Abstract
Throughout adulthood and ageing our brains undergo structural loss in an average pattern resembling faster atrophy in Alzheimer’s disease (AD). Using a longitudinal adult lifespan sample (aged 30-89; 2–7 timepoints) and four polygenic scores for AD, we show that change in AD-sensitive brain features correlates with genetic AD-risk and memory decline in healthy adults. We first show genetic risk links with more brain loss than expected for age in early Braak regions, and find this extends beyond APOE genotype. Next, we run machine learning on AD-control data from the Alzheimer’s Disease Neuroimaging Initiative using brain change trajectories conditioned on age, to identify AD-sensitive features and model their change in healthy adults. Genetic AD-risk linked with multivariate change across many AD-sensitive features, and we show most individuals over age ~50 are on an accelerated trajectory of brain loss in AD-sensitive regions. Finally, high genetic risk adults with elevated brain change showed more memory decline through adulthood, compared to high genetic risk adults with less brain change. Our findings suggest quantitative AD risk factors are detectable in healthy individuals, via a shared pattern of ageing- and AD-related neurodegeneration that occurs along a continuum and tracks memory decline through adulthood.
Subject terms: Cognitive ageing, Neural ageing, Alzheimer's disease
This study shows that among healthy adults tracked across their lives, brain atrophy rates correlate with genetic risk for Alzheimer’s and memory decline, and that degeneration in Alzheimer’s-vulnerable brain regions is a continuous phenomenon.
Introduction
Advanced age is the primary risk factor for Alzheimer’s disease (AD) – the leading form of dementia. Across healthy adult life, our brains undergo gradual and widespread structural changes1–4. Many of these changes are qualitatively similar to atrophy patterns seen in AD, suggesting shared vulnerability of brain systems in ageing and AD5–7. For example, medial temporal lobe regions including hippocampus and entorhinal cortex are amongst the earliest regions to show atrophy and tau deposition in AD8,9, and each exhibits accelerated structural loss from around ~50 years of age. Prior to this, many brain structures exhibit slow but steady average volume reduction from early adulthood2,10–13. Whole-cortical atrophy patterns are also largely shared between ageing and AD4,6, with characteristic temporo-parietal atrophy patterns in AD also found to a lesser degree in healthy people, including those at low AD risk5,14. This parallel pattern is critical to understand4, because reported AD incidence increases exponentially after 65 years of age15,16.
If brain regions vulnerable in AD exhibit gradual change through ageing, healthy individuals at higher AD risk may show faster atrophy over extended age spans. Polygenic risk scores for AD (PRS-AD) calculated from risk variants in genome-wide association studies (GWAS) provide a marker to test this; in AD patients, genetic AD risk links with longitudinal outcomes including faster brain and cognitive decline, earlier AD onset, and clinical progression17–19. In healthy adults, however, attempts to link genetic AD risk to alterations in brain structure have typically been cross-sectional and yielded mixed results20–26. For example, although many studies report no effect of APOE-ε4 on cross-sectional hippocampal volumes20–26, recent large-scale studies found smaller hippocampal volumes in older ε4 carriers27,28. However, evidence suggests smaller hippocampal volume as a function of genetic AD risk may be evident in neonates29, children2,30 and young adults31,32, and longitudinal work suggests the effect of genetic risk upon lower hippocampal volume is roughly equivalent from childhood to old age2. Further, many 70 year-olds have similar size brain measures to many 30 year-olds, and individual differences in brain structure at any age typically exceed the magnitude of change effects through ageing2,33. Hence, brain differences observed in older at-risk individuals may be ascribable to preexisting differences from early life. Consequently, only longitudinal designs are suited to examine whether elevated genetic AD risk confers a direct genetic effect on the slope of brain ageing across adult life.
Longitudinal studies attempting to link brain changes to genetic AD risk in healthy adults have been inconclusive, often restricted to small samples of older adults34–36, and adult lifespan samples with extensive follow-up are lacking. Small studies have reported group-level effects34,35 or no effect of APOE-ε4 upon hippocampal change in healthy older adults37. Another study found evidence PRS-AD related to hippocampal and entorhinal thinning in an older sample enriched for APOE-ε4 and memory concerns, though did not report polygenic effects beyond APOE36. Additionally, in a large sample of healthy older individuals, hippocampal change was found to be greater in APOE-ε4 carriers (N = 748)38. However, a recent GWAS39 (N = 15,640) observed that an association between APOE and faster hippocampal and amygdala change in ageing disappeared when accounting for disease (notably, the sample included many AD cases). Thus, the effect of APOE upon brain change in candidate AD regions was seemingly driven by disease-related processes and not detected in healthy brains39. Moreover, the trajectories of genetically high-risk versus low-risk groups provide little evidence that genetic AD risk affects the slope of brain decline across adult life2,24,25. Individualized estimates of the degree to which a healthy person’s brain is changing more or less than expected for their age may be better suited to answer whether genetic AD risk impacts the slope of brain ageing in healthy adults.
Regions with greater brain atrophy in AD are encompassed within the Braak staging scheme8,40. This describes the spatiotemporal sequence of tau deposition9,41—from a cortical entorhinal epicentre (Stage I) to hippocampus (Stage II), amygdala and inferior temporal cortex (Stage III), and later the rest of cortex8,40. This “AD signature”9 is not specific to AD but found to a lesser degree in normal ageing5,6,42. Beyond this core set of regions with seemingly shared vulnerability in ageing and AD, many other brain features exhibit faster atrophy in AD. Applying a data-driven approach to delineate these in AD patients—combined with multivariate analyses of individualized brain change estimates—may reveal new insights into whether genetic AD risk influences the slope of brain ageing in a select few or across many AD-relevant features in healthy adults.
Finally, several studies suggest that genetic AD risk is subtly related to longitudinal memory decline in healthy older adults43–45, and one adult lifespan study reported a weak association with decline in cognition on average46. Thus, AD risk variants may influence differences in memory decline trajectories that are protracted through life and begin in early adulthood45–48. However, the extent to which one’s genetic predisposition influences brain and cognitive outcomes probably differs also between individuals at high genetic risk, which may explain why genetic risk alone is not highly predictive of cognitive change46,49. Given that individualized approaches to risk assessment are predicated on assessing the conjunction of risks, considering genetic risk together with an individualized marker of relative brain ageing may improve identification of individuals at higher AD risk in healthy adult lifespan data.
Here, we aimed to the test the hypothesis that brain change trajectories in healthy adults correlate with AD-related genetic variation and memory decline across adult life. We hypothesized that neurodegeneration in ageing and AD is linked on a continuum, and that individuals changing more than their age would predict in AD-sensitive features across adult life would have quantitatively higher genetic AD risk. We further hypothesized that individuals with both higher genetic risk and faster atrophy for their age would exhibit more memory decline detectable across adulthood. Using brain trajectories in a healthy adult lifespan sample with frequent longitudinal follow-up (2-7 timepoints, 1430 scans from 420 individuals aged 30–89 years), and genome-wide significant single nucleotide polymorphisms (SNPs) from four AD GWAS, we (1) show that PRS-AD associates with change in several early Braak Stage regions in healthy adults. Next, we empirically identify brain features with faster atrophy in AD using machine learning (ML) on the individual-specific slopes in longitudinal AD-control data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI; scans = 4410, N = 978, 2–9 timepoints). Modelling multivariate change in these in our adult lifespan sample, we (2) show that PRS-AD is associated with change in many AD-sensitive brain features in healthy adults. In an independent sample with notably less follow-up (2-3 timepoints), we corroborate some of the observed PRS-AD associations with brain change in healthy adults. Last, we (3) show that high PRS-AD individuals who are also high on a multivariate brain change marker show greater drop-off in memory over adult life, compared to high PRS-AD individuals with less brain change. Thus, the conjunction of a multivariate brain change marker and known genetic risk identified a subset of comparatively high-risk individuals showing more memory decline over healthy adult life (30–89 years).
Results
Univariate associations between PRS-AD and brain change
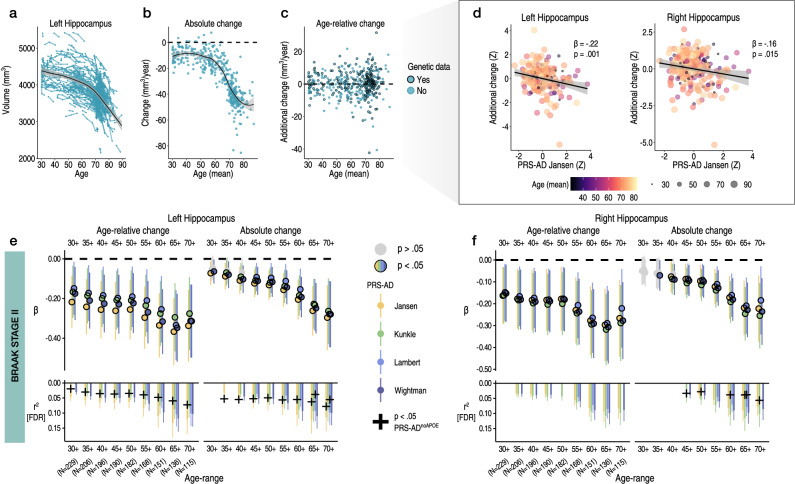
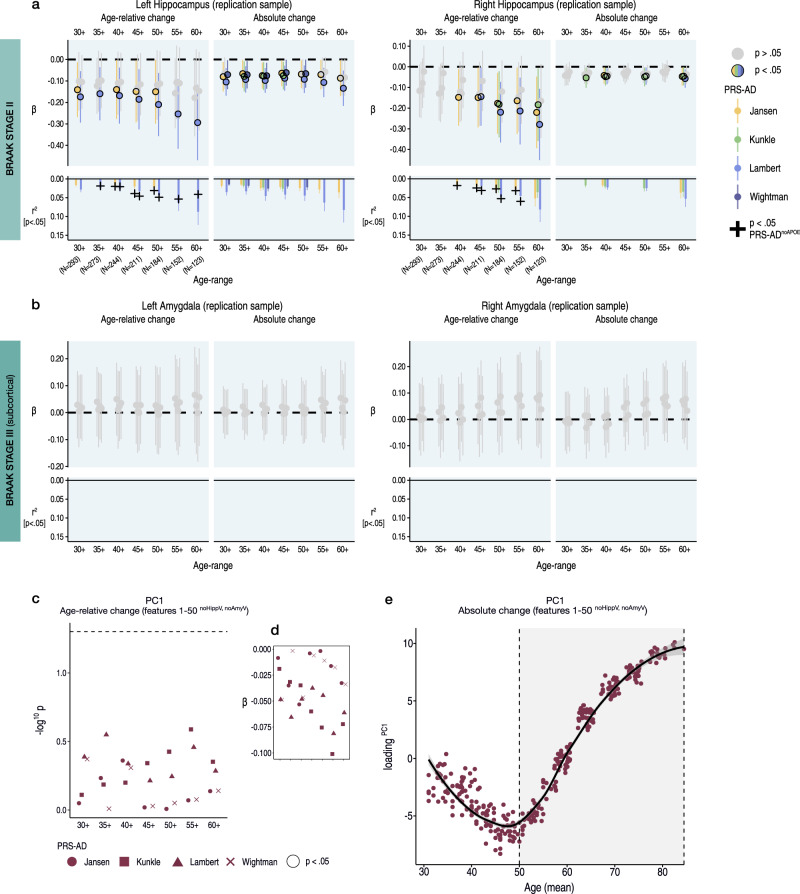
To estimate brain change relative to a person’s age in adult lifespan data, we used all longitudinal scans fitting age-range and inclusion criteria (≥30 years of age; Methods). This allowed us to obtain the best-fitting age trajectory models from which we could subsequently estimate how much an individual’s change trajectory deviated from the population-average (i.e., from the level of change predicted given age). Change was estimated via the individual-specific random slopes in a Generalized Additive Mixed Model (GAMM) of age (sex, scanner, and intracranial volume [ICV] corrected; Methods). We first explored change in initial hippocampal ROI’s – Braak Stage II50. Fig. 1a–c shows the longitudinal lifespan trajectory, and individual-specific degree of absolute and age-relative change for the left hippocampus (see Supplementary Fig. 1 for right). As expected2, almost all individuals aged ≥ 30 years were estimated to exhibit hippocampal loss, but to differing degrees, and very few individual-specific slopes were estimated to show growth over time (Fig. 1a–c). As also expected, the degree of absolute hippocampal change accelerated on average between the ages of 50 and 60 years. Note that negative absolute change values reflect hippocampal loss, whereas positive would indicate an estimated growth. Positive values on age-relative change then correspond to less hippocampal loss than expected given age, whereas negative values reflect more hippocampal loss than expected given age. Fig. 1b, c thus provides the context that higher hippocampal change values correspond to less decline, not growth. The degree of age-relative change was significantly associated with PRS-AD in the hypothesized negative direction: on average across the adult lifespan (30–89 years), individuals losing more hippocampus than expected for their age had significantly higher PRS-AD. This genetic association was probed separately for the bilateral hippocampi (left: β = −0.22, t(212) = −3.3, p = 0.001; right: β = −0.16, t = 2.4, p = 0.015; [PRS-AD Jansen]) and was significant using all four GWAS-derived scores (covariates: mean age, sex, N timepoints, interval between first and last timepoint, and 10 genetic ancestry factors [GAFs]). To ensure we were capturing ageing-specific effects at some point (see Supplementary Fig. 1), we tested the association using change rates extracted from progressively older age-ranges (i.e., progressively discarding data from comparatively younger individuals; Methods). This also ensured the analysis outcome was not based on a single arbitrary decision such as the age range to test the association across51,52. FDR-correction was applied across all 576 PRS-AD tests in this analysis. We then tested whether surviving associations remained statistically significant at p < 0.05 using polygenic scores computed without APOE (PRS-ADnoAPOE), assuming a 5% chance false positive rate per structure. Despite the progressively smaller sample size, all tested PRS-AD associations with age-relative hippocampal change (left and right) were significant at p < 0.05 [uncorrected] using all four scores (coloured points in Fig. 1e, f depict associations at p < 0.05 [uncorrected]). 31 of the 36 tests (86%) with age-relative left hippocampal change, and 25/36 (69%) with age-relative right hippocampal change, survived FDR-correction (see lower panels in Fig. 1e, f; partial r2 effect size is shown for associations surviving correction). Using PRS-AD to predict absolute hippocampal change instead in comparable statistical models (i.e., also correcting for mean age), PRS-AD associations were also mostly significant (47/72 [65%] survived correction). Probing whether these surviving associations remained after discounting the effect of APOE per structure, 19/58 (33%) PRS-ADnoAPOE associations with left hippocampal change (age-relative or absolute) remained significant at p < 0.05, surpassing the 5% false positive rate expected by chance (black crosses in Fig. 1e, f depict partial r2 of PRS-ADnoAPOE where significant [p < 0.05]). For right hippocampus, 6/45 (13%) associations remained significant at p < 0.05 with PRS-ADnoAPOE, also surpassing the chance false positive rate (Fig. 1f). Post-hoc tests confirmed the impression that the estimated regression coefficients became more negative as the age subset steadily comprised only older individuals (Fig. 1e, f); on average across change metrics, each increasing age subset was associated with a reduction in the negative beta coefficient of −.026 for left hippocampus (t = −14.1; pperm = 9.9e−4), and −.023 for right hippocampus (t = −15.4; pperm = 9.9e−4). Alternative post-hoc analyses dependent on power across the full age-range (30–89 years) found significant PRS-AD × age (mean) interactions upon age-relative change in left and right hippocampi for all four scores but these did not survive multiple comparison correction (Supplementary Table 6; Supplementary Fig. 5).
