Abstract
In bacteria the formation of disulphide bonds is facilitated by a family of enzymes known as the disulphide bond forming (Dsb) proteins, which, despite low sequence homology, belong to the thioredoxin (TRX) superfamily. Among these enzymes is the disulphide bond-forming protein A (DsbA); a periplasmic thiol oxidase responsible for catalysing the oxidative folding of numerous cell envelope and secreted proteins. Pathogenic bacteria often contain diverse Dsb proteins with distinct functionalities commonly associated with pathogenesis. Here we investigate FtDsbA1, a DsbA homologue from the Gram-negative bacterium Francisella tularensis. Our study shows that FtDsbA1 shares a conserved TRX-like fold bridged by an alpha helical bundle showcased by all DsbA-like proteins. However, FtDsbA1 displays a highly unique variation on this structure, containing an extended and flexible N-terminus and secondary structural elements inserted within the core of the TRX fold itself, which together twist the overall DsbA-like architecture. Additionally, FtDsbA1 exhibits variations to the well conserved active site with an unusual dipeptide in the catalytic CXXC redox centre (CGKC), and a trans configuration for the conserved cis-proline loop, known for governing DsbA-substrate interactions. FtDsbA1’s redox properties are comparable to other DsbA enzymes, however, consistent with its atypical structure, functional analysis reveals FtDsbA1 has a high degree of substrate specificity suggesting a specialised role within F. tularensis’ oxidative folding pathway. Overall, this work underscores the remarkable malleability of the TRX catalytic core, a ubiquitous and ancestral protein fold. This not only contributes to broadening the structural and functional diversity seen within proteins utilising this core fold but will also enhance the accuracy of AI-driven protein structural prediction tools.
Keywords: Thioredoxin protein, Disulphide bond: Oxidoreductase, Crystal structure, Redox biology, AlphaFold
Graphical Abstract
1. Introduction
Members of the thioredoxin (TRX) protein superfamily are widespread in nature where they facilitate essential processes including protein folding and redox signalling as well as contributing to oxidative stress responses and detoxification [47], [59]. TRX-like proteins are defined by a common architecture centred on a core βαβ-α-ββα scaffold, where different inserts between the βαβ and ββα motifs along with extensions to the N and C termini expand the functional capabilities of this protein family (reviewed in [2], [49], [64]). The TRX fold is particularly prevalent among redox active proteins that participate in the introduction of disulphide bonds into substrates, a process known as oxidative folding [24], [43]. In Gram-negative bacteria these oxidative folding reactions occur in the periplasm and are carried out by a group of enzymes known as the disulphide bond forming proteins (Dsb) [4], [43]. Owing to the abundance of disulphide bonds in secreted virulence factors and cell envelope proteins, these Dsb molecular machines are implicated in a wide array of pathogenesis processes and antimicrobial resistance mechanisms across a range of pathogens [20], [24], [75].
The canonical bacterial model for oxidative folding comes from Escherichia coli K-12 which comprises the DsbA/DsbB oxidative pathway [24], [33], where monomeric DsbA introduces disulphide bonds into substrate proteins and is then re-oxidised by the membrane protein DsbB [5]. The additional DsbC/DsbD isomerase pathway corrects non-native bonds via the periplasmic dimer DsbC [92], which is kept reduced by the membrane embedded DsbD [43], [73].
DsbA is at the spearhead of the oxidative folding pathway and is the best characterised Dsb protein in bacteria [41], [42], [63] with DsbA from E. coli (EcDsbA) serving as the prototypical model for unravelling the structural and functional properties of this protein family. The highly oxidising EcDsbA encompasses a core TRX-like domain which is interrupted by a long alpha helical insertion embedded between the βαβ and ββα motifs, and contains an active site utilising a CPHC redox motif and GVcisP loop - all highly conserved features of the DsbA family [24], [50], [75].
While the structure and catalytic residues of EcDsbA are highly conserved across DsbAs from many bacterial species [78] it is increasingly apparent that more diverse catalytic machinery exists, notably within pathogenic bacteria [84], [14], [24], [43].
Here, we report the crystal structure of DsbA1 from Francisella tularensis (FtDsbA1); a highly infectious Gram-negative coccobacillus responsible for the condition tularaemia [71], [74]. The overall structure reveals a distorted TRX-like architecture marked by an extended N-terminus, insertions within the fold core motifs, and a highly unusual active site, all atypical features that deviate from canonical DsbA-like proteins. However, biophysical and biochemical analyses of FtDsbA1 reveals redox properties comparable to other DsbA proteins although, attributed to its unique structural elements, this protein features narrowed substrate specificity suggesting a specialised function within F. tularensis’ oxidative pathway. These findings provide insights into the structural plasticity of the ubiquitous TRX fold that enables a remarkable functional diversity within the TRX superfamily.
2. Experimental procedures
2.1. Cloning, expression, and purification
Codon optimised FtDsbA1 (locus FTT_0507) with the signal sequence removed and an N terminal TRX-His6 tag was cloned into a modified version of a pMCSG7 vector [17], [26] to create pMCSG7::TRX-His6-FtDsbA1 (Epoch Life Science). FtDsbA1 was then expressed in E. coli BL21(DE3) using autoinduction media (24 h at 30 °C) in the presence of ampicillin (100 μg/mL) [81]. Cells were then harvested, resuspended in Tris buffer (20 mM Tris, 150 mM NaCl, 20 mM imidazole, pH 7) and lysed by sonication (Misonix S-4000 Ultrasonic Liquid Processor (Qsonica)). N-terminal His6-tagged FtDsbA1 was purified from cytoplasmic extracts by nickel affinity chromatography followed by His6-tag cleavage by TEV protease and reverse nickel affinity chromatography. The protein was oxidised using 10 molar equivalents of oxidised glutathione (GSSG; Sigma–Aldrich, USA) and was subsequently purified to homogeneity by size exclusion chromatography using a HiLoad® 16/600 Superdex® column (GE Healthcare) equilibrated in HEPES buffer (25 mM HEPES, 150 mM NaCl, pH 7). Protein purity was assessed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
For cell-based assays, a second plasmid construct was utilised. Prior to cloning, the EcDsbAss-∆ss-CcScsC cassette was subcloned from pSC108 into a pWSK29 vector. For periplasmic expression in the previously described E. coli K-12 strains JCB817 [5] and PL263 (MC4100 ΔdsbC ΔmdoG)[46], the mature form of FtDsbA1 was cloned into pWSK29::EcDsbAss-∆ss-CcScsC vectors such that the FtDsbA1 gene exchanges the CcScsC gene at the NcoI/HindIII cut sites. This results in a mature FtDsbA1 construct which carries an N-terminal EcDsbA signal sequence for periplasmic translocation (pWSK29:: EcDsbAss-∆ss-FtDsbA1). For simplicity, this plasmid will be referred to as pWSK29-FtDsbA1.
