Abstract
Host restriction factors play key roles in innate antiviral defense, but it remains poorly understood which of them restricts HIV-1 in vivo. Here, we used single-cell transcriptomic analysis to identify host factors associated with HIV-1 control during acute infection by correlating host gene expression with viral RNA abundance within individual cells. Wide sequencing of cells from one participant with the highest plasma viral load revealed that intracellular viral RNA transcription correlates inversely with expression of the gene PTMA, which encodes prothymosin α. This association was genome-wide significant (Padjusted < 0.05) and was validated in 28 additional participants from Thailand and the Americas with HIV-1 CRF01_AE and subtype B infections, respectively. Overexpression of prothymosin α in vitro confirmed that this cellular factor inhibits HIV-1 transcription and infectious virus production. Our results identify prothymosin α as a host factor that restricts HIV-1 infection in vivo, which has implications for viral transmission and cure strategies.
INTRODUCTION
Multiple human innate factors that inhibit HIV-1 during its replication cycle have been identified. Most of these restriction factors, such as APOBEC3G, BST-2 (tetherin), and TRIM5α, were discovered in vitro by comparative gene expression analyses and by screening complementary DNA (cDNA) libraries in restrictive versus susceptible cell line models (1). The importance of these restriction factors is evident by the evolution of HIV-1 accessory proteins that counter their effects; for example, HIV-1 Vif counteracts APOBEC3G (2), and Vpu antagonizes BST-2 (3, 4). However, it is currently unknown which factors are most relevant in restricting present-day circulating HIV-1 strains in vivo.
Genome-wide studies of bulk transcriptomics in CD4+ T cells from an HIV-1 infection cohort have identified host genes up-regulated during chronic infection (5). Expression of interferon (IFN)–stimulated genes (ISGs), including known antiretroviral restriction factors, correlated with higher, rather than lower, viral loads. The observed changes in global RNA expression therefore likely reflect responses to viral replication, rather than viral control. To identify host factors affecting HIV-1 gene expression, it is important to combine cellular transcriptomics and quantification of viral transcripts in the same cell, as has been recently reported (6). The frequency of productively infected cells during suppressive antiretroviral therapy (ART) is extremely low, and characterizing gene expression in such cells without stimulation is not possible (7). We thus reasoned that analysis of infected and uninfected cells during acute HIV-1 infection (AHI), when viral replication is at its peak, may provide an opportunity to discover cellular factors that modulate viral transcription. Understanding which cellular factors are capable of restricting HIV-1 in infected cells in vivo is critical because such cells may present a source of rebounding virus after analytic treatment interruption (ATI) (8, 9). Knowledge about mechanisms of viral inhibition during acute infection may also facilitate the rational design of drugs or vaccines to prevent reservoir seeding.
Next-generation sequencing allows integration of data from thousands of individual single cells and enables detection of host gene expression (10) and viral RNA (vRNA). For example, more than 20,000 human genes can be assessed using droplet-based single-cell RNA sequencing (scRNA-seq) (11–13), which can also be used to detect viral transcripts. Using this technology, we confirmed previous findings showing that memory CD4+ T cells expressing cytotoxic markers exhibit the highest abundance of HIV-1 RNA transcripts (6). We then used an unbiased omics approach to identify host genes that correlate with vRNA expression within individual peripheral blood mononuclear cells (PBMCs) as early as 2 weeks after HIV-1 infection and then again 48 weeks after ART initiation, when viremia was suppressed. We found that expression of PTMA host gene inversely correlated with vRNA expression within individual infected cells in vivo. Because prothymosin α, a small protein encoded by the PTMA gene, also inhibits HIV-1 transcription and infectious virus production in cell culture, it represents a host restriction factor identified by unbiased screening of infected cells in vivo.
RESULTS
Multiomics data were generated using samples collected from individuals during AHI
We selected 14 participants enrolled in an AHI cohort to identify host factors that restrict HIV-1, because samples from these individuals allowed us to map the immune landscape of single cells during peak viremia. Furthermore, participants immediately started treatment upon diagnosis, thus providing an opportunity to study the same individuals at a later time point during ART-induced viral suppression (14, 15). We generated and analyzed single-cell data including scRNA-seq and T cell receptor (TCR) results from PBMCs as well as protein expression from plasma proteomics to define differences in host gene expression and protein composition between the AHI and ART time points. In addition, vRNA expression was assessed in the same single cells and compared both within individuals and across multiple participants (Fig. 1A). Demographic characteristics including age, ethnicity, sex, and infecting viral subtype (CRF01_AE) were similar between participants (table S1) and were staged on the basis of the emergence of virological and immunological markers after HIV-1 infection as described by Fiebig and colleagues (16). All 14 participants started ART during Fiebig stage III, when HIV-1 antibodies are first detected in the blood by enzyme-linked immunosorbent assay (ELISA), within 20 days after HIV infection and when plasma viral loads (VLs) were reaching peak viremia (16, 17). All participants became virally suppressed within 48 weeks of treatment and continued to be suppressed, as exemplified by undetectable plasma VL for up to 288 weeks of follow-up (fig. S1A). As expected, CD4+ T cell counts increased, and plasma VL markedly decreased between the AHI (0-week) and ART (48-week) time points (fig. S1B). Additional participants from the same and other cohorts (6, 18) were assessed to validate findings of host and viral gene expression (Fig. 1A). Last, longitudinal single-cell multimodal omics data, including transcriptomics, surface protein expression, and chromatin accessibility, were generated for seven participants from the same study diagnosed at Fiebig stage III with samples collected at the preinfection, AHI, and ART time points. These data were used to identify transcription factors that may regulate host genes of interest.
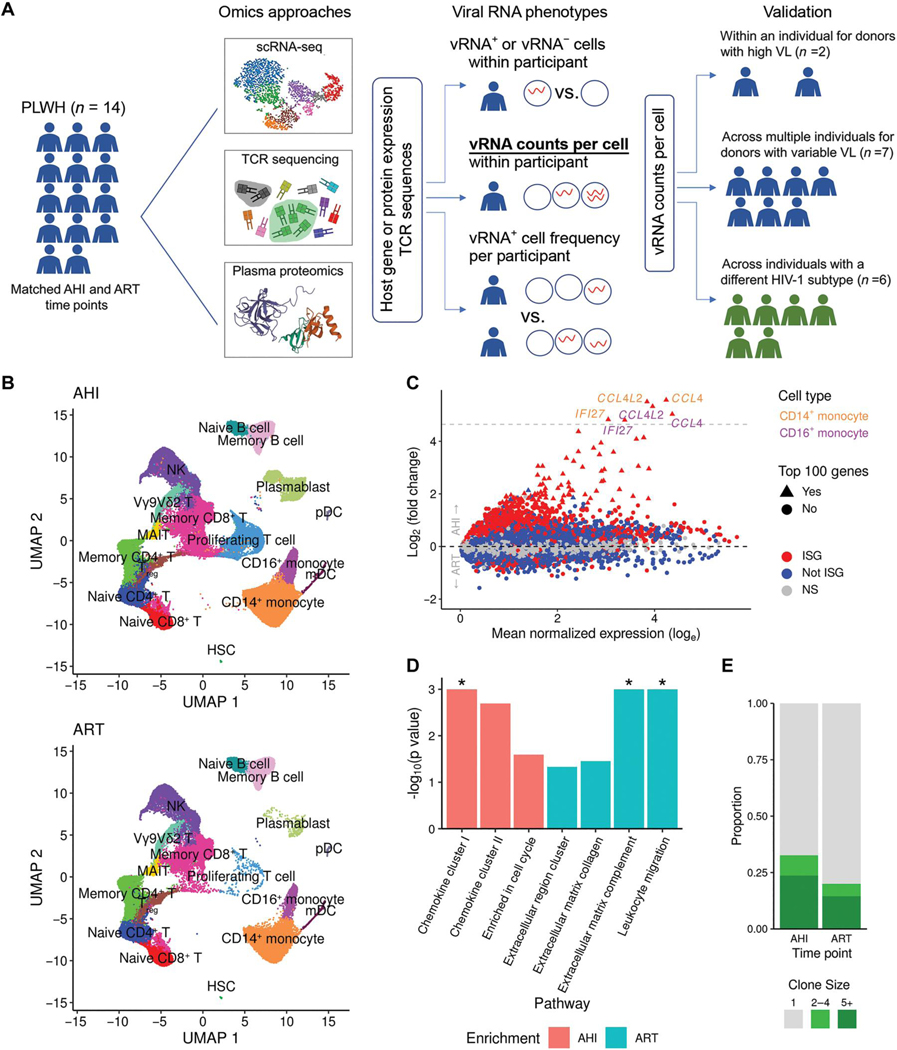
Fig. 1. Longitudinal single-cell multiomics confirm that IFN responses dominate differences between the AHI and ART time points in an acutely treated HIV cohort.
(A) scRNA-seq, TCR sequencing, and plasma proteomics data were generated from samples collected from 14 people living with HIV (PLWH) who initiated treatment during AHI. Data were generated for samples from all 14 participants at two time points, including AHI and ART. Viral RNA phenotypes from single cells within participants and across participants that were used to assess correlations with host gene expression are indicated. Analytical approaches are also shown, and the method used to validate findings in individuals with different VLs and subtypes is highlighted in bold and underlined font. (B) Unsupervised clustering of single-cell transcriptomic phenotypes from the 14 PLWH who were treated during acute infection at two time points (AHI and ART). HSC, hematopoietic stem cell; mDC, myeloid dendritic cells; NK, natural killer; pDC, plasmacytoid DC. (C) DEGs between the two time points from the scRNA-seq dataset showing significant ISGs as red symbols. Gray dashed line indicates genes up-regulated more than 25-fold. (D) Differentially enriched protein pathways between the two time points from the plasma proteomics datasets. Significant results [false discovery rate (FDR) < 0.05] are shown with an asterisk. (E) Differences in TCR expansion between the two time points. Clone size indicates the number of cells in which the same TCR was detected.
Elevated IFN responses during acute infection confound longitudinal analyses of HIV-1 restriction factors
Paired PBMC samples from 14 participants at the AHI and ART time points were subjected to scRNA-seq analyses on the 10x Genomics platform. A total of 95,872 and 61,539 single cells passed quality filters at the AHI and ART time points, respectively, and 23,102 genes were collectively expressed across all cell types from all participants. Cell clustering and annotation based on gene expression of variable genes or lineage markers revealed 17 discrete immune populations (Fig. 1B). All major canonical cell populations in PBMCs were detected, including cells from the innate and adaptive arms of the immune system (data file S1). There were significant differences in cell frequencies between the AHI and ART time points for most of the 16 immune cell populations that had frequencies of greater than 0.1% (P < 0.05, q < 0.01). The largest differences included a higher frequency of proliferating T cells at AHI, whereas frequencies of memory and naïve B cells, memory CD4+ T cells, mucosal-associated invariant T (MAIT) cells, and regulatory T cells (Tregs) were higher at the ART time point, indicating varying cellular expansion of immune cell populations at both time points (fig. S2A). For differentially expressed genes (DEGs) between AHI and ART time points across all cell subsets, an increase of up to 48-fold was observed at the AHI time point, with ISGs representing 95 of the top 100 genes that were significantly (Padjusted < 0.05) up-regulated (fig. S2B and data file S2). In addition, all genes up-regulated more than 25-fold at AHI were ISGs expressed in monocytes (Fig. 1C). Proteomics analyses of corresponding plasma samples independently confirmed enrichment of ISG chemokine pathways (chemokine clusters I and II) at AHI compared with the ART time points (Fig. 1D). Clonal T cell expansion was also significantly higher at the AHI than the ART time point (P = 0.011) (Fig. 1E and fig. S2C). That host factors at the AHI time point are mainly ISG-driven, in both infected and uninfected cells, is consistent with previous observations of elevated gene expression, cytokines, and IFN-resistant viral phenotypes during acute infection (19–21). This analysis did not allow us to examine gene expression within individual cells in response to HIV-1 infection, because it was impossible to differentiate global IFN-mediated effects dominated by uninfected cells from the influence of antiviral factors expressed in cells containing replicating HIV-1. We thus aimed to identify single cells with viral transcripts to search for host factors that restrict HIV-1 expression in vivo.
