion development and the ability to recover viable organisms in the resulting lesions, both from control rabbits and from rabbits immunized with the purified His-tagged hemolysin protein.
(i) Immunization with hemolysin does not affect lesion development.
There was no significant difference between lesions induced by strain 35000 in the immunized and control animals and between lesions induced by strain 35000ΔAPC in the immunized and control animals (Table 4). The average ulcer scores for inocula of 107, 106, and 105 CFU were 4.0, 3.7, and 3.6 for strain 35000 in control rabbits and 4.0, 3.8, and 3.0 for strain 35000 in immunized rabbits. Similar lesion scores were observed for strain 35000ΔAPC (Table 4).
(ii) Immunization with hemolysin decreases survival of wild-type organisms in lesions.
We analyzed the ability of strains 35000 and 35000ΔAPC to survive in lesions in both immunized and control rabbits (Table 5). Again these experiments were performed with aspirates of pustular material withdrawn from the center of the lesions (experiment 1, 2, and 3) and from biopsies obtained from the sides of the lesions (experiments 2 and 3). We found that immunization with hemolysin affected the recovery rate of viable bacteria from lesions sampled by both biopsy and aspiration. Thus, viable H. ducreyi 35000 was recovered from aspirates from 64% (9 of 14) of the lesions from unimmunized rabbits but from none of the 13 lesions from immunized rabbits. Similarly, viable H. ducreyi 35000 was recovered from 90% (9 of 10) of biopsies obtained from unimmunized rabbits but from only 33% (3 of 9) of biopsies from immunized rabbits. These differences in recovery of strain 35000 from immunized and unimmunized animals were significant (P = 0.003 for aspirates, P = 0.04 for biopsies; Table 5).
Strain 35000ΔAPC, which produces no hemolysin protein, was expected to be unaffected by antibodies specific to HhdA and was used as an internal control in the unimmunized and immunized rabbits. We found in these experiments that there were no significant differences in the recovery of strain 35000ΔAPC from the unimmunized rabbits compared to the immunized rabbits. Thus, strain 35000ΔAPC was recovered by aspiration from 15% (2 of 13) and 23% (3 of 13) of the lesions in the control and immunized rabbits, respectively, and by biopsy, 70% (7 of 10) and 78% (7 of 9), respectively.
Combining the results from both aspirates and biopsies, we found a significant difference in recovery of strain 35000 from unimmunized compared to immunized rabbits (18 of 24 lesions versus 3 of 22 lesions with viable organisms, respectively; P = 0.0004) but no significant difference in the recovery of strain 35000ΔAPC from unimmunized versus immunized rabbits (9 of 23 versus 10 of 22 lesions with viable organisms, respectively; P = 0.6942). Thus, we conclude that specific immunity to hemolysin mediates the killing of the hemolysin-expressing strain 35000 in immunized animals.
DISCUSSION
In this study, we show that the hemolysin genes are present and expressed in vitro in all strains of H. ducreyi in our large collection of diverse isolates. In addition, humans and animals infected with H. ducreyi produce antibodies to this protein, suggesting that hemolysin is also expressed in vivo. We found that hemolysin had no effect on the development and severity of lesions induced by this organism in the temperature-dependent rabbit model and that immunization with hemolysin also had no effect on the severity of lesions induced by H. ducreyi infection. However, immunized rabbits were better able to clear this organism from infected lesions, suggesting that hemolysin may be an important immunogen that could be exploited for the development of a vaccine for chancroid.
