Abstract
Aim
To develop a multiclass non‐clinical screening tool for periodontal disease and assess its accuracy for differentiating periodontal health, gingivitis and different stages of periodontitis.
Materials and Methods
A cross‐sectional diagnostic study on a convenience sample of 408 consecutive subjects was conducted by applying three non‐clinical index tests estimating different features of the periodontal health–disease spectrum: a self‐administered questionnaire, an oral rinse activated matrix metalloproteinase‐8 (aMMP‐8) point‐of‐care test (POCT) and determination of gingival bleeding on brushing (GBoB). Full‐mouth periodontal examination was the reference standard. The periodontal diagnosis was made on the basis of the 2017 classification of periodontal diseases and conditions. Logistic regression and random forest (RF) analyses were performed to predict various periodontal diagnoses, and the accuracy measures were assessed.
Results
Four‐hundred and eight subjects were enrolled in this study, including those with periodontal health (16.2%), gingivitis (15.2%) and stage I (15.9%), stage II (15.9%), stage III (29.7%) and stage IV (7.1%) periodontitis. Nine predictors, namely ‘gum disease’ (Q1), ‘a rating of gum/teeth health’ (Q2), ‘tooth cleaning’ (Q3a), the symptom of ‘loose teeth’ (Q4), ‘use of floss’ (Q7), aMMP‐8 POCT, self‐reported GBoB, haemoglobin and age, resulted in high levels of accuracy in the RF classifier. High accuracy (area under the ROC curve > 0.94) was observed for the discrimination of three (health, gingivitis and periodontitis) and six classes (health, gingivitis, stages I, II, III and IV periodontitis). Confusion matrices showed that the misclassification of a periodontitis case as health or gingivitis was less than 1%–2%.
Conclusions
Machine learning‐based classifiers, such as RF analyses, are promising tools for multiclass assessment of periodontal health and disease in a non‐clinical setting. Results need to be externally validated in appropriately sized independent samples (ClinicalTrials.gov NCT03928080).
Keywords: artificial intelligence, multiclass prediction, periodontitis, random forest, screening
Clinical Relevance.
Scientific rationale for study: Gingivitis and periodontitis are two of humanity's most common non‐communicable diseases and remain undetected worldwide. The development of an accurate screening tool for non‐clinical settings is essential for improving their early diagnosis and effective care.
Principal findings: The developed machine learning model based on age, self‐reported measures, gingival bleeding on brushing and activated matrix metalloproteinase‐8 yielded satisfactory accuracy in differentiating periodontal health, gingivitis and different stages of periodontitis.
Practical implications: The model based on self‐reported parameters and salivary biomarkers may be a valuable tool to screen for periodontal disease at the population level once validated in multiple populations.
1. INTRODUCTION
Non‐clinical approaches for the detection and differential diagnosis of periodontal health status hold great promise for better prevention and management of periodontal diseases (gingivitis and periodontitis), which remain a significant health burden and the most common non‐communicable condition of humanity (Tonetti et al., 2017). Self‐reported measures proposed by the Centers for Disease Control and Prevention (CDC) and the American Academy of Periodontology (AAP) have shown potential in different populations (Carra et al., 2018; Deng et al., 2021b; Eke & Genco, 2007). Recent efforts have also focused on using biomarkers and their combinations (Arias‐Bujanda et al., 2020; Deng et al., 2021a, 2022; Grant et al., 2022). Despite persisting limitations, recent progress has been substantial (Gürsoy & Kantarci, 2022). Combining information from multiple variables has shown the potential to improve accuracy. For example, adding demographics to biomarker data has improved the performance of multivariate diagnostic models in periodontal health and disease (Deng et al., 2021a, 2021b; Grant et al., 2022).
Previous work from our group had identified the differential utility of specific questions in the Chinese version of the AAP/CDC questionnaire, gingival bleeding on brushing (GBoB) and levels of activated matrix metalloproteinase‐8 (aMMP‐8) in discriminating periodontal health, gingivitis and periodontitis (Deng et al., 2021a, 2021b, 2021c, 2022). The Chinese version of the CDC/AAP questionnaire showed good accuracy in the screening for severe periodontitis (Deng et al., 2021b). However, this questionnaire was less helpful in detecting gingivitis and incipient periodontitis. Recent studies have indicated that GBoB and haemoglobin (Hb) concentrations in oral rinses could be a sentinel sign of gingival inflammation with great potential for the discrimination of periodontal health and disease, especially for the detection of gingival inflammation (Deng et al., 2021c; Tonetti et al., 2020). Despite its valuable role in periodontal diagnostics, GBoB has low to moderate accuracy for periodontitis, probably because this feature is shared by both gingivitis and periodontitis. Point‐of‐care diagnostics based on salivary biomarkers could give an instant indication of the probable disease status, allowing periodontal health monitoring outside the clinic (Aro et al., 2017). Matrix metalloproteinases (MMPs) are a family of proteinases that regulate the cell‐matrix composition, and MMP‐8 is the primary type of collagenase associated with collagen degradation in periodontal disease (Birkedal‐Hansen et al., 1993; Sorsa et al., 2016). A consumer version of point‐of‐care testing (POCT) for aMMP‐8 has shown promising potential for detecting or excluding periodontitis (Deng et al., 2021a, 2022). The accuracy of aMMP‐8 alone, however, has been shown to be limited.
A fundamental limitation of this approach has been the use of conventional diagnostic accuracy analyses developed for binary conditions. Applying these analyses to multiple diagnostic scenarios, such as the discrimination between periodontal health, gingivitis and periodontitis, requires the artificial dichotomization of the diagnostic question: health and gingivitis compared to periodontitis, or health against gingivitis and periodontitis. Identifying the spectrum of periodontal case diagnoses might be valuable for optimal case management. This problem is not unique to periodontal health and is typical for multistage diseases, and specific analytical approaches have been proposed (Lorena et al., 2009). Machine learning, a subset of artificial intelligence (AI) and computer science, enables disease diagnosis and differentiation by constructing a model in the form of input–output variable analysis, thereby allowing the selection of an optimal feature subset and automatic classification (Sajda, 2006). Methods based on machine learning have been used to identify diagnostic patterns with increased accuracy (Maity & Das, 2017). Systematic reviews have identified their potential benefits for diabetes and its complications as well as for cardiovascular disease (Baashar et al., 2022; Shin et al., 2022). Initial methodological reports have shown potential for applying AI to the screening of a periodontally healthy population (Bashir et al., 2022).
We hypothesize that combining specific non‐clinical features of the periodontal health–disease spectrum can improve the discrimination of periodontal health, gingivitis and periodontitis and its various stages with a single algorithm. The specific aims of this work are the following: (i) preliminary assessment of candidate diagnostics using multivariate logistic regression; (ii) development and internal validation of screening tools based on random forests (RFs), a machine learning algorithm, to correctly classify subjects into three case definitions (periodontal health, gingivitis, and periodontitis or periodontal health, gingivitis and stage I periodontitis, stages II–IV periodontitis) and six case definitions (periodontal health, gingivitis, stage I, stage II, stage III and stage IV periodontitis).
2. MATERIALS AND METHODS
2.1. Study design and population
This was a cross‐sectional diagnostic accuracy study involving a convenience sample of consecutive patients seeking dental care in the Prince Philip Dental Hospital, Hong Kong, between July 2019 and August 2020. Details of the design and methods have been described elsewhere (Deng et al., 2021b). All adults aged 18 or older with the willingness to participate were eligible; edentulous patients, pregnant females, subjects using antibiotics within the previous 3 months and those having a history of periodontal treatment (other than supragingival cleaning) within the last 12 months were excluded. The study protocol was registered on ClinicalTrials.gov (NCT03928080) and HKU Clinical Trials Registry (HKUCTR‐2631). The ethical approval for the study was obtained from the local Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (reference: UW19‐188). Written informed consent was obtained from all participants. This study was conducted following the Declaration of Helsinki and followed the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) guidelines (Collins et al., 2015).