The frequency of HIV-1 RNA+ cells correlates with clinical parameters
In addition to obtaining host transcripts at the single-cell resolution, we identified vRNA transcripts in the same individual cells by aligning sequences to an HIV-1 reference genome. Cells that were vRNA+ were detected at both AHI (n = 164) and ART (n = 2) time points (Fig. 2A). Each single cell was classified on the basis of the presence (vRNA+ or vRNA−) or quantity (normalized counts) of intracellular viral transcripts as determined by unique molecular identifiers (UMIs), which are random oligonucleotide barcodes used to tag individual transcripts before polymerase chain reaction (PCR) amplification. Cells were considered high viral expressors on the basis of the presence of 10 or more vRNA UMIs (n = 17). As expected, most vRNA+ cells with high viral counts were found in CD4+ T cell subsets (64.7%). These cell subsets were generated by unsupervised analysis of all mRNA expression and annotated by expression of lineage markers, including the CD4 mRNA, which, in contrast to cell surface expression of the CD4 protein (22), is not down-regulated during HIV-1 infection (Fig. 2B). Among CD4+ T cells, the memory subset had the largest number of high vRNA–expressing cells (n = 7), followed by naïve CD4+ T cells (n = 2) and Tregs (n = 2). Flow cytometry sorting of cell subsets based on protein surface expression of lineage markers (CD3, CD4, CD8, CD14, CD19, CD45RO, and CD56) in one participant before single cell sequencing showed that CD4+ T cells were the only cell subsets with high vRNA detection (fig. S3, A and B). We aligned HIV-1 transcripts to a reference sequence that matched the viral subtype of the participants and showed that obtained reads spanned the entire viral genome, including spliced transcripts observed at the AHI time point (Fig. 2C). These data indicate that the scRNA-seq approach could detect viral transcripts in all of the expected CD4+ T cell subsets (23) and that these sequences were indicative of intact proviruses and active viral gene expression.
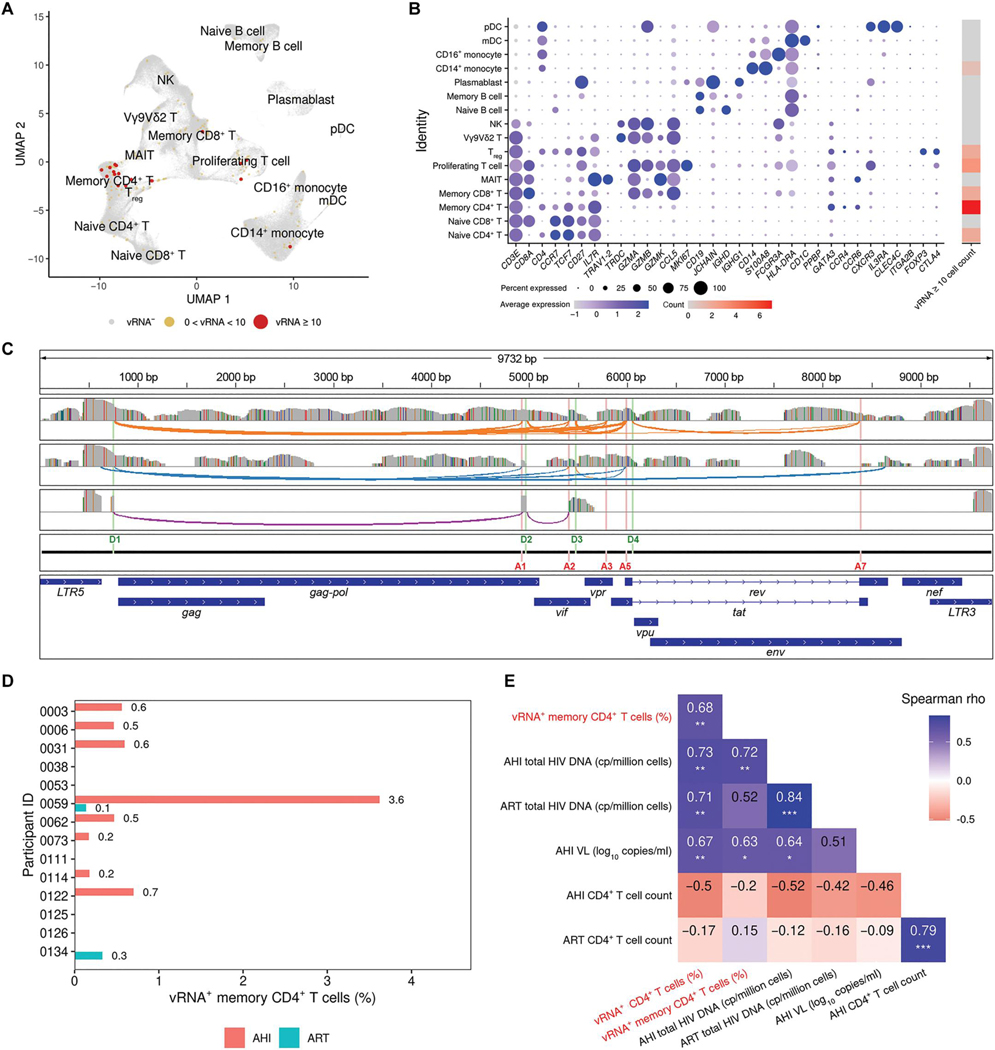
Fig. 2. HIV-1 vRNA transcripts identified during AHI span full viral transcriptomes and correlate with clinical parameters.
(A) High dimensionality reduction plot of single cells from PBMCs showing high viral reads mainly in the memory CD4+ T cell cluster in 14 participants at both AHI and ART time points. (B) For the PBMC clusters determined in (A), the expression of host genes identifying these populations is shown, as well as the number of cells with ≥10 HIV-1 transcripts. (C) Three representative cells of viral transcripts were mapped to the HIV-1 genome, showing detection of both full-length virus and active splicing. Gray bars represent coverage of viral sequences from three individual cells, colored bars in these coverage tracks represent single-nucleotide changes from the reference sequence, and colored arches show splicing events. Donor and acceptor splice sites and gene annotation of the HIV-1 reference genome are shown as separate tracks. (D) Frequency of vRNA+ cells (%) in memory CD4+ T cells is shown for each participant during AHI and ART time points. (E) Correlation of frequencies of vRNA+ CD4+ or memory CD4+ T cells obtained from scRNA-seq (red font) with significant clinical parameters (*P < 0.05, **P < 0.01, and ***P < 0.001).
The fraction of vRNA+ cells in total CD4+ T cells varied between participants but was proportional to the frequency of the vRNA+ memory subset, which represented the most commonly infected cell type (fig. S4). We thus focused our analyses on vRNA+ cells belonging to the larger CD4+ memory immune cell subset. Among 14 participants analyzed at the AHI time point, individual 0059 exhibited the highest fraction of vRNA+ memory CD4+ T cells (Fig. 2D). To ensure that the detection of vRNA in single cells was a biologically meaningful marker, we examined potential correlations of the frequency of vRNA+ single CD4+ T cells at the AHI time point with clinical parameters, including overall CD4+ T cell counts, total plasma HIV-1 RNA, and cell-associated DNA (AHI and ART). We found a significant positive correlation between the frequency of vRNA+ CD4+ T cells and the quantity of total cell–associated HIV-1 DNA detected both at the ART (rho = 0.71, P = 0.004) and AHI (rho = 0.73, P = 0.003) time points as well as plasma VL at AHI (rho = 0.67, P = 0.009) and the vRNA+ memory population (rho = 0.68, P = 0.008) (Fig. 2E). Thus, the correlation of the frequency of vRNA+ cells with HIV-1 clinical parameters such as plasma viremia and total cell–associated HIV-1 DNA validated our single-cell vRNA detection approach as physiologically relevant.
Markers of T cell activation and persistence are associated with the presence of vRNA
To characterize cells expressing vRNA in greater detail, we sequenced 10 times more cells from participant 0059 (originally 826 CD4+ T cells), who had the highest systemic plasma VL at the AHI time point. Using this “wide” sequencing approach, we obtained a total of 8228 CD4+ T cells, of which 223 were vRNA+. HIV-1 RNA was detected in all three CD4+ T cell types (naïve, memory, and Tregs) at varying frequencies. To identify host gene expression profiles, we focused on the memory CD4+ T cells, because these represented most of the cells with detectable vRNA. Unsupervised clustering based on gene expression of the top 2000 most variable genes identified four subpopulations within the memory CD4+ T population (Fig. 3A). Clusters 2 and 3 had the highest frequencies of cells with viral transcripts (Fig. 3B). The cells comprising one or both of these clusters also collectively expressed markers of T cell activation (GZMA, GZMK, NKG7, GNLY, PRF1, CCL5, and CXCR3) and persistence or exhaustion (PDCD1, TIGIT, CTLA4, and IFI27) (Fig. 3C and data file S3). TCR analyses revealed expanded clonotypes in cluster 2, which had the greatest number and highest percentage of vRNA+ cells (Fig. 3D). Combined single-cell transcriptomics and surface protein expression using CITE-seq (cellular indexing of transcriptomes and epitopes) data from seven additional AHI donors from the same cohort confirmed that the predicted phenotypes of all four memory CD4+ T cell clusters were CD45RO+ and CD45RA− based on surface protein markers (fig. S5, A to C). Cells from predicted clusters from participant 0059 mainly belonged to central memory (clusters 1 and 3) and effector memory (clusters 0 and 2) types based on expression of CCR7, CD62L, and CD27 (Fig. 3C and fig. S5, D and E). Overall, higher expression of CD278 (ICOS), an immune checkpoint protein in cluster 3, CD195 (CCR5) in cluster 2, and other cluster-specific markers was also observed.
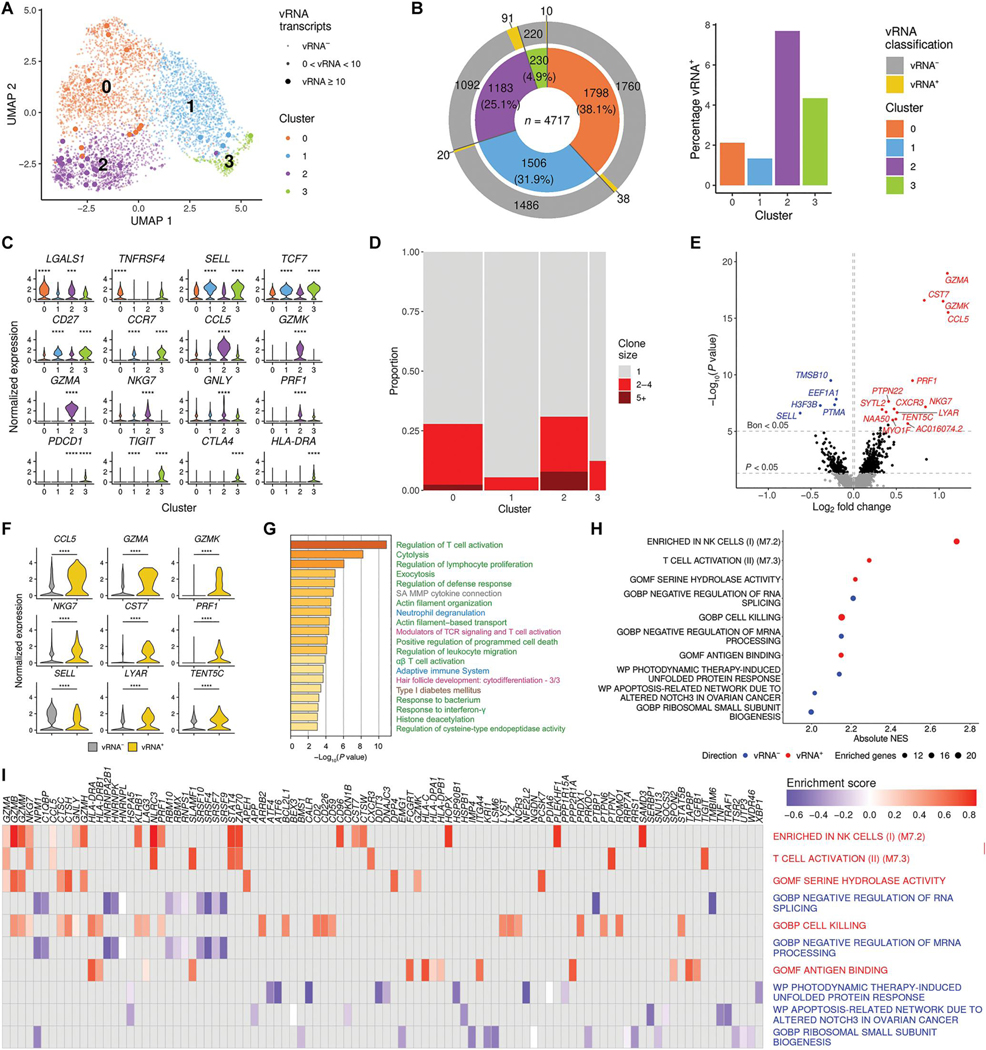
Fig. 3. Host and TCR diversity in memory CD4+ T cells with viral transcripts.
