Abstract
Enterococcus faecalis has become a pervasive clinical problem due to the emergence of resistance to most antibiotics. The cytolysin of E. faecalis is a novel bacterial toxin that contributes to the severity of disease. It consists of two structural subunits, which together possess both hemolytic and bactericidal activity. Both toxin subunits are encoded in a complex operon frequently harbored on pheromone-responsive plasmids. E. faecalis strains lacking such plasmids are susceptible to the bactericidal effects of the cytolysin. A novel cytolysin immunity determinant at the 3′ end of the pAD1 cytolysin operon is described in the present study. Deletion analysis and specific mutagenesis isolated the immunity function to a single open reading frame. Specific mutagenesis experiments demonstrate that cytolysin immunity is unrelated to cytolysin activator (CylA) expression as previously proposed. Cytolysin immunity is, however, encoded on the same transcript as and 3′ to CylA, and previous associations between immunity and CylA can be ascribed to the polar behavior of Tn917 insertion.
Enterococcus faecalis is a leading cause of nosocomial bacteremias, urinary tract infections, and subacute endocarditis. E. faecalis has become a pervasive clinical problem due to the emergence of resistance to most, and in some cases all, clinically relevant antibiotics (17, 24). As many as 60% of clinical isolates produce a cytolysin (22), which by multivariate analysis is associated with acutely terminal outcome (18). Further, utilizing isogenic strains of E. faecalis differing only in production of the cytolysin, three animal models of infection demonstrate unambiguously that the cytolysin contributes to the severity and lethality of infection (7, 21, 25).
Early studies of the cytolysin demonstrated its ability to act both as a hemolysin and as a bacteriocin active against a wide range of gram-positive bacteria (3, 4). The cytolysin is typically encoded by large, pheromone-responsive plasmids, the prototype of which is pAD1 (8). The genetic organization of the pAD1 cytolysin operon was ascertained by transposon and site-directed mutagenesis, followed by intracellular and extracellular complementation (13, 14, 19). The complete sequence of five structural genes identified as sufficient for expression of the cytolysin in the naturally bacteriocin-resistant host E. coli has been reported (13, 14, 41). The cytolysin is heterodimeric, consisting of a large subunit and a small subunit, both of which are required for hemolytic and bactericidal activity (14). Recent studies have demonstrated that both subunits possess lanthionine residues, placing the cytolysin as a uniquely toxic relative of the lantibiotic class of bacteriocins (2). Lantibiotics, which are produced by a number of gram-positive bacteria, possess the unusual amino acids lanthionine and β-methyllanthionine, as well as other modified amino acids (26). The lantibiotics are divided into two subgroups. Subgroup A lantibiotics are elongated, amphiphilic peptides, while those in subgroup B are globular in nature (36). The cytolysin formally fits the definition of subgroup A, which also includes nisin (16), subtilin (15), epidermin (1), and gallidermin (27). Where the mechanism of bactericidal activity is known, subgroup A lantibiotics form voltage-dependent pores that dissipate the bacterial membrane potential and interfere with energy transduction (36).
Lanthionine modifications are posttranslationally introduced into the structures of both nonidentical subunits, encoded by the cylLL and cylLS genes, a process that is believed to involve the product of the cylM gene (14). Both cytolysin subunits are secreted through a dedicated ATP-binding cassette transporter, encoded by the cylB gene product (13). Secretion of each subunit is accompanied by a proteolytic processing event (2). Once extracellular, both subunits require an additional proteolytic removal of six residues from the amino terminus (2). The final activating cleavage, which is accomplished by a subtilisin-class serine protease encoded by the cylA gene product (41), renders the cytolysin subunits active against prokaryotic and eukaryotic cells.
Because the cytolysin is unique among both bacterial toxins and lantibiotics in consisting of two nonidentical lanthionine-containing subunits, it was of interest to determine whether the immunity mechanism was also unique. The results demonstrate that cytolysin immunity is in fact unrelated to any known mechanism. Moreover, immunity can be ascribed to a single open reading frame (ORF) at the 3′ end of the cytolysin operon and is unrelated to CylA activity as previously suspected. The immunity gene is, however, cotranscribed with cylA, the gene encoding the extracellular cytolysin activator, allowing previously detected associations to be ascribed to the polar effect of Tn917 insertion.
MATERIALS AND METHODS
Bacteria, media, and reagents.
The main characteristics of the relevant bacterial strains and plasmids used in this study are listed in Table 1. E. faecalis FA2-2, a plasmid-free derivative of JH2 (10), was utilized to express the various transposon insertion and deletion derivatives of the cytolysin immunity determinant. In the absence of these derivatives, FA2-2 is noncytolytic and cytolysin susceptible. E. coli DH5α (Bethesda Research Laboratories, Inc., Gaithersburg, Md.) and XL1-Blue (Stratagene, La Jolla, Calif.), which are intrinsically cytolysin resistant, as are all gram-negative bacteria tested (23), were used for cloning and generation of deletion constructs. E. faecalis strains were cultivated routinely in brain heart infusion (BHI) (Difco Laboratories, Detroit, Mich.), whereas Luria-Bertani broth (38) was used for the cultivation of E. coli strains. Blood agar plates were used for the qualitative detection of hemolytic activity. These plates contained BHI and 1.5% Bacto Agar (Difco), to which washed human or bovine erythrocytes were added to a final concentration of 5%. Antibiotics (Sigma, St. Louis, Mo.) used for selection of recombinant E. faecalis strains included 25 μg of rifampin per ml, 10 μg of fusidic acid per ml, 50 μg of erythromycin per ml, and 500 μg of spectinomycin per ml. Antibiotics (Sigma) utilized for selection of recombinant E. coli strains included 100 μg of ampicillin per ml, 12.5 μg of tetracycline per ml, and 150 μg of spectinomycin per ml. Restriction enzymes and other molecular biological reagents were purchased from Gibco BRL and used according to the manufacturer’s instructions.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | recA1 lacZΔM15 | Bethesda Research Laboratories |
| E. coli XL-1 Blue | recA1 lacZΔM15 Tn10 Tetr | Stratagene |
| E. faecalis FA2-2 | Rifr Fusr | 10 |
| Plasmids | ||
| pBluescript SK(+/−) | Ampr | Stratagene |
| pAM714 | Tn917 inserted into pAD1 3′ to the cytolysin operon within EcoRI G | 21 |
| pAM9055 | Tn917 inserted into pAD1 within cylA | 19 |
| pAM9054 | Tn917 inserted into pAD1 91 bp 3′ to cylI | 19 |
| pAT28 | Spcr | 44 |
| pERN101 | pAT28 containing the pAD1 4.1-kb EcoRI D fragment (encodes the 3′ end of cylB, cylA, and cylI and 1.6 kb of sequence 3′ to cylI) | This work |
| pATMB | Derivative of pXMA9AT (14) containing the 3′ end of cylB and functional cylA | This work |
| pERN101 Δ XbaI | pAT28 containing the EcoRI D fragment truncated at the first XbaI site (encodes the 3′ end of cylB, cylA, and cylI truncated at the first XbaI site) | This work |
| pERN101 Δ SalI | pAT28 containing the EcoRI D fragment truncated at the first SalI site (encodes the 3′ end of cylB, cylA, and cylI truncated at the first SalI site) | This work |
| pPSC101 | pAT28 containing an EcoRI/SstI fragment encoding the 3′ end of cylB, cylA, and cylI | This work |
| pPSC102 | pAT28 containing an EcoRI/SstI fragment encoding the 3′ end of cylB, nonfunctional cylA, and cylI | This work |
Transposon mutagenesis.