Fig. 1. Hippocampal change in healthy adults associates with genetic AD risk.
Longitudinal data was used to estimate individual-specific age-relative and absolute change in hippocampus (Braak Stage II), modelling the adult lifespan trajectories using GAMMs with random slopes. a Adult lifespan trajectory for left hippocampus from 30–89 years (data corrected for sex and scanner). b Estimated absolute change per individual (datapoints) in left hippocampus as a function of their mean age (across timepoints). This contextualizes change values in terms of an estimated loss. c Estimated age-relative change per individual in left hippocampus (individual-specific slopes) as a function of their mean age. Units are interpretable in terms of additional hippocampal volume loss per individual, above or below the mean level of loss expected for their age. Black stroke indicates whether or not genetic data was available per participant, and thus whether the datapoint was included in the PRS-AD association tests. d Linear models found more hippocampal loss than expected given age was associated with higher PRS-AD, on average across the full adult lifespan sample with genetic data (30–89 years; N = 229; association visualized for one score [Jansen]; colour and size depicts mean age). The association is shown for both the left and right hippocampus; however, note that across the full age-range only the left hippocampus survived FDR-correction, as depicted in panels (e, f). Models and datapoints are corrected for mean age and other covariates (Methods). e-f Linear PRS-AD associations with age-relative (left facet) and absolute change (right facet) in left (E) and right (F) hippocampus, using four GWAS-derived scores, tested for progressively older age-ranges to ensure capture of ageing-specific effects (i.e., moving from left to right on the X-axis, the leftmost age-range represents tests across the full adult lifespan [30–89 years; N = 229], whereas the rightmost age-range depicts associations tested in only the oldest adults [70–89 years]; standardized β). Significant associations at p < 0.05 (uncorrected) are depicted in colour (upper panels), with colours corresponding to the GWAS used to derive the four scores (Jansen, Kunkle, Lambert, Wightman). For associations surviving FDR-correction (p[FDR] < 0.05 applied across 576 two-sided tests), partial r2 of PRS-AD is shown (lower panels). Where the association survived correction, we retested it after removing APOE (PRS-ADnoAPOE). Partial r2 of PRS-ADnoAPOE is depicted by a black cross if the FDR-corrected association remained significant at p < 0.05. Trajectories depict mean measures. Error bands and error bars depict 95% CI. Summary-level source data are provided as a Source Data file.
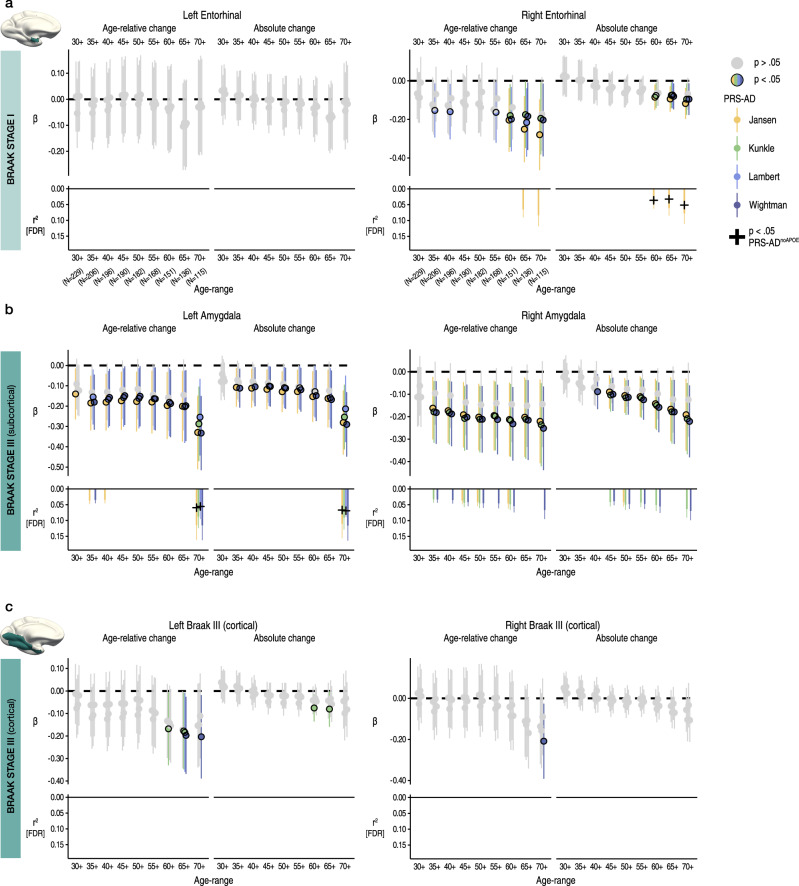
We then repeated the procedure for Braak Stage I (entorhinal) and III regions (subcortical and cortical ROIs; Methods). For Stage I, we observed no significant PRS-AD associations with change (age-relative or absolute) in left entorhinal cortex, but observed several significant associations with each in right entorhinal cortex, 5 of which survived correction (Fig. 2a). 3 of these surviving associations remained significant at p < 0.05 with PRS-ADnoAPOE (lower panels in Fig. 2a), surpassing the false positive rate. Post-hoc tests confirmed that the estimated regression coefficients became more negative as the age subset comprised only older individuals for right (beta reduction = −0.013, t = −7.8, pperm = 0.018) but not for left entorhinal cortex (beta reduction = −0.008, t = −5.0, pperm = 0.10). However, alternative post-hoc analyses across the full age-range (30–89 years) found no PRS-AD × age (mean) interactions upon age-relative entorhinal change, suggesting our data may have been underpowered to detect a two-way continuous interaction using change estimates (Supplementary Fig. 5; Supplementary Table 6).
Fig. 2. Change in early Braak stage regions in healthy adults associates with genetic AD risk.
Linear PRS-AD associations with age-relative and absolute change in brain regions encompassed within (a) Braak Stage I (entorhinal) and (b-c) Braak Stage III regions (amygdala and inferior temporal cortical ROI), using the four GWAS-derived scores, tested for progressively older age-ranges to ensure capture of ageing-specific effects (i.e., moving from left to right on the X-axis, the leftmost age-range represents tests across the full adult lifespan [30–89 years; N = 229], whereas the rightmost age-range shows the associations tested in only the oldest adults [70–89 years]; standardized β). Significant associations at p < 0.05 [uncorrected] are depicted in colour (upper panels), with colours corresponding to the GWAS used to derive the four scores (Jansen, Kunkle, Lambert, Wightman). For associations surviving FDR-correction (p[FDR] < 0.05 applied across 576 two-sided tests), partial r2 of PRS-AD is shown (lower panels; lower panels in A [left] and C [left and right] are correctly empty because no association survived correction). Where the association survived correction, we retested it after removing APOE (PRS-ADnoAPOE). Partial r2 of PRS-ADnoAPOE is depicted by a black cross if the FDR-corrected association remained significant at p < 0.05. Error bars depict 95% CI. Summary-level source data are provided as a Source Data file.
For the subcortical Stage III region (amygdala), we similarly observed negative associations between age-relative change in left and right amygdala and PRS-AD (Fig. 2b), 21 of which were significant after FDR-correction, and using absolute change yielded similar results (15 surviving associations). 4/11 (36%) PRS-ADnoAPOE associations remained significant at p < 0.05 for left amygdala (surpassing the false positive rate), whereas no associations with right amygdala change remained after excluding APOE. The estimated regression coefficients became stronger as the age subset comprised only older individuals (beta reduction left amygdala = −0.018, t = −7.6, pperm = 0.002; right amygdala = −0.019, t = 10.0 pperm = 0.004), though alternative analyses dependent on power across the full age-range found no post-corrected significant PRS-AD × age (mean) interactions upon age-relative change in amygdala (Supplementary Fig. 5; Supplementary Table 6). For the cortical Stage III region, none of the tested PRS-AD associations with change in left or right cortex survived correction (Fig. 2c), the regression coefficients became stronger as the age subset comprised only older individuals in each (beta reduction left cortex = −0.011, t = −6.2, pperm = 0.013; right cortex = −0.013, t = −6.3; pperm = 0.004), and we found no significant PRS-AD × age (mean) interactions in alternative analyses across the full age-range (Supplementary Fig. 5; Supplementary Table 6).
Across the 576 PRS-AD tests, conditioning brain change estimates on age conferred a 91% average strengthening in the observed PRS-AD regression coefficients, compared to using absolute change estimates and correcting for mean age (t = −26.9, p = 2.0e−80; Figs. 1, 2). This was also evident in the subset of results where both change estimates were FDR-corrected significant (54%, t = −20.6, p = 1.1e−28).
Sensitivity analysis
The PRS-AD results were not driven by outlying observations (see Supplementary Fig. 3). Furthermore, post-hoc analysis supported our choice of a genome-wide significant threshold for constructing PRS-AD scores (see Discussion); at more liberal SNP inclusion thresholds, the four scores became less comparable, dropping from a median correlation between scores of R = 0.73 at our chosen threshold, to R = 0.29 at the most liberal threshold. Moreover, the data suggested including more SNP’s was not beneficial, but may be detrimental to finding PRS-AD effects on brain change in healthy adults, at least in hippocampus (Supplementary Fig. 4).
Identifying features with faster atrophy in AD
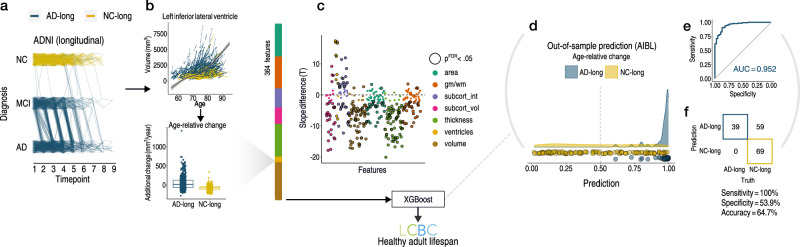
Given the univariate results, we expected that multivariate measures of change would be better suited to detect PRS-AD associations with brain change in healthy adults. Thus, we sought to empirically obtain a list of brain features with faster atrophy in AD, then test whether multivariate change across these features relates to PRS-AD in the LCBC healthy adult lifespan sample (Methods). First, in longitudinal AD patient-control data from ADNI (Supplementary Table 3), we defined two longitudinal groups we could be maximally confident consisted of healthy individuals and those succumbing to AD based on diagnosis: NC-long consisted of normal controls consistently classed as healthy over time, whereas AD-long comprised all individuals with an AD diagnosis by their final timepoint (Fig. 3a; Methods). Then, in 364 features we modelled a GAMM of age (irrespective of group), and entered the individual-specific slopes into ML binary classification (Fig. 3b; sex, scanner field strength, and ICV corrected; note that the ADNI sample did not change field strength over time; Methods). Group differences in slopes (age-relative change) were in the expected direction (Fig. 3c). The top features deemed most important for separating AD-long from NC-long individuals based on age-relative change in ADNI included many well-known AD brain vulnerabilities (e.g., ventricles, medial temporal and temporo-parietal regions; see Fig. 4a). Though our intention was not to refine prediction of AD cases, we note the model achieved an area under the curve (AUC) of 0.952 in independent data from the Australian Imaging Biomarker & Lifestyle Flagship Study of Ageing (AIBL; Fig. 3d–f; Supplementary Fig. 8; precision-recall AUC [AUC-PR] = 0.883).
