Abstract
Background & Aims:
Chronic viral infections present serious public health challenges; however, direct-acting antivirals (DAAs) are now able to cure nearly all patients infected with hepatitis C virus (HCV), representing the only cure of a human chronic viral infection to date. DAAs provide a valuable opportunity to study immune pathways in the reversal of chronic immune failures in an in vivo human system.
Methods:
To leverage this opportunity, we used plate-based single-cell RNA-seq to deeply profile myeloid cells from liver fine needle aspirates in patients with HCV before and after DAA treatment. We comprehensively characterised liver neutrophils, eosinophils, mast cells, conventional dendritic cells, plasmacytoid dendritic cells, classical monocytes, non-classical monocytes, and macrophages, and defined fine-grained subpopulations of several cell types.
Results:
We discovered cell type-specific changes post-cure, including an increase in MCM7+STMN1+ proliferating CD1C+ conventional dendritic cells, which may support restoration from chronic exhaustion. We observed an expected down-regulation of interferon-stimulated genes (ISGs) post-cure as well as an unexpected inverse relationship between pre-treatment viral load and post-cure ISG expression in each cell type, revealing a link between viral loads and sustained modifications of the host’s immune system. We found an upregulation of PD-L1/L2 gene expression in ISG-high neutrophils and IDO1 expression in eosinophils, pinpointing cell subpopulations crucial for immune regulation. We identified three recurring gene programmes shared by multiple cell types, distilling core functions of the myeloid compartment.
Conclusions:
This comprehensive single-cell RNA-seq atlas of human liver myeloid cells in response to cure of chronic viral infections reveals principles of liver immunity and provides immunotherapeutic insights.
Clinical Trial registration:
This study is registered at ClinicalTrials.gov (NCT02476617).
Keywords: Chronic infections, Viral infections, Hepatitis C virus, Direct-acting antiviral, Immune cells, Myeloid cells, Neutrophils, Eosinophils, PD-L1, Innate immunity, Single-cell RNA-sequencing, Fine needle aspiration, Liver
Introduction
Chronic viral infections, including hepatitis C virus (HCV) and hepatitis B virus (HBV) infections, remain serious public health challenges.1-4 These diseases arise from a failure to clear the virus by the host’s immune system and often result in persistent and aberrant immune activation. The liver, in particular, is an organ predisposed to chronic viral infections potentially because of its immune tolerance and sampling of the external environment.5 Direct-acting antivirals (DAAs) are the only curative therapies for a chronic viral infection, virtually curing all patients infected with common strains of HCV.6-9 DAAs, which have been available since 2011, target and inhibit key HCV life-cycle proteins, including the NS3/4A protease, NS5A, and the RNA polymerase NS5B.9 However, the host’s immune state post-cure of chronic infection is not well understood.10 A detailed understanding of the host’s immune response during chronic infections and post-cure can provide insights into designing HCV vaccines for low-resource communities, providing optimal care for post-DAA-cured individuals, and developing effective treatments for other chronic infections and immune-mediated diseases.11
Myeloid cells are among the first responders to pathogens, key constituents of the innate immune system and regulators of adaptive immunity and tissue repair. Myeloid cells include neutrophils, eosinophils, monocytes, macrophages, dendritic cells (DCs), and plasmacytoid dendritic cells (pDCs), each of which comprises a number of subsets with diverse functions that adapt their phenotype to different environmental cues.12 Myeloid cell involvement in viral disease pathogenesis is well recognised. In HCV, studies have shown that myeloid cells in the liver play a critical role in controlling the virus and resolving attributable liver damage.13-15 Therapeutic strategies targeting myeloid cells have shown promise in treating chronic immune-mediated diseases.16 However, the biological and clinical significance of different myeloid cell types and subsets is far from being elucidated. Understanding the state of myeloid cells in chronic infections and cures can shed light on novel myeloid cell-targeting therapies.
With advances in single-cell RNA sequencing (scRNA-seq) technologies, we can now characterise heterogeneous biological systems at a high resolution. Moreover, the development of fine needle aspiration (FNA) enables longitudinal, minimally invasive access to the tissue site of infection, including, in the case of HCV, the liver. These advances allow for transcriptomic characterisation of heterogeneous liver cells longitudinally at the single-cell resolution. Although single-cell maps of the liver have been developed,17-19 an atlas of human liver immune cells in chronic infections and recovery is not yet available. An atlas of cells during cure can reveal pathways involved in the reversal of chronic immune failures in humans, providing important insights for numerous diseases marked by a chronically dysfunctional immune response. Additionally, several key knowledge gaps remain. (1) Many infectious diseases are monitored via peripheral blood, which do not accurately reflect the activities in tissues where the disease is localised. (2) Single-cell transcriptomic data of human granulocytes (e.g. neutrophils and eosinophils) in tissues are rare because of limitations of droplet-based scRNA-seq technologies. The present lack of high-resolution data on these critical cell types in tissues hinders our ability to understand the innate immune system in detail. (3) DAA is a relatively new therapy and the host’s response to DAA has not been completely characterised. Some studies have revealed persistent liver abnormalities after elimination of HCV infections; therefore, understanding the host’s response to DAA can provide insight into the long-term health and prognosis of patients treated with such therapy.10 These knowledge gaps call for a high-resolution characterisation of liver myeloid cells from patients with HCV treated with DAA.
In this study, we leveraged scRNA-seq technology and the liver FNA procedure to comprehensively dissect the myeloid compartment in the liver from chronic patients with HCV pre- and post-DAA treatment. We chose to use the Smart-seq2 scRNA-seq technology as it provides robust detection of mRNA transcripts and enables profiling of human neutrophils and other granulocytes with a high sensitivity.20,21 We created a transcriptomic landscape of liver myeloid cells, from which we uncovered subsets of liver myeloid cells and identified molecular signatures associated with a cure for the infection. We further longitudinally studied serum dynamics of immune activation markers associated with liver virus removal. Altogether, our single-cell atlas presents a map of in vivo human response to curable chronic infections, shedding light on principles of liver immunity.
Materials and methods
For details regarding the materials and methods used, please refer to the CTAT table and supplementary information.
Results
Characteristics of patients with HCV treated with DAA
We studied 23 patients chronically infected with HCV (genotype 1a) who received 12 weeks of DAA treatment and were followed up to 24 weeks post-treatment (PTW24) (Supplementary methods). Patients were enrolled into a previously described phase IIIb open-label study of all-oral DAAs.22,23 We performed scRNA-seq on 18 of these patients and serum measurements on all patients. Table S1 summarises their baseline demographics and clinical characteristics. The majority were male, white, and treatment naïve. Liver stiffness (by transient elastography) values ranged from 4.2 to 11 kPa (median: 5.9 kPa; >12.8 kPa threshold for cirrhosis).24 Accordingly, the studied patients had no cirrhosis, and most had no to mild fibrosis. HCV RNA levels significantly decreased at treatment week 2 (W2) (p <0.0001) and became undetectable at W4 (Fig. 1A). Alanine transaminase (ALT) levels significantly decreased at W4 (p <0.001) (median, range: 15 IU/L, 8–40) and remained low throughout the study (PTW12, median, range: 12 IU/L, 8–17).
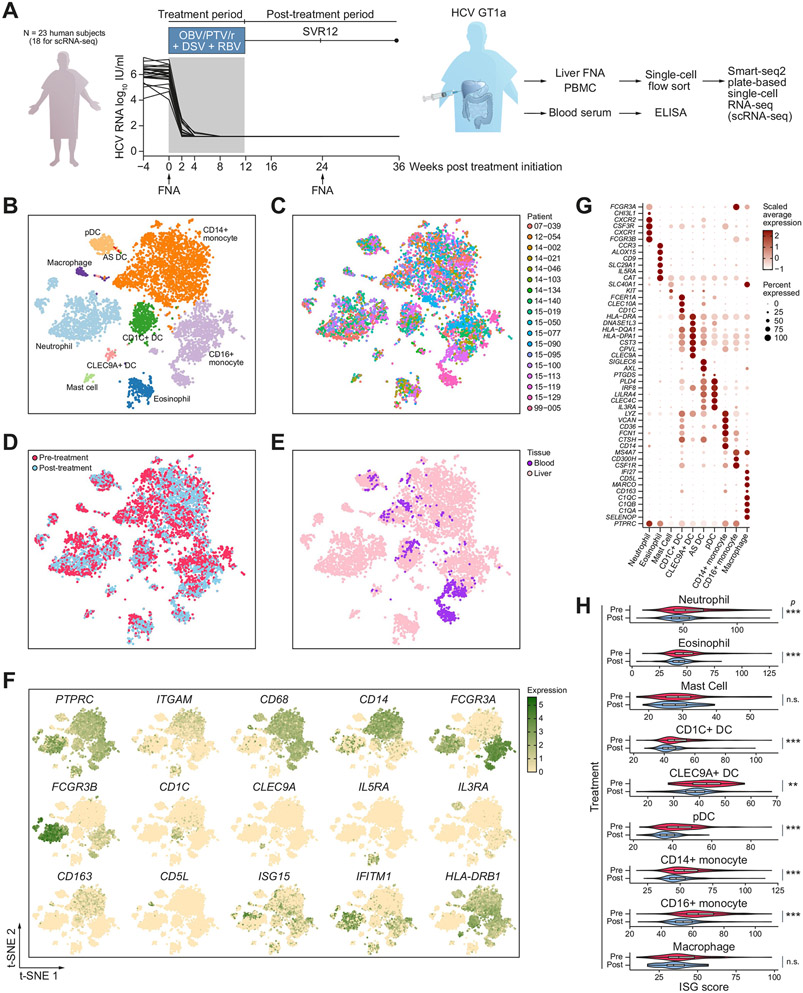
Fig. 1. Experimental design and single-cell RNA-sequencing of myeloid cells in chronic viral hepatitis and post-DAA-cure.