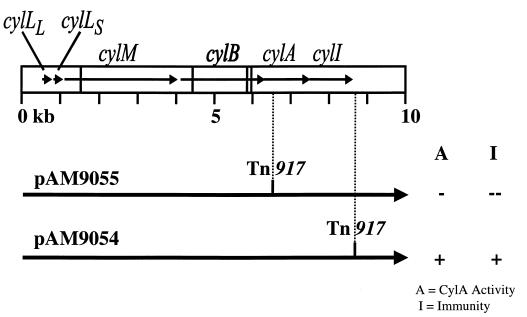
Previously generated Tn917 insertion mutations of pAD1 (19) used to assess cytolysin immunity are illustrated in Fig. 1.
FIG. 1.
Map of the cytolysin operon showing the transposon insertions which localized immunity to the 3′ end of the cytolysin operon. Insertion mutations in the cytolysin operon were characterized with respect to immunity to the cytolysin and CylA production. The two Tn917 insertion mutations of plasmid pAD1 were generated previously (19). The corresponding phenotype of E. faecalis FA2-2 harboring each pAD1 derivative is indicated on the right. Horizontal arrows within the boxed area represent the individual genes of the cytolysin operon, and the vertical lines transversing the boxed area represent EcoRI sites.
Mapping of the location of the Tn917 insertion in pAM9054.
To localize the Tn917 insertion in pAM9054 (Fig. 1), which was known to be near the putative immunity ORF, but not to affect either cytolysin activation by CylA or cytolysin immunity, PCR was conducted with primer 3 (5′-TCA ACT TTC TTA AAT ACA ACT GAG G-3′), which is complementary to the sequence 450 bp 5′ to the end of the putative immunity ORF, and primer 4 (5′-CTT ATC GAT ACA AAT TCC-3′), which is complementary to the end of Tn917. The resulting 581-bp PCR product was cloned into the TA cloning vector, pKRX (40), and sequenced.
Construction of deletion derivatives of the cytolysin operon.
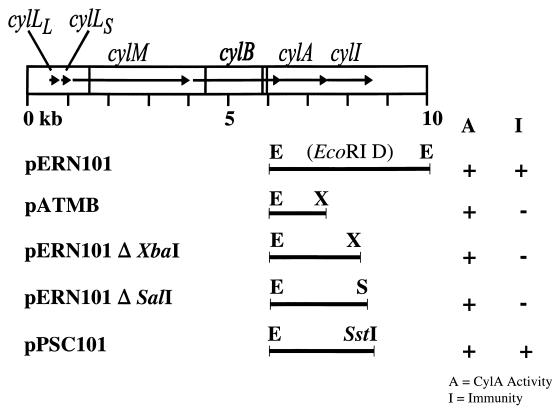
To elucidate the region of the cytolysin operon necessary for immunity, various deletions were generated. To localize the cytolysin immunity functions to sequences contained within the 4.1-kb pAD1 EcoRI D fragment, this fragment was inserted into the 6.7-kb shuttle vector pAT28 (44) by using standard recombinant DNA techniques (38), generating pERN101 (10.8 kb) (Fig. 2). Vector pAT28 carries the origin of replication from pUC18 that allows replication in E. coli and the origin of replication from pAMβ1 that allows replication in E. faecalis (44). In addition, it contains the multiple cloning site and the lacZα reporter gene of pUC18 (44). To determine whether the only previously known complete ORF within the EcoRI D fragment, cylA, is sufficient to confer immunity, sequences 5′ and 3′ to cylA were deleted. pXMA9AT, a previously generated construction lacking sequences 3′ to cylA (14), but containing the genes cylLL, cylLS, cylM, cylB, and cylA for cytolysin biosynthesis, was digested with EcoRI and recircularized by ligation with T4 DNA ligase. The ligated product contained functional cylA, with cylLL, cylLS, and cylM deleted and cylB partially deleted. Since pXMA9AT was derived by Bal 31 exonuclease digestion up to the 3′ end of cylA (14), nucleotide sequence analysis was performed at the junction between the 3′ end of cylA and the vector to demonstrate the presence of a complete cylA. Sequencing information revealed that cylA was fused in frame with the β-galactosidase gene of the vector. To interrupt this gene fusion, a self-complementary oligonucleotide of sequence 5′-CTA GGT TAA GGA TCC TTA AC-3′, which encodes a BamHI site, a stop codon, and XbaI compatible ends, was inserted into the XbaI site located within the previously inserted linker (14). Digestion with BamHI was performed to eliminate multiple linker insertions; this was followed by religation, generating the plasmid pATMB (Fig. 2). Finally, confirmation of the presence of a single BamHI/XbaI linker was obtained by BamHI digestion.
FIG. 2.
Map of the cytolysin operon showing deletion constructs generated to analyze cytolysin immunity. Deletion mutations in the cytolysin operon were characterized with respect to immunity to the cytolysin and CylA production. Each of the constructions was introduced into the noncytolytic, cytolysin-susceptible strain FA2-2 (that does not possess pAD1). The corresponding phenotype of FA2-2 transformants harboring each plasmid is indicated on the right. The single-letter restriction enzyme designations are as follows: E, EcoRI; X, XbaI; and S, SalI. EcoRI D, 4.1-kb pAD1 EcoRI D fragment. Horizontal arrows within the boxed area represent the individual genes of the cytolysin operon, and the vertical lines transversing the boxed area represent EcoRI sites.
To determine the involvement in immunity of the ORF located immediately 3′ to cylA, two deletion derivatives of the 4.1-kb pAD1 EcoRI D fragment were generated. pERN101ΔXbaI was generated by restricting pERN101 with XbaI and recircularizing the resultant plasmid, minus a 1.7-kb XbaI fragment, with T4 DNA ligase. This plasmid contains the EcoRI D fragment truncated at the first XbaI site (Fig. 2). The deletion clone pERN101ΔSalI was constructed in a fashion similar to that for pERN101ΔXbaI, except pERN101 was first restricted with SalI. pERN101ΔSalI contains the EcoRI D fragment truncated at the first SalI site (Fig. 2).
