Abstract
Pod dehiscence facilitates seed dispersal in wild legumes while indehiscence is a key domestication trait in cultivated ones. However, the evolutionary genetic mechanisms underlying its diversity are largely unclear. In this study, we compared transcriptomes of two warm-season (Glycine spp. and Phaseolus spp.) and two cool-season (Pisum spp. and Medicago ruthenica) legumes in analysis of dehiscent and indehiscent pod genotypes. Differentially expressed genes in AP2/ERF-like transcription factors and seven structural gene families, including lactoperoxidase, laccase, and cellulose synthase-interactive proteins, which are involved in secondary cell wall component accumulation, were identified to exert key roles in pod dehiscence variation. In accordance with this, higher lignin and cellulose contents were observed in pod secondary cell wall of dehiscent accessions of soybean and pea; however, the variation patterns of lignin polymers in soybean (accumulation) and pea (proportion) differed between dehiscent and indehiscent pods. Moreover, genome-wide comparative analysis revealed that orthogroups represented <1% of all identified differentially expressed genes could be traced among the four genera of legumes, while recruiting paralogous members may constitute the genetic robustness of legume pod dehiscence. This study compared the genetic mechanism among several legumes in pod dehiscence formation and revealed a compensating role of paralogous redundancy of involved gene families in seed dispersal, which can guide crop breeding.
Keywords: domestication, genetic basis, gene expression, lignin, legumes, pod dehiscence
Significance.
Pod dehiscence across several legume species was related to paralogous instead of orthologous genes from multiple gene families including AP2/ERF-like and genes affecting secondary cell wall formation and lignin/cellulose content, which were differentially expressed between pod dehiscent and indehiscent accessions.
Introduction
Seed dispersal is an essential driver of the diversity affecting the structure, composition, and spatial arrangement of plant populations in natural environments (Aslan et al. 2019). Changes in seed dispersal are also one of the most visible domestication syndromes (Hammer 1984) in crops including cereals such as rice (Oryza sativa) or legumes as soybean (Glycine max) and pea (Pisum sativum) (Fuller and Allaby 2009). Pod dehiscence or seed shattering during harvest is one of the major challenges in agricultural mechanization resulting in yield losses, especially under high temperatures and arid conditions (Maity et al. 2021). Therefore, varieties with dehiscence/shattering resistance have been a common goal (Ogutcen et al. 2018; Di Vittori et al. 2019; Parker, Lo, and Gepts 2021). Leguminosae (legumes) is one of the most important angiosperm plant family in the global ecosystem and the second largest in the case of economic crops (Azani et al. 2017; Zhao et al. 2021). Pod dehiscence is suggested to be under convergent evolution (Lenser and Theißen 2013; Fuller et al. 2014; Chamberlain-Irwin and Hufford 2022), in which independent selections lead to similar changes in different regions of domestication and historical periods among multiple legumes (Parker, Lo, and Gepts 2021). Elucidation of the genetic basis and evolutionary mechanisms underlying pod dehiscence diversity is important to understand the adaption of wild legume species and also in the breeding of pod dehiscence-resistant crop varieties.
Seed dispersal via pod dehiscence is a result of interactions between multiple pod tissues. The pod wall of legumes consists of the exocarp (epidermal cells), mesocarp (parenchyma cells), and endocarp (sclerenchyma and inner epidermal layer), and the dehiscence zone at the ventral suture terminates in the fiber cap-mesocarp boundary formed by the sclerenchyma at the pod-shell boundary (Ogutcen et al. 2018; Parker, Lo, and Gepts 2021). When the pod is desiccating and dried, differences in the cellular water content of the pod wall lead to differences in shrinkage and twist, resulting in pod dehiscence (Armon et al. 2011; Ogutcen et al. 2018; Parker, Lo, and Gepts 2021). Furthermore, developmental anatomical changes in endocarp sclerenchyma and fiber cap cells (FCCs) of the ventral suture have been identified as major drivers of pod dehiscence in various legumes (Hradilová et al. 2017; Di Vittori et al. 2021; Parker, de Sousa, et al. 2021; Guo et al. 2022; Yong et al. 2023). Pod wall twists show a strong positive correlation with the thickness of sclerenchyma in legumes (Takahashi et al. 2020). Moreover, the number variation of FCC can influence pod wall bond strength and pod dehiscence in soybean (Dong et al. 2014). The identification of key genetic components and regulations of anatomical differences is essential and lacking.
Various domesticated legumes including soybean, common bean (Phaseolus vulgaris), and pea have been studied in relation to pod development and dehiscence (Dong et al. 2014; Funatsuki et al. 2014; Hradilová et al. 2017; Lo et al. 2018; Rau et al. 2019; Di Vittori et al. 2021; Parker, de Sousa, et al. 2021; Guo et al. 2022; Yong et al. 2023). Soybean Pod dehiscence 1 (Pdh1, encoding a dirigent-like protein that regulates lignin deposition and biosynthesis) and SHATTERING 1-5 (SHAT1-5, a homolog of Arabidopsis NST1/2 encoding a NAC protein) were identified to contribute to pod dehiscence by modulating lignin deposition in sclerenchyma and FCC, respectively (Dong et al. 2014; Funatsuki et al. 2014). In common bean, besides the soybean Pdh1 orthologous gene PvPdh1 (Moghaddam et al. 2016; Parker, de Sousa, et al. 2021), a major quantitative trait locus (QTL) qPD5.1 was predicted to be PvMYB26, an ortholog of AtMYB26 transcription factor (TF). This gene is upregulated in early dehiscence pods and regulates the abscission zone formation of ventral suture (Rau et al. 2019; Di Vittori et al. 2021). Homologous VuMYB26 and VaMYB26 genes were found to affect pod dehiscence by regulating the development of sclerenchyma in phylogenetically related cowpea (Vigna unguiculata) and adzuki bean (Vigna angularis) (Yang and Wang 2016; Yang et al. 2017; Lo et al. 2018). Pod dehiscence in pea has been identified to be regulated by Dpo1 locus (Weeden et al. 2002; Weeden 2007), yet to be identified at gene level. Cell wall modification genes located at the same position as Dpo1 are differentially expressed between dehiscence-prone and dehiscence-resistant pea varieties (Hradilová et al. 2017). Similarly, two QTL governing pod dehiscence were identified in lentil (Lens spp.) (Cao et al. 2024). In addition, the Arabidopsis bHLH homologous TF genes INDEHISCENCE (IND) and ALACATRAS (ALC) are associated with pod dehiscence in soybean and common bean (Hu et al. 2019; Parker et al. 2020). Altogether, orthologous members of genes that regulate pod dehiscence variability seem to have often been selected in parallel during the legume domestication (Parker, Lo, and Gepts 2021; Yong et al. 2023). However, the soybean Pdh1 paralogous instead of orthologous gene in chickpea (Cicer arietinum), named CaPdh1, was recruited to regulate pod dehiscence (Aguilar-Benitez et al. 2020; Yong et al. 2023). Orthologous genes derived from a single ancestral gene could have similar functions, but paralogous genes via duplication events provide more genetic resources due to functional differentiation (Koonin 2005).
Although the anatomy of pod dehiscence has been associated with sclerenchyma and fiber cap in various legumes, underlying genetic mechanisms remain largely unknown. In the present study, we comparatively analyzed transcriptomes related to pod dehiscence in two warm-season legumes (soybean and common bean) and two cool-season legumes (pea and Medicago ruthenica). As a result, we identified that multiple gene families such as AP2/ERF-like TFs and those involved in lignin metabolic process form the common genetic mechanism regulating pod dehiscence. In corroboration with this, we found that lignin composition was altered in both soybean and pea, resulting in modified physical properties of the pod wall. Our results revealed the genetic diversity and compensatory mechanisms recruiting paralogous genes for genetic robustness in seed dispersal of legumes, providing a guide for breeding pod dehiscence-resistant varieties of legume crops.
Results
Intraspecific DEGs Between Pod Dehiscent and Indehiscent Legumes
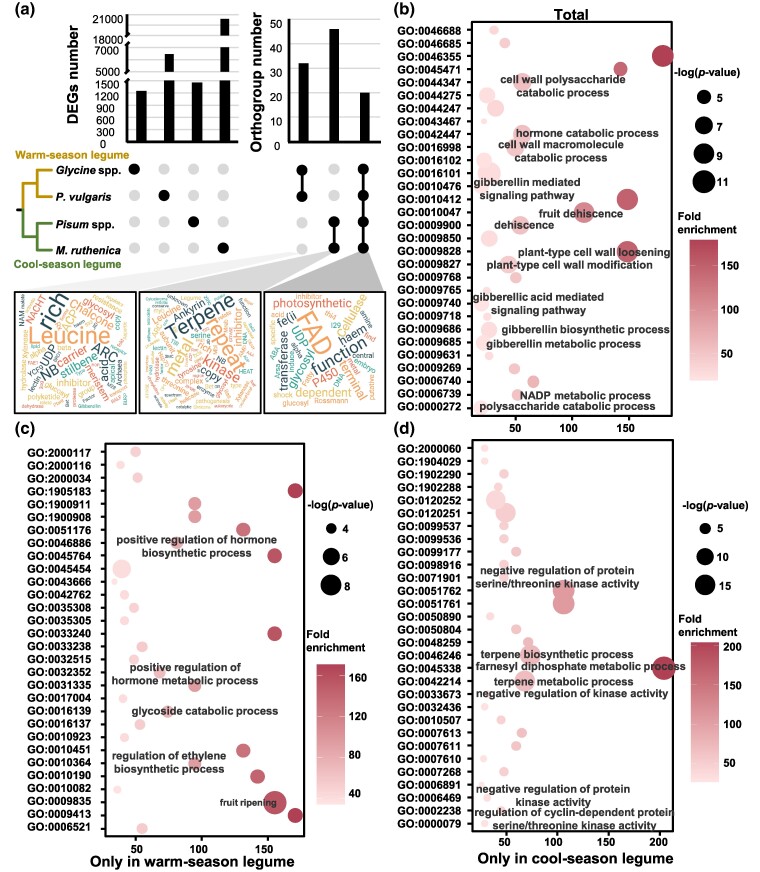
To reveal transcriptomic differences between dehiscent and indehiscent pods in studied legumes, we compared pod dehiscence-related transcriptome data from soybean (Glycine spp.: Glycine soja and G. max), common bean (P. vulgaris), pea (Pisum spp.: Pisum elatius and P. sativum), and M. ruthenica, encompassing six species across four genera (supplementary fig. S1a, Supplementary Material online). Differentially expressed genes (DEGs) between dehiscent and indehiscent pods in each species identified candidate genes, and interspecific comparison provided the subset of conserved genes. In this scenario, the number of DEGs ranged from 84 to 14,687 in different subgroups of four studied genera of legume species (supplementary fig. S1b, Supplementary Material online). After combing and de-duplicating the DEGs in different subgroups of each species, these revealed 1,252, 6,453, 1,453, and 20,391 candidate DEGs for Glycine spp., P. vulgaris, Pisum spp., and M. ruthenica, respectively (Fig. 1a). These DEGs were enriched for photosynthesis, secondary metabolism related to cell wall composition, and other biological processes like hormone synthesis/metabolism (supplementary table S1, Supplementary Material online). Interestingly, the known genes SHAT1-5, MYB26, and a bHLH gene (homolog of Arabidopsis IND and ALC) were DEGs in common bean and M. ruthenica but not in soybean and pea, whereas Pdh1 (Yong et al. 2023) was only differentially expressed in soybean and common bean, both warm-season legumes (supplementary fig. S2, Supplementary Material online). These identified DEG families thus provide gene regulatory network involved in pod dehiscence in a wider set of legumes.