2.2. Crystallisation
FtDsbA1 crystals were grown via the hanging drop vapour diffusion method at 293 K over the course of 2 weeks. Crystallisation conditions were determined following initial high-throughput commercial screening. Initial hits were obtained via the sitting drop diffusion method with 0.2 μL droplets at 293 K in the 36th PACT premiere condition (Molecular Dimensions, UK) (0.01 M ZnCl2, 0.1 M HEPES (pH 7), 20 % v/v PEG 6000). Diffraction quality crystals were produced in 2 μL droplets containing 1 μL of reservoir solution consisting of 0.1 M HEPES (pH 7.58), 0.01 M ZnCl2 and 20% PEG 3350 (v/v), and 1 μL of protein at 58 mg/mL in 25 mM HEPES, 150 mM NaCl, pH 7. Crystals were transferred to a cryoprotectant solution containing crystallisation solution supplemented with 20 % glycerol (v/v) and cryocooled in liquid nitrogen prior to data collection.
2.3. Structural determination and refinement
Diffraction data was collected at the Australian synchrotron using the MX2 beamline (100 K, 13 kEV) using an EIGER x 16 M detector. Data was collected covering 360° with 0.1° per frame with 0.02 s exposure (average x-ray dose for the exposed region 35.6 MGy (calculated using Raddose-3D [93])). The data was indexed and integrated using XDS [37] and scaled using POINTLESS [18] and SCALA [37]. Crystals belonged to spacegroup P212121 with cell dimensions of a = 43.9 Å, b = 57.5 Å, c = 168.8 Å and α = β = γ = 90.0°, diffracted at a resolution of 1.95 Å and contained two molecules per asymmetric unit [52]. The crystal structure of FtDsbA1 was solved using Phaser by molecular replacement [70] using an AlphaFold2 [85], [36], [55] partial computational model of FtDsbA1. The resulting model was refined using REFMAC5 [57] and further built using COOT [15]. The quality of the model was monitored during refinement by the Rfree value, which represented 5 % of the data. The structure was validated by the MolProbity server [11] and Figures were created with PyMOL [12]. Details of data-processing and final refinement statistics are summarised in Table 1.
Table 1.
Data collection and refinement statistics.
| PDB ID: 9EDL | |
|---|---|
| Data Collection | |
| Resolution range (Å) | 47.39 – 1.95 (2.06 – 1.95) |
| Wavelength (Å) | 0.95373 |
| Space group | P212121 |
| Unit cell parameters (Å, °) | a= 43.74, b= 57.28, c= 168.81 α = β = γ = 90.00 |
| Number of molecules per asymmetric unit | 2 |
| Total number of reflections | 137047 (16557) |
| Number of unique reflections | 31035 (3729) |
| Completeness (%) | 99.2 (94.3) |
| Redundancy | 4.4 (4.4) |
| I/σ(I) | 8.2 (1.8) |
| Rmerge | 0.108 (0.685) |
| Rmeas | 0.123 (0.778) |
| Rpim | 0.058 (0.364) |
| CC(1/2) | 0.996 (0.802) |
| Refinement | |
| Resolution (Å) | 47.39 – 1.95 (2.06 – 1.95) |
| Rwork/Rfree | 18.36/22.58 (26.14/28.44) |
| Ccwork | 0.926 (0.88) |
| No. of atoms (excluding H) | |
| Protein | 3494 |
| Solvent | 266 |
| Zn2+ | 6 |
| PEG | 7 |
| Protein Residues | 448 |
| R.M.S.D from ideal geometry | |
| Bonds (Å) | 0.006 |
| Angles (°) | 0.73 |
| Ramachandran (%) | |
| Favoured | 98.87 |
| Allowed | 1.13 |
| Outliers | 0.00 |
| Rotamer Outliers (%) | 0.53 |
| Clashscore (%) | 2.70 |
| Average B-factors | |
| Overall | 32.95 |
| Protein | 32.55 |
| Solvent | 37.24 |
*Values in parentheses represent the highest resolution shell
2.4. Sedimentation velocity analytical ultracentrifugation (SV-AUC)
SV-AUC using an Optima XL-A analytical ultracentrifuge (Beckman) was used to determine the quaternary structure of FtDsbA1 in both its oxidised and reduced forms. FtDsbA1 was loaded into double sector quartz cells (370 μL) at 2.5, 1 and 0.25 mg/mL in HEPES buffer (25 mM HEPES, 150 mM NaCl, pH 7) using a An50-Ti rotor. Initial scans were performed at 3000 rpm to determine the optimal wavelength and radial positions. Absorbance readings were collected at 270 nm and 40,000 rpm at 20 °C. Solvent density, solvent viscosity and estimates of the partial specific volume of FtDsbA1 (0.7371 mL g−1) at 20 °C were calculated with SEDNTERP [44]. Data was fitted using SEDFIT [76] to determine the c(s) and c(M) values with Z test values < 40 and RMSD < 0.008 for all data fitting.
2.5. Determination of redox potential
The redox equilibrium of FtDsbA1 was determined as described previously [31]. Briefly, oxidised FtDsbA1 was diluted to 12 μM in 100 mM sodium phosphate (pH 7), 1 mM GSSG (Sigma–Aldrich, USA) and 32 different concentrations of reduced glutathione (GSH; Sigma-Aldrich, USA) (7.1 μM to 3.8 mM). After overnight incubation, the redox state of FtDsbA1 was analysed in a Spectra max M5e plate reader (Bio-strategy) by measuring the fluorescence emission at 330 nm after excitation at 280 nm. The equilibrium constant Keq and redox potential were determined using the standard equations [56]. Briefly, the data was then fitted using GraphPad Prism (GraphPad Software, Inc., San Diego, CA, USA) and the equilibrium constant Keq estimated according to the equation below:
| Yobs=( Yox+ (M/Keq)Yred) / 1+ (M/Keq), |
where Yobs is the fraction of reduced protein at equilibrium, Yox and Yred are the signals for the oxidised and reduced proteins, respectively, and M is the ratio of [GSH]2/[GSSG].
The redox potential was determined from the Nernst equation:
E0’= E0’GSH/GSSG(RT/2 F) × ln Keq, where E0’GSH/GSSG is the standard potential of −240 mV, R is the universal gas constant 8.314 J K−1 mol−1, T is the absolute temperature in K, F is the Faraday constant 9.648 × 104 C mol−1, and Keq is the equilibrium constant. Keq was calculated as mean ± standard error of the mean (SEM) with three independent replicates.