To eliminate the possibility of the involvement of additional genes in immunity and to demonstrate conclusively the role of the ORF immediately 3′ to cylA in conferring immunity to the cytolysin, a pERN101 derivative possessing the 3′ end of cylB, the cylA coding sequence, and the putative immunity ORF was constructed. PCR, utilizing pERN101 as template DNA, was performed with primer 1 (5′-AGC GGA TAA CAA TTT CAC ACA GG-3′) and primer 2 (5′-GTG TGA GCT CTT TTA CGT CGA CAG TCT AGC TGG-3′). Primer 1 is complementary to the pAT28 multiple cloning site sequence immediately 5′ to the EcoRI site demarcating the 5′ end of the EcoRI D fragment. Primer 2 contains an SstI site at its 5′ end to facilitate cloning and is complementary to the sequence beginning 32 bp 3′ to the stop codon of the putative immunity ORF. PCR generated a 2,668-bp product with an EcoRI site at the 5′ end and an SstI site at the 3′ end. The product was restricted with EcoRI and SstI and then ligated to EcoRI/SstI-digested pAT28 with T4 DNA ligase. The resulting plasmid was termed pPSC101 (Fig. 2). All deletion derivatives then were introduced into E. faecalis FA2-2 by electroporation (11). Since pAT28 is not an expression vector, expression of the above-described cloned inserts in E. faecalis was dependent on the presence of the native cylA promoter, located in the 358 bp of sequence 5′ to cylA. This 358-bp sequence was included in all pAT28 derivatives described.
Complementation of FA2-2(pAM9055) to detect CylA activity.
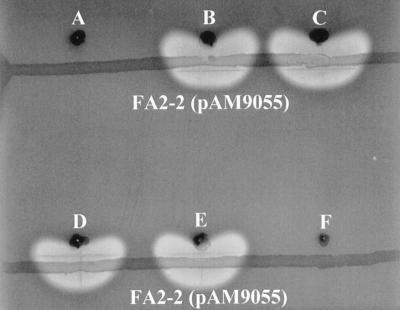
In order to detect CylA activity expressed from various derivatives, sterile swabs were used to make horizontal streaks of FA2-2(pAM9055) on blood agar plates. Strains to be tested for CylA activity were stabbed into the agar 2 to 3 mm above the horizontal FA2-2(pAM9055) streak. The plate was then incubated at 37°C overnight. FA2-2(pAM9055) does not produce active CylA but does secrete the preactivated cytolysin components. If the test strain produces and secretes active CylA, a zone of hemolysis is detected between the FA2-2(pAM9055) streak and the stab of the test strain (Fig. 3).
FIG. 3.
Extracellular complementation assay to detect CylA activity. Functional CylA production was assayed by stabbing single colonies of test clones into the agar above a horizontal streak of FA2-2(pAM9055). A zone of hemolysis is observed between clones secreting functional CylA and FA2-2(pAM9055), which secretes both preactivated cytolysin subunits. A, FA2-2(pAT28); B, FA2-2(pERN101); C, FA2-2(pERN101ΔXbaI); D, FA2-2(pATMB); E, FA2-2(pPSC101); F, FA2-2(pPSC102).
Cytolysin immunity assay.
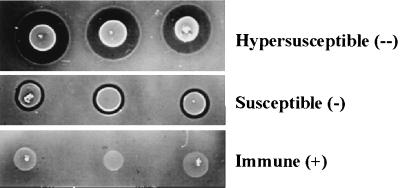
To test the various strains for immunity to cytolysin, a soft agar assay was performed essentially as described previously (20). Briefly, 50 μl of an overnight culture of a test strain grown in BHI was added to 3 ml of molten BHI soft agar (0.75% [wt/vol]) and poured onto a BHI agar plate. After solidification, 3 μl of a overnight culture of the cytolysin-producing strain FA2-2(pAM714) was spotted in triplicate onto the plate. To control for the production of other potentially inhibitory metabolites other than cytolysin, 3 μl of an overnight culture of plasmid-free strain FA2-2 was also spotted in triplicate on the plate. The plate was then incubated at 37°C overnight. Three phenotypes—hypersusceptible, susceptible, and immune—were defined as follows. The plasmid-free strain FA2-2, as well as FA2-2 that possesses the vector pAT28, were utilized to define the susceptible phenotype. These strains do not possess any genes related to cytolysin or immunity. Any strain qualitatively exhibiting zones of inhibition around the spots of FA2-2(pAM714) that were comparable to those on lawns of FA2-2 or FA2-2(pAT28) was also deemed susceptible. FA2-2(pAM9055) was the only strain that consistently exhibited zones of inhibition around the spots of FA2-2(pAM714) that were qualitatively larger than FA2-2 alone or FA2-2(pAT28); FA2-2(pAM9055) was therefore assigned the hypersusceptible phenotype. A strain was considered immune if no zones of inhibition were detected around the spots of FA2-2(pAM714) (Fig. 4).
FIG. 4.
Cytolysin immunity assay. The transposon insertion mutants pAM9055 and pAM9054 and the deletion constructs pERN101, pATMB, pERN101ΔXbaI, pERN101ΔSalI, pPSC101, and pPSC102 were introduced into the cytolysin-susceptible strain FA2-2, and the resulting transformants were tested for immunity as described in Materials and Methods. Three phenotypes were defined: hypersusceptible, susceptible, and immune. The strains tested (cytolysin-related genotypes in parentheses) are as follows. FA2-2(pAM9055) (cylLL+ cylLS+ cylM+ cylB+ cylA::Tn917 cylI+) demonstrated the hypersusceptible phenotype; FA2-2, FA2-2(pAT28), FA2-2(pATMB) (cylA+), FA2-2(pERN101ΔXbaI) (cylA+ cylIΔXbaI), and FA2-2(pERN101ΔSalI) (cylA+ cylIΔSalI) demonstrated the susceptible phenotype; FA2-2(pAM9054) (cylLL+ cylLS+ cylM+ cylB+ cylA+ cylI+), FA2-2(pERN101) (cylA+ cylI+), FA2-2(pPSC101) (cylA+ cylI+), and FA2-2(pPSC102) (cylAΩ cylI+) demonstrated the immune phenotype. Each of the three phenotypes is shown in triplicate.
Construction of a frameshift mutation in cylA.
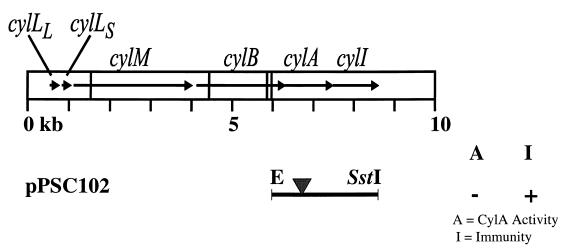
To rule out the possibility that both cylA and the putative immunity ORF, termed cylI, are required for immunity, or that cylI is dependent in any way on cylA translation or activity, the effect of introducing a frameshift mutation into cylA was examined. Utilizing pPSC101, a unique BclI site located at position 6877 of the cytolysin sequence was restricted, the cohesive ends were filled with the Klenow fragment of DNA polymerase I, and the blunt ends were ligated. This eliminated the BclI site, transforming it into a new ClaI site. The resultant plasmid, designated pPSC102 (see Fig. 6) was introduced into FA2-2 by electroporation (11). To confirm the elimination of the BclI site and introduction of the ClaI site, which was predicted to result in a frameshift, PCR was conducted by using primer CYLAF (5′-GAG AGA ATT CCC ATA GCT CTA ATT GAC TCG GG-3′), which has an EcoRI site at the 5′ end, and primer CYLAR (5′-GAG AAA GCT TCC ATA TCC TCC TGC GGG ACC AT-3′), which has a HindIII site at the 5′ end. The resulting 555-bp PCR product, encompassing the region 235 bp 5′ and 294 bp 3′ to the unique BclI site within cylA, was cloned into pBluescript SK(+/−) (Stratagene) and sequenced.