2.2. Sample size considerations
There are no similar screening and diagnostic studies for differentiating periodontal health, gingivitis and different stages of periodontitis. Therefore, the sample size was primarily calculated according to the existing model for screening periodontitis and using the current approach to detecting gingival inflammation as a secondary consideration. The sample size was estimated based on the observed screening accuracy of the CDC/AAP questionnaire for periodontitis in the 2009–2010 National Health and Nutrition Examination survey (sensitivity, 59.3%; specificity, 57.0%) (Eke et al., 2013) and the expected sensitivity and specificity of 70.0%. As the most recent systematic review and meta‐analysis showed an overall synthesis of 70% sensitivity and specificity of MMP‐8 alone for the diagnosis of periodontitis (Arias‐Bujanda et al., 2020), the minimum acceptable level was expected to be 70% for the combined model. With a 5% significance level and 80% power, 159 periodontitis patients and 109 non‐periodontitis (periodontal health and gingivitis) participants were required. Furthermore, there is no previous investigation on the screening accuracy of gingivitis. A study assessing the utility of self‐reported GBoB for detecting gingival inflammation (defined by ≥10% of bleeding on probing [BOP]) determined the sensitivity to 47% (Romano et al., 2020). By setting the acceptable sensitivity of the screening/diagnostic model of gingivitis as 70%, at least 35 gingivitis cases were needed.
A total of 400 subjects were recruited, considering 5% missing data and the prevalence of periodontitis in Hong Kong ranging from 40% to 59% (Department of Health, Hong Kong Government SAR, 2011). Based on the rule that the ratio of the number of subjects to the number of predictor items should be at least 3 (Mundfrom et al., 2005), the sample size in the present study was sufficient for internal validation: the ratio of the number of subjects with different periodontal case definitions to the number of predictor items ranged between 3.4 and 17.5.
2.3. Predictors: Index tests
The study procedures comprised three consecutive screening tests (index tests) and a full‐mouth periodontal examination (reference standard). Demographic characteristics, including age (years) and sex (male/female), lifestyle factors including tobacco smoking status (current, never/former) and personal medical history (e.g., presence of cardiovascular disease and diabetes) were collected by a trained nurse through face‐to‐face interviews. The screening tests consisted of a self‐administered questionnaire, a subsequent oral rinse aMMP‐8 POCT and the final GBoB test.
The development and validation of the Chinese version of the CDC/AAP questionnaire followed a standardized forward–backwards translation procedure and a cognitive assessment before it was administered to patients (Deng et al., 2021b). The complete set of the questionnaire includes a self‐assessment of ‘gum disease’ (Q1), ‘a rating of gum/teeth health’ (Q2), the self‐reported experience of ‘supragingival scaling’ (Q3a), ‘deep scaling’ (Q3b), self‐perceived tooth mobility (Q4), self‐reported professionally diagnosed bone loss (Q5), self‐awareness of the change of ‘tooth appearance’ (Q6), use of dental floss (Q7) and mouth rinse (Q8).
A commercial aMMP‐8 lateral flow immunoassay system (PerioSafe PRO, Dentognostics GmbH, Jena, Germany) and its digital analysis device (ORALyzer, Dentognostics) were used in the POCT. The aMMP‐8 test was administered to the participants by a trained and calibrated nurse according to the manufacturer's instructions: (i) a 30‐s pre‐rinse with tap water; (ii) a 60‐s wait; (iii) a 30‐s rinse with 5 mL purified water; (iv) pouring the oral rinse and drawing up 3 mL into a syringe and (v) placing a filter into the syringe and adding 3–4 droplets of the filtered samples to the test system. The value of aMMP‐8 concentration was shown on the digital reader within 5 min. A positive test result was considered if the aMMP‐8 concentration was >10 ng/mL; a concentration below the detection level was considered 10 ng/mL for the statistical analysis.
The GBoB test was carried out in a separate room with a mirror and washbasin by the same nurse in a standard way as follows: (i) all participants were asked to perform their routine without any disturbance; (ii) their saliva/toothpaste slurries (TPS) were collected and stored for further quantitative analysis of Hb at the end of each session and (iii) participants were asked to self‐assess the presence or absence of blood in the TPS (GBoB). The quantification of Hb in the TPS samples by ultraviolet–visible (UV–vis) spectroscopy has been described in detail previously (Deng et al., 2021c).
2.4. Periodontal examination and clinical diagnoses: The reference standard
A single trained and calibrated examiner (DK), who was blind to the results of all index tests, performed the comprehensive periodontal examination, including probing pocket depth (PPD), BOP and clinical attachment level (CAL) at six sites per tooth with a standardized periodontal probe (UNC‐15, Hu Friedy, Chicago, USA); furcation involvement (FI) according to the Hamp's classification (Hamp et al., 1975); tooth mobility according to the Miller scale and number of teeth lost attributed to periodontitis.
Case definitions of periodontal health, gingivitis and periodontitis were based on the 2017 World Workshop on the Classification of Periodontal and Peri‐implant Diseases and Conditions (Chapple et al., 2018; Papapanou et al., 2018; Tonetti et al., 2018; Trombelli et al., 2018). In this paper, ‘periodontal disease’ refers to plaque‐induced gingivitis and periodontitis, to avoid misunderstanding by including a broader spectrum of gingival diseases and other forms of periodontitis. A periodontally healthy case was defined as the absence of gingival inflammation (BOP < 10%) and attachment loss attributed to periodontitis (Chapple et al., 2018). A gingivitis case was defined as the presence of gingival inflammation (BOP ≥ 10%) and absence of detectable attachment loss resulting from periodontitis (Trombelli et al., 2018). A periodontitis case was defined by detectable inter‐dental attachment loss at a minimum of two non‐adjacent teeth. The periodontitis stage was determined by the severity and complexity of the disease (Tonetti et al., 2018).
2.5. Quality control
To ensure data quality, all investigators were trained and calibrated before the implementation of the study; all standard procedures were rigorously conducted, and the blinded interpretation of study results was ensured. In particular, the self‐administered questionnaire was pre‐tested during a pilot phase and adapted accordingly before validation. In the aMMP‐8 test, subjects were required to avoid eating, drinking, brushing or using a mouthwash at least 30 min before the test (Deng et al., 2021a). The methodology for quantifying Hb was validated in a pilot study with good validity and reproducibility before sample analysis. The clinical examiner was trained in standardized diagnostic criteria and calibrated with the reference examiner with good reliability. The intra‐examiner reliability assessed by the intraclass correlation coefficient (ICC) for PPD was >0.90, and the ICC for CAL was >0.83 (Deng et al., 2021b).
2.6. Data analysis
The analytical approach consisted of two steps: (i) a preliminary assessment with multivariate logistic regression and (ii) assessment of a multiclass screening tool incorporating different features of the periodontal health–disease spectrum using forest plots. IBM SPSS statistics, version 26 (IBM Corp., Armonk, NY, USA), was used for descriptive and logistic regression analysis. To explore the discriminative performance of the combined index tests for differentiating various periodontal cases, multivariable logistic regression models were used for predicting ‘periodontal health’, ‘gingivitis’, ‘periodontitis’, ‘stages I/II periodontitis’ and ‘stages III/IV periodontitis,’ according to the disease distribution in the study population. The algorithm for detecting each scenario was developed using the best predictors in the logistic regression model. A bootstrap approach was used for internal validation. One thousand bootstrap samples were generated with replacement. The final logistic regression models incorporated significant predictors using backward stepwise selection. The parameter associated with each predictor was examined using the Wald test and removed if non‐significant at the 95% confidence level. The model was re‐run and the process repeated until no more predictors were removed. Model 1 was the combination of the questionnaire, aMMP‐8 POCT and GBoB test. Model 2 was the combined index tests in conjunction with demographic and lifestyle behavioural factors. Receiver operating characteristic (ROC) curves, the area under the ROC curve (AUROC), sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were estimated on the basis of the predicted probability from the logistic regression model. The predicted probability cut‐off value was chosen as the point by maximizing the sum of the sensitivity and specificity across the ROC curve.