(A) Experimental design and HCV patient viral load before, during, and after DAA treatment. Patients received 12 weeks of DAA treatment. Liver FNAs and blood were collected before and 24 weeks after the initiation of DAA (12 weeks after the last dose of DAA). Myeloid cells were sorted and scRNA-seq was performed using the plate-based Smart-seq2 scRNA-seqmethod. For comparison, PBMC samples from seven patients were also profiled by scRNA-seq. Key immune cell activation markers were measured longitudinally by ELISA from the 252 Journal patient serum. (B–E) t-SNE maps of single cells colour-coded for (B) cell type, (C) individual patients, (D) pre-vs. post-treatment samples, and (E) liver vs. blood. (F) t-SNE maps coloured by the expression level of cell type and activation markers. (G) Dot plot of expressions of marker genes in each cell type. Shade represents the average expression. Size represents the percentage of cells that express the gene. (H) Violin plot of ISG scores in pre- or post-treatment samples in each cell type. The Wilcoxon rank-sum test was used to determine statistical significance between pre-treatment and post-treatment ISG scores. *p <0.05, **p <0.01, ***p <0.001. n.s., non-significant; DAA, direct-acting antiviral; DC, dendritic cell; FNAs, fine needle aspirates; ISG, interferon-stimulated gene; PBMCs, peripheral blood mononuclear cells; scRNA-seq, single cell RNA sequencing; SVR12, sustained virologic response after 12 weeks post-treatment; t-SNE, t-distributed stochastic neighbour embedding.
A landscape of intrahepatic myeloid cells defined by scRNA-seq
To obtain a high-resolution landscape of intrahepatic myeloid cell populations in patients with HCV, we used the Smart-seq2 scRNA-seq method to deeply profile liver FNA cells from study patients pre- and 12 weeks post-DAA when the viral load became undetectable in all patients (Fig. 1A). scRNA-seq data generated from three flow-cytometry sorted myeloid cell populations (CD14 high, dim, and negative, all negative for T/B/NK cell lineage markers) were pooled (Fig. S1A). To enrich dendritic cells, HLA-DR+ cells were selected for the CD14-negative population. For comparison, scRNA-seq data of peripheral blood mononuclear cells (PBMCs) from seven patients were included and sorted using the same approach. After quality control (Supplementary methods), we obtained a total of 8,515 high-quality single-cell transcriptomes (7,759 liver FNA cells and 756 PBMCs). These cells highly expressed housekeeping genes, such as ACTB and B2M, confirming the quality of our data (Fig. S1B).
Our data showed diverse myeloid cell populations in the liver and the blood. We visualised these cells on a 2D t-distributed stochastic neighbour embedding (t-SNE) plot, and coloured them by cell type, patient, treatment status, or tissue site (Fig. 1B-E). We performed clustering and found 15 coarse clusters of cells with distinct expressions of known cell type markers (Fig. S1C,D). These clusters were identified as neutrophils, eosinophils, mast cells, basophils, CD1C+ DCs, CLEC9A+ DCs, AXL+SIGLEC6+ (AS) DCs, pDCs, CD14+ monocytes, CD16+ monocytes, and macrophages (Fig. 1B, F, G, Table S2). We further performed subclustering to confirm the identity of each cell type and removed low quality subclusters (Supplementary methods). We found that the cells clustered by cell type rather than by patient, suggesting a conservation of immune cell phenotype across patients with HCV and an absence of technical artefacts such as patient-to-patient batch effects (Fig. 1B,C). Furthermore, we identified heterogeneous cell types in nearly all patients and in both the liver and the blood (Fig. S1E,F). On average, we measured ~1,000 genes per cell for granulocytes and ~4,000 genes per cell for other myeloid cell types, consistent with prior findings that Smartseq2 measures a much higher number of genes than most other scRNA-seq platforms (Fig. S1G,H).21
We found that pre- and post-treatment samples were separated within each cell type, suggesting that the DAA treatment led to a transcriptomic response in myeloid cells (Fig. 1D, Table S3). To study whether our data accurately reflected changes related to the treatment, we performed a global analysis on all major cell types to examine the known pathways related to antiviral therapy. Consistent with prior literature, we observed a significant decrease in interferon-stimulated gene (ISG) expression post-DAA, which reflected a decrease in antiviral activity after virus removal (Fig. 1H).22,25 Each cell type showed a strong decrease in ISG expression, suggesting that the antiviral programme was highly and broadly regulated by interferons (IFNs) produced by infected cells.
We found that blood myeloid cells only partially overlapped with intrahepatic myeloid cells (in the cell types identified in both tissues), indicating that blood samples did not capture the full spectrum of heterogeneous states of intrahepatic immune cells (Fig. 1E). Furthermore, we found a higher magnitude of treatment-induced changes in the liver compared with the blood (Fig. S2A-D). For example, we observed a mean 16.1% decrease in the ISG score in CD16+ monocytes in the liver and a mean 7.0% decrease in the blood (Fig. S2A). We also measured protein markers of immune activation, including CD163, HLA-DR, and CD86, which consistently showed that the changes between pre- and post-treatment were much more pronounced in the liver than in the blood (Fig. S2E). Studies on HBV have also shown that the composition and immune responses differ in the liver vs. the blood.26 Although peripheral blood is frequently used to monitor infectious disease activities, our data highlight the importance of studying immune cells from diseased tissues for both RNA and protein measurements as the peripheral immune cells may have a different phenotype.
Liver neutrophil subsets revealed distinct functions and segregation between ISG-high and MHC-II-high populations
We recovered 1,410 high-quality neutrophils from the liver FNAs from all patient samples, providing a transcriptomic atlas of tissue neutrophils (Fig. 2A). The data allowed us to comprehensively characterise liver neutrophil heterogeneity and their response to successful antiviral therapy. Subclustering of all neutrophils revealed their heterogeneity and identified four transcriptomically distinct cell subclusters, named Neutro_C1 through Neutro_C4 (Fig. 2A). Each of these subclusters encompassed cells from pre- and post-treatment conditions (Fig. 2B). We examined published markers for pro-neutrophils, pre-neutrophils, and mature neutrophils, and found that all four clusters robustly expressed markers for mature neutrophils including FCGR3B and not markers for immature cells (Fig. S3A).27,28 An analysis of differentially expressed genes (DEGs) between subclusters revealed that the four neutrophil subclusters were marked by distinct functions (Fig. 2C-E). The most abundant cluster, Neutro_C1, overexpressed chemokine and chemokine receptor genes, including CXCR4, CXCL8, CCR3, as well as genes associated with adhesion, including SIGLEC10. Pathway enrichment analysis showed that this cluster was enriched for granulocyte migration and cell chemotaxis (Fig. 2E). Neutro_C2 overexpressed alarmin genes including S100A4, S100A6, S100A12, and highly expressed signatures associated with the granulocytic myeloid-derived suppressor cells (G-MDSCs) (Fig. S3B).29 Neutro_C3 was marked by ISG genes, including ISG15, MX1, IFI6, and IFIT1, which represent a broad antiviral response. Neutro_C4 was associated with major histocompatibility complex II (MHC-II) complex and antigen presentation, including HLA-DPA1, HLA-DPB1, and HLA-DRA.
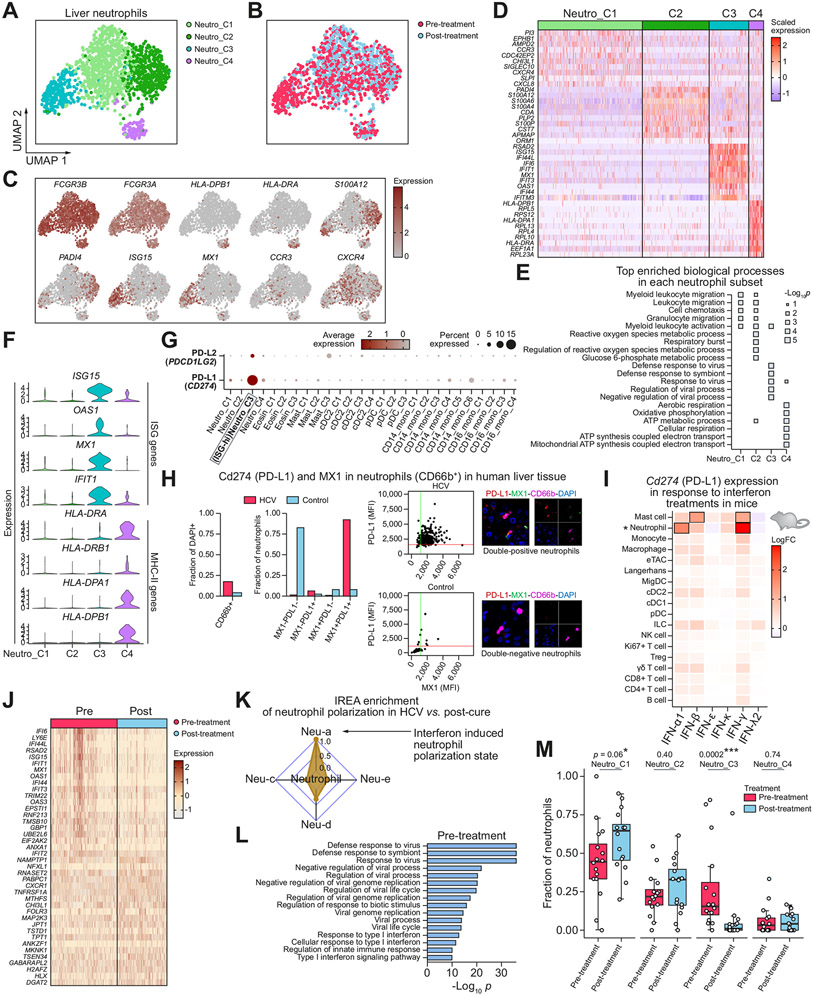
Fig. 2. Liver neutrophil subsets and phenotypic changes after DAA-cure.
(A,B) UMAPs of high-quality liver neutrophil cells, coloured by (A) neutrophil subcluster, and (B) treatment status. (C) UMAPs coloured by expression of marker genes. (D) Heatmap showing the overexpressed genes between each subcluster and all other neutrophil subclusters. (E) Pathways enriched for each subcluster. (F) Expressions levels of key ISG and MHC-II genes, showing segregation between ISG-high and MHC-II-high cells. (G) Dot plot showing expression levels of PD-L1 (CD274) and PD-L2 (PDCD1LG2) genes in each cell subcluster, some of which are defined later in the manuscript. (H) Immunofluorescent validation of PD-L1 expression in ISG+ (MX1+) neutrophils in human livers, HCV+ (top) and HCV− control (bottom). Shown as neutrophil fraction of cells, coexpression fraction of neutrophils, and as scatter plots of MFIs for PD-L1 and MX1, as well as composite and individual immunofluorescent channels. (I) Heatmap showing fold change in average expression of PD-L1 gene (CD274) in each immune cell type in response to interferon treatments in vivo in mice. Results are aggregated from three independent mice for each treatment condition. Boxes annotate most significant changes relative to PBS controls (log fold change >1 and FDR <0.01). Neutrophils are marked by *. (J) Heatmap showing the top differentially expressed genes between all pre- and post-treatment neutrophils. (K) IREA analysis shows an enrichment of interferon-mediated neutrophil polarisation state in pre-treatment neutrophils compared to post-treatment. (L) Pathway enrichment of genes overexpressed in pre-treatment neutrophils. (M) Box plot showing the distribution of the fraction of each neutrophil subset in pre- and post-treatment samples across patients. Each dot represents a patient. *p <0.05, **p <0.01, ***p <0.001 (Wilcoxon rank-sum test). DAA, direct-acting antiviral; FDR, false discovery rate; IREA, immune response enrichment analysis; ISG, interferon-stimulated gene; MFIs, mean fluorescent intensities; MHC-II, major histocompatibility complex II; UMAPs, uniform manifold approximation and projections.