Fig. 3. Visualization of longitudinal AD analysis pipeline.
a Longitudinal grouping in ADNI data. X-axis shows the scan observations across timepoints in the sample. Each line represents a participant. Single-timepoint ADNI diagnoses (Y-axis; NC normal controls, MCI mild cognitive impairment, AD Alzheimer’s disease) were used to define two longitudinal groups of AD and NC individuals (AD-long; N = 606, obs = 2730; NC-long, N = 372; obs = 1680). NC-long individuals were classified as healthy at every timepoint whereas AD-long individuals were diagnosed with AD by their final timepoint (Methods). Single-timepoint MCI diagnoses were considered only for the purpose of defining the longitudinal AD group. Because the grouping used all diagnosis observations (i.e., not only scan observations), trajectories of AD-long individuals that appear to end with a NC or MCI diagnosis also correspond to individuals with an AD diagnosis by their final timepoint, as do those seemingly reverting. b GAMMs of Age (across groups; upper plot) were used to model age-relative change (individual-specific slopes) in 364 brain features, shown for one example feature (lower plot). The ADNI-derived slopes were then used as input to machine learning binary classification using XGBoost115. c Most features exhibited significant group-differences in age-relative change as expected (datapoints depict t-statistics for t-tests); black stroke indicates significant differences after FDR-correction (p[FDR] < 0.05 applied across 364 two-sided tests). d–f Out-of-sample prediction for the binary classifier (AIBL data; Supplementary Fig. 8) including receiver operator curve (d), confusion matrix and performance metrics (e). The purpose of the classification procedure was to empirically derive brain features with accelerated change in AD, to use these in healthy adult lifespan data. Subcort subcortical, vol volume, int intensity, gm/wm grey/white matter contrast. Error bands depict 95% confidence intervals, while the boxplot displays the median as the measure of centre with the box spanning from the 25th to the 75th percentiles.
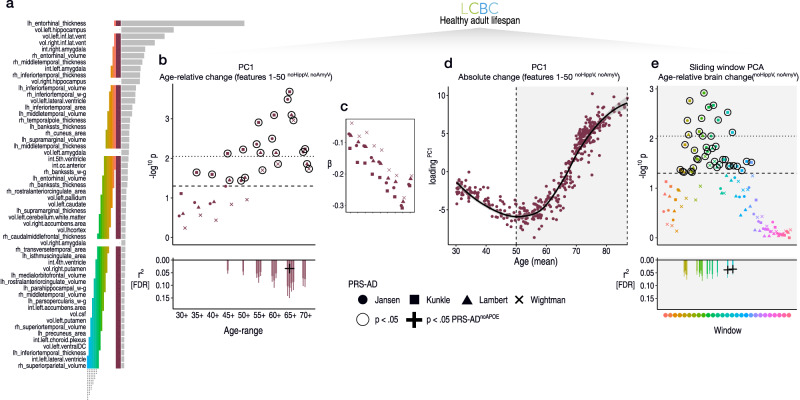
Fig. 4. ADNI-derived features applied to the healthy adult lifespan.
a Top brain features for classifying AD-long from NC-long individuals in ADNI data based on age-relative change. Coloured bars indicate feature selections across which we calculated PC1, and link with the subsequent plotted data in (b–e). b Linear PRS-AD associations in the LCBC healthy adult lifespan sample using PC1 of age-relative change across the top 50 features with accelerated change in AD (excluding hippocampal and amygdala volumes); PC1relChange; maroon bar in (a). Datapoints show -log10 p-values for PRS-AD associations with PC1relChange, tested at progressively older age-ranges, for all four scores. Dashed line indicates p = 0.05, and black stroke depicts significant PRS-AD associations at p < 0.05 (uncorrected). Datapoints above the dotted line are significant at p(FDR) < 0.05. Datapoint symbol corresponds to the GWAS used to derive the four scores (Jansen, Kunkle, Lambert, Wightman). For associations surviving FDR-correction (across 144 two-sided tests), partial r2 of PRS-AD is shown (lower panel). Where the association survived correction, we retested it after removing APOE (PRS-ADnoAPOE). Partial r2 of PRS-ADnoAPOE is depicted by a black cross if the FDR-corrected association remained significant at p < 0.05. c Standardized PRS-AD betas in b as a function of age-range (inversed to be negative due to the non-directional nature of PCA). d PC1 of absolute change across the top 50 brain features with accelerated change in AD (excluding hippocampal and amygdala volumes); maroon bar in (a) as a function of mean age (across timepoints). Accelerated brain change in AD-accelerated features was evident between ages 50–60. Note that since the y-axis represents change, the slope of the curve represents acceleration (see also Supplementary Fig. 14). e Linear PRS-AD-change associations using a PCA-based sliding window analysis within the age-range 50–89 years. Colours and order correspond to the coloured bars in (a). Dashed line indicates p = 0.05, and datapoints with black stroke depict significant PRS-AD associations at p < 0.05 (uncorrected). Datapoints above the dotted line are significant at p(FDR) < 0.05. For associations surviving FDR-correction, partial r2 of PRS-AD is shown (lower panel). Error bands and error bars depict 95% CI. lh left hemisphere, rh right hemisphere, vol volume (subcortical), int intensity (subcortical), w–g grey/white matter contrast, cc corpus callosum, DC diencephalon, csf cerebrospinal fluid. Subcortical features (aseg atlas) are delineated with “.”, whereas cortical features (aparc atlas) are delineated with “_”. Summary-level source data are provided as a Source Data file.
Multivariate associations between PRS-AD and brain change
In the LCBC healthy adult lifespan sample, we then modelled adult lifespan change in all 364 features used to train the AD-control model, and estimated the individual-specific slopes as before. Then, we calculated a multivariate marker of change based on the list of features the model found most important for classifying AD patients from controls, and related this to PRS-AD. Specifically, we calculated the principal component (PC1) of age-relative change across the first 50 features with model-implied importance (PC1relChange). Note that hippocampal and amygdala volumes were not included in PC1relChange to ensure these did not drive the multivariate effect (see the maroon bar in Fig. 4a; explaining 13% variance). As hypothesized, 14 of the 36 tested associations relating PC1relChange to PRS-AD were FDR-corrected significant (Fig. 4b; correction applied across all 144 PRS-AD tests in this analysis). Again, post-hoc tests confirmed the estimated regression coefficients became stronger as the age subset comprised only older individuals (beta reduction = −0.023, t = −9.9, pperm = 0.002) and alternative analyses across the full age-range found post-corrected significant PRS-AD × age (mean) interactions upon PC1relChange using all four scores (Supplementary Table 7). Next, to determine the age at which brain change in AD-sensitive features starts increasing in healthy adults, we calculated the principal component of absolute change across the same set of 50 features (PC1absChange; explaining 45% variance) plotted as a function of mean age (Fig. 4c). The results suggested that all healthy individuals were on a trajectory of change in AD features that showed onset of accelerated change around age ~50 (Fig. 4c; see Supplementary Fig. 14 for derivative plots). Further, change trajectories were steepest in features that were most important for separating AD-patients from controls (Supplementary Fig. 10). To ensure that the multivariate associations were not driven by one or a few brain features, we ran a sliding window PCA within the 50–89 year age-range (Methods). PRS-AD associations with age-relative change were evident when calculating PC1 across many combinations of features (coloured bars in Fig. 4a depict feature windows for the PCA and link with the coloured points denoting p-values for the PRS-AD associations in Fig. 4d; see Supplementary Fig. 11 for correlations between features). 13 of the tested associations were significant after FDR-correction, illustrating that change across many AD-sensitive features relates to PRS-AD in healthy adults (Fig. 4d). The data suggested that PRS-AD associations derived via this method were largely though not entirely driven by APOE (3 of the 27 [11%] FDR-corrected tests remained significant at p < 0.05 using PRS-ADnoAPOE, surpassing the 5% false positive rate; lower panels in Fig. 4).
As proof-of-principle, we directly applied the AD-control model weights to the LCBC healthy adult lifespan change estimates. This prediction incorporates information from the weights of all 364 features. The dependent variable was the model-implied log odds of having AD (probADrelChange; Methods). Importantly, because the model was trained on an index of relative brain change conditioned on age, the logistic prediction applied to the healthy adult lifespan data cannot be interpreted in terms of its implied binary outcome (i.e., AD/no-AD). This is because the model could assign the same probability of having AD to a hypothetical 30 year-old with an estimated additional brain loss of 10 mm3/year as to a 60 year-old with the same additional brain loss, despite the change being higher in the 60 year-old, because it exceeds the mean brain loss anticipated at age 60 (see Fig. 1c). Note that this clarifies why the model had high sensitivity but lower specificity in separating AD from controls in AIBL; it is a characteristic of the measures we used for model training more than the model itself. Still, we hypothesized the learned model weights would be useful, and would relate to PRS-AD similar to the relative change values in specific features. As expected, almost all of the tested PRS-AD associations with probADrelChange were significant at p < 0.05, 14 of which survived correction; (see Supplementary Fig. 9B). Repeating all steps of the model estimation procedure using absolute change instead (from hyperparameter estimation to prediction; AUC = 0.933 in unseen data from AIBL; AUC-PR = 0.864; Supplementary Fig. 8), we found fewer significant PRS-AD associations with probADabsChange (7 survived correction; Supplementary Fig. 9C, D). This suggests age-relative change may be a superior marker for capturing differences in brain ageing. Again, the data indicated PRS-AD associations derived by this method were largely though not entirely driven by APOE (8/21 [38%] of the FDR-corrected tests with change remained significant using PRS-ADnoAPOE; Supplementary Fig. 9; FDR-correction applied across all 72 PRS-AD tests in this analysis).
Replication analysis
To reduce the number of tests, in an independent adult lifespan replication sample with fewer follow-up observations (2-3 timepoints; Lifebrain sample), we tested PRS-AD associations with hippocampal and amygdala change, and PC1 of age-relative change across the first 50 AD-sensitive features, not including hippocampal or amygdala volumes (i.e., PC1relChange; Fig. 4a). For hippocampus (Supplementary Fig. 12), we observed similarly negative effects, 22 of which were significant for age-relative change (p < 0.05 [uncorrected]; 31 for absolute change; Fig. 5a). Similar to the discovery sample, PRS-AD effects on age-relative hippocampal change were larger than absolute change, and often remained significant after discounting APOE (black crosses in Fig. 5a depict partial r2 for PRS-ADnoAPOE where this remained significant at p < 0.05). For amygdala, we observed no significant PRS-AD associations within any age-range, and we also observed no significant associations with PC1relChange (Fig. 5b, c). However, like the discovery sample, the principal component of absolute change across the same set of 50 features (PC1absChange; explaining 54%) revealed that all healthy individuals lay on a trajectory of accelerated change in AD features, with a similar onset of acceleration around the age of 50 years (Fig. 5d; Supplementary Fig. 14).
Fig. 5. Replication.
a-b Linear PRS-AD associations with age-relative and absolute brain change in an independent adult lifespan sample (Lifebrain replication sample), using the four GWAS-derived scores, tested for progressively older age-ranges to ensure capture of ageing-specific effects (i.e., moving from left to right on the X-axis, the leftmost age-range represents the association across the full adult lifespan on average [30–88 years; N = 293], whereas the rightmost age-range shows the associations tested in only the oldest adults [60–88 years]). Univariate linear associations were tested for (a) left and right hippocampus, and (b) left and right amygdala. Significant associations at p < 0.05 (uncorrected) are depicted in colour (upper panels), with colours corresponding to the GWAS used to derive the four scores (Jansen, Kunkle, Lambert, Wightman). For associations that were significant at p < 0.05 (uncorrected), partial r2 of PRS-AD is shown (lower panels). For these, we retested the association after removing APOE (PRS-ADnoAPOE). Partial r2 of PRS-ADnoAPOE is depicted by a black cross if the association remained significant at p < 0.05 (uncorrected). c Multivariate linear PRS-AD association tests using PC1 of age-relative change across the top 50 brain features with accelerated change in AD (excluding hippocampal and amygdala volumes; PC1relChange; as in Fig. 4a, b). Datapoints show −log10 p-values for the association with PC1relChange, tested at progressively older age-ranges, for all four scores. Datapoint symbol corresponds to the GWAS used to derive the four scores (Jansen, Kunkle, Lambert, Wightman). d Standardized Betas in c as a function of age-range (inversed to be negative due to the non-directional nature of PCA). Dashed line indicates p = 0.05. e PC1 of absolute change across the top 50 brain features with accelerated change in AD (excluding hippocampal and amygdala volumes; maroon bar in Fig. 4a), plotted as a function of mean age across timepoints. Accelerated brain change in AD-accelerated features was evident around age 50–60. Note that since the y-axis represents change, the slope of the curve represents acceleration (see also Supplementary Fig. 14). Error bands and error bars depict 95% CI. Summary-level source data are provided as a Source Data file.