2.6. Thermal unfolding by circular dichroism (CD)
Conformation of the secondary structure and temperature induced stability curves of the protein were investigated by circular dichroism (CD) spectroscopy using a JASCO Model J-1100 CD spectrophotometer (JASCO, USA). For analysis, 0.3 mg/mL of protein was oxidised or reduced with a 10 times molar excess of GSSG or DTT respectively in a 100 mm sodium phosphate buffer (pH 7) supplemented with 1 mM EDTA. Wavelength scans were performed in triplicate measuring between 190 nm and 250 nm in 0.5 nm intervals at 20 °C while thermal unfolding was monitored at 222 nm between 20 °C and 90 °C with a heating rate of 0.5 °C/min. Three replicates were taken for both oxidised and reduced protein. The melting temperature was calculated according to the below equation assuming a two-state model using GraphPad Prism (GraphPad Software, Inc., San Diego, CA, USA):
where Y is CD signal (mdeg), x is temperature (°C), t is temperature x in Kelvin (K), Tm is the midpoint of the thermal unfolding curve (K), yf is the intercept of the fully folded baseline pre-transition, mf is the gradient of the fully folded baseline pre-transition, yu is the intercept of the fully unfolded baseline post-transition, mu is the gradient of the fully unfolded baseline post-transition, h is the change in enthalpy for unfolding at Tm [58].
2.7. Thiol-oxido-reductase activity
The thiol-oxidase activity was measured in vitro using a Fluorescence Resonance Energy Transfer (FRET) based assay against a substrate peptide derived from the arylsulfate sulfotransferase (ASST) enzyme [89]. Oxidation of the cysteine pair in the peptide ASST (CNENGLCK) causes peptide cyclisation that results in the N-terminal 1,4,7,10-tetraazacyclododecane-1,4,7,10- tetraacetic acid (DOTA) - europium (Eu3+) to come into close proximity with a methylcoumarin amide linked to a C-terminal lysine. This causes excitation of the coumarin moiety at 340 nm with a fluorescence emission that was detected at 615 nm.
To perform the assays, 50 μL reaction mixtures containing 500 nM of the protein, 2 mM GSSG and 15 µM ASST peptide in 50 mM MES (pH 5.5), 50 mM NaCl, 2 mM EDTA, were added into a PerkinElmer 384-well white opaque microplate (OptiPlate−384). The time-resolved fluorescence (excitation λ = 340 nm and emission λ = 615 nm) was monitored over the course of 30 min using a CLARIOstar plate reader (BMG Labtech) fitted with a TR-FRET module. Measurements were carried out in triplicate and data were analysed and mean ± standard error of the mean (SEM) was plotted using GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA).
The ability of FtDsbA1 to reduce disulphide bonds in vitro in the presence of DTT was analysed using the insulin reduction assay [30]. Briefly, reaction samples were prepared by adding 131 μM insulin to 10 μM FtDsbA1 (or controls EcDsbA and EcDsbC) in 100 mM sodium phosphate (pH 7), 2 mM EDTA and 0.35 mM DTT. Reduction of insulin was monitored by measuring the change in optical density at 650 nm over 80 min
2.8. Kinetics of FtDsbA1 mediated oxidation
The kinetics of oxidation of DTT, PapD peptide [89], [35] (FICNGSRCSV) and FtPilA peptide (TCNIDGVSASQVPSNCN) by FtDsbA1 was monitored following the change in intrinsic fluorescence of FtDsbA1 upon reduction using a SX20 Stopped-flow Spectrometer (Applied Photophysics). In preparation for the analysis, 20 μM FtDsbA1 and EcDsbA were fully oxidised with an excess of GSSG (Sigma–Aldrich, USA) for 1 h and buffer exchanged to remove oxidising agent. The oxidised proteins (2 μM) were individually mixed in increasing concentrations of DTT (Sigma), FtPilA or PapD peptide (2–10 μM) at 30 °C in 25 mM Tris (pH 7), 50 mM NaCl, 1 mM EDTA. The change in fluorescence upon reduction was monitored using a λem 330 nm emission filter (λex 295 nm). Data was processed and analysed using Pro-Data SX (Applied photophysics), and the initial rates (r) of oxidation were determined from the linear portion of the fluorescence data. Initial rates calculated as mean ± standard error of the mean (SEM) with three independent replicates.
2.9. In vivo swimming motility assay
The ability of FtDsbA1 to oxidatively fold a component of the E. coli flagellar motor, FlgI [28], was assessed. The pWSK29-FtDsbA1 construct was transformed into JCB817 (∆dsbA) cells to assess its ability to reduce bacterial swimming motility, indicative of thiol-oxidase activity, on a 24 well Nunc plate as previously described [86]. Briefly, overnight cultures were normalised to an OD600 of 1.0 and inoculated on the left side of wells containing 0.25 % w/v LB-Lennox agar (Gibco™ Bacteriological agar) supplemented with IPTG (500 μM) and chloramphenicol (17 μg/mL). As bacteria migrate across the agar surface, a radius of motility corresponding to an increased OD600 was observed and monitored over 18 hrs using a CLARIOstar pate reader (BMG, Australia).
2.10. Mucoidal morphology assay
To assess the in vivo disulphide reductase activity (indicative of isomerase activity) of FtDsbA1, the production of colanic acid resulting in a distinctive mucoidal plate morphology was assessed as previously described [46]. Briefly, PL263 (∆dsbC, ΔmdoG) cells carrying pWSK29-FtDsbA1 were patched onto 1.5 % (w/v) solid agar containing M9 media (2 mM MgSO4, 100 μM CaCl2, 0.4 % v/v glycerol, 0.1 % w/v casamino acid, 1x M9 salt) supplemented with IPTG (500 μM) and chloramphenicol (17 μg/mL). Following 48 h of incubation at 29 °C, plates were imaged using a ChemiDoc MP Imaging System (Bio-Rad) and visually inspected for changes in mucoidal morphology.
3. Results
3.1. FtDsbA1 contains DsbA-like architecture
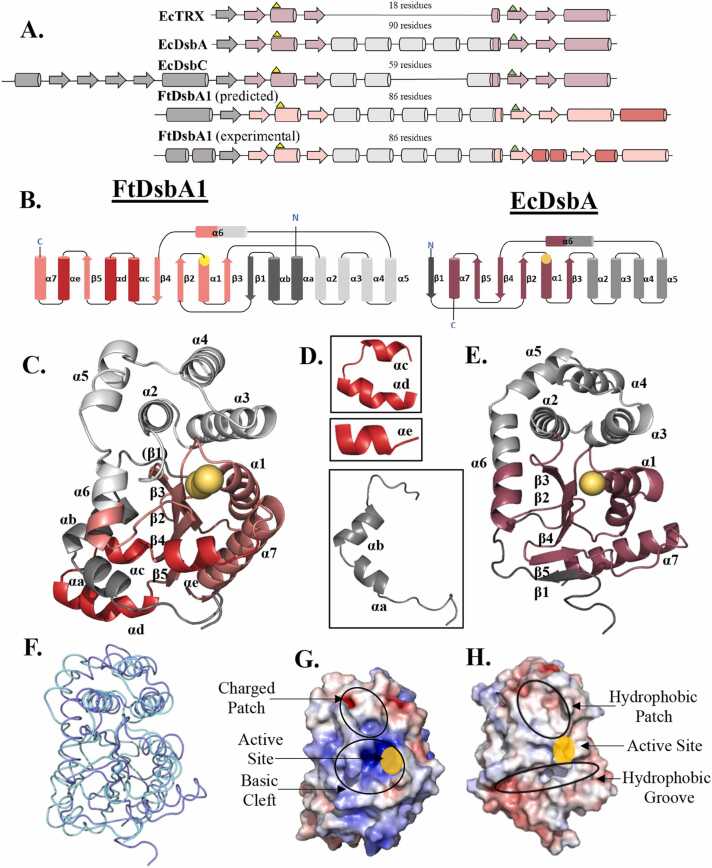
Sequence analysis of FtDsbA1 (Supplementary Figure A1) anticipated the presence of a TRX DsbA-like architecture, identified by the prediction of the βαβ and ββα motifs linked by a 90 residue long alpha helical insertion domain, a CXXC catalytic motif (C57G58K59C60), and a proline loop (Q172G173P174). However, this analysis also proposed 20- and 30-residue N and C terminal extensions, respectively. This predicted domain organisation differs both from previously characterised DsbA-like proteins, which typically lack both N or C terminal elements, and oligomeric Dsb proteins like DsbC, which have an N-terminal extension to allow oligomerisation but lack extensions at the C-terminus (Fig. 1A).