Our analysis revealed that MHC-II-high neutrophils (Neutro_C4) and ISG-high neutrophils (Neutro_C3) were distinct populations found in the liver (Fig. 2D,F). This is in contrast to previous studies in other cell types which showed that both MHC-II and ISG expressions were induced by type I and II IFNs in DCs.30,31 Our data showed the neutrophils expressing a high level of ISGs tended to have a low expression of MHC-II and vice versa (Fig. 2F). This suggests that neutrophil responses to IFNs are distinct from the known IFN-MHC-II association previously observed in DCs. A recent study showed that FcγR engagement is involved in upregulating MHC-II programmes in neutrophils and turning them into professional antigen presenting cells, which provides a potential mechanism for the segregation we observed.32 Furthermore, it has been shown that human circulating neutrophils do not express MHC-II under normal conditions, and MHC-II-high neutrophil clusters were not observed in blood single-cell data collected from infectious diseases.33,34 However, our data revealed that the MHC-II-high neutrophil populations were present in the liver in both pre- and post-treatment conditions. Altogether this showed heterogeneous neutrophil populations with distinct functions in the liver.
ISG-high neutrophils expressed PD-L1/L2 and contracted post-cure
PD-L1+ neutrophils have been shown to have suppressive functions on T cells as well as on their own cytotoxicity in COVID-19, HIV, and cancer.33,35-37 By examining all the neutrophil subsets, we found that PD-L1 (CD274) and PD-L2 (PDCD1LG2) genes were predominantly expressed in the ISG-high Neutro_C3 subcluster and very scantly expressed in other neutrophil subclusters (Fig. 2G). To validate this finding we performed immunofluorescence staining in the human liver with untreated HCV infection compared with an uninfected control (Supplementary methods). We confirmed that ISG+/MX1+ neutrophils were also positive for PD-L1 and were abundant in the HCV-infected liver (Fig. 2H and Fig. S9). This suggests that a neutrophil population most responsive to IFNs mounts a regulatory programme to potentially limit T cell activation in the presence of the virus. Furthermore, PD-L1/L2 genes were very minimally expressed in monocytes, DCs, and eosinophils including ISG-high subclusters in these cell types (defined later in the manuscript) (Fig. 2G). Our data thus reveal that neutrophils are a major cell type likely to perform an immunoregulatory role in the liver under chronic infections.
To further validate in mice and study immune responses to IFNs in vivo, we injected type I, II, and III IFNs into mice and collected scRNA-seq profiles of immune cells using an optimal sample processing strategy.38 We studied PD-L1 gene expression in response to each IFN in each immune cell type (Fig. 2I). We found that in response to type I and type II IFNs, multiple immune cell types had mild upregulation of PD-L1. However, neutrophils are the cell type with the highest magnitude of upregulation in response to IFN-α and IFN-γ, having at least twofold larger changes than in any other cell types. Thus our in vivo data of both humans and mice revealed that neutrophils are the cell type with the highest potential for immunoregulation via the PD-1/PD-L1 pathway in an active antiviral process where IFNs are involved.
We then analysed changes in neutrophils in response to DAA therapy. We performed a differential gene expression analysis of all neutrophils collected pre- vs. post-treatment (Fig. 2J). Immune response enrichment analysis (IREA) on the differential gene expression shows a strong enrichment of neutrophil polarisation towards an IFN-induced polarisation state in chronic HCV relative to post-cure (Fig. 2K).38 Pathway enrichment shows the most remarkable changes were associated with reduced antiviral activities post-cure, consistent with the removal of the virus (Fig. 2L). We then computed the fraction of cells in each neutrophil subcluster to detect changes in relative fractions (Fig. 2M). After treatment, the ISG-high Neutro_C3 population significantly contracted though remained present (p < 0.01, Wilcoxon rank-sum test). Because the Neutro_C3 subset had a high expression of ISGs and PD-L1/PD-L2 genes (Fig. 2D,F,G), this suggests that neutrophil-mediated immune exhaustion was reduced once the virus was removed, promoting a homeostatic state. Our atlas thus pinpoints the specific neutrophil populations involved in immune regulation and resolution.
ISG-high liver eosinophils upregulated IDO1 and contracted after virus removal
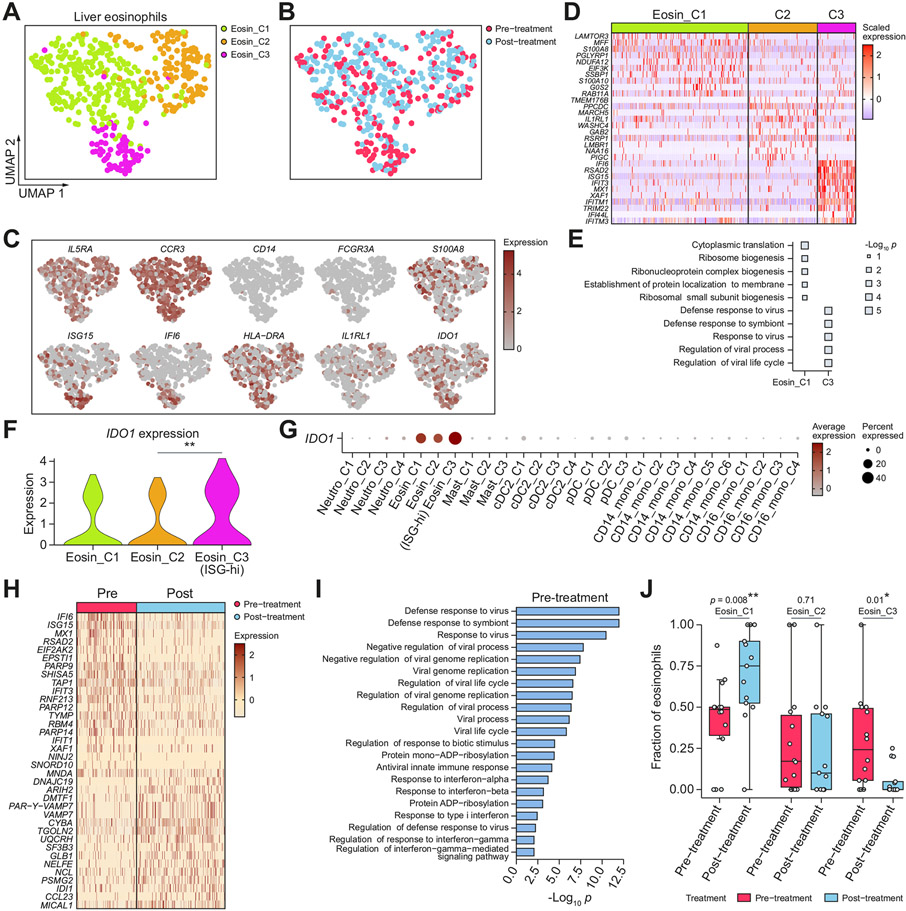
We identified three subclusters from 447 high-quality eosinophils from the liver, which we named Eosin_C1 through Eosin_C3 (Fig. 3A,B). Eosin_C1 increased in frequency after cure, and overexpressed S100A8, which have recently been suggested to promote eosinophil-mediated tissue healing39 (Fig. 3C). The Eosin_C3 subset strongly expressed ISGs and was enriched for antiviral defence programmes (Fig. 3C-E). Among the differentially expressed genes, we found that IDO1 was significantly overexpressed in the ISG-high Eosin_C3 subcluster (Fig. 3F). IDO1 is well established in its role in T cell immunosuppression and tolerance during viral infections, but the regulation of IDO1 expression is less clear.40 Consistent with prior findings, we found that IDO1 was constitutively expressed in eosinophils (Fig. 3G).41 The over-expression of IDO1 in the ISG-high Eosin_C3 subcluster showed that the antiviral programme induced a higher level of IDO1 expression, which could mediate T cell regulation. Next, we analysed the differential expression of all eosinophils in pre-vs. post-treatment (Fig. 3H). We found that DAA treatment was associated with a decrease in IFN response, and the IDO1-high ISG-high eosinophil cluster significantly decreased in fraction after cure (p = 0.01) (Fig. 3I,J).
Fig. 3. Liver eosinophil subsets and phenotypic changes after DAA-cure.
(A,B) UMAPs of high-quality liver eosinophils cells, coloured by (A) eosinophil subcluster, and (B) treatment status. (C) UMAPs coloured by expression of marker genes. (D) Heatmap showing the overexpressed genes between each subcluster and all other eosinophil subclusters. (E) Pathways enriched for each subcluster. (F) Violin plot showing IDO1 expression across eosinophil subclusters. *p <0.05, **p <0.01, ***p <0.001 (Wilcoxon rank-sum test). (G) IDO1 expression across all myeloid cell subclusters, some of which are defined later in the manuscript. (H) Heatmap showing top differentially expressed genes between all pre- and post-treatment eosinophils. (I) Pathway enrichment of the top overexpressed genes in pre-treatment eosinophils. (J) Box plot showing the distribution of the fraction of each eosinophil cluster in pre- and post-treatment samples across patients. Each dot represents a patient. *p <0.05, **p <0.01, ***p <0.001 (Wilcoxon rank-sum test). DAA, direct-acting antiviral; DC, dendritic cell; pDC, plasmacytoid dendritic cell; UMAPs, uniform manifold approximation and projections.
Aside from neutrophils and eosinophils, we uncovered mast cells, a type of rare granulocyte (Fig. S3C-I). A total of 79 profiled liver mast cells split into three subclusters: the ISG-low Mast_C1 upregulated FCER1A, a critical receptor for mast cell function, whereas the ISG-high Mast_C2 down-regulated FCER1A and upregulated ANXA1 (Annexin-A1), a potent negative regulator of mast cell activation.42 Collectively, our data on neutrophils, eosinophils, and mast cells revealed that granulocytes mount immunoregulatory programmes such as PD-L1/L2, IDO1, and ANXA1 during chronic infections in the cell subpopulations with the strongest ISG antiviral response.