Fig. 1.
DEGs for pod dehiscence in legumes. a) Orthogroups of DEGs in legumes. Left histogram: DEGs were identified in pod dehiscent and indehiscent accessions in Glycine spp. (including wild and cultivar), P. vulgaris, Pisum spp. (including wild and cultivar), and M. ruthenica. Right histogram: the orthogroup number of the coexpressed genes. The word cloud represents major protein families of the identified orthogroups. b) Top 30 GO terms of orthogroups shared by the four genera of legume species. c) Top 30 GO terms of warm-season-specific orthogroups. d) Top 30 GO terms of cool-season-specific orthogroups. Annotations of GO terms associated with pod dehiscence are given.
We identified orthologous genes within the DEGs to explore the conservation and specificity of the regulatory mechanism of pod dehiscence. Thirty-two, 46, and 20 DEGs orthogroups were found in warm-season legumes (Glycine spp. and P. vulgaris), cool-season legumes (Pisum spp. and M. ruthenica), and all four genera of legumes (six species), which accounted for <1% of the identified DEGs (Fig. 1a). Among these, the orthologous genes of flavin adenine dinucleotide-dependent oxidoreductase, fatty acid desaturase, and cellulase were shared by all legumes, suggesting a common mechanism involved in cell wall synthesis-related metabolism, hormone metabolism, and biological processes related to cell wall composition (Fig. 1a and b). The orthologous genes encoding leucine-rich repeat proteins and chalcone and stilbene synthases participating in the processes related to fruit ripening, hormone synthesis, and glucose metabolism were specifically recruited in the warm-season legumes (Fig. 1a and c). On the other hand, orthologous genes in cool-season legumes were dominated by terpene synthase and protein kinase and those related to biological processes such as primary metabolism and phosphorylation (Fig. 1a and d).
Coexpression Modules Associated with Pod Dehiscence and Indehiscence in Legumes
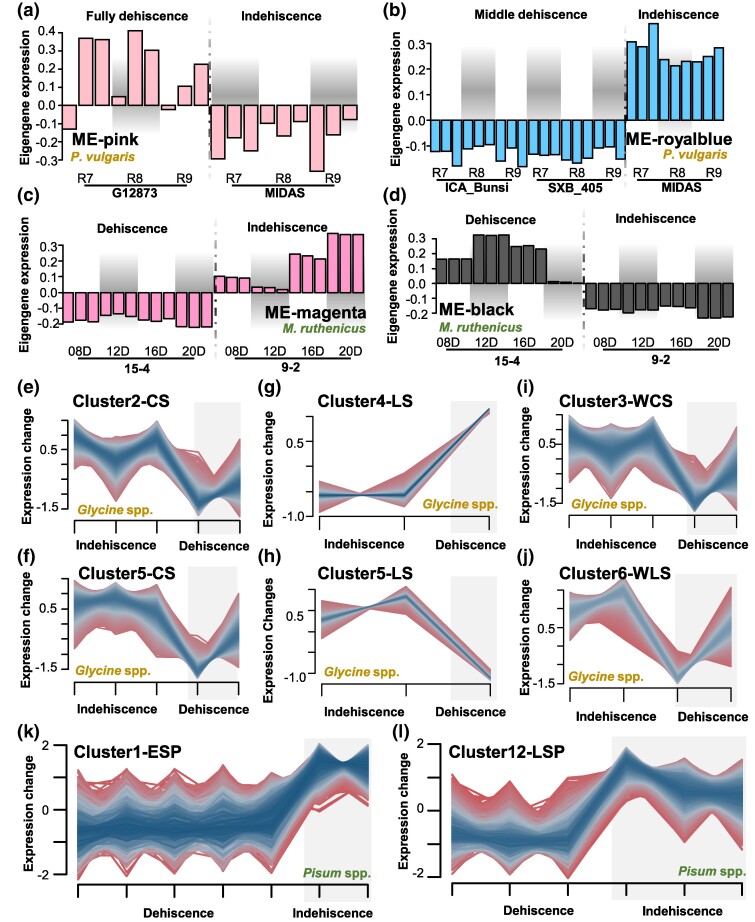
The coexpression module analysis was performed to verify the results of DEGs analysis and to reveal regulatory mechanisms controlling pod dehiscence in legumes. In common bean, weighted gene coexpression network analysis (WGCNA) revealed 22 coexpression modules (ME) when clustering completely dehiscent pods with indehiscent ones, with the pink module (ME-pink) specifically associated with the dehiscent pod phenotype, and corresponding genes implicated in cell wall composition (Fig. 2a; supplementary fig. S3a and b, Supplementary Material online). Meanwhile, the ME-royal blue from 25 coexpressed modules was associated with pod dehiscence when clustering the middle/intermediate dehiscent pods with the indehiscent pods, and genes within these modules were involved in hormonal responses and cell wall loosening (Fig. 2b; supplementary fig. S3c and d, Supplementary Material online).
Fig. 2.
Identification of expression clusters for pod dehiscence in four genera of legumes. a) Coexpression modules between fully dehiscent and indehiscent pods in common bean by WGCNA analysis. G12873, fully dehiscent pod; MIDAS, indehiscent pod. b) Coexpression modules between middle dehiscent and indehiscent pod in common bean by WGCNA analysis. ICA_Bunsi and SXB_405, middle dehiscent pod; MIDAS, indehiscent pod. In a) and b), R7, R8, and R9 represent the R7, R8, and R9 stages in the development of common bean pods. c, d) Coexpression modules of ME-magenta c) and ME-black d) between dehiscent and indehiscent pods in M. ruthenica by WGCNA analysis. 08D, 8-day pods; 12D, 12-day pods; 16D, 16-day pods; 20D, 20-day pods. 15-4, dehiscent pod; 9-2, indehiscent pod. e to j) Coexpression modules between dehiscent and indehiscent pod in cultivated e, f), landrace g, h), and wild soybean compared with cultivar i) or landrace j) by Mfuzz analysis. CS, comparison of indehiscent and dehiscent pod in cultivated soybean; LS, comparison of indehiscent and dehiscent pod in soybean landrace; WCS, comparison of indehiscent pod in cultivar and dehiscent pod in wild soybean; WLS, comparison of indehiscent pod in landrace and dehiscent pod in wild soybean. k, l) Coexpression modules between dehiscent pod and indehiscent pod in pea at early k) and late l) stage by Mfuzz analysis. ESP, comparison of indehiscent and dehiscent pods of pea at early developmental stages; LSP, comparison of indehiscent and dehiscent pods of pea at late developmental stages.
In M. ruthenica, WGCNA identified two of the 23 coexpression modules associated with pod dehiscence, specifically ME-magenta and ME-black (Fig. 2c and d; supplementary fig. S4a, Supplementary Material online). The genes within the ME-magenta were implicated in a diverse array of metabolic processes, whereas the genes within the ME-black were mainly involved in cell growth and cellular morphogenesis (supplementary fig. S4b and c, Supplementary Material online).
In soybean, two coexpression modules were found to be associated with pod dehiscence in both cultivars (Cluster2-CS and Cluster5-CS) and landraces (Cluster4-LS and Cluster5-LS). Genes within Cluster2-CS and Cluster4-LS were mainly involved in histone modification processes, while genes within Cluster5-CS and Cluster5-LS participated in cell wall composition-related metabolic processes (Fig. 2e to h; supplementary figs. S5 and S6, Supplementary Material online). When clustering with wild soybean, six and eight coexpression modules were respectively identified in cultivars and landraces, and only Cluster3-WCS and Cluster6-WLS were associated with pod dehiscence (Fig. 2i and j; supplementary fig. S7a and b, Supplementary Material online). Gene ontology (GO) enrichment analysis showed that genes within Cluster3-WCS were mostly involved in the regulatory processes such as glycan metabolism, protein phosphorylation, and microtubule cytoskeleton organization. At the same time, genes within Cluster6-WLS were also enriched in relevant cell wall biosynthesis and composition-related biological processes (supplementary fig. S7c and d, Supplementary Material online).
In pea, we analyzed two groups of early and late developmental stages for gene coexpression, and as a result, 16 coexpression modules were identified (Fig. 2k and l; supplementary figs. S8 and S9, Supplementary Material online). The Cluster1-ESP at early stage and Cluster12-LSP at late stage were associated with the pod dehiscence (Fig. 2k and l), and the GO enrichments indicated that most of the coexpressed genes at early and late stages were involved in polysaccharide metabolism and cell wall composition or synthesis (supplementary figs. S8b and S9b, Supplementary Material online).
Altogether, the above analyses further support the secondary cell wall-related metabolic processes playing a crucial role in legume pod dehiscence. The results of orthologous genes identification showed that the orthogroups of coexpressed genes in the warm-season legumes, cool-season legumes, and all four genera of legumes were 38, 10, and 8, respectively, which accounted for <1% of the number of coexpressed genes in each species (supplementary fig. S10a, Supplementary Material online). In addition to cell wall composition, GO enrichments also revealed that RNA splicing and protein phosphorylation/dephosphorylation modification associated with the abscisic acid (ABA) signaling pathways, such as pyrabactin resistance 1-like proteins (PYLs) in warm-season legumes and cyclin-dependent kinases (CDKs) in cool-season legumes, were different between dehiscent and indehiscent pods (supplementary fig. S10b to d, Supplementary Material online), implying that RNA splicing and epigenetic modification were involved in the regulating pod dehiscence.
Candidate Gene Families Involved in Pod Dehiscence
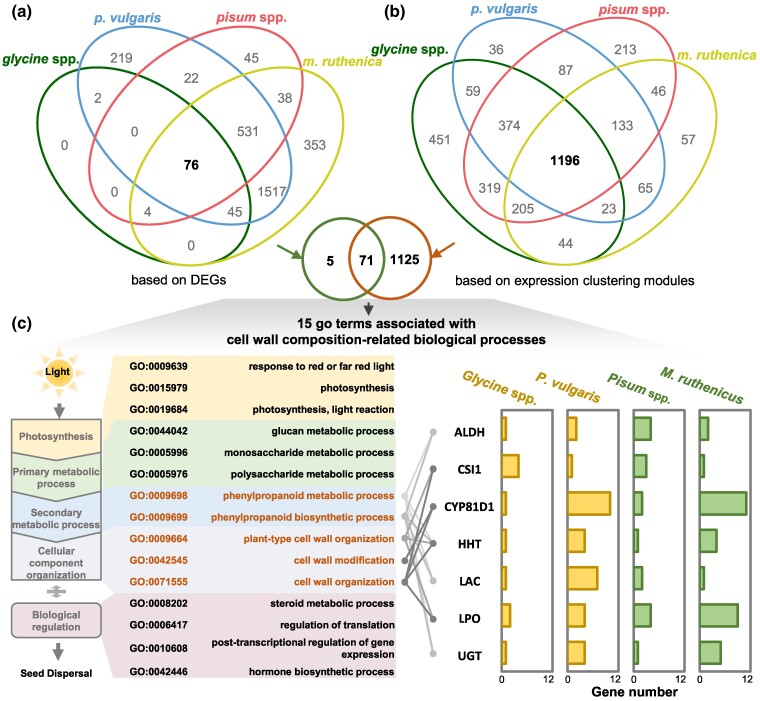
As a relatively small number of orthologous genes were identified in all four genera of studied legumes, we integrated the GO enrichment of DEGs and coexpressed genes to determine conserved biological processes regulating pod dehiscence. A total of 76 conserved GO terms were identified in DEGs (Fig. 3a) and 1,196 GO terms were shared by the four genera of legumes in coexpression genes (Fig. 3b), revealing 71 conserved GO terms. Fifteen of which were found to be involved in cell wall composition, including photosynthesis-related, glucose metabolism-related, phenylpropane metabolism pathway, cellular component organization, and hormonal regulation (Fig. 3c). We therefore focused on the five GO terms involved in secondary cell wall formation, consisting of phenylpropanoid metabolic pathway (GO: 0009698), phenylpropanoid synthesis pathway (GO: 0009699), plant-type cell wall organization (GO: 0009664), cell wall modification (GO: 0042545), and cell wall organization (GO: 0071555) (Fig. 3c). We found that members of only seven gene families were involved in these five GO terms within the four genera of legumes. These encoded lactoperoxidase (LPO), ω-hydroxypalmitate O-feruloyl transferase (HHT), UDP-glycosyltransferase (UGT), aldehyde dehydrogenase (ALDH), Cytochrome P450/isoflavone 2′-hydroxylase (CYP81D1), laccase (LAC), and cellulose synthase-interactive protein 1 (CSI1) (Fig. 3c). Different paralogous members of each gene family were differentially expressed in pods of the different legume species (supplementary fig. S11, Supplementary Material online). These seven gene families and their potential regulatory genes might be essential for pod dehiscence of legume species.
