Abstract
Our study aimed to identify the bacterial source of a previously detected mobile antibiotic-resistant gene, mecA, found in a lake that serves as a source to a water treatment plant operated by a First Nation reserve. Three methicillin-resistant presumptive Staphylococcus spp. isolated from the sample using selective media were verified as mecA positive by PCR. MALDI-TOF and whole-genome sequencing of each isolate confirmed that all three were Mammaliicoccus fleurettii. Antibiotic-resistant gene analysis of the assembled genomes predicted mecA with 99.7% sequence identity, and phylogenetic analysis grouped our three mecA genes with the mecA allele from a methicillin-resistant strain of Staphylococcus aureus. Identifying microbial species known to harbour mobile antibiotic-resistant elements can provide greater depth of information about drinking water, an especially essential need in First Nation reserves where water quality too frequently is poor.
Keywords: Mammaliicoccus fleurettii, methicillin resistance, mobile genetic elements, water quality
Impact Statement
The global presence of antibiotic-resistant bacteria is a major public health concern. The transmission of antibiotic-resistant genes via mobile genetic elements is primarily talked about within pathogenic bacteria; however, non-pathogenic bacteria can also act as carriers and promote the movement of these resistant genes. The mecA gene confers methicillin resistance and is one of the most significant mobile resistance elements to β-lactam antibiotics (e.g., penicillin). Surface waters – including lakes, rivers and streams –are ecosystems that can act as a vehicle for bacterial movement. We discovered the presence of mecA-harbouring Mammaliicoccus fleurettii, a non-pathogenic bacterium, in a lake that provides drinking water to a remote First Nation reserve in Manitoba, Canada. This finding raises concern as the mecA mobile genetic element could potentially be transferred to staphylococcal pathogens within the water supply or distribution system. Our study highlights the need to proactively monitor potable water sources for non-pathogenic bacteria harbouring harmful mobile antibiotic-resistant genes.
Data Summary
All data and protocols for the three presumptive Staphylococcus spp. confirmed as Mammaliicoccus fleurettii have been provided within the article or are available upon request. NCBI assembly accession numbers: SF 001: GCA_016808275.1; SF 002: GCA_018310175.1; SF 003: GCA_018310195.1.
Introduction
Access to safe drinking water has been deemed a basic human right by the United Nations, essential for people’s well-being and dignity [1]. Poor water conditions in developing countries are a long-fought problem, but there is little public awareness of the poor water quality that affects people in developed countries. Canadian drinking water supplies are generally considered to be excellent [2]; however, water quality across many First Nation reserves is extremely poor [3], often prompting long-term drinking water advisories [4].
One of the major reasons for water advisories is the poor microbiological quality of the water [5]. We already documented this problem in First Nation reserves in Manitoba, Canada, where drinking water was found to have high levels of Escherichia coli despite the communities having access to water treatment plants; we also revealed the presence of antibiotic-resistant genes – considered an emerging water contaminant [6] – within the water supply of these communities [7]. One of the antibiotic-resistant genes identified was mecA, a clinically significant methicillin-resistant gene exemplified by methicillin-resistant Staphylococcus aureus (MRSA). The mecA gene encodes for a penicillin-binding protein and makes bacteria harbouring the gene less vulnerable to eradication upon exposure to most β-lactam antibiotics [8]. The mecA gene is commonly located on the staphylococcal cassette chromosome mec (SCCmec), a mobile genetic element that can also carry other antimicrobial-resistant genes, virulence determinants and genes that confer survival under stress [9].
In this study, we aimed to identify the bacterial source of the mecA gene that we previously discovered in a First Nation reserve’s lake water [7] from an area used as a source of water and for recreational activities. Such information can help guide and improve water monitoring protocols and assess potential health risks to the members of the community.
