Abstract
Background
The catenin delta 2 (CTNND2) gene has been implicated in the progression of various cancers, but its specific role in melanoma has not yet been thoroughly investigated. This study sought to explore the expression and biological function of CTNND2 in malignant melanoma tissues to identify new targets or biomarkers for melanoma diagnosis and treatment.
Methods
Immunohistochemistry was used to examine the levels of CTNND2 in melanoma and adjacent non-tumor tissues. A Western blot analysis was performed to quantify the expression levels of CTNND2 in human immortalized keratinocytes and melanoma cell lines. The Cell Counting Kit-8 (CCK-8) assay, plate colony formation assay, cell adhesion assay, scratch test, and Transwell assay were used to assess the effects of CTNND2 knockdown on the proliferation, adhesion, migration, and invasion of melanoma cells. The Harmonizome database was used to research the biological processes (BPs) involved in CTNND2.
Results
In the melanoma tissues, CTNND2 expression was substantially upregulated and its levels were closely linked with the pathological features of patients. The CTNND2 levels were notably more increased in the melanoma cell lines than the immortalized keratinocytes. The suppression of the CTNND2 gene substantially impeded the capacity of the melanoma cells to proliferate, migrate, and invade, and also significantly decreased their potential to attach to collagen I and IV, and fibronectin. The Harmonizome database results revealed a strong correlation between the BPs controlled by CTNND2 and the focal adhesion signaling pathway of the cells. The inhibition of the CTNND2 gene in melanoma cells resulted in a significant decrease in the phosphorylation of focal adhesion kinase (FAK) and the production of paxillin protein. In the melanoma cells, the reduction of CTNND2 did not have a significant effect on the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway. However, it did considerably prevent the activation of mitogen-activated extracellular signal-regulated kinase 1/2 (MEK1/2) and its downstream molecule extracellular signal-regulated protein kinase 1/2 (ERK1/2).
Conclusions
The expression of the CTNND2 gene is increased in melanoma tissues, which enhances the ability of melanoma cells to proliferate both in vivo and in vitro. Additionally, the CTNND2 gene is crucial in controlling the adhesion process of melanoma cells. This mechanism is associated with the regulation of the FAK and MEK1/2/ERK1/2 signaling pathways. Based on our findings, CTNND2 could be used as an oncogene target for melanoma and a new treatment target or diagnostic biomarker.
Keywords: Melanoma, catenin, catenin delta 2 (CTNND2), focal adhesion kinase (FAK), biomarker
Highlight box.
Key findings
• Catenin delta 2 (CTNND2) expression is significantly upregulated in melanoma tissues compared to adjacent non-tumor tissues.
• The suppression of the CTNND2 gene in melanoma cells reduces their proliferation, migration, and invasion capabilities, and impairs their adhesion to key extracellular matrix components.
• CTNND2 modulates melanoma cell behavior through the focal adhesion kinase and mitogen-activated extracellular signal-regulated kinase 1/2/extracellular signal-regulated protein kinase 1/2 signaling pathways, but not the phosphatidylinositol 3-kinase/protein kinase B pathway.
What is known, and what is new?
• Research has been conducted on the role of CTNND2 in other cancers, but little is known about its specific function and effect in melanoma.
• This study identified CTNND2 as a potential oncogene in melanoma, highlighting its involvement in key signaling pathways that regulate melanoma cell adhesion and proliferation.
What is the implication, and what should change now?
• Our findings suggest that CTNND2 could serve as a novel target for therapeutic interventions and as a biomarker for melanoma diagnosis.
• Further research and clinical trials need to be conducted to explore the potential of targeting CTNND2 in melanoma treatment, which may lead to the development of more effective therapeutic strategies.
Introduction
Melanoma, while comprising less than 1% of skin cancers, causes up to 90% of skin cancer-related deaths, making it the deadliest form of skin cancer (1-3). The prognosis and treatment outcome of melanoma are closely related to the time of diagnosis. Stage I–II melanomas are often superficial, can be surgically removed, and have a 5-year survival rate of 82–98% (4). However, currently, there are no established treatment guidelines or effective drugs available for advanced-stage metastatic melanoma. In recent years, the introduction of targeted immunotherapies has extended survival times and increased survival rates for many cancer patients. However, due to drug resistance and toxicity, these advanced treatments are not beneficial in about 60% of patients with metastatic melanoma (5,6), and only 23% of these patients achieve 5-year survival (7). Thus, current understandings of the mechanisms of melanoma metastasis and prognostic factors are limited, and further research needs to be conducted to explore the mechanisms of melanoma development and metastasis, and to identify more effective treatment targets and strategies.
In epithelial or neuronal cells, catenin delta 2 (CTNND2) is highly expressed and plays a role in morphological control and cell adhesion. Functionally, CTNND2 interacts with various intracellular calcium-regulated cytoskeletal proteins that participate in signal transduction or cell shape regulation (8,9). Previous research has shown that the levels of the CTNND2 gene are abnormal in several tumor tissues and are associated with both treatment resistance and the malignant advancement of diverse types of cancer (10). Research by Shimizu et al. (11) indicated that CTNND2 overexpression increased the invasiveness of glioma cells in vivo and in vitro, and linked CTNND2 to the ability of tumor cells to invade and migrate. Based on these studies, we think that CTNND2 may function as an oncogene and play a critical role in cancer formation and progression. However, the precise function of CTNND2 in melanoma is still uncertain. This study focused on melanoma cells, using short hairpin RNA (shRNA) to construct CTNND2 knockdown cell lines. Through in vivo and in vitro functional experiments, it is the first to explore the role of the CTNND2 gene in the malignant progression and pathogenesis of melanoma. This research seeks to provide new perspectives on the treatment and prognosis of melanoma patients. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2159/rc).
Methods
Clinical sample collection
Clinical data, excised cancerous tissues, and corresponding tissues surrounding the cancerous area were obtained from 46 melanoma patients who underwent surgical treatment at the Dermatology Department of the Affiliated Yixing Clinical School of Medical School of Yangzhou University from January 2018 to December 2019. This research included patients: (I) who were aged between 40 and 70 years; (II) who had not undergone radiotherapy or chemotherapy before surgery; (III) who signed an informed consent form; and (IV) whose tumor and adjacent tissues were examined by more than two pathologists. The tissues collected during surgery were rinsed with sterile saline, dried, and then rapidly frozen in liquid nitrogen. The clinical data collected included lymph node metastasis, age, ulceration level, sex, and tumor-node-metastasis (TNM) staging data. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of the Affiliated Yixing Clinical School of Medical School of Yangzhou University (approval No. 2023-159-01), and informed consent was obtained from all the patients.