Fig. 3.
Integrative analysis of DEGs and coexpressed genes for legume pod dehiscence. a) Venn of GO enrichment of DEGs. b) Venn of GO enrichment for coexpression modules. c) GO terms related to cell wall composition. Five GO terms and seven structural gene families closely related to pod dehiscence are highlighted, and their gene numbers in DEGs and coexpression modules are given in different legume species. LPO, lactoperoxidase; HHT, ω-hydroxypalmitate O-feruloyl transferase; UGT, UDP-glycosyltransferase; ALDH, aldehyde dehydrogenase; CYP81D1, Cytochrome P450/isoflavone 2′-hydroxylase; LAC, laccase; CSI1, cellulose synthase-interactive protein 1.
AP2/ERF-Like TFs Act Potential Regulators for Pod Dehiscence
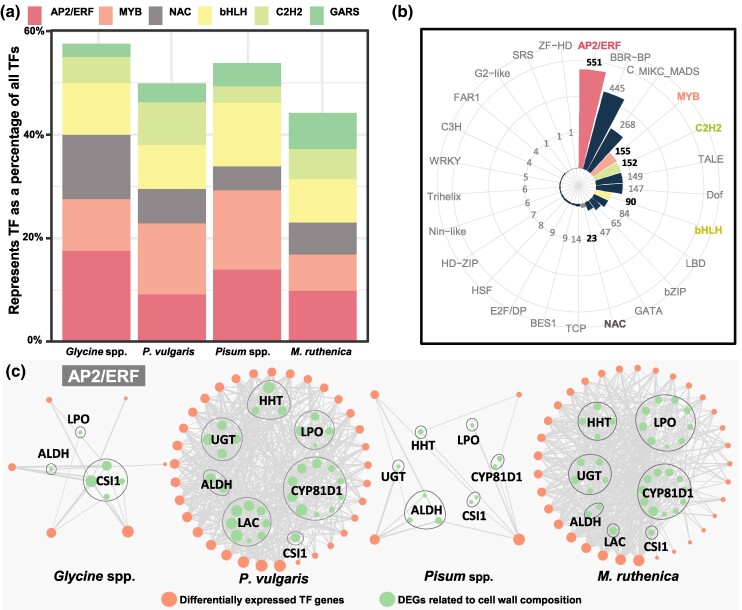
To determine the hypothesized regulators, we identified TF genes within DEGs. Multiple regulatory gene families were identified, with a high proportion of AP2/ERF, bHLH, NAC, and MYB-like TFs (Fig. 4a; supplementary fig. S12, Supplementary Material online). Next, we analyzed the cis-motifs on the promoters of seven candidate gene families to discern which TFs may have been involved in their regulations (supplementary fig. S13, Supplementary Material online). We found that their promoter regions contained the binding sites of MYB-, bHLH-, and NAC-like TFs, previously described as involved in pod dehiscence regulation (Dong et al. 2014; Hu et al. 2019; Rau et al. 2019; Parker et al. 2020; Di Vittori et al. 2021). Interestingly, the highest proportion of AP2/ERF-like TF binding sites (24.5%) was found in promoter regions of candidate genes (Fig. 4b), implying that AP2/ERF-like TFs may act as key regulators for pod dehiscence in legumes.
Fig. 4.
Identification of potential key regulators of pod dehiscence in legumes. a) Percentage of Top6 representative TFs. The proportion of each TF family to the total differentially expressed TF genes was calculated in each species. b) Statistics on the number of promoter binding sites. Important differentially expressed TF genes are highlighted in bold. c) Expression correlation between AP2/ERF-like TF genes and structural genes. The circle size indicates the count of the genes associated with its expression. The line width represents the correlation level. LPO, lactoperoxidase; HHT, ω-hydroxypalmitate O-feruloyl transferase; UGT, UDP-glycosyltransferase; ALDH, aldehyde dehydrogenase; CYP81D1, Cytochrome P450/isoflavone 2′-hydroxylase; LAC, laccase; CSI1, cellulose synthase-interactive protein 1. All correlations are significant at P < 0.05.
To further establish the association of TFs with the candidate genes and already reported genes involved in pod dehiscence formation, we assessed expression levels (fragments per kilobase million [FPKM]) using Pearson's correlation coefficient (Fig. 4c; supplementary figs. S14 to S19, Supplementary Material online). We found significant expression correlations between AP2/ERF-like TFs and candidate genes, like ALDH, CSI1, and CYP81D1, in both common bean and M. ruthenica, less so in soybean and pea (Fig. 4c; supplementary fig. S14, Supplementary Material online). Moreover, some AP2/ERF-like TFs showed significant expression correlations with reported pod dehiscence genes in soybean, common bean, pea, and M. ruthenica (supplementary figs. S15 to S18, Supplementary Material online). Furthermore, significant expression correlations between MYB26, Pdh1, and SHAT1-5 and some members of the candidate genes were found in the four genera of legume species (supplementary fig. S19, Supplementary Material online). Altogether, these suggested that AP2/ERF family TFs might act as regulators for pod dehiscence.
Evolution of Candidate Gene Families in Legumes
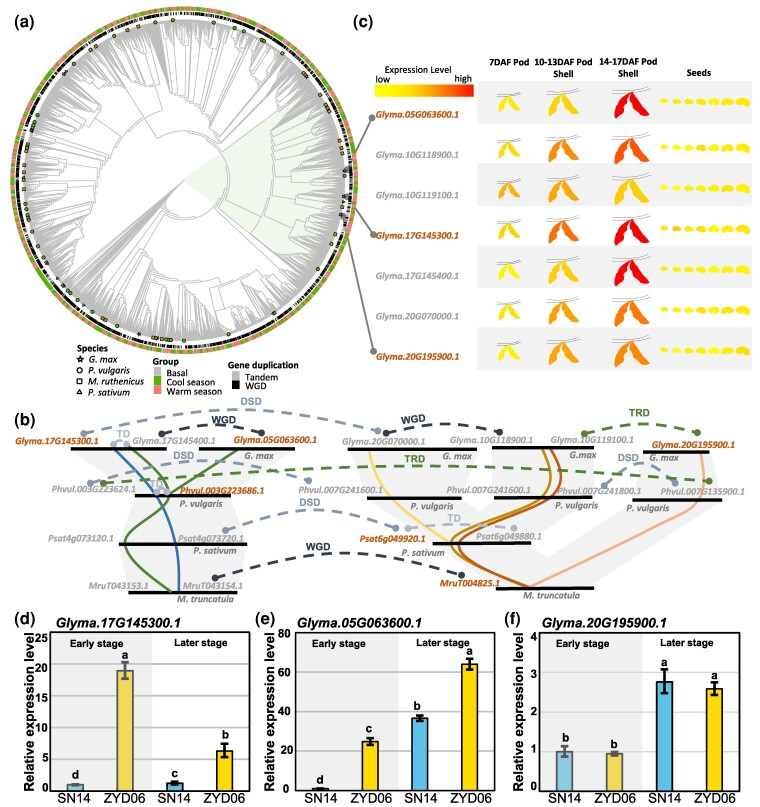
To reveal the evolution of pod dehiscence variation, we conducted phylogenetic analyses of the identified eight gene families in 31 legumes belonging to 18 genera. Multiple paralogous branches or subfamilies were observed in each of these candidate gene families (Fig. 5a; supplementary figs. S20 and S21a, Supplementary Material online). We mapped the identified DEGs in these legume species on these phylogenetic trees and found that the DEGs from ALDH, CSI1, CYP81D1, LAC, LPO, HHT, and UGT families in different legumes were mainly distributed in one or two paralogous branches or subfamilies within their own families (supplementary figs. S20 and S21a, Supplementary Material online). For example, DEGs of ALDH family were distributed in two branches, which belonged to two orthogroups and existed as tandem duplicates in legumes (supplementary fig. S21b, Supplementary Material online). Moreover, through microsynteny and ancestral state reconstruction, we found that warm-season- or cool-season-specific DEGs were detected in the gene families of CYP81D1, LAC, LPO, HHT, and UGT in legumes (supplementary figs. S22 and S23, Supplementary Material online). However, phylogenetic reconstruction indicated that the identified DEGs within the AP2/ERF family were randomly dispersed among most branches (Fig. 5a).
Fig. 5.
Evolution pattern and expression of AP2/ERF-like TFs. a) Phylogenetic analysis of legume AP2/ERF-like TFs. Members of DEGs are labeled on the tree using different shapes. b) Analysis of microsynteny and gene duplication events of candidate AP2/ERF-like TFs among the indicated four genera of legume species. DSD, dispersed duplication; TRD, transposed duplication; TD, tandem duplication; WGD, whole-gene duplication. c) Tissue expression patterns of candidate AP2/ERF-like TFs in soybean. Expression data are from ePlants (https://bar.utoronto.ca). In b) and c), highlighted and bold genes are DEGs. d to f) Relative expression level of Glyma.17G145300.1 d), Glyma.05G063600.1 e), and Glyma.20G195900.1 f) in soybean pods at early and later stages. Total RNA from cultivar SN14 and wild soybean ZYD06 was subjected to qRT-PCR. In d) to f), different letters (a to c) indicate significant difference (P < 0.05) in one-way ANOVA. Each bar corresponds to the mean value ± SD of three biological replicates.
Orthologous members of four DEGs from the soybean AP2/ERF family Glyma.02G016100.1, Glyma.05G063600.1, Glyma.13G236600.1, and Glyma.17G145300.1 were identified in three other species (Fig. 5b; supplementary fig. S24a and b, Supplementary Material online). We found that orthologous Phvul.003G223686.1 of Glyma.05G063600.1 was also differentially expressed in common bean, whereas its orthologous in pea (Psat4g073720.1) and M. ruthenica (MruT043154.1) were not; however, the paralogous genes Psat6g049920.1 and MruT004825.1 of Psat4g073720.1 and MruT043154.1 were DEGs between dehiscent and indehiscent pods in pea and M. ruthenica, respectively (Fig. 5b; supplementary table S2, Supplementary Material online). In addition, Glyma.20G195900.1, the orthologous gene of MruT004825.1, was also differentially expressed between dehiscent and indehiscent pods (Fig. 5b; supplementary table S2, Supplementary Material online). The orthologous genes of Glyma.02G016100.1 and Glyma.13G236600.1 and their paralogous genes were not differentially expressed between dehiscent and indehiscent pea (supplementary fig. S24a and b and table S2, Supplementary Material online). We further found that all genes associated with Glyma.05G063600.1 were highly expressed in pod walls and less in developing seeds, whereas only Glyma.05G063600.1, Glyma.17G145300.1, and Glyma.20G195900.1 were DEGs between dehiscent and indehiscent pods (Fig. 5c). However, the expression of Glyma.02G016100.1 was relatively low in both pods and seeds, while Glyma.13G236600.1 exhibited high expression levels in both tissues (supplementary fig. S24c and d, Supplementary Material online). The expression variation of these DEGs between dehiscent and indehiscent pods was consistent with that between ZYD06 (dehiscent pod) and SN14 (indehiscent pod), except Glyma.20G195900.1 (Fig. 5d to f; supplementary fig. S24e and f, Supplementary Material online). Integrating evolutionary and expression patterns, the orthologous genes of Glyma.05G063600.1 and Glyma.17G145300.1 and closely related paralogous genes might play a regulatory role in pod dehiscence formation.