Liver cDCs displayed heterogeneity and had enhanced proliferative programmes after virus removal
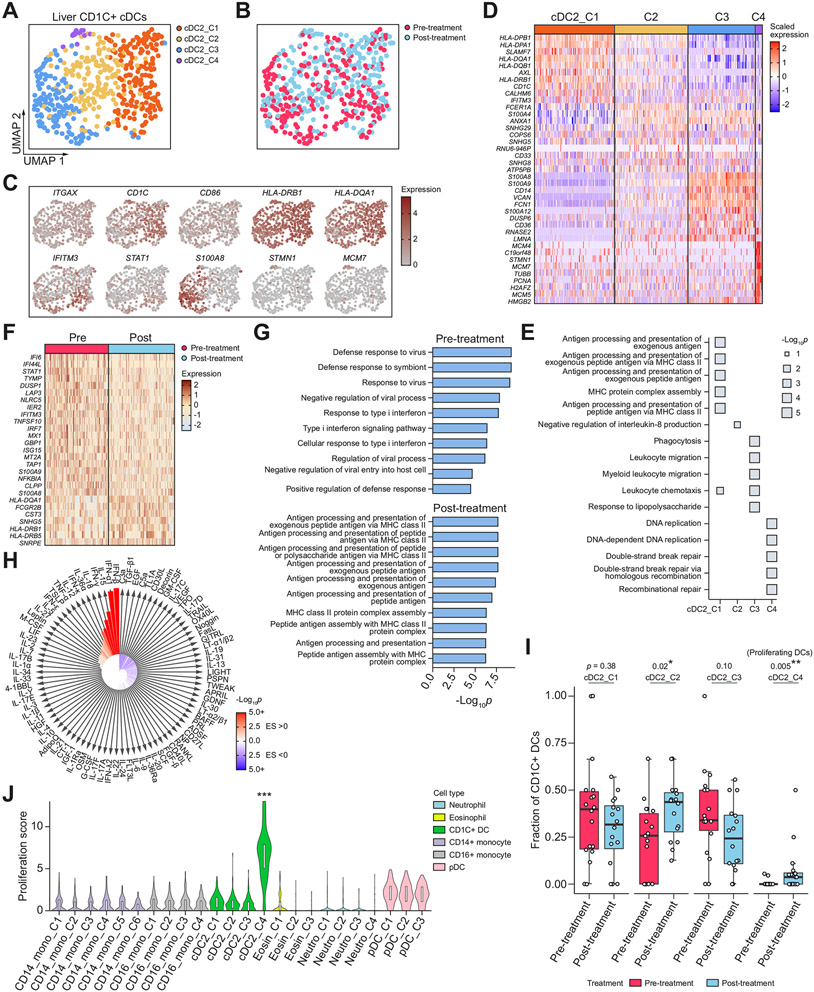
We observed three broad clusters of conventional DCs corresponding to CD1C+ DCs (cDC2; 568 cells), CLEC9A+ DCs (cDC1; 81 cells), and AS DCs we previously described (33 cells)43 (Fig. 1B). The most abundant CD1C+ DCs displayed heterogeneity and split into four subclusters (Fig. 4A,B), each of which expressed CD1C+ DC cell type markers, including ITGAX, CD1C, and HLA-DRB1 (Fig. S4A). We performed differentially expressed gene analysis to identify functional enrichments of each subcluster (Fig. 4C-E). cDC2_C1 was enriched for MHC-II-mediated antigen presentation, including HLA-DPA1, HLA-DPB1, and HLA-DRA. cDC2_C2 was an intermediate state and was enriched for FCER1A. cDC_C3 was enriched for inflammatory markers, including CD14, S100A8, S100A9, and S100A12. cDC2_C4 was marked by proliferation signatures including CDK1, MCM4, MCM5, MCM7, and STMN1. In contrast to neutrophils where ISG-high cells formed a distinct cluster and were separated from MHC-II-high cells, the cDC2_C1 cluster expressed both MHC-II genes and ISGs highly (Fig. 4C,D).
Fig. 4. Liver CD1C+ conventional dendritic cell subsets and phenotypic changes after DAA-cure.
(A,B) UMAPs of high-quality liver CD1C+ DCs, coloured by (A) CD1C+ DC subcluster, and (B) treatment status. (C) UMAPs coloured by expression of marker genes. (D) Heatmap showing the overexpressed genes between each subcluster and all other CD1C+ DC subclusters. (E) Pathways enriched for each subcluster. (F) Heatmap showing top differentially expressed genes between all preand post-treatment CD1C+ DCs. (G) Pathway enrichment of the top overexpressed genes in pre- and post-treatment CD1C+ DCs. (H) IREA cytokine analysis showing enrichment of cytokine signatures in pre-treatment relative to post-cure. Red indicates an enrichment in pre-treatment; blue indicates an enrichment in post-cure. (I) Box plot showing the distribution of the fraction of each CD1C+ DC subset in pre- and post-treatment samples across patients. Each dot represents a patient. *p <0.05, **p <0.01, ***p <0.001 (Wilcoxon rank-sum test). (J) Violin plot showing distribution of proliferation score of each cell subset across myeloid cell types. ***p <0.001 (Wilcoxon rank-sum test) relative to the reference group (CD14_Mono_C1). DAA, direct-acting antiviral; DCs, dendritic cells; IREA, immune response enrichment analysis; pDC, plasmacytoid dendritic cell; UMAPs, uniform manifold approximation and projections.
A comparison between pre- and post-treatment CD1C+ DCs revealed that DCs in the post-cure state had enhanced antigen presentation and proliferation programmes. In the differential expression analysis between all pre- and post-treatment CD1C+ DCs, aside from the expected down-regulation of ISGs post-cure, we also observed upregulation of MHC-II mediated antigen presentation programmes (Fig. 4F,G). IREA cytokine analysis shows the strongest enrichment for IFN-α1 and IFN-β signatures in pre-treatment relative to post-cure (Fig. 4H). Furthermore, when we analysed the changes in the fraction of each subcluster, we found that the subcluster cDC2_C4 had a significant increase in fraction (p = 0.005, Wilcoxon rank sum test) (Fig. 4I). Although this population was relatively rare, most of the patients had an increased fraction of cells in this population post-cure. As cDC2_C4 was enriched for genes associated with proliferation, this suggests that dendritic cells had an enhanced proliferative ability following successful removal of chronic infections. Next, we studied whether other myeloid cell types also acquired such properties. We found that the proliferation markers STMN1 and MCM7 were only elevated in cDC2_C4 and not in other liver myeloid cell types (Fig. S4B,C). We created a proliferation score for each cell (Supplementary methods) and again found that this proliferation property was unique to cDC2_C4 (Fig. 4J). To ensure this observation was not caused by clustering (i.e. the proliferating cells being clustered together with other cells), we computed the fraction of cells with a high proliferation score and found that the increase in proliferating cells was only present in DCs (Fig. S4D). Notably, the proliferating DCs were only observed in the liver, and not in the blood. Altogether, our findings suggest that tissue CD1C+ DCs gaining enhanced antigen presentation and proliferative abilities is associated with a reversal of chronic immune failure. We hypothesised that the reason for this property being unique to DCs and not other myeloid cells is that DCs are the most powerful antigen presenting cells and can constantly activate T cells in the presence of a chronic virus; thus, a reduction in their function leads to a homeostatic state in controlling the virus and preventing tissue damage in the setting of chronic infections.
CD14+ monocytes in the liver formed diverse subsets, all of which were detected in both pre- and post-treatment
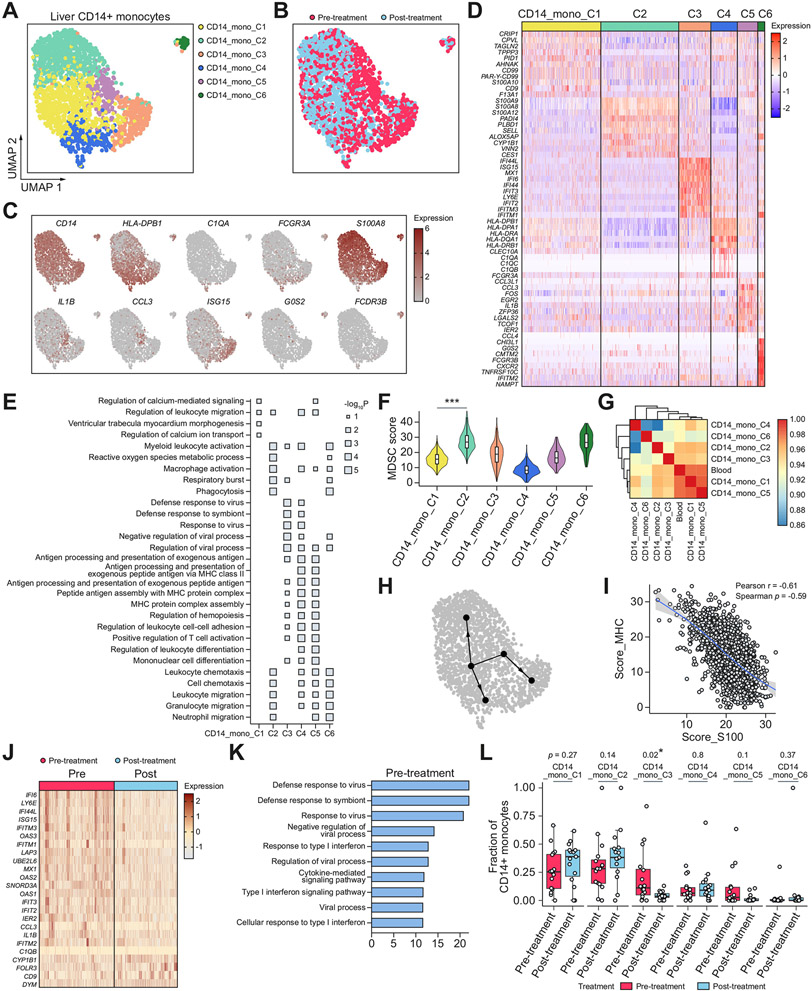
We obtained a total of 2,012 high-quality CD14+ monocytes from liver FNAs from all patient samples, from which we identified six subclusters, each of which included both pre- and post-treatment samples (Fig. 5A,B). Through DEG and pathway enrichment analysis, we found some of these subclusters resembled the subclusters we previously defined in neutrophils and DCs, whereas others were distinct (Fig. 5C-E). CD14_Mono_C2 over-expressed alarmins including S100A8, S100A9, and S100A12, and was highly enriched for signatures of monocytic myeloid derived suppressor cells (M-MDSCs) (Fig. 5F).44 CD14_Mono_C3 was enriched for ISGs, and CD14_Mono_C4 was enriched for MHC-II molecules. The cells expressing the highest levels of MHC-II also overexpressed C1q molecules, which are components of the complement pathway and markers of macrophages, indicating that this population was differentiating into a macrophage-like phenotype (Fig. 5C). Similar to neutrophils and in contrast to DCs, we found that the ISG-high monocytes and MHC-II-high monocytes were distinct populations of cells (Figs 2D,F, 4D, and 5D). This segregated expression was confirmed in the HCV-infected human liver by immunofluorescence staining (Fig. S10). CD14_Mono_C5 over-expressed IL1B, CCL3, and TMEM176B and was enriched for cytokine activities and chemotaxis. CD14_Mono_C6 cells, which were separated from other clusters, were enriched for granulocytic molecules and likely to be granulocytic monocytes. CD14_Mono_C4 and CD14_Mono_C6 also overexpress FCGR3A (CD16), which may correspond to the CD14+CD16+ double-positive populations that were found to be enriched in liver pathology.45-47
Fig. 5. Liver CD14+ monocyte subsets and phenotypic changes after DAA-cure.