Memory change analysis
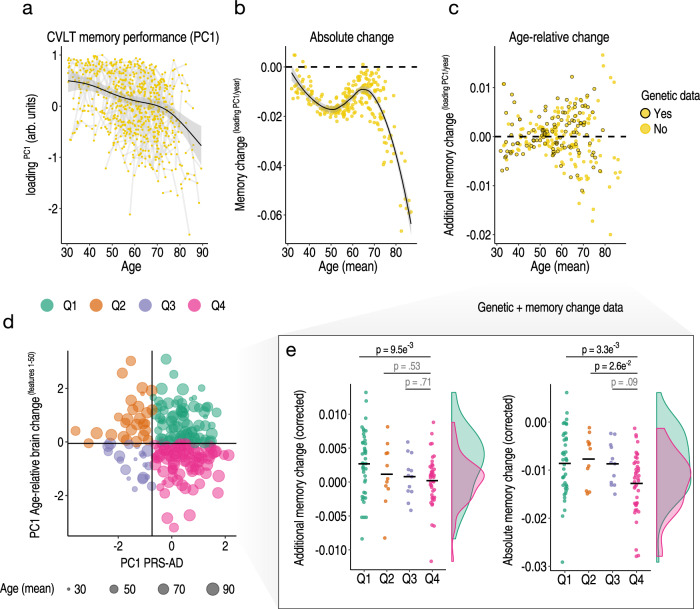
Finally, in the LCBC adult lifespan discovery sample, we separated individuals into discrete groups based on the conjunction of brain and genetic risk factors. We hypothesized that high PRS-AD individuals who are also high on a multivariate marker of relative change in AD-sensitive features would show more pronounced longitudinal memory decline across adult life (pink quadrant 4 in Fig. 6d; Methods). For this, we used the partial association between PC1 of age-relative change calculated across the first 50 AD-sensitive features—here including hippocampal and amygdala volumes (PC1relChange1-50)—and PC1 calculated across the four PRS-AD scores (PC1PRS-AD; explaining 87%; Methods). Akin to the brain analysis, memory-change estimates were derived via the individual-specific random slopes in a GAMM of age, and we used longitudinal memory observations from the full adult lifespan sample to optimize memory-change estimates in the subset of participants that also had genetic data (Methods). Fig. 6a–c shows the longitudinal lifespan trajectory, and estimated individual-specific degree of absolute and age-relative change in memory performance on the California Verbal Learning Test (CVLT; PC1 across subtests; Methods). Absolute memory change was estimated to be predominantly negative, with memory decline occurring gradually across the adult lifespan and accelerating around the mid ~60 s (Fig. 6b; though we also observed a trend towards steeper slopes in mid-life prior to this; see Supplementary Fig. 15 and Discussion). High PRS-AD individuals who were also high on a multivariate marker of relative brain change showed significantly more age-relative (p = 0.01) and absolute memory decline (p = 0.003) on average across the adult lifespan, compared to high PRS-AD individuals with less brain change. These group differences in memory change were not driven by differences in APOE-ε4 carriership (Fig. 6e; main models corrected for carriership, mean age, sex, N timepoints, interval between first and last timepoint), and persisted in alternative models controlling for the number of APOE-ε4 alleles (p = 0.009; p = 0.003) and baseline memory performance (p = 0.008; p = 0.002). In the main model, we also observed a significant difference in absolute memory change between the high PRS-AD-high brain change group and the low PRS-AD-low brain change group (p = 0.026; Fig. 6e). These group differences survived multiple comparison correction (FDR-correction applied across six one-sided group tests; Methods). Importantly, the reported group differences in memory-change persisted when correcting for differences in genetic risk (PC1PRS-AD) but not for differences in multivariate brain change (Supplementary Fig. 16). Finally, the main group difference in memory decline was not driven by the oldest adults or by residual group-differences in age, but persisted using change estimates from alternative age subsets (see Supplementary Fig. 17). These data suggest the conjunction of risk markers – a multivariate marker of change in AD-sensitive features and known PRS-AD – helped identify a subset of comparatively higher-risk individuals showing more longitudinal memory decline through heathy adult life.
Fig. 6. Longitudinal memory change analyses.
Exclusively longitudinal data was used to estimate individual-specific age-relative and absolute change in CVLT task performance (PC1 across subtests), modelling the adult lifespan trajectories using GAMMs with random individual-specific slopes. a Adult lifespan trajectory analysis for CVLT memory performance from 30–89 years. Lines connect longitudinal observations. b Estimated absolute memory change per individual (datapoints) in CVLT task performance plotted as a function of their mean age (across timepoints). c Estimated age-relative change per individual in CVLT task performance (individual-specific slopes) as a function of mean age. For each participant with memory change data, black stroke indicates whether or not genetic data was available. d The linear association between the principal component across the four PRS-AD scores and the principal component of age-relative change across the first 50 ADNI-derived features (listed in Fig. 4a) was used to define four quadrant groups representing the conjunction of brain and genetic risk factors. e Memory change for individuals with both memory change and genetic data within the quadrant groups (colours in d-e depict groups). Linear models found that individuals with higher PRS-AD who also exhibited more age-relative brain change in AD-sensitive features (in pink) showed significantly more age-relative (left plot) and absolute (right plot) change in memory across the healthy adult lifespan, relative to high PRS-AD individuals estimated to show less relative brain change. These significant group differences survived FDR-correction for multiple comparisons (applied across six one-sided tests; Methods; two-sided p-values shown). The distributions are visualized for these two groups; datapoints corrected for covariates including mean age and APOE-ε4 carriership [Methods] (see also Supplementary Fig. 17). Error bands depict 95% CI. Summary-level source data are provided as a Source Data file.
Discussion
Variation in brain ageing trajectories in healthy adults links with AD-related genetic variation and memory decline outcomes through adulthood. Specifically, we found that healthy individuals who are losing more brain than expected for their age in early Braak regions—bilateral hippocampus, amygdala, and right entorhinal cortex – are at significantly higher genetic AD risk. Some of these polygenic associations extend beyond the risk conferred by APOE alone, most notably in hippocampus. In multivariate analyses, we then show that faster-than-expected brain ageing across many AD-sensitive features associates with PRS-AD in healthy adults, and that accelerated atrophy in AD features is evident in most healthy individuals over age ~50. This latter finding suggests that neurodegeneration in ageing and AD occurs on a continuum. Accordingly, we find that ML models trained on longitudinal AD-control data can be applied to brain change estimates in healthy adults and the prediction relates to PRS-AD. Finally, genetically high-risk individuals showing faster-than-expected brain change exhibited more longitudinal memory decline compared to genetically high-risk individuals with less brain change, across the adult lifespan, and independent of APOE-ε4. Thus, the conjunction of a multivariate brain change marker and known genetic risk found a subset of comparatively high-risk individuals exhibiting more memory decline through adulthood.
Univariate analyses using change in early Braak regions found many PRS-AD associations in healthy adults, illustrating accelerated brain ageing in genetically at-risk individuals. The clearest genetic effects upon faster atrophy were in hippocampus; healthy adults at higher genetic AD risk lose hippocampal volume faster than their age would predict – observed consistently using all four scores. Particularly for left hippocampus, the association was evident after discounting APOE, suggesting differences in left hippocampal loss also arise from genetic factors beyond APOE. However, we also observed PRS-ADnoAPOE associations with right hippocampal change, and confirmed these in independent data. Shrinkage of the hippocampus—a critical structure underpinning episodic memory and spatial navigation operations—is a well-known AD risk marker in patients9,32,53, with atrophy rates predicting clinical conversion54. However, most studies in healthy adults have not linked genetic AD risk to hippocampal change2,20–26,39,55, including in large adult lifespan samples24,25 and our previous report2. And since AD risk variants may influence hippocampal differences early in life2,29,30, cross-sectional findings in older adults27,28,56 cannot attribute genetic effects to accelerated brain ageing57. By isolating genetic effects on change, our study confirms quantitative genetic AD risk influences variation in hippocampal change rates in healthy adults.
This aligns with a study by Harrison et al.36 finding a longitudinal relationship between hippocampal change and PRS-AD in older adults. However, that study recruited participants with memory complaints and a family AD history via memory clinics. In contrast, our sample comprised healthy adults in longitudinal studies which are known to be biased towards retaining high performers58,59, as seems evident in the cognitive scores of the older adults here (see Supplementary Table 2; Supplementary Fig. 18). It also agrees with a study finding more hippocampal atrophy in healthy older APOE-ε4 carriers38. Yet, we also found AD risk SNP’s beyond APOE predict hippocampal ageing trajectories in healthy adults, which to our knowledge has not been shown. Previously, we did not find evidence PRS-AD or APOE-ε4 alters the slope of hippocampal ageing, but found an offset effect suggesting the difference between high- and low-risk groups in hippocampal volume was as large at age ~25 as at age ~802. However, that study used a PRS-AD incorporating many more SNP’s (p < 0.0560), and did find some, albeit inconsistent, evidence for a slope effect using the same SNP-inclusion threshold as here. Taking an individual-centric approach to estimate change, we found genome-wide significant SNPs could explain up to ~13% variance in hippocampal change rates (effect sizes after discounting APOE were ~5%; Fig. 1e, f). This longitudinal marker of relative brain ageing consistently excelled, exhibiting stronger relationships to PRS-AD than absolute change that were detectable over wider age-spans. This suggests that conditioning change estimates on age may help uncover signal in comparatively younger adults. Still, while the data suggest PRS-AD associations were not driven by only the oldest adults, comparatively older adults likely contributed more to the differences in brain change signal (Supplementary Fig. 7). This fits with the tendency we observed towards stronger genetic effects upon slopes in older adults, consistent with theories suggesting genetic effects become amplified in older age when neural resources are depleted61.
PRS-AD also linked with faster atrophy in right entorhinal cortex and bilateral amygdala. This also aligns with Harrison et al.36, where entorhinal change linked with PRS-AD in older adults in memory clinics. It may also fit with a study finding right entorhinal cortex shows among the largest differences in APOE-ε4 carriers28. However, we also found evidence SNPs beyond APOE predict entorhinal change. Similarly, faster amygdala loss was related to PRS-AD in healthy adults, and there was some evidence to suggest SNP’s beyond APOE predict amygdala trajectories, at least in left amygdala. PRS-AD associations in right amygdala were seemingly driven by APOE. These data contradict a recent GWAS, which found the effect of APOE upon amygdala and hippocampal change in ageing disappeared after accounting for disease39. In contrast, we found faster amygdala atrophy in healthy adults with higher PRS-AD. However, while amygdala effects were clear in the discovery sample—one of the most densely sampled MRI datasets for lifespan follow-up—these did not replicate in a sample with less follow-up, hence this awaits replication. Nevertheless, in healthy ageing and AD, medial temporal lobe structures show early vulnerability to structural loss5, highest expression of top AD risk genes62–64, and our study provides evidence PRS-AD influences faster atrophy in some of these structures in healthy adults. Speculatively, faster atrophy may co-occur with faster tau accumulation, consistent with higher tau in risk-allele carriers64,65. Critical questions remain concerning which mechanisms drive the shared vulnerability of these structures to lifespan influences and AD, and why AD risk variants speed up their age-related neurodegeneration. One candidate shared characteristic may be a high degree of plasticity66–68.
Yet many other brain features exhibit faster atrophy in AD. Through data-driven analyses to delineate these, we found faster change across many AD-sensitive features relates to PRS-AD in healthy adults. These associations with multivariate change measures were largely though not entirely driven by APOE (Fig. 4; Supplementary Fig. 9). We also found replicable evidence that almost all individuals above age ~50 are on an accelerated trajectory of neurodegenerative ageing in brain features showing faster atrophy in AD (see also Supplementary Fig. 10). This agrees with work documenting overlapping atrophy patterns in ageing and AD4,5,14. These individualized estimates suggest that neurodegeneration occurs along a continuum from normal ageing to AD. Further, since it is unlikely most healthy adults here would be amyloid positive, this may run counter to the amyloid cascade hypothesis, which posits plaque build-up as an initial triggering event for neurodegeneration69–71. However, amyloid may be associated with differences in its degree. Our approach to link neurodegenerative changes in AD to ageing likely benefitted from multivariate analyses using change in healthy adults. We also found that ML models trained on AD-control data can be applied to healthy adults and the prediction relates to PRS-AD. This seemed to work best when the model was trained on estimates of change conditional on age, possibly because this places often extreme change values in AD on a scale more comparable across ages. Modelling relative change in AD vs. controls may also enable better identification of features exhibiting a quantitative difference in change despite the presence of a similar qualitative pattern. That our patient-control groups were based on two extremes (consistently healthy versus becoming AD) further suggests the difference may lie more in degree than kind, as does the fact that our ML model still captured 100% of independent AD cases (Fig. 3). Together, these findings suggest genetic AD risk elicits a widespread impact on faster brain ageing in healthy adults, and that the border between neurodegeneration in ageing and AD is unclear.