Variation in Gene Pathways Correlated to Altered Lignin Composition Between Dehiscent and Indehiscent Pods
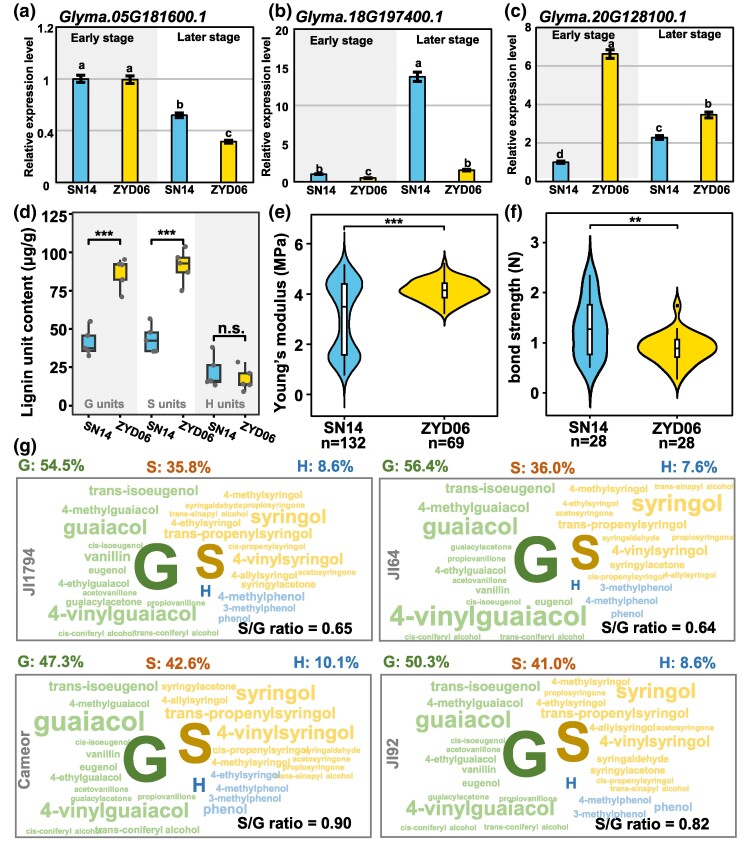
To determine the potential consequence of recruiting these DEGs, we analyzed the changes of candidate gene expression and metabolite composition in the secondary cell wall between dehiscent and indehiscent pods in soybean and pea. At first, we examined the expression variation of some DEGs from the candidate gene families, including Glyma.05G181600.1 (CSI1), Glyma.18G197400.1 (LAC), and Glyma.20G128100.1 (UDGT), in pods of cultivar ‘SN14’ and wild soybean ‘ZYD06’. Glyma.05G181600.1 was not differentially expressed at the early stage in SN14 and ZYD06 but was significantly higher in SN14 than in ZYD06 at a later stage (Fig. 6a). The expression of Glyma.18G197400.1 in SN14 was significantly higher than that of ZYD06 at both developmental stages, whereas Glyma.20G128100.1 showed an opposite pattern (Fig. 6b and c). These genes were indeed differentially expressed between dehiscent and indehiscent pods in soybean. Since most of the identified structural gene families were related to cellulose and lignin accumulation, the observed difference may result in variations in cell wall components.
Fig. 6.
Secondary cell wall composition in soybean and pea. a to c) Relative expression level of CSI1 (Glyma.05G181600.1), LAC (Glyma.18G197400.1), and UGT (Glyma.20G128100.1) in soybean pods at early and later stages. Total RNA from cultivar SN14 and wild soybean ZYD06 was subjected to qRT-PCR. Different letters (a to c) indicate significant difference (P < 0.05) in one-way ANOVA. Each bar corresponds to the mean value ± SD of three biological replicates. d) Lignin polymer content of soybean 30-day pods. Each box corresponds to the mean value ± SD of five biological replicates. e) Young's modulus of endocarp of soybean pods at 30 days. In d) and e), ***P < 0.001. f) Bond strength of mature soybean pods. **P < 0.01. g) Lignin polymer content of pea. JI1794 and JI64 are wild peas with pod dehiscence; Cameor and JI92 are cultivated peas with pod indehiscence. G units, guaiacyl units; H units, p-hydroxyphenyl units; S units, syringyl units.
The analysis of the secondary cell wall components of pod walls showed that the cellulose and lignin contents in ZYD06 (dehiscent pod) were significantly higher than those in SN14 (indehiscent pod) at the late developmental stage (supplementary fig. S25, Supplementary Material online). Then, we evaluated the composition of lignin polymers in SN14 and ZYD06 and found that the content of guaiacyl (G) and syringyl (S)-lignin units in ZYD06 was significantly higher than that of SN14, while the difference in p-hydroxyphenyl (H) units was not significant (Fig. 6d). Consistently, a significantly lower Young's modulus of pod walls in SN14 than in ZYD06 was observed (Fig. 6e), showing that the mature pods of wild soybean are stiffer than cultivated soybean. Moreover, the mature dry SN14 pods showed a stronger binding strength than ZYD06 (Fig. 6f). Therefore, the variation in the accumulation of cellulose and lignin correlated with pod dehiscence or indehiscence in soybeans.
This presumption in soybean was supported by the observation in pea (Fig. 6g; supplementary fig. S26 and table S3, Supplementary Material online). The predominant components of lignin polymer in the secondary cell walls of the pod walls were G- and S-lignin units derived from coniferyl and sinapyl alcohol, respectively. Lignin composed mainly of G units has more resistance linkages than S units (Boerjan et al. 2003; Vanholme et al. 2019). G-type phenolic compounds were detected in higher abundance from the dehiscent pods in wild “JI64” and “JI1794” accessions than from the indehiscent pods in cultivated pea “Cameor” and “JI92” accessions. In general terms, the lignin composition of the dehiscent pods of wild accessions showed higher content of G units and, therefore, lower S/G ratios (0.65) than the indehiscent pods of cultivated, conversely having higher abundances of S units and higher S/G ratios (0.86) (Fig. 6g; supplementary table S3, Supplementary Material online).
Taken together, the variation in accumulation and composition (including the ratio of different units) of secondary cell wall components, especially lignin, contributes to pod dehiscence or indehiscence in legumes.
Discussion
Pod dehiscence (pod shattering) is an adaptive trait as a mode of legume seed dispersal under natural conditions, whereas it leads to yield losses of domesticated legumes during the harvest (Ogutcen et al. 2018; Di Vittori et al. 2019; Maity et al. 2021). The anatomical basis of this trait and the several regulatory genes of pod dehiscence have been reported in several legumes (Lee et al. 2017; Han et al. 2019; Hu et al. 2019; Miranda et al. 2019; Kang et al. 2020; Seo et al. 2020; Ali et al. 2023; Yong et al. 2023); however, its genetic components remain elusive. In this study, we analyzed transcriptome data related to pod dehiscence in soybean, common bean, pea, and M. ruthenica. We found that AP2/ERF-like TFs and their putative target genes, including LAC, CSI1, and ALDH, were associated with pod dehiscence formation. Paralogous genes of these gene families showed differential expression between dehiscent and indehiscent pods, which was correlated with the lignin and cellulose content variation of the secondary cell wall of pod walls, ultimately leading to pod dehiscence variation in legumes.
Secondary metabolism plays an essential role in biotic or abiotic interactions and stress response in plants (Pichersky et al. 2006; Kessler and Kalske 2018; Yuan and Grotewold 2020), as well as development (Tohge et al. 2014). Recent studies have shown that efficient seed dispersal in plants is also highly dependent on secondary metabolism in fruit traits, including color, odor, and pod wall (Rajani and Sundaresan 2001; Dong et al. 2014, 2024; Funatsuki et al. 2014; Hofhuis et al. 2016; Nelson and Whitehead 2021; Lyu et al. 2023; Nelson et al. 2023; Smýkal and Parker 2023). Tension resulting from the differential mechanical properties of lignified and nonlignified pod tissues is important to pod dehiscence (Hofhuis et al. 2016; Ballester and Ferrándiz 2017). Changes in the mode of lignin deposition and the secondary cell wall thickness affect the mechanical properties of pod wall, which results in pod dehiscence variation (Liljegren et al. 2000, 2004; Mitsuda and Ohme-Takagi 2008; Dong et al. 2014, 2024; Zhang et al. 2018). In the present study, we identified DEGs and coexpressed genes associated with pod dehiscence in two warm-season and two cool-season legumes and found the gene families shared by the four genera of legume species participated in the cell wall formation, which related to secondary metabolisms, gibberellin signaling pathway, and epigenetic modification.
Multiple Gene Families Related to Secondary Cell Walls Act in Pod Dehiscence
Lignin and cellulose are important components of the secondary cell wall (Kumar et al. 2016), and their deposition and synthesis have been demonstrated to be critical for silique dehiscence in Brassicaceae and seed shattering in Poaceae (Liljegren et al. 2000, 2004; Mitsuda and Ohme-Takagi 2008; Steigemann and Gerlich 2009; Pourkheirandish et al. 2015; Di Vittori et al. 2019; Wu, He, and Wang 2023). Asymmetric lignification of endocarp b cells controlled by LACs (LAC4/11/7) is required for explosive seed dispersal in Cardamine hirsuta (Pérez-Antón et al. 2022). SLENDER RICE1 encodes DELLA protein, a key repressor of gibberellin signaling, which represses the expression of lignin biosynthesis gene 4-coumarate-CoA ligase 3 and consequently increases the lignin deposition in the abscission layer, thus resulting in reduced seed shattering in rice (Wu, He, et al. 2023). In our work, we found that seven gene families, including LAC, related to the phenylpropane metabolic pathway and cell wall composition and modification were associated with pod dehiscence variation in legumes. Moreover, we found that lignin content and subunit composition differed between dehiscent and indehiscent pods. Cellulose content was also significantly different between dehiscent and indehiscent pod in soybeans. Thus, the variation in cellulose and lignin accumulation may be a major factor contributing to pod dehiscence in soybeans at the later developmental stage. The same was observed in common bean (Murgia et al. 2017). This likely influences the composition or deposition pattern of secondary cell walls of the endocarp, leading to alterations in the physical properties of pod wall and ultimately to differential contraction between different pod tissues of the pods in response to pod maturation and drying leading to pod dehiscence. Similarly, the asymmetric deposition of lignin in the secondary cell wall of endocarp b cells drives explosive energy release for seed dispersal in C. hirsuta (Hofhuis et al. 2016). Interestingly, we found different patterns of lignin composition in pea. The predominance of G units in wild pea dehiscent pods produced a more condensed and branched lignin structure than the lignin of the pods with higher S/G ratios (such as found in the nondehiscent domesticated ones). This is in agreement with the finding in common bean (Murgia et al. 2017). Similarly, alternations in lignin deposition were shown to be responsible for a novel fruit morphology and dispersal strategy in Medicago (Fourquin et al. 2013). Thus, measurement and quantification of lignin deposition, especially G units, may rapidly identify pod dehiscence characteristics in legume crops. Differently modifying lignin polymerization in pod secondary wall via multiple mechanisms and recruiting different paralogous genes may contribute to species-specific adaptations in pod dehiscence formation.