In constructing multiclass models, an RF algorithm was trained using unique features describing the periodontal health–disease spectrum and, in case of multiple measurements, the most significant predictors obtained in the logistic regression analyses as input data. Multiclass decision algorithms for differentiating subjects with periodontal health, gingivitis and different stages of periodontitis were applied to the selected variables to train and construct the final model. The RF was generated using 100 estimators (‘trees’) for each forest with a maximum depth value of 6. Node‐splitting was determined using the Gini coefficient. Such parameters have been set based on our experience and after a few initial tests, to verify that they could produce reliable results. The mode of the response across the 100 trees was used as the estimate. The RF models were validated by running 100 independent trainings on the whole set of data and comparing the results to ensure the internal consistency of the model. RF models were fitted using the Python (version 3.9.13, www.python.org) Scikit‐learn package (Pedregosa et al., 2011). The average and deviation of the model performance, including sensitivity, specificity, PPV, NPV and accuracy, were reported. Sensitivity and specificity values were defined to be low (<60%), moderate (60%–79%) or high (≥80%) (Nelson et al., 2001). The accuracy results derived from the AUROC values were interpreted as low (0.50–0.70), moderate (0.71–0.90) and high (>0.90) (Swets, 1988). The package ‘Yellowbrick’ in Python was used to compute the micro‐ and macro‐averaged multiclass AUROC values as well as the per‐class AUROC value.
Micro‐averaging computes a global average by summing up the true positives and false positives across all classes. Macro‐averaging computes an average of metrics across all classes by taking the average of curves across all classes. In addition to the micro‐ and macro‐average curves for each class, a curve for each class was plotted to assess the trade‐off between sensitivity and specificity on a per‐class basis. Accuracy was calculated based on the following definition: accuracy = sensitivity × prevalence + specificity × (1 − prevalence). 95% confidence intervals were obtained using the MedCalc tool (version 20.115, MedCalc Software bv, Ostend, Belgium; https://www.medcalc.org). RF models were also run by giving different weights to the multiple trees to specifically train the model to avoid misclassification of a periodontitis case as health or gingivitis. Weights of 0.1 (light weights) and 0.2 (heavy weights) for the trees predicting the more severe diagnosis were added in these additional analyses.
3. RESULTS
After assessing the eligibility criteria from 501 consecutive patients seeking dental care at the reception clinic, 408 eligible Chinese subjects were included in this study, resulting in a participation rate of 81.4% (Figure 1). The demographic and clinical characteristics of the study population have been reported and are also shown in Tables S1 and S2 (Deng et al., 2021a, 2021b). In brief, 66 (16.2%) subjects were periodontally healthy cases and 62 (15.2%) had gingivitis; 280 (68.6%) subjects were diagnosed with periodontitis, including stage I (15.9%), stage II (15.9%), stage III (29.7%) and stage IV (7.1%). There were 189 (46.3%) males and 219 (53.7%) females aged between 18 and 86 (41 ± 18) years. Thirty‐two (7.8%) subjects were current smokers, and 20 (4.9%) suffered from diabetes. All subjects completed the questionnaire and the aMMP‐8 and GBoB tests except for one invalid sample for Hb measurement. Therefore, the entire dataset of self‐reported parameters and salivary biomarker profiles from 407 subjects was used to create the logistic regression and RF models for differentiating various periodontal diagnoses.
FIGURE 1.
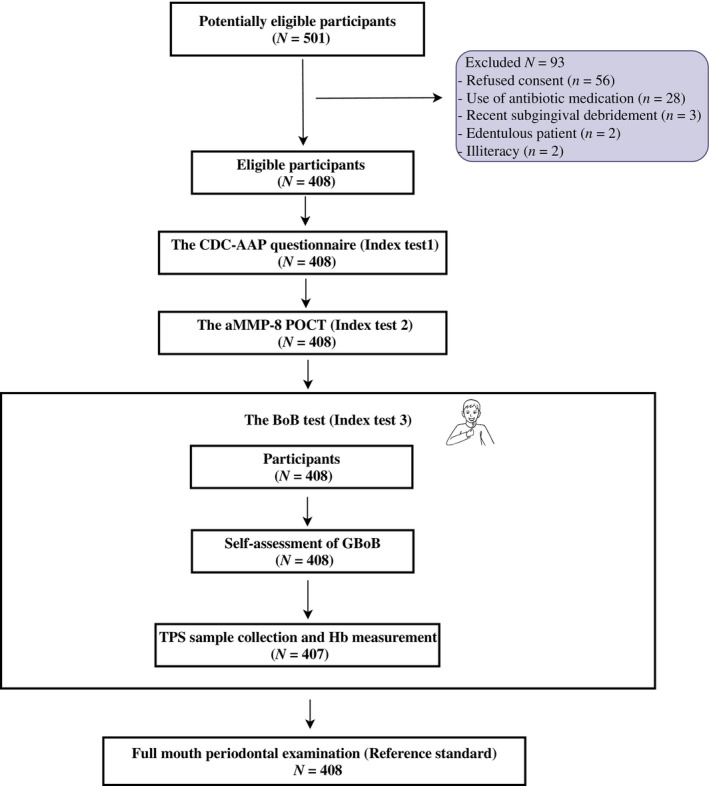
Flow‐chart of inclusion of participants and study procedures. CDC‐AAP, Centers for Disease Control and Prevention‐American Academy of Periodontology; aMMP‐8, activated matrix metalloproteinase‐8; POCT, point‐of‐care test; BoB, bleeding on brushing; GBoB, gingival bleeding on brushing; TPS, toothpaste slurries; Hb, haemoglobin.
3.1. Binary classifiers and predictive accuracy from logistic regression
The performance of self‐reported non‐clinical parameters, salivary biomarkers and Models 1 and 2 for predicting periodontal health, gingivitis and different stages of periodontitis is shown in Table 1. The results from conventional logistic regression analyses indicate that (i) a combination of self‐reported non‐clinical parameters and salivary biomarkers (Model 1) could achieve high accuracy for the discrimination of periodontal health and disease and moderate accuracy for detecting gingivitis; (ii) joint use of the aMMP‐8 POCT and the CDC/AAP questionnaire could slightly improve the screening accuracy for periodontitis compared with the questionnaire alone; (iii) the well‐accepted risk factors/indicators (age and smoking) show moderate/high accuracy in detecting periodontitis/stages III/IV periodontitis; (iv) the predictor alone or in combination performed poorly for identifying stages I/II periodontitis and (v) the CDC/AAP tool had moderate to high accuracy in screening stage III/IV periodontitis and its combination, with age and smoking strongly predicting stages III/IV periodontitis with excellent performance.
TABLE 1.