FIG. 6.
Map of the cytolysin operon indicating the frameshift mutation introduced into the cylA gene. The derivative pPSC102, possessing the frameshift mutation within cylA, was introduced into FA2-2, and the transformant was characterized with respect to cytolysin activator activity and immunity. The corresponding phenotype of the FA2-2 transformant harboring this plasmid is indicated on the right. The frameshift mutation introduced into the cylA gene is indicated by the inverted triangle. Horizontal arrows within the boxed area represent the individual genes of the cytolysin operon, and the vertical lines transversing the boxed area represent EcoRI sites.
RNA isolation.
E. faecalis FA2-2(pAM714) and E. faecalis FA2-2(pAM9055) were cultivated in 10 ml of BHI for 18 h. Total RNA was then isolated from these cultures according to the method of Cheung et al. (6), but with minor modifications as described by Shepard and Gilmore (42). This technique entails centrifugation of the cultures followed by resuspension in 1.5 ml of Tri Reagent (Sigma) and rapid disruption of the bacterial cells with 0.1-mm-diameter zirconia-silica beads (BioSpec Products, Bartlesville, Okla.) in a reciprocating shaker (BioSpec Products) at 5,000 rpm. Subsequent total RNA extraction and purification was conducted essentially as described by Shepard and Gilmore (42).
Transcriptional analysis of cylA and cylI by RT-PCR.
To determine whether cylA and cylI are cotranscribed, reverse transcription-PCR (RT-PCR) was performed. The RETROscript kit (Ambion, Inc., Austin, Tex.), which uses Moloney murine leukemia virus (MMLV) reverse transcriptase, was used for RT-PCR according to the manufacturer’s instructions. For first-strand cDNA synthesis, 1 μg of total RNA isolated from the cytolysin-producing strain FA2-2(pAM714) cultivated in BHI for 18 h and 50 pmol of primer 5 (5′-CCA AAA GCT AAG TGC CCT AAC TCA TG-3′), which is complementary to sequence 137 bp 3′ to the initiation codon of cylI, were utilized. To control for genomic DNA contamination, identical reactions were established except that MMLV reverse transcriptase was excluded. PCR amplification of the first-strand cDNA was conducted utilizing Taq DNA polymerase (Promega, Madison, Wis.), 50 pmol of primer 5, and 50 pmol of primer 6 (5′-CCT GGA TGA TAG TGA TAG GAA G-3′), which is complementary to sequence 201 bp 3′ to the initiation codon of cylA. PCR was performed at 94°C for 20 s, 55°C for 30 s, and 72°C for 60 s for a total of 25 cycles. Reactions were analyzed on an 1% native agarose gel (see Fig. 7).
FIG. 7.
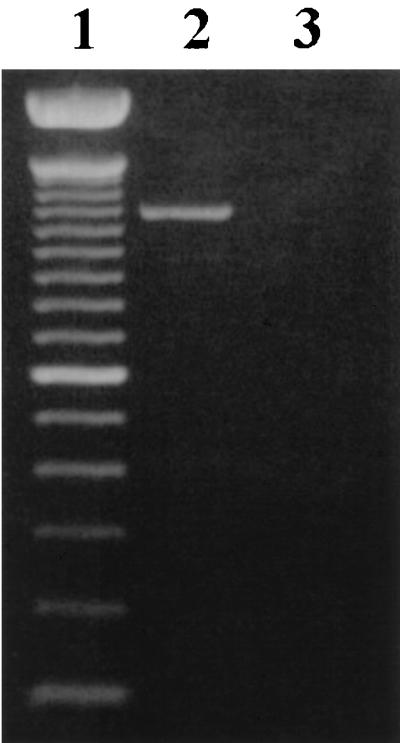
Transcriptional analysis of cylA and cylI by RT-PCR. Total RNA isolated from FA2-2(pAM714) grown in BHI was utilized in RT-PCR with primers specific to cylI and cylA. Lane 1 is a 100-bp DNA marker. Lane 2 shows a 1,176-bp product which results only if the two primer binding sites lie on the same transcript. Lane 3 is a minus reverse transcriptase control.
To confirm that the hypersusceptible phenotype observed with FA2-2(pAM9055) is related to exertion of a polar effect on cylI transcription by Tn917 insertion within cylA, quantitative RT-PCR was performed at positions on each side of the Tn917 insertion in FA2-2(pAM9055). Primers complementary to sequence 5′ (cylA specific) and 3′ (cylI specific) to the Tn917 insertion in FA2-2(pAM9055) were used. Primer cylAleft (5′-GGA GGT ACA AAA TGA AAA AAA GAG G-3′) corresponds to the ribosome binding site of cylA, and primer cylAright (5′-CCT ATA AAC TTC CTA TCA CTA TCA TC-3′) is complementary to sequence beginning 230 bp 3′ to the start codon of cylA. Primer cylIleft (5′-CCG ATG ATC TTT ACG TTA CTT CTC G-3′) is complementary to sequence beginning 13 bp 3′ to the initiator codon of cylI, and primer cylIright (5′-CCT CAG TTG TAT TTA AGA AAG TTG ACT-3′) is complementary to sequence beginning 556 bp 3′ to the initiator codon of cylI. In addition, to allow for normalization of the cylA and cylI derived products, RT-PCR was conducted with FA2-2(pAM9055) and FA2-2(pAM714) RNAs and two control primers, 23left (5′-GGT TGC ACA GAC AAC TAG GA-3′) and 23right (5′-GGA CAG GAT TCT CAC GTG TCT T-3′), which amplify a 23S rRNA sequence. For first-strand cDNA synthesis, 1 μg of total RNA isolated from FA2-2(pAM9055) or FA2-2(pAM714) cultivated in BHI for 18 h was used a template with 50 pmol of either primer cylAright, primer cylIright, or primer 23right. To control for genomic DNA contamination, identical reactions were prepared except that MMLV reverse transcriptase was excluded. PCR amplification of the first-strand cDNA was conducted with Taq DNA polymerase (Promega) and 50 pmol each of either primers cylAleft and cylAright, primers cylIleft and cylIright, or primers 23left and 23right. PCR was performed at 94°C for 20 s, 55°C for 30 s, and 72°C for 60 s for a total of 25 cycles. Reactions were analyzed on a 1% native agarose gel following normalization based on the 23S rRNA control product amplified in parallel (see Fig. 8). Relative quantities of each RT-PCR product amplified from both FA2-2(pAM714) and FA2-2(pAM9055) were determined by image analysis as previously described (42) (see Fig. 8).
FIG. 8.