The development of the cell wall is subjected to epigenetic regulation in response to environments (McCahill and Hazen 2019; Tang et al. 2020), and this also includes the pod dehiscence plasticity in legumes as a reaction to precipitation changes (Parker, Lo, and Gepts 2021; Yong et al. 2023). In this work, we revealed that some DEGs, like ABA receptors PYLs in warm-season legumes and CDKs in cool-season legumes between dehiscent and indehiscent pods, might be associated with protein phosphorylation. Both PYLs and CDKs could regulate protein phosphatases activity associated with ABA signaling and/or metabolism (Yang et al. 2002; Cao et al. 2017), where ABA is not only involved in abiotic stress such as water-deficient conditions but may also participate in expression regulation of genes controlling pod dehiscence in legumes (Cao et al. 2017; Marsh et al. 2023; Wang et al. 2024). However, epigenetic modifications in pod dehiscence in legumes involving PYLs or CDKs and related protein phosphatases in ABA signaling need further investigations.
Potential Roles of AP2/ERF-Like Genes in Seed Dispersal
Fruit development and ripening are preconditions for seed dispersal in angiosperms (Forlani et al. 2019, 2021). TFs involved in regulating fruit development and ripening include MADS-box, NAC, MYB, and AP2/ERF (Zeng et al. 2015; Cao et al. 2020; Fu et al. 2020; Sharma et al. 2020; Forlani et al. 2021; Zhai et al. 2022). MYB, NAC, and bHLH-like TFs are involved in secondary metabolism, cell wall modification, and fruit development (Liljegren et al. 2004; Zhong et al. 2006; Dong et al. 2014; Di Vittori et al. 2021), impacting fruit dehiscence. Indeed, some of their homologous genes were found among DEGs between dehiscent and indehiscent legume pods. Particularly, the abovementioned cell wall composition-related genes, including CSI1, LPO, and ALDH genes, contained a large number of AP2/ERF-type TF-binding sites in their promoters, and their expression showed significant correlation. In Glycine spp., Glyma.05G063600.1 and Glyma.17G145300.1 are candidate genes for regulating the expression of the identified structural genes associated with plant cell wall formation. The proposed regulations need further investigation; nonetheless, these results implied that AP2/ERF-like may be involved in the regulation of pod dehiscence in legumes. This agrees with findings in Arabidopsis and rice, where the abscission zone differentiation and secondary cell wall modification of fruit were shown to be regulated by AP2/ERF-like genes affecting seed dispersal (Ripoll et al. 2011; Zhou et al. 2012). Moreover, seed dispersal can be influenced by other various fruit-related traits. The color and aroma of mature fruits significantly affect the efficiency of natural fruit and seed dispersal (Rodríguez et al. 2013; Nelson et al. 2023), as well as facilitate human-mediated transport and dispersal to more distant areas (Shi et al. 2022; Wang and Seymour 2022). All these traits associated with seed dispersal are partially regulated by AP2/ERF-like TFs (Gu et al. 2017; Zhai et al. 2022; Li et al. 2023). Furthermore, AP2/ERF-like TFs are involved in the regulation of fruit and seed size (Liu et al. 2022; Wang et al. 2022; Qi et al. 2023), which may also assist in seed dispersal. Taking together, AP2/ERF-like may be essential players in seed dispersal mechanisms in angiosperms.
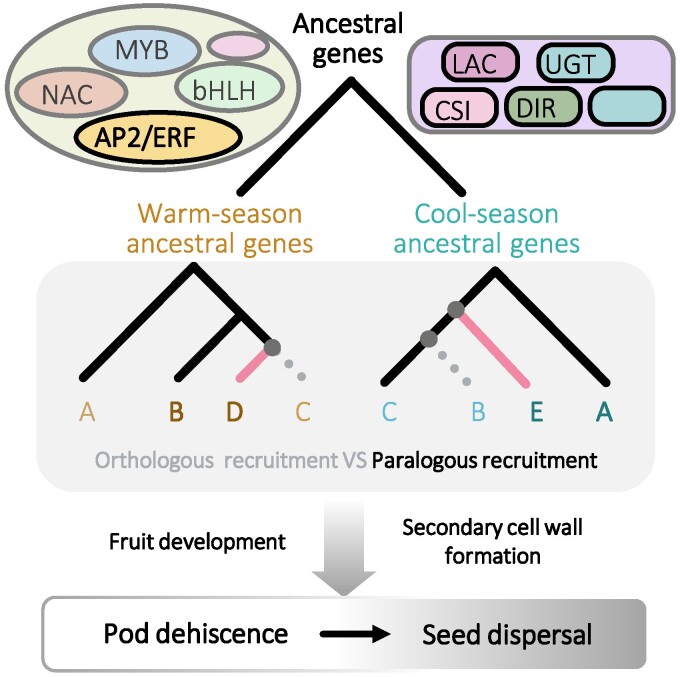
Recruiting Paralogous Genes Is a Predominant Evolutionary Event in Legumes
Phenotypes are influenced by both genetics and environments, and genetic robustness could help organisms maintain phenotypic stability in complex and variable environments (Félix and Wagner 2008). Genetic robustness and environmental adaptation often rely on similar regulatory networks and influence each other (Meiklejohn and Hartl, 2002; Masel and Siegal 2009; Diss et al. 2014). Expanded gene families and functional redundancy among paralogous genes could help organisms achieve their genetic robustness (Akazome et al. 2010; Diss et al. 2014; Suzuki et al. 2018; Bailon-Zambrano et al. 2022; Dai et al. 2023). Generating paralogous genes through gene family duplication may provide a compensatory mechanism for maintaining the robustness of regulatory mechanisms of seed dispersal in legumes, and similar compensatory mechanisms are present in triterpenoid metabolism, peptide hormones, and the MADS-like genes that regulate flower development (Cárdenas et al. 2019; Selby and Jones 2023). In the present study, we found that the identified DEGs and coexpressed genes had similar biological functions in different legumes, but only <1% of their orthologous genes were recruited in different species. The same was observed for seven structural gene families and the AP2/ERF regulatory gene family for potential pod dehiscence formation. Interestingly, the differentially expressed candidate structural genes were exclusively distributed in one or two branches of the phylogenetic tree of their gene families, whereas the differentially expressed AP2/ERF-like genes were distributed throughout the major branches of the phylogenetic tree. This implies that, unlike these structural genes, whose function might be specific to cell wall formation, regulatory genes may be involved in multiple aspects of fruit development.
Pod indehiscence, one of the important domestication syndromes, has been regulated by a few genes or loci in different legume crops, such as Pdh1 in soybean (Funatsuki et al. 2014; Yong et al. 2023), St and qPD5.1 in common bean (Parker, Lo, and Gepts 2021), and Dpo or Dpo1 in pea (Weeden et al. 2002). Certainly, incorporating wild legumes or diverse legumes would help to gain a deeper understanding of the regulatory and evolutionary mechanisms. Our evolutionary analyses showed that although orthologous members of candidate genes in other species did not show differential expression in accessions with different pod dehiscence, their paralogs usually were differentially expressed. Therefore, the functional redundancy of these paralogs might act as a major driver to maintain the genetic robustness of the pod dehiscence in legumes (Fig. 7). The widespread existence of genetic compensatory mechanisms in plants illustrates their importance in plant evolution and development (Iohannes and Jackson 2023). Thus, understanding the genetic compensatory mechanisms through gene family expansion, particularly the regulatory gene families, will provide new insights into plant morphological and molecular evolutionary innovations. In future molecular breeding for pod dehiscence-resistant legumes, AP2/ERF-like genes could be primarily targeted. Particularly, Glyma.05G063600.1 and Glyma.17G145300.1 from this regulatory gene family were highly expressed in dehiscent wild soybean ZYD06 compared to indehiscent soybean SN14. Therefore, gene editing of orthologous and paralogous genes will be promising for breeding new legume varieties or de novo domestication from wild legumes. However, the compensating roles of paralogous genes in pod dehiscence may bring potential limitations or challenges in translating these findings from wild to cultivated legumes.
Fig. 7.
Evolution of genetic architecture of pod dehiscence in legumes. Various TF gene families including AP2/ERF-like and structural gene families such as LAC, CSI1, UGT, and DIR might be involved in pod dehiscence formation in legumes. Members in these gene families participate in fruit development and secondary cell wall formation, thus affecting seed dispersal. During evolution, recruiting either orthologous or paralogous members in these gene families to be differentially expressed likely constitutes the genetic architecture and robustness of legume pod dehiscence formation. In legume, paralogous recruitment was the major evolutionary event compared to orthologous recruitment. Orthologous genes are represented by the same letters and paralogous genes are represented by different letters. Moreover, specific gene loss or gene gain might occur, which represented additional evolutionary mechanisms for pod dehiscence variation. Gray dotted line indicates gene loss and pink line represents genes that were specifically originated. CSI1, cellulose synthase-interactive protein 1; DIR, dirigent; LAC, laccase; UGT, UDP-glycosyltransferase.
In conclusion, this work identified DEGs and coexpressed genes associated with pod dehiscence formation, and these were involved in secondary cell wall-related metabolism, hormone metabolism, and epigenetics. We suggest that seven structure gene families (including LPO, ALDH, LAC, CSI1, CYP81D1 HHT, and UGT), which are implicated in the biosynthesis, accumulation, and deposition of secondary cell wall components, represent potential candidate genes for pod dehiscence formation and may be primarily regulated by AP2/ERF-like TFs. In soybean, lignin and cellulose contents of pod dehiscent accessions were higher than those of pod indehiscent accessions. Lignin, G unit, and S unit composition in soybean or the G/S ratio in pea was lower in pod indehiscent accessions than pod dehiscent accessions, implying that this affects the physical properties of the pod walls. Genome-wide evolutionary analyses revealed that recruiting paralogous genes is a predominant evolutionary event, which may provide a compensatory mechanism to maintain genetic and developmental robustness of pod dehiscence in legumes. These findings shed new light in the genetic control of pod dehiscence formation and provide the framework for breeding dehiscence-resistant varieties in legumes.
Materials and Methods
Plant Materials and Growth Conditions
The soybean accessions of G. max “Suinong14” (SN14, a cultivar with indehiscent pods) and G. soja “ZYD00006” (ZYD06, wild soybean displaying dehiscent pods) were grown in greenhouse in Beijing (Institute of Botany, Beijing, China, latitude 39.9, longitude 116.3). Two wild pea (P. elatius) “JI1794” and “JI64” with dehiscent pods and two cultivated pea (P. sativum) “Cameor” and “JI92” with indehiscent pods used previously (Zablatzká et al. 2021; Balarynová et al. 2022) were grown in the greenhouse (as described in Zablatzká et al. 2021) at Palacký University in Olomouc (the Czech Republic, latitude 49.6, longitude 17.3).