Diagnostic accuracy of self‐reported non‐clinical parameters and salivary biomarkers alone and in combination to detect periodontal health, gingivitis, periodontitis and different stages of periodontitis from the study sample (N = 408) using conventional logistic regression analyses.
| Diagnostic performance | CDC‐AAP questionnaire | aMMP‐8 POCT | Haemoglobin or (and) self‐reported GBoB | Model 1 | Model 2 |
|---|---|---|---|---|---|
| Periodontal health (n = 62) | |||||
| Sensitivity | 91.1% | 95.5% | 93.5% | 86.7% | 93.3% |
| Specificity | 61.4% | 28.9% | 52.5% | 78.8% | 80.8% |
| PPV | 29.7% | 20.6% | 21.9% | 42.3% | 46.7% |
| NPV | 97.3% | 97.1% | 93.4% | 97.0% | 98.5% |
| Accuracy | 65.9% | 39.0% | 58.7% | 80.0% | 82.7% |
| Misclassification | 34.1% | 61.0% | 41.3% | 20.0% | 17.3% |
| AUROC (95% CI) | 0.837 (0.783, 0.891) | 0.622 (0.557–0.687) | 0.754 (0.705–0.803) | 0.908 (0.872, 0.944) | 0.904 (0.869, 0.939) |
| Gingivitis (n = 66) | |||||
| Sensitivity | 54.1% | 9.7% | 61.3% | 75.7% | 86.5% |
| Specificity | 80.9% | 95.5% | 67.1% | 77.0% | 83.4% |
| PPV | 35.4% | 66.7% | 25.0% | 34.1% | 45.1% |
| NPV | 95.6% | 52.9% | 90.6% | 95.3% | 97.5% |
| Accuracy | 76.6% | 81.6% | 66.2% | 76.8% | 83.9% |
| Misclassification | 23.4% | 18.4% | 33.8% | 23.2% | 16.1% |
| AUROC (95% CI) | 0.692 (0.623, 0.762) | 0.590 (0.520–0.661) | 0.642 (0.566–0.718) | 0.768 (0.707, 0.830) | 0.893 (0.855, 0.931) |
| Diagnostic performance | CDC‐AAP questionnaire | aMMP‐8 POCT | Haemoglobin or (and) self‐reported GBoB | Risk factors/indicators | Model 1 | Model 2 |
|---|---|---|---|---|---|---|
| Periodontitis (n = 280) | ||||||
| Sensitivity | 67.9% | 33.2% | 50.2% | 71.4% | 64.5% | 80.1% |
| Specificity | 83.5% | 93.0% | 68.0% | 92.2% | 89.4% | 92.9% |
| PPV | 90.0% | 91.2% | 76.9% | 95.4% | 93.8% | 96.6% |
| NPV | 69.8% | 38.9% | 38.2% | 84.4% | 50.3% | 65.3% |
| Accuracy | 72.8% | 52.0% | 56.1% | 77.9% | 72.3% | 84.1% |
| Misclassification | 27.2% | 48.0% | 43.9% | 22.1% | 27.7% | 15.9% |
| AUROC (95% CI) | 0.803 (0.758, 0.849) | 0.631 (0.576–0.685) | 0.579 (0.521–0.638) | 0.853 (0.814, 0.891) | 0.811 (0.767, 0.855) | 0.898 (0.866, 0.929) |
| Stage I/II periodontitis (n = 130) | ||||||
| Sensitivity | 86.8% | 34.6% | NA | 87.7% | 82.4% | 55.9% |
| Specificity | 35.3% | 93.0% | NA | 36.3% | 40.2% | 74.5% |
| PPV | 38.6% | 44.1% | NA | 39.2% | 37.0% | 48.3% |
| NPV | 53.8% | 72.2% | NA | 54.9% | 84.2% | 79.8% |
| Accuracy | 51.7% | 74.4% | NA | 52.7% | 53.7% | 68.6% |
| Misclassification | 48.3% | 25.6% | NA | 47.3% | 46.3% | 31.4% |
| AUROC (95% CI) | 0.608 (0.550, 0665) | 0.638 (0.570–0.706) | NA | 0.597 (0.542,0.653) | 0.668 (0.609, 0.727) | 0.688 (0.632, 0.743) |
| Stages III/IV periodontitis (n = 150) | ||||||
| Sensitivity | 72.5% | 32.0% | NA | 72.8% | 72.5% | 96.1% |
| Specificity | 84.1% | 79.1% | NA | 84.1% | 84.1% | 89.4% |
| PPV | 73.3% | 47.1% | NA | 72.7% | 73.3% | 84.5% |
| NPV | 83.6% | 66.7% | NA | 90.1% | 83.6% | 96.8% |
| Accuracy | 80.8% | 61.8% | NA | 79.9% | 79.8% | 91.9% |
| Misclassification | 19.2% | 38.2% | NA | 20.1% | 20.2% | 9.1% |
| AUROC (95% CI) | 0.870 (0.830, 0.910) | 0.555 (0.497–0.614) | NA | 0.921 (0.895–0.947) | 0.870 (0.830, 0.910) | 0.953 (0.930, 0.977) |
Note: Model 1 represents the combination of CDC‐AAP questionnaire, aMMP‐8 POCT and gingival bleeding on brushing. Model 2 is Model 1 with the addition of age and smoking status.
Abbreviations: 95% CI, confidence interval of 95%; AUROC, area under receiver operator characteristic curve; CDC‐AAP, Centers for Disease Control and Prevention‐American Academy of Periodontology; GBoB, gingival bleeding on brushing; NA, not applicable as the predictor was not significant in the model; NPV, negative predictive value; OR, odds ratio; PPV, positive predictive value.
Based on Model 2 (Tables S3–S7), the fitted linear predictor for estimating log‐odds of periodontal health was = −1.750 × Q2 + 0.844 × Q3a + 0.951 × Q7 − 0.027 × Hb (total amount) − 1.471 × aMMP‐8 POCT − 0.050 × Age + 2.112; the fitted linear predictor for estimating log‐odds of gingivitis was = −0.992 × Q3a + 1.396 × self‐reported GBoB − 0.160 × Age − 14.719; the fitted linear predictor for estimating log‐odds of periodontitis was = 1.407 Q2 + 1.845 × aMMP‐8 POCT + 0.102 × Age − 4.203; the fitted linear predictor for estimating log‐odds of stage I/II periodontitis was = −1.540 × Q4 + 1.022 × aMMP‐8 POCT + 0.521 × Gender − 0.963; and the fitted linear predictor for estimating log‐odds of stage III/IV periodontitis was = 1.729 × Q2 + 1.756 × Q6 + 0.115 × Age + 2.453 × Smoking − 8.024.
3.2. Multiclass categorization and performance using RF
Eleven parameters estimating different features of the periodontal health–disease spectrum were included in the RF: ‘gum disease’ (Q1), ‘rating of gum/ teeth health’ (Q2), ‘tooth cleaning’ (Q3a), the symptom of ‘loose teeth’ (Q4), ‘tooth appearance’ (Q6), ‘use of floss’ (Q7), aMMP‐8 POCT, self‐reported GBoB, Hb, smoking and age. The robustness of the RF models was evaluated by (i) analysis of the patterns of misclassification and (ii) the leave‐one‐out or add‐one‐in approach. The leave‐one‐out or add‐one‐in approach means that a predictor was removed from or (and) added in the model one by one to assess whether the accuracy was improved or not. Patterns of misclassification refer to the misclassification of different case definitions (e.g., the underestimation of stages III/IV periodontitis to periodontal health cases or overestimation of a periodontal health case to a periodontitis case). Focusing on the more serious misclassification (declaring as healthy a subject with periodontitis, N = 17), the pattern of misclassification showed that all errors occurred in younger subjects (<35 years of age), reporting that teeth looked alright and use of dental floss, without bleeding on brushing and with a negative aMMP‐8 test. In general, leave‐one‐out analyses resulted in a decrease in precision except for smoking (only 7.8% of the population) and ‘tooth appearance’ (Q6), which did not significantly affect the estimates and were therefore removed. The best combination of the predictors for differentiating periodontal health, gingivitis and different stages of periodontitis were ‘gum disease’ (Q1), ‘rating of gum/teeth health’ (Q2), ‘tooth cleaning’ (Q3a), symptom of ‘loose teeth’ (Q4), ‘use of floss’ (Q7), aMMP‐8 POCT, self‐reported GBoB, Hb and age.
Figures 2, 3, 4 and Tables S7–S19 show the diagnostic characteristics of each final model (sensitivity, specificity, positive and negative predictive value and accuracy), their confusion matrix, ROC, AUROC and the relative influence of the multiple parameters in the model.
FIGURE 2.