Fig. 1.
Comparison of FtDsbA1 to EcDsbA. (A.) Domain organisation of model TRX-like proteins compared to predicted and experimental domain organisation of FtDsbA1. EcTRX consists of a canonical thioredoxin motif containing the βαβ and ββα domains linked by an 18-residue α helical motif. Monomeric Dsb oxidases like EcDsbA contain a long ∼90-residue α helical insertion linking the βαβ and ββα elements, while oligomeric Dsb isomerases like EcDsbC have a N-terminal oligomerisation domain preceding the TRX fold and contain a ∼59-residue alpha helical insertion between the βαβ and ββα elements. FtDsbA1 was predicted to contain both an N- and C- terminal extension. The experimental domain organisation of FtDsbA1 however includes a 45-residue N-terminal extension encompassing two alpha helices flanked by loops, and three α helical insertions into the core of the TRX fold. (B.) Topology schematics of the FtDsbA1 (left) and EcDsbA (PDB ID 1FVK [22] (right) structures with cylinders representing α-helices and arrows representing β-sheets. Catalytic cysteines are shown as yellow circles. FtDsbA1 features a Class II β-sheet topology (1−3-2−4-5) while EcDsbA features a Class I β-sheet topology (3−2-4−5-1) [54]. (C). Cartoon depictions of FtDsbA1. Secondary elements are labelled and coloured according to B. FtDsbA1 features an extended N terminus with two helices (αa-b), two additional helices between β4 and β5 (αc-d), and a third additional helix between β5 and α7 (αe). (D.) Cut-outs of the additional helices αa-e of FtDsbA1. (E.) Cartoon depictions of reference protein EcDsbA (PDB ID 1FVK [22]). Secondary elements are labelled and coloured according to B. (F.) Superimposition of FtDsbA1 (light blue) and EcDsbA (dark blue). The two proteins superimpose with a high RMSD value of 3.13 Å over 118 equivalent Cα atoms indicating significant structural differences. (G&H.) Electrostatic surface representation of FtDsbA1 (left) and EcDsbA (right). EcDsbA features an overall hydrophobic surface with a hydrophobic patch above the active site and a hydrophobic groove below. FtDsbA1 features an overall basic surface with a positively charged patch above the active site and a broad basic cleft enveloping the catalytic cysteines. The electrostatic potential was calculated with APBS [3] in PyMOL [12] showing positive charges in blue (saturating at 5 kT/e) and negative charges in red (saturating at −5 kT/e).
To gain insight into the structure of this unusual Dsb-like protein we determined the crystal structure of FtDsbA1 at 1.95 Å resolution by molecular replacement and refined it to Rfactor and Rfree values of 0.19 and 0.24, respectively. Two symmetrically independent FtDsbA1 molecules were built in the asymmetric unit, with monomer A and B covering residues 12–233 and 8–233, respectively (Table 1). Both monomers are structurally very similar with a root-mean-square deviation (RMSD) of 0.49 Å over 221 Cα atoms. The structural alignment deviates slightly in the C- terminal alpha helices (residues 205–233) and more considerably at the N-terminus (residues 12–17) where chain A and B adopt different configurations (Supplementary Figure A2). While FtDsbA1 was oxidised prior to crystallisation, the catalytic cysteines appear reduced in both monomers. The reduction of the previously oxidised catalytic cysteines may be attributed to radiation damage caused during data collection or may have been caused by the coordination of the Zn2+ cation. Since overall, no significant differences were observed between the two molecules in the asymmetric unit subsequent structural analyses were performed with monomer A.
As predicted from the sequence analysis (Fig. 1A), the structure of FtDsbA1 contains a TRX-like domain encompassing motifs β2α1β3 (residues 46–83) and β4β5α7 (residues 169–212) which are bridged by an inserted alpha-helical domain comprised of a three helical bundle, α2α3α4 (residues 90–143), one additional α-helix, α5 (residues 147–157) and a short α6 (residues 161–165) (Fig. 1B and C). As predicted bioinformatically, FtDsbA1 also features a 45-residue extended N-terminus, consisting of helices αa and αb (residues 17–23 and 25–33 respectively) and several connecting unstructured loops preceding the β1 strand (Fig. 1C and D). Although FtDsbA1 was anticipated to have a C-terminal extension, the crystal structure revealed that it instead contains two insertions embedded within the TRX-like domain. Specifically, it incorporates a 19-residue insertion in the loop connecting β4 and β5, which features two short alpha helices αc and αd (residues 182–186 and 189–198, respectively). The second additional inserted element is a 6-residue long αe alpha helix (residues 204–210), which connects β5 and α7 (Fig. 1C and D). Altogether, these insertions twist the overall TRX-like structure of FtDsbA1.
3.2. Comparison of FtDsbA1 to DsbA homologues
Although FtDsbA1 contains the structural features of DsbA-like proteins, the overall structure diverges from canonical EcDsbA (PDB ID 1FVK [22]) (Fig. 1E) with the two protein structures aligning with a root-mean-square deviation value of 3.13 Å over 118 equivalent Cα atoms (Fig. 1F). An area of significant difference between the two proteins is the N-terminus, with FtDsbA1 harbouring the additional αa and αb helices prior to the first β-strand (β1), in contrast to the shorter and unstructured N-terminus preceding the EcDsbA β1. Furthermore, the topology of their respective N-terminal regions differ, whereby in FtDsbA1 β1 forms hydrogen bonds to β3 on the non-catalytic side of the protein mimicking the β-sheet topology of Class II DsbA proteins, whereas in EcDsbA β1 is hydrogen bonded to β5 on the catalytic side, which is the β-sheet topology of Class I DsbA proteins [54] (Fig. 1B,C and E).