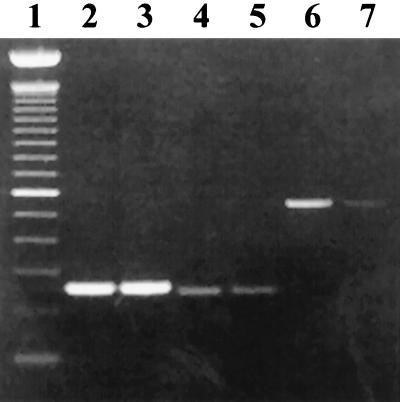
Analysis of the effect of Tn917 insertion into cylA on expression of cylI by quantitative RT-PCR. Total RNA isolated from FA2-2(pAM9055) and FA2-2(pAM714) grown in BHI was utilized in RT-PCR by using primers specific to cylI and cylA. RT-PCR products generated with primers specific to cylI and cylA were normalized based on a control product generated with primers specific to a 23S rRNA sequence. The relative quantities of each of the RT-PCR products were determined by image analysis. Lane 1 is a 100-bp DNA marker. Lanes 2 and 3 show normalized 254-bp control products generated with RNA from FA2-2(pAM714) (lane 2) and FA2-2(pAM9055) (lane 3). Lanes 4 and 5 show a 244-bp product generated with primers specific to cylA and RNA from FA2-2(pAM714) (lane 4) and FA2-2(pAM9055) (lane 5). Lanes 6 and 7 show a 544-bp product generated with primers specific to cylI and RNA from FA2-2(pAM714) (lane 6) and FA2-2(pAM9055) (lane 7).
DNA sequencing.
DNA sequence information was obtained by using standard chain termination reactions employing a fluorescein-labeled primer and a T7 DNA polymerase-based Autoread Sequencing Kit, with a Pharmacia LKB A.L.F. DNA Sequencer. GCG utilities on a VAX mainframe were employed for BLAST searches (46).
Nucleotide sequence accession number.
The nucleotide sequence of cylI has been deposited in GenBank under accession number AF102550.
RESULTS
CylA, the cytolysin activator, does not confer immunity to the cytolysin.
It was previously shown that Tn917 insertion into cylA of pAD1 results in a noncytolytic, cytolysin-hypersusceptible phenotype (pAM9055) (19, 41) (Fig. 1), suggesting that the cytolysin activator protease, CylA, may be involved at some level in immunity. It was hypothesized that cytolysin activation resulted from CylA-mediated cleavage of the cytolysin subunits at an activation site and that immunity could result from cleavage at a low-affinity site secondary to the activation cleavage. However, subsequently no direct secondary cleavage could be shown when both cytolysin subunits were incubated for an extended time with excess CylA (2), leaving the enigmatic association between CylA and immunity unexplained. To directly test whether CylA conferred immunity, a recombinant plasmid containing only the cylA coding sequence was generated. This plasmid, designated pATMB, was introduced into E. faecalis FA2-2, which lacks pAD1, and is thus natively noncytolytic and cytolysin sensitive (Fig. 2). CylA production by FA2-2(pATMB) was confirmed by extracellular complementation of FA2-2(pAM9055), in that a zone of hemolysis was detected between the horizontal streak of FA2-2(pAM9055) and the stab of FA2-2(pATMB) (Fig. 3). pATMB was then tested for the ability to confer immunity to the cytolysin, as described in Materials and Methods. FA2-2(pATMB) was found to be as susceptible to the cytolysin as plasmid-free FA2-2, indicating that CylA by itself is insufficient for protection of the producing strain from the bactericidal effects of the cytolysin (Fig. 4).
Role of pAD1 sequences adjacent to cylA.
Previous work had shown that a Tn917 insertion into the cylM coding sequence exerted a strong polar effect in abolishing expression of the immediately downstream gene, cylB (14). The observation that Tn917 insertions into the cytolysin operon exert strong polar effects on downstream genes, combined with a lack of observed direct contribution to immunity by CylA in the previous experiments, indicated that the loss of immunity in the pAD1 derivative possessing a Tn917 insertion in cylA (pAM9055) may have resulted from a polar effect on the expression of gene(s) 3′ to cylA. Another Tn917 insertion into the same general region of pAD1, designated pAM9054, was found to retain activator activity and immunity (Fig. 1). Based on restriction mapping, the Tn917 insertion in pAM9054 was localized grossly within the EcoRI D fragment at a point approximately 1.1 kb 3′ to the cylA stop codon (19). Since Tn917 insertion within cylA abrogated immunity, whereas a Tn917 insertion 1.1 kb 3′ to the cylA reading frame did not, these two insertions appeared to bracket the immunity-specifying region of pAD1 (19, 41). The 4.1-kb EcoRI D fragment from pAD1, which contains the cylA coding sequence, 358 bp 5′ to cylA, and approximately 2.5 kb of sequence 3′ to cylA, was therefore cloned into pAT28 (44). The resultant plasmid, pERN101 (Fig. 2), was introduced into the naturally sensitive strain FA2-2. FA2-2(pERN101) was tested for activator activity and immunity to the cytolysin. This strain produced and secreted functional CylA (Fig. 3) and was immune to the cytolysin. This result demonstrated that all coding information for cytolysin immunity was contained within the EcoRI D fragment. Moreover, based on the observations that cylA by itself was insufficient to confer immunity, whereas a Tn917 insertion approximately 1.1 kb 3′ to cylA did not disrupt immunity, cytolysin immunity includes sequences 3′ to the cylA gene. In addition, because of the known polarity of the Tn917 insertion (14), cytolysin immunity probably does not include sequences 3′ to the Tn917 insertion within pAM9054.
The cylI gene.
Sequencing of the region immediately 3′ to cylA revealed an ORF initiating at position 7472 and terminating at position 8455 of the cytolysin sequence (Fig. 5). This ORF, which encodes a polypeptide of 327 amino acids, is pioneer sequence as database searches revealed no homologs.
FIG. 5.
The cylI reading frame initiates at position 7472 and terminates at position 8455 of the cytolysin sequence, encoding a polypeptide of 327 amino acids. The initiator codon of cylI immediately follows the stop codon of cylA. The ribosome binding site and start codon are highlighted in boldface. The first XbaI site was utilized to generate pERN101ΔXbaI, and the SalI site was utilized to generate pERN101ΔSalI.
The observation that this ORF, termed cylI, is immediately adjacent to the reading frame encoding cylA, with only an intervening stop codon, supported the prospect that the transcription of cylI could be dependent on the expression of cylA (Fig. 5). Moreover, a potential ribosome binding site occurs relatively far upstream of the putative cylI initiator, compared to consensus spacing among genes of gram-positive bacteria (32, 33, 45). This observation prompted the hypothesis that in addition to transcriptional dependence, translation of the immunity ORF may be wholly or partially dependent on translation of cylA as a means of concentrating ribosomes in the microenvironment of the translation initiation region. These hypotheses were tested as described below.
Analysis of the predicted amino acid sequence of cylI revealed a hydrophobic N terminus and a hydrophilic C terminus, findings globally similar to the pepI gene product, which is related to immunity to the lantibiotic Pep5 (34). Examination of the N terminus did not reveal a lipoprotein consensus signal sequence, cleavage (L-X-G/A↓C) site, or membrane attachment site (cysteine), which are features of the immunity proteins, NisI and SpaI (12, 28). The central portion of the amino acid sequence possesses alternating hydrophilic and hydrophobic stretches, such as are found among transmembrane proteins (30).