Immunohistochemistry
The melanoma tissues were fixed with 4% paraformaldehyde (RO1321; Jiangsu Dingguo Technology Co., Nanjing, China), embedded in paraffin, and then sectioned, baked, deparaffinized, and hydrated. The sections underwent depigmentation using the potassium permanganate/oxalic acid method. After depigmentation, the sections were subjected to antigen retrieval under high temperature and pressure in citrate buffer for 3 minutes, then cooled to room temperature and rinsed with phosphate-buffered saline (PBS). The sections were treated for 10 minutes with freshly prepared hydrogen peroxide (3%) and rinsed two times with PBS (for 5 minutes each time). Next, 0.5% Triton X-100 (P0096; Beyotime, Shanghai, China) was applied on the permeabilized cell membranes for 15 minutes. A proper boundary was drawn on the slides using an immunohistochemistry pen, and an appropriate amount of 5% goat serum blocking solution (C0265; Beyotime) was added. The slides were then placed in a humidified box for 2 hours at 37 °C. The samples were then treated overnight with the primary antibody solution (1:150; LS-C677610-50; LSBio, Seattle, WA, USA) at 4 °C, rinsed three times with PBS (for 5 minutes each time), incubated with the secondary antibody (1:1,000) solution at room temperature (RT) for 90 minutes, and rinsed three times with PBS (for 5 minutes each time). 3,3'-diaminobenzidine (DAB) (P0203; Beyotime) was used for color development, and hematoxylin was then applied for counterstaining, followed by differentiation in hydrochloric acid ethanol, bluing in tap water, dehydration in graded ethanol, clearing in xylene, and mounting with neutral resin. The samples were visualized via a microscope (DMI 40008; Leica, Wetzlar, Germany). Five fields of view were randomly selected from each slide. Scoring was done based on staining intensity (0 points for no staining, 1 point for light yellow, 2 points for brown-yellow, and 3 points for tan) and the percentage of positive cells (0 points for ≤5%, 1 point for 5%< to ≤25%, 2 points for 25%< to ≤50%, 3 points for 50%< to ≤75%, and 4 points for >75%). The final score was determined by multiplying the two scores together. A score ≥2, was considered positive (+), and a score <2 was considered negative (−).
Western blot
Total cellular protein was isolated using radioimmunoprecipitation assay (RIPA) lysis buffer (P0013C; Beyotime). The protein concentration was quantified using a bicinchoninic acid protein test kit (P0010; Beyotime). Samples were then mixed with protein loading buffer and denatured for 5 minutes at 100 °C. Next, the samples were subjected to sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which involved a 90-minute transfer to a polyvinylidene fluoride (PVDF) membrane (IPVH00010; Millipore Corp., Atlanta, GA, USA) in an ice bath (80 V until the marker entered the separating gel, followed by 120 V until completion). The PVDF membrane was blocked with 5% bovine serum albumin solution (P0007; Beyotime) at RT for 2 hours. The membrane was then incubated overnight on a shaker at 4 °C with primary antibodies (12), including CTNND2 (1:1,000; LS-C677610-50; LSBio), p-extracellular signal-regulated protein kinase 1/2 (p-ERK1/2) (phospho T202/Y204; 1:800; ab278538; Abcam, Cambridge, UK), ERK1/2 (1:1,000; ab184699; Abcam), p-mitogen-activated extracellular signal-regulated kinase 1 (p-MEK1) (phospho T286; 1:800; ab307509; Abcam), MEK1 (1:1,000; ab32091; Abcam), p-protein kinase B (p-AKT) (phospho T308; 1:800; ab38449; Abcam), AKT (1:1,000, ab8805; Abcam), p-focal adhesion kinase (p-FAK) (phospho Y576; 1:800; ab76120; Abcam), p-FAK (phospho Y397; 1:800; ab81298; Abcam), FAK (1:1,000; ab40794; Abcam), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1,000; ab263962; Abcam), vimentin (1:1,000; EPR3776; Abcam), N-cadherin (1:1,000; ab18203; Abcam), and E-cadherin (1:800; ab314063; Abcam). The membranes were rinsed three times with PBS (for 5 minutes each time), and then incubated for 2 hours with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:5,000; ab6728; Abcam) at RT. Proteins were visualized using an enhanced chemiluminescence detection reagent (34095; Pierce, Rockford, IL, USA) and then rinsed three more times with Tris-buffered saline with Tween 20 (TBST) (PPB002; Sigma-Aldrich, St. Louis, MO, USA) for 5 minutes each time. The relative expression levels of the target proteins were evaluated using ImageJ software (version 1.53; NIH, Bethesda, MD, USA), with β-actin serving as an endogenous control.
Cell transfection
The HaCat cell line, derived from human immortalized keratinocytes, and the A375, A875, A2058, and M14 cell lines, derived from human melanoma, were supplied by the Shanghai Cell Bank at the Chinese Academy of Sciences (Shanghai, China). The cell lines M14, A2058, and A875 were maintained in the Roswell Park Memorial Institute (RPMI)-1640 medium (11875-093; Invitrogen, Carlsbad, CA, USA). The A375 and HaCat cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (D0422; Sigma-Aldrich) containing 10% fetal bovine serum (FBS) (F0193; Sigma-Aldrich) in an incubator containing 5% CO2 at 37 °C.
The A375 cells demonstrating logarithmic growth were separated into a negative control group (shRNA-control) and several transfection groups [shRNA-CTNND2-1 (5'-CCACACGAAAUUAUGAUGA-3') and shRNA-CTNND2-2 (5'-CUUACAGUGAACUGAACUA-3')], and the cell density was adjusted to 2×105/mL. The cells (5×105/well) were propagated in a 6-well plate. For each group, three replicate wells were designated. The lentivirus dilution was prepared, and the transfection groups were then added to the diluted lentiviral plasmids containing a low expression of CTNND2, while the negative control group received an equal concentration of diluted empty vector lentiviral plasmids. The cells were incubated overnight in a 5% CO2 incubator at 37 °C. After 72 hours, the lentiviral fluid was removed and replaced with 500 µL of complete medium containing 2.5 ng/mL puromycin, and then transferred back into a 5% CO2 incubator at 37 °C, for continued culturing until the confluency reached 80–90% for passaging. To evaluate the transfection efficiency, fluorescent protein expression was assessed using a fluorescence microscope (IX71; Olympus, Tokyo, Japan).
Cell Counting Kit-8 (CCK-8) assay
The shRNA-control-A375 cells and shRNA-CTNND2-1-A375 cells (1×104/well) were grown in a 96-well plate at 37 °C in a 5% CO2 incubator. CCK-8 solution (C0039; Beyotime) (10 µL) was added to each well, and the cells were incubated for 30 minutes at 37 °C in 5% CO2 for 24, 48, 72, and 96 hours. Six replicate wells were examined at each time point, and the absorbance was measured at 450 nm using a microplate reader (ELx800; BioTek Instruments Inc., Winosky, VE, USA) (13).
Plate colony formation assay
The shRNA-control-A375 cells and shRNA-CTNND2-1-A375 cells (1×104/well) were grown in a 6-well plate at 37 °C in 5% CO2 for 48 hours. After media removal, 500 µL of 4% formaldehyde solution (RO1321; Jiangsu Dingguo Technology Co.) was added to each well to cover the bottom layer of cells, and the cells were fixed at RT overnight. The formaldehyde solution was aspirated, and the bottoms of the wells were rinsed with low-pressure tap water for 10 minutes. Next, 200 µL of 1% crystal violet solution (C0121; Beyotime) was introduced to each well, and the cells were stained in the dark at RT for 30 minutes. The wells were then rinsed using low-pressure tap water until the rinse water was clear and colorless, and the excess water was removed. The plates were dried and placed under a light source for counting cell colonies.
Adhesion assay
The adhesion capability of each cell group was assessed using the CytoSelect™ 48-well Cell Adhesion Assay Kit (CBA-071; Cell Biolabs, San Diego, CA, USA). Specifically, 0.25% trypsin (C0201; Beyotime) was used to digest the shRNA-control-A375 cells and shRNA-CTNND2-1-A375 cells for 1 minute, and the digestion was stopped by adding complete medium (3 mL). The cells were then washed and transferred to sterile Eppendorf tubes. A final concentration of 1×105 cells/mL was achieved by centrifuging the cells for three minutes at 600 rpm (5810 R; Eppendorf, Hamburg, Germany), after which the supernatant was discarded, and the cells were diluted with a suitable volume of serum-free DMEM. Following propagation in a 48-well plate pre-coated with laminin, fibrinogen, fibronectin, and collagen types I and IV, the cells were cultured at 37 °C for 1 hour in a 5% CO2 incubator. The bottoms of the wells were washed with sterile PBS after the medium was aspirated. Each well was treated with 200 µL of crystal violet solution (C0121; Beyotime). The cells were then stained at room temperature for 10 minutes. Next, the staining solution was removed, and the cells were washed with sterile PBS five times. Using an Olympus IX71, microscope, the cells were observed, photographed, and counted.