RNA Extraction, qRT-PCR, and RNA-seq
RNA was extracted from soybean pods at 15 (early stage) and 30 (late stage) days after flowering using the SV Total RNA Isolation System (Promega, Madison, WI, USA). An M-MLV complementary DNA (cDNA) synthesis kit (Invitrogen, Carlsbad, CA, USA) was used to synthesize the first-strand cDNA. Quantitative reverse-transcription PCR (qRT-PCR) was performed on a Stratagene Mx3000P qPCR system (Agilent, Santa Clara, CA, USA) using SYBR Premix Ex Taq (Takara, Kusatsu, Shiga, Japan) and gene-specific primers (supplementary table S4, Supplementary Material online). The soybean gene GmActin11 (Glyma.18G290800.1) was used as an internal control. Total RNA from sutures of pea pods at early (11 to 17 days after pollination [DAP]) and late (23 to 27 DAP) stages was isolated as described in Balarynová et al. (2022). Total RNA was isolated using PureLink Plant RNA Reagent (Thermo Fisher Scientific, Waltham, USA) and the traces of residual DNA were removed by a DNase I (TOP-BIO Ltd., Czech Republic) treatment followed by phenol-chloroform extraction. The RNA sequencing was performed at the Novogene (Cambridge, UK) using an Illumina NovaSeq platform.
Transcriptomic Data and Gene Expression Quantification
The transcriptome raw data related to pod dehiscence traits in soybean (Glycine spp., including five genotypes: three cultivars and two landraces exhibiting indehiscent pods, alongside two cultivars, one landrace, and two wild soybeans displaying dehiscent pods) (Kang et al. 2020) and common bean (P. vulgaris, including four genotypes: G12873 with fully dehiscent pods; ICA_Bunsi and SXB_405 with intermediate pod dehiscence; and MIDAS displaying indehiscent pods) were downloaded from the NCBI database (PRJNA533590 and PRJNA746142). Those for M. ruthenica (including two genotypes: 15-4 with dehiscent pods; 9-2 with indehiscent pods) were collected from previous work (Guo et al. 2022) and those for pea were generated in this study. The quality control of these data was performed using FastQC v0.11.4, and then the reads were aligned with reference genome by using software fastp and hisat2 after removing low-quality sequence bases (Chen et al. 2018; Kim et al. 2019). Gene expression was quantitatively conversed to FPKM using featureCounts software (Liao et al. 2014), and DEGs were identified for each species using R package DEseq2 with 1.5-fold differentially expressed level (https://github.com/thelovelab/DESeq2).
Expression Pattern Clustering and Enrichment Analysis
For species with <15 sequenced samples (soybean and pea), coexpression module analysis was performed using the R package Mfuzz (http://mfuzz.sysbiolab.eu/). For species with sequencing sample sizes >15 (common bean and M. ruthenica), weighted correlation network analysis (WGCNA) was performed using the R package WGCNA (Langfelder and Horvath 2008). GO annotation files of soybean, common bean, M. ruthenica, and pea were constructed using eggnog-mapper v2 software (Cantalapiedra et al. 2021). The identified DEGs and coexpressed genes were analyzed for functional enrichment using R package clusterProfiler (Wu et al. 2021).
Gene Family Identification
Genomic data of legumes used in this work were downloaded from public databases (supplementary table S5, Supplementary Material online). Gene family was identified based on the conserved hidden Markov model. The LAC gene family was determined based on three conserved structural domains of Cu-oxidase (PF00394), Cu-oxidase2 (PF07731), and Cu-oxidase3 (PF07732). The cellulose synthase-interactive protein family was identified based on C2 domain (PF00168) and armadillo/beta-catenin-like repeat (Arm; PF00514). The gene families of AP2/ERF, ALDH, cytochrome P450/isoflavone 2′-hydroxylase, LPO, HHT, and UGT were identified according to PF00847 (AP2 domain), PF00171, PF00067 (cytochrome P450 domain), PF00141, PF02458, and PF00201, respectively.
Evolutionary Analysis
OrthoFinder was used to construct species trees and identify orthogroups with a default algorithm (Emms and Kelly 2019). All gene families were searched in whole genomes using HMMER version 3.3.2 software (Johnson et al. 2010). MUSCLE v3.8.31 was used for multiple sequence alignment (Edgar 2004). Phylogenetic trees were reconstructed in IQ-TREE using the maximum-likelihood method with default parameters (Nguyen et al. 2015) and visualized using the R package ggtree (https://github.com/YuLab-SMU/ggtree). Microsyntenic analysis was performed using MCScan-Python (Tang et al. 2008). Ancestral state reconstruction of candidate genes was performed using pamlX version 1.3.1 (Xu and Yang 2013) and Mesquite version 3.70 (http://www.mesquiteproject.org). The TF-binding sites in the promoters were predicted using PlantTFDB (Jin et al. 2017). Expression correlations were calculated based on gene expression FPKM values and visualized using Cytoscape (Shannon et al. 2003). Gene duplication was analyzed using DupGen-finder (Qiao et al. 2019).
Determination of Cell Wall Components
Matured soybean pod walls were dried at 60 °C, and then cell wall components and lignin units were assayed using spectrophotometry and LC-MS by BiotechPack Analytical Co. (Beijing, China). The lignin composition of mature dry pea pod walls was determined by Pyrolysis coupled to gas chromatography and mass spectrometry. Pyrolysis of the pea pod samples (ca. 1 mg) was performed at 500 °C in an EGA/PY-3030D microfurnace pyrolyzer (Frontier Laboratories Ltd., Fukushima, Japan) connected to a GC 7820A (Agilent Technologies, Inc., Santa Clara, CA, USA) and an Agilent 5975 mass-selective detector (electron impact at 70 eV). The column used was a 30 m × 0.25 mm internal diameter, 0.25-μm film thickness, DB-1701 (J&W Scientific, Folsom, CA, USA). The oven temperature was programmed from 50 °C (1 min) to 100 °C at 20 °C min−1 and then ramped to 280 °C (5 min) at 6 °C min−1. Helium was the carrier gas (1 mL min−1). Peak molar areas were calculated for the H, G, and S-lignin degradation products, and the summed areas were normalized and expressed as percentages.
Young's Modulus Test
The 30-day soybean pods shell were observed under FastScan Bio Atomic Force Microscope (Bruker, Karlsruhe, Germany) and analyzed for Young's modulus using NanoScope (http://nanoscaleworld.bruker-axs.com/nanoscaleworld/).
Statistical Analysis
Significance analyses were performed by using SPSS Statistics 17.0 software (International Business Machines Corporation, New York, USA). Significant differences among samples were determined using one-way analysis of variance (ANOVA). Pearson correlation coefficient was estimated using the R function “cor. test.” (R Core Team 2020).
Supplementary Material
Acknowledgments
The technical assistance of Cui Song (Institute of Process Engineering, Chinese Academy of Sciences) in histological experiment is acknowledged. Thanks to Jun Li and Lin Zhu (Grassland Research Institute, Chinese Academy of Agricultural Sciences) for transferring the transcriptomic data of M. ruthenica.
Contributor Information
Bin Yong, State Key Laboratory of Plant Diversity and Specialty Crops/State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jana Balarynová, Department of Botany, Faculty of Sciences, Palacky University, Olomouc 773 71, Czech Republic.
Bingbing Li, State Key Laboratory of Plant Diversity and Specialty Crops/State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Denisa Konečná, Department of Botany, Faculty of Sciences, Palacky University, Olomouc 773 71, Czech Republic.
Jorge Rencoret, Instituto de Recursos Naturales y Agrobiología de Sevilla, CSIC, 41012 Seville, Spain.
José C del Río, Instituto de Recursos Naturales y Agrobiología de Sevilla, CSIC, 41012 Seville, Spain.
Petr Smýkal, Department of Botany, Faculty of Sciences, Palacky University, Olomouc 773 71, Czech Republic.
Chaoying He, State Key Laboratory of Plant Diversity and Specialty Crops/State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; The Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
C.H., P.S., and B.Y. conceived and designed the project. B.Y. performed all analyses and experiments unless otherwise noted. J.B. and D.K. conducted the pea transcriptomic analysis. B.L. and D.K. participated in the histological experiments. J.R. and J.C.d.R. performed the lignin polymer determination of the pea pod wall. B.Y., C.H., J.B., and P.S. analyzed the data and wrote the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (31930007 and 31525003) to C.H. P.S. acknowledges support from Grant Agency of the Czech Republic (19-07155S and 24-10730S), Palacky University Grant Agency (PrF-2023-001 and PrF-2024-001), and the Ministry of Education, Youth and Sports (LUAUS25035). J.R. and J.C.d.R. acknowledge support from the project PID2023-152543OB-I00 funded by the Spanish MICIU/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”.
Data Availability
All data are available in the manuscript and the Supplementary materials are provided in Supplementary material online. Transcriptomic data of pea pod generated for this research have been submitted to NCBI (PRJNA1138002).