(A,B) UMAPs of high-quality liver CD14+ monocytes, coloured by (A) CD14+ monocyte subcluster, and (B) treatment status. (C) UMAPs coloured by expression of marker genes. (D) Heatmap showing the overexpressed genes between each subcluster and all other CD14+ monocyte subclusters. (E) Pathway enrichment of the top overexpressed genes in each subcluster. (F) Violin plot showing M-MDSC scores in each subcluster. ***p <0.001 (Wilcoxon rank-sum test) relative to the reference group (CD14_Mono_C1). (G) Heatmap showing correlation coefficients between transcriptomic profiles of blood CD14+ monocytes and each subcluster of liver CD14+ monocytes. (H) Inferred cell maturation trajectories. (I) Scatter plot showing the relationship between MHC-II score and S100 score. Each dot is an individual cell. Line of best fit and 95% CIs are shown. (J) Heatmap showing top differentially expressed genes between all pre- and post-treatment CD14+ monocytes. (K) Pathway enrichment of the top overexpressed genes in pre-treatment CD14+ monocytes. (L) Box plot showing the distribution of the fraction of each CD14+ monocyte subset in pre- and post-treatment samples across patients. Each dot represents a patient. *p <0.05, **p <0.01, ***p <0.001 (Wilcoxon rank-sum test). DAA, direct-acting antiviral; M-MDSC, monocytic myeloid-derived suppressor cells; UMAPs, uniform manifold approximation and projections.
Given the diverse liver CD14+ monocyte subsets with distinct functional enrichments, we then analysed their differentiation trajectories. A correlation analysis between blood monocytes and each liver monocyte subcluster showed that the blood monocytes most resembled CD14_Mono_C1 and CD14_Mono_C5 (Fig. 5G). As monocytes are known to infiltrate from the blood to tissues, we performed a trajectory analysis originating from the CD14_Mono_C1 cluster on all the non-granulocytic monocyte subclusters (Fig. 5H). Our trajectory analysis supported the theory that upon entering the liver, monocytes could be differentiated into either S100-high, MHC-II-high, or ISG-high phenotype. As S100-high and MHC-II-high were on the opposite end of the spectrum, we hypothesised that these gene programmes were mutually antagonising at the individual cell level. To test this, we calculated the expression of S100 genes and MHC-II genes for each cell, and indeed found a strong inverse correlation between them (Pearson r = −0.61), which is consistent with a recent observation from protein measurements of these markers in COVID-19 samples (Fig. 5I).48 Our analysis supports the notion that S100 and MHC-II were mutually antagonising gene programmes in CD14+ monocytes: CD14+ monocytes could acquire S100 programmes to become a more MDSC-like phenotype, or MHC-II programmes to become a more matured or macrophage-like phenotype.
We then compared the changes in each subset after DAA treatment (Fig. 5J-L). As expected, the antiviral CD14_Mono_C3 cluster significantly decreased in proportion after treatment, and the inflammatory CD14_Mono_C5 also decreased, although not significantly. We found that there was a trend, although non-significant, towards an increase in fraction in blood-resembling CD14_Mono_C1 and MDSC-like CD14_Mono_C2 clusters post-treatment. Future study should explore whether this CD14_Mono_C2 population were pathologically induced MDSCs that remained in the liver post-treatment. Our analyses concluded that liver CD14+ monocytes formed subsets with diverse functions, all of which persisted post-treatment albeit there were significant changes in relative fractions compared to pre-treatment.
Liver CD16+ monocytes showed distinct features compared to CD14+ monocytes
The 1,071 high-quality liver CD16+ monocytes (FCGR3A+, CD14-/dim) partitioned into four subclusters (Fig. S6A-E). CD16_Mono_C1 was most closely related to blood monocytes and likely reflected baseline CD16+ monocyte states (Fig. S6F). CD16_Mono_C2 over-expressed MHC-II genes, and CD16_Mono_C3 overexpressed ISGs. Unlike CD14+ monocytes, the cluster enriched for alarmins and MDSC signatures was much smaller (CD16_Mono16_C4) (Fig. S6D,G), and there was no correlation between S100 and MHC-II gene scores (Fig. S6H). This suggests that the gene programmes involved in maturation or activation are distinct between CD14+ and CD16+ monocytes. CD16_Mono_C4 also overexpresses CD14, which may correspond to the double positive population similar to CD14_Mono_C4.
The DAA treatment was associated with a decrease in IFN response as revealed by differential gene expression between all pre-vs. post-treatment CD16+ monocytes, and the ISG-high CD16_Mono_C3 subset significantly decreased in fraction after cure (Fig. S6I-K). In contrast, CD16_Mono_C1, which likely represented the baseline CD16+ monocyte state, increased from an average of 30% of CD16+ monocytes in pre-treatment to 70% after treatment (Fig. S6K). Altogether this suggests a return to the homeostatic state in CD16+ monocytes post-DAA.
Macrophage activation markers, sCD163 and sCD5L, were found in serum pre- and post-DAA and followed different dynamics
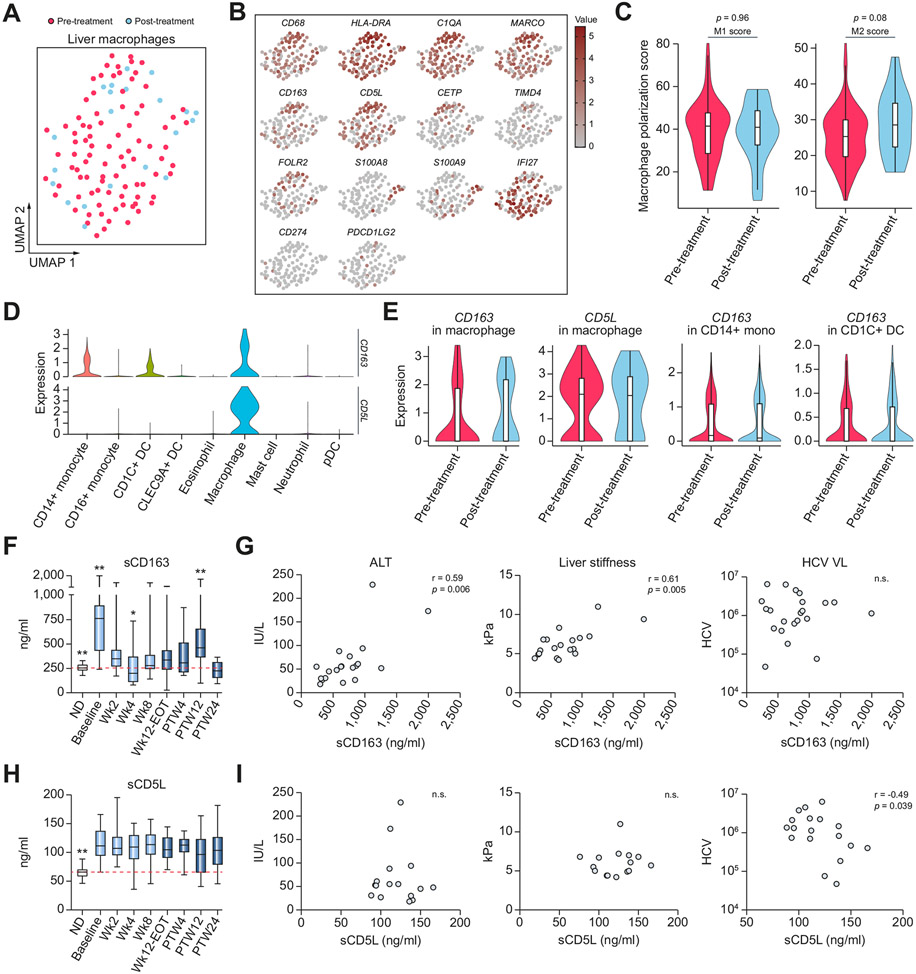
We studied macrophage states using both single-cell transcriptomics of liver cells and protein measurements of activation markers in the serum. Although the number of liver macrophages that could be collected by the FNA procedure in the scRNA-seq was limited, we were able to uncover their heterogeneity. Some cells overexpressed embryonic-derived macrophage markers, including CETP, TIMD4, and FOLR2, whereas others overexpressed monocyte-derived macrophage markers, including S100A8 and S100A9 (Fig. 6A,B). Using transcriptomic signatures of M1-like and M2-like macrophages, we found an elevation in the M2-like macrophage signature post-treatment, consistent with the role of this macrophage state in immune resolution (Fig. 6C).49
Fig. 6. Liver macrophage phenotypic changes upon DAA and serum macrophage activation marker dynamics during and after treatment.