Scratch test
After being seeded into a 6-well plate, the shRNA-control-A375 and shRNA-CTNND2-1-A375 cells were cultured to 90% confluence. A micropipette tip (200 µL) was used to linearly wound the confluent monolayer, and the cell debris was then washed off with PBS. After a 24-hour incubation period, cell migration was examined using a 100× magnification microscope (IX71; Olympus). In five fields of view selected randomly from each sample, the number of cells that moved across the black lines was recorded. After normalization, which involved setting the amount for the control group to 100%, the inhibitory effect was reported.
Transwell assay
The migratory and invasive abilities of the cells were evaluated using the Transwell insert for 8 µm pores in 24-well plates. The shRNA-control-A375 and shRNA-CTNND2-1-A375 cells were seeded into the Transwell insert (upper chamber) for the Transwell migration assays using a serum-free culture medium. In the lower chamber, a chemo-attractant culture medium containing 10% FBS was applied. The invasion tests were conducted in the same manner, but 200 µg/mL of Matrigel (35234; BD Biosciences, Franklin Lakes, NJ, USA) was applied to the upper chambers of 24-well cell culture inserts. Following the 48-hour incubation period, the inserts were removed, and the cells were fixed with 4% paraformaldehyde for 30 minutes and stained with 1% crystal violet. The non-invading cells were carefully removed from the upper side of the insert membrane using cotton swabs. The quantification of cell migration to the lower surface of the membrane was conducted by counting 10 randomly chosen fields of view using the Olympus IX71 microscope.
Immunofluorescence experiment
The shRNA-control-A375 and shRNA-CTNND2-1-A375 cells were fixed for 2 hours with 1 mL of 4% paraformaldehyde solution. The cell membranes were then permeabilized by adding 500 µL of 0.1% Triton X-100 for 10 minutes. After permeabilization, 200 µL of paxillin antibody solution (ab32115; 1:1,000; Abcam) was applied, and the cells were incubated overnight at 4 °C. The next day, 200 µL of 488 fluorescent antibody solution (1:500) was added to the cells, which were incubated in the dark at room temperature for 1 hour. Following this, 200 µL of an oversaturated 4',6-diamidino-2-phenylindole (DAPI) (C1005; Beyotime) was used to stain the nuclei, with staining performed in the dark for 5 minutes. A laser confocal imaging system (DMI 40008; Leica) was employed to observe the cytoskeletal protein paxillin and cell nuclei in the 488 channels and 405 channels, respectively. Throughout the process, 500 µL of PBS buffer was used to maintain cell integrity.
Tumor-bearing experiment in mice
For this study, nude specific pathogen free (SPF) Balb/c mice (aged 3–4 weeks) were acquired from the Shanghai Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China). The mice were relocated to the experimental site and given 1 week to acclimate to their new surroundings. During this time, the mice had free access to food and water and were kept on a 12-hour light-dark cycle. The mice were weighed the day before the experiment and randomly allocated into the following two groups: the shRNA-CTNND2-1 group and the shRNA-control group (n=5 for both groups). The shRNA-control-A375 cells and shRNA-CTNND2-1-A375 cells were digested, and the cell concentration was diluted with normal saline to 2.0×107 cells/mL and temporarily stored at 4 °C. The mice were subcutaneously injected with 0.1 mL of the cell suspension in their flanks. Starting from the day of injection as day 1, the animals were weighed daily. Starting on the 15th day, the dimensions of each tumor mass were recorded every 3 days to determine the volume. The following formula was used to calculate the volume of the tumor block: volume = (length × width2)/2. After 4 weeks, the mice were euthanized, and their tumors were dissected and weighed. The animal experiment was approved by the Medical Ethics Committee of the Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College (No. 2023-KY-055), in compliance with national guidelines for the care and use of animals.
Statistical analysis
All the analyses were repeated three times. The statistical analysis and visualization procedures were conducted using GraphPad Prism software (version 9.0; GraphPad Software, San Diego, CA, USA). The count data were presented as the number (n, %), and the Chi-squared test was used for the intergroup comparisons. An independent sample t-test was used to compare the measurement data to a normal distribution. The Mann-Whitney rank-sum test was used when the measurement data deviated from a normal distribution. The survival curve was established by Kaplan-Meier. A P value of less than 0.05 was considered statistically significant.
Results
CTNND2 is highly expressed in melanoma cell lines and tissues
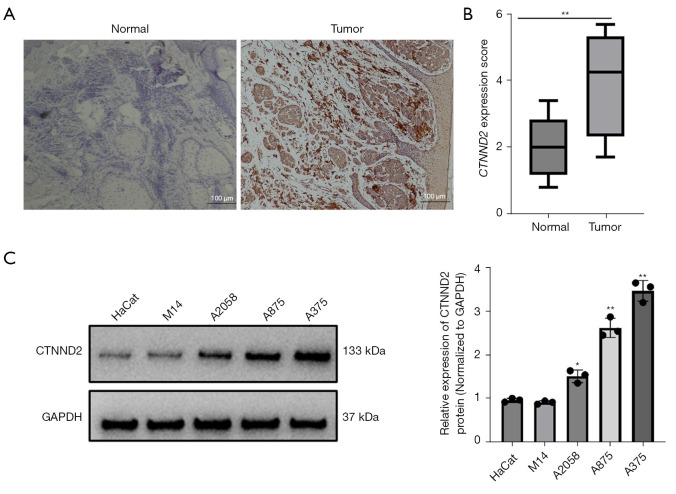
Previously, we performed RNA-sequencing to predict the differential expression of mRNAs in clinical melanoma and paired paracancerous tissues (n=5). Following screening criteria (fold change ≥1.5, P value <0.05, false discovery rate <0.05), the results showed that 865 genes were up-regulated while 631 genes were down-regulated. We identified that CTNND2 was significantly upregulated in melanoma tissues. The immunohistochemical results further showed that the CTNND2 was primarily localized in the cytoplasm of keratinocytes and melanocytes. The CTNND2 levels were markedly increased in the melanoma tissues (n=46) compared to the normal nevus tissues (n=10) (Figure 1A). The CTNND2 score for the melanoma tissues was higher than that for the pericancerous tissues (Figure 1B). Further statistical analysis of the clinical samples revealed that in the melanoma tissues, 34 cases (73.91%) showed positive staining for CTNND2, while 12 cases (26.09%) showed negative staining. Conversely, in the pericancerous tissues, 21 cases (45.65%) showed positive staining and 25 cases (54.35%) showed negative staining. The positive CTNND2 expression rate was notably higher in the melanoma tissues than the pericancerous tissues (P=0.01) (Table 1).
Figure 1.
Expression of CTNND2 in melanoma cell lines and tissues. (A) Immunohistochemical analysis of CTNND2 expression in melanoma tissues (hematoxylin staining). (B) Scoring of the immunohistochemical results. (C) Western blot assay was used to assess CTNND2 protein expression in melanoma cell lines. *, P<0.05; **, P<0.01. CTNND2, catenin delta 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Table 1. Staining of CTNND2 in tissues of clinical samples.
| Tissue samples | N | CTNND2 expression, n (%) | P | |
|---|---|---|---|---|
| Positive | Negative | |||
| Melanoma | 46 | 34 (73.91) | 12 (26.09) | 0.01 |
| Paracancerous | 46 | 21 (45.65) | 25 (54.35) | |
CTNND2, catenin delta 2.