Another notable difference between FtDsbA1 and EcDsbA lies in the helical domain integrated within the TRX fold. In EcDsbA, this region comprises of a three helical bundle (α2–α4), an additional helix (α5) and an extension of the connecting helix α6. The helical domain of the FtDsbA1 protein also adopts the three helical bundle (α2–α4) but exhibits a longer loop connecting α4 and α5 and lacks the extended helix α6 seen in EcDsbA, resulting in a different topology and a poor overlap between the helical domains of the two proteins (Fig. 1B-F and Supplementary Figure 3A). Other differences between the two proteins are the insertions of the α c-e helices between β4-β5 and β5-α7 in FtDsbA1 in place of the short loops present in EcDsbA and other DsbA proteins [78], [50]. In EcDsbA, the β4β5α7 motif forms one side of a peptide-binding groove adjacent to the catalytic cysteines, which has been shown to be involved in binding the partner oxidase EcDsbB and substrate proteins [34], [60]. The insertion of α e between β5α7 changes the configuration of this motif in FtDsbA1 resulting in a twisted and truncated groove (Fig. 1C, E, G, H and Supplementary Figure A3). In accordance with the low sequence identity between FtDsbA1 and EcDsbA (17 %), their surface properties differ markedly (Fig. 1F and G). Neighbouring the catalytic site in EcDsbA is a hydrophobic substrate binding patch [60] and a long hydrophobic groove required for interaction with the cognate oxidase [34]. Conversely, FtDsbA1 incorporates a charged path in the region corresponding to the EcDsbA substrate binding site and a truncated basic groove adjacent to the active site (Fig. 1F and G).
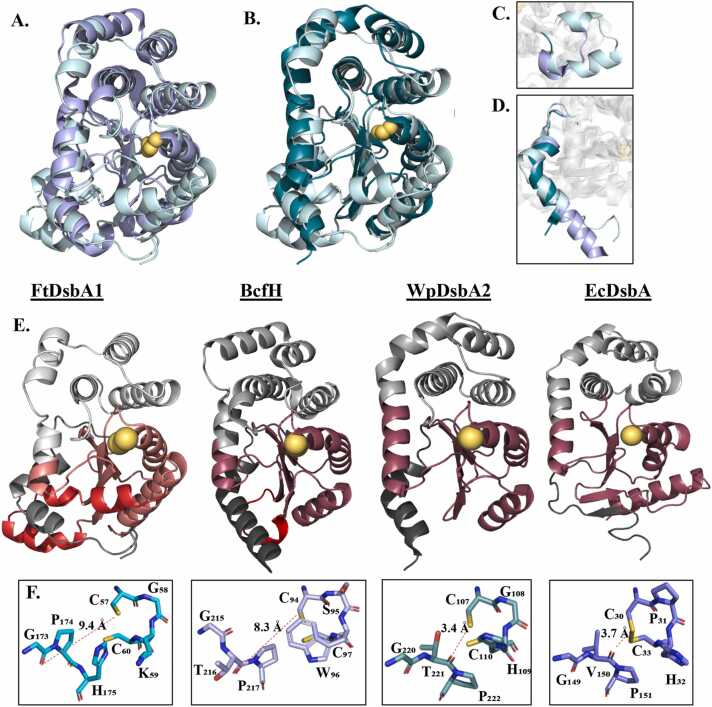
A Dali search [29] identified the protomers of two trimeric DsbA-like proteins as the closest structural homologues of FtDsbA1, Salmonella Typhimurium BcfH (PDB ID 7JVE [83]) (31 % sequence identity, Z-score 22.1 and RMSD of 1.8 Å across 161 equivalent Cα atoms), and Wolbachia pipientis WpDsbA2 (PDB ID 6EEZ [88]) (21 % sequence identity, Z-score 18.3 and RMSD of 2.3 Å across 140 equivalent Cα atoms) (Fig. 2A and B). In addition to the anticipated similarities in the TRX domain motifs, the FtDsbA1 N-terminus bears a resemblance to the N-terminal regions of BcfH and WpDsbA2 through which they both oligomerise [83], [88] (Fig. 2A and B, D, Supplementary Figure A4). Additionally, similar to FtDsbA1, BcfH has an insertion in the C-terminus of the TRX-like domain linking β4 and β5, however this insertion is comparatively smaller with only one short helix present (Fig. 2C).
Fig. 2.
Comparison of FtDsbA1 Structure and Active Site with Structural Homologues. (A.) Superimposition of FtDsbA1 and BcfH (PDB ID 7JVE [83]). BcfH (lavender) and FtDsbA1 (light cyan) superimpose with a RMSD of 1.8 Å across 161 equivalent Cα atoms indicative of broad structural similarities across the N terminus, α-helical insertion domain and additional TRX-like elements. (B.) Superimposition of FtDsbA1 and WpDsbA2 (PDB ID 6EEZ [88]). WpDsbA2 (dark cyan) and FtDsbA1 (light cyan) superimpose with a RMSD of 2.3 Å across 140 equivalent Cα atoms reflecting larger differences in their α-helical insertion domains and WpDsbA2’s lack of additional inserted TRX helices. (C&D.) Comparison of the (C.) β4 β5 loop and (D.) N-terminus in FtDsbA1 (light cyan), BcfH (lavender) and WpDsbA2 (dark cyan). Like other DsbA-like proteins, WpDsbA2 lacks any secondary structural elements in the TRX ββα motif, rather has a short loop to connect β4 and β5. In contrast, BcfH uniquely features a half helical turn in this region and FtDsbA1 features two helices. All three DsbA-like proteins feature an elongated and crooked N terminus which, in BcfH and WpDsbA2, facilitate their trimerisation. (E.) Cartoon representation of, from left to right, FtDsbA1, BcfH (PDB ID 7JVE [83]), WpDsbA2 (PDB ID 6EEZ [88]) and EcDsbA (PDB ID 1FVK [22]) with domains coloured as per Fig. 1. (F.) Comparison of the active sites of, from left to right, FtDsbA1, BcfH (PDB ID 7JVE [83]), WpDsbA2 (PDB ID 6EEZ [88]) and EcDsbA (PDB ID 1FVK [22]). FtDsbA1 and BcfH feature trans-proline loops which orient the carbonyl of the pro-1 residue (G173 in FtDsbA1 and T216 in BcfH) away from the catalytic cysteines. In comparison to EcDsbA and WpDsbA2 which harbour cis-proline loops where the Pro-1 residue carbonyl group (V150 in EcDsbA and T221 in α-WpDsbA2) is oriented towards the catalytic cysteines.
3.3. FtDsbA1 features a unique active site
The CXXC motif is the redox catalytic centre in Dsb proteins, mapping at the N-terminus of α1 in the TRX-like domain adopting a righthand hook configuration [32], [69]. The specific amino acids forming the XX dipeptide between the catalytic cysteines are important in controlling the redox properties of Dsb proteins with over 80 % of DsbAs containing a histidine in the C-terminal position (CXHC) [24], [69]. Conversely, FtDsbA1 lacks this conserved histidine and instead contains a C57G58K59C60 motif, which represent a rare dipeptide, occurring only within other Francisella species [24] (Fig. 2F).