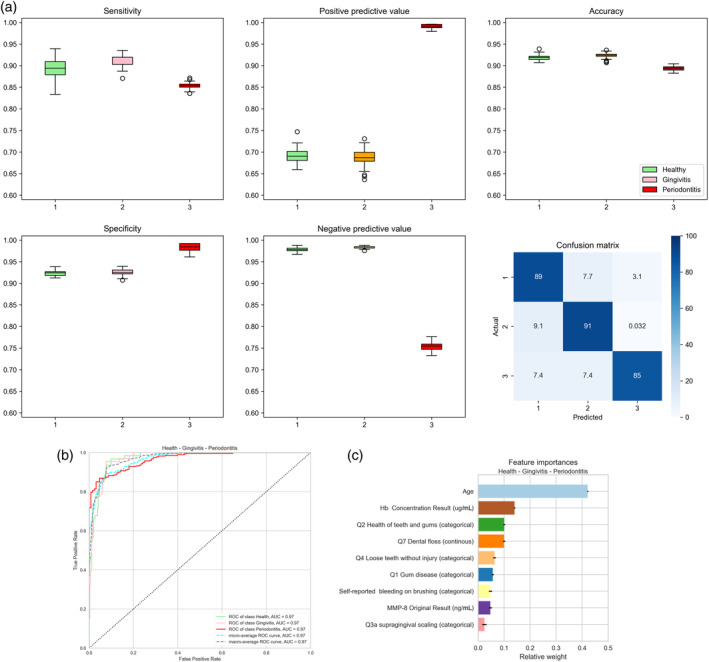
Accuracy of the three‐class prediction of periodontal health, gingivitis and periodontitis using the random forest classifier. (a) Periodontal health (1, green), gingivitis (2, pink) and periodontitis (3, red). The figure shows box plots of sensitivity (upper left diagram), specificity (lower left), positive predictive value (upper centre), negative predictive value (lower centre) and accuracy (upper right). The lower right diagram shows the confusion matrix plotting the frequency of predicted versus actual diagnosis. Values were obtained by averaging 100 runs. (b) Receiver‐operating‐curves plotting true positive rates against false positive rates and the area under the ROC (AUROC) for the three‐class random forest diagnosis obtained using the ‘yellowbrick’ package in Python (see text for details). (c) Diagrammatic representation of the relative influence of the multiple factors in the random forest classification. Values are obtained by averaging 100 runs. Hb, haemoglobin; MMP‐8, matrix metalloproteinase‐8.
FIGURE 3.
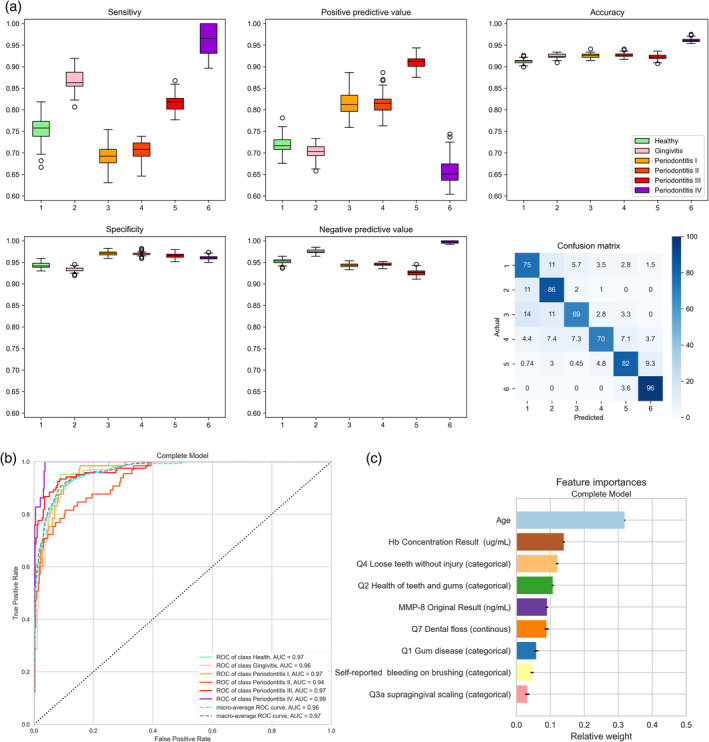
Accuracy of the six‐class prediction of periodontal health, gingivitis and stages I–IV periodontitis using the random forest classifier. (a) Periodontal health (1, green), gingivitis (2, pink), stage I periodontitis (3, orange), stage II periodontitis (4, light red), stage III periodontitis (5, darker red) and stage IV periodontitis (6, purple). The figure shows box plots of sensitivity (upper left diagram), specificity (lower left), positive predictive value (upper centre), negative predictive value (lower centre) and accuracy (upper right). The lower right diagram shows the confusion matrix plotting the frequency of predicted versus actual diagnosis. Values were obtained by averaging 100 runs. (b) Receiver‐operating‐curves plotting true positive rates against false positive rates and the area under the ROC (AUROC) for the six‐class random forest diagnosis obtained using the ‘yellowbrick’ package in Python (see text for details). (c) Diagrammatic representation of the relative influence of the multiple factors in the six‐class random forest classification. Values were obtained by averaging 100 runs. Hb, haemoglobin; MMP‐8, matrix metalloproteinase‐8.
FIGURE 4.
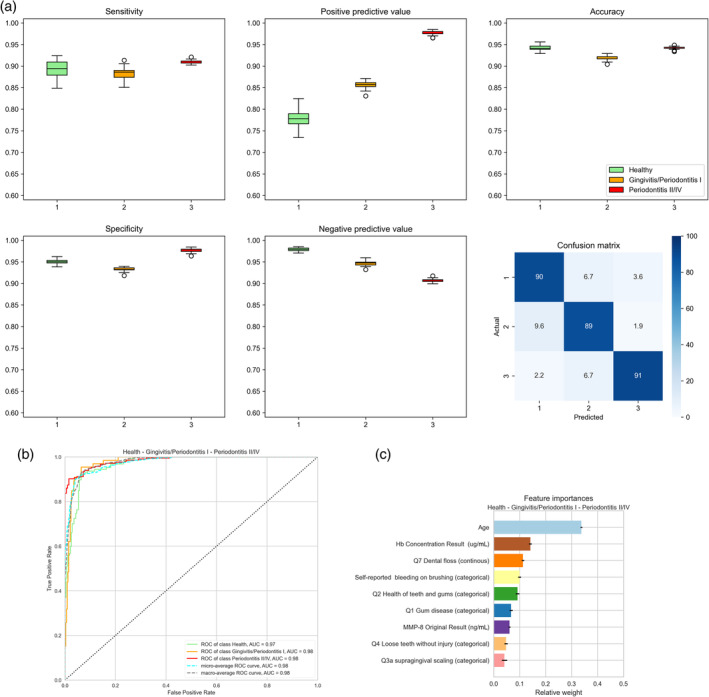
Accuracy of the three‐class prediction of periodontal health, gingivitis + stage I periodontitis and stages II–IV periodontitis using the random forest classifier. (a) Periodontal health (1, green), gingivitis and stage 1 periodontitis (2, orange) and stages II–IV periodontitis (3, red). The figure shows box plots of sensitivity (upper left diagram), specificity (lower left), positive predictive value (upper centre), negative predictive value (lower centre) and accuracy (upper right). The lower right diagram shows the confusion matrix plotting the frequency of predicted versus actual diagnosis. Values were obtained by averaging 100 runs. (b) Receiver‐operating‐curves plotting true positive rates against false positive rates and the area under the ROC (AUROC) for the three‐class random forest diagnosis obtained using the ‘yellowbrick’ package in Python (see text for details). (c) Diagrammatic representation of the relative influence of the multiple factors in the three‐class random forest classification. Values were obtained by averaging 100 runs. Hb, haemoglobin; MMP‐8, matrix metalloproteinase‐8.