In the melanoma cell lines, CTNND2 expression was examined by Western blot assay. The CTNND2 levels were more elevated in the melanoma cell lines (M14, A2058, A875, and A375) than the human immortalized keratinocyte cell lines (HaCat), and the highest expression level was observed in the A375 (Figure 1C) cell line. Therefore, the A375 cell line was selected for the subsequent functional experiments.
Correlation analysis between patients’ clinical pathological characteristics and CTNND2 expression
Based on the immunohistochemical results, the patients were divided into CTNND2 positive and CTNND2 negative groups. The examination of the relationship between the clinical pathological features of the expression of CTNND2 revealed that in patients with melanoma, the expression of CTNND2 was associated with tumor stage, lymph node metastasis, and tumor ulceration (Table 2).
Table 2. Correlation analysis between patients’ clinicopathological features and CTNND2 expression.
| Clinicopathological features | N | CTNND2 expression, n | P | |
|---|---|---|---|---|
| Positive (n=34) | Negative (n=12) | |||
| Age (years) | 0.18 | |||
| <55 | 22 | 14 | 8 | |
| ≥55 | 24 | 20 | 4 | |
| Sex | 0.51 | |||
| Female | 19 | 13 | 6 | |
| Male | 27 | 21 | 6 | |
| Tumor ulcer | 0.02 | |||
| Yes | 29 | 25 | 4 | |
| No | 17 | 9 | 8 | |
| Lymphatic metastasis | 0.003 | |||
| Yes | 19 | 18 | 1 | |
| No | 27 | 16 | 11 | |
| TNM stage | 0.049 | |||
| I | 11 | 6 | 5 | |
| II | 16 | 10 | 6 | |
| III | 8 | 7 | 1 | |
| IV | 11 | 11 | 0 | |
| Tumor thickness (mm) | 0.54 | |||
| <1 | 8 | 6 | 2 | |
| 1–4 | 25 | 17 | 8 | |
| >4 | 13 | 11 | 2 | |
CTNND2, catenin delta 2; TNM, tumor-node-metastasis.
The impact of CTNND2 expression on overall survival (OS)
A 3-year follow-up was conducted for all patients, and the results showed that the survival rate of the CTNND2-negative expression group was 91.7%, with a mean survival time of 33.90±1.99 months [95% confidence interval (CI): 30.00–37.81]. In contrast, the survival rate of the CTNND2-positive expression group was 52.9%, with a mean survival time of 26.48±1.73 months (95% CI: 23.10–29.86). The survival rate of patients with positive CTNND2 expression was significantly lower than that of patients with negative CTNND2 expression (P=0.043) (Figure 2).
Figure 2.
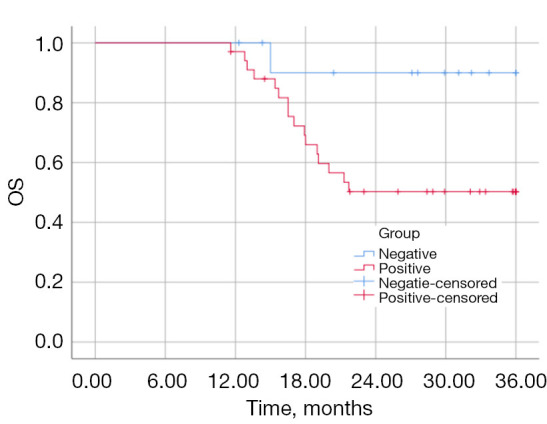
OS curves for patients with different CTNND2 expression levels. OS, overall survival; CTNND2, catenin delta 2.
Establishment of a CTNND2 knockdown melanoma cell line
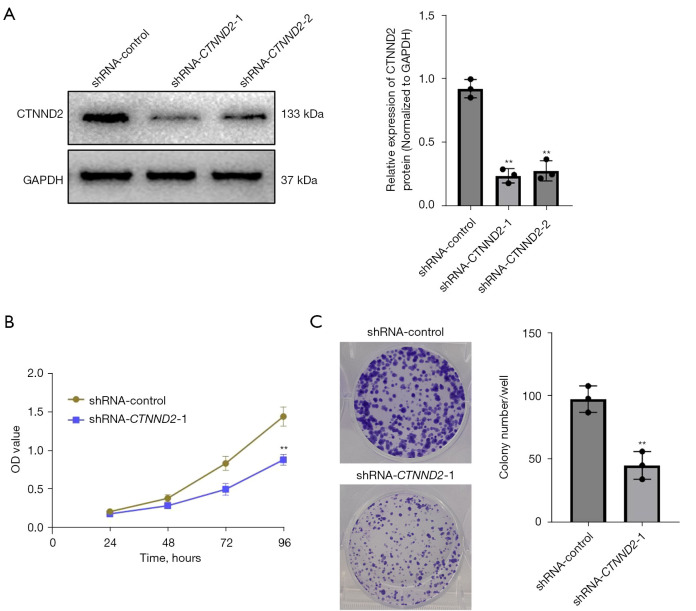
After transfection, the CTNND2 expression levels of the cells were measured by Western blot analysis. CTNND2 expression was significantly more reduced in the cells transfected with two interfering sequences targeting CTNND2 (shRNA-CTNND2-1 and shRNA-CTNND2-2) than in the cells in the blank control group (shRNA-control) (Figure 3A). Thus, the results indicated that the CTNND2 knockdown melanoma cell line had been successfully established. As the knockdown efficiency of shRNA-CTNND2-1 was higher than that of shRNA-CTNND2-2, shRNA-CTNND2-1 was selected for the subsequent knockdown experiments.
Figure 3.
The effects of downregulating CTNND2 expression on melanoma cell growth. (A) Following cell transfection, western blot was used to examine CTNND2 mRNA and protein expression in melanoma cell lines. (B) CCK-8 assay was used to evaluate the effects of CTNND2 knockdown on the proliferation capacity of melanoma cells. (C) Plate colony formation tests were used to evaluate the effects of CTNND2 knockdown on colony-dependent proliferation in melanoma cells (crystal violet staining). **, P<0.01. CTNND2, catenin delta 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; shRNA, short hairpin RNA; OD, optical density; mRNA, messenger RNA; CCK-8, Cell Counting Kit-8.
CTNND2 knockdown inhibits the proliferative ability of melanoma cells
The CCK-8 assay results showed that transfection with shRNA-CTNND2-1 substantially suppressed the proliferation of the melanoma A375 cells compared to that of the cells in the blank control group. Notably, after 24 and 48 hours, transfection had no discernible effect on cell proliferation. However, as the culture time extended, cell proliferation became inhibited (starting from 72 hours) with the most significant effect observed at 96 hours (Figure 3B). The colony formation assay showed that shRNA-CTNND2-1 transfection considerably decreased the total number of colonies generated by the melanoma A375 cells, compared to that of the cells in the shRNA-control group. This suggests that suppressing CTNND2 expression significantly hinders the capacity of melanoma cells to proliferate in a colony-dependent manner (Figure 3C).