(A) UMAP of high-quality liver macrophages, coloured by treatment status. (B) UMAPs coloured by expression of marker genes. (C) Violin plot showing scores of M1-like and M2-like macrophage signatures in pre- or post-treatment samples. Values of p were calculated using the Wilcoxon rank-sum test. (D) Violin plot showing gene expressions of CD163 and CD5L in each liver myeloid cell type. (E) Violin plot showing gene expressions of CD163 and CD5L in pre- or post-treatment samples in each cell type where they are expressed. (F) Box plots showing longitudinal protein expressions of serum sCD163. Baseline was significantly higher than all study timepoints (p <0.009). PTW12 was significantly higher than all study timepoints except baseline and Wk2 (p <0.002). Wk4 was significantly lower than all study timepoints except PTW24 (p <0.03). Normal donors were significantly lower than baseline, Wk2, and PTW12 (p <0.01). *p <0.05, **p <0.01 (Wilcoxon rank-sum test). (G) Scatter plots showing the relationship between sCD163 expression and ALT, liver stiffness, and HCV VL at the pre-treatment baseline. Values of p were obtained from Spearman correlation analysis. (H) Box plots showing longitudinal protein expressions of serum sCD5L. Normal donors were significantly lower than all other study time points (**p <0.01, Wilcoxon rank-sum test). (I) Scatter plots showing the relationship between sCD5L expression and ALT, liver stiffness, and HCV VL at the pre-treatment baseline. Values of p were obtained from Spearman correlation analysis. ALT, alanine transaminase; DAA, direct-acting antiviral; ND, normal donor; PTW12, post-treatment week 12; PTW24, post-treatment week 24; UMAPs, uniform manifold approximation and projections; VL, viral load.
In macrophages, CD163 protein and CD5L glycoprotein can be shed or secreted, respectively, and both can be measured as soluble (s)CD163 or sCD5L in the serum. Serum levels of sCD163 and sCD5L have been linked to inflammatory conditions, in particular sCD163 levels are strongly associated with liver inflammation and fibrosis in chronic HCV and are decreased upon successful antiviral therapy.50,51 We confirmed that both CD163 and CD5L transcripts were highly present in liver macrophages (Fig. 6D); CD5L was almost exclusively expressed in macrophages, whereas CD163 was also expressed in CD14+ monocytes and CD1C+ DCs, and both genes were expressed in both pre- and post-treatment conditions (Fig. 6E). To monitor the immune response to DAA longitudinally, we then measured protein levels of these markers in the serum at eight time points during or after DAA-therapy (Fig. 6F-I). As expected, serum levels of sCD163 and sCD5L at the pre-treatment baseline from HCV-infected patients were both significantly higher than those in healthy controls, and sCD163 levels were significantly correlated with liver inflammation and fibrosis markers. However, the dynamics of protein expression across the study time points differed between the two markers. Levels of sCD163 initially rapidly normalised by W4 of treatment when the viral load became undetectable for most of the patients, likely reflecting reduced monocyte/macrophage activation because of virus removal. After viral clearance, however, sCD163 levels progressively rose and peaked at PTW12 (p <0.01), although levels remained significantly lower than baseline, and were normalised by PTW24. This second wave increase may reflect mild activation of macrophages as part of the resolution phase of the chronic inflammation. Levels of sCD5L did not follow the same dynamics and continued to be higher than the levels observed in healthy controls. Surprisingly, the pre-treatment baseline sCD5L inversely correlated with the HCV viral load (r = −0.49, p = 0.04), probably reflecting the ability of sCD5L to bind to and aggregate viral damage-associated molecular patterns (DAMPs) (HCV ssRNA and dsRNA). It is noteworthy that at baseline, expression of S100 genes in macrophages also inversely correlated with HCV viral load (r = −0.58) (Fig. S7C,D). These elevations of sCD163 and sCD5L levels post-treatment potentially reflect macrophage-mediated ongoing regulation of inflammatory and healing responses months after viral clearance.
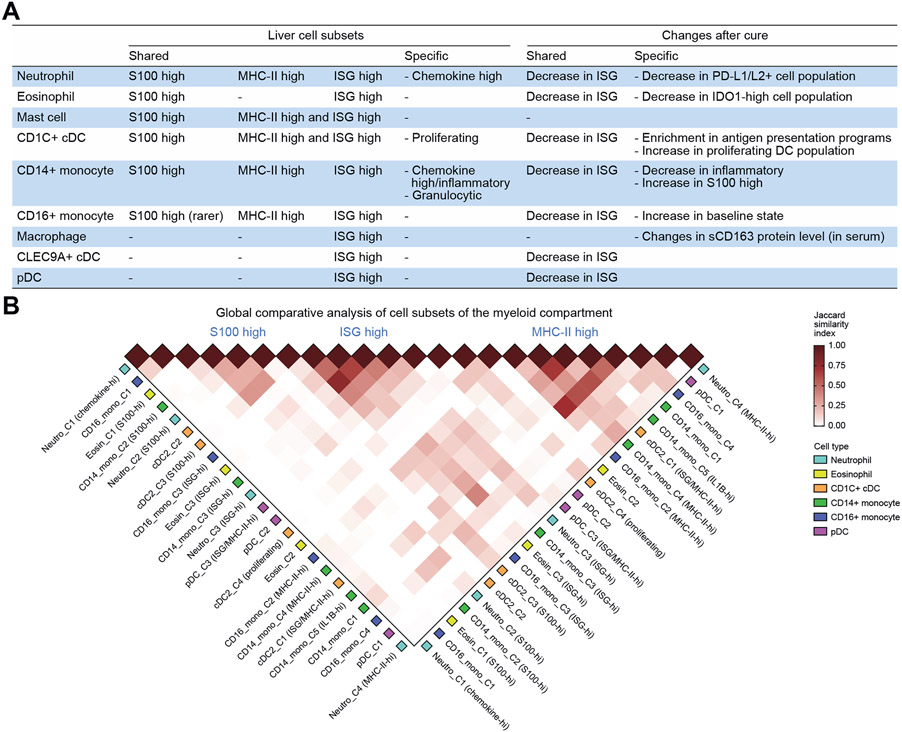
Global view of myeloid cell subsets and changes post-cure across cell types identifies shared gene programmes
From the analyses of each myeloid cell type, we noticed that some subset features were similar between multiple cell types, while others were specific to a cell type. To gain a comprehensive view of each myeloid cell type, we summarised all subsets we identified in Fig. 7. We found that nearly all cell types had a cluster of cells corresponding to ISG-high population. This population was always more abundant in pre-treatment samples, but was also present in post-cure samples in every cell type analysed. We found that neutrophils, cDC2s, and CD14+ monocytes all contained large subsets marked by either MHC-II-high, ISG-high, or S100-high expression. CD16+ monocytes also contained the MHC-II-high population and ISG-high population, but a smaller number of S100-high cells. Our observation of a convergence across cell types suggests that myeloid cell types share core programmes important for myeloid cell function.
Fig. 7. A global view of cell subsets in the liver myeloid compartment and their changes post-DAA.
(A) A summary of myeloid cell subsets identified in the liver (left) and their changes in response to DAA-induced cure (right). (B) Jaccard similarity index between each pair of myeloid cell subsets across cell types. cDC, conventional dendritic cell; DAA, direct-acting antiviral; ISG, interferon-stimulated gene; MHC-II, major histocompatibility complex II; pDC, plasmacytoid dendritic cell.
We also summarised the main changes post-DAA treatment in each cell type. A major change that was observed across all cell types is the decrease in ISG expression, consistent with reduced antiviral activities upon virus removal. The major feature of granulocytes was that the ISG-high cell population overexpressed immune checkpoint molecules. Specifically, the subset of ISG-high neutrophils upregulated PD-L1/L2 genes, whereas ISG-high eosinophils upregulated IDO1, both of which decreased post-cure. Another major cell-type-specific change was the increase in proliferating DCs after virus removal, suggesting that the proliferating activities may have been enhanced after treatment. Altogether these findings suggest a level of commonality across myeloid cell types as well as cell-type-specific responses in chronic immune activation and resolution.
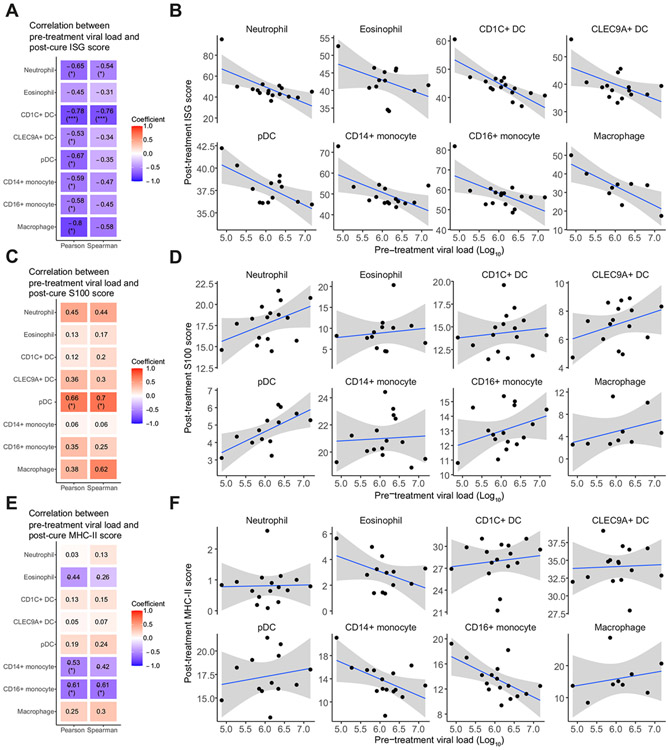
A higher pre-treatment viral load was associated with a lower post-cure ISG expression in host’s immune cells
In our global view, we identified a number of recurring gene programmes across cell types, namely ISG, S100, and MHC-II. We next investigated how these gene programmes related to viral load. We first analysed the relationship with ISG expression in each cell type. Consistent with prior studies, we did not observe a strong correlation between pre-treatment viral load and pre-treatment ISG expression (Fig. S7A,B).52,53 We also did not observe strong associations with S100 gene expression or with MHC-II gene expression (Fig. S7C-F). However, we found a strong inverse correlation between post-cure ISG expression and pre-treatment viral load in every cell type (Fig. 8A,B). Additionally, post-cure S100 gene programmes positively correlated with pre-treatment viral load, consistent with our prior finding that S100 gene programmes might have an antagonistic relationship with ISG (Fig. 8C,D). We did not find a strong correlation between antigen presentation and virus load aside from CD14+ monocytes (Fig. 8E,F). We performed the same analyses on differential expressions of these gene programmes between post-treatment and pre-treatment (Fig. S8A). Differential ISG expression showed a similar trend as post-cure ISG expression. We also tested the correlations with other clinical parameters, and did not observe a strong correlation between gene module expressions and bilirubin level or liver stiffness (Fig. S8B-E). Altogether this suggests that pre-treatment viral load has a profound role in shaping gene expressions in the host that would persist even after the patients were cured of the infections. Our analyses thus provide a link between viral load and sustained modifications of the host’s immune system.
Fig. 8. Post-cure ISG expression is inversely related to pre-treatment viral load.