3.2.1. Periodontal health, gingivitis and periodontitis
The performance of three‐class discrimination of periodontal health, gingivitis and periodontitis is shown in Figure 2 and Table S17. Overall, the RF classifier provided high accuracy for multiclass discrimination compared with those derived from the logistic regression algorithms for binary classification. Across the 100 runs of the RF algorithm, the classifier achieved a sensitivity of 89.3 ± 2.2%, a specificity of 92.3 ± 0.4% and an accuracy of 91.8 ± 0.5% for periodontal health; a sensitivity of 91.0 ± 1.1%, a specificity of 92.6 ± 0.6% and an accuracy of 92.4 ± 0.6% for gingivitis; and a sensitivity of 91.0 ± 1.1%, a specificity of 92.6 ± 0.6% and an accuracy of 92.4 ± 0.6% for periodontitis in the three‐class classification. The confusion matrix shows the major limitation of the model: 15% of periodontitis cases are misclassified as health or gingivitis. The AUROC for each diagnosis and their micro‐ and macro‐averaging were 0.97 (Figure 2b). Figure 2c shows the relative importance of the different features used in the model. Age of the subject, concentration of Hb in the toothbrushing slurry, response to the questions related to the health of the teeth and gum and the number of episodes of use of dental floss in a week were the most important features.
3.2.2. Periodontal health, gingivitis and periodontitis stages I, II, III and IV
Figure 3 and Table S18 show the performance of six‐class discrimination (periodontal health, gingivitis, stages I–IV periodontitis). In Figure 3a, the best predictions were obtained for stage IV periodontitis (3.6% of cases were misclassified as stage III) and gingivitis (11% of cases misclassified as healthy and 3% as stage I or II). AUROC values ranged from 0.94 to 0.99 (Figure 3b). In the six‐class analysis, the most important features were the age of the subject, Hb concentration in the toothbrushing slurry and the response to the questions related to the presence of lose teeth and the health of the teeth and gums (Figure 3c).
3.2.3. Periodontal health, gingivitis + periodontitis stage I, and periodontitis stages II, III and IV
The performance of three‐class discrimination combining gingivitis and stage I periodontitis (periodontal health, gingivitis + stage I periodontitis, stages II–IV periodontitis) is displayed in Figure 4 and Table S19. The confusion matrix (Figure 4a) shows the major limitation of the model: 9% of periodontitis subjects were misclassified as being healthy or gingivitis/stage I periodontitis. Nonetheless, high AUROC values of 0.97–0.98 were observed (Figure 4b). The most important features in this model were the age of the subject, concentration of Hb in the toothbrushing slurry and the number of weekly flossing and self‐reported bleeding on brushing (Figure 4c).
3.2.4. Insertion of different weights into the RF predictions
As the clinical consequences of misclassification are not symmetric, an analysis was performed to add additional weights to the trees in the RF with the more advanced disease predictions. Results are shown in Figures S1–S3. In the three‐class diagnostic model (Figure S1), adding light weights to more severe predictions decreased the misclassification of periodontitis cases from 15% to 2%. This, however, carried the price of classifying as periodontitis 29% of healthy subjects and 17% of gingivitis subjects. Adding different weights did not significantly improve the six‐class model or the three‐class model combining gingivitis and stage I in a single entity.
4. DISCUSSION
This study showed that combining non‐clinical parameters reflecting the unique features of periodontal health, gingivitis and periodontitis in a machine learning algorithm enables the development of multiclass prediction/diagnostic models of periodontal health status with high accuracy. RF models had high accuracy (AUROC > 0.94) for three‐class predictions (health, gingivitis and periodontitis or health, gingivitis/stage I periodontitis and stages II, III and IV periodontitis) and six‐class predictions (health, gingivitis, stages I, II, III and IV periodontitis). Accuracy levels achieved by the RF multiclass classifier were significantly better than those obtained for multivariate logistic regression analyses, particularly for the diagnoses that were more difficult to discriminate: gingivitis, stage I and stage II periodontitis. With RF, the probability of misclassification was low (Figures 2a, 3a and 4a). Adding specific weights to trees predicting more severe disease during the training of the model enabled a decrease from 15% to 2% in the misclassification of periodontitis subject as being healthy or gingivitis, arguably the most severe mistake for a screening test. Further studies in independent populations need to be performed to validate the model and approach. The results are notable because the multiclass screening of periodontal health status in non‐clinical settings may provide specific utility and added efficiency to manage preventive and therapeutic services/care pathways. Ideally, the screening results must be able to identify subjects requiring primary prevention services alone (Tonetti et al., 2015), those who need management of gingivitis before the onset of periodontitis and those who should avail periodontitis treatment either in primary settings (stage I and II periodontitis) or in specialist settings (stage III and IV periodontitis) (Tonetti et al., 2017). The clinical relevance of multiclass screening of periodontal health status includes referring the subject to the appropriate level of care, preventive, primary or specialist care, for subsequent confirmation and management.
Nine predictors, namely ‘gum disease’ (Q1), ‘rating of gum/teeth health’ (Q2), ‘tooth cleaning’ (Q3a), the symptom of ‘loose teeth’ (Q4), ‘use of floss’ (Q7), aMMP‐8 POCT, self‐reported GBoB, Hb and age, resulted in high levels of accuracy. RF is a machine learning algorithm widely used in classification by generating multiple decision trees and using a randomly selected subset of variables, thereby reducing overfitting and bias (Breiman, 2001). The RF model was stable, leaving one parameter out and using multiple seeds to build/train the model. Mechanistically, the predictors in the model provide information on different underlying features of the spectrum of periodontal disease (gingivitis and the various stages of periodontitis), which adds biological credibility. Interestingly, the analysis of the influencing factors (Figures 2c, 3c and 4c) gave slightly different rankings for the three tested scenarios. This appears to be mechanistically in line with the current understanding of the differential features of periodontal health, gingivitis and the different stages of periodontitis.
Notably, the set of predictors in the machine learning tool for multiclass identification performed better than the logistic regression models did in bivariate analyses. Improved accuracy has been reported for machine learning algorithms in the multiclass identification of complex diseases such as diabetes or cardiovascular diseases (Baashar et al., 2022; Shin et al., 2022). In the present study, the machine learning algorithm improved the detection accuracy of stages I and II periodontitis from an AUROC of 0.68 for logistic regression to an AUROC of 0.97 for predicting stage I periodontitis and 0.94 for predicting stage II periodontitis with the RF classifier. Discriminating incipient periodontitis offers the most significant challenge, even for experienced clinicians performing a complete periodontal examination. The application of AI based on non‐clinical parameters may be handy for this diagnostic question. As expected, the prediction accuracy of the more severe forms of the disease was high, even with the use of the CDC/AAP questionnaire alone. From the 6 × 6 (3 × 3) confusion matrix for the RF models, the value of adding additional parameters lies in the ability to better discriminate among the more subtle/initial stages of the disease spectrum.
The logistic regression analysis also uncovered additional findings. Despite the minimal added value of aMMP‐8 and GBoB for periodontitis screening, the combination of the predictors performed best for discrimination of periodontal health from disease and detection of gingivitis, compared with the questionnaire or biomarker testing alone. Although slightly more resources and medical costs are required, there is a vast array of potential public health benefits from the combined predictors: nearly 80% of suspected cases with periodontal diseases, including those with gingivitis, stages I/II periodontitis and stage III/IV periodontitis, can be identified for early referral, further professional diagnosis and timely intervention. The detection of gingivitis and incipient periodontitis particularly contributes to a more favourable treatment outcome and less healthcare cost, thereby alleviating the socio‐economic consequences and promoting periodontal health and general well‐being. Consistent with logistic regression analysis, the feature importance plots from RF also reinforced the rationale of incorporating biomarker testing (e.g., Hb), in line with our current medical understanding. The differential significance of GBoB in the screening for gingivitis and aMMP‐8 POCT for periodontitis highlights the key features of these different diseases: superficial inflammation captured by bleeding on brushing and the periodontal breakdown captured by MMP activation. Diagnostic strategies combining them may show increased overall accuracy.