Predicting the biological functions of CTNND2 in melanoma cells
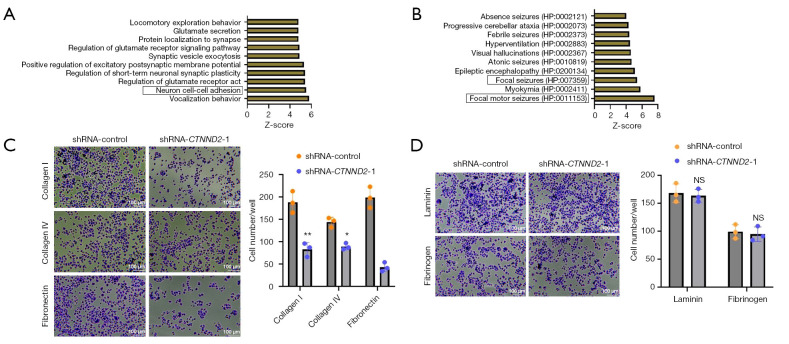
To further investigate the regulatory mechanisms by which CTNND2 exerts its biological functions, an initial prediction of the biological processes (BPs) [Gene Ontology (GO)] regulated by CTNND2 and its phenotypes in human tissues and cells was conducted using the online website Harmonizome (https://maayanlab.cloud/Harmonizome/). Based on the Z-scores ranking, the top 10 ranked processes and phenotypes were selected for presentation and ordered according to their score. The analysis of the BPs (GO) revealed that CTNND2 is primarily associated with vocalization behavior and neuron cell-cell adhesion (Figure 4A). Additionally, further analysis indicated that CTNND2 is also related to the focal pathway, such as focal motor seizures and focal seizures (Figure 4B). These results indicated that CTNND2 may be linked with the focal adhesion signaling pathway of the cells.
Figure 4.
The effects of CTNND2 knockdown on the adhesion ability of melanoma cells. (A) The BPs regulated by CTNND2 and (B) human phenotypes in tissue cells were predicted through the online website Harmonizome (https://maayanlab.cloud/Harmonizome/). (C) The effects of the CTNND2 knockdown on the adhesion ability of melanoma cells to adhere to the ECM components collagen I, collagen IV, and fibronectin (crystal violet staining). (D) The effects of CTNND2 suppression on the capacity of melanoma cells to adhere to fibrinogen and laminin, two components of the ECM (crystal violet staining). *, P<0.05; **, P<0.01. NS, not significant; CTNND2, catenin delta 2; shRNA, short hairpin RNA; BP, biological process; ECM, extracellular matrix.
CTNND2 knockdown inhibits the adhesion ability of melanoma cells
Given that CTNND2 is associated with cadherin proteins, we hypothesized that it might affect the adhesion ability of cells, thereby influencing the abilities of melanoma cells to invade and migrate. The effects of CTNND2 knockdown on the ability of the melanoma cells to adhere to the extracellular matrix (ECM), which includes fibronectin, collagen types IV and I, laminin, and fibrinogen, were studied using the CytoSelect™ 48-well Cell Adhesion Assay Kit. The results showed that transfection with interfering sequences targeting CTNND2 significantly inhibited the ability of the melanoma cells to adhere to collagen I, collagen IV, and fibronectin compared to that of the cells in the shRNA-control group (Figure 4C). However, no substantial effect on the ability of the cells to adhere to fibrinogen and laminin was observed (Figure 4D).
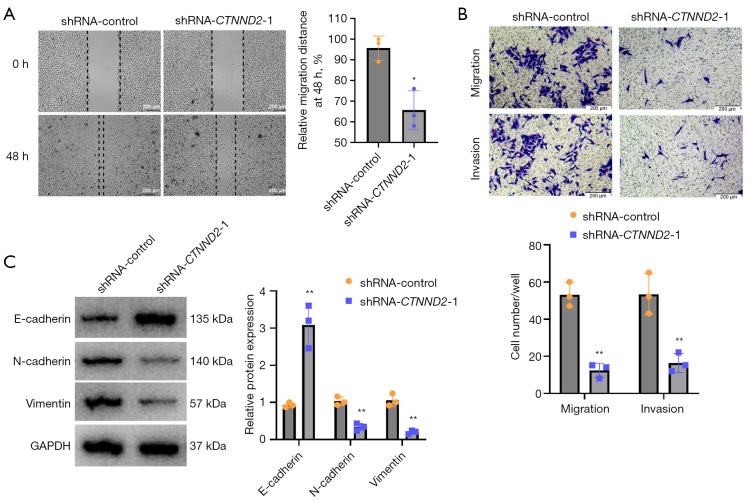
CTNND2 suppression inhibits melanoma cell invasion and migration
Scratch and Transwell tests were used to assess the effects of CTNND2 knockdown on melanoma cell migration and invasion. According to the findings of the scratch test, transfection with shRNA-CTNND2-1 substantially decreased the capacity of the melanoma A375 cells to migrate compared to that of the cells in the blank control group (Figure 5A). The Transwell experiment showed that in comparison to the shRNA-control cells, transfection with shRNA-CTNND2-1 substantially lowered the invasion and migration abilities of the melanoma A375 cells (Figure 5B). The effects of CTNND2 knockdown on the expression of the proteins linked to melanoma cell motility and invasion, including vimentin, N-cadherin, and E-cadherin, were examined by Western blot analysis. The results showed that the transfection of shRNA-CTNND2-1 substantially inhibited the epithelial-mesenchymal transformation process of the melanoma A375 cells compared to that of the cells in the blank control group (shRNA-control); that is, it increased the expression of E-cadherin protein and decreased the expression of N-cadherin and vimentin proteins (Figure 5C).
Figure 5.
Melanoma cell invasion and migration are inhibited by CTNND2 knockdown. (A) Scratch assay was used to assess the effects of CTNND2 knockdown on melanoma cell migration. (B) The Transwell test was used to examine the effects of CTNND2 knockdown on melanoma cell migration and invasion (crystal violet staining). (C) Western blot assay was used to assess the effects of CTNND2 knockdown on the expression of proteins involved in melanoma cell invasion and migration. *, P<0.05; **, P<0.01. shRNA, short hairpin RNA; CTNND2, catenin delta 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
The effects of CTNND2 on the focal adhesion signaling pathway in melanoma cells
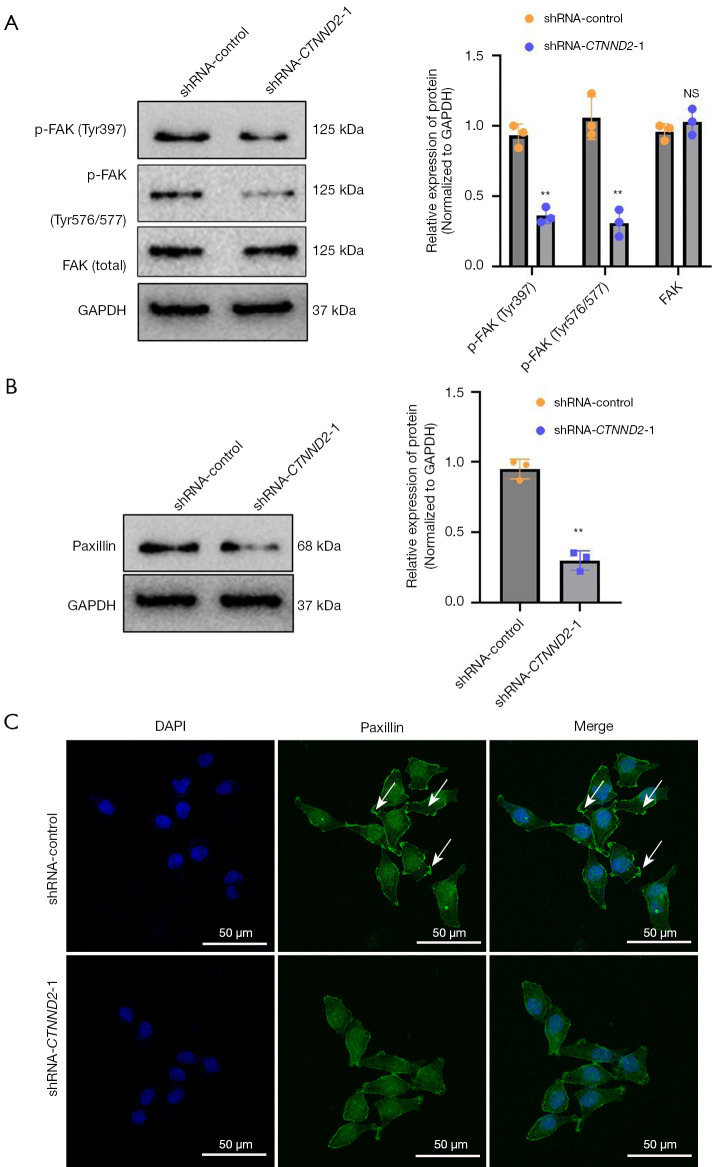
Additionally, Western blot assay was used to investigate the effects of CTNND2 suppression on the focal adhesion signaling pathway in the melanoma cells. The main component of the focal adhesion signaling pathway is FAK, which is a member of the protein tyrosine kinase superfamily (14). The results showed that transfection with the interfering sequence targeting CTNND2 (shRNA-CTNND2-1) markedly suppressed the FAK phosphorylation in the melanoma cells compared to that of the cells in the blank shRNA-control group. Tyr397 is the main phosphorylation site of FAK, while Tyr576/577 are two major phosphorylation sites that enhance kinase activity; however, no marked alteration in the total FAK levels was observed (Figure 6A).