Previous studies by Palmer and Munson (35) indicated that all of 10 H. ducreyi strains tested contained genes for hemolysin. However, the ability of these strains to express hemolysin was not evaluated in this study. The possible existence of hemolysin-negative strains of H. ducreyi was suggested in studies by Alfa et al. (3) which showed that 3 of the 33 H. ducreyi isolates tested failed to produce a detectable fibroblast contact cytotoxin. Because this toxin was later identified as the H. ducreyi hemolysin (1, 34), these results suggested that these H. ducreyi strains produced reduced or no hemolytic activity, similar to strain CIP A75, which produces low levels of hemolysin (50). We tested the three H. ducreyi strains, 78226, 6V, and 35199, previously reported as negative for the fibroblast contact cytotoxin (3) and found that they contained DNA which amplified with hhdA- and hhdB-specific PCR primers but produced less than 20% of the hemolytic activity of strain 35000. Thus, we conclude that while all strains produced hemolysin, they differed in the amount of hemolysin activity produced, possibly reflecting differences either in the regulation of hemolysin, efficiency of secretion, or activities of different hemolysin structural genes. Factors that contribute to the expression, secretion, and regulation of hemolysin are poorly understood and need to be further studied.
The fact that strain ATCC 27721 grows only anaerobically suggests that the atmospheric requirements for growth of H. ducreyi strains may differ. Since all known strains have been acquired from human lesions, it is interesting to speculate that chancroidal ulcers can support an anaerobic environment. In addition, if other strains of H. ducreyi that grow preferentially under anaerobic conditions exist, perhaps recovery of H. ducreyi from lesions could be improved by anaerobic incubation as well as the traditional incubation in candle jars. Interestingly, some of the earliest methods used to recover H. ducreyi from human lesions included the use of tubes containing freshly clotted human or rabbit blood (11), which might have provided an environment suitable for isolation of bacteria under more anaerobic conditions. Identifying virulence factors and studying their expression becomes even more complex when in vivo and in vitro culture conditions differ so widely.
Our studies showing that animals and humans infected with H. ducreyi mount an immune response to hemolysin indicate that hemolysin is expressed in vivo and is immunogenic. Thus, primates infected genitally with H. ducreyi produce antibodies that react with hemolysin as do rabbits infected on their backs in the temperature-dependent rabbit model. Patients with chancroid also produced antibodies to hemolysin, indicating hemolysin is expressed in humans during natural infection. Because hemolysin is a universal feature of all H. ducreyi strains, is expressed in vivo, and is immunogenic, our results suggest further studies to evaluate the usefulness of the hemolysin-specific antibodies as a serologic test for chancroid, either independently or in conjunction with the standard absorption EIA (13) and the LOS EIA (2) assays.
We found that production of hemolysin in H. ducreyi 35000 had no effect on virulence in the temperature-dependent rabbit model, as measured by comparison of lesion development and severity in H. ducreyi 35000 and isogenic mutants lacking the hemolysin genes. Similar results were obtained for an isogenic hemolysin mutant in the human model of infection (37). However, both of these models have strengths and drawbacks for the evaluation of the contribution of H. ducreyi virulence factors. The human model, while requiring the inoculation of only 30 CFU to induce lesions, is limited to evaluation of lesions on the upper arms less than 14 days old that have not ulcerated (43). In contrast, the temperature-dependent rabbit model requires 105 CFU for lesion induction and can detect only viable organisms up to 5 to 10 days postinoculation. However, this model has been used to show that mutants defective in acquisition of heme, and thus growth, in vitro, are also less virulent in vivo (39, 45). Undoubtedly, H. ducreyi pathogenesis is multifactorial and requires the expression of several different virulence factors to allow this organism to obtain nutrients, damage host tissue, and evade the host immune response. Because the H. ducreyi hemolysin is able to lyse human fibroblasts and keratinocytes as well as inflammatory cell types such as macrophages, T cells, and B cells (54), hemolysin may contribute to ulcer development as well as survival in the presence of inflammatory cells. We plan to evaluate the effect of the hemolysin on lesion development and bacterial survival in the primate model. In this model, viable bacteria can be recovered for up to 21 days postinfection, genital, rather than peripheral sites are infected, and later stages of ulcer development can be studied than is possible in humans (49).