(A) Clustering based on scRNA-seq host transcriptomes of memory CD4+ T cells from one donor identifies four distinct subpopulations. (B) For each cluster in (A), the total number of cells, as well as the number and percentage of cells with viral transcripts, is shown. (C) Gene expression values are plotted for the top DEGs discriminating the four memory CD4+ T cell clusters (***P < 0.001 and ****P < 0.0001). (D) Proportion of expanded TCR clonotypes in the four clusters. Bar widths reflect the number of cells in each cluster. Clone size indicates the number of cells in which the same TCR was detected. (E) Categorical DEGs performed between cells with and without viral transcript in memory CD4+ T cells from all four clusters. Statistical significance and fold change are displayed as a volcano plot, with Bonferroni significant genes indicated in blue (vRNA− direction) or red (vRNA+ direction). Black dots are nominally significant genes. (F) Violin plots showing the expression of some top DEGs from the categorical analysis (all adjusted P < 0.05; ****P < 0.0001). (G and H) The top 100 DEGs shown in (E) were used in pathway analysis identifying associations with the presence of viral transcripts in memory CD4+ T cells (G) and preranked GSEA based on fold change of the genes (H). Pathways in (G) are shown in different colored font representing their source, including Gene Ontology (GO) terms (green), Canonical Pathways (gray), Reactome (blue), WikiPathways (pink), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (brown). In (H) are the top five significant gene sets in both directions. NES, normalized enrichment score. (I) Leading edge analysis with the 10 gene sets from (H).
To further identify cellular markers associating with the presence of vRNA, we compared gene expression between vRNA+ and vRNA− cells within the memory CD4+ T cell cluster initially in a categorical analysis. The top DEGs with higher expression in the vRNA+ versus vRNA− cells included markers associated with T cell activation (CCL5) and cytotoxicity (GZMA, GZMK, PRF1, NKG7, and CST7) (Fig. 3, E and F, and data file S4). Pathway analyses of the 100 top DEGs between memory CD4+ T cells with and without viral transcripts showed that these genes were associated with regulating T cell activation in the memory CD4+ T cell subset (Fig. 3G and data file S4). Gene set enrichment analysis (GSEA) using all DEGs confirmed that the top pathways enriched in the vRNA+ group were activation and cytolytic pathways, whereas the vRNA− group was enriched for pathways related to negative regulation of RNA splicing and mRNA processing (Fig. 3H). The top overlapping co-enriched genes were related to cytolytic activity (GZMA, GZMB, GZMM, and NKG7) or histone binding (NPM1) (Fig. 3I). Overall, both cluster markers and vRNA differential analyses indicated that the highest frequencies of viral reads were in memory CD4+ T cells defined by expression of cytotoxic markers.
High PTMA expression is associated with low HIV-1 transcription
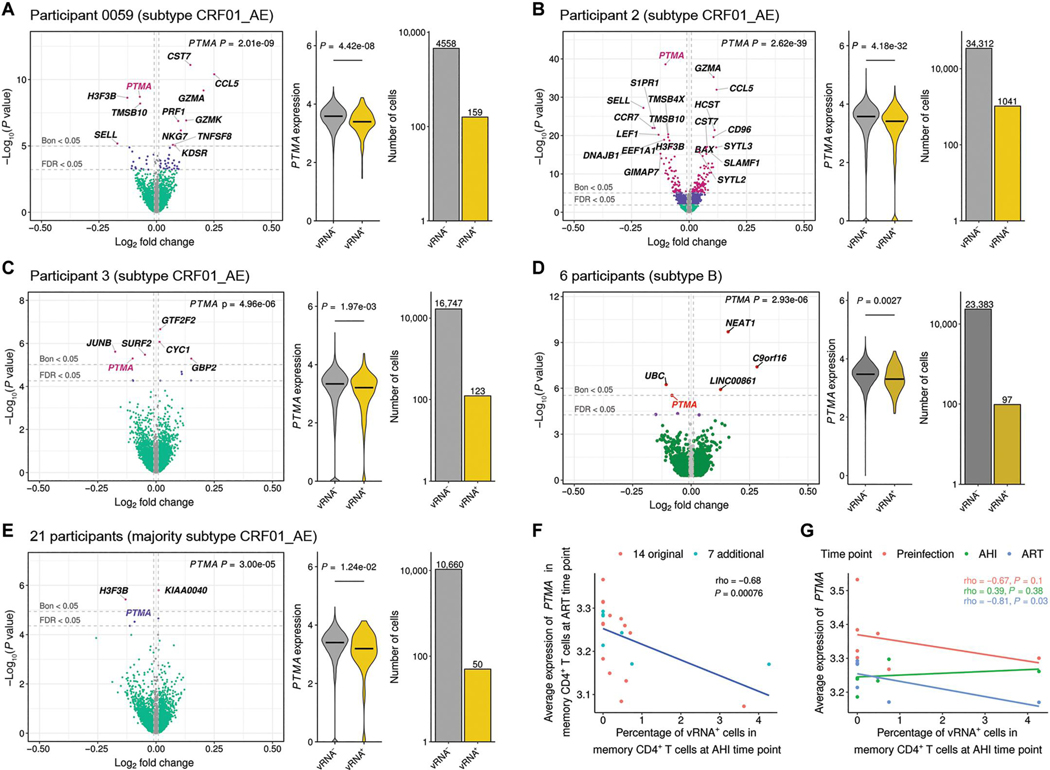
To identify cellular factors associated with HIV-1 transcription, we correlated all 4782 expressed host genes at AHI with the normalized number of viral transcripts detected as a continuous variable in vRNA+ memory CD4+ T cells from participant 0059. This analysis identified the gene encoding prothymosin α (PTMA) as the top genome-wide hit associating with lower intracellular vRNA counts (Fig. 4A). We validated this finding in two additional AHI participants sampled at peak viremia from a different prospectively followed early AHI cohort (18), who were not part of the initial 14 participants but were selected because of their high peak VL: participant 2 (7.58 log10 copies/ml) and participant 3 (6.82 log10 copies/ml). For these participants, we first enriched for memory CD4+ T cells by cell sorting using flow cytometry and then performed droplet-based scRNA-seq analyses. Because HIV-1 down-regulates CD4 on the cell surface, we captured all memory T cells expressing CD3 that were negative for CD8 and included both CD4+ and CD4− cells. Again, we identified PTMA as being associated with lower intracellular vRNA+ counts with genome-wide significance (Padjusted < 0.05; Fig. 4, B and C). To include participants with lower plasma VL and fewer sequenced cells, we next tested host gene expression for association with viral transcription in the same single cells in the remaining 13 participants from the initial cohort and observed an association that was nominally significant (β = −0.09, P = 0.007). To assess whether a similar relationship could also be observed in participants infected with other HIV-1 subtypes, we took advantage of a recently published single CD4+ T cell dataset, which included six participants infected with subtype B viruses (6). Combining cells from all six AHI participants and performing single gene analysis with vRNA expression as a continuous variable, we confirmed a genome-wide significant association between PTMA expression and lower vRNA counts within memory CD4+ T cells (β = −0.08, P = 2.93 × 10−6) (Fig. 4D). Thus, the observed relationship of PTMA expression and reduced intracellular vRNA transcription was not a unique feature of one particular HIV-1 (CRF01_AE) subtype. Last, to gain mechanistic insight, we performed pathway analyses of the top 100 genes associating with vRNA counts but did not find enriched biological pathways that also included PTMA (fig. S6A). We also observed that coexpressed genes based on our in vivo single-cell data with PTMA differed depending on the infecting viral subtype (data file S5). To identify potential transcription factor binding sites, we performed a coaccessibility analysis using single-cell chromatin data from memory CD4+ T cells of seven additional participants who were part of the same AHI cohort but also had samples before infection as well as at AHI and ART time points (18). We found that the binding sites for CCCTC-binding factor (CTCF), Myc-associated zinc finger protein (MAZ), SP3, and zinc finger protein 148 (ZNF148) were all enriched in potential enhancer regions within 500 kb of the PTMA gene, thus revealing variation in chromatin accessibility over time and pointing to a possible role of epigenetic modification in PTMA regulation (fig. S6, B and C).
Fig. 4. Individual cell and participant-specific PTMA expression at AHI and ART associate with decreased vRNA transcripts identified in memory CD4+ T cells at the single-cell resolution.
(A) Correlation of PTMA with abundance of vRNA detected in the same individual memory CD4+ T cells from participant 0059. The color of the dots indicates degree of significance for each gene at P < 0.05: green, nominal; purple, FDR; pink, Bonferroni. Gray dots denote genes with absolute log fold change (logFC) of <0.01. (B and C) Correlation of PTMA with abundance of vRNA detected in sorted memory CD4+ T cells from each of two additional participants, participant 2 (B) and participant 3 (C), with samples at peak viremia and with high viral loads. (D and E) Similarly, correlation of PTMA with intracellular vRNA counts from the same single cells in a combined analysis of cells from different participants with varying viral loads from 6 participants infected with HIV-1 subtype B (D) and 21 participants of whom most were infected with subtype CRF01_AE (E). For (A) to (E), the horizontal bars in the violin plots indicate median normalized expression of PTMA. (F and G) Correlation of frequency of vRNA+ memory CD4+ T cells at the AHI time point and participant-specific PTMA expression at the ART time point in 21 individuals from whom longitudinal data were available (F) or among the 7 participants having gene expression at three longitudinal time points, including preinfection, AHI, and ART (G).