Figure 6.
The effects of CTNND2 knockdown on paxillin and FAK phosphorylation levels in melanoma cells. (A) Western blot assay was used to examine the effects of CTNND2 knockdown on FAK phosphorylation and (B) paxillin levels in melanoma cells. (C) Immunofluorescence assay was used to detect paxillin expression levels in melanoma cells after CTNND2 knockdown (DAPI staining). The white arrows indicate pseudopodia. **, P<0.01. NS, not significant; FAK, focal adhesion kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; shRNA, short hairpin RNA; CTNND2, catenin delta 2; DAPI, 4',6-diamidino-2-phenylindole.
To further validate the above experimental results, this study examined the expression of downstream paxillin by Western blot and immunofluorescence. Transfection with the interfering sequence targeting CTNND2 (shRNA-CTNND2-1) notably suppressed the paxillin levels in the melanoma cells (Figure 6B). Immunofluorescence detection indicated that in the blank shRNA-control group, the melanoma cells had a large number of pseudopodial spots formed by paxillin at their edges, while in the group transfected with the interfering sequence targeting CTNND2 (shRNA-CTNND2-1), the number of pseudopodial spots formed by paxillin at the edges of the melanoma cells was significantly reduced (Figure 6C).
CTNND2 knockdown substantially suppresses the ERK1/2 and MEK1/2 signaling pathway in melanoma cells
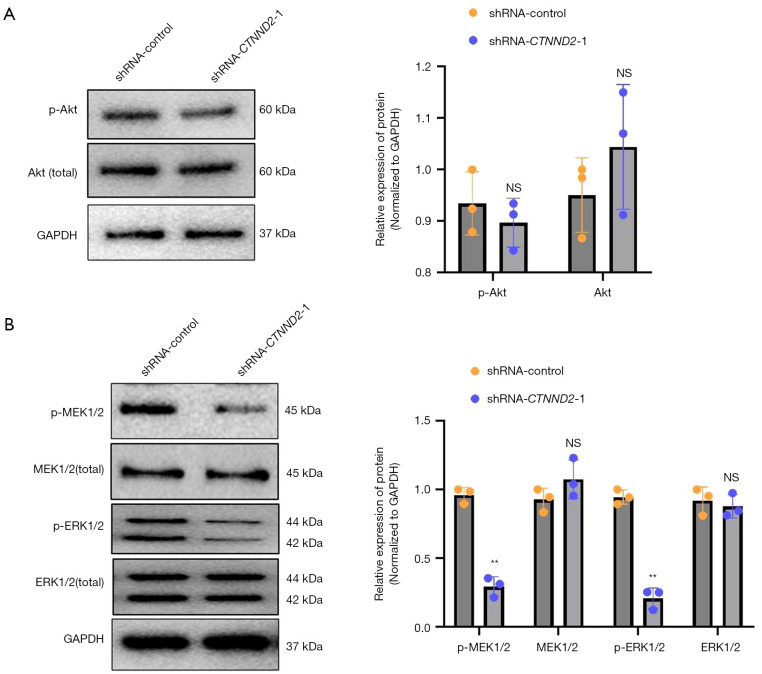
The data obtained from the Western blot analysis showed that transfection with the interfering sequence targeting CTNND2 (shRNA-CTNND2-1) did not substantially change the levels of AKT and phosphorylated AKT in the melanoma cells compared to those in the cells in the blank shRNA-control group (Figure 7A). Thus, the knockdown of CTNND2 may not have a significant effect on the PI3K-AKT signaling pathway in cells.
Figure 7.
CTNND2 knockdown significantly inhibits the MEK1/2 and ERK1/2 signaling pathways in melanoma cells. (A) Western blot assay was used to determine the effects of CTNND2 knockdown on AKT, phosphorylated AKT, and AKT expression levels and (B) MEK1/2, p-MEK1/2, ERK1/2, and p-ERK1/2 in melanoma cells. **, P<0.01. NS, not significant; AKT, protein kinase B; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; shRNA, short hairpin RNA; CTNND2, catenin delta 2; MEK1/2, mitogen-activated extracellular signal-regulated kinase 1/2; ERK1/2, extracellular signal-regulated protein kinase 1/2.
Further investigation of the MEK1/2, ERK1/2 signaling pathway revealed that transfection with the interfering sequence targeting CTNND2 (shRNA-CTNND2-1) significantly reduced the p-ERK1/2and p-MEK1/2 levels in melanoma cells, compared to those in the cells in the blank shRNA-control group. However, the expression levels of total ERK1/2 (ERK1/2 total) and MEK1/2 (MEK1/2 total) were not significantly affected (Figure 7B). Thus, reducing the expression of CTNND2 can significantly hinder the activation of MEK1/2 and its subsequent molecule ERK1/2 in melanoma cells.
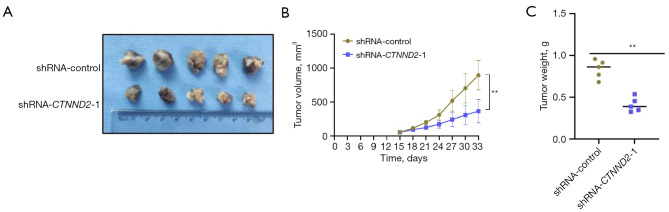
CTNND2 suppression substantially inhibits the growth of melanoma cells in vivo
To further verify the effects of the CTNND2 knockdown on the proliferation ability of the melanoma cells in vivo, melanoma cells in the transfected shRNA-CTNND2 and CTNND2-1 group (shRNA-CTNND2-1) were subcutaneously inoculated into nude mice. The results indicated that the volume of the tumor-bearing tissue in the shRNA-control group was higher than that in the CTNND2-low (shRNA-CTNND2-1) group (Figure 8A). The results also showed a considerable reduction in the volume of the tumor-bearing tissue created by melanoma cells in the shRNA-CTNND2-1 group compared to the shRNA-control group, as indicated by the growth curve of the tumor-bearing tissue (Figure 8B). Further weighing of tumor-bearing tissues showed that the weight of the tumor-bearing tissues formed by the melanoma cells in the shRNA-CTNND2-1 group was substantially lower than that in the shRNA-control group (Figure 8C).
Figure 8.