Methods
Community profile and water sampling
The water sample was collected in June 2019 (congruent with the sampling location and time of year in the initial study [7]) from a natural lake within a remote First Nation reserve in Manitoba, Canada, at an area frequented by the community. This area of the lake is also used as a source of water for the water treatment plant in the First Nation reserve. The water sample was collected using a standard method [10]. Sterile sampling bottles were opened just prior to filling; the sample was preserved with 0.003% sodium thiosulfate and stored at 4 °C or on ice packs. The water sample was processed immediately upon arrival at the laboratory, within 24 h of collection.
Isolation of methicillin-resistant presumptive Staphylococcus spp.
Oxacillin screening for methicillin-resistant presumptive Staphylococcus spp. followed published guidelines [11] and was performed with the lake water sample, autoclaved tap water (negative control) and an S. aureus clinical isolate HA-MRSA 100697 (positive control). Briefly, 100 ml of each water sample was filtered through a 0.45 µm sterile polyethersulfone membrane (Pall Corporation). Each membrane was placed on a lysogeny broth (LB) agar plate [BD Difco LB Agar (Lennox); Fisher Scientific] supplemented with 6 µg ml−1 oxacillin (Millipore Sigma) [11] and incubated at 37 °C for 48 h. Colonies from the LB+oxacillin plates containing the lake water sample were presumptively identified as methicillin-resistant staphylococci and were sub-cultured onto mannitol salt agar (MSA; Oxoid) supplemented with 6 µg ml−1 oxacillin for 48 h at 37 °C. Three yellow-pigmented colonies from the MSA+oxacillin plate – designated SF 001, SF 002 and SF 003 – were selected for further propagation and analysis, including MIC testing using the Gram-positive GPALL1F AST plate on the Sensititre system (Thermo Fisher Scientific).
Genotypic analysis of the bacterial isolates
Colony PCR [12] was used to screen the isolates for three genes: Staphylococcus species-specific rpoB [13], methicillin-resistant mecA [14] and S. aureus-specific nuc [15] (Table S1, available in the online version of this article). The PCR products were resolved on 1.3% agarose gels and visualized using an Axygen BL Gel Documentation System (Corning).
MALDI-TOF analysis of the bacterial isolates
The peptide profile of each of the isolates was determined using the Bruker Daltonics Microflex MALDI-TOF mass spectrometer. Chemical extraction of each bacterial isolate was performed following the manufacturer’s protocol; S. aureus was included as an extraction control. The Bruker Bacterial Test Standard (#8255343) was used to calibrate the system. One microlitre of each extract was spotted onto an MSP 96-well polished steel plate in triplicate, dried at room temperature and then overlaid with 1 μl of a saturated solution of α-cyano-4-hydroxycinnamic acid (Millipore Sigma). Using flexControl software (v.3.4), 240 laser shots (60 Hz N2-Cartridge-Laser) were accumulated for each spectrum. Spectra were analysed using MALDI BioTyper Compass Explorer software (v.4.1) and compared against the latest spectral database (BDAL DB, 8468 MSPs). Scoring was based on Bruker Daltonic’s BioTyper algorithm.
Whole-genome sequencing of the bacterial isolates and genomic analysis
Whole-genome sequencing was performed using genomic DNA purified from each of the three isolates using the Epicentre MasterPure Complete DNA and RNA Purification kit (Illumina) and prepared using the Nextera XT DNA Library Preparation kit (Illumina). Sequencing was performed using the Illumina MiSeq (150 bp paired-end reads). Sequence quality was assessed via FastQC [16], and de novo assembly was conducted using SPAdes (v.3.14.0) [17]. The quality of the assembled genomes was assessed using QUAST v.5.0.2 [18], and annotation was carried out using the NCBI Prokaryotic Genome Annotation Pipeline [19]. Each assembled genome sequence was submitted to the NCBI GenBank database. Antibiotic-resistant genes were predicted from the assembled genomes using the Comprehensive Antibiotic Resistance Database using RGI main under strict (>95% identity) and perfect (100% identity) parameters [20]. Phylogenetic relationships of the mecA genes were analysed in mega X [21] using the neighbour-joining method [22]; pairwise deletion removed ambiguous positions [23]. Genomic alignments were performed using Geneious Prime [24], BLASTn [25] and Easyfig [26]. All analyses were performed using default settings to ensure consistency across each analysis.