Deletion analysis of the pAD1 EcoRI D fragment.
To demonstrate the role of cylI in immunity, several deletion derivatives of the pAD1 4.1-kb EcoRI D fragment were constructed. The pERN101ΔXbaI and pERN101ΔSalI derivatives (Fig. 2), which truncate the C terminus of cylI at the first XbaI site at position 8287 and at the first SalI site at position 8413, respectively, were introduced into FA2-2, and the resulting transformants were again tested for production of functional CylA and cytolysin immunity. While functional CylA production was demonstrated for both FA2-2(pERN101ΔXbaI) (Fig. 3) and FA2-2(pERN101ΔSalI), neither was found to retain immunity to the cytolysin. This suggested that the complete coding sequence of cylI may be required for immunity.
Despite a lack of polar effect on the expression of immunity of the Tn917 insertion in pAM9054, the location of which was shown by sequence analysis to be 91 bp 3′ to the stop codon of cylI, it remained a formal though unlikely possibility that downstream ORFs, contained within the 1.5-kb sequence constituting the remainder of the EcoRI D fragment, were required for immunity. To rule out the involvement of any ORFs 3′ to cylI and to demonstrate conclusively the involvement of cylI in conferring protection from the cytolysin, PCR was used to amplify the sequence encoding the 3′ end of cylB, all of cylA, and cylI ending 32 bp 3′ to cylI’s stop codon. The coding sequence of cylA was included for several reasons. First, although CylA was shown not to be directly involved in cytolysin immunity, it remained a formal possibility that CylA may be required to cleave, and thus activate, the product of cylI. Second, because of the nonconsensus spacing between the Shine-Dalgarno sequence and the initiator codon, translation of cylI was hypothesized to be dependent on translation of cylA. Finally, Tn917 insertion into cylA abrogates cytolysin immunity, suggesting that since cylA was insufficient to confer immunity, cylA and cylI may be cotranscribed. The PCR product spanning this region was cloned into pAT28, and the resultant plasmid, designated pPSC101 (Fig. 2), was introduced into FA2-2. FA2-2(pPSC101) was tested for extracellular complementation of FA2-2(pAM9055) and demonstrated to secrete functional CylA (Fig. 3). Moreover, this strain also demonstrated immunity to the cytolysin. Based on these results, immunity could be concluded to require cylI and not sequences 3′ to it.
Construction of a frameshift mutation within cylA.
To unambiguously determine whether CylA is involved indirectly in immunity, perhaps by cleaving the product of cylI to an active form, and/or to determine whether translation of cylI is dependent on cylA translation, a frameshift mutation was introduced into the coding sequence of cylA. This was accomplished by restricting pPSC101 with BclI, filling in the overhangs with Klenow, and religating the blunt ends, thus generating pPSC102 (Fig. 6). This eliminates the unique BclI site found at position 6877 within the cylA sequence but in the process introduces a ClaI site and generates a frameshift resulting in premature termination of translation of cylA. The cylA reading frame is shifted immediately following amino acid 216 into a frame specifying 13 additional amino acids not found in the cylA coding sequence. These amino acids are immediately followed by a stop codon. Thus, CylA is predicted to be truncated by 196 residues. The truncated CylA polypeptide is predicted to possess the first two members of the serine protease catalytic triad (Asp-142 and His-173) but lacks the third member of the triad, namely, Ser-359, and the entire oxyanion binding site (Ala-261, Gly-262, and Asn-263). The resulting derivative pPSC102 was introduced into FA2-2, and the transformant was tested for CylA production and cytolysin immunity. As expected, this strain failed to complement FA2-2(pAM9055), indicating lack of CylA production (Fig. 3). However, this strain retained full immunity to the cytolysin. Two deductions can be made from this finding. First, CylA is not involved in immunity to the cytolysin even indirectly, and second, cylI is not translationally dependent on cylA translation beyond the first half of the gene. Although the spacing between the Shine-Dalgarno sequence and the initiator codon of cylI is nonconsensus, the ribosome binding site is apparently functional.
Analysis of the transcriptional dependence of cylI on cylA.
As discussed above, it had been observed previously that a Tn917 insertion within cylA (pAM9055) abrogates both cytolysin activator activity and immunity. Based on the findings of the previous section, it was hypothesized that this was due to a polar effect on cylI, the next gene 3′ to the Tn917 insertion. To address this hypothesis and to provide evidence for the cotranscription of cylA and cylI, RT-PCR was performed with a primer complementary to sequence 140 bp 3′ to the initiation codon of cylI, a primer complementary to sequence 204 bp 3′ to the initiation codon of cylA, and RNA from FA2-2(pAM714), which both produces and is resistant to the cytolysin. The position for the primer complementary to cylA was chosen to ensure that the primer binding site would lie well within the cylA structural gene. RT-PCR with these primers gave rise to a 1,176-bp product, the generation of which was possible only if the two primer binding sites lie on the same transcript (Fig. 7, lane 2).
To confirm that the hypersusceptible phenotype observed with FA2-2(pAM9055) is related to the polar effect on cylI expression of Tn917 insertion within cylA, quantitative RT-PCR was performed at positions on each side of the Tn917 insertion in FA2-2(pAM9055). Primers complementary to sequence 5′ (cylA specific) and 3′ (cylI specific) to the Tn917 insertion in FA2-2(pAM9055) and RNA isolated from FA2-2(pAM9055), as well as control RNA derived from cytolysin-expressing and fully immune FA2-2(pAM714), were utilized. The cylA and cylI derived products were normalized based on results obtained for a control product from a 23S rRNA sequence (42). After normalization, RT-PCR products were visualized by electrophoresis and analyzed by image analysis to determine relative quantities (Fig. 8). Normalized 254-bp control products, generated by using primers specific to a 23S rRNA sequence and RNA from FA2-2(pAM714) (Fig. 8, lane 2) and FA2-2(pAM9055) (Fig. 8, lane 3), are shown. Although the primers specific to cylA were designed 5′ to the previously mapped location of Tn917 within pAM9055 (20), and thus the quantities of the 244-bp cylA derived RT-PCR products generated by using RNA from FA2-2(pAM714) and FA2-2(pAM9055) were predicted to be equivalent, the amount of the cylA derived product generated with RNA from FA2-2(pAM9055) (Fig. 8, lane 5) is reduced 2.8-fold relative to the amount of the cylA derived product generated with RNA from FA2-2(pAM714) (Fig. 8, lane 4). Conceivably, unexpected slight reduction in RNA 5′ to the point of Tn917 insertion could result from an increased instability of the altered cylA transcript. The amount of the 544-bp RT-PCR product (cylI specific) derived from sequences downstream of Tn917 insertion in FA2-2(pAM9055) (Fig. 8, lane 7) is reduced 35.8-fold, relative to the amount of the cylI derived RT-PCR product derived from FA2-2(pAM714) RNA (Fig. 8, lane 6). These data, as well as the results demonstrating that cylA and cylI are transcribed on a polycistronic message, indicate that Tn917 insertion within the cylA gene exerts a strong polar effect on the expression of cylI, the gene immediately 3′ to cylA, and confirm that the hypersusceptible phenotype observed with FA2-2(pAM9055) is related to abrogation of cylI transcription. The hypersusceptibility of FA2-2(pAM9055) thus results from the contribution of two factors. First, Tn917 insertion within cylA impairs expression of cylI. Second, this strain still produces both preactivated cytolysin precursors. This increases the concentration of precursors available for activation by exogenous CylA from the cytolysin-producing strain, FA2-2(pAM714), that is used as the source of cytolysin in the immunity assays, and this results in an increased zone of inhibition relative to that observed with a susceptible strain.