Of note, while PRS-AD effects were not solely driven by APOE, APOE nevertheless accounted for much of the predictive power of PRS-AD, as associations often disappeared or were attenuated using PRS-ADnoAPOE. This fits with studies finding PRS-AD associations with cognitive and metabolic factors in adults are largely driven by APOE72, and limited utility of SNP’s beyond APOE to predict AD markers18. Most associations after excluding APOE were with scores derived from the genome-wide significant SNP’s/weightings reported by Jansen et al.73, possibly suggesting these better capture differences in brain ageing (though PRS-ADnoAPOE effects were also evident using scores from two other GWAS60,74). We chose a conservative PRS-AD threshold based on studies indicating this shows highest discrimination of patients75,76, and an assumption that scores would be less comparable at more liberal thresholds, due to including different sets of genetic variants and less consistent effect size estimates (see77 for why simply deferring to the latest AD GWAS estimates is also not without assumption). Indeed, we found no evidence that incorporating more SNPs in the PRS increased sensitivity to detect genetic effects upon brain ageing. Rather, it may be detrimental to this goal (Supplementary Fig. 4). PRS-AD scores also correlated more poorly when including more variants. The implication is that the choice of GWAS and PRS will affect the outcome of any PRS-AD study, possibly because different AD GWAS capture signals that become less comparable across the wider genome.
Individuals at higher genetic risk who also showed more atrophy for their age in AD-sensitive features exhibited more memory decline across adult life, compared to genetically at-risk individuals with less atrophy. Hence, knowing one’s genetic risk was insufficient, as it was not necessarily reflected in brain and cognitive outcomes. However, considered together with a multivariate marker of brain change, we found a subset of high PRS-AD individuals whose brain status over time was reflected in a greater drop-off in memory. Thus, our results speak to the importance of considering overlapping risk factors rather than only each in isolation, as we found substantial variation in risk also within genetically high-risk individuals—highlighting that genetic AD risk neither determines nor sufficiently predicts cognitive and brain outcomes. Rather, group differences in memory decline were more driven by brain change differences than by genetic differences, as they persisted when controlling for PRS-AD and APOE-ε4 but not atrophy. Thus, brain change may be crucial for detecting comparatively at-risk individuals in adult lifespan data. Further, memory decline differences were protracted through adulthood, as they were evident in different age subsets, including comparatively younger adults (e.g., within the age-range 30–65; Supplementary Fig. 17). This indicates neurodegeneration in AD-sensitive regions tracks with memory decline differences that are detectable through adulthood. It also emphasizes that memory decline is a gradual phenomenon that is not confined to old age. This perspective may be obscured in studies that use clinical tests that do not capture subtle cognitive variations, and estimates of decline relative to a group rather than one’s earlier capacity. Our findings extend previous studies finding PRS-AD43,44,46 or APOE-ε445 relates to memory decline across adult life, and possibly shed light on why such genetic associations are often weak43–46 or absent49. Whether these brain trajectory differences are relevant for later AD outcomes will require follow-up and biomarker assessment, but our results show these neurodegenerative changes are not benign. They also underscore the need for follow-up data over extended age-spans for prediction or prevention of AD, and suggest a continuous view on lifespan brain health may aid understanding of AD78,79. Multivariate atrophy measures may help assess AD risk and improve selection into clinical trials. Future research should examine why some high PRS-AD individuals decline more in brain and memory where others remain resilient, as well as combine multivariate change with other biomarkers (e.g., tau, amyloid, inflammation) as we move towards a future of individualized risk assessment.
Our study has several strengths. First, our brain change estimation circumvents the drawbacks of other approaches attempting to capture individual differences in brain ageing—such as brain age models80—which may not necessarily reflect change81. Second, we used all available longitudinal data to estimate brain and memory change, each in a single model. This likely optimized the change estimates for all, including the relatively modest subset with genetic data, in part due to improved age trajectory modelling from which one can estimate the deviation of an individual’s change trajectory. The mixed-model change estimation is equivalent to estimating factor scores, and psychometrically superior to more manual calculations of change82,83. It should also be less influenced by outliers due to the shrinkage effect. This limits the influence of extreme data points by estimating random effects from a probability distribution, the parameters of which are derived from the data. As more longitudinal measures are incorporated, the distribution becomes more robust, reducing the influence of extreme slopes and pulling them closer to the mean82,83. This is exemplified in Supplementary Fig. 13, where we show that PRS-AD-change associations in the same individuals in the BETULA study improved when their slopes were estimated together with NESDA data, compared to using BETULA data alone. Further, to ensure we were capturing ageing-specific processes at some point (Supplementary Fig. 1), we allowed the data to be increasingly comprised of only older adults and repeatedly tested PRS-AD associations. As inferences based on significance are affected by arbitrary analysis choices, we took inspiration from multiverse methods to define a defensible set of choices to perform analyses across51,52. In our case, the main arbitrary covariate was the age-range to test the association across. Despite accounting for age- and time-related covariates, the influence of this choice on statistical significance is clear in Figs. 2, 4b, and 5. This clarifies why we used multiple scores; using a single PRS could have obscured the results, as significance fluctuated across scores, or using the same score across age-range specifications. Adopting this approach, we could ensure capture of ageing-specific processes, document the stability of PRS-AD-change associations in healthy adults, and ensure the results were independent of a single arbitrary decision51,52.
There are also limitations. First, characteristics of lifespan data will affect the mixed-model change estimation. Our study had more timepoints in the older age-ranges (Supplementary Fig. 6), likely resulting in more accurate estimates in older adults. Hence, alongside mean age, we corrected for timepoints and the interval between first and last visit to ensure the results were not driven by residual age-related variation. Similarly, normalizing change estimates by age does not reduce variability along the age variable. Because more variability in older age is a known phenomenon84, adjusting for mean age in all association tests further ensured the results were not driven by higher dispersion in older adults. Conversely, estimates in younger, less-sampled age-ranges may be biased by the magnitude of change in older adults. This may help explain why the memory slopes of also younger adults were estimated as negative. Further, selection bias and attrition vary by age-group, which alongside data density differences may explain why adults in their 60’s were estimated with less negative memory slopes compared to middle age (Fig. 6b). Caution is thus advised around overinterpreting change estimates in terms of their absolute values, hence we refer to “estimated change”. Relatedly, scanner parameters changed over time for some samples. While we made efforts to correct for or reduce scanner variation (Methods), this will influence estimates. Second, our approach disregards heterogeneity in ageing or AD-related atrophy, treating all individuals with an AD diagnosis as one group compared to all normal controls. This was reasonable for our goal of identifying features with faster average change in AD, given there may be a predominant AD atrophy pattern85 that overlaps with the average ageing pattern5,6,42. But since there are AD subtypes85–87, an important question is whether AD variability traces to brain change heterogeneity in adults. Third, we relied on FreeSurfer-derived measures. While these are well-validated and reliable88–90, some measures may be less so90. Indeed, that we observed no PRS-AD associations with left entorhinal change was surprising. When we quantified the proportion of individuals estimated to show positive absolute change cortex-wide (i.e., “growth”), entorhinal measures were clear outliers, with ~16% estimated to be growing (Supplementary Fig. 2). The median across cortical regions was 0.2%. This may reflect poorer reliability of entorhinal measures, as suggested by others89,91,92. Possibly, manual entorhinal tracing or alternative tools may have led to different results90. Our results also point to the advantage of multivariate change measures over univariate measures. Fourth, while the discovery sample screened out participants with mental disorders, the replication sample included individuals with disorders (Methods). This decision aimed to increase power to estimate change in the less-powered replication sample, but potentially influenced the results. Fifth, the adult samples consisted mainly of homogenous white ethnic populations from their respective countries, as did the GWAS on which PRS scores were based, possibly limiting result generalizability. Sixth, alongside the more limited longitudinal coverage which will negatively impact change estimates, sampled or geographic differences in APOE genotype may account for the lack of full replication in the independent adult lifespan cohort (Supplementary Fig. 19; Supplementary Table 8). Seventh, longitudinal studies inevitably recruit and culminate in unrepresentatively high-performing samples58. Our data also suggest this, as we observed a tendency for better memory in older adults with more repeat visits (Supplementary Fig. 18), and higher average IQ scores in those older than 60 (Supplementary Table 2). Since even in these we find variation in brain ageing slopes that correlates with AD-related genetic variation and memory decline, the population effect-sizes may be larger. Eighth, we used only structural measures. While these are sensitive to detecting subtle changes in brain structure that ultimately reflect a continuous, lifelong process of change, other biomarkers are necessary to refine detection of AD-risk in healthy adult samples. Finally, we do not know which individuals here will be diagnosed with AD later in life, or have other AD biomarkers suggesting a biological trajectory to AD93. While our analyses suggest one could assign differential transition probabilities to healthy individuals, only time and follow-up data will tell.
In conclusion, brain change trajectories in healthy adults are accelerated by the presence of AD risk variants, in many brain features, and also beyond APOE. We show that brain features most susceptible to faster deterioration in AD are on a trajectory of accelerated change from age ~50 in most healthy individuals, and that models trained on AD patients can be applied to adult lifespan data and the prediction relates to genetic AD risk. Finally, genetically at-risk individuals with more brain change showed more memory decline through adulthood, compared to genetically at-risk individuals with less brain change. Thus, tracking change in AD-sensitive regions enhanced the value of knowing a person’s genetic risk, and atrophy predicted memory decline more than the genetics. Our findings show that brain ageing slopes in healthy adults correlate with AD-related genetic variation and memory decline through adulthood, and that neurodegeneration occurs along a continuum from normal ageing to AD.
Methods
Studies conducted at the Center for Lifespan Changes in Brain and Cognition (LCBC) were approved by the Regional Ethical Committee of South-East Norway (2017/653). Ethical approval for the other datasets was granted by the relevant authorities and all participants provided informed consent. LCBC participants received compensation.
Samples
Age-relative change estimation
Adult lifespan discovery sample
After applying exclusion criteria (see below), an exclusively longitudinal adult lifespan sample (minimum two timepoints) comprising 1430 scans from 420 healthy individuals aged 30–89 years (248 females; mean age [SD] = 63.7 [14.4]; 2–7 timepoints [median = 3]; follow-up range = 0.15–11.1 years) was drawn from the Center for Lifespan Changes in Brain and Cognition database (LCBC; Department of Psychology, University of Oslo; see Supplementary Note 1). Observations were collected across 3 scanners. Prior to participation, all individuals were screened via health and neuropsychological assessments. Generally, the LCBC sample is comprised of cognitively high-performing individuals (see summary of cognitive scores in Supplementary Table 2; Supplementary Fig. 18). The following exclusion criteria were applied across LCBC studies: evidence of neurodegenerative or neurologic disorders, conditions or injuries known to affect central nervous system (CNS) function (e.g., hypothyroidism, stroke, serious head injury), and MRI contraindications as assessed by a clinician. At baseline, participants were thoroughly screened for evidence of cognitive deficits, and excluded based on lifetime presence of psychiatric disorders and/or use of medication known to affect the CNS (e.g., benzodiazepines, antidepressants or other central nervous agents). Additionally, to guard against including participants with incipient AD in our sample, we here excluded adults whose scores on the Mini Mental State Exam (MMSE)94 suggested longitudinal cognitive deficit with no later recovery (MMSE < 25 at their final timepoint; 2 participants; 4 scans), and adults aged 40+ whose scores on the Beck Depression Inventory (BDI)95 or Geriatric Depression Scale (GDS)96 suggested depression symptoms over time with no later recovery (BDI > 21 or GDS > 10 at their final timepoint; 7 participants; 32 scans). All LCBC studies were approved by the Norwegian Regional Committee for Medical and Health Research Ethics, complied with ethical regulations, and all participants provided informed consent.
Adult lifespan replication sample
To test replication, we used the two remaining longitudinal adult cohorts from the Lifebrain consortium that had up to three MRI timepoints available: the BETULA project97 and the Netherlands Study of Depression and Anxiety (NESDA)98. BETULA participants underwent dementia assessment by a clinician using cognitive data and medical records, and those reporting neurological disorders (stroke, AD, other dementias, MS), or presenting with severe memory deficits or MRI contraindications at any timepoint were excluded. Because it is a population-based sample, BETULA employs no screening/inclusion criteria for mental disorders. NESDA participants reporting neurological disorders (stroke, AD, other dementias, MS), or presenting with severe memory deficits or MRI contraindications were excluded. Although neurologically normal, 97 of the NESDA participants were diagnosed with a current or remitted depressive and/or anxiety disorder, whereas 41 had no history of mental health disorders. One extreme outlier in the change data of each sample was also detected and excluded here (see Supplementary Fig. 12). In all, we collated the data from 449 scans from 182 individuals aged 31–8 from BETULA (mean age = 64.3 [11.9], 2-3 timepoints, follow-up = 3.5–7.7 years; 85 females), with 331 scans from 138 individuals from NESDA aged 30–65 (mean age = 45.1 [7.9], 2-3 timepoints, follow-up = 1–10 years; 91 females), into a single adult lifespan replication sample (Supplementary Table 1).
Polygenic risk associations
To test associations with PRS-AD we used the subset of participants with both quality-controlled genetic data (European ancestry) and longitudinal change estimates, as estimated from the full adult lifespan models with all participants (also those without genetic data). For the discovery sample, 229 participants had genetic and brain change data. For the replication sample, 175 participants from BETULA and 118 from NESDA (92 diagnosed) had genetic and brain change data.