Literature Cited
- Aguilar-Benitez D, Rubio J, Millán T, Gil J, Die JV, Castro P. Genetic analysis reveals PDH1 as a candidate gene for control of pod dehiscence in chickpea. Mol Breeding. 2020:40(4):40. 10.1007/s11032-020-01117-9. [DOI] [Google Scholar]
- Akazome Y, Kanda S, Okubo K, Oka Y. Functional and evolutionary insights into vertebrate kisspeptin systems from studies of fish brain. J Fish Biol. 2010:76(1):161–182. 10.1111/j.1095-8649.2009.02496.x. [DOI] [PubMed] [Google Scholar]
- Ali S, Kucek LK, Riday H, Krom N, Krogman S, Cooper K, Jacobs L, Mehta P, Trammell M, Bhamidimarri S, et al. Transcript profiling of hairy vetch (Vicia villosa Roth) identified interesting genes for seed dormancy. Plant Genome. 2023:16(2):e20330. 10.1002/tpg2.20330. [DOI] [PubMed] [Google Scholar]
- Armon S, Efrati E, Kupferman R, Sharon E. Geometry and mechanics in the opening of chiral seed pods. Science. 2011:333(6050):1726–1730. 10.1126/science.1203874. [DOI] [PubMed] [Google Scholar]
- Aslan C, Beckman NG, Rogers HS, Bronstein J, Zurell D, Hartig F, Shea K, Pejchar L, Neubert M, Poulsen J, et al. Employing plant functional groups to advance seed dispersal ecology and conservation. AoB Plants. 2019:11(2):plz006. 10.1093/aobpla/plz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azani N, Babineau M, Bailey CD, Banks H, Barbosa AR, Pinto RB, Boatwright JS, Borges LM, Brown GK, Bruneau A, et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny: the legume phylogeny working group (LPWG). Taxon. 2017:66(1):44–77. 10.12705/661.3. [DOI] [Google Scholar]
- Bailon-Zambrano R, Sucharov J, Mumme-Monheit A, Murry M, Stenzel A, Pulvino AT, Mitchell JM, Colborn KL, Nichols JT. Variable paralog expression underlies phenotype variation. eLife. 2022:11:e79247. 10.7554/eLife.79247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balarynová J, Klčová B, Sekaninová J, Kobrlová L, Cechová MZ, Krejčí P, Leonova T, Gorbach D, Ihling C, Smržová L, et al. The loss of polyphenol oxidase function is associated with hilum pigmentation and has been selected during pea domestication. New Phytol. 2022:235(5):1807–1821. 10.1111/nph.18256. [DOI] [PubMed] [Google Scholar]
- Ballester P, Ferrándiz C. Shattering fruits: variations on a dehiscent theme. Curr Opin Plant Biol. 2017:35:68–75. 10.1016/j.pbi.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003:54(1):519–546. 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 2021:38(12):5825–5829. 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Chen J, Yue M, Xu C, Jian W, Liu Y, Song B, Gao Y, Cheng Y, Li Z. Tomato transcriptional repressor MYB70 directly regulates ethylene-dependent fruit ripening. Plant J. 2020:104(6):1568–1581. 10.1111/tpj.15021. [DOI] [PubMed] [Google Scholar]
- Cao M-J, Zhang Y-L, Liu X, Huang H, Zhou XE, Wang W-L, Zeng A, Zhao C-Z, Si T, Du J, et al. Combining chemical and genetic approaches to increase drought resistance in plants. Nat Commun. 2017:30(1):1183. 10.1038/s41467-017-01239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Socquet-Juglard D, Daba K, Vandenberg A, Bett KE. Understanding genome structure facilitates the use of wild lentil germplasm for breeding: a case study with shattering loci. Plant Genome. 2024:17(2):e20455. 10.1002/tpg2.20455. [DOI] [PubMed] [Google Scholar]
- Cárdenas PD, Almeida A, Bak S. Evolution of structural diversity of triterpenoids. Front Plant Sci. 2019:10:1523. 10.3389/fpls.2019.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain-Irwin HN, Hufford MB. Convergent domestication: finding the genes that make crops. Curr Biol. 2022:32(12):R585–R588. 10.1016/j.cub.2022.05.003. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):884–890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Luo L, Zhao Z. Genetic robustness control of auxin output in priming organ initiation. Proc Natl Acad Sci U S A. 2023:120(28):e2221606120. 10.1073/pnas.2221606120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diss G, Ascencio D, DeLuna A, Landry CR. Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J Exp Zool B Mol Dev Evol. 2014:322(7):488–499. 10.1002/jez.b.22555. [DOI] [PubMed] [Google Scholar]
- Di Vittori V, Bitocchi E, Rodriguez M, Alseekh S, Bellucci E, Nanni L, Gioia T, Marzario S, Logozzo G, Rossato M, et al. Pod indehiscence in common bean is associated with the fine regulation of PvMYB26. J Exp Bot. 2021:72(5):1617–1633. 10.1093/jxb/eraa553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Vittori V, Gioia T, Rodriguez M, Bellucci E, Bitocchi E, Nanni L, Attene G, Rau D, Papa R. Convergent evolution of the seed shattering trait. Genes (Basel). 2019:10(1):68. 10.3390/genes10010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XM, Chen JW, Zhou Q, Luo D, Fang LF, Liu WX, Liu ZP. Pod-shattering characteristic differences between shattering-resistant and shattering-susceptible common vetch accessions are associated with lignin biosynthesis. J Integr Agr. 10.1016/j.jia.2024.03.032, 2024, preprint: not peer reviewed. [DOI] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang B-H, Liu B-L, Wang Y-Z. Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun. 2014:5(1):3352. 10.1038/ncomms4352. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004:5(1):113. 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Bio. 2019:20(1):238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A, Wagner A. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity (Edinb). 2008:100(2):132–140. 10.1038/sj.hdy.6800915. [DOI] [PubMed] [Google Scholar]
- Forlani S, Masiero S, Mizzotti C. Fruit ripening: the role of hormones, cell wall modifications, and their relationship with pathogens. J Exp Bot. 2019:70(11):2993–3006. 10.1093/jxb/erz112. [DOI] [PubMed] [Google Scholar]
- Forlani S, Mizzotti C, Masiero S. The NAC side of the fruit: tuning of fruit development and maturation. BMC Plant Biol. 2021:21(1):238. 10.1186/s12870-021-03029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourquin C, del Cerro C, Victoria FC, Vialette-Guiraud A, de Oliveira AC, Ferrándiz C. A change in SHATTERPROOF protein lies at the origin of a fruit morphological novelty and a new strategy for seed dispersal in Medicago genus. Plant Physiol. 2013:162(2):907–917. 10.1104/pp.113.217570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Chen H, Gao H, Lu Y, Han C, Han Y. Two papaya MYB proteins function in fruit ripening by regulating some genes involved in cell-wall degradation and carotenoid biosynthesis. J Sci Food Agric. 2020:100(12):4442–4448. 10.1002/jsfa.10484. [DOI] [PubMed] [Google Scholar]
- Fuller DQ, Allaby R. Seed dispersal and crop domestication: shattering, germination and seasonality in evolution under cultivation. In: Østergaard L, editor. Annual plant reviews volume 38: fruit development and seed dispersal. Oxford: Wiley-Blackwell; 2009. p. 238–295. 10.1002/9781444314557.ch7. [DOI] [Google Scholar]
- Fuller DQ, Denham T, Arroyo-Kalin M, Lucas L, Stevens CJ, Qin L, Allaby RG, Purugganan MD. Convergent evolution and parallelism in plant domestication revealed by an expanding archaeological record. Proc Natl Acad Sci U S A. 2014:111(17):6147–6152. 10.1073/pnas.1308937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsuki H, Suzuki M, Hirose A, Inaba H, Yamada T, Hajika M, Komatsu K, Katayama T, Sayama T, Ishimoto M, et al. Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci U S A. 2014:111(50):17797–17802. 10.1073/pnas.1417282111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Guo Z-H, Hao P-P, Wang G-M, Jin Z-M, Zhang S-L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot Stud. 2017:58(1):6. 10.1186/s40529-016-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MW, Zhu L, Li HY, Liu WP, Wu ZN, Wang CH, Liu L, Li ZY, Li J. Mechanism of pod shattering in the forage legume Medicago ruthenica. Plant Physiol Biochem. 2022:185:260–267. 10.1016/j.plaphy.2022.06.013. [DOI] [PubMed] [Google Scholar]
- Hammer K. Das Domestikationssyndrom. Kulturpflanze. 1984:32(1):11–34. 10.1007/BF02098682. [DOI] [Google Scholar]
- Han J, Han D, Guo Y, Yan H, Wei Z, Tian Y, Qiu L. QTL mapping pod dehiscence resistance in soybean (Glycine max L. Merr.) using specific-locus amplified fragment sequencing. Theor Appl Genet. 2019:132(8):2253–2272. 10.1007/s00122-019-03352-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhuis H, Moulton D, Lessinnes T, Routier-Kierzkowska A-L, Bomphrey RJ, Mosca G, Reinhardt H, Sarchet P, Gan X, Tsiantis M, et al. Morphomechanical innovation drives explosive seed dispersal. Cell. 2016:166(1):222–233. 10.1016/j.cell.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hradilová I, Trněný O, Válková M, Cechová M, Janská A, Prokešová L, Aamir K, Krezdorn N, Rotter B, Winter P, et al. A combined comparative transcriptomic, metabolomic, and anatomical analyses of two key domestication traits: pod dehiscence and seed dormancy in pea (Pisum sp.). Front Plant Sci. 2017:8:542. 10.3389/fpls.2017.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Kan G, Hu W, Li Y, Hao D, Li X, Yang H, Yang Z, He X, Huang F, et al. Identification of loci and candidate genes responsible for pod dehiscence in soybean via genome-wide association analysis across multiple environments. Front Plant Sci. 2019:10:811. 10.3389/fpls.2019.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iohannes SD, Jackson D. Tackling redundancy: genetic mechanisms underlying paralog compensation in plants. New Phytol. 2023:240(4):1381–1389. 10.1111/nph.19267. [DOI] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017:45(D1):1040–1045. 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010:11(1):431. 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Cai J, Chen Y, Yan Y, Yang S, He R, Wang D, Zhu Y. Pod-shattering characteristics differences between two groups of soybeans are associated with specific changes in gene expression. Funct Integr Genomics. 2020:20(2):201–210. 10.1007/s10142-019-00702-2. [DOI] [PubMed] [Google Scholar]
- Kessler A, Kalske A. Plant secondary metabolite diversity and species interactions. Annu Rev Ecol Evol Syst. 2018:49(1):115–138. 10.1146/annurev-ecolsys-110617-062406. [DOI] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8):907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005:39(1):309–338. 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Kumar M, Campbell L, Turner S. Secondary cell walls: biosynthesis and manipulation. J Exp Bot. 2016:67(2):515–531. 10.1093/jxb/erv533. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008:9(1):559. 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kim KR, Ha B-K, Kang S. Identification of SNPs tightly linked to the QTL for pod shattering in soybean. Mol Breed. 2017:37(4):54. 10.1007/s11032-017-0656-2. [DOI] [Google Scholar]
- Lenser T, Theißen G. Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 2013:18(12):704–714. 10.1016/j.tplants.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Li T, Liu Z, Lv T, Xu Y, Wei Y, Liu W, Wei Y, Liu L, Wang A. Phosphorylation of MdCYTOKININ RESPONSE FACTOR4 suppresses ethylene biosynthesis during apple fruit ripening. Plant Physiol. 2023:191(1):694–714. 10.1093/plphys/kiac498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014:30(7):923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000:404(6779):766–770. 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004:116(6):843–853. 10.1016/S0092-8674(04)00217-X. [DOI] [PubMed] [Google Scholar]
- Liu C, Ma T, Yuan D, Zhou Y, Long Y, Li Z, Dong Z, Duan M, Yu D, Jing Y, et al. The OsEIL1-OsERF115-target gene regulatory module controls grain size and weight in rice. Plant Biotechnol J. 2022:20(8):1470–1486. 10.1111/pbi.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S, Muñoz-Amatriaín M, Boukar O, Herniter I, Cisse N, Guo Y-N, Roberts PA, Xu S, Fatokun C, Close TJ. Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci Rep. 2018:8(1):6261. 10.1038/s41598-018-24349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X, Li Y-H, Li Y, Li D, Han C, Hong H, Tian Y, Han L, Liu B, Qiu L-J. The domestication-associated L1 gene encodes a eucomic acid synthase pleiotropically modulating pod pigmentation and shattering in soybean. Mol Plant. 2023:16(7):1178–1191. 10.1016/j.molp.2023.06.003. [DOI] [PubMed] [Google Scholar]
- Maity A, Lamichaney A, Joshi DC, Bajwa A, Subramanian N, Walsh M, Bagavathiannan M. Seed shattering: a trait of evolutionary importance in plants. Front Plant Sci. 2021:12:657773. 10.3389/fpls.2021.657773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JI, Nestor BJ, Petereit J, Tay Fernandez CG, Bayer PE, Batley J, Edwards D. Legume-wide comparative analysis of pod shatter locus PDH1 reveals phaseoloid specificity, high cowpea expression, and stress responsive genomic context. Plant J. 2023:115(1):68–80. 10.1111/tpj.16209. [DOI] [PubMed] [Google Scholar]
- Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009:25(9):395–403. 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCahill IW, Hazen SP. Regulation of cell wall thickening by a medley of mechanisms. Trends Plant Sci. 2019:24(9):853–866. 10.1016/j.tplants.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL. A single mode of canalization. Trends Ecol Evol. 2002:17(10):468–473. 10.1016/S0169-5347(02)02596-X. [DOI] [Google Scholar]
- Miranda C, Culp C, Škrabišová M, Joshi T, Belzile F, Grant D, Bilyeu K. Molecular tools for detecting Pdh1 can improve soybean breeding efficiency by reducing yield losses due to pod shatter. Mol Breeding. 2019:39(2):27. 10.1007/s11032-019-0935-1. [DOI] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2008:56(5):768–778. 10.1111/j.1365-313X.2008.03633.x. [DOI] [PubMed] [Google Scholar]
- Moghaddam SM, Mamidi S, Osorno JM, Lee R, Brick M, Kelly J, Miklas P, Urrea C, Song Q, Cregan P, et al. Genome-wide association study identifies candidate loci underlying agronomic traits in a middle American diversity panel of common bean. Plant Genome. 2016:9(3):1–21. 10.3835/plantgenome2016.02.0012. [DOI] [PubMed] [Google Scholar]
- Murgia ML, Attene G, Rodriguez M, Bitocchi E, Bellucci E, Fois D, Nanni L, Gioia T, Albani DM, Papa R, et al. A comprehensive phenotypic investigation of the “pod-shattering syndrome” in common bean. Front Plant Sci. 2017:8:251. 10.3389/fpls.2017.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AS, Gelambi M, Morales-M E, Whitehead SR. Fruit secondary metabolites alter the quantity and quality of a seed dispersal mutualism. Ecology. 2023:104(5):e4032. 10.1002/ecy.4032. [DOI] [PubMed] [Google Scholar]
- Nelson AS, Whitehead SR. Fruit secondary metabolites shape seed dispersal effectiveness. Trends Ecol Evol. 2021:36(12):1113–1123. 10.1016/j.tree.2021.08.005. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogutcen E, Pandey A, Khan MK, Marques E, Penmetsa RV, Kahraman A, Von Wettberg EJB. Pod shattering: a homologous series of variation underlying domestication and an avenue for crop improvement. Agronomy. 2018:8(8):137. 10.3390/agronomy8080137. [DOI] [Google Scholar]
- Parker TA, Berny Mier Y Teran JC, Palkovic A, Jernstedt J, Gepts P. Pod indehiscence is a domestication and aridity resilience trait in common bean. New Phytol. 2020:225(1):558–570. 10.1111/nph.16164. [DOI] [PubMed] [Google Scholar]
- Parker TA, de Sousa LL, de Oliveira Floriani T, Palkovic A, Gepts P. Toward the introgression of PvPdh1 for increased resistance to pod shattering in common bean. Theor Appl Genet. 2021:134(1):313–325. 10.1007/s00122-020-03698-7. [DOI] [PubMed] [Google Scholar]
- Parker TA, Lo S, Gepts P. Pod shattering in grain legumes: emerging genetic and environment-related patterns. Plant Cell. 2021:33(2):179–199. 10.1093/plcell/koaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Antón M, Schneider I, Kroll P, Hofhuis H, Metzger S, Pauly M, Hay A. Explosive seed dispersal depends on SPL7 to ensure sufficient copper for localized lignin deposition via laccases. Proc Natl Acad Sci U S A. 2022:119(24):e2202287119. 10.1073/pnas.2202287119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N. Biosynthesis of plant volatiles: nature's diversity and ingenuity. Science. 2006:311(5762):808. 10.1126/science.1118510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkheirandish M, Hensel G, Kilian B, Senthil N, Chen G, Sameri M, Azhaguvel P, Sakuma S, Dhanagond S, Sharma R, et al. Evolution of the grain dispersal system in barley. Cell. 2015:162(3):527–539. 10.1016/j.cell.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Qi X, Liu L, Liu C, Song L, Dong Y, Chen L, Li M. Sweet cherry AP2/ERF transcription factor, PavRAV2, negatively modulates fruit size by directly repressing PavKLUH expression. Physiol Plant. 2023:175(6):e14065. 10.1111/ppl.14065. [DOI] [PubMed] [Google Scholar]
- Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Bio. 2019:20(1):38. 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2020. https://www.R-project.org/. [Google Scholar]
- Rajani S, Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol. 2001:11(24):1914–1922. 10.1016/S0960-9822(01)00593-0. [DOI] [PubMed] [Google Scholar]
- Rau D, Murgia ML, Rodriguez M, Bitocchi E, Bellucci E, Fois D, Albani D, Nanni L, Gioia T, Santo D, et al. Genomic dissection of pod shattering in common bean: mutations at non-orthologous loci at the basis of convergent phenotypic evolution under domestication of leguminous species. Plant J. 2019:97(4):693–714. 10.1111/tpj.14155. [DOI] [PubMed] [Google Scholar]
- Ripoll JJ, Roeder AH, Ditta GS, Yanofsky MF. A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development. 2011:138(23):5167–5176. 10.1242/dev.073031. [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Alquézar B, Peña L. Fruit aromas in mature fleshy fruits as signals of readiness for predation and seed dispersal. New Phytol. 2013:197(1):36–48. 10.1111/j.1469-8137.2012.04382.x. [DOI] [PubMed] [Google Scholar]
- Selby R, Jones DS. Complex peptide hormone signaling in plant stem cells. Curr Opin Plant Biol. 2023:75:102442. 10.1016/j.pbi.2023.102442. [DOI] [PubMed] [Google Scholar]
- Seo J-H, Kang B-K, Dhungana SK, Oh J-H, Choi M-S, Park J-H, Shin S-O, Kim H-S, Baek I-Y, Sung J-S, et al. QTL mapping and candidate gene analysis for pod shattering tolerance in soybean (Glycine max). Plants. 2020:9(9):1163. 10.3390/plants9091163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003:13(11):2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Koul A, Kaul S, Dhar MK. Tissue-specific transcriptional regulation and metabolite accumulation in tomato (Solanum lycopersicum L.). Protoplasma. 2020:257(4):1093–1108. 10.1007/s00709-020-01492-2. [DOI] [PubMed] [Google Scholar]
- Shi Y, Li BJ, Su G, Zhang M, Grierson D, Chen KS. Transcriptional regulation of fleshy fruit texture. J Integr Plant Biol. 2022:64(9):1649–1672. 10.1111/jipb.13316. [DOI] [PubMed] [Google Scholar]
- Smýkal P, Parker T. Domestication-related changes in seed dispersal and pigmentation: visual selection and functional trait? Mol Plant. 2023:16(8):1240–1242. 10.1016/j.molp.2023.07.007. [DOI] [PubMed] [Google Scholar]
- Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 2009:19(11):606–616. 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Suzuki IK, Gacquer D, Van Heurck R, Kumar D, Wojno M, Bilheu A, Herpoel A, Lambert N, Cheron J, Polleux F, et al. Human-specific NOTCH2NL genes expand cortical neurogenesis through Delta/Notch regulation. Cell. 2018:173(6):1370–1384.e16. 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Kongjaimun A, Muto C, Kobayashi Y, Kumagai M, Sakai H, Satou K, Teruya K, Shiroma A, Shimoji M, et al. Same locus for non-shattering seed pod in two independently domesticated legumes, Vigna angularis and Vigna unguiculata. Front Genet. 2020:11:748. 10.3389/fgene.2020.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Gallusci P, Lang Z. Fruit development and epigenetic modifications. New Phytol. 2020:228(3):839–844. 10.1111/nph.16724. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008:320(5875):486–488. 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tohge T, Alseekh S, Fernie AR. On the regulation and function of secondary metabolism during fruit development and ripening. J Exp Bot. 2014:65(16):4599–4611. 10.1093/jxb/ert443. [DOI] [PubMed] [Google Scholar]
- Vanholme R, De Meester B, Ralph J, Boerjan W. Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol. 2019:56:230–239. 10.1016/j.copbio.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Wang D, Seymour GB. Molecular and biochemical basis of softening in tomato. Mol Hortic. 2022:2(1):5. 10.1186/s43897-022-00026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu X, Li Q, Xu N, He C. A lineage-specific arginine in POS1 is required for fruit size control in Physaleae (Solanaceae) via gene co-option. Plant J. 2022:111(1):183–204. 10.1111/tpj.15786. [DOI] [PubMed] [Google Scholar]
- Wang W-N, Wei Y-T, Zhao S-T, Yu F-H, Wang J-W, Gu C-Y, Liu X-R, Sai N, Zhu J-L, Wang QM, et al. ABSCISIC ACID-INSENSITIVE 5-KIP-RELATED PROTEIN 1-SHOOT MERISTEMLESS modulates reproductive development of Arabidopsis. Plant Physiol. 2024:195(3):2309–2322. 10.1093/plphys/kiae146. [DOI] [PubMed] [Google Scholar]
- Weeden NF. Genetic changes accompanying the domestication of Pisum sativum: is there a common genetic basis to the ‘domestication syndrome’ for legumes? Ann Bot. 2007:100(5):1017–1025. 10.1093/aob/mcm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden NF, Brauner S, Przyborowski JA. Genetic analysis of pod dehiscence in pea (Pisum sativum L.). Cell Mol Biol Lett. 2002:7(2B):657–663. https://api.semanticscholar.org/CorpusID:13872236. [PubMed] [Google Scholar]
- Wu H, He Q, He B, He S, Zeng L, Yang L, Zhang H, Wei Z, Hu X, Hu J, et al. Gibberellin signaling regulates lignin biosynthesis to modulate rice seed shattering. Plant Cell. 2023:35(12):4383–4404. 10.1093/plcell/koad244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, He Q, Wang Q. Advances in rice seed shattering. Int J Mol Sci. 2023:24(10):8889. 10.3390/ijms24108889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2021:2(3):100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Yang Z. PAMLX: a graphical user interface for PAML. Mol Biol Evol. 2013:30(12):2723–2724. 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- Yang C, Song J, Ferguson AC, Klisch D, Simpson K, Mo R, Taylor B, Mitsuda N, Wilson ZA. Transcription factor MYB26 is key to spatial specificity in anther secondary thickening formation. Plant Physiol. 2017:175(1):333–350. 10.1104/pp.17.00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Wang H. Molecular mechanisms for vascular development and secondary cell wall formation. Front Plant Sci. 2016:7:356. 10.3389/fpls.2016.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Jin E, Chung IK, Kim WT. Cell cycle-dependent regulation of telomerase activity by auxin, abscisic acid and protein phosphorylation in tobacco BY-2 suspension culture cells. Plant J. 2002:29(5):617–626. 10.1046/j.0960-7412.2001.01244.x. [DOI] [PubMed] [Google Scholar]
- Yong B, Zhu W, Wei S, Li B, Wang Y, Xu N, Lu J, Chen Q, He C. Parallel selection of loss-of-function alleles of Pdh1 orthologous genes in warm-season legumes for pod indehiscence and plasticity is related to precipitation. New Phytol. 2023:240(2):863–879. 10.1111/nph.19150. [DOI] [PubMed] [Google Scholar]
- Yuan L, Grotewold E. Plant specialized metabolism. Plant Sci. 2020:298:110579. 10.1016/j.plantsci.2020.110579. [DOI] [PubMed] [Google Scholar]
- Zablatzká L, Balarynová J, Klčová B, Kopecký P, Smýkal P. Anatomy and histochemistry of seed coat development of wild (Pisum sativum subsp. elatius (M. Bieb.) Asch. et Graebn. and domesticated pea (Pisum sativum subsp. sativum L.). Int J Mol Sci. 2021:22(9):4602. 10.3390/ijms22094602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng JK, Li X, Xu Q, Chen JY, Yin XR, Ferguson IB, Chen KS. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol J. 2015:13(9):1325–1334. 10.1111/pbi.12351. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Fan Z, Cui Y, Gu X, Chen S, Ma H. APETALA2/ethylene responsive factor in fruit ripening: roles, interactions and expression regulation. Front Plant Sci. 2022:13:979348. 10.3389/fpls.2022.979348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QY, Tu BJ, Liu CK, Liu XB. Pod anatomy, morphology and dehiscing forces in pod dehiscence of soybean (Glycine max (L.) Merrill). Flora. 2018:248:48–53. 10.1016/j.flora.2018.08.014. [DOI] [Google Scholar]
- Zhao Y, Zhang R, Jiang K-W, Qi J, Hu Y, Guo J, Zhu R, Zhang T, Egan AN, Yi T-S, et al. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol Plant. 2021:14(5):748–773. 10.1016/j.molp.2021.02.006. [DOI] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye Z-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006:18(11):3158–3170. 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lu D, Li C, Luo J, Zhu B-F, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B, et al. Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell. 2012:24(3):1034–1048. 10.1105/tpc.111.094383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript and the Supplementary materials are provided in Supplementary material online. Transcriptomic data of pea pod generated for this research have been submitted to NCBI (PRJNA1138002).