Neighbouring the CXXC motif is the FtDsbA1 proline loop (Q172G173transP174) bridging α6 and β4 (Fig. 2F). Unlike most Dsb proteins, which contain a similar loop with proline in a cis configuration (e.g EcDsbA features a G149V150cisP151 loop), both FtDsbA1 protomers in the asymmetric unit showcase this proline in a trans-configuration (Fig. 2F). When proline is in the cis-conformation, the carbonyl of the residue preceding the proline (Pro-1) is oriented toward the CXXC motif, within hydrogen bond distance of the N-terminal cysteine (3.7 Å in EcDsbA, Fig. 2F). Additionally, the sidechain of this Pro-1 residue (typically either a Val or Thr in DsbAs) has been demonstrated to interact with the N-terminal catalytic cysteine and regulate redox properties in TRX proteins [72]. In FtDsbA1, the trans-Pro174 loop maps far from the catalytic cysteines, with the carbonyl of the Pro-1 residue oriented away (9.4 Å) from the CGKC motif. Additionally, since this Pro-1 residue is a glycine (Gly173), it lacks a functional group side chain, further preventing any interactions with the catalytic cysteines [72]. Notably, one of the protomers in BcfH, the closest FtDsbA1 structural homolog, has been shown to also contain proline in a trans-configuration [83] (G215T216transP217), while WpDsbA2 has a more conserved G220T221cisP222 loop (Fig. 2F).
3.4. FtDsbA1 is a monomeric highly oxidising protein
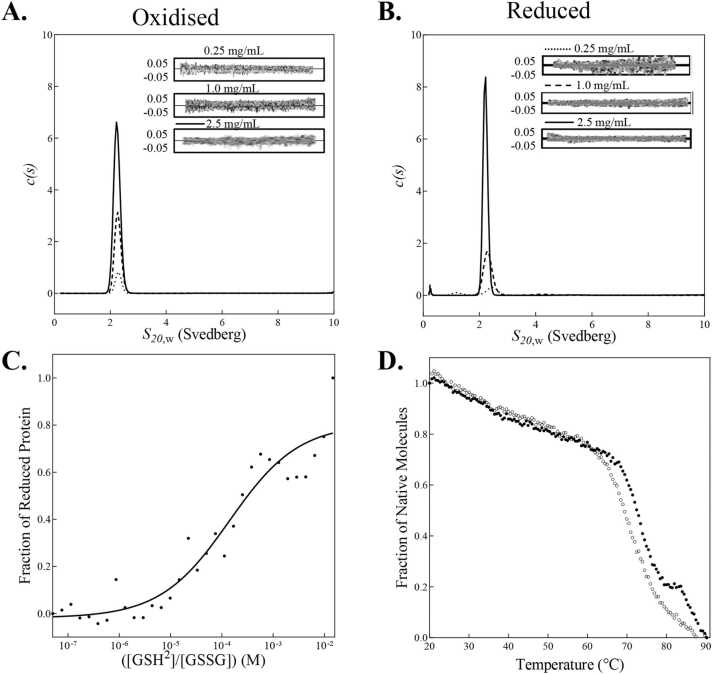
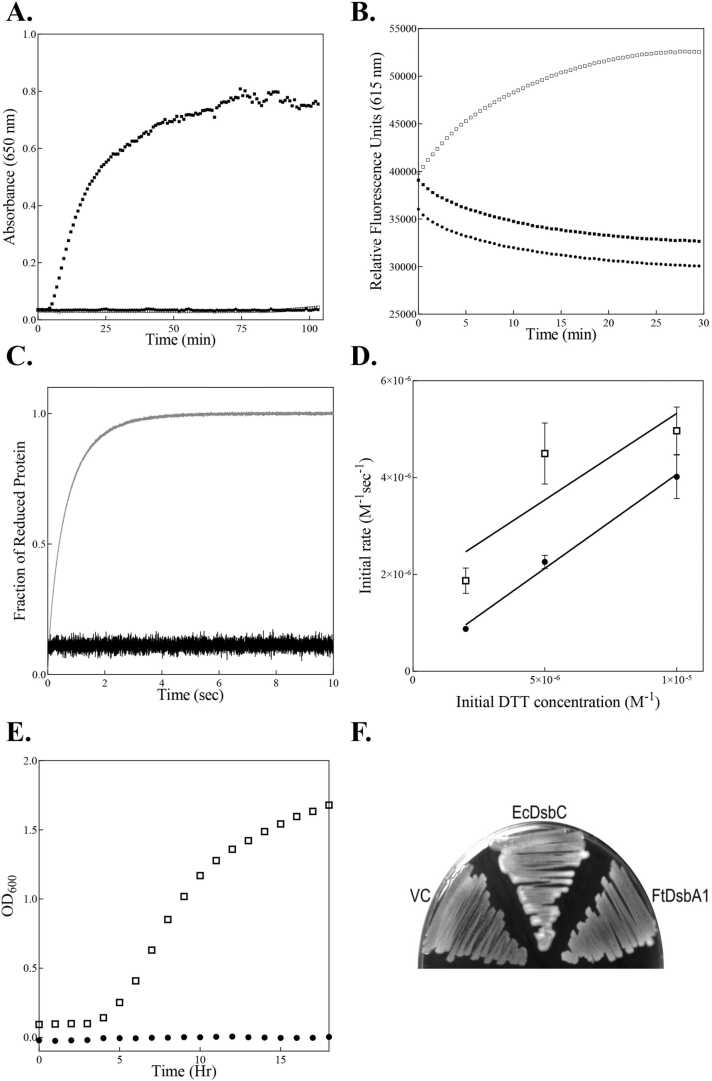
To date, monomeric Dsbs have been found to be oxidases, whereas oligomeric Dsbs are primarily isomerases [94]. Given the presence of an extended N terminus in FtDsbA1, its oligomeric state in solution state was determined in an initial approach to define the putative function of this protein. Analytical ultracentrifugation was performed on FtDsbA1 at three concentrations (0.25 mg/mL, 1 mg/mL, 2.5 mg/mL) in its oxidised and reduced forms. At all concentrations FtDsbA1 formed a single peak at 2.5 S, corresponding to a 26 kDa monomer (Fig. 3A and B), confirming FtDsbA1 is a monomer regardless of redox state or concentration, which is consistent with typical DsbA oxidases.
Fig. 3.
Redox Characterisation of FtDsbA1. (A&B.) Overlay of AUC continuous standardised sedimentation coefficient (s20,w) distribution of (A.) oxidised and (B.) reduced FtDsbA1 at 2.5 (solid line), 1 (dashed line) and 0.25 (dotted line) mg/mL. Residuals from the c(s) distribution best fits plotted as a function of radial position from the axis of rotation. (C.) FtDsbA1 redox potential. FtDsbA1 was incubated with various concentrations of reduced glutathione (GSH) overnight and was analysed fluorometrically. The fraction of reduced protein was plotted against log([GSH]²/[GSSG]) to determine the Keq (1.85 ×10⁻⁴ M) and redox potential was then calculated (−129 mV). Data represents one biological replicate with three technical replicates that is representative of three independent replicates. (D.) Thermal melt of oxidised (open circles ○) and reduced (closed circles •) FtDsbA1. CD thermal melts are plotted as a fraction of α-helical content (based on molar ellipticity [θ] at 222 nm) and temperature (°C). Thermal melts represent three technical replicates.