As previously suggested, individual questions of the CDC/AAP questionnaire seem to be better suited to confirm/exclude specific diagnoses (Deng et al., 2021b). Combining them in specific sets may be helpful for multiclass classification of periodontal health status. Additionally, models incorporating well‐accepted risk indicators (such as age or smoking status) were generally more accurate. In this respect, it must be emphasized that this portion of the model is particularly prone to error in populations with divergent characteristics. Extensive validation will be required to better estimate their significance and contribution to the models.
This study has several limitations, which are implicit in the model development nature of the study. The original sample consists of Chinese subjects seeking care at a Hong Kong Dental Hospital. Although they seem to reflect well the distribution of periodontal health and disease in the local population, a selection bias is likely. Therefore, the external validity of this study needs to be addressed in an ongoing validation study using a representative sample of households in Hong Kong generated by systematic random sampling from the Census and Statistics Department of Hong Kong. Additionally, model validation will have to be extended to different populations around the world. Second, no robust sample size calculation could be done, as this was the first investigation on the multiclass classification of periodontal health status. Third, despite the conservative approach used to insert and maintain predictors in the RF model, it is unclear whether a more conservative model/approach may result in a similar level of accuracy. Further calibration of the model will have to be performed based on a much larger sample from multicentre validation. The reported feature importance data may be particularly useful in the future analyses. Fourth, the incorporation of Hb concentration in the model due to its high importance in the feature analyses requires additional development of a simple quantitative method that could be effectively used by subjects in a screening context. Investigations are ongoing in this area. Lastly, a misclassification analysis showed that the model did not perform well in younger periodontitis subjects presenting with features generally associated with health. In these subjects, the biomarkers did not manage to detect the disease. Additional validation in multiple populations is necessary, ideally in a multicentre study.
Within the limitations of this initial study, AI prediction based on non‐clinical data appears to be a promising approach to classify periodontal health status and can potentially open new avenues to manage periodontitis better.
AUTHOR CONTRIBUTIONS
Ke Deng and Francesco Zonta contributed to protocol development, data collection, analysis and interpretation, and manuscript preparation. Huan Yang contributed to data analysis and manuscript preparation. George Pelekos contributed to data collection and manuscript preparation. Maurizio S. Tonetti devised this study and contributed to protocol development, data interpretation and manuscript preparation.
FUNDING INFORMATION
This study was supported by the Hong Kong Human Medical Research Fund (HMRF) grant no. 07182796, the Shanghai Innovative Research Team Award of High‐Level University (SHSMU‐ZDCX202125000), the National Clinical Research Center for Oral Diseases (19411950100), Clinical Research Program of Ninth People's Hospital affiliated Shanghai Jiao Tong University School of Medicine (JYLJ201909) and the European Research Group on Periodontology, Switzerland.
CONFLICT OF INTEREST STATEMENT
Maurizio Tonetti, Ke Deng and Francesco Zonta have applied for a patent based on the present work. Otherwise, all authors declare no conflict of interest.
ETHICS STATEMENT
The study was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (reference: UW19‐188). Written informed consent was obtained from all participants. All study procedures comply with data protection regulations (fully anonymized data). The study protocol was prospectively registered in ClinicalTrials.gov (NCT03928080) and HKU Clinical Trials Registry (HKUCTR‐2631).
Supporting information
Data S1. Supplementary material.
Deng, K. , Zonta, F. , Yang, H. , Pelekos, G. , & Tonetti, M. S. (2024). Development of a machine learning multiclass screening tool for periodontal health status based on non‐clinical parameters and salivary biomarkers. Journal of Clinical Periodontology, 51(12), 1547–1560. 10.1111/jcpe.13856
Ke Deng and Francesco Zonta contributed equally to this work.
DATA AVAILABILITY STATEMENT
The machine learning algorithm will be made available upon reasonable request in the context of the required international validation studies.
REFERENCES
- Arias‐Bujanda, N. , Regueira‐Iglesias, A. , Balsa‐Castro, C. , Nibali, L. , Donos, N. , & Tomás, I. (2020). Accuracy of single molecular biomarkers in saliva for the diagnosis of periodontitis: A systematic review and meta‐analysis. Journal of Clinical Periodontology, 47(1), 2–18. 10.1111/jcpe.13202 [DOI] [PubMed] [Google Scholar]
- Aro, K. , Wei, F. , Wong, D. T. , & Tu, M. (2017). Saliva liquid biopsy for point‐of‐care applications. Frontiers in Public Health, 5, 77. 10.3389/fpubh.2017.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baashar, Y. , Alkawsi, G. , Alhussian, H. , Capretz, L. F. , Alwadain, A. , Alkahtani, A. A. , & Almomani, M. (2022). Effectiveness of artificial intelligence models for cardiovascular disease prediction: Network meta‐analysis. Computational Intelligence and Neuroscience, 2022, 5849995. 10.1155/2022/5849995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir, N. Z. , Rahman, Z. , & Chen, S. L. (2022). Systematic comparison of machine learning algorithms to develop and validate predictive models for periodontitis. Journal of Clinical Periodontology, 49, 958–969. 10.1111/jcpe.13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkedal‐Hansen, H. , Moore, W. G. , Bodden, M. K. , Windsor, L. J. , Birkedal‐Hansen, B. , DeCarlo, A. , & Engler, J. A. (1993). Matrix metalloproteinases: A review. Critical Reviews in Oral Biology and Medicine, 4(2), 197–250. 10.1177/10454411930040020401 [DOI] [PubMed] [Google Scholar]
- Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Carra, M. C. , Gueguen, A. , Thomas, F. , Pannier, B. , Caligiuri, G. , Steg, P. G. , Zins, M. , & Bouchard, P. (2018). Self‐report assessment of severe periodontitis: Periodontal screening score development. Journal of Clinical Periodontology, 45(7), 818–831. 10.1111/jcpe.12899 [DOI] [PubMed] [Google Scholar]
- Chapple, I. L. C. , Mealey, B. L. , Van Dyke, T. E. , Bartold, P. M. , Dommisch, H. , Eickholz, P. , Geisinger, M. L. , Genco, R. J. , Glogauer, M. , Goldstein, M. , Griffin, T. J. , Holmstrup, P. , Johnson, G. K. , Kapila, Y. , Lang, N. P. , Meyle, J. , Murakami, S. , Plemons, J. , Romito, G. A. , … Yoshie, H. (2018). Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: Consensus report of workgroup 1 of the 2017 World Workshop on the Classification of Periodontal and Peri‐Implant Diseases and Conditions. Journal of Clinical Periodontology, 45(Suppl 20), S68–s77. 10.1111/jcpe.12940 [DOI] [PubMed] [Google Scholar]
- Collins, G. S. , Reitsma, J. B. , Altman, D. G. , & Moons, K. (2015). Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. BMC Medicine, 13(1), 1. 10.1186/s12916-014-0241-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, K. , Pelekos, G. , Jin, L. , & Tonetti, M. S. (2021a). Diagnostic accuracy of a point‐of‐care aMMP‐8 test in the discrimination of periodontal health and disease. Journal of Clinical Periodontology, 48(8), 1051–1065. 10.1111/jcpe.13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, K. , Pelekos, G. , Jin, L. , & Tonetti, M. S. (2021b). Diagnostic accuracy of self‐reported measures of periodontal disease: A clinical validation study using the 2017 case definitions. Journal of Clinical Periodontology, 48(8), 1037–1050. 10.1111/jcpe.13484 [DOI] [PubMed] [Google Scholar]