Participant-specific variations in PTMA expression correlate with HIV-1 inhibition
The robust inverse correlation between host PTMA and vRNA expression raised the question of whether higher PTMA expression inhibited HIV-1 transcription or whether HIV-1 was down-regulating PTMA expression. We thus asked whether there was natural variation in PTMA expression among the AHI cohort participants that was indicative of different cell infection rates. This could not be addressed at the AHI time point because of IFN-driven host gene expression in both infected and uninfected cells as shown earlier (Fig. 1C). Thus, we used our scRNA-seq gene expression data from the ART time point when viremia was suppressed to quantify average PTMA expression for each participant and investigated whether expression correlated with the frequency of vRNA+ cells at the AHI time point. To increase statistical power, we included 7 additional participants from the same cohort as the 14 participants for whom scRNA-seq analyses were performed and confirmed a significant inverse correlation of PTMA expression and intracellular vRNA counts by analyzing single cells in all 21 participants (β = −0.09, P = 3.0 × 10−5) (Fig. 4E). We next used the PTMA expression data from each participant and correlated it with the frequency of vRNA+ memory CD4+ T cells identified at AHI. These analyses revealed that a lower frequency of vRNA+ memory CD4+ T cells during acute infection correlated significantly with higher PTMA expression at the ART time point (rho = −0.68, P = 0.00076) (Fig. 4F). This association remained significant when the seven additional participants were analyzed separately (rho = −0.81, P = 0.03) (Fig. 4G). Because these seven participants had also been sampled before HIV-1 infection, we generated scRNA-seq for their preinfection time points. Higher PTMA expression before infection was inversely correlated with lower frequencies of vRNA+ memory CD4+ T cells at AHI (rho = −0.67). As expected, there was no correlation with PTMA at AHI (rho = 0.39, P = 0.38). These findings suggest that higher PTMA expression inhibits HIV-1 transcription, rather than the virus down-regulating PTMA. Although PTMA expression at ART correlated inversely with intracellular vRNA counts measured at AHI, it did not correlate with either plasma VL or CD4+ T cell counts (fig. S7), indicating that this finding was independent of baseline clinical parameters.
PTMA overexpression inhibits HIV-1 in vitro
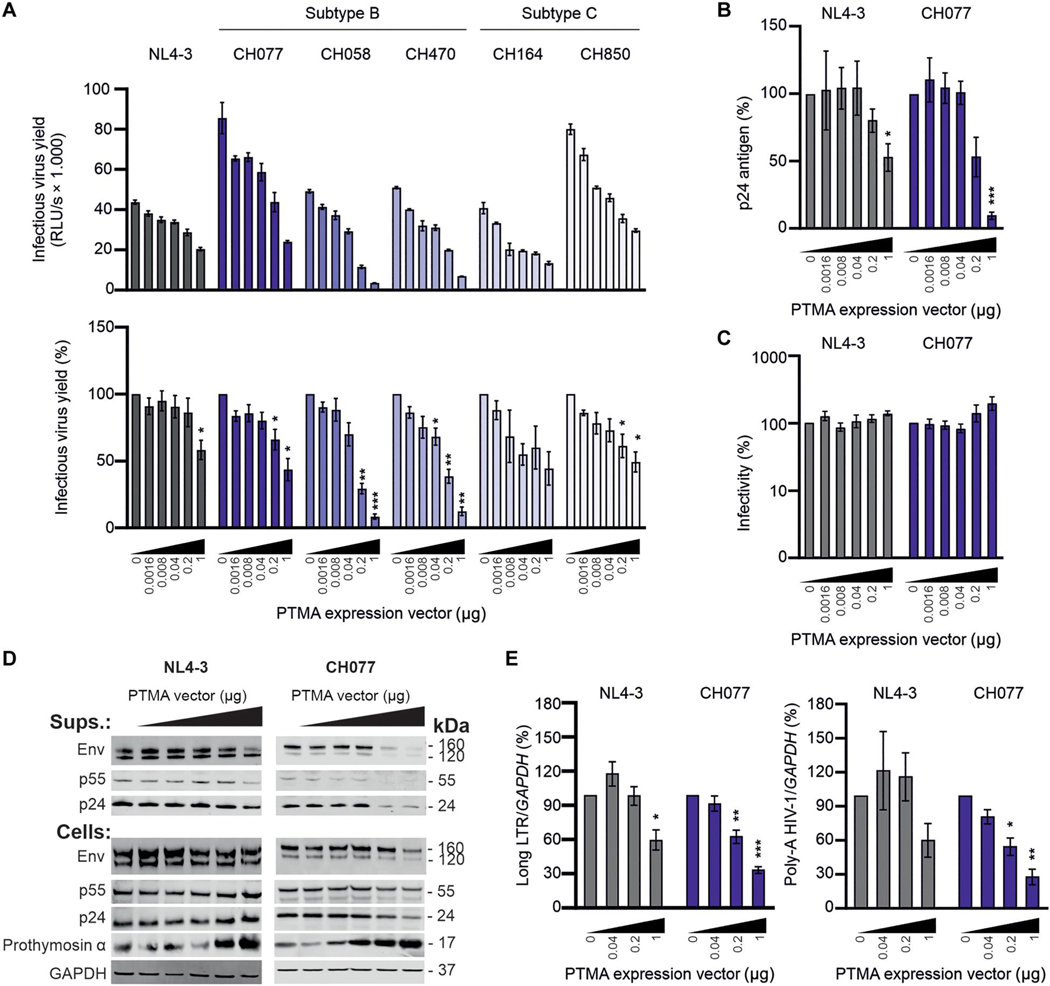
To assess the potential antiviral activity of PTMA directly, we examined its in vitro activity using a panel of infectious molecular clones (IMCs). To determine the susceptibility of these IMCs to PTMA, we measured infectious virus yield from human embryonic kidney (HEK) 293T cells after cotransfection of proviral constructs with increasing amounts of a PTMA expression vector. Control experiments showed that PTMA overexpression was not cytotoxic (fig. S8). All IMCs produced high quantities of infectious virus and were inhibited by PTMA in a dose-dependent manner (Fig. 5A). The susceptibility of the HIV-1 strains to PTMA varied to some extent. The subtype B CH058 and CH470 HIV-1 IMCs were more susceptible to PTMA than the subtype C CH164 and CH850 IMCs, the clade B CH077 IMC, and the T cell line–sadapted NL4–3 IMC (Fig. 5A). PTMA expression reduced the quantity of p24 capsid antigen in the cell culture supernatants for both NL4–3 and the transmitted-founder CH077 IMC (Fig. 5B) but had no significant effect (P > 0.05) on normalized viral infectivity (Fig. 5C), indicating an effect on the quantity rather than quality of virus produced. Western blot analysis supported that PTMA overexpression was associated with reduced viral protein expression (Fig. 5D). Quantitative real-time PCR (qRT-PCR) revealed that PTMA suppressed the production of initial [Long LTR (long terminal repeat)–containing mRNA] and fully completed poly-A HIV-1 CH077 RNA transcripts in transfected HEK293T cells (Fig. 5E). At the lowest concentrations of PTMA expression constructs, the quantity of NL4–3 RNA transcripts fluctuated around the respective empty vector control, but at a higher PTMA expression, it was reduced by about 40% and reached significance for Long LTR transcription (P < 0.05). Together, these results indicate that PTMA restricts transcription of primary HIV-1 strains. Thus, multiple orthogonal approaches performed in vivo and in vitro provided direct evidence that PTMA exerts an antiviral effect.
Fig. 5. PTMA overexpression inhibits transcription of HIV-1 in vitro.
(A) Proviral HIV-1 constructs and increasing amounts of a plasmid expressing PTMA (fivefold incremental change ranging from 0.0016 to 1 μg) were cotransfected into HEK293T cells. Infectious virus yield was measured using the TZM-bl reporter cell infectivity assay. The top panel shows absolute infectivity, and the lower panel shows values normalized to the infectious virus yield obtained in the absence of PTMA overexpression (100%). (B and C) p24 antigen measured by ELISA (B) and normalized infectivity produced by HEK293T cells (C) were measured in the presence of increasing quantities of PTMA. (D) Expression of HIV-1 proteins, PTMA, and GAPDH in viral particles (Sups.) or cellular extracts (Cells) was determined by Western blot. (E) Abundance of viral transcripts determined by qRT-PCR in multiplex reactions with GAPDH as control. Bar diagrams show mean values (±SEM) derived from three or four independent experiments performed in technical triplicates. *P < 0.05, **P < 0.01, and ***P < 0.001.
DISCUSSION
Innate immune defense mechanisms are of enormous interest because antiviral factors restrict viral pathogens, including HIV-1 (24, 25). However, the cellular factors that are most relevant for restricting HIV-1 replication, transmission, and rebound after treatment interruption in vivo remain to be determined. Addressing this question has been challenging in part because of the difficulty of characterizing the transcriptional profile of individual cells infected with HIV-1 in vivo. Here, we used single-cell transcriptomic analyses to identify individual vRNA+ memory CD4+ T cells during AHI and confirmed that they expressed markers of persistence and cytotoxicity (6, 26), thus supporting our approach. Moreover, by analyzing both viral and host gene expression in the same cells, we were able to correlate intracellular vRNA measurements with corresponding host factors, rather than comparing systemic plasma VL and IFN-driven responses across cellular populations. Our investigation showed that high intracellular expression of PTMA was associated with low vRNA expression, which was confirmed for individuals infected with two different HIV-1 subtypes (CRF01_AE and CRF01_B). This correlation could have been caused by HIV-1 down-regulating PTMA. However, support for PTMA actually inhibiting HIV-1 was provided by the observation that the frequency of vRNA+ cells during acute infection inversely correlated with PTMA expression more than 40 weeks later when virus was suppressed by ART. These data suggested that interindividual variation in PTMA expression influences the frequency of infected cells during AHI, an observation that was further corroborated when preinfection samples were compared for a subset of individuals. Direct evidence that PTMA inhibits HIV-1 was obtained by overexpression in vitro. At the highest dose, PTMA reduced infectious virus yields of clade B HIV-1 CH077 and CH058 IMCs by ≥90%, although subtype C CH164 and CH850 IMCs were less affected. It is expected that not all pandemic HIV-1 strains are similarly susceptible to PTMA because they counteract or evade essentially all restriction factors (27). It has recently been shown that an additional nuclear factor κB (NF-κB) site allows HIV-1 subtype C to evade restriction by nuclear Pyrin and hematopoietic IFN-inducible nuclear (HIN) domain–containing (PYHIN) proteins (28), and it will be interesting to determine whether subtype C viruses have also evolved mechanisms to avoid restriction by PTMA. Together, the strength of the inhibitory effect of PTMA was similar to those observed in overexpression assays of tetherin, zinc-finger antiviral protein (ZAP), and guanylate binding protein 5 (GBP5) (29–31). Genes correlating with PTMA expression varied depending on the infecting viral subtype, which could also be attributed to differences in ethnicity, showing that varying host responses can affect viral inhibition in vivo. Because the PTMA effect was discovered by an unbiased analysis of infected patient cells and inhibited primary HIV-1 IMCs in vitro, it likely represents an important factor restricting HIV-1 transcription in vivo.
We analyzed matched peripheral blood samples collected from participants during AHI and after viral suppression by ART using single-cell transcriptomics. AHI cohorts have previously been used to investigate clinical and immunological characteristics of early HIV-1 infection (14, 15, 18, 32), which provided previously unknown insights into viral dynamics and host responses (21, 33–39). Our current analyses of host gene expression using single-cell data are consistent with previous findings, indicating that ISGs and genes involved in the chemokine response are strongly induced during HIV infection. However, we also observed that responses of uninfected cells to IFN drive expression of a number of host genes. Thus, it is clear that the identification of host factors that restrict HIV-1 in vivo requires an assessment of host and viral expression profiles within the same individual infected cell.
We were able to detect viral transcripts in single cells from AHI participants during peak viremia without stimulation. This approach allowed us to compare the genome-wide expression of host genes with vRNA counts in the same individual cells. We found that PTMA expression correlated with lower vRNA measurements by single-cell analysis of three CRF01_AE–infected participants and by combining vRNA+ CD4+ T cell data from 20 additional participants. Last, similar observations were made by reanalyzing an independent dataset of six participants infected with subtype B virus (6). In agreement with DEG analyses from that study, we observed higher expression of cytotoxic markers in cells with vRNA. However, although Collora and colleagues (6) did not identify PTMA as genome-wide significant in their cohort, we observed a significant inverse association of PTMA expression and vRNA counts (Padjusted < 0.05). This difference is most likely explained by the fact that the previous study examined only the presence or absence of vRNA, whereas we measured the quantity of intracellular vRNA counts when testing for an association. Thus, our quantitative analyses show that PTMA expression correlates inversely with intracellular vRNA counts in vivo across multiple cohorts and subtypes.