Results and discussion
Contaminated drinking water is a commonly recognized source of gastrointestinal infections [27]. Previous studies by us have highlighted the presence of antibiotic-resistant genes in lakes as well as post-treated tap and cistern water samples from First Nation reserves in Manitoba, Canada [7,28]. Families in these communities rely on lake water for their livelihood as well as spiritual practices. The presence of antibiotic-resistant microbes and their genes in these waters warrants further collaboration with the community to assess risk. The goal here was to determine the bacterial source of the mecA gene that we previously found in a water sample from a First Nation reserve [7].
We isolated three colonies from MSA plates supplemented with oxacillin, a standard screening media for pathogenic Staphylococcus spp. [11]. The smooth, elevated colonies were yellow, indicating the fermentation of mannitol [29]. The three isolates showed resistance towards oxacillin (resistance breakpoint ≥4 µg ml−1) and penicillin (resistance breakpoint 0.254 µg ml−1) (Table 1) according to Clinical and Laboratory Standards Institute guidelines [30]. A positive PCR reaction for the Staphylococcus species-specific rpoB gene and the mecA gene highly suggested that the isolates were staphylococci; however, the nuc gene was absent, meaning that the isolates were not specifically S. aureus (Fig. S1).
Table 1. MICs of antimicrobials in the GPALL1F AST plate for the three M. fleurettii isolates from this study.
| Antimicrobials | SF 001 | SF 002 | SF 003 |
| Ampicillin | 0.5 | 0.5 | 0.5 |
| Chloramphenicol | 8.0 | 8.0 | 8.0 |
| Ciprofloxacin | ≤1.0 | ≤1.0 | ≤1.0 |
| Clindamycin | ≤0.5 | ≤0.5 | ≤0.5 |
| Daptomycin | ≤0.5 | ≤0.5 | ≤0.5 |
| Dtest1 | ≤4.0 | ≤4.0 | ≤4.0 |
| Dtest2 | ≤8.0 | ≤8.0 | ≤8.0 |
| Erythromycin | ≤0.25 | ≤0.25 | ≤0.25 |
| Gentamicin | ≤2.0 | ≤2.0 | ≤2.0 |
| Levofloxacin | 0.5 | 0.5 | 0.5 |
| Linezolid | 2.0 | 2.0 | 2.0 |
| Moxifloxacin | 0.5 | 0.5 | 0.5 |
| Nitrofurantoin | ≤32.0 | ≤32.0 | ≤32.0 |
| Oxacillin+2% NaCl | >4.0 | >4.0 | >4.0 |
| Penicillin | 0.5 | 1.0 | 0.5 |
| Quinupristin/dalfopristin | ≤0.5 | ≤0.5 | ≤0.5 |
| High-level streptomycin (1000 µg ml−1) | ≤1000 | ≤1000 | ≤1000 |
| Tetracycline | ≤2.0 | ≤2.0 | ≤2.0 |
| Tigecycline | ≤0.03 | 0.06 | ≤0.03 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | ≤0.5 | ≤0.5 |
| Vancomycin | 1.0 | 1.0 | 1.0 |
We then performed MALDI-TOF mass spectrometric analysis on protein extracts from each isolate and confirmed that the bacterium was not S. aureus. Whole-genome sequencing and 16S rRNA annotation of each assembled genome allowed us to identify our isolates as M. fleurettii. M. fleurettii is a recently reclassified species, once called Staphylococcus fleurettii [31], and believed to be the first point of emergence of the mecA gene [32,33]. It is most commonly associated with cattle, sheep, goat and camelids [34]; has been isolated from wild animals (rabbits [35], shrews, voles and mice [36]) and companion animals (dogs and cats [37]) and has also been linked with bovine mastitis [38]. Most animal-associated M. fleurettii isolates showed the presence of mecA [34,35, 37]. To our knowledge, our samples are the first Canadian lake-water-derived, mecA-carrying M. fleurettii isolates; superficial waters and wastewater have also been documented as sources of mecA-harbouring M. fleurettii [39].