Consistent with our finding that hemolysin does not affect lesion development in the rabbit model, we observed no difference in survival of hemolysin-negative mutant and wild-type strains, as measured by culture of biopsies obtained from these lesions. In contrast, in lesion aspirates, wild-type strain 35000 was recovered more often than the hemolysin-negative mutant. The pustular material was collected from the center of the lesion whereas biopsies sample the tissue on the edges of the lesion, indicating possible differences in the local environment within these lesions which might favor the survival of bacteria expressing hemolysin. Palmer et al. (37) found that the infiltrate of lesions in the human model contained macrophages, CD4+ T lymphocytes, and a few B cells. Because hemolysin is able to lyse T cells, B cells, and macrophages (54), expression of this protein might help H. ducreyi survive in the pustular material. Further studies of the histology of different regions in these lesions, as well as studies including complemented hemolysin mutants, might clarify the role of hemolysin in survival in these different locations.
The ability of rabbits immunized with hemolysin protein to clear hemolysin-producing organisms in infected lesions suggests that hemolysin is an important target for the immune system and may have implications for future vaccine development. These experiments were internally controlled by the inoculation of both hemolysin-producing and hemolysin-negative strains in each rabbit. Because the lesions induced by these two strains progressed similarly, these differences in survival were not a consequence of differences in lesion development. Similarly, Hansen et al. (20) showed that prior immunization with cellular envelopes affected the recovery of viable bacteria more significantly than it affected lesion development. Desjardins et al. (12) also showed that immunization with a pilus preparation induced protective immunity in rabbits with both homologous and heterologous strains. Perhaps reflecting the heterogeneity of outer membrane proteins between strains, both methods of immunization reported differences in the protective effect of homologous rather than a heterologous strains (20). Because hemolysin appears to be a universal feature of H. ducreyi strains, immunization with hemolysin may be effective against most, if not all, strains of H. ducreyi. However, further work on the conservation of the hemolysin sequence and immunogenic epitopes between strains is clearly needed before the potential role of hemolysin as an H. ducreyi vaccine can be considered. In addition, the role of immunization in protection from lesion development might be further enhanced by evaluation in alternative animal or human models for disease. Because lesion development in rabbits requires an inoculum of 104 to 106 organisms (39), a dose at which purified LOS exhibits a significant toxicity (7), the elimination of lesion development by immunization may be an unrealistic goal in this model.
The existence and molecular and cellular nature of protective immunity in naturally acquired chancroidal infection is not known. However, both in the experimental human model and in naturally acquired chancroid, an immune response typical of cell-mediated delayed hypersensitivity, rather than a humoral immunity, is predominant (25, 36). Furthermore, the induction of CD8+ as well as CD4+ cells in chancroidal lesions suggests that this organism may exist intracellularly in lesions (36). In our experiments, rabbits immunized with the H. ducreyi hemolysin produced hemolysin antibodies, but the contribution of these antibodies to protective immunity is not known. Further studies on the cellular and molecular nature of the protective immunity induced in rabbits by immunization with hemolysin as well as the possible induction of similar protective responses in primates are further research directions that will be pursued by this laboratory.
ACKNOWLEDGMENTS
We thank Andrew Haydock for performing dot blot analyses and Jane Kuypers, Steve Lory, Steve Moseley, and Sheila Lukehart for scientific discussions and review of the manuscript. We also thank Eric Hansen (University of Texas, Dallas) for the pooled rabbit serum and Chen-Yen Chen (CDC, Atlanta, Ga.) for the human sera from Mississippi chancroid patients.
This work was supported by grant R29 AI33533 to P.A.T. and NIH STD/AIDS research training grant T32 AI07140 to G.E.W.
REFERENCES
- 1.Alfa M J, DeGagne P, Totten P A. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect Immun. 1996;64:2349–2352. doi: 10.1128/iai.64.6.2349-2352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfa M J, Olson N, Degagne P, Plummer F, Namaara W, MacLean I, Ronald A R. Humoral immune response of humans to lipooligosaccharide and outer membrane proteins of Haemophilus ducreyi. J Infect Dis. 1993;167:1206–1210. doi: 10.1093/infdis/167.5.1206. [DOI] [PubMed] [Google Scholar]
- 3.Alfa M J, Stevens M K, DeGagne P, Klesney-Tait J, Radolf J D, Hansen E J. Use of tissue culture and animal models to identify virulence-associated traits of Haemophilus ducreyi. Infect Immun. 1995;63:1754–1761. doi: 10.1128/iai.63.5.1754-1761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfa M J, Yang C L, Slaney L A, Kwok A Y C, Ronald A R, Jay F T. Identification of highly conserved and species-specific polypeptides of Haemophilus ducreyi. J Med Microbiol. 1992;37:413–419. doi: 10.1099/00222615-37-6-413. [DOI] [PubMed] [Google Scholar]
- 4a.Barreau, C. Personal communication.