Although the observed in vivo correlation of PTMA with low vRNA counts could be due to HIV-1 down-regulating PTMA, multiple lines of evidence suggest that PTMA inhibits HIV-1. PTMA expression differed in participant 0059 when using the presence or absence of vRNA as a categorical variable, but the association between PTMA and vRNA was much stronger when the number of viral transcripts was used as a continuous variable in this and other participants. This suggests that inhibition by PTMA was not occurring during viral entry of target cells (as assessed by vRNA presence) but that PTMA played a role in inhibiting HIV-1 transcription (as assessed by vRNA counts). Additional evidence for PTMA expression inhibiting HIV-1 was provided by a correlation of intracellular vRNA transcription at AHI with PTMA expression on ART across 21 participants, suggesting that inherent differences in PTMA expression among individuals resulted in varying degrees of HIV-1 inhibition. Last, overexpression of PTMA in HEK293T cells inhibited transcription of a variety of transmitted-founder subtype B and C HIV-1 IMCs. Together, our findings suggest that PTMA is expressed in CD4+ T cells and that higher expression silences viral transcription.
PTMA is a small highly acidic protein of 109 to 111 amino acids. Previous studies have shown that PTMA is widely expressed and seemingly involved in a multitude of cellular functions, including protection against oxidative stress and apoptosis, DNA remodeling, and cellular transcription (40–42). PTMA is abundant in the nucleus but may be released into the cytosol upon necrosis-inducing stress and subsequently processed into several peptides, which have been proposed to exert immunomodulatory, antiviral, and anticancer activity (43). One of the peptides, thymosin α1 (thymalfasin), was reported to have a beneficial effect in people with HIV-1 infection (44), but these findings have not been independently confirmed. In addition, PTMA and resulting peptides released from CD8+ T cells were reported to inhibit HIV-1 through Toll-like receptor 4–mediated type 1 IFN induction (45). Thus, although it is currently unclear whether PTMA has potential as an antiviral agent in vivo, our findings that it inhibited virus transcription may prompt efforts to examine this possibility more rigorously.
We show that higher PTMA expression correlates with lower vRNA expression in vivo. This agrees with our finding that overexpression of PTMA inhibits the transcription of primary HIV-1 strains in vitro. It has been reported that PTMA inhibits HIV-1 LTR-driven gene expression in macrophages (46). In addition, PTMA binds to histone H1, increases the accessibility of chromatin, and promotes cell proliferation (42, 47). Our analyses of single-cell chromatin accessibility in longitudinal samples of AHI cohort participants showed that accessible chromatin regions in PTMA in memory CD4+ T cells contain binding sites for DNA binding transcription factors with a Cys2His2 (C2H2) zinc finger binding domain (48, 49). One transcription factor, ZNF148, has been shown to be negatively associated with HIV-1 replication in lymphoid tissue (50). It is thus tempting to speculate that the interaction between PTMA and histone H1 may allow transcription factors that inhibit HIV-1 transcription to access the LTR promoter. Furthermore, unlike the other three transcription factors, CTCF enrichment decreased during AHI and increased at the ART time point. This transcriptional repressor has been shown to play a role in the induction of anti–HIV-1 responses and the promotion of latency, pointing to another potential mechanism through which PTMA could be inhibiting HIV transcription (51–53). However, further investigation is required to define the exact mechanism.
There are some limitations to this study. One is that in vivo it assesses host restriction factors in the context of only two HIV-1 subtypes (CRF01_AE and CRF01_B) and is restricted to two ethnic populations (Thai and American) and a limited number of participants. Additional research using a greater number of participants, viral subtypes, and geographic populations will be required to thoroughly elucidate the negative association between PTMA and HIV-1 transcript accumulation. A second limitation of our study is that we do not have insight into the precise molecular mechanism of viral suppression by PTMA. Further work will show whether the interaction between PTMA and HIV-1 is direct or indirect and whether inhibition of vRNA accumulation occurs transcriptionally or posttranscriptionally. Last, this study focused on characterizing host factors in memory CD4+ T cells from the periphery at AHI. Future work investigating the relationship between PTMA and HIV-1 in additional tissue compartments, including lymph nodes and mucosa, as well as other biological situations, such as chronic infection and time to rebound after ATI, is warranted. In conclusion, we have identified a candidate antiviral host factor on the basis of unbiased analyses of transcriptomes of individual vRNA+ cells in people living with HIV-1. Our findings illustrate that new unbiased omics technologies enable identification of antiviral factors that may have utility for HIV-1 prevention and cure strategies.
MATERIALS AND METHODS
Study design
Participants from an AHI cohort were selected for omics analyses to identify host factors that restricted HIV-1 at the single-cell resolution. Samples were selected from the RV254 cohort enrolling participants with AHI (Fiebig stages I to V) in Bangkok, Thailand exploring the impact of early ART on immune responses and HIV-1 disease progression (14, 15). Most participants in the cohort are men who have sex with men and are followed prospectively to gather longitudinal data, including HIV-1 VL and CD4+ T cell counts at baseline (2 to 3 days after diagnosis) before and after ART initiation. Sample size was based on availability of samples at Fiebig stage III from an AHI cohort with known clinical parameters. We selected 21 male participants at Fiebig stage III with variable VL at peak viremia and with age ranging between 20 and 46 years (table S1). Further validation was performed in one participant with the highest VL sequenced 10-fold more than the other 20 participants. In addition, further biological replication of findings was performed in two participants from an independent AHI cohort (RV217) with high VL at peak viremia by scRNA-seq wide sequencing analyses of sorted memory CD4+ T cells (18). A previously published scRNA-seq dataset from six participants at AHI from the Sabes cohort in South America was mined to validate scRNA-seq findings in a third independent study (6). Last, longitudinal samples were used to generate simultaneous single-cell surface protein and transcriptome, as well as chromatin accessibility data to support findings using orthogonal methods in 7 of the 21 participants who in addition to the AHI and ART time points had PBMCs collected before infection.
All participants from the aforementioned human studies from Thailand provided informed consent, and use of samples for research was approved by ethical review boards at the Walter Reed Army Institute of Research, the Thai Ministry of Public Health, the Royal Thai Army Medical Department, Mahidol University Faculty of Tropical Medicine, and Chulalongkorn University Faculty of Medicine.
HIV-1 reservoir measurements
Total HIV-1 DNA and integrated DNA were measured by quantitative PCR as described previously (54). Briefly, pellets of PBMCs were digested for 12 hours at 55°C using a proteinase K lysis buffer. Total HIV-1 DNA was quantified using primers and a probe situated in the 5′-LTR, whereas the primers and probe used for integrated DNA were situated in Alu and the 5′-LTR. ACH-2 cells, which carry a single copy of the integrated HIV-1 genome, were used to generate a standard curve for both assays. The lower limit of detection of this assay was 10 copies per 106 cells. Samples with HIV DNA values below the lower limit of detection were assigned a numerical value of 0 for downstream quantitative analyses.
Isolation of memory CD4+ T cells by flow sorting
Cryopreserved PBMCs from two participants at AHI time point were thawed in medium containing 10% fetal bovine serum. Cell counts and viabilities were assessed using Guava ViaCount reagent and Muse Cell Analyzer (Millipore Sigma). Cells were washed and stained with Aqua Live/Dead stain (Molecular Probes), washed, and blocked using normal mouse immunoglobulin G (IgG) (Caltag). The cells were surface-stained for the following antibodies [all Becton Dickinson (BD)]: CD3 fluorescein isothiocyanate (FITC) (UCHT1, 555916, 1:10, AB_396217), CD4 allophycocyanin (APC) (RPA-T4, 555349, 1:20. AB_398593), CD14 BV421 (MϕP9, 563743, 1:320, AB_2744289), CD8 BV786 (RPA-T8, 563823, 1:160, AB_2687487), CD45RA BUV395 (5H9, 740315, 1:160, AB_2740052), CD16 BUV496 (3G8, 612944, 1:80, AB_2870224), CD19 BUV737 (SJ25C1, 612756, 1:80, AB_2870087), CD45RO phycoerythrin (PE) (UCHL1, 555493, 1:5, AB_395884), and CD56, PE-Cy7 (B159, 557747, 1:320, AB_396853). The cells were then washed, sorted, and analyzed on the FACSAria II SORP Cell Sorter (BD) or the BigFoot Spectral Cell Sorter (Thermo Fisher Scientific).
scRNA library generation and sequencing
Total PBMCs from longitudinal AHI and ART time points of 14 participants were washed, resuspended in phosphate-buffered saline (PBS), and processed using the Chromium NextGEM Single Cell 5′ v1.1 Kit and the Chromium Controller (both 10x Genomics) per the manufacturer’s instructions. About 21,000 or 13,900 PBMCs from each participant from the AHI or ART time points, respectively, were loaded into individual wells of Chromium G chips for cell partitioning and subsequent gene expression (GEX) library construction. In addition, flow-sorted cell suspensions of memory CD4+ T cells, monocytes, and CD8+ T cells from one participant, and memory CD4+ T cells from a second participant, at the AHI time point were processed similarly. Briefly, 19,000 to 21,000 sorted cells were loaded into separate wells of Chromium chips, and amplified cDNA was used to make GEX and TCR libraries, which were assessed for quality and concentration using the High Sensitivity DNA Assay with the 2100 BioAnalyzer (both Agilent) and pooled. Library component ratios were determined with the MiSeq Reagent Nano v2 Kit (300 cycles) (Illumina) sequencing run per the manufacturer’s recommendations. Final normalized library pools were sequenced using the NovaSeq 6000 S4 Reagent Kit (300 cycles) on a NovaSeq 6000 instrument (both Illumina) per the manufacturer’s instructions.
Single-cell transcriptome, surface protein, and chromatin accessibility sequencing
Longitudinal samples from seven additional donors at three time points were processed using the CITE-seq method (55, 56). Samples were stained with unique hashtag oligo-conjugated antibodies (HTO) targeting ubiquitously expressed surface proteins to enable sample pooling, and other antibody-derived tag antibodies (ADTs) targeting specific surface-expressed proteins using a manually constructed pool of 63 TotalSeq-C antibodies or the 137 target TotalSeq-C Human Universal Cocktail V1.0 (all BioLegend), before cell partitioning on the Chromium Controller. Around 40,000 cells were loaded into each well of Chromium chips G, and GEX and protein (HTO/ADT) libraries were constructed as previously described (21).
Cells from these seven donors were also processed for transposase-accessible chromatin with sequencing (ATAC-Seq) with hashing (57). For ATAC-Seq, the Chromium Next GEM scATAC Library and Gel Bead v1.1 Kit (10x Genomics) was used per the manufacturer’s protocol, and about 15,300 nuclei from each donor were loaded into different wells of Chromium chips H. DNA (transposase fragmentation) libraries were pooled and quantitated using the MiSeq Reagent Nano Kit before sequencing on the NovaSeq 6000 S4 Reagent Cartridge v1.5 kit (300 cycles) (all Illumina). For cell hashing, donor PBMCs were stained with different HTO, washed, and pooled into batches before formaldehyde fixation. About 33,000 fixed cells from each batch were loaded into multiple wells of Chromium chips H, and DNA (transposase fragmentation) and HTO libraries were constructed. The pooled libraries were quantitated and sequenced as described above.