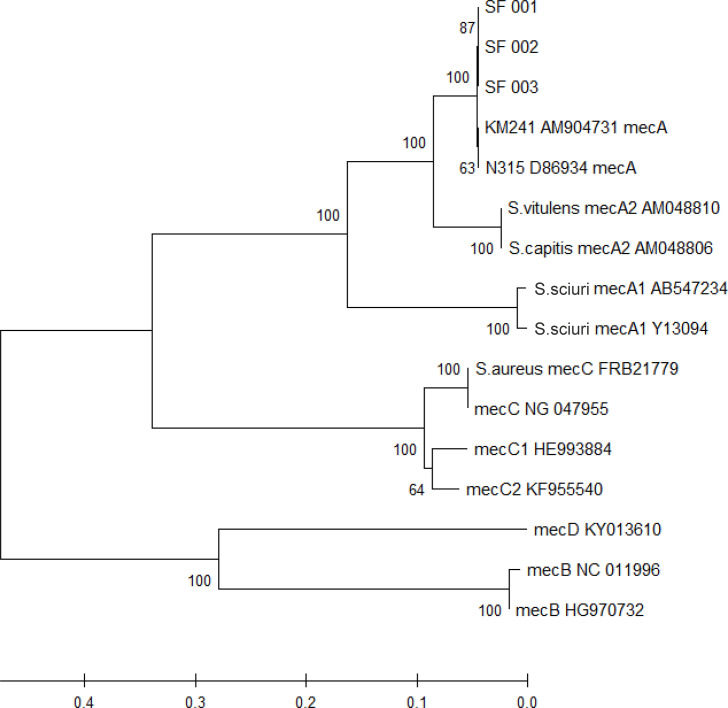
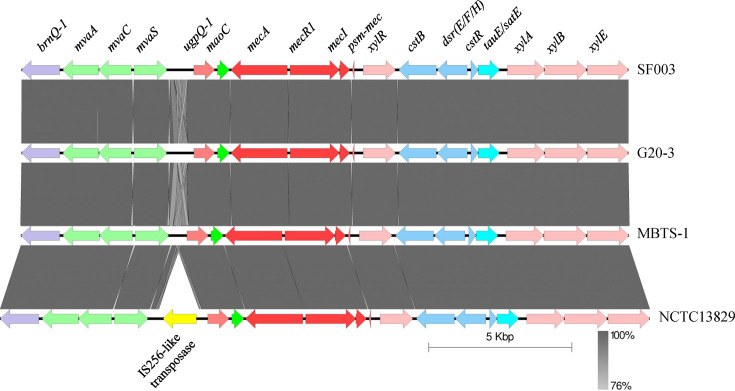
Sequencing data were used to generate genome assemblies (total length: >2.47 Mbp; length N50: >113,000; %GC: 31%) (Fig. S2), identifying >2400 protein-encoding genes in each of the samples. Antibiotic-resistant genes were predicted from the assemblies using the Comprehensive Antibiotic Resistance Database; under ‘strict’ parameters, a hit yielding 99.7% sequence identity with mecA was found. No further antibiotic-resistant genes were identified under ‘loose’ parameters (<95% identity). The per cent nucleotide identity of the mec gene complex of our three isolates against reference genomes (MBTS-1, D86934.2MBTS-1D86934.2) showed at least 98.8% identity (Table 2). Phylogenetic tree analysis linked our three mecA genes to the mecA allele type from the SCCmec cassette of Staphylococcus pseudintermedius KM241 (AM904731.1AM904731.1) and methicillin-resistant S. aureus N315 (D86934.2D86934.2) (Fig. 1). Furthermore, the mec gene complex flanking region – a 21 881 bp sequence comprising brnQ-1-mvaA-mvaC-mvaS-ugpQ-1-maoC-mecA-mecR1-mecI-psm-xylR-cstB-DsrE/DsrF/DsrH-cstR-TauE/SafE-xylA-xylB-xylE – showed 100% nucleotide coverage and identity with a representative M. fleurettii isolate and three other M. fleurettii genomes (MBTS-1, G20-3MBTS-1G20-3 and NCTC18329NCTC18329) (Fig. 2).