The propensity of a DsbA protein to donate its disulphide bond to substrates, and thus its function, is in part dictated by its redox potential and the stability of its oxidised and reduced form. The redox potential of FtDsbA1 was determined using a fluorescence-based assay measuring the reduction potential of FtDsbA1 against the standard redox potential of the GSH/GSSG redox pair. This resulted in a Keq of 1.48 ( ± 0.4) x10-4 M equating to an intrinsic redox potential of −127 mV ( ± 3 mV) (Fig. 3C). This redox potential is comparable to that of EcDsbA (−122 mV) [91], [33] and is slightly more oxidising than FtDsbA2 (−131 mV) [71]. Thermal stability of FtDsbA1 was assessed in both redox states at 222 nm from 20 to 90 °C. Interestingly, the reduced state was found to be equally stable to the oxidised state with Tm values of 71.8 ± 0.8 °C and 70.7 ± 1.13 °C, respectively (Fig. 3D). This may indicate that FtDsbA1 would act as a comparatively weaker oxidase to DsbAs with a larger difference in thermostability.
3.5. FtDsbA1 displays high substrate specificity
Next, we sought to investigate FtDsbA1’s oxidoreductase activity. First we determined the disulphide reductase activity of FtDsbA1 by measuring changes in the optical density of a solution caused by the precipitation of insulin due to disulphide reduction [30]. FtDsbA1 did not show reductase activity against insulin (Fig. 4A). The thiol oxidase activity of the protein was first assessed using a peptide oxidation assay with a fluorescently labelled peptide (CNENGLCK) derived from the EcDsbL substrate ASST in the presence of GSSG to maintain the oxidised form of the protein [89], [45] (Fig. 4B). In a parallel approach, the ability of FtDsbA1 to oxidise a peptide derived from PapD (FICNGSRCSV), an EcDsbA substrate [35], was measured using stopped-flow fluorescence, monitoring the change in tryptophan fluorescence of FtDsbA1 upon reduction. FtDsbA1 did not show activity against either substrate (Supplementary figure A5). This lack of oxidative activity may suggest that FtDsbA1 displays heightened substrate specificity and is thus unable to interact with peptides derived from E. coli proteins. To investigate this further, a peptide derived from a F. tularensis PilA homologue, FtPilA (TCNIDGVSASQVPSNCN) was tested against FtDsbA1 using stopped-flow fluorescence. FtDsbA1 showed no activity against the FtPilA peptide while EcDsbA reacted rapidly, highlighting the substrate specificity of the former enzyme and the promiscuity of the latter (Fig. 4C). Given the possibility that FtDsbA1 shows narrow substrate specificity for an unknown substrate, we next used stopped-flow fluorescence to monitor the oxidation of DTT and assess redox activity. EcDsbA catalysed oxidation of DTT displays second order kinetics and proceeds at a much more rapid rate (3.57 (± 1.11) x105 M-1sec-1) than the uncatalysed reaction. Importantly, we found a similar rate for FtDsbA1 oxidation of DTT (3.88 (± 0.45) x105 M-1sec-1) supporting an oxidative role for this enzyme (Fig. 4D and Supplementary Figure A5).
Fig. 4.
Disulphide Oxidase Activity of FtDsbA1. (A.) In vitro disulphide reductase activity. Increase in absorbance (650 nm) indicates the dissolution of insulin following disulphide cleavage. Neither FtDsbA1 (closed circles •) nor EcDsbA (open squares □) show disulphide reductase activity to insulin while EcDsbC (an isomerase) (closed squares ■) shows activity. A buffer control containing GSH only, showing no activity, was omitted for clarity. (B.) In vivo thiol oxidase activity. Fluorescence curves taken at 340 nm indicate oxidation of the peptide substrate ASST by EcDsbA (open squares □), FtDsbA1 (closed circles •) or buffer control (closed squares ■). FtDsbA1 shows little catalytic activity towards ASST and reacts comparably to the buffer control. (C.) Real time oxidation of the FtPilA peptide by EcDsbA and FtDsbA1. Change in fluorescence (λex 295 nm, λem 330 nm) indicates the reduction of DsbA enzymes (2 μM) resulting from the oxidation of the peptide substrate (2 μM). EcDsbA (light grey) reacts rapidly with the FtPilA peptide, becoming fully reduced within 4 s while FtDsbA1 (black) remains fully oxidised, indicating no activity towards the peptide. (D.) Initial rates of reaction of FtDsbA1 (closed circles •) and EcDsbA (open squares □) against the reductant DTT plotted to give rate constants. Both DsbA enzymes catalyse the oxidation of DTT with second order kinetics at comparable rates, indicating FtDsbA1 plays an oxidative role. (E.) Swimming motility assay. Optical density curves taken at 600 nm indicate the migration of cells over time. Cells containing FtDsbA1 (closed circles •) did not migrate due to flagella mediated motility and displayed migration comparable to an empty vector control (omitted from graph for clarity) while EcDsbA transformed cells (open squares □) did display flagella mediated motility. (F.) Mucoidal morphology assay. The production of colanic acid results in morphological changes which can be seen as a haze surrounding cells. Cells expressing EcDsbC display this haze, indicating in vivo isomerase activity while both isolates of FtDsbA1 transformed cells (only one shown for clarity) show no difference in phenotype compared to an empty vector control (VC).
To determine the in vivo activity of FtDsbA1, a pWSK29 vector harbouring the EcDsbA signal sequence followed by the mature FtDsbA1 gene was constructed to facilitate its translocation into the periplasmic space of E. coli. The plasmid was then transformed into E. coli strains lacking either EcDsbA (JCB817 [5]) or EcDsbC (PL263 [46]). A swimming motility assay was performed to determine the FtDsbA1 thiol-oxidase activity [86], given that surface migration required the disulphide bond dependant folding of Flgl [28]. However, FtDsbA1 failed to restore the motility in the ΔdsbA strain, showing migration levels similar to the negative vector control (VC), suggesting an inability to fold FlgI (Fig. 4E). This lack of activity could also be attributed to the inability of EcDsbB to re-oxidize FtDsbA1. This seems less likely, as EcDsbB has been demonstrated to oxidise heterologous DsbA proteins [40] and can be complemented by FtDsbB [65] suggesting a conserved mechanism that is supported by their significant (40 %) sequence identity. PL263 (ΔdsbC, ΔmdoG) was used to assess the disulphide isomerase activity of FtDsbA1 in vivo as previously described [46]. The strain is unable to synthesise membrane-derived oligosaccharides, which consequently triggers RcsF dependant colanic acid synthesis, resulting in a distinctive mucoidal phenotype. RcsF contains non-consecutive cysteine bridges that demand disulfide isomerisation, as such, only Dsb-like proteins with disulphide isomerase activity can restore this mucoidal phenotype. Cells expressing FtDsbA1 showed no difference in mucoidal morphology compared to the empty VC and exhibited a significantly diminished phenotype compared to the EcDsbC expressing control, as visually assessed by the absence of a haze surrounding the cell mass on the agar surface (Fig. 4F).