Plasma proteomics
Plasma protein was measured from 14 participants at two time points using the SOMAscan V4 Assay (SomaLogic). A total of 5284 SOMAmers covering 4791 proteins were interrogated across log10 dynamic range (fM to μM) as previously described (58, 59). Briefly, SOMAmer reagents were mixed with the plasma samples at different dilutions, forming SOMAmer-protein complexes bound to streptavidin-coated beads. SOMAmer-bound proteins were washed, labeled with biotin, and released from the beads. The complexes went through a second capture and wash step before SOMAmer release, quantification, and normalization using adaptive normalization by maximum likelihood. The SomaLogic proteomics data were imported into R (v3.6) and processed with the SomaDataIO library. Proteomics data were ranked on the basis of log2 fold change of the mean protein expression of AHI and ART samples and corrected P values from a two-sided Student’s t test. The preranked dataset of 4660 proteins was then analyzed in the GSEA 4.2.0 software using the preranked method (60). Protein names were converted to gene names and analyzed with gene sets belonging to the blood transcription modules (61). Unannotated gene sets are not shown.
Cell culture
All cells were cultured at 37°C in a 5% CO2 atmosphere. HEK293T cells (American Type Culture Collection) and TZM-bl cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), L-glutamine (2 mM), streptomycin (100 μg/ml), and penicillin (100 U/ml). TZM-bl cells were provided and authenticated by the National Institutes of Health (NIH) AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH from J. C. Kappes, X. Wu, and Tranzyme Inc. (62).
Expression and proviral constructs
Codon-optimized isoform 1 of prothymosin α (UniProt, P06454–1) was synthetized by Integrated DNA Technologies. The region encoding for PTMA was PCR-amplified and subcloned in pCG-based backbone using flanking restriction sites Xba I and Mlu I. HIV-1 proviral constructs were previously described (63–65).
Transfections
HEK293T cells were transiently transfected using TransIT-LT1 (Mirus) according to the manufacturer’s protocol. To test the antiviral effect of PTMA, pCG-based expression constructs were cotransfected with the proviral constructs in 24-well plates. The amounts of pCG expression vectors were increased incrementally by fivefold ranging from 0.0016 to 1 μg, and empty vector control plasmids were used to keep the total DNA amount constant. Transfected cells were incubated for 8 to 16 hours before the medium was replaced by fresh supplemented DMEM.
Cytotoxicity
HEK293T cells were transiently transfected using TransIT-LT1 (Mirus) according to the manufacturer’s protocol, and the medium was replaced 24 hours after transfection. To test the cell viability, CellTiter-Glo Luminescent Cell Viability Assay (Promega) was performed according to the manufacturer’s protocol. Briefly, cells were lysed with 300 μl of 5× passive lysis buffer, and 25 μl was transferred into a fresh plate in duplicates. Twenty-five microliters of the CellTiter-Glo Reagent was added to the lysates and incubated for 5 min at room temperature. Luminescent signal was read using an Orion microplate luminometer (Berthold).
Viral infectivity
To determine infectious virus yield, 10,000 TZM-bl reporter cells per well were seeded in 96-well plates and infected with cell culture supernatants in triplicate on the following day. Three days after infection, cells were lysed. β-Galactosidase reporter gene expression was determined using the GalScreen Kit (Applied Bioscience) according to the manufacturer’s instructions with an Orion microplate luminometer (Berthold).
ELISA p24 and virion infectivity analysis
The amount of HIV-1 p24 in cell culture supernatants was determined by ELISA (66). Briefly, 96-well MaxiSorp microplates were coated with anti–HIV-1 p24 antibody (0.5 mg/ml; EXBIO, 11-CM006-BULK) and incubated in a wet chamber at room temperature overnight. The plates were then washed three times with PBS-T (PBS and 0.05% Tween 20) and incubated with blocking solution [PBS and 10% (v/v) FCS] for 2 hours at 37°C. After washing, the plates were loaded with 100 μl of serial dilutions of HIV-1 p24 protein (Abcam, 43037) as standard and dilutions of virus supernatants were lysed with 1% (v/v) Triton X-100 and incubated overnight in a wet chamber at room temperature. After washing away unbound capsid, 100 μl per well of polyclonal rabbit antiserum against p24 antigen [1:1000 in PBS-T with 10% (v/v) FCS; Eurogentec] was added for 1 hour at 37°C. After washing, 100 μl of goat anti-rabbit horseradish peroxidase–coupled antibody (1:2000; AB_2337937, Dianova, 111–035-008) was loaded on the plates and incubated for 1 hour at 37°C. Last, the plates were washed, and 100 μl of SureBlue TMB 1-Component Microwell Peroxidase Substrate (Medac, 52–00-04) was added. After shaking for 20 min at room temperature, the reaction was stopped with 0.5 M H2SO4 (100 μl per well). The optic density was determined by comparing with a standard curve and measured at 450 and 650 nm with the Thermo Max microplate reader (Molecular Devices). The virion infectivity values were obtained by normalizing the raw infectivity values to the amount of p24 for a specific volume of supernatant.
Supernatants and whole-cell lysates
To determine expression of cellular and viral proteins, cells were washed in PBS and subsequently lysed in transmembrane lysis buffer [150 mM NaCl, 5 mM Hepes, 5 mM EDTA, and 1% Triton X-100 (pH 7.5)] supplemented with protease inhibitor (1:500; Roche). After 10 min of incubation on ice, samples were centrifuged (4°C for 20 min at 14,000 rpm) to remove cell debris. The supernatant was transferred to a fresh tube, and the protein concentration was measured with the Pierce Rapid Gold BCA Protein Assay Kit (Thermo Fisher Scientific) and adjusted using transmembrane lysis buffer. Virus-containing supernatants (750 μl) were centrifuged on top of a 20% sucrose layer (250 μl) at 21,000g for 2 hours. The viral pellet was then lysed in transmembrane lysis buffer with 4× protein sample loading buffer (LI-COR) supplemented with 10% β-mercaptoethanol (Sigma-Aldrich) and heated at 95°C for 5 min.
SDS–polyacrylamide gel electrophoresis and immunoblotting
Whole-cell lysates were mixed with 4× protein sample loading buffer (LI-COR, at a final dilution of 1×) supplemented with 10% β-mercaptoethanol (Sigma-Aldrich), heated at 95°C for 10 min, separated on NuPAGE 4 ± 12% bis-tris gels (Invitrogen) for 90 min at 110 V, and blotted onto Immobilon-FL polyvinylidene difluoride membranes (Merck Millipore) at a constant voltage of 30 V for 30 min. For blotting of PTMA, the transfer was performed using an acidic transfer buffer (20 mM sodium acetate, pH 5.2) as previously described (67). Briefly, after transfer, the membrane was fixed in 0.5% glutaraldehyde for 5 min, transferred to a fresh 0.5% glutaraldehyde solution for 10 min, and transferred to a 500 mM glycine solution for 10 min to stop the cross-linking. Afterward, the membrane was blocked in casein (Thermo Fisher Scientific) for 1 hour at room temperature. Proteins were stained using primary antibodies against PTMA (1:500; LSBio, LS-C358258–100), HIV-1 p24 (1:1000; Abcam, ab9071,AB_306981), HIV-1 Env (1:1000; NIH, ARP-12559), and anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1000; BioLegend, 607902,AB_2734503).
Quantitative real-time PCR
Viral transcript quantity was determined as previously described (68). Briefly, HEK293T cells were cotransfected with proviral constructs of either NL4–3, CH077 TF, or CH077 CC and increasing doses of expression constructs for PTMA. At 40 hours after transfection, cells were washed with PBS and lysed in RLT Plus buffer containing 1% β-mercaptoethanol. Total RNA was isolated using the RNeasy Plus Mini Kit (QIAGEN) according to the manufacturer’s instructions. Residual genomic DNA was removed from the RNA preparations using the DNA-free DNA Removal Kit (Thermo Fisher Scientific). qRT-PCR was performed according to the manufacturer’s instructions using TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific) and the OneStepPlus Real-Time PCR System (96-well format, fast mode) using viral primer/probe sets (Biomers/TIB Molbiol) in multiplex reactions with GAPDH (Thermo Fisher Scientific) as control. Viral primers and probes were designed as previously described (69) to measure R-U5/Gag mRNA as indicator of proximal elongation and U3-polyA mRNA as indicator for completed transcription.
Single-cell gene expression analysis
Single-cell gene expression data from PBMCs were generated using the 10x Genomics Cell Ranger pipeline (10x Genomics, CA, v3.1.0) (13). The human reference genome GRCh38 with Ensembl gene annotations v93/98 was combined with the sequence of the HIV-1 subtype CRF01-AE virus (GenBank, AB070352.1). We used this concatenated reference as input for Cell Ranger mkref, and the FASTQ files were aligned to the reference according to the manufacturer’s recommendations. A sequencing depth of 42,000 to 235,000 mean reads per cell was obtained, and a median of 1607 genes and 4066 transcripts (UMI) was detected in each cell, with median Q30 bases greater than 90%. The fraction of reads mapped to the genome was ≥86% for all participants, and mean sequencing saturation was 82.4%. Quality filtering, normalization, multisample integration, visualization, and differential gene expression were performed using the R package Seurat (v3.1.5 to v4.0.5) (70, 71). Cells with greater than 10% of transcripts derived from mitochondrial genes were removed from the analysis. Gene expression was normalized with the Seurat NormalizeData function, and participant-specific Seurat objects were prepared for integration using FindVariableFeatures, ScaleData, and RunPCA (rpca). Integration features were selected from the top 2000 variable features using the first 30 principal components analysis (PCA) dimensions, and the gene expression data were integrated across participants using rpca. Cells were clustered by the Shared Nearest Neighbor method with a resolution of 0.5, and clusters were visualized by Uniform Manifold Approximation and Projection (UMAP). Cluster marker genes were determined using Seurat FindAllMarkers, and cluster identities were manually annotated using DEGs between the clusters and known lineage cell markers. Differential gene expression analysis within each cell type subset comparing cells at the AHI and ART time point was performed within Seurat using a logistic regression framework with participant as the latent variable and Bonferroni correction (n = 23,102). Genes that were not expressed in at least 10% of cells in either group were excluded from consideration. Further analysis of wide sequencing data of participant 0059 was performed by selecting all memory CD4+ T cells classified on the basis of transcriptome data and then reclustered.
Identification and differential analyses of vRNA+ cells
HIV RNA+ cells were defined as those having one or more UMI-tagged transcripts that mapped to viral sequences from the hybrid reference genome. Differential analysis on cells using the normalized HIV-1 count data and the number of genes expressed in the cell as continuous covariates was performed using the MAST software. Reported P values implement the “Hurdle” model for continuous analyses (72). Genes that were not expressed in at least 10% of cells were excluded from the analysis, and participant information was included as a categorical latent variable when multiple participants were analyzed. All memory CD4+ T cell DEG analyses were corrected for multiple testing, with the number of genes ranging between 4782 and 5551 per participant. In addition, for both the categorical and continuous analyses, genes excluded from further consideration for analyses included mitochondrial, ribosomal, variable Ig/TCR, non–protein coding, and pseudogenes (Ensembl v93, 98 and National Center for Biotechnology Information annotations). DEGs comparing vRNA+ and vRNA− memory CD4+ T cells from participant 0059 were analyzed using Metascape to identify enriched pathways (73). Similar single-cell vRNA detection was performed on an additional two Thai participants with CRF01_AE infection and a previously published single-cell dataset as described above (6). For the B subtype samples, FASTQ files for RNA-seq experiments performed on unstimulated CD4+ T cells from six viremic individuals were downloaded from the Gene Expression Omnibus (GEO) database (GSE187515) and aligned to a reference genome containing the human genome GRCh38 [Ensembl gene annotations v97 and the subtype B HIV-1 genome (GenBank, accession NC_001802.1)] using the Cell Ranger count program. Cells were clustered on the basis of gene expression patterns using Seurat, and memory CD4+ T cells were selected on the basis of protein expression data obtained from Cell Ranger matrix files provided as supplementary data for the GEO submission in GSE187515 processed data.tar.gz files.