Mice tumor development was used to assess the effects of CTNND2 knockdown on the ability of melanoma cells to proliferate in vivo. (A) The tumor-bearing tissue morphology of melanoma cells with CTNND2 knockdown was formed after 33 days of subcutaneous inoculation in nude mice. (B) The growth curve of melanoma cells with CTNND2 knockdown was measured and analyzed after subcutaneous inoculation for 15 days. (C) The weight of the inoculated tumor-bearing tissue was analyzed. **, P<0.01. shRNA, short hairpin RNA; CTNND2, catenin delta.
Discussion
This study first used immunohistochemistry and Western blot to show that the CTNND2 protein expression level was higher in melanoma tissues and cell lines. This suggests that CTNND2 may have a key function in the development and advancement of melanoma. Further, this study uncovered a remarkable association between the positive expression of CTNND2 and the clinical pathological characteristics of individuals with melanoma. Tumor ulceration, which is considered an indicator of tumor aggressiveness and progression, is strongly correlated with the presence of CTNND2 expression (15). This suggests that CTNND2 may be related to the invasive behavior of melanoma. The study also found a substantial correlation between the positive expression of CTNND2 and lymph node metastasis, which is a crucial prognostic factor for poor outcomes in melanoma. The study suggests that CTNND2 is likely to have a substantial effect on the mobility and proliferation of cancer cells. Moreover, the presence of CTNND2 is strongly correlated with the classification of tumors, suggesting that the level of CTNND2 expression could be linked to the advancement of tumors. The OS survival rate of patients with positive CTNND2 expression was significantly lower than that of patients with negative CTNND2 expression. Thus, CTNND2 could serve as a potential candidate biomarker for the detection and monitoring of melanoma.
The elevated expression levels of CTNND2 across various melanoma cell lines and the establishment of CTNND2 knockdown melanoma cell lines highlighted the significant role of CTNND2 in melanoma cell proliferation and adhesion. The knockdown of CTNND2 significantly inhibited the proliferative capacity of the A375 cells, which might be achieved by affecting the expression or activity of cell cycle regulation-related proteins (16). Additionally, the knockdown of CTNND2 altered the adhesion ability of melanoma cells to various ECM components, especially collagen I, collagen IV, and fibronectin. This may affect the interaction between tumor cells and their surrounding microenvironment (17), thus regulating tumor growth and spread.
To investigate the regulatory mechanisms by which CTNND2 exerts the aforementioned biological functions, the present study initially used the online website Harmonizome to predict the BPs regulated by CTNND2 and its phenotypes in human tissues and cells. The results indicated that CTNND2 might be related to the focal adhesion signaling pathway of the cells. Focal adhesions are complex multiprotein structures that serve not only as physical links but also as nodes for signal transduction. Key components of the focal adhesion signaling pathway include integrins, FAK, Src family kinases, paxillin, and vinculin. FAK controls vital BPs such as the adhesion, migration, proliferation, and survival of cells (18,19). It plays a significant role in promoting key malignant traits during cancer progression, such as cancer stemness, epithelial-mesenchymal transition (EMT), tumor angiogenesis, chemotherapy resistance, and stromal fibrosis (20,21). Due to its involvement in these processes, FAK is considered a promising target for cancer therapy (22,23). FAK is frequently overexpressed in many cancer types, and the majority of research has examined whether decreases in FAK levels or suppressing FAK activity can impede tumor development and metastasis (24,25). Previous research has shown that FAK can control the production of the subsequent scaffolding protein paxillin (26). To confirm these experimental results, this study used Western blot and immunofluorescence techniques to investigate the expression of paxillin, and found that transfection with the interfering sequence targeting CTNND2 substantially inhibited the expression of paxillin in the melanoma cells as compared to the shRNA-control cells.
Paxillin is a multifunctional adaptor protein located primarily at the adhesions between cells and the ECM. Functioning as a scaffolding protein, it facilitates the arrangement and positioning of diverse signaling molecules at adhesions, exerting a pivotal influence on cellular migration, multiplication, and survival (27-29). Paxillin interacts with a wide array of proteins, such as kinases and structural proteins, and is involved in regulating the actin cytoskeleton. Changes in the expression and function of paxillin in various physiological processes and diseases, particularly cancer, can affect tumor cell behavior (30). The results of this study suggest that CTNND2 may control cellular activities by directly or indirectly influencing paxillin via the focal adhesion signaling pathway.
FAK is capable of activating several downstream signaling pathways, including the PI3K-AKT and MEK1/2-ERK pathways (31-33). The knockdown of CTNND2 in the melanoma cells did not significantly affect AKT and its phosphorylation levels, which suggests that CTNND2 may not directly regulate the PI3K-AKT signaling pathway. However, the knockdown of CTNND2 significantly reduced the phosphorylation levels of MEK1/2 and ERK1/2, without significantly affecting the overall expression levels of MEK1/2 and ERK1/2. These results suggest that CTNND2 regulates the MEK1/2-ERK signaling pathway, and that by controlling this pathway, CTNND2 may affect the biological properties of melanoma cells. However, it is still unclear how CTNND2 lowers the phosphorylation levels of ERK1/2 and MEK. CTNND2, a protein associated with adhesive junctions in the armadillo/β-catenin superfamily, is thought to facilitate the disruption of E-cadherin-based adherens junctions. This disruption promotes cell proliferation when stimulated by hepatocyte growth factor. CTNND2 then affects paxillin through the focal adhesion signaling pathway.
Conclusions
This research showed for the first time that CTNND2 is elevated in melanoma tissues, promotes the in vitro proliferative ability of melanoma cells, participates in regulating the adhesion process of melanoma cells, and its mechanism is related to regulating the FAK and MEK1/2/ERK1/2 signaling pathways. Clinically, patients who are CTNND2-positive with advanced tumor stage, lymph node metastasis, and tumor ulceration can be considered a high-risk group. For high-risk patients, personalized treatment strategies targeting CTNND2 may be considered, in combination with existing therapeutic methods to improve treatment efficacy. Future studies will further explore the specific application of CTNND2 as a target for risk stratification and personalized therapy through larger clinical sample sizes. Based on our findings, CTNND2 could be used as an oncogene target for melanoma, and a new treatment target or diagnostic biomarker. However, some limitations of this study should be mentioned. First, the study exclusively used a single cell line to assess the impact of CTNND2 knockdown on cellular function. Future research will incorporate multiple melanoma cell lines representing diverse subtypes to validate the findings and ensure the generalizability of the results. Second, our findings lack support from in vivo animal experiments (e.g., knockdown mouse models). Future studies will include the development of CTNND2 knockdown or overexpression mouse models to examine the gene’s functional role in melanoma progression and metastasis in a more physiological context.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank all the investigators, including the physicians and laboratory technicians, for their work in this study.
Funding: The present research project was funded by the General project of the Yangzhou Health Commission (No. 2023-2-15) and the Natural Science Foundation of Jiangsu Province (No. BK20231115).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Affiliated Yixing Clinical School of Medical School of Yangzhou University (approval No. 2023-159-01), and informed consent was obtained from all the patients. The animal experiment was approved by the Medical Ethics Committee of the Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College (No. 2023-KY-055), in compliance with national guidelines for the care and use of animals.
Footnotes
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2159/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2159/coif). The authors have no conflicts of interest to declare.