Table 2. Per cent identity matrix of mecA, mecR1 and mecI nucleotide sequences from an M. fleurettii reference genome (MBTS-1MBTS-1), a methicillin-resistant S. aureus N315 (D86934.2D86934.2) and the three M. fleurettii isolates from this study (SF 001, SF 002 and SF 003) (Geneious Prime [24]).
| Strains | MBTS-1 (%) | N315 (%) | SF 001 (%) | SF 002 (%) | SF 003 (%) |
| MBTS-1 | 98.82 | 98.96 | 98.96 | 98.96 | |
| N315 | 98.82 | 99.86 | 99.86 | 99.86 | |
| SF 001 | 98.96 | 99.86 | 100% | 100% | |
| SF 002 | 98.96 | 99.86 | 100% | 100% | |
| SF 003 | 98.96 | 99.86 | 100% | 100% |
Fig. 1. Phylogenetic analysis of the mecA gene from M. fleurettii (SF 001, SF 002 and SF 003) was inferred using the neighbour-joining method [22]. The optimal tree with the sum of branch length=1.373 is shown. Evolutionary distances were computed using the maximum composite likelihood method; branch lengths represent the number of base substitutions per site (scale shown), and units on the branches indicate branch support values; pairwise deletion removed ambiguous positions [23]. A total of 2042 positions comprised the final dataset (mega X [21]).
Fig. 2. Genomic alignment of the mec gene complex and its flanking region using a representative M. fleurettii isolate (SF 003) along with a M. fleurettii reference genome (MBTS-1MBTS-1) and two other NCBI M. fleurettii genomes (G20-3 and NCTC18329G20-3NCTC18329) (Easyfig [26]).
Our study had a defined purpose and so did not investigate peripheral questions. The source of M. fleurettii in this First Nation reserve’s lake water is unknown, but given the vastness and remoteness of the lake, it would be difficult for this to be precisely determined. One could speculate that M. fleurettii was of animal origin, wild or companion [35,37]. Given that we isolated M. fleurettii years after discovering the mecA gene does suggest, though, that there is a consistent source of contamination. Broader water sampling around the lake may be a beneficial future collaboration with the First Nation community.
Conclusion
In this study, we showed that M. fleurettii was present in a lake water sample taken from a First Nation reserve in Manitoba, Canada, and established that this bacterium was a source of the mecA gene previously identified in the same water. The mobility of the mecA gene means that M. fleurettii can potentially transfer the gene to other known staphylococcal pathogens [40]. Our study underscores the importance of understanding the full bacterial composition of the lake and drinking waters to identify seemingly harmless bacteria harbouring transferable antibiotic-resistant genes. Such precautions may reduce potentially serious human health issues.
supplementary material
Acknowledgements
We thank the members of the First Nation reserve for their valuable research partnership and Anita Murdock for her indispensable assistance; without all of their help, this study would not have been possible. The authors acknowledge that the University of Manitoba campuses are located on the original lands of Anishinaabeg [ah-nish-in-ah-bek], Ininewuk [in-i-nuh-wuk], Anisininewuk [un-shin-i-wuk], Dakota Oyate [oh-yah-tay] and Denesuline [deh-nay-sue-li-nay] and on the National Homeland of the Red River Métis.
Abbreviations
- LB
lysogeny broth
- MRSA
methicillin-resistant Staphylococcus aureus
- MSA
mannitol salt agar
- SCCmec
staphylococcal cassette chromosome mec
Footnotes
Funding: This research was supported by a project grant from the Canadian Institutes of Health Research to AK and AF.
Accession No: NCBI assembly accession numbers: SF 001: GCA_016808275.1; SF 002: GCA_018310175.1; SF 003: GCA_018310195.1.