Surface protein CITE-seq data analysis
Paired single-cell gene and surface protein expression data from PBMCs were generated using the 10x Genomics Cell Ranger pipeline (10x Genomics, CA, v3.1.0) (13) Surface protein data were analyzed in Seurat v4 as described previously (21). HTO data were centered log ratio (CLR)–normalized across features and demultiplexed using HTODemux with a positive quantile threshold of 0.99 (55). After removal of doublets and negative droplets, cells were split and combined into participant-specific Seurat objects, which were then used for the default single-cell gene expression analysis described above. The surface protein measurements were additionally CLR-normalized and used for protein-based dimensionality reduction, clustering, and differential expression analysis.
Single-cell ATAC-seq data analysis
Single-cell reads for hashed and ATAC libraries were demultiplexed using Cellranger arc (version 2.0.1), followed by alignment to the human genome reference (GRCh38, GENCODE v32/Ensembl 98) and quantification using Cellranger-atac count (version 2.0.0). All samples were subsequently analyzed using Signac (v1.4.0) to call peaks from cells and perform quality control on the basis of thresholds for peak region fragments (3 to 20 kb), percentage of reads in peaks (>15%), black-list ratio (>0.05), nucleosome signal score (<4), and transcription start site (TSS) enrichment score (>2) (74). Cells were normalized and subjected to dimensionality reduction using Signac’s latent semantic indexing (LSI) method. Only singlet cells (identifying with a unique hash) were retained. Cell clusters were identified and annotated using gene activity based on lineage markers. Memory CD4+ T cells from preinfection, AHI, and ART time points were integrated into one Seurat object using a common set of peaks as described in Signac’s integration vignette (71). Scores for co-accessibility of peaks in the region 500 kb upstream and downstream of PTMA were calculated by Cicero (v1.3) (75). Peaks with coaccessibility score < 0.1 with any of the PTMA peaks were removed from analysis. Signac’s FindMotifs method was used to calculate fold enrichments of motifs present in co-accessible peaks using JASPAR (76). Gene expression in the scRNA-seq data for these seven participants was used to restrict the set of motifs analyzed to transcription factors for which there was expression in at least 10% of memory CD4+ T cells.
TCR sequence analysis
TCR clonotype identification, alignment, and annotation were performed using the 10x Genomics Cell Ranger pipeline (v3.1.0, 10x Genomics) according to the manufacturer’s recommendations. Clonotype alignment was performed against the Cell Ranger human V(D)J reference library 3.1.0 (GRCh38 and Ensembl GTF v94). TCR clonotypes were defined as unique TCR-β complementarity determining region (CDR3) nucleotide sequences, and clonal expansion was determined using either a binary or a three-group classification. For the binary classification, TCR clonotypes were defined as either expanded (detected in at least two cells) or unique (detected in no more than one cell) in all T cells. In the three-group classification method, TCR clonotypes observed among these cells were classified as singlets, medium clones (detected in two to four cells), and large clones (detected in five or more cells). The percentage of cells in singlet, medium, and large clones was determined for cells within each of the four memory CD4+ T cell clusters.
Statistical analysis
All raw, individual-level data for experiments where n < 20 are presented in data file S6. All paired comparisons were performed by Wilcoxon signed-rank test or Mann-Whitney U test. Correlations were performed by Spearman’s rank correlation coefficient. A two-sided P value of <0.05 was considered statistically significant with multiple testing as appropriate for all statistical analyses described above. All descriptive and inferential statistical analyses were performed using R 3.6.0 and higher. For the boxplots, the lower and upper bounds corresponding to the first and third quartiles (the 25th and 75th percentile) and the median (the 50th percentile) are shown. All data points representing samples are plotted individually when possible. Pairs of data points are linked using gray solid lines. For the violin plots, the center line is plotted at the median. Unless otherwise stated, data for the overexpression assays are shown as the mean of at least three independent experiments ± SEM.
Supplementary Material
Acknowledgments:
We would like to thank the volunteers and staff of the RV254/SEARCH010 and RV217 cohorts. We thank R. Gottardo for advice on analysis. We thank M. Rolland and D. Bolton for critical review. Assistance using NGS instrumentation from various members of the Viral Disease branch at WRAIR is acknowledged. The views expressed are those of the authors and should not be construed to represent the positions or views of the U.S. Army, the U.S. Department of Defense (DOD), the U.S. Centers for Disease Control and Prevention, the U.S. Public Health Service, or the U.S. government.
Funding:
This work was supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine Inc. and the DOD. This research was also funded in part by the U.S. National Institute of Allergy and Infectious Disease (AAI20052001 to J.A.A., 5UM1AI126603-05 to S.V., and R01AI16246 and UM1AI164570 to B.H.H.). RV254/SEARCH 010 is supported by cooperative agreements (W81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine Inc. and the U.S. Department of Defense (DOD). F.K. was supported by the German Research Foundation (DFG, Collaborative Research Centre 1279) and the Baden-Württemberg Foundation (BWST_ISF2018-032). C.P.B. and A.L. are part of the International Graduate School in Molecular Medicine Ulm (IGradU). We are grateful to the Thai Government Pharmaceutical Organization (GPO), ViiV Healthcare, Gilead Sciences, and Merck for providing the antiretroviral medications for this study. Material has been reviewed by the Walter Reed Army Institute of Research, and there is no objection to its presentation or publication. The investigators have adhered to the policies for protection of human participants as prescribed in AR 70-25.
Footnotes
Competing interests:
The authors declare that they have no competing interests.
Data and materials availability:
All data associated with this study are in the paper or the Supplementary Materials. Single-cell gene expression data generated in this study are submitted to the National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository under accession number GSE233747. Source data files have been provided for plasma proteomics and chromatin accessibility datasets and are available on Figshare: https://doi.org/10.6084/m9.figshare.c.6288354. The previously published scRNA-seq dataset was accessed from GEO accession number GSE187515. Scripts for data analysis were written in the R programming language. Code used for the main figure analyses is available in R markdown format at the Figshare repository: https://doi.org/10.6084/m9.figshare.c.6288354.
REFERENCES AND NOTES
- 1.Malim MH, Bieniasz PD, HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med 2, a006940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheehy AM, Gaddis NC, Choi JD, Malim MH, Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J, The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neil SJ, Zang T, Bieniasz PD, Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, Shianna KV, Ge D, Günthard HF, Goldstein DB, Telenti A; Swiss HIV Cohort Study; Center for HIV/AIDS Vaccine Immunology, Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLOS Pathog. 6, e1000781 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collora JA, Liu R, Pinto-Santini D, Ravindra N, Ganoza C, Lama JR, Alfaro R, Chiarella J, Spudich S, Mounzer K, Tebas P, Montaner LJ, van Dijk D, Duerr A, Ho YC, Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones. Immunity 55, 1013–1031.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, Massanella M, Brassard N, Alsahafi N, Delgado GG, Routy JP, Walker BD, Finzi A, Chomont N, Kaufmann DE, Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe 20, 368–380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colby DJ, Trautmann L, Pinyakorn S, Leyre L, Pagliuzza A, Kroon E, Rolland M, Takata H, Buranapraditkun S, Intasan J, Chomchey N, Muir R, Haddad EK, Tovanabutra S, Ubolyam S, Bolton DL, Fullmer BA, Gorelick RJ, Fox L, Crowell TA, Trichavaroj R, Connell RO, Chomont N, Kim JH, Michael NL, Robb ML, Phanuphak N, Ananworanich J; RV411 study group, Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat. Med 24, 923–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colby DJ, Sarnecki M, Barouch DH, Tipsuk S, Stieh DJ, Kroon E, Schuetz A, Intasan J, Sacdalan C, Pinyakorn S, Grandin P, Song H, Tovanabutra S, Shubin Z, Kim D, Paquin-Proulx D, Eller MA, Thomas R, de Souza M, Wieczorek L, Polonis VR, Pagliuzza A, Chomont N, Peter L, Nkolola JP, Vingerhoets J, Truyers C, Pau MG, Schuitemaker H, Phanuphak N, Michael N, Robb ML, Tomaka FL, Ananworanich J, Safety and immunogenicity of Ad26 and MVA vaccines in acutely treated HIV and effect on viral rebound after antiretroviral therapy interruption. Nat. Med 26, 498–501 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Stuart T, Satija R, Integrative single-cell analysis. Nat. Rev. Genet 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, Farland CM, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananworanich J, Fletcher JL, Pinyakorn S, van Griensven F, Vandergeeten C, Schuetz A, Pankam T, Trichavaroj R, Akapirat S, Chomchey N, Phanuphak P, Chomont N, Michael NL, Kim JH, de Souza M; RV254/SEARCH 010 Study Group, A novel acute HIV infection staging system based on 4th generation immunoassay. Retrovirology 10, 56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Souza MS, Phanuphak N, Pinyakorn S, Trichavaroj R, Pattanachaiwit S, Chomchey N, Fletcher JL, Kroon ED, Michael NL, Phanuphak P, Kim JH, Ananworanich J; RV254SEARCH 010 Study Group, Impact of nucleic acid testing relative to antigen/antibody combination immunoassay on the detection of acute HIV infection. AIDS 29, 793–800 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP, Dynamics of HIV viremia and antibody seroconversion in plasma donors: Implications for diagnosis and staging of primary HIV infection. AIDS 17, 1871–1879 (2003). [DOI] [PubMed] [Google Scholar]
- 17.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF, The immune response during acute HIV-1 infection: Clues for vaccine development. Nat. Rev. Immunol 10, 11–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S, Kroon E, Sawe FK, Sinei S, Sriplienchan S, Jagodzinski LL, Malia J, Manak M, de Souza MS, Tovanabutra S, Sanders-Buell E, Rolland M, Dorsey-Spitz J, Eller MA, Milazzo M, Li Q, Lewandowski A, Wu H, Swann E, O’Connell RJ, Peel S, Dawson P, Kim JH, Michael NL; RV 217 Study Team, Prospective study of acute HIV-1 infection in adults in East Africa and Thailand. N. Engl. J. Med 374, 2120–2130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer SS, Bibollet-Ruche F, Sherrill-Mix S, Learn GH, Plenderleith L, Smith AG, Barbian HJ, Russell RM, Gondim MV, Bahari CY, Shaw CM, Li Y, Decker T, Haynes BF, Shaw GM, Sharp PM, Borrow P, Hahn BH, Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc. Natl. Acad. Sci. U.S.A 114, E590–E599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, Lebedeva M, DeCamp A, Li D, Grove D, Self SG, Borrow P, Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol 83, 3719–3733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li SS, Hickey A, Shangguan S, Ehrenberg PK, Geretz A, Butler L, Kundu G, Apps R, Creegan M, Clifford RJ, Pinyakorn S, Eller LA, Luechai P, Gilbert PB, Holtz TH, Chitwarakorn A, Sacdalan C, Kroon E, Phanuphak N, de Souza M, Ananworanich J, O’Connell RJ, Robb ML, Michael NL, Vasan S, Thomas R, HLA-B *46 associates with rapid HIV disease progression in Asian cohorts and prominent differences in NK cell phenotype. Cell Host Microbe 30, 1173–1185.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levesque K, Finzi A, Binette J, Cohen EA, Role of CD4 receptor down-regulation during HIV-1 infection. Curr. HIV Res. 2, 51–59 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Okoye AA, Picker LJ, CD4+ T-cell depletion in HIV infection: Mechanisms of immunological failure. Immunol. Rev 254, 54–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauter D, Kirchhoff F, Evolutionary conflicts and adverse effects of antiviral factors. ELife 10, e65243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duggal NK, Emerman M, Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol 12, 687–695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sékaly RP, Chomont N, CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLOS Pathog. 