Data Sharing Statement
Available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-2159/dss
References
- 1.Subramani P, Das RK. Possible therapeutic role of short-chain fatty acids from skin commensal bacteria in UVB-induced skin carcinogenesis. Biocell 2023;47:2195-205. 10.32604/biocell.2023.030383 [DOI] [Google Scholar]
- 2.Arnold M, Singh D, Laversanne M, et al. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol 2022;158:495-503. 10.1001/jamadermatol.2022.0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12-49. 10.3322/caac.21820 [DOI] [PubMed] [Google Scholar]
- 4.Limonta P, Queirolo P. New insights in melanoma biology: Running fast towards precision medicine. Semin Cancer Biol 2019;59:161-4. 10.1016/j.semcancer.2019.09.019 [DOI] [PubMed] [Google Scholar]
- 5.Boutros A, Croce E, Ferrari M, et al. The treatment of advanced melanoma: Current approaches and new challenges. Crit Rev Oncol Hematol 2024;196:104276. 10.1016/j.critrevonc.2024.104276 [DOI] [PubMed] [Google Scholar]
- 6.Huang B, Han W, Sheng ZF, et al. Identification of immune-related biomarkers associated with tumorigenesis and prognosis in cutaneous melanoma patients. Cancer Cell Int 2020;20:195. 10.1186/s12935-020-01271-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebecca VW, Somasundaram R, Herlyn M. Pre-clinical modeling of cutaneous melanoma. Nat Commun 2020;11:2858. 10.1038/s41467-020-15546-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maity AK, Stone TC, Ward V, et al. Novel epigenetic network biomarkers for early detection of esophageal cancer. Clin Epigenetics 2022;14:23. 10.1186/s13148-022-01243-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abu-Elneel K, Ochiishi T, Medina M, et al. A delta-catenin signaling pathway leading to dendritic protrusions. J Biol Chem 2008;283:32781-91. 10.1074/jbc.M804688200 [DOI] [PubMed] [Google Scholar]
- 10.Lu Q, Aguilar BJ, Li M, et al. Genetic alterations of δ-catenin/NPRAP/Neurojungin (CTNND2): functional implications in complex human diseases. Hum Genet 2016;135:1107-16. 10.1007/s00439-016-1705-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu T, Ishida J, Kurozumi K, et al. δ-Catenin Promotes Bevacizumab-Induced Glioma Invasion. Mol Cancer Ther 2019;18:812-22. 10.1158/1535-7163.MCT-18-0138 [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Yang H, Liu J, et al. Metabolomics-based Approach to Analyze the Therapeutic Targets and Metabolites of a Synovitis Ointment for Knee Osteoarthritis. Curr Pharm Anal 2023;19:222-34. 10.2174/1573412919666221223152915 [DOI] [Google Scholar]
- 13.Tian X, He X, Qian S, et al. Immunoregulatory effects of human amniotic mesenchymal stem cells and their exosomes on human peripheral blood mononuclear cells. BIOCELL 2023;47:1085-93. 10.32604/biocell.2023.027090 [DOI] [Google Scholar]
- 14.Pang XJ, Liu XJ, Liu Y, et al. Drug Discovery Targeting Focal Adhesion Kinase (FAK) as a Promising Cancer Therapy. Molecules 2021;26:4250. 10.3390/molecules26144250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chopra A, Sharma R, Rao UNM. Pathology of Melanoma. Surg Clin North Am 2020;100:43-59. 10.1016/j.suc.2019.09.004 [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Yao B, Niu Y, et al. Corrigendum to hypoxia-induced lncRNA EIF3J-AS1 accelerates hepatocellular carcinoma progression via targeting miR-122-5p/CTNND2 axis. Biochem Biophys Res Commun 2021;565:101-2. 10.1016/j.bbrc.2021.04.109 [DOI] [PubMed] [Google Scholar]
- 17.Fujisaki H, Futaki S. Epithelial-Mesenchymal Transition Induced in Cancer Cells by Adhesion to Type I Collagen. Int J Mol Sci 2022;24:198. 10.3390/ijms24010198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan X, Yan Y, Song B, et al. Focal adhesion kinase: from biological functions to therapeutic strategies. Exp Hematol Oncol 2023;12:83. 10.1186/s40164-023-00446-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panera N, Crudele A, Romito I, et al. Focal Adhesion Kinase: Insight into Molecular Roles and Functions in Hepatocellular Carcinoma. Int J Mol Sci 2017;18:99. 10.3390/ijms18010099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee BY, Timpson P, Horvath LG, et al. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther 2015;146:132-49. 10.1016/j.pharmthera.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Liu S, Zhou S, et al. Focal adhesion kinase: Insight into its roles and therapeutic potential in oesophageal cancer. Cancer Lett 2021;496:93-103. 10.1016/j.canlet.2020.10.005 [DOI] [PubMed] [Google Scholar]
- 22.Mohanty A, Pharaon RR, Nam A, et al. FAK-targeted and combination therapies for the treatment of cancer: an overview of phase I and II clinical trials. Expert Opin Investig Drugs 2020;29:399-409. 10.1080/13543784.2020.1740680 [DOI] [PubMed] [Google Scholar]
- 23.Dawson JC, Serrels A, Stupack DG, et al. Targeting FAK in anticancer combination therapies. Nat Rev Cancer 2021;21:313-24. 10.1038/s41568-021-00340-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chauhan A, Khan T. Focal adhesion kinase-An emerging viable target in cancer and development of focal adhesion kinase inhibitors. Chem Biol Drug Des 2021;97:774-94. 10.1111/cbdd.13808 [DOI] [PubMed] [Google Scholar]
- 25.Quispe PA, Lavecchia MJ, León IE. Focal adhesion kinase inhibitors in the treatment of solid tumors: Preclinical and clinical evidence. Drug Discov Today 2022;27:664-74. 10.1016/j.drudis.2021.11.025 [DOI] [PubMed] [Google Scholar]
- 26.Mitsos P, Anastasiou I, Constantinides C, et al. Clinical Importance of Focal Adhesion Kinase (FAK)-Src and Paxillin Expression in Renal Cell Carcinoma. Cureus 2024;16:e62706. 10.7759/cureus.62706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ripamonti M, Wehrle-Haller B, de Curtis I. Paxillin: A Hub for Mechano-Transduction from the β3 Integrin-Talin-Kindlin Axis. Front Cell Dev Biol 2022;10:852016. 10.3389/fcell.2022.852016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz AUR, Deng S, Jin Y, et al. The explorations of dynamic interactions of paxillin at the focal adhesions. Biochim Biophys Acta Proteins Proteom 2022;1870:140825. 10.1016/j.bbapap.2022.140825 [DOI] [PubMed] [Google Scholar]
- 29.Vann K, Weidner AE, Walczyk AC, et al. Paxillin knockout in mouse granulosa cells increases fecundity†. Biol Reprod 2023;109:669-83. 10.1093/biolre/ioad093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Huang X, Luo W, et al. The Role of Paxillin Aberrant Expression in Cancer and Its Potential as a Target for Cancer Therapy. Int J Mol Sci 2023;24:8245. 10.3390/ijms24098245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Gao Y, Ren C. Focal adhesion kinase inhibitor BI 853520 inhibits cell proliferation, migration and EMT process through PI3K/AKT/mTOR signaling pathway in ovarian cancer. Discov Oncol 2021;12:29. 10.1007/s12672-021-00425-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Zhao D, Wang Y, et al. Focal adhesion kinase (FAK) inhibitor-defactinib suppresses the malignant progression of human esophageal squamous cell carcinoma (ESCC) cells via effective blockade of PI3K/AKT axis and downstream molecular network. Mol Carcinog 2021;60:113-24. 10.1002/mc.23273 [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Zhang M, Tian J, et al. WNT1-inducible signalling pathway protein 1 stabilizes atherosclerotic plaques in apolipoprotein-E-deficient mice via the focal adhesion kinase/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase pathway. J Hypertens 2022;40:1666-81. 10.1097/HJH.0000000000003195 [DOI] [PubMed] [Google Scholar]