4. Discussion
The periplasm of Gram-negative bacteria harbour enzymes known as Dsb proteins, which catalyse the formation of disulphide bonds in a wide range of cell envelope and secreted proteins that facilitate bacterial colonisation, proliferation and persistence [79], [20], [24], [38], [4]. The prototypical Dsb system includes thiol oxidases DsbA, DsbB and disulfide isomerases/reductases DsbC and DsbD, although there is significant diversity in the Dsb pathways among bacterial species [43]. This diversity is often manifested through the presence of multiple DsbA homologues or modifications to the shared thioredoxin fold which confer distinct functionalities [8], [77], [83], [82], [84], [25], [43].
One bacterium that showcases variations to the oxidative folding system is Francisella tularensis, causative agent of tularaemia and one of the most infectious bacterial pathogens known [80], [13]. Owing to its infectivity and ability to be transmitted through multiple routes including insect bites, contact with infected animals or water, and inhalation of aerosolised organisms, this zoonotic bacterium has the potential to be misused as an agent of biological warfare [9], [10]. The ability of this bacteria to infect hosts and to evade current antibiotic agents have been tied to its distinct disulphide catalysis pathway [68], [71], which encompasses two DsbA proteins; FtDsbA1 [74] and FtDsbA2 [66], and a DsbB homologue; FtDsbB [65]. Notably its catalysis pathway lacks any distinct isomerase machinery, a crucial function accommodated for by the bifunctional FtDsbA2 [7], [67], [71].
The present study aimed to investigate the mostly uncharacterised FtDsbA1. Through structural analysis we show that this protein has a novel DsbA-like architecture, which retains the identifiable TRX-like domain (β2α1β3 - β4β5α7) and the alpha helical insertion domain (α2 – α5) characteristic of DsbA-like oxidases [78], but includes several non-canonical structural elements. For example, FtDsbA1 has an extended N-terminus with a long unstructured loop and two alpha helices that wrap around the TRX-fold. Its closest structural homologues, the trimeric BcfH and WpDsbA2 [83], [19], [62], or dimeric Dsb proteins like EcDsbC [53] and EcDsbG [23], use their long N-terminal extensions for oligomerisation. However, FtDsbA1 is monomeric as shown by AUC and X-ray crystallography. As FtDsbA1 is predicted to be membrane-associated due to the presence of a putative lipid-binding motif in the signal sequence [48], the extended N-terminus may provide conformational flexibility to better orient the enzyme's catalytic face when attached to the membrane.
A notable observation in the structure of FtDsbA1 is that it features three helices inserted within the β4β5α7 motif, two between the β strands and a third preceding the last α7 helix (Fig. 1C). Long insertions in this core component of the TRX motif are highly atypical, with some DsbAs only featuring a short α-helix turn directly preceding β5 (BcfH [83] and α-WpDsbA1 (PDB ID 3F4R [41])) or prior to α7 (Class Ia and Ib DsbAs such as S. enterica DsbA (PDB ID 3L9S [25]) and Vibrio cholerae DsbA (PDB ID 4DVC [87]). Instead, FtDsbA1 has a 17-residue αc-loop-αd motif linking β4 and β5 and a two turn α-helix (αe) preceding α7. These insertions completely change the overall TRX topology of FtDsbA1 (Fig. 1G and H), particularly regions adjacent to the CXXC catalytic centre. Structural changes compounded by the sequence divergence also alter the surface properties of FtDsbA1. Unlike the canonical EcDsbA, which features a hydrophobic patch and a long hydrophobic peptide binding groove [78], [50] to facilitate the binding of cognate oxidase and substrate proteins [34], [60], FtDsbA1 has a mostly basic surface and lacks the deep peptide-binding hydrophobic channel near the active site. This suggests that FtDsbA1 has different substrate specificity compared to typical DsbAs.
FtDsbA1 also possesses a distinctive active site characterised by a CGKC motif, which lacks the histidine residue commonly found in DsbA enzymes [78], [69], and an atypical Pro-1 residue (Gly173). In EcDsbA, a C-terminal histidine (CXHC) has been shown to stabilise the thiolate group of the reduced N-terminal catalytic cysteine through electrostatic interaction with its sidechain and a hydrogen bond via its amide backbone [22]. These interactions, absent in the oxidised form, contribute to the higher stability of the reduced form compared to the oxidized form [5], [46] and promotes oxidative activity by making the reduction of DsbA, through the oxidation of its substrates, thermodynamically favourable [22], [21], [69]. However, the crystal structure of FtDsbA1 reveals that the equivalent residue, Lys59, likely does not interact with the N-terminal cysteine thiolate and rather is oriented firmly towards the solvent exposed surface of the enzyme. Consequently, FtDsbA1 exhibits similar stability in both its oxidised and reduced forms, as demonstrated by thermal unfolding experiments (Fig. 3D). Since DsbA homologues frequently have a destabilising disulphide bond which facilitates the donation of the disulphide bond to substrates [91], this may result in FtDsbA1 being a less efficient oxidase activity.
Another characteristic property of TRX-like proteins is a conserved cisPro loop, which precedes the ββα motif and is adjacent to the CXXC site in the three-dimensional structure. This loop was previously shown to be critical for substrate binding and modulating the redox properties of TRX proteins by interacting with the N-terminal catalytic cysteine [38], [60], [72]. In the case of FtDsbA1 the equivalent proline is in a trans configuration,which rarely seen, with examples limited to one monomer of the trimeric DsbA-like protein BcfH [83], and in Eps1p- a TRX-like protein from Naumovozyma dairenensis [6]. This altered conformation results in the Q172G173-transP174 loop mapping away from the C57G58K59C60 motif, preventing any interactions between the loop residues and the catalytic cysteines. However, while Gly173 is unable to interact with the N terminal cysteine (Cys57) the sidechain of His175 is only 4 Å away from this cysteine, suggesting it may- albeit weakly, interact. Confounding this however is a Zn2+ ion present within the active site of FtDsbA1 (an artifact from the crystallisation solution) which His175 is only 2.3 Å away from, obscuring this residue’s true orientation in the FtDsbA1 active site.
Although the function of the trans-proline loop is unclear, it has been speculated that this conformation may slow-down the catalytic rate by influencing substrate retention [83]. Additionally, a consequence of the unique sequence elements utilised by FtDsbA1 is completely altering the outline of the groove that in DsbA proteins in involved in substrate binding. However, despite the numerous structural differences, FtDsbA1 appears to function as a thiol-oxidase, being monomeric in solution – characteristic of DsbA oxidases, having a comparable redox potential to EcDsbA [