12, e1005761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauter D, Kirchhoff F, Key viral adaptations preceding the AIDS pandemic. Cell Host Microbe 25, 27–38 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Bosso M, Stürzel CM, Kmiec D, Badarinarayan SS, Braun E, Ito J, Sato K, Hahn BH, Sparrer KMJ, Sauter D, Kirchhoff F, An additional NF-κB site allows HIV-1 subtype C to evade restriction by nuclear PYHIN proteins. Cell Rep. 36, 109735 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauter D, Schindler M, Specht A, Landford WN, Münch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F, Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kmiec D, Nchioua R, Sherrill-Mix S, Stürzel CM, Heusinger E, Braun E, Gondim MVP, Hotter D, Sparrer KMJ, Hahn BH, Sauter D, Kirchhoff F, CpG frequency in the 5′ third of the env gene determines sensitivity of primary HIV-1 strains to the zinc-finger antiviral protein. MBio 11, e02903–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Stürzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, Hornung V, Sauter D, Telenti A, Kirchhoff F, Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19, 504–514 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Ndhlovu ZM, Kamya P, Mewalal N, Kløverpris HN, Nkosi T, Pretorius K, Laher F, Ogunshola F, Chopera D, Shekhar K, Ghebremichael M, Ismail N, Moodley A, Malik A, Leslie A, Goulder PJR, Buus S, Chakraborty A, Dong K, Ndung’u T, Walker BD, Magnitude and kinetics of CD8+ T cell activation during hyperacute HIV infection impact viral set point. Immunity 43, 591–604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Townsley SM, Donofrio GC, Jian N, Leggat DJ, Dussupt V, Mendez-Rivera L, Eller LA, Cofer L, Choe M, Ehrenberg PK, Geretz A, Gift S, Grande R, Lee A, Peterson C, Piechowiak MB, Slike BM, Tran U, Joyce MG, Georgiev IS, Rolland M, Thomas R, Tovanabutra S, Doria-Rose NA, Polonis VR, Mascola JR, McDermott AB, Michael NL, Robb ML, Krebs SJ, B cell engagement with HIV-1 founder virus envelope predicts development of broadly neutralizing antibodies. Cell Host Microbe 29, 564–578.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ndhlovu ZM, Kazer SW, Nkosi T, Ogunshola F, Muema DM, Anmole G, Swann SA, Moodley A, Dong K, Reddy T, Brockman MA, Shalek AK, Ndung’u T, Walker BD, Augmentation of HIV-specific T cell function by immediate treatment of hyperacute HIV-1 infection. Sci. Transl. Med 11, eaau0528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kazer SW, Walker BD, Shalek AK, Evolution and diversity of immune responses during acute HIV infection. Immunity 53, 908–924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kløverpris HN, Kazer SW, Mjösberg J, Mabuka JM, Wellmann A, Ndhlovu Z, Yadon MC, Nhamoyebonde S, Muenchhoff M, Simoni Y, Andersson F, Kuhn W, Garrett N, Burgers WA, Kamya P, Pretorius K, Dong K, Moodley A, Newell EW, Kasprowicz V, Karim SSA, Goulder P, Shalek AK, Walker BD, Ndung’u T, Leslie A, Innate lymphoid cells are depleted irreversibly during acute HIV-1 infection in the absence of viral suppression. Immunity 44, 391–405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krebs SJ, Kwon YD, Schramm CA, Law WH, Donofrio G, Zhou KH, Gift S, Dussupt V, Georgiev IS, Schätzle S, McDaniel JR, Lai YT, Sastry M, Zhang B, Jarosinski MC, Ransier A, Chenine AL, Asokan M, Bailer RT, Bose M, Cagigi A, Cale EM, Chuang GY, Darko S, Driscoll JI, Druz A, Gorman J, Laboune F, Louder MK, McKee K, Mendez L, Moody MA, O’Sullivan AM, Owen C, Peng D, Rawi R, Sanders-Buell E, Shen CH, Shiakolas AR, Stephens T, Tsybovsky Y, Tucker C, Verardi R, Wang K, Zhou J, Zhou T, Georgiou G, Alam SM, Haynes BF, Rolland M, Matyas GR, Polonis VR, McDermott AB, Douek DC, Shapiro L, Tovanabutra S, Michael NL, Mascola JR, Robb ML, Kwong PD, Doria-Rose NA, Longitudinal analysis reveals early development of three MPER-directed neutralizing antibody lineages from an HIV-1-infected individual. Immunity 50, 677–691.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rolland M, Tovanabutra S, Dearlove B, Li Y, Owen CL, Lewitus E, Sanders-Buell E, Bose M, O’Sullivan AM, Rossenkhan R, Labuschagne JPL, Edlefsen PT, Reeves DB, Kijak G, Miller S, Poltavee K, Lee J, Bonar L, Harbolick E, Ahani B, Pham P, Kibuuka H, Maganga L, Nitayaphan S, Sawe FK, Eller LA, Gramzinski R, Kim JH, Michael NL, Robb ML; The RV217 Study Team, Molecular dating and viral load growth rates suggested that the eclipse phase lasted about a week in HIV-1 infected adults in East Africa and Thailand. PLOS Pathog. 16, e1008179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gantner P, Buranapraditkun S, Pagliuzza A, Dufour C, Pardons M, Mitchell JL, Kroon E, Sacdalan C, Tulmethakaan N, Pinyakorn S, Robb ML, Phanuphak N, Ananworanich J, Hsu D, Vasan S, Trautmann L, Fromentin R, Chomont N, HIV rapidly targets a diverse pool of CD4+ T cells to establish productive and latent infections. Immunity 56, 653–668.e5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Kim HE, Shu H, Zhao Y, Zhang H, Kofron J, Donnelly J, Burns D, Ng SC, Rosenberg S, Wang X, Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science 299, 223–226 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Su BH, Tseng YL, Shieh GS, Chen YC, Shiang YC, Wu P, Li KJ, Yen TH, Shiau AL, Wu CL, Prothymosin α overexpression contributes to the development of pulmonary emphysema. Nat. Commun 4, 1906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez-Marquez J, Rodriguez P, Prothymosin α is a chromatin-remodelling protein in mammalian cells. Biochem. J 333, 1–3 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matteucci C, Grelli S, Balestrieri E, Minutolo A, Argaw-Denboba A, Macchi B, Sinibaldi-Vallebona P, Perno CF, Mastino A, Garaci E, Thymosin α 1 and HIV-1: Recent advances and future perspectives. Future Microbiol. 12, 141–155 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Garaci E, Rocchi G, Perroni L, D’Agostini C, Soscia F, Grelli S, Mastino A, Favalli C, Combination treatment with zidovudine, thymosin α1 and interferon-α in human immunodeficiency virus infection. Int. J. Clin. Lab. Res 24, 23–28 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Mosoian A, Teixeira A, Burns CS, Sander LE, Gusella GL, He C, Blander JM, Klotman P, Klotman ME, Prothymosin-α inhibits HIV-1 via toll-like receptor 4-mediated type I interferon induction. Proc. Natl. Acad. Sci. U.S.A 107, 10178–10183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosoian A, Teixeira A, High AA, Christian RE, Hunt DF, Shabanowitz J, Liu X, Klotman M, Novel function of prothymosin alpha as a potent inhibitor of human immunodeficiency virus type 1 gene expression in primary macrophages. J. Virol 80, 9200–9206 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karetsou Z, Sandaltzopoulos R, Frangou-Lazaridis M, Lai CY, Tsolas O, Becker PB, Papamarcaki T, Prothymosin alpha modulates the interaction of histone H1 with chromatin. Nucleic Acids Res. 26, 3111–3118 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert SA, Jolma A, Campitelli LF, das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT, The human transcription factors. Cell 175, 598–599 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, Greenblatt J, Frey BJ, Hughes TR, C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol 33, 555–562 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Smith AJ, Li Q, Wietgrefe SW, Schacker TW, Reilly CS, Haase AT, Host genes associated with HIV-1 replication in lymphatic tissue. J. Immunol 185, 5417–5424 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rheinberger M, Costa AL, Kampmann M, Glavas D, Shytaj IL, Sreeram S, Penzo C, Tibroni N, Garcia-Mesa Y, Leskov K, Fackler OT, Vlahovicek K, Karn J, Lucic B, Herrmann C, Lusic M, Genomic profiling of HIV-1 integration in microglia cells links viral integration to the topologically associated domains. Cell Rep. 42, 112110 (2023). [DOI] [PubMed] [Google Scholar]
- 52.Jefferys SR, Burgos SD, Peterson JJ, Selitsky SR, Turner AMW, James LI, Tsai YH, Coffey AR, Margolis DM, Parker J, Browne EP, Epigenomic characterization of latent HIV infection identifies latency regulating transcription factors. PLOS Pathog. 17, e1009346 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Li G, Ivanova A, Aaron S, Simm M, The critical role of human transcriptional repressor CTCF mRNA up-regulation in the induction of anti-HIV-1 responses in CD4+ T cells. Immunol. Lett 117, 35–44 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandergeeten C, Fromentin R, Merlini E, Lawani MB, DaFonseca S, Bakeman W, McNulty A, Ramgopal M, Michael N, Kim JH, Ananworanich J, Chomont N, Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol 88, 12385–12396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P, Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck III WM, Smibert P, Satija R, Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mimitou EP, Lareau CA, Chen KY, Zorzetto-Fernandes AL, Hao Y, Takeshima Y, Luo W, Huang TS, Yeung BZ, Papalexi E, Thakore PI, Kibayashi T, Wing JB, Hata M, Satija R, Nazor KL, Sakaguchi S, Ludwig LS, Sankaran VG, Regev A, Smibert P, Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol 39, 1246–1258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams SA, Kivimaki M, Langenberg C, Hingorani AD, Casas JP, Bouchard C, Jonasson C, Sarzynski MA, Shipley MJ, Alexander L, Ash J, Bauer T, Chadwick J, Datta G, DeLisle RK, Hagar Y, Hinterberg M, Ostroff R, Weiss S, Ganz P, Wareham NJ, Plasma protein patterns as comprehensive indicators of health. Nat. Med 25, 1851–1857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D, Aptamer-based multiplexed proteomic technology for biomarker discovery. PLOS ONE 5, e15004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, Kasturi S, Carlone GM, Quinn C, Chaussabel D, Palucka AK, Mulligan MJ, Ahmed R, Stephens DS, Nakaya HI, Pulendran B, Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol 15, 195–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D, Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol 72, 2855–2864 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P, Relative resistance of HIV-1 founder viruses to control by interferon-α. Retrovirology 10, 146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr., Bjorkman PJ, Wilen CB, Doms RW, O’Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH, Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. U.S.A 110, 6626–6633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM, Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med 206, 1273–1289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konvalinka J, Litterst MA, Welker R, Kottler H, Rippmann F, Heuser AM, Kräusslich HG, An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J. Virol 69, 7180–7186 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsunaga H, Ueda H, Stress-induced non-vesicular release of prothymosin-α initiated by an interaction with S100A13, and its blockade by caspase-3 cleavage. Cell Death Differ. 17, 1760–1772 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Bosso M, Bozzo CP, Volcic M, Kirchhoff F, IFI16 knockdown in primary HIV-1 target cells. STAR Protoc. 2, 100236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK, HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med 10, eaap9927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck III WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao Y, Hao S, Andersen-Nissen E, Mauck III WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stuart T, Srivastava A, Madad S, Lareau CA, Satija R, Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, Adey AC, Steemers FJ, Shendure J, Trapnell C, Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, Santana-Garcia W, Tan G, Chèneby J, Ballester B, Parcy F, Sandelin A, Lenhard B, Wasserman WW, Mathelier A, JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or the Supplementary Materials. Single-cell gene expression data generated in this study are submitted to the National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository under accession number GSE233747. Source data files have been provided for plasma proteomics and chromatin accessibility datasets and are available on Figshare: https://doi.org/10.6084/m9.figshare.c.6288354. The previously published scRNA-seq dataset was accessed from GEO accession number GSE187515. Scripts for data analysis were written in the R programming language. Code used for the main figure analyses is available in R markdown format at the Figshare repository: https://doi.org/10.6084/m9.figshare.c.6288354.