AD samples
We used exclusively longitudinal data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI99), and the single-timepoint ADNI diagnosis (normal controls [NC]; mild cognitive impairment [MCI]; AD) to define two longitudinal groups based on final-timepoint diagnosis (2–9 timepoints): NC-long consisted of subjects classed as NC at every diagnosed timepoint; AD-long consisted of all subjects where the final diagnosed timepoint was AD7. After grouping, for subjects where scanner field strength changed over time (from 1.5 T to 3 T), we used only the observations from the field strength with the most timepoints (or where equal used the 3 T scans). This was to ensure the validity of change estimates in the multiscanner, multisite ADNI data100. In all, NC-long consisted of 1680 scans from 372 subjects, and AD-long consisted of 2730 scans from 606 subjects (Supplementary Table 3). The ADNI (PI: Michael W. Weiner, MD) was launched in 2003, with a goal of testing whether serial MRI can be used to measure the progression of MCI and early AD (see https://adni.loni.usc.edu/about/). An independent AD-control sample consisting of 107 scans from 39 AD-long subjects and 435 scans from 128 NC-long subjects was used for validation of ML models (AIBL dataset; data collected by the AIBL study group101; Supplementary Fig. 8).
Genotyping and polygenic scores
In the LCBC dataset, buccal swab and saliva samples were collected for DNA extraction, followed by genome-wide genotyping using the Global Screening Array (Illumina, Inc., San Diego, CA). For a full description of genotyping, post-genotyping, and quality control and imputation methods applied to the genetic samples here, see refs. 2,102,103. We used the summary statistics from four previous large-scale GWAS of AD60,74 two of which included AD-by-proxy subjects based on parental status73,104. We then computed polygenic risk scores based on the genome-wide significant SNPs reported in each (p < 5 × 10−8), weighted by their allelic effect sizes. Prior to this, shared SNPs between each GWAS and our data were pruned to be nearly independent using PLINK105 with the following parameters --clump-p1 0.9999 --clump-p2 0.9999 –clump-r2 0.1 –clump-kb 500. The linkage disequilibrium structure was based on the European subpopulation of the 1000 Genomes Project Phase 3106. Because of the complexity of the major histocompatibility complex region (build hg19; chr6: 25,652,429-33,368,333), we removed SNPs in this region except the most significant one prior to pruning. We computed the four PRS-AD both with and without SNPs from the APOE region (build hg19; chr19: 44,909,011-45,912,650). We chose a genome-wide significant SNP threshold based on recent studies showing highest discrimination ability between patients and controls75,76. We also reasoned PRS’ constructed with more relaxed p-value thresholds are more likely to contain different sets of genetic variants, and hence would be less comparable across the four scores. As an exploratory analysis, we tested eleven other p-value thresholds. This confirmed the four scores became less correlated at more liberal thresholds (see Results). Furthermore, adding more SNPs was detrimental to finding PRS-AD associations with hippocampal change in healthy adults (Supplementary Fig. 4). From the summary files, we removed SNPs not in the reference data, with minor allele frequencies <0.05, or with low imputation scores. Genetic ancestry factors (GAFs) were computed using established principal components methods. For the discovery sample analyses, we used the first 10 as covariates in genetic analyses107. For genetic analyses in the combined Lifebrain replication sample, the first 4 were used as covariates (NESDA data was prepared using ENIGMA protocols requiring 4 GAFs102).
MRI acquisition and pre-processing
T1-weighted (T1w) anatomical scans from each MRI dataset (acquisition parameters in Supplementary Table 4) were processed using FreeSurfer’s longitudinal stream108 (v.7.1 for LCBC, BETULA, ADNI and AIBL, v6.0 for NESDA), yielding a reconstructed cortex and subcortex for each participant and timepoint109,110. Data for the main discovery sample comprised T1w magnetization prepared rapid gradient echo (MPRAGE) sequences collected on 3 scanners at Oslo University Hospital; a 1.5 T Avanto (599 scans), a 3 T Skyra (769 scans), and a 3 T Prisma (62 scans; Siemens Medical Solutions, Germany).
A priori ROIs
We first analyzed subcortical and cortical volumes for a priori defined ROI’s based on known AD vulnerability. These were based on the Braak staging scheme, initially defined using post-mortem measures of tau50 and subsequently applied to in vivo imaging111. Similar to others111,112, we used FreeSurfer regions from the aseg and Desikan-Killiany (DK) atlas113 that anatomically approximate the various stages (see https://jagustlab.neuro.berkeley.edu/s/Braak_ROI-3l2g.pdf). ROIs were constructed separately per hemisphere7. After initial analyses with our main hippocampal ROI’s—corresponding to Braak Stage II50—we analyzed ROI’s corresponding to Stages I (entorhinal) and Stage III50, the latter we subdivided into a subcortical (amygdala) and a composite cortical ROI (parahippocampal, fusiform, lingual).
Data-driven ROIs
To empirically derive brain features with accelerated change in AD, we used machine learning in ADNI data (below) on a total of 364 features from the aseg and DK atlas113, comprising measures of cortical volume, area, thickness, grey matter/white matter contrast, subcortical volume and intensity (Fig. 3). This set of 364 features was also extracted and modelled within the discovery and replication samples.
Statistics and reproducibility
Age-relative brain change across the adult lifespan
We used Generalized Additive Mixed Models (GAMMs, gamm4 v 0.2-6114) to estimate age models for each of the 364 brain features, fitting a nonlinear term for age, with covariates added for sex, scanner, and intracranial volume (knots = 8). No statistical method was used to predetermine sample size: we used all longitudinal MRI data from those meeting the inclusion criteria. We specified random intercepts and slopes for each participant. This enabled fitting an individual-specific linear model (level and slope) across all of their timepoints, to estimate how each person’s slope as a function of age deviates from the average nonlinear estimation. For an age model of e.g., hippocampus, random slopes are interpretable as the extent of additional (or reduced) hippocampal change an individual exhibits relative to the predicted change given their age (taking other covariates into consideration). Hence, we refer to this as an estimate of “age-relative change”. To partition unique variance associated with individual-specific slope, the estimation requires that a number of participants have three or more timepoints, although estimates are also produced for participants with fewer, but then are drawn from a population distribution more skewed towards the sample mean82. This estimation is equivalent to estimating factor scores, and as such is psychometrically superior to manual calculations of change. Absolute change was calculated by adding the random slopes to the first derivative of the GAMM average age trajectory.
Polygenic risk associations
Univariate associations between PRS-AD and brain change in healthy adults
For each of our a priori ROIs, we used the random slopes as response variable in linear models with a PRS-AD predictor and the following covariates: mean age (across timepoints), sex, GAFs, the number of timepoints, and the interval between first and last timepoint. There was a tendency for older adults to have more timepoints (Supplementary Fig. 6). Hence, the latter two covariates helped ensure the effects were not driven by an uneven timepoint distribution across age. No statistical method was used to predetermine the genetic sample size: we used all quality-controlled genetic data from those in the brain change analysis. We tested the associations between PRS-AD (4 scores; tested separately) and age-relative change with progressively older age-ranges (i.e., 30-89, 35-89, 40-89 … 70-89). The reasons for this were threefold. First, because some brain features were estimated to have more negative individual-specific slopes in younger adults compared with middle-age (Supplementary Fig. 1), we could not test the association across the entire age-range (30–89) and ensure we were capturing only ageing-specific processes. Second, it enabled assessing the stability of PRS-AD associations detectable in adult lifespan data (note that older age-ranges correspond to smaller sample sizes). Third, because empirical outcomes are influenced by arbitrary analysis decisions, we took inspiration from multiverse methods that attempt to reduce such bias by testing associations across a set of theoretically justified alternatives51,52. We also tested each association with absolute change, and FDR correction was applied across all 576 PRS-AD tests (8 structures × 4 scores × 9 age-ranges × 2 change metrics; significance considered at p[FDR] < 0.05). For surviving PRS-AD associations, we tested whether the FDR-corrected association including APOE remained significant at p < 0.05 using PRS-ADnoAPOE, and determined whether the number of significant hits exceeded the 5% false positive rate per structure. We also ran post-hoc tests to confirm that the PRS-AD-change estimates became more negative as the age subset steadily comprised only older individuals (see Fig. 1e, f). Here, we used the pre-computed beta estimates from all PRS-AD-change models (age-relative and absolute; all four scores) as response variable, and the age-range as predictor (coded 0-8), and tested the linear effect of age-range upon the PRS-AD beta estimates (main effect across change models). The observed coefficient thus represents the strengthening of the negative PRS-AD-change association for each increasing age subset. Next, we permuted the empirical p-value for this observed association, by generating a null distribution across 1000 random permutations of the age variable (mean age) in the PRS-AD change associations, then recalculating the effect of age-range (randomized) upon the PRS-AD beta estimates.
Identifying features with faster atrophy in AD
We repeated the procedure to estimate age-relative change in ADNI data, fitting a GAMM of age across NC-long and AD-long groups (Fig. 3a; covariates: sex, field strength, ICV). To guard against overfitting the age trajectories and account for the roughly three-decade drop in age coverage in the AD datasets (Supplementary Table 3), we reduced the number of knots in the GAMM to 5. Next, we ran machine learning binary classification with XGBoost (https://xgboost.readthedocs.io115), using the random individual-specific slopes (age-relative change) across all 364 features as input. No statistical method was used to predetermine sample size: we used all longitudinal MRI data from those meeting our criteria (see AD samples). Hyperparameters were chosen using 10-fold cross validation across 500 random combinations of the following possible parameter values: nrounds (100–600, step = 50), eta (0.01, 0.05, 0.1, 0.15, 0.2), max_depth (2–8, step = 1), gamma (0.5–1.5, step = 0.5), min_child_weight (1–4, step = 1). To reduce the risk of overfitting to the training data and increase generalizability, we selected the final hyperparameters based on the mean AUC obtained across the 500 iterations of 10-fold cross-validation, where each iteration logged the maximum AUC achieved across folds (mean = 0.927; final hyperparameters: nrounds = 500, eta = 0.2, max_depth = 5, gamma = 1, min_child_weight = 1). This approach ensures a more robust and stable estimate of model performance across diverse data subsets while also avoiding potential overfitting to a single hyperparameter combination. For comparison, we also computed a classification model using absolute brain change as input following the same procedure (hyperparameters: nrounds = 600, eta = 0.01, max_depth = 7, gamma = 0.5, min_child_weight = 2). Model performance was evaluated in AIBL data (Fig. 3; Supplementary Fig. 8).
Multivariate associations between PRS-AD and brain change in healthy adults
First, we extracted the feature matrix to derive a list of brain features important for classifying AD-long from NC-long individuals based on age-relative change in ADNI. Then, in the LCBC healthy adult lifespan discovery sample, we calculated the principal component of age-relative change (PC1relChange) across the top 50 features, not including hippocampal and amygdala volumes (to ensure these did not drive the effect). We then used PC1relChange to test for PRS-AD associations with change in our healthy adult lifespan sample, at progressively older age-ranges, for all four scores. Next, we aimed to ensure the observed multivariate associations were not disproportionately driven by one or a few brain features. To do this, we first calculated the age at which absolute brain change accelerates, reasoning analyses within this age-range would give maximal chance of detecting PRS-AD effects upon individual ageing trajectories. Here, we took the principal component of absolute change across the same set of features (PC1absChange), plotted as a function of mean age. Then, within the 50–89 years age-range (Fig. 4c), we ran a sliding window PCA, iteratively calculating PC1 across 20 features with a step size of 3, across the first ~100 features (complete windows of 20 up to 98 features; 27 windows), and tested PC1 associations with PRS-AD within each window. FDR-correction was applied across all 144 PRS-AD tests in this analysis, and surviving associations were tested with PRS-ADnoAPOE.
As a final proof-of-principle, we applied the weights from the binary classification procedure in AD-control data directly to the healthy adult lifespan data (i.e., LCBC as test data). This prediction uses information from the weights of all 364 features. Here, the dependent variable was calculated as log[p/(1-p)], where p is the model-implied probability of having AD (probADrelChange). The aim of this was not to classify healthy individuals as AD or not, but rather test our hypothesis that the learned model weights would nevertheless prove useful, and would relate to PRS-AD in healthy adult lifespan data. We also tested whether predictions derived from the ML model based on absolute change were related to PRS-AD. Again, FDR-correction was applied across all 72 PRS-AD tests in this analysis, and surviving associations were tested with PRS-ADnoAPOE.
Replication analysis
We first ran a GAMM separately in each of the replication cohorts, revealing a strong outlier for each in the hippocampal change data (−7.4 SD in BETULA; +5.5 SD in NESDA; see Supplementary Fig. 12). Then, we collated the data from the two cohorts, ran a GAMM comparable to the main analysis estimating the random slopes, and excluded these two outliers. Note that since each of the two cohorts originated from a single scanner, the scanner covariate indexed study cohort. No statistical method was used to predetermine sample size: we used all longitudinal MRI data from those meeting the inclusion criteria. Similar to the main analysis, we expected including as many longitudinal observations as possible in the GAMM would optimize the change estimates for all. Testing this assumption post-hoc, we found that in the same individuals with genetic data from BETULA, beta estimates with left hippocampal change were significantly lower when their random slopes were estimated together with NESDA data, relative to only using BETULA data (p = 0.009; Supplementary Fig. 13). To reduce the number of tests, we tested PRS-AD associations with change in hippocampus and amygdala, and with PC1relChange (top 50 AD-accelerated features excluding hippocampal and amygdala volumes). PRS-AD models matched the discovery sample, except for an added cohort covariate. We tested the model at progressively older age-ranges for all four scores (here until a lower age-bound of 60, above which the genetic sample was comprised entirely of BETULA subjects). Where the association was significant (p < 0.05 [uncorrected]), we tested whether it remained significant with PRS-ADnoAPOE. We considered it a replication where the number of significant tests per structure exceeded the 5% false positive rate. Lastly, we assessed whether the trajectory of accelerated brain ageing in AD features mirrored the discovery sample (i.e., modelled PC1absChange as a function of mean age).