DISCUSSION
The E. faecalis cytolysin is a novel bacterial toxin distantly related to lantibiotics. Among lantibiotics it is unique in several respects. First, it is the only lantibiotic which is toxic to eukaryotic cells, having been demonstrated to contribute to the virulence of enterococcal disease in a number of animal models (7, 21, 25), as well as in humans (18, 22, 31). Second, it is the only lantibiotic that possesses two components, both of which are required for bactericidal activity (14).
Because the cytolysin acts as a bacteriocin capable of inhibiting the growth of gram-positive bacteria, cytolysin-producing strains, similar to other bacteriocin producers, must possess a self-protection mechanism. The exact mechanism of immunity to conventional lantibiotics is somewhat controversial, as a number of immunity factors have been described, and models for how these factors confer immunity to various lantibiotic-producing strains have been proposed (39). The putative lipoproteins, NisI and SpaI, have been shown to contribute to immunity to nisin and subtilin, respectively (12, 28, 29). Both NisI and SpaI contain the consensus cleavage and attachment site, Leu-X-Gly/Ala↓Cys (12, 28), typical of bacterial lipoproteins attached to the extracellular face of the cytoplasmic membrane. This suggests that they may contribute to immunity by interacting with the cognate lantibiotic to prevent insertion of the lantibiotic into the membrane, and/or to prevent formation of a transmembrane pore (12, 28, 29, 39).
Members of the ABC transporter family found to be encoded within a number of the lantibiotic operons also appear to contribute to conventional lantibiotic immunity (28, 35, 39, 43). It is hypothesized that these ABC transporters may protect the lantibiotic-producer organism by extruding the lantibiotic from the cytoplasmic membrane in a manner similar to the eukaryotic multidrug-resistant pumps (39). Presumably, this mechanism would maintain a concentration of lantibiotic in the membrane that is lower than that required to form a functional channel (39). In the case of nisin, which requires a transnegative potential to form pores (37) and thus can only form pores from outside of the cell, the ABC transporter may function to pump the active nisin species into the cytoplasm, where it is degraded (43).
In the present study, the region of the E. faecalis cytolysin operon related to cytolysin immunity was characterized, and a previously undefined ORF immediately adjacent to the gene encoding the cytolysin activator, cylA, was shown to be necessary and sufficient. It had previously been speculated that the cylA gene product might itself be involved in protecting cytolysin-producing strains by cleavage of one or both of the cytolysin subunits at a secondary, low-affinity site (19, 41). However, in the present study, direct involvement of CylA was ruled out when (i) the cloned cylA gene failed to confer immunity even though cytolysin activator, CylA, activity could be detected, and (ii) a frameshift mutation introduced into cylA failed to abrogate immunity. Moreover, RT-PCR experiments demonstrate that cylA and cylI are transcribed on a polycistronic message, providing an explanation for the observation that Tn917 insertion into the gene for the cytolysin activator also inactivates immunity.
Seemingly contradictory evidence to the cotranscription of cylA and cylI was provided by Ike et al. (20), who characterized pAD1::Tn916 derivatives altered in cytolysin expression and found that these derivatives fell into five classes. Four of the five classes of Tn916 insertion mapped to a similar site at the 3′ end of cylA. While these four classes altered cytolysin expression to different extents, all four classes maintained the immune phenotype (20). This lack of polar effect would suggest that cylI is transcribed independently of cylA in these derivatives. However, based on the results of the current study, which demonstrated that cylA and cylI are cocistronic, the lack of polar effect of the Tn916 insertions most likely is the result of transcription proceeding out the distal end of Tn916 into cylI, thus preserving the immune phenotype. Outward transcription from Tn916 has been advanced by Clewell et al. to explain the observation that E. faecalis mutants possessing Tn916 insertions adjacent to the cytolysin operon of pAD1 frequently exhibit hyperexpression of the cytolysin (9). These authors have shown that in a number of these hyperhemolytic mutants, Tn916 is inserted such that a potential promoter element is placed proximal to the cytolysin determinant (9). Using Northern analysis, Celli and Trieu-Cuot (5) have recently demonstrated that outward reading transcripts, initiated from promoters located in the end of Tn916 containing the tetM gene, encoding tetracycline resistance, and the xis and int genes, encoding excisionase and integrase, respectively, can extend into the host chromosome. These authors were able to detect transcripts encompassing a host gene, yufQ, located immediately adjacent to the Tn916 insertion (5).
The exact mechanism by which cytolysin immunity is achieved is not known. Database searches of the predicted amino acid sequence of cylI revealed no homologs. The predicted cylI gene product, which confers immunity, is enriched in hydrophobic amino acids, with a hydrophobic N terminus and alternating hydrophobic/hydrophilic regions in the remainder of the sequence. This suggests a membrane location for the predicted polypeptide. The high degree of hydrophobicity may relate to an adsorbent property for the activated cytolysin species. The polypeptide appears not to be a lipoprotein, as no obvious lipoprotein consensus signal sequence could be detected. Cytolysin immunity is specific to the cytolysin and does not confer cross-immunity to nisin (data not shown). This lack of cross-immunity is common among lantibiotic immunity mechanisms (39).
E. faecalis infections are becoming increasingly refractory to therapy because of the proliferation of multiply antibiotic-resistant strains. As many as 60% of E. faecalis strains isolated from sites of infection elaborate the cytolysin (22). The cytolysin has been shown to lower the 50% lethal dose in a intraperitoneal mouse model (21) and to contribute to toxicity in experimental endocarditis and endophthalmitis (7, 25). In addition, the cytolysin is associated with a fivefold-increased risk of acutely terminal outcome in patients with nosocomial enterococcal bacteremia (18). Targeting expression, modification, activation, or immunity to the cytolysin may be of value in deriving new therapeutics for treating refractory enterococcal infection.
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Agriculture grant 96-35201-3459, the U.S. Public Health Service grants EY08289 and AI41108, and by an unrestricted grant from Research to Prevent Blindness.
We gratefully acknowledge Viswanathan Shankar, Wolfgang Haas, Brett Shepard, Pravina Srinivas, and Michelle Callegan for helpful discussions.