- 5.Caillaud F, Courvalin P. Nucleotide sequence of the ends of the conjugative shuttle transposon Tn1545. Mol Gen Genet. 1987;209:110–115. doi: 10.1007/BF00329844. [DOI] [PubMed] [Google Scholar]
- 6.Cameron D W, Simonsen J N, D’Costa L J, Ronald A R, Maitha G M, Gakinya M N, Cheang M, Nydinya-Achola J O, Piot P, Brunham R C, Plummer F A. Female to male transmission of human immunodeficiency virus type 1: risk factors for seroconversion in men. Lancet. 1989;ii:403–407. doi: 10.1016/s0140-6736(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 7.Campagnari A A, Wild L M, Griffiths G E, Karalus R J, Wirth M A, Spinola S M. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect Immun. 1991;59:2601–2608. doi: 10.1128/iai.59.8.2601-2608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Mertz K J, Spinola S M, Morse S A. Comparison of enzyme immunoassays for antibodies to Haemophilus ducreyi in a community outbreak of chancroid in the United States. J Infect Dis. 1997;175:1390–1395. doi: 10.1086/516471. [DOI] [PubMed] [Google Scholar]
- 9.Clewell D B, Flannagan S E, Ike Y, Jones J M, Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988;170:3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cope L D, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson R S, Jr, Lagergard T, Radolf J D, Hansen E J. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deacon W E, Albritton D C, Olansky S, Kaplan W. V.D.R.L. chancroid studies. I. A simple procedure for the isolation and identification of Haemophilus ducreyi. J Investig Dermatol. 1956;26:399–406. doi: 10.1038/jid.1956.51. [DOI] [PubMed] [Google Scholar]
- 12.Desjardins M, Filion L G, Robertson S, Cameron D W. Inducible immunity with a pilus preparation booster vaccination in an animal model of Haemophilus ducreyi infection and disease. Infect Immun. 1995;63:2012–2020. doi: 10.1128/iai.63.5.2012-2020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjardins M, Thompson C E, Filion L G, Ndinya-Achola J O, Plummer F A, Ronald A R, Piot P, Cameron D W. Standardization of an enzyme immunoassay for human antibody to Haemophilus ducreyi. J Clin Microbiol. 1992;30:2019–2024. doi: 10.1128/jcm.30.8.2019-2024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiCarlo R P, Armentor B S, Martin D H. Chancroid epidemiology in New Orleans men. J Infect Dis. 1995;172:446–452. doi: 10.1093/infdis/172.2.446. [DOI] [PubMed] [Google Scholar]
- 15.Elkins C, Chen C, Thomas C E. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson B W, Campagnari A A, Melaugh W, Phillips N J, Apicella M A, Grass S, Wang J, Palmer K L, Munson R S., Jr Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J Bacteriol. 1997;179:5062–5071. doi: 10.1128/jb.179.16.5062-5071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond G W, Lian C J, Wilt J C, Ronald A R. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob Agents Chemother. 1978;13:608–612. doi: 10.1128/aac.13.4.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning, a practical approach. Vol. 1. Washington, D.C: IRL Press; 1985. pp. 108–135. [Google Scholar]
- 18a.Hansen, E. Personal communication.