Author contributions: S.B. – conceptualization, investigation, data curation, formal analysis, writing-original draft, visualization; R.P. – conceptualization, methodology; A.W. – formal analysis, software, writing-reviewing and editing; D.W. – writing-reviewing and editing, formal analysis; G.R.G. – supervision, resources, writing-reviewing and editing; A.F. – funding acquisition, supervision, writing-reviewing and editing; A.K. – funding acquisition, conceptualization, supervision, writing-reviewing and editing.
Contributor Information
Sabrin Bashar, Email: sbashar@ualberta.ca.
Rakesh Patidar, Email: rakeshk.patidar82@gmail.com.
Alvan Wai, Email: alvanwai@outlook.com.
Dawn White, Email: Dawn.White@umanitoba.ca.
George R. Golding, Email: George.golding@phac-aspc.gc.ca.
Annemieke Farenhorst, Email: Annemieke.Farenhorst@umanitoba.ca.
Ayush Kumar, Email: ayush.kumar@umanitoba.ca.
References
- 1.UN Office of the High Commissioner for Human Rights . United Nations, Geneva; 2010. Fact sheet no. 35: the right to water. [Google Scholar]
- 2.Government of Canada Canadian Drinking Water Guidelines. 2022. [13-April-2024]. https://www.canada.ca/en/health-canada/services/environmental-workplace-health/water-quality/drinking-water/canadian-drinking-water-guidelines.html accessed.
- 3.Patrick RJ. Uneven access to safe drinking water for First Nations in Canada: connecting health and place through source water protection. Health Place. 2011;17:386–389. doi: 10.1016/j.healthplace.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Harbison M. ENSC 501 – independent environmental study project; 2012. [13-April-2024]. An analysis of water quality and human health issues in First Nations communities in Canada.https://qspace.library.queensu.ca/server/api/core/bitstreams/5f5fe8f8-e964-44e4-ad93-9d8ce6992d87/content accessed. [Google Scholar]
- 5.Galway LP. Boiling over: a descriptive analysis of drinking water advisories in First Nations Communities in Ontario, Canada. Int J Environ Res Public Health. 2016;13:505. doi: 10.3390/ijerph13050505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivalingam P, Poté J, Prabakar K. Extracellular DNA (eDNA): neglected and potential sources of Antibiotic Resistant Genes (ARGs) in the aquatic environments. Pathogens. 2020;9:874. doi: 10.3390/pathogens9110874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernando DM, Tun HM, Poole J, Patidar R, Li R, et al. Detection of antibiotic resistance genes in source and drinking water samples from a first nations community in Canada. Appl Environ Microbiol. 2016;82:4767–4775. doi: 10.1128/AEM.00798-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierre J, Williamson R, Bornet M, Gutmann L. Presence of an additional penicillin-binding protein in methicillin-resistant Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, and Staphylococcus simulans with a low affinity for methicillin, cephalothin, and cefamandole. Antimicrob Agents Chemother. 1990;34:1691–1694. doi: 10.1128/AAC.34.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice EW, Baird RB, Eaton AD, Clesceri LS. Standard Methods for the Examination of Water and Wastewater, 22nd edn. Washington, DC: American Public Health Association, American Water Works, Water Environment Federation; 2012. [Google Scholar]
- 11.Brown DFJ, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, et al. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA) J Antimicrob Chemother. 2005;56:1000–1018. doi: 10.1093/jac/dki372. [DOI] [PubMed] [Google Scholar]
- 12.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 13.Mellmann A, Becker K, von Eiff C, Keckevoet U, Schumann P, et al. Sequencing and staphylococci identification. Emerg Infect Dis. 2006;12:333–336. doi: 10.3201/eid1202.050962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martineau F, Picard FJ, Grenier L, Roy PH, Ouellette M, et al. Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. The ESPRIT trial. J Antimicrob Chemother . 2000;46:527–534. doi: 10.1093/jac/46.4.527. [DOI] [PubMed] [Google Scholar]
- 15.Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 17.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A . 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geneious Geneious Prime 2023.0.1. 2023. https://www.geneious.com
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Health Canada Guidance on waterborne pathogens in drinking water. 2022. [13-April-2024]. https://www.canada.ca/content/dam/hc-sc/documents/services/environmental-workplace-health/reports-publications/water-quality/guidance-waterborne-pathogens-drinking-water/guidance-waterborne-pathogens-drinking-water.pdf accessed.