REFERENCES
- 1.Allgaier H, Jung G, Werner R G, Schneider U, Zahner H. Epidermin: sequencing of a heterodet tetracyclic 21-peptide amide antibiotic. Eur J Biochem. 1986;160:9–22. doi: 10.1111/j.1432-1033.1986.tb09933.x. [DOI] [PubMed] [Google Scholar]
- 2.Booth M C, Bogie C P, Sahl H-G, Siezen R J, Hatter K L, Gilmore M S. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol Microbiol. 1996;21:1175–1184. doi: 10.1046/j.1365-2958.1996.831449.x. [DOI] [PubMed] [Google Scholar]
- 3.Brock T D, Davie J M. Probable identity of a group D hemolysin with a bacteriocine. J Bacteriol. 1963;86:708–712. doi: 10.1128/jb.86.4.708-712.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brock T D, Peacher B, Pierson D. Survey of the bacteriocins of enterococci. J Bacteriol. 1963;86:702–707. doi: 10.1128/jb.86.4.702-707.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celli J, Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading though the attachment site. Mol Microbiol. 1998;28:103–117. doi: 10.1046/j.1365-2958.1998.00778.x. [DOI] [PubMed] [Google Scholar]
- 6.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 7.Chow J W, Thal L A, Perri M B, Vazquez J A, Donabedian S M, Clewell D B, Zervos M J. Plasmid-associated hemolysin and aggregation substance production contributes to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewell D B. Bacterial sex pheromone-induced plasmid transfer. Cell. 1993;73:9–12. doi: 10.1016/0092-8674(93)90153-h. [DOI] [PubMed] [Google Scholar]
- 9.Clewell D B, Flannagan S E, Ike Y, Jones J M, Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988;170:3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clewell D B, Tomich P K, Gawron-Burke M C, Franke A E, Yagi Y, An F Y. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J Bacteriol. 1982;152:1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz-Rodz A, Gilmore M S. High-efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet. 1990;224:152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- 12.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian K-D. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol. 1994;60:814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmore M S, Segarra R A, Booth M C. An HlyB-type function is required for expression of the Enterococcus faecalis hemolysin/bacteriocin. Infect Immun. 1990;58:3914–3923. doi: 10.1128/iai.58.12.3914-3923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmore M S, Segarra R A, Booth M C, Bogie C P, Hall L R, Clewell D B. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol. 1994;176:7335–7344. doi: 10.1128/jb.176.23.7335-7344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross E, Kiltz H, Nebelin E. Subtilin. VI. Die Struktur des Subtilins. Hoppe-Seyler’s Z Physiol Chem. 1973;354:810–812. [PubMed] [Google Scholar]
- 16.Gross E, Morell J L. The structure of nisin. J Am Chem Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 17.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huycke M M, Spiegel C A, Gilmore M S. Bacteremia caused by hemolytic, high level gentamicin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ike Y, Clewell D B, Segarra R A, Gilmore M S. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J Bacteriol. 1990;172:155–163. doi: 10.1128/jb.172.1.155-163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ike Y, Flannagan S E, Clewell D B. Hyperhemolytic phenomena associated with insertions of Tn916 into the hemolysin determinant of Enterococcus faecalis plasmid pAD1. J Bacteriol. 1992;174:1801–1809. doi: 10.1128/jb.174.6.1801-1809.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ike Y, Hashimoto H, Clewell D B. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immun. 1984;45:528–530. doi: 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ike Y, Hashimoto H, Clewell D B. High incidence of hemolysin production by Enterococcus (Streptococcus) faecalis strains associated with human parenteral infections. J Clin Microbiol. 1987;25:1524–1528. doi: 10.1128/jcm.25.8.1524-1528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jett B D, Gilmore M S. The growth-inhibitory effect of the Enterococcus faecalis bacteriocin encoded by pAD1 extends to the oral streptococci. J Dent Res. 1990;69:1640–1645. doi: 10.1177/00220345900690100301. [DOI] [PubMed] [Google Scholar]
- 24.Jett B D, Huycke M M, Gilmore M S. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jett B D, Jensen H G, Nordquist R E, Gilmore M S. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect Immun. 1992;60:2445–2452. doi: 10.1128/iai.60.6.2445-2452.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung G. Lantibiotics—ribosomally synthesized biologically active polypeptides containing sulfide bridges and α,β-didehydroamino acids. Angew Chem Int Ed Engl. 1991;30:1051–1068. [Google Scholar]
- 27.Kellner R, Jung G, Horner T, Zahner H, Schnell N, Entian K-D, Gotz F. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur J Biochem. 1988;177:53–59. doi: 10.1111/j.1432-1033.1988.tb14344.x. [DOI] [PubMed] [Google Scholar]
- 28.Klein C, Entian K-D. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl Environ Microbiol. 1994;60:2793–2801. doi: 10.1128/aem.60.8.2793-2801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPRK of Lactococcus lactis and evidence for the involvement of expression of the nisA and nisI genes in product immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 30.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 31.Libertin C R, Dumitru R, Stein D S. The hemolysin/bacteriocin produced by enterococci is a marker of pathogenicity. Diagn Microbiol Infect Dis. 1992;15:115–120. doi: 10.1016/0732-8893(92)90033-p. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin J R, Murray C L, Rabinowitz J C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus β-lactamase gene. J Biol Chem. 1981;256:11283–11291. [PubMed] [Google Scholar]
- 33.Moran C P, Lang N, LeGrice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 34.Reis M, Eschbach-Bludau M, Iglesias-Wind M I, Kupke T, Sahl H-G. Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl Environ Microbiol. 1994;60:2876–2883. doi: 10.1128/aem.60.8.2876-2883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rince A, Dufour A, Uguen P, Le Pennec J-P, Haras D. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl Environ Microbiol. 1997;63:4252–4260. doi: 10.1128/aem.63.11.4252-4260.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahl H-G, Jack R W, Bierbaum G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 37.Sahl H-G, Kordel M, Benz R. Voltage-dependent depolarization of bacterial membranes and artificial lipid bilayers by the peptide antibiotic nisin. Arch Microbiol. 1987;149:120–124. doi: 10.1007/BF00425076. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Saris P E, Immonen T, Reis M, Sahl H-G. Immunity to lantibiotics. Antonie Leeuwenhoek. 1996;69:151–159. doi: 10.1007/BF00399420. [DOI] [PubMed] [Google Scholar]
- 40.Schutte B C, Ranade K, Pruessner J, Dracopoli N. Optimized conditions for cloning PCR products into an XcmI T-vector. BioTechniques. 1997;22:40–44. doi: 10.2144/97221bm06. [DOI] [PubMed] [Google Scholar]
- 41.Segarra R A, Booth M C, Morales D A, Huycke M M, Gilmore M S. Molecular characterization of the Enterococcus faecalis cytolysin activator. Infect Immun. 1991;59:1239–1246. doi: 10.1128/iai.59.4.1239-1246.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepard B D, Gilmore M S. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed polymerase chain reaction. Appl Environ Microbiol. 1999;65:1470–1476. doi: 10.1128/aem.65.4.1470-1476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegers K, Entian K-D. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl Environ Microbiol. 1995;61:1082–1089. doi: 10.1128/aem.61.3.1082-1089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vellanweth R L, Rabinowitz J C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 46.Warren G, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]