(A,C,E) Pearson and Spearman correlation coefficients between pre-treatment viral load and post-cure gene set scores for (A) ISG genes, (C) S100 genes, and (E) MHC-II genes for each cell type. Colours correspond to correlation coefficients. Statistically significant correlations are marked. *p <0.05, **p <0.01, ***p <0.001. (B,D,F) Scatter plots between pre-treatment viral load and post-cure gene set scores for (B) ISG genes, (D) S100 genes, and (F) MHC-II genes, for each cell type. Line of best fit and 95% confidence interval are shown. DC, dendritic cell; ISG, interferon-stimulated gene; MHC-II, major histocompatibility complex II; pDC, plasmacytoid dendritic cell.
Discussion
The introduction of DAAs has made HCV the first curable chronic viral infection of humans. However, for other chronic viral infections and immune disorders, a complete cure remains extremely difficult. In diseases where viral triggers are unknown such as the recent childhood hepatitis, immunotherapies can offer a feasible treatment approach. A more complete elucidation of the evolution of the myeloid compartment in chronic viral infections and recovery can enhance our understanding of innate immune biology and offer insights into design of more effective immunotherapeutic strategies.
Basic biology studies of chronic infections and recovery have led to immunotherapy breakthroughs as cancer checkpoint inhibitor therapies were originally inspired by discoveries from mouse chronic lymphocytic choriomeningitis virus infections.54 However, to date only animal models exist for treatable chronic viral infections, which do not fully recapitulate human disease pathology. Here, we study an in vivo human system of cure, revealing multiple layers of innate immune regulation. We observe that granulocytes, in response to IFNs, simultaneously upregulate checkpoint molecules that could suppress lymphocyte activation. Specifically, we found that ISG-high neutrophils expressed PD-L1/PD-L2 and ISG-high eosinophils overexpressed IDO1. Indeed, we found that the chronically stimulated T cells collected from the same cohort of patients displayed a phenotype of exhaustion, and the expression levels of T cell inhibitory molecules were significantly decreased post-cure.23 Our atlas therefore pinpoints the specific cell populations that contribute to immune checkpoint regulation. CD1C+ DCs, in contrast, had a more active proliferation programme post-cure. DCs, as powerful antigen-presenting cells, can constantly activate T cells. We therefore hypothesise that a lower level of DC proliferation is one way for the immune system to adapt to persistent infection. Our study shows that innate immune cells create multiple layers of regulatory mechanisms, promoting a homeostatic state in chronic infections. After a successful cure of the chronic infection, this homeostatic state is shifted to favour the host. Thus, our atlas reveals specific populations of neutrophils, eosinophils, and DCs that can be targets of therapeutic intervention for chronic immune failures where a cure is not yet available.
ISGs are critical for a variety of host defence activities, and a lower response to IFNs is known to be associated with worse outcome for many types of infectious diseases and malignancies.55,56 Moreover, ISG expression has been shown to be higher in end-of-treatment liver biopsies of patients who had sustained virologic response than in patients who later relapsed.57 However, the factors that contribute to patient-to-patient variability in IFN response remain largely unknown. Understanding the factors that contribute to an ISG response can help stratify patients, predict disease prognosis, and suggest more targeted therapies. Here, we studied the association between pre-treatment viral load and ISG response. Consistent with prior findings, we did not observe an association between pre-treatment viral load and pre-treatment ISG expression. However, we discovered a strong inverse relationship between pre-treatment viral load and post-cure ISG expression (i.e. patients with a higher viral load pre-treatment tended to have a lower level of ISG expression 12 weeks after DAA treatment). Therefore, our study shows that chronic high viral load is a factor that could contribute to a lower level of basal ISG expression in the host when the virus is suppressed. A possible mechanism for this observation is that high viral load induces stable expression of negative regulators for ISGs, and such modifications are sustained post-DAA-cure. Indeed, several studies have shown sustained epigenetic modifications post-cure of chronic HCV infections.58,59 Future studies should explore whether these patients also had a lower level of ISG activities when freshly treated with IFNs, as well as the duration for which such a lower basal ISG expression was sustained post-cure.
Tissue immune cell heterogeneity is continuously being revealed, especially with the use of high-throughput profiling methods.12 Here, we comprehensively analysed subsets of each major liver myeloid cell type and found a high level of heterogeneity in each cell type. As expected, because every cell type is known to have unique functions, we found cell-type-specific cell subclusters, such as an inflammatory cluster found in CD14+ monocytes. However, at the same time, we found a surprising level of convergence across several myeloid cell types. In particular, neutrophils, DCs, and monocytes each displayed cell populations marked by the overexpression of (1) ISG, (2) S100, and (3) MHC-II. The ISG and MHC-II programmes could be co-regulated, as observed in DCs, or segregated, as observed in neutrophils and monocytes. S100 programmes, in many cases, have an antagonising relationship with ISG/MHC-II programmes. These observations suggest that the myeloid compartment, despite having evolved into cell types with distinct specialties, still retains core programmes that are critical for shared innate immune functions. They also reinforce the importance of deep profiling of cell types at the site of infection, as made possible through scRNA-seq and FNA sampling from the liver.
Our study also enhanced the clinical understanding of DAA therapy. Recent studies have pointed out that after DAA-induced cure of HCV, a number of perturbations persist, including risk for hepatocellular carcinoma in certain patients with more advanced hepatic fibrosis.10 Indeed, we previously reported that the T cells collected from the same cohort of cured HCV patients stop short of achieving full memory function.23 These findings call for a comprehensive analysis of post-HCV immunopathogenesis to provide optimal care for DAA-cured individuals. Our study provided two biological insights for these clinical observations. First, through our longitudinal study of patient serums, we found that immune cell activation markers, sCD163 and sCD5L, were upregulated in HCV patients even after treatment. sCD163 levels fluctuated during the course of the study, suggesting an ongoing immune response in some patients even at 6 months post-treatment. Second, we found that the patients with high viral load had lower basal ISG expression post-DAA. As ISGs are involved in controlling numerous types of infections and malignancies, our study suggests that patients should be stratified based on pre-treatment viral load for optimal post-DAA care.
In conclusion, we created a scRNA-seq atlas of intrahepatic myeloid cells in patients with HCV pre-treatment and post-cure. We comprehensively characterised each major myeloid cell type and studied their association with a curative response, which uncovered principles of myeloid cell gene programmes, multiple types of innate immune regulations, residual immune responses post-cure, and associations between pre-treatment viral load and post-cure immune state. Through a deeper understanding of the basic biology of chronic immune failure and resolution in humans, our atlas provides a roadmap to a cure for currently uncured chronic infections and immune-mediated disorders.
Supplementary Material
Acknowledgements
We thank Michael Waring and Nathalie Bonheur for additional assistance with cell sorting at the Immunology Core Facility of the Ragon Institute, which is part of the Harvard University Center for AIDS Research (CFAR), an NIH funded programme (P30 AI060354). We thank Dr Matthew P. Salomon and Dr Matthew A. Burchill for technical assistance. We acknowledge the Akoya scientist, Aniket Gad, for his valuable input with the Opal 3-plex tissue immunostaining used for validation. We also thank members of the Hacohen, Chung, and Lauer Laboratories for helpful discussions. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Financial support
This work was supported by grants from the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases U19AI082630 to Raymond T. Chung, Nir Hacohen, Georg M. Lauer, and Nadia Alatrakchi, NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK098079 to Raymond T. Chung and Nadia Alatrakchi, NIH/NIDDK R56DK134251 to Nadia Alatrakchi and Raymond T. Chung, NIH/NIDDK R01DK108370 to Raymond T. Chung. We acknowledge the MGH Flow and Mass Cytometry Research Core for their assistance with the single-cell sort, which was supported by NIH Shared Instrumentation programme grants 1S10OD012027-01A1, 1S10OD016372-01, 1S10RR020936-01, and 1S10RR023440-01A1.
Abbreviations
- ALT
alanine transaminase
- ANXA1
Annexin-A1
- AS
AXL+SIGLEC6+
- cDC
conventional dendritic cell
- DAA
direct-acting antiviral
- DAMPs
damage-associated molecular patterns
- DCs
dendritic cells
- DEG
differentially expressed gene
- FDR
false discovery rate
- FNA
fine needle aspirate
- G-MDSC
granulocytic myeloid-derived suppressor cells
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- IFN
interferon
- IREA
immune response enrichment analysis
- ISG
interferon-stimulated gene
- LCMV
lymphocytic choriomeningitis virus
- MFIs
mean fluorescent intensities
- MHC-II
major histocompatibility complex II
- M-MDSC
monocytic myeloid-derived suppressor cells
- ND
normal donor
- PBMC
peripheral blood mononuclear cells
- pDC
plasmacytoid dendritic cell
- PTW12
post-treatment week 12
- PTW24
post-treatment week 24
- scRNA-seq
single-cell RNA sequencing
- SVR12
Sustained virologic response after 12 weeks post-treatment
- t-SNE
t-distributed stochastic neighbour embedding
- UMAP
uniform manifold approximation and projection
- W2
week 2
- W4
week 4
Footnotes
Conflicts of interest
AC was a consultant for Foresite Capital and Altimmune Inc. for unrelated work. NH holds equity in BioNTech and is an advisor for Related Sciences/Danger Bio, Repertoire Immune Medicines and CytoReason. RTC received research grants to the institution from Abbvie, Gilead Sciences, Merck, Boehringer, Janssen, and BMS. NA received a research grant to the institution from Boehringer for unrelated work. The remaining authors declare no conflicts of interest that pertain to this work. Please refer to the accompanying ICMJE disclosure forms for further details.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2023.02.040.