- Deng, K. , Pelekos, G. , Jin, L. , & Tonetti, M. S. (2021c). Gingival bleeding on brushing as a sentinel sign of gingival inflammation: A diagnostic accuracy trial for the discrimination of periodontal health and disease. Journal of Clinical Periodontology, 48(12), 1537–1548. 10.1111/jcpe.13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, K. , Wei, S. , Xu, M. , Shi, J. , Lai, H. , & Tonetti, M. S. (2022). Diagnostic accuracy of active matrix metalloproteinase‐8 point‐of‐care test for the discrimination of periodontal health status: Comparison of saliva and oral rinse samples. Journal of Periodontal Research, 57, 768–779. 10.1111/jre.12999 [DOI] [PubMed] [Google Scholar]
- Department of Health, Hong Kong Government SAR . (2011). Oral Health Survey 2011. Accessed September 3, 2023. https://www.toothclub.gov.hk/en/en_pdf/Oral_Health_Survey_2011/Oral_Health_Survey_2011_WCAG_20141112_(EN_Full).pdf
- Eke, P. I. , Dye, B. A. , Wei, L. , Slade, G. D. , Thornton‐Evans, G. O. , Beck, J. D. , Taylor, G. W. , Borgnakke, W. S. , Page, R. C. , & Genco, R. J. (2013). Self‐reported measures for surveillance of periodontitis. Journal of Dental Research, 92(11), 1041–1047. 10.1177/0022034513505621 [DOI] [PubMed] [Google Scholar]
- Eke, P. I. , & Genco, R. J. (2007). CDC periodontal disease surveillance project: Background, objectives, and progress report. Journal of Periodontology, 78(7s), 1366–1371. 10.1902/jop.2007.070134 [DOI] [PubMed] [Google Scholar]
- Grant, M. M. , Taylor, J. J. , Jaedicke, K. , Creese, A. , Gowland, C. , Burke, B. , Doudin, K. , Patel, U. , Weston, P. , Milward, M. , Bissett, S. M. , Cooper, H. J. , Kooijman, G. , Rmaile, A. , de Jager, M. , Preshaw, P. M. , & Chapple, I. L. C. (2022). Discovery, validation, and diagnostic ability of multiple protein‐based biomarkers in saliva and gingival crevicular fluid to distinguish between health and periodontal diseases. Journal of Clinical Periodontology, 49(7), 622–632. 10.1111/jcpe.13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürsoy, U. K. , & Kantarci, A. (2022). Molecular biomarker research in periodontology: A roadmap for translation of science to clinical assay validation. Journal of Clinical Periodontology, 49(6), 556–561. 10.1111/jcpe.13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamp, S. E. , Nyman, S. , & Lindhe, J. (1975). Periodontal treatment of multirooted teeth. Results after 5 years. Journal of Clinical Periodontology, 2(3), 126–135. 10.1111/j.1600-051x.1975.tb01734.x [DOI] [PubMed] [Google Scholar]
- Lorena, A. C. , de Carvalho, A. C. P. L. F. , & Gama, J. M. P. (2009). A review on the combination of binary classifiers in multiclass problems. Artificial Intelligence Review, 30(1), 19–37. 10.1007/s10462-009-9114-9 [DOI] [Google Scholar]
- Maity, N. G. , & Das, S. (2017). Machine learning for improved diagnosis and prognosis in healthcare. Paper presented at the 2017 IEEE Aerospace Conference.
- Mundfrom, D. J. , Shaw, D. G. , & Ke, T. L. (2005). Minimum sample size recommendations for conducting factor analyses. International Journal of Testing, 5(2), 159–168. 10.1207/s15327574ijt0502_4 [DOI] [Google Scholar]
- Nelson, D. E. , Holtzman, D. , Bolen, J. , Stanwyck, C. A. , & Mack, K. A. (2001). Reliability and validity of measures from the Behavioral Risk Factor Surveillance System (BRFSS). Sozial‐ und Präventivmedizin, 46(Suppl 1), S3–S42. [PubMed] [Google Scholar]
- Papapanou, P. N. , Sanz, M. , Buduneli, N. , Dietrich, T. , Feres, M. , Fine, D. H. , Flemmig, T. F. , Garcia, R. , Giannobile, W. V. , Graziani, F. , Greenwell, H. , Herrera, D. , Kao, R. T. , Kebschull, M. , Kinane, D. F. , Kirkwood, K. L. , Kocher, T. , Kornman, K. S. , Kumar, P. S. , … Tonetti, M. S. (2018). Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri‐Implant Diseases and Conditions. Journal of Periodontology, 89(Suppl 1), S173–s182. 10.1002/jper.17-0721 [DOI] [PubMed] [Google Scholar]
- Pedregosa, F. , Varoquaux, G. , Gramfort, A. , Michel, V. , Thirion, B. , Grisel, O. , Blonde, M. , Prettenhofer, P. , Weiss, R. , Dubourg, V. , Vanderplas, J. , Passos, A. , Cournapeau, D. , Brucher, M. , Perrot, M. and Duchesnay, E. 2011. Scikit‐learn: Machine Learning in Python. Journal of Machine Learning Research 12, 2825–2830. [Google Scholar]
- Romano, F. , Perotto, S. , Bianco, L. , Parducci, F. , Mariani, G. M. , & Aimetti, M. (2020). Self‐perception of periodontal health and associated factors: A cross‐sectional population‐based study. International Journal of Environmental Research and Public Health, 17(8). 10.3390/ijerph17082758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajda, P. (2006). Machine learning for detection and diagnosis of disease. Annual Review of Biomedical Engineering, 8, 537–565. 10.1146/annurev.bioeng.8.061505.095802 [DOI] [PubMed] [Google Scholar]
- Shin, H. , Schneeweiss, S. , Glynn, R. J. , & Patorno, E. (2022). Cardiovascular outcomes in patients initiating first‐line treatment of type 2 diabetes with sodium‐glucose cotransporter‐2 inhibitors versus metformin: A cohort study. Annals of Internal Medicine, 175(7), 927–937. 10.7326/m21-4012 [DOI] [PubMed] [Google Scholar]
- Sorsa, T. , Gursoy, U. K. , Nwhator, S. , Hernandez, M. , Tervahartiala, T. , Leppilahti, J. , Gursoy, M. , Könönen, E. , Emingil, G. , Pussinen, P. J. , & Mäntylä, P. (2016). Analysis of matrix metalloproteinases, especially MMP‐8, in gingival creviclular fluid, mouthrinse and saliva for monitoring periodontal diseases. Periodontology 2000, 70(1), 142–163. 10.1111/prd.12101 [DOI] [PubMed] [Google Scholar]
- Swets, J. A. (1988). Measuring the accuracy of diagnostic systems. Science, 240(4857), 1285–1293. 10.1126/science.3287615 [DOI] [PubMed] [Google Scholar]
- Tonetti, M. S. , Chapple, I. L. , Jepsen, S. , & Sanz, M. (2015). Primary and secondary prevention of periodontal and peri‐implant diseases: Introduction to, and objectives of the 11th European Workshop on Periodontology consensus conference. Journal of Clinical Periodontology, 42(Suppl 16), S1–S4. 10.1111/jcpe.12382 [DOI] [PubMed] [Google Scholar]
- Tonetti, M. S. , Deng, K. , Christiansen, A. , Bogetti, K. , Nicora, C. , Thurnay, S. , & Cortellini, P. (2020). Self‐reported bleeding on brushing as a predictor of bleeding on probing: Early observations from the deployment of an internet of things network of intelligent power‐driven toothbrushes in a supportive periodontal care population. Journal of Clinical Periodontology, 47(10), 1219–1226. 10.1111/jcpe.13351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonetti, M. S. , Greenwell, H. , & Kornman, K. S. (2018). Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. Journal of Clinical Periodontology, 45(Suppl 20), S149–S161. 10.1111/jcpe.12945 [DOI] [PubMed] [Google Scholar]
- Tonetti, M. S. , Jepsen, S. , Jin, L. , & Otomo‐Corgel, J. (2017). Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. Journal of Clinical Periodontology, 44(5), 456–462. 10.1111/jcpe.12732 [DOI] [PubMed] [Google Scholar]
- Trombelli, L. , Farina, R. , Silva, C. O. , & Tatakis, D. N. (2018). Plaque‐induced gingivitis: Case definition and diagnostic considerations. Journal of Clinical Periodontology, 45(Suppl 20), S44–S67. 10.1111/jcpe.12939 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary material.
Data Availability Statement
The machine learning algorithm will be made available upon reasonable request in the context of the required international validation studies.