- 19.Hansen E J, Latimer J L, Thomas S E, Helminen M, Albritton W L, Radolf J D. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J Bacteriol. 1992;174:5442–5449. doi: 10.1128/jb.174.16.5442-5449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen E J, Lumbley S R, Richardson J A, Purcell B K, Stevens M K, Cope L D, Datte J, Radolf J D. Induction of protective immunity to Haemophilus ducreyi in the temperature-dependent rabbit model of experimental chancroid. J Immunol. 1994;152:184–192. [PubMed] [Google Scholar]
- 21.Haydock A K, Martin D H, Morse S A, Cammarata C, Mertz K J, Totten P A. Molecular characterization of Haemophilus ducreyi strains from Jackson, MS and New Orleans, LA. J Infect Dis. 1999;179:1423–1432. doi: 10.1086/314771. [DOI] [PubMed] [Google Scholar]
- 22.Hirono I, Tange N, Aoki T. Iron-regulated haemolysin gene from Edwardsiella tarda. Mol Microbiol. 1997;24:851–856. doi: 10.1046/j.1365-2958.1997.3971760.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan W, Deacon W E, Olansky S, Albritton D C. V.D.R.L. chancroid studies. II. Experimental chancroid in the rabbit. J Investig Dermatol. 1956;26:407–414. doi: 10.1038/jid.1956.52. [DOI] [PubMed] [Google Scholar]
- 24.Kilian M, Theilade J. Cell wall ultrastructure of strains of Haemophilus ducreyi and Haemophilus piscium. Int J Syst Bacteriol. 1975;25:351–356. [Google Scholar]
- 25.King R, Gough J, Ronald A, Nasio J, Ndinya-Achola J O, Plummer F, Wilkins J A. An immunohistochemical analysis of naturally occurring chancroid. J Infect Dis. 1996;174:427–430. doi: 10.1093/infdis/174.2.427. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 28.McEntegart M G, Hafiz S, Kinghorn G R. Haemophilus ducreyi infections—time for reappraisal. J Hyg (Cambridge) 1982;89:467–478. doi: 10.1017/s0022172400071035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mertz K J, Weiss J B, Webb R M, Levine W C, Lewis J S, Orle K A, Totten P A, Overbaugh J, Morse S A, Currier M M, Fishbein M, St Louis M E. An investigation of genital ulcers in Jackson, Mississippi: high prevalence of chancroid and human immunodeficiency virus infection. J Infect Dis. 1998;178:1060–1066. doi: 10.1086/515664. [DOI] [PubMed] [Google Scholar]
- 30.Moseley S L, Falkow S. Nucleotide sequence homology between the heat-labile enterotoxin gene of Escherichia coli and Vibrio cholerae deoxyribonucleic acid. J Bacteriol. 1980;144:444–446. doi: 10.1128/jb.144.1.444-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odumeru J A, Wiseman G M, Ronald A R. Virulence factors of Haemophilus ducreyi. Infect Immun. 1984;43:607–611. doi: 10.1128/iai.43.2.607-611.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ondraczek R, Hobbie S, Braun V. In vitro activation of the Serratia marcescens hemolysin through modification and complementation. J Bacteriol. 1992;174:5086–5094. doi: 10.1128/jb.174.15.5086-5094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paik G. Reagents, stains, and miscellaneous test procedures. In: Lennette E H, Balows A, Hausler J W J, Truant J P, editors. Manual of clinical microbiology. 3rd ed. Washington, D.C: American Society for Microbiology; 1980. [Google Scholar]
- 34.Palmer K L, Goldman W E, Munson R S., Jr An isogenic haemolysin-deficient mutant of Haemophilus ducreyi lacks the ability to produce cytopathic effects on human foreskin fibroblasts. Mol Microbiol. 1996;21:13–19. doi: 10.1046/j.1365-2958.1996.00615.x. [DOI] [PubMed] [Google Scholar]
- 35.Palmer K L, Munson J R S. Cloning and characterization of the genes encoding the haemolysin of Haemophilus ducreyi. Mol Microbiol. 1995;18:821–830. doi: 10.1111/j.1365-2958.1995.18050821.x. [DOI] [PubMed] [Google Scholar]