Data availability statement
The human RNA-sequencing (scRNA-seq) count data can be visualized and downloaded at: https://singlecell.broadinstitute.org/single_cell/study/SCP2407/single-cell-atlas-of-the-liver-myeloid-compartment-before-and-after-cure-of-chronic-viral-hepatitis. Raw scRNA-seq data are also deposited in the dbGaP portal (ID phs002510.v1.p1)
References
- [1].Polaris Observatory HCV Collaborators. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2017;2:161–176. [DOI] [PubMed] [Google Scholar]
- [2].Yuen M-F, Chen D-S, Dusheiko GM, Janssen HLA, Lau DTY, Locarnini SA, et al. Hepatitis B virus infection. Nat Rev Dis Primers 2018;4:18035. [DOI] [PubMed] [Google Scholar]
- [3].Martínez I, Ryan P, Valencia J, Resino S. The challenging road to hepatitis C virus eradication. J Clin Med 2021;10:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hill AM, Nath S, Simmons B. The road to elimination of hepatitis C: analysis of cures versus new infections in 91 countries. J Virus Erad 2017;3:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zheng M, Tian Z. Liver-mediated adaptive immune tolerance. Front Immunol 2019;10:2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen T, Terrault N. Treatment of chronic hepatitis C in patients with cirrhosis. Curr Opin Gastroenterol 2016;32:143–151. [DOI] [PubMed] [Google Scholar]
- [7].Götte M, Feld JJ. Direct-acting antiviral agents for hepatitis C: structural and mechanistic insights. Nat Rev Gastroenterol Hepatol 2016;13:338–351. [DOI] [PubMed] [Google Scholar]
- [8].Wirth TC, Manns MP. The impact of the revolution in hepatitis C treatment on hepatocellular carcinoma. Ann Oncol 2016;27:1467–1474. [DOI] [PubMed] [Google Scholar]
- [9].Li DK, Chung RT. Overview of direct-acting antiviral drugs and drug resistance of hepatitis C virus. Methods Mol Biol 2019;1911:3–32. [DOI] [PubMed] [Google Scholar]
- [10].Polyak SJ, Crispe IN, Baumert TF. Liver abnormalities after elimination of HCV infection: persistent epigenetic and immunological perturbations post-cure. Pathogens 2021;10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roingeard P, Beaumont E. Hepatitis C vaccine: 10 good reasons for continuing. Hepatology 2020;71:1845–1850. [DOI] [PubMed] [Google Scholar]
- [12].Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC, Schultze JL. The myeloid cell compartment-cell by cell. Annu Rev Immunol 2019;37:269–293. [DOI] [PubMed] [Google Scholar]
- [13].Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 2017;17:306–321. [DOI] [PubMed] [Google Scholar]
- [14].Triantafyllou E, Woollard KJ, McPhail MJW, Antoniades CG, Possamai LA. The role of monocytes and macrophages in acute and acute-on-chronic liver failure. Front Immunol 2018;9:2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ritz T, Krenkel O, Tacke F. Dynamic plasticity of macrophage functions in diseased liver. Cell Immunol 2018;330:175–182. [DOI] [PubMed] [Google Scholar]
- [16].Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer 2016;16:447–462. [DOI] [PubMed] [Google Scholar]
- [17].Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019;572:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Poch T, Krause J, Casar C, Liwinski T, Glau L, Kaufmann M, et al. Single-cell atlas of hepatic T cells reveals expansion of liver-resident naive-like CD4 T cells in primary sclerosing cholangitis. J Hepatol 2021;75:414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014;9:171–181. [DOI] [PubMed] [Google Scholar]
- [21].Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND, et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 2020;38:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Holmes JA, Carlton-Smith C, Kim AY, Dumas EO, Brown J, Gustafson JL, et al. Dynamic changes in innate immune responses during direct-acting antiviral therapy for HCV infection. J Viral Hepat 2019;26:362–372. [DOI] [PubMed] [Google Scholar]
- [23].Tonnerre P, Wolski D, Subudhi S, Aljabban J, Hoogeveen RC, Damasio M, et al. Differentiation of exhausted CD8+ T cells after termination of chronic antigen stimulation stops short of achieving functional T cell memory. Nat Immunol 2021;22:1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Afdhal NH, Bacon BR, Patel K, Lawitz EJ, Gordon SC, Nelson DR, et al. Accuracy of fibroscan, compared with histology, in analysis of liver fibrosis in patients with hepatitis B or C: a United States multicenter study. Clin Gastroenterol Hepatol 2015;13:772–779.e1–3. [DOI] [PubMed] [Google Scholar]
- [25].Burchill MA, Salomon MP, Golden-Mason L, Wieland A, Maretti-Mira AC, Gale M Jr, et al. Single-cell transcriptomic analyses of T cells in chronic HCV-infected patients dominated by DAA-induced interferon signaling changes. PLoS Pathog 2021;17:e1009799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang C, Li J, Cheng Y, Meng F, Song J-W, Fan X, et al. Single-cell RNA sequencing reveals intrahepatic and peripheral immune characteristics related to disease phases in HBV-infected patients. Gut 2023;72:153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kwok I, Becht E, Xia Y, Ng M, Teh YC, Tan L, et al. Combinatorial single-cell analyses of granulocyte-monocyte progenitor heterogeneity reveals an early uni-potent neutrophil progenitor. Immunity 2020;53:303–318.e5. [DOI] [PubMed] [Google Scholar]
- [28].Ng LG, Ostuni R, Hidalgo A. Heterogeneity of neutrophils. Nat Rev Immunol 2019;19:255–265. [DOI] [PubMed] [Google Scholar]
- [29].Veglia F, Hashimoto A, Dweep H, Sanseviero E, De Leo A, Tcyganov E, et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J Exp Med 2021;218:e20201803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Steimle V, Siegrist C-A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-γ mediated by the transactivator gene CIITA. Science 1994;265:106–109. [DOI] [PubMed] [Google Scholar]
- [31].Simmons DP, Wearsch PA, Canaday DH, Meyerson HJ, Liu YC, Wang Y, et al. Type I IFN drives a distinctive dendritic cell maturation phenotype that allows continued class II MHC synthesis and antigen processing. J Immunol 2012;188:3116–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mysore V, Cullere X, Mears J, Rosetti F, Okubo K, Liew PX, et al. FcγR engagement reprograms neutrophils into antigen cross-presenting cells that elicit acquired anti-tumor immunity. Nat Commun 2021;12:4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schulte-Schrepping J, Reusch N, Paclik D, Baßler K, Schlickeiser S, Zhang B, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 2020;182:1419–1440.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pylaeva E, Lang S, Jablonska J. The essential role of type I interferons in differentiation and activation of tumor-associated neutrophils. Front Immunol 2016;7:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bowers NL, Helton ES, Huijbregts RPH, Goepfert PA, Heath SL, Hel Z. Immune suppression by neutrophils in HIV-1 infection: role of PD-L1/PD-1 pathway. PLoS Pathog 2014;10:e1003993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cheng Y, Li H, Deng Y, Tai Y, Zeng K, Zhang Y, et al. Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death Dis 2018;9:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yajuk O, Baron M, Toker S, Zelter T, Fainsod-Levi T, Granot Z. The PD-L1/PD-1 axis blocks neutrophil cytotoxicity in cancer. Cells 2021;10:1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cui A, Huang T, Li S, Ma A, Pérez JL, Sander C, et al. Dictionary of immune responses to cytokines at the single-cell resolution. Nature 2023. 10.1038/s41586-023-06816-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Reichman H, Moshkovits I, Itan M, et al. Transcriptome profiling of mouse colonic eosinophils reveals a key role for eosinophils in the induction of s100a8 and s100a9 in mucosal healing. Sci Rep 2017;7:7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schmidt SV, Schultze JL. New Insights into IDO biology in bacterial and viral infections. Front Immunol 2014;5:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Odemuyiwa SO, Ghahary A, Li Y, Puttagunta L, Lee JE, Musat-Marcu S, et al. Cutting edge: human eosinophils regulate T cell subset selection through indoleamine 2,3-dioxygenase. J Immunol 2004;173:5909–5913. [DOI] [PubMed] [Google Scholar]
- [42].Sinniah A, Yazid S, Bena S, Oliani SM, Perretti M, Flower RJ. Endogenous annexin-A1 negatively regulates mast cell-mediated allergic reactions. Front Pharmacol 2019;10:1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017;356:eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mastio J, Condamine T, Dominguez G, Kossenkov AV, Donthireddy L, Veglia F, et al. Identification of monocyte-like precursors of granulocytes in cancer as a mechanism for accumulation of PMN-MDSCs. J Exp Med 2019;216:2150–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liaskou E, Zimmermann HW, Li K-K, Oo YH, Suresh S, Stamataki Z, et al. Monocyte subsets in human liver disease show distinct phenotypic and functional characteristics. Hepatology 2013;57:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Seidler S, Zimmermann HW, Bartneck M, Trautwein C, Tacke F. Age-dependent alterations of monocyte subsets and monocyte-related chemokine pathways in healthy adults. BMC Immunol 2010;11:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dhanda AD, Williams EL, Yates E, Lait PJP, Schewitz-Bowers LP, Hegazy D, et al. Intermediate monocytes in acute alcoholic hepatitis are functionally activated and induce IL-17 expression in CD4+ T cells. J Immunol 2019;203:3190–3198. [DOI] [PubMed] [Google Scholar]
- [48].Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020;369:1210–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS−) vs. alternatively activated macrophages. Front Immunol 2019;10:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lidofsky A, Holmes JA, Feeney ER, Kruger AJ, Salloum S, Zheng H, et al. Macrophage activation marker soluble CD163 is a dynamic marker of liver fibrogenesis in human immunodeficiency virus/hepatitis C virus coinfection. J Infect Dis 2018;218:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sanjurjo L, Aran G, Roher N, Valledor AF, Sarrias M-R. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J Leukoc Biol 2015;98:173–184. [DOI] [PubMed] [Google Scholar]
- [52].Hedegaard DL, Tully DC, Rowe IA, Reynolds GM, Bean DJ, Hu K, et al. High resolution sequencing of hepatitis C virus reveals limited intrahepatic compartmentalization in end-stage liver disease. J Hepatol 2017;66:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wieland S, Makowska Z, Campana B, Calabrese D, Dill MT, Chung J, et al. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology 2014;59: 2121–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006;439:682–687. [DOI] [PubMed] [Google Scholar]
- [55].McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol 2015;15:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer 2016;16:131–144. [DOI] [PubMed] [Google Scholar]
- [57].Meissner EG, Wu D, Osinusi A, Bon D, Virtaneva K, Sturdevant D, et al. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. J Clin Invest 2014;124(8):3352–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Perez S, Kaspi A, Domovitz T, Davidovich A, Lavi-Itzkovitz A, Meirson T, et al. Hepatitis C virus leaves an epigenetic signature post cure of infection by direct-acting antivirals. PLoS Genet 2019;15:e1008181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hamdane N, Jühling F, Crouchet E, El Saghire H, Thumann C, Oudot MA, et al. HCV-induced epigenetic changes associated with liver cancer risk persist after sustained virologic response. Gastroenterology 2019;156:2313–2329.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human RNA-sequencing (scRNA-seq) count data can be visualized and downloaded at: https://singlecell.broadinstitute.org/single_cell/study/SCP2407/single-cell-atlas-of-the-liver-myeloid-compartment-before-and-after-cure-of-chronic-viral-hepatitis. Raw scRNA-seq data are also deposited in the dbGaP portal (ID phs002510.v1.p1)