Memory change analysis
Finally, we tested differences in memory change between groups of individuals defined by the conjunction of brain and genetic risk markers. We hypothesized higher PRS-AD individuals also high on a multivariate marker of brain change would show more memory decline across the adult lifespan. This analysis proceeded in two parts. First, we took the principal component across the four PRS-AD scores (PC1PRS-AD; explaining 87%), and used the partial association between PC1PRS-AD and the principal component across the first 50 AD-accelerated features (here including hippocampal and amygdala volumes), to divide individuals into quadrant groups (Fig. 6d; pink group depicts individuals high on both risk factors; covariates: mean age, sex, GAFs, N timepoints, and interval between first and last timepoint). Second, from the full adult lifespan discovery sample described above (N = 420; scans = 1430), we identified those with observations on the California Verbal Learning Test (CVLT)116,117. Of these, we discarded individuals with non-usable memory data (due to being part of on-off memory training projects at LCBC; see Supplementary Note 1 for information on the projects that comprised the LCBC sample). In the resulting data (713 observations from 267 individuals), we took the principal component across the three main CVLT subtests (learning, immediate, and delayed free recall; scaled) to index general memory, expressed the loadings as a proportion of the maximum loading, and kept only those with longitudinal memory observations (707 observations from 261 individuals). Then, we ran a GAMM of age on Memory (sex corrected, knots = 8). Akin to the brain analysis, age-relative memory change was estimated via the random slopes, and absolute memory change was calculated by adding the slopes to the first derivative of the GAMM average age trajectory. Having estimated memory change using as many longitudinal CVLT observations as possible—108 individuals had both memory change and genetic data (i.e., were included in the quadrant-groups). Finally, we tested our hypothesis that the high brain change-high PRS-AD group would exhibit more adult lifespan memory decline, setting this group to the intercept, in linear models of quadrant-group on memory change, correcting for group differences in mean age, sex, N timepoints, interval between first and last timepoint, and APOE-ε4 carriership (main model). These were tested using both age-relative and absolute memory change. For these tests—where the intercept group was hypothesized to show the most negative change—we corrected for multiple comparisons applying FDR-correction across the six group comparisons in the two main change models using one-sided tests. Alternative models correcting for the number of APOE-ε4 alleles, baseline memory, PC1PRS-AD, and PC1relChange1-50, were also run, and the main model was retested using change estimates within different age subsets.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
Scripts were run on the Colossus processing cluster at the University of Oslo, and on resources provided by UNINETT Sigma2 (project NN9769K). LCBC funding: grant 302854 (FRIPRO; to Y.W.), European Research Council under grants 283634, 725025 (to A.M.F.), and 313440 (to K.B.W.); Norwegian Research Council (to A.M.F. and K.B.W.) under grants 249931 (TOPPFORSK) and, The National Association for Public Health’s dementia research program, Norway (to A.M.F). The Lifebrain project is funded by the EU Horizon 2020 Grant: “Healthy minds 0–100 years: Optimising the use of European brain imaging cohorts (Lifebrain).” Grant agreement number: 732592. The Betula project was supported by a Scholar grant from Knut and Alice Wallenberg’s (KAW) foundation to L.N. The infrastructure for the NESDA study (www.nesda.nl) is funded through the Geestkracht program of the Netherlands Organisation for Health Research and Development (ZonMw, grant number 10-000‐1002) and financial contributions by participating universities and mental health care organizations (VU University Medical Center, GGZ inGeest, Leiden University Medical Center, Leiden University, GGZ Rivierduinen, University Medical Center Groningen, University of Groningen, Lentis, GGZ Friesland, GGZ Drenthe, Rob Giel Onderzoekscentrum). Some of the data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. ADNI investigators contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. Some of the data used in the preparation of this article were obtained from the Australian Imaging Biomarkers and Lifestyle Flagship Study of Ageing (AIBL) funded by the Commonwealth Scientific and Industrial Research Organisation (CSIRO), which was made available at the ADNI database (www.loni.usc.edu/ADNI). AIBL investigators contributed to the design and implementation of AIBL and/or provided data but did not participate in analysis or writing of this report.
Author contributions
J.M.R., Y.W., D.V.-P., Ø.S., K.B.W. and A.M.F. conceived the study. J.M.R. designed, developed and performed the analysis. Ø.S., D.V.-P, H.G., E.H.L., R.A.K. and P.G. contributed statistical expertise. Ø.S. contributed to code development. A.M. managed, created and curated cross-site data infrastructure. Y.W., L.B., M.A., Y.M. and M.P. managed/curated/transferred genetic data. L.Nawijn, Y.M. and B.P. procured/managed/curated/transferred NESDA data. L.Nyberg, M.A. and S.P. procured/managed/curated/transferred BETULA data. A.C.S.B., J.K., E.M.F., K.Ø. procured recent MRI data for LCBC included here. D.V.-P., Ø.S., H.G., E.H.L., O.I., M.S., L.Nawijn, Y.M., M.A., S.P., A.C.S.B., K.Ø., R.A.K., K.P.E., U.L., P.G., N.D., C-J.B., C.A.D., B.P., L.B., L.Nyberg, K.B.W., A.M.F. and Y.W. reviewed/edited paper drafts. ADNI investigators (M.W., P.A., R.P.) contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. AIBL investigators (C.L.M., C.C.R.) contributed to the design and implementation of AIBL and/or provided data but did not participate in analysis or writing of this report. Y.W. provided supervision. Y.W., K.B.W. and A.M.F. procured project funding. J.M.R. created figures and wrote the paper, with substantive input from all authors.
Peer review
Peer review information
Nature Communications thanks Vijaya Kolachalama, who co-reviewed with Varuna JasodanandDenise Park, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The individual-level data supporting the results of the current study may be available upon request, given appropriate ethical and data protection approvals. Different limitations on data access apply to different samples. Participants in LCBC, BETULA and NESDA have not consented to share their data publicly online. Requests for the raw data can be submitted to the relevant principal investigator of each contributing study. Contact details are provided in Supplementary Note 2. ADNI and AIBL data are available at https://adni.loni.usc.edu/data-samples/access-data/ pending application approval and compliance with the data usage agreement. Summary-level source data are provided with this paper. Source data are available as a source data file. Source data are provided with this paper.
Code availability
Code for statistical analyses is available at https://github.com/jamesmroe/ADchangeRisk (archived at 10.5281/zenodo.13844701).
Competing interests
CAD is a consultant, board member, and stock owner in the analytical laboratory Vitas Ltd, Oslo, Norway. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lists of authors and their affiliations appears at the end of the paper.
Contributor Information
James M. Roe, Email: j.m.roe@psykologi.uio.no
Alzheimer’s Disease Neuroimaging Initiative:
Michael Weiner, Paul Aisen, and Ronald Petersen
the Australian Imaging Biomarkers and Lifestyle Flagship Study of Ageing:
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53548-z.
References
- 1.Sexton, C. E. et al. Accelerated changes in white matter microstructure during aging: a longitudinal diffusion tensor imaging study. J. Neurosci.34, 15425–15436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walhovd, K. B. et al. Genetic risk for Alzheimer disease predicts hippocampal volume through the human lifespan. Neurol. Genet.6, e506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidal-Pineiro, D. et al. Cellular correlates of cortical thinning throughout the lifespan. Sci. Rep.10, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fjell, A. M., McEvoy, L., Holland, D., Dale, A. M. & Walhovd, K. B. What is normal in normal aging? Effects of aging, amyloid and Alzheimer’s disease on the cerebral cortex and the hippocampus. Prog. Neurobiol.117, 20–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fjell, A. M., McEvoy, L., Holland, D., Dale, A. M. & Walhovd, K. B. Brain changes in older adults at very low risk for Alzheimer’s disease. J. Neurosci.33, 8237–8242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fjell, A. M. et al. One-year brain atrophy evident in healthy aging. 29, 15223–15231 (2009). [DOI] [PMC free article] [PubMed]
- 7.Roe, J. M. et al. Asymmetric thinning of the cerebral cortex across the adult lifespan is accelerated in Alzheimer’s disease. Nat. Commun.12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braak, H. & Braak, E. Staging of Alzheimer-related cortical destruction. Rev. Clin. Neurosci.33, 403–408 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci.19, 687–700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bethlehem, R. A. I. et al. Brain charts for the human lifespan. Nature604, 525–533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutherford, S. et al. Charting brain growth and aging at high spatial precision. Elife11, 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walhovd, K. B. et al. Neurodevelopmental origins of lifespan changes in brain and cognition. Proc. Natl Acad. Sci. USA.113, 9357–9362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raz, N. et al. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cereb. Cortex15, 1676–1689 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Fjell, A. M. et al. Accelerating cortical thinning: unique to dementia or universal in aging? Cereb. Cortex24, 919–934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corrada, M. M., Brookmeyer, R., Paganini-Hill, A., Berlau, D. & Kawas, C. H. Dementia incidence continues to increase with age in the oldest old the 90+ study. Ann. Neurol.67, 114–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorm, A. & Jolley, D. The incidence of dementia: a meta-analysis. Neurology51, 728–733 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Desikan, R. S. et al. Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med.14, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altmann, A. et al. A comprehensive analysis of methods for assessing polygenic burden on Alzheimer’s disease pathology and risk beyond APOE. Brain Commun.2, fcz047 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logue, M. W. et al. Use of an Alzheimer’s disease polygenic risk score to identify mild cognitive impairment in adults in their 50 s. Mol. Psychiatry24, 421–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyall, D. M. et al. Association between APOE e4 and white matter hyperintensity volume, but not total brain volume or white matter integrity. Brain Imaging Behav.14, 1468–1476 (2020). [DOI] [PMC free article] [PubMed]
- 21.Machulda, M. M. et al. Effect of APOE? 4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch Neurol.68, 1131–1136 (2011). [DOI] [PMC free article] [PubMed]
- 22.Habes, X. M. et al. Relationship between APOE genotype and structural MRI measures throughout adulthood in the study of health in Pomerania population-based cohort. AJNR Am. J. Neuroradiol.37, 1636–42 (2016). [DOI] [PMC free article] [PubMed]
- 23.Bunce, D. et al. APOE genotype and entorhinal cortex volume in non-demented community-dwelling adults in midlife and early old age. J. Alzheimer’s Dis.30, 935–942 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Henson, R. N. et al. Effect of apolipoprotein E polymorphism on cognition and brain in the Cambridge Centre for Ageing and Neuroscience cohort. Brain Neurosci. Adv.4, 2398212820961704 (2020). [DOI] [PMC free article] [PubMed]
- 25.Jack, C. R. et al. Age, sex, and APOEε4 effects on memory, brain structure, and β-amyloid across the adult life span. JAMA Neurol.72, 511-9 (2022). [DOI] [PMC free article] [PubMed]
- 26.Protas, H. D. et al. Posterior cingulate glucose metabolism, hippocampal glucose metabolism, and hippocampal volume in cognitively normal, late-middle-aged persons at 3 levels of genetic risk for Alzheimer disease. JAMA Neurol.70, 320–325 (2013). [DOI] [PMC free article] [PubMed]
- 27.Foo, H. et al. Associations between Alzheimer’s disease polygenic risk scores and hippocampal subfield volumes in 17, 161 UK biobank participants. Neurobiol. Aging98, 108–115 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Du, J. et al. Exploration of Alzheimer’s disease MRI biomarkers using APOE4 carrier status in the UK biobank. medRxiv10.1101/2021.09.09.21263324 (2021).