- 28.Murdock A, Bashar S, White D, Uyaguari-Diaz M, Farenhorst A, et al. Bacterial diversity and resistome analysis of drinking water stored in cisterns from two First Nations communities in Manitoba, Canada. Microbiol Spectr . 2024;12:e03141–23. doi: 10.1128/spectrum.03141-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp SE, Searcy C. Comparison of mannitol salt agar and blood agar plates for identification and susceptibility testing of Staphylococcus aureus in specimens from cystic fibrosis patients. J Clin Microbiol. 2006;44:4545–4546. doi: 10.1128/JCM.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CLSI . CLSI Supplement M100. 30th edn. Clinical and Laboratory Standards Institute; 2020. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 31.Madhaiyan M, Wirth JS, Saravanan VS. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int J Syst Evol. 2020;70:5926–5936. doi: 10.1099/ijsem.0.004498. [DOI] [PubMed] [Google Scholar]
- 32.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 2010;54:4352–4359. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwenderen S, Perreten V. The bla and mec families of β-lactam resistance genes in the genera Macrococcus, Mammiliicoccus and Staphylococcus: an in-depth analysis with emphasis on Macrococcus. J Antimicrob Chemother. 2020;77:1796–1827. doi: 10.1093/jac/dkac107. [DOI] [PubMed] [Google Scholar]
- 34.Schauer B, Szostak MP, Ehricht R, Monecke S, Feßler AT, et al. Diversity of methicillin-resistant coagulase-negative Staphylococcus spp. and methicillin-resistant Mammaliicoccus spp. isolated from ruminants and New World camelids. Vet Microbiol. 2021;254:109005. doi: 10.1016/j.vetmic.2021.109005. [DOI] [PubMed] [Google Scholar]
- 35.Sousa M, Silva V, Silva A, Silva N, Ribeiro J, et al. Staphylococci among wild European rabbits from the azores: a potential zoonotic issue? J Food Prot. 2020;83:1110–1114. doi: 10.4315/0362-028X.JFP-19-423. [DOI] [PubMed] [Google Scholar]
- 36.Hauschild T, Sliżewski P, Masiewicz P. Species distribution of staphylococci from small wild mammals. Syst Appl Microbiol. 2010;33:457–460. doi: 10.1016/j.syapm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Loncaric I, Tichy A, Handler S, Szostak MP, Tickert M, et al. Prevalence of methicillin-resistant Staphylococcus sp. (MRS) in different companion animals and determination of risk factors for colonization with MRS. Antibiotics. 2019;8:36. doi: 10.3390/antibiotics8020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naushad S, Barkema HW, Luby C, Condas LAZ, Nobrega DB, et al. Comprehensive phylogenetic analysis of bovine non-aureus Staphylococci species based on whole-genome sequencing. Front Microbiol. 2016;7:1990. doi: 10.3389/fmicb.2016.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gómez P, Casado C, Sáenz Y, Ruiz-Ripa L, Estepa V, et al. Diversity of species and antimicrobial resistance determinants of staphylococci in superficial waters. FEMS Microbiol Ecol. 2017;93:fiw208. doi: 10.1093/femsec/fiw208. [DOI] [PubMed] [Google Scholar]
- 40.Lienen T, Schnitt A, Hammerl JA, Maurischat S, Tenhagen B-A. Mammaliicoccus spp. from german dairy farms exhibit a wide range of antimicrobial resistance genes and non-wildtype phenotypes to several antibiotic classes. Biology. 2022;11:1525. doi: 10.3390/biology11020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.