- 36.Palmer K L, Schnizlein-Bick C T, Orazi A, John K, Chen C Y, Hood A F, Spinola S M. The immune response to Haemophilus ducreyi resembles a delayed-type hypersensitivity reaction throughout experimental infection of human subjects. J Infect Dis. 1998;178:1688–1697. doi: 10.1086/314489. [DOI] [PubMed] [Google Scholar]
- 37.Palmer K L, Thornton A C, Fortney K R, Hood A F, Munson R S, Jr, Spinola S M. Evaluation of an isogenic hemolysin-deficient mutant in the human model of Haemophilus ducreyi infection. J Infect Dis. 1998;178:191–199. doi: 10.1086/515617. [DOI] [PubMed] [Google Scholar]
- 38.Plummer F A, Simonsen J N, Cameron D W, Ndinya-Achola J O, Kreiss J K, Gakinya M N, Waiyaki P, Cheang M, Piot P, Ronald A R, Ngugi E N. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163:233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- 39.Purcell B K, Richardson J A, Radolf J D, Hansen E J. A temperature-dependent rabbit model for production of dermal lesions by Haemophilus ducreyi. J Infect Dis. 1991;164:359–367. doi: 10.1093/infdis/164.2.359. [DOI] [PubMed] [Google Scholar]
- 40.Purven M, Lagergard T. Haemophilus ducreyi, a cytotoxin-producing bacterium. Infect Immun. 1992;60:1156–1162. doi: 10.1128/iai.60.3.1156-1162.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 42.Simmons N A. Isolation of campylobacters. Br Med J. 1977;2:707. doi: 10.1136/bmj.2.6088.707-a. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spinola S M, Wild L M, Apicella M A, Gaspari A A, Campagnari A A. Experimental human infection with Haemophilus ducreyi. J Infect Dis. 1994;169:1146–1150. doi: 10.1093/infdis/169.5.1146. [DOI] [PubMed] [Google Scholar]
- 44.Stamm W E, Handsfield H H, Rompalo A M, Ashley R L, Roberts P L, Corey L. The association between genital ulcer disease and acquisition of HIV infection in homosexual men. JAMA. 1988;260:1429–1433. [PubMed] [Google Scholar]
- 45.Stevens M K, Porcella S, Klesney-Tait J, Lumbley S, Thomas S E, Norgard M V, Radolf J D, Hansen E J. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect Immun. 1996;64:1724–1735. doi: 10.1128/iai.64.5.1724-1735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Totten P A, Handsfield H H, Peters D, Holmes K K, Falkow S. Characterization of ampicillin resistance plasmids from Haemophilus ducreyi. Antimicrob Agents Chemother. 1982;21:622–627. doi: 10.1128/aac.21.4.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Totten P A, Kuypers J M, Morse S A. Haemophilus ducreyi. Detection by PCR. In: Peeling R, Sparling P F, editors. Sexually transmitted diseases. Methods and protocols. Vol. 20. Totowa, N.J: Humana Press; 1999. pp. 47–65. [DOI] [PubMed] [Google Scholar]
- 48.Totten P A, Lara J C, Norn D V, Stamm W E. Haemophilus ducreyi attaches to and invades cultured human foreskin epithelial cells in vitro. Infect Immun. 1994;62:5632–5640. doi: 10.1128/iai.62.12.5632-5640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Totten P A, Morton W R, Knitter G H, Clark A M, Kiviat N B, Stamm W E. A primate model for chancroid. J Infect Dis. 1994;169:1284–1290. doi: 10.1093/infdis/169.6.1284. [DOI] [PubMed] [Google Scholar]
- 50.Totten P A, Norn D V, Stamm W E. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect Immun. 1995;63:4409–4416. doi: 10.1128/iai.63.11.4409-4416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Totten P A, Stamm W E. Clear broth and plate media for culture of Haemophilus ducreyi. J Clin Microbiol. 1994;32:2019–2023. doi: 10.1128/jcm.32.8.2019-2023.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Towbin H, Staehelm J G T. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wasserheit J N. Epidemiological synergy. Interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sex Transm Dis. 1992;19:61–77. [PubMed] [Google Scholar]
- 54.Wood, G. E., S. M. Dutro, and P. A. Totten. Target cell range of the Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]