- 29.Knickmeyer, R. C. et al. Common variants in psychiatric risk genes predict brain structure at birth. Cerebral Cortex.24, 1230–1246 (2014). [DOI] [PMC free article] [PubMed]
- 30.Axelrud, L. K. et al. Polygenic risk score for Alzheimer’s disease: implications for memory performance and hippocampal volumes in early life. Am. J. Psychiatry175, 555–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foley, S. F. et al. Multimodal brain imaging reveals structural differences in Alzheimer’s disease polygenic risk carriers: a study in healthy young adults. Biol. Psychiatry81, 154–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mormino, E. C. et al. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology87, 481–488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fjell, A. M. et al. Self-reported sleep relates to hippocampal atrophy across the adult lifespan—results from the lifebrain consortium. Sleep43, zsz280 (2019). [DOI] [PMC free article] [PubMed]
- 34.Donix, M. et al. Longitudinal changes in medial temporal cortical thickness in normal subjects with the APOE-4 polymorphism. Neuroimage53, 37–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, P. H. et al. Apolipoprotein e genotype is associated with temporal and hippocampal atrophy rates in healthy elderly adults: a tensor-based morphometry study. J. Alzheimer’s Dis.23, 433–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison, T. M., Mahmood, Z., Lau, E. P., Karacozoff, A. M. & Alison, C. An Alzheimer’s disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro3, ENEURO.0098-16.2016 (2016). [DOI] [PMC free article] [PubMed]
- 37.Taylor, J. L. et al. Neurobiology of Aging APOE-epsilon4 and aging of medial temporal lobe gray matter in healthy adults older than 50 years. NBA35, 2479–2485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorbach, T. et al. Longitudinal association between hippocampus atrophy and episodic-memory decline in non-demented APOE ε4 carriers. Alzheimer’s Dement. Diagnosis. Assess. Dis. Monit.12, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brouwer, R. M. et al. Genetic variants associated with longitudinal changes in brain structure across the lifespan. Nat. Neurosci.25, 421–432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braak, H. & Braak, E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging16, 271–284 (1995). [DOI] [PubMed] [Google Scholar]
- 41.La Joie, R. et al. Prospective longitudinal atrophy in Alzheimer’s disease correlates with the intensity and topography of baseline tau-PET. Sci. Transl. Med.12, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roe, J. M. et al. Asymmetric thinning of the cerebral cortex across the adult lifespan is accelerated in Alzheimer’s disease. bioRxiv10.1101/2020.06.18.158980 (2020). [DOI] [PMC free article] [PubMed]
- 43.Marioni, R. E. et al. Genetic stratification to identify risk groups for Alzheimer’s disease. J. Alzheimer’s Dis.57, 275–283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayden, K. M., Lutz, M. W., Kuchibhatla, M., Germain, C. & Plassman, B. L. Effect of APOE and CD33 on cognitive decline. PLoS ONE10, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caselli, R. J. et al. Longitudinal modeling of age-related memory decline and the APOE ε4 Effect. N. Engl. J. Med.361, 255–263 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kauppi, K., Rönnlund, M., Nordin Adolfsson, A., Pudas, S. & Adolfsson, R. Effects of polygenic risk for Alzheimer’s disease on rate of cognitive decline in normal aging. Transl. Psychiatry10, 250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salthouse, T. A. What and when of cognitive aging. Curr. Dir. Psychol. Sci.13, 140–144 (2004). [Google Scholar]
- 48.Salthouse, T. A. Why are there different age relations in cross-sectional and longitudinal comparisons of cognitive functioning? Curr. Dir. Psychol. Sci.23, 252–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris, S. E. et al. Polygenic risk for Alzheimer’s disease is not associated with cognitive ability or cognitive aging in non-demented older people. J. Alzheimer’s Dis.39, 565–574 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol.82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 51.Simonsohn, U., Simmons, J. P. & Nelson, L. D. Specification curve analysis. Nat. Hum. Behav.4, 1208–1214 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Steegen, S., Tuerlinckx, F., Gelman, A. & Vanpaemel, W. Increasing transparency through a multiverse analysis. Perspect Psychol. Sci.11, 702–712 (2016). [DOI] [PubMed]
- 53.Heijer, T. D. et al. Magnetic resonance imaging in early dementia and cognitive decline. Cochrane Database Syst. Rev.3, CD009628 (2010).
- 54.Jack, C. R. et al. Comparison of different MRI brain athrophy rate measures with clinical disease progression in AD. Neurology62, 591–600 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lupton, M. K. et al. The effect of increased genetic risk for Alzheimer’s disease on hippocampal and amygdala volume. Neurobiol. Aging40, 68–77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chauhan, G. et al. Association of Alzheimer’s disease GWAS loci with MRI markers of brain aging. Neurobiol. Aging36, 1765.e7–1765.e16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walhovd, K. B., Lövden, M. & Fjell, A. M. Timing of lifespan influences on brain and cognition. Trends Cogn. Sci. 27, 901–915 (2023). [DOI] [PubMed]
- 58.Storsve, A. B. et al. Differential longitudinal changes in cortical thickness, surface area and volume across the adult life span: regions of accelerating and decelerating change. J. Neurosci.34, 8488–8498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salthouse, T. A. Attrition in longitudinal data is pimarily selective with respect to level rather than rate of change. J. Int. Neuropsychol. Soc.25, 618–623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lambert, J. C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet.45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papenberg, G., Lindenberger, U. & Bäckman, L. Aging-related magnification of genetic effects on cognitive and brain integrity. Trends Cogn. Sci.19, 506–514 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei, Y. et al. Statistical testing in transcriptomic-neuroimaging studies: A how-to and evaluation of methods assessing spatial and gene specificity. Hum. Brain Mapp.43, 885–901 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franzmeier, N. et al. The BIN1 rs744373 SNP is associated with increased tau-PET levels and impaired memory. Nat. Commun.10, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Therriault, J. et al. Association of apolipoprotein e ϵ4 with medial temporal tau independent of amyloid-β. JAMA Neurol.77, 470–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mesulam, M. A. Plasticity-based theory of the pathogenesis of Alzheimer’s disease. Ann. N. Y. Acad. Sci. 924, 42–52 (2000). [DOI] [PubMed]
- 67.Walhovd, K. B. et al. Premises of plasticity—and the loneliness of the medial temporal lobe. Neuroimage131, 48–54 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Douaud, G. et al. A common brain network links development, aging, and vulnerability to disease. Proc. Natl Acad. Sci. USA. 111, 17648–17653 (2014). [DOI] [PMC free article] [PubMed]
- 69.Jack, C. R. et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol.12, 207–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hardy, J. A., Higgins, G. A., Hardy, J. A. & Higgins, G. A. Alzheimer’s disease: the amyloid cascade hypothesis. Science256, 184–185 (1992). [DOI] [PubMed] [Google Scholar]
- 71.Herrup, K. The case for rejecting the amyloid cascade hypothesis. Nat. Neurosci.18, 794–799 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Korologou-Linden, R. et al. The causes and consequences of Alzheimer’s disease: phenome-wide evidence from mendelian randomization. Nat. Commun.13, 4726 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet.51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet.51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Q. et al. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat. Commun.11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Rojas, I. et al. Common variants in Alzheimer’s disease and risk stratification by polygenic risk scores. Nat. Commun.12, 3417 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Escott-Price, V. & Hardy, J. Genome-wide association studies for Alzheimer’s disease: bigger is not always better. Brain Commun.4, 1–7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Korczyn, A. D. & Grinberg, L. T. Is Alzheimer disease a disease? Nat. Rev. Neurol.20, 245–251 (2024). [DOI] [PubMed] [Google Scholar]
- 79.Tucker-Drob, E. M. Cognitive aging and dementia: a life-span perspective. Annu. Rev. Dev. Psychol.1, 177–196 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cole, J. H. & Franke, K. Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci.40, 681–690 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Vidal-Pineiro, D. et al. Individual variations in ‘brain age’ relate to early-life factors more than to longitudinal brain change. Elife10, 1–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Demidenko, E. Mixed Models: Theory and Applications With R. (John Wiley & Sons, 2013).
- 83.Pinheiro, J. & Bates, D. Mixed-Effects Models in S and S-PLUS 1st edn, Vol. 528 (Springer-Verlag, 2000).
- 84.Nelson, E. A. & Dannefer, D. Aged heterogeneity: fact or fiction? the fate of diversity in gerontological research. Gerontologist32, 17–23 (1992). [DOI] [PubMed] [Google Scholar]
- 85.Ferreira, D., Nordberg, A. & Westman, E. Biological subtypes of Alzheimer disease a systematic review and meta-analysis. Neurology. 94, 436–448 (2020). [DOI] [PMC free article] [PubMed]
- 86.Vogel, J. W. et al. Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nat. Med.27, 871–881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohanty, R., Ferreira, D., Nordberg, A. & Westman, E. Associations between different tau ‑ PET patterns and longitudinal atrophy in the Alzheimer’s disease continuum: biological and methodological perspectives from disease heterogeneity. Alzheimers. Res. Ther. 15, 37 (2023). [DOI] [PMC free article] [PubMed]
- 88.Cardinale, F. et al. Validation of FreeSurfer-estimated brain cortical thickness: comparison with histologic measurements. Neuroinformatics12, 535–542 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Han, X. et al. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage32, 180–194 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Leng, Y., Ng, K. E. T., Vogrin, S. J., Meade, C. & Ngo, M. Comparative utility of manual versus automated segmentation of hippocampus and entorhinal cortex volumes in a memory clinic sample. J. Alzheimers Dis.68, 159–171 (2019). [DOI] [PubMed]
- 91.Hedges, E. P. et al. Reliability of structural MRI measurements: the effects of scan session, head tilt, inter-scan interval, acquisition sequence, FreeSurfer version and processing stream. Neuroimage246, 118751 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McGuire, S. A. et al. Reproducibility of quantitative structural and physiological MRI measurements. Brain Behav.7, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ossenkoppele, R. et al. Amyloid and tau PET-positive cognitively unimpaired individuals are at high risk for future cognitive decline. Nat. Med.28, 2381–2387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Folstein, M. F., Folstein, S. E. & McHugh, P. R. Mini-mental state’. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res.12, 189–198 (1975). [DOI] [PubMed] [Google Scholar]
- 95.Beck, A. T., Ward, C., Mendelson, M., Mock, J. & Erbaugh, J. An inventory for measuring depression. Arch. Gen. Psychiatry4, 561 (1961). [DOI] [PubMed] [Google Scholar]
- 96.Yesavage, J. A. et al. Development and validation of a geriatric depression screening scale: a preliminary report. J. Psychiatr. Res.17, 37–49 (1982). [DOI] [PubMed] [Google Scholar]
- 97.Nilsson, L. G. et al. The Betula prospective cohort study: memory, health, and aging. Aging Neuropsychol. Cogn.4, 1–32 (1997). [Google Scholar]
- 98.Penninx, B. W. J. H. et al. Cohort profile of the longitudinal Netherlands study of depression and anxiety (NESDA) on etiology, course and consequences of depressive and anxiety disorders. J. Affect. Disord.287, 69–77 (2021). [DOI] [PubMed] [Google Scholar]
- 99.Weiner, M. W. et al. The Alzheimer’s disease neuroimaging initiative: a review of papers published since its inception. Alzheimer’s Dement.8, S1–S68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chow, N. et al. Comparing 3T and 1.5T MRI for mapping hippocampal atrophy in the Alzheimer’s disease neuroimaging initiative. Am. J. Neuroradiol.36, 653–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ellis, K. A. et al. The Australian imaging, biomarkers and lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int. Psychogeriatr.21, 672–687 (2009). [DOI] [PubMed] [Google Scholar]
- 102.Grasby, K. L. et al. The genetic architecture of the human cerebral cortex. Science367, eaay6690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hong, S. et al. TMEM106B and CPOX are genetic determinants of cerebrospinal fluid Alzheimer’s disease biomarker levels. Alzheimer’s Dement.17, 1628–1640 (2021). [DOI] [PubMed] [Google Scholar]
- 104.Wightman, D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet.53, 1276–1282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Auton, A. et al. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet.2, 2074–2093 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reuter, M., Rosas, H. D. & Fischl, B. Highly accurate inverse consistent registration: a robust approach. Neuroimage53, 1181–1196 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fischl, B., Sereno, M. I., Tootell, R. B. & Dale, A. M. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum. Brain Mapp.8, 272–284 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fischl, B., Sereno, M. I. & Dale, A. M. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage9, 195–207 (1999). [DOI] [PubMed] [Google Scholar]
- 111.Schöll, M. et al. PET imaging of Tau deposition in the aging human brain. Neuron89, 971–982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Franzmeier, N. et al. Functional brain architecture is associated with the rate of tau accumulation in Alzheimer’s disease. Nat. Commun.11, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage31, 968–980 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Wood, S. & Scheipl, F. gamm4: Generalized Additive Mixed Models Using ‘mgcv’ and ‘lme4’. R Package Version 0.2-5.https://cran.r-project.org/web/packages/gamm4/gamm4.pdf (2017).
- 115.Chen, T. & Guestrin, C. XGBoost: A scalable tree boosting system. Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min. 10.1145/2939672.2939785 (2016).
- 116.Delis, D. C., Kramer, J. H., Kaplan, E. & Thompkins, B. A. O. CVLT: California Verbal Learning Test-Adult Version: Manual, Vol. 91 (Psychological Corporation, 1987).
- 117.Roe, J. M. Brain Change Trajectories in Healthy Adults Correlate with Alzheimer’s Related Genetic Variation and Memory Decline Across Life.https://github.com/jamesmroe/ADchangeRisk (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The individual-level data supporting the results of the current study may be available upon request, given appropriate ethical and data protection approvals. Different limitations on data access apply to different samples. Participants in LCBC, BETULA and NESDA have not consented to share their data publicly online. Requests for the raw data can be submitted to the relevant principal investigator of each contributing study. Contact details are provided in Supplementary Note 2. ADNI and AIBL data are available at https://adni.loni.usc.edu/data-samples/access-data/ pending application approval and compliance with the data usage agreement. Summary-level source data are provided with this paper. Source data are available as a source data file. Source data are provided with this paper.
Code for statistical analyses is available at https://github.com/jamesmroe/ADchangeRisk (archived at 10.5281/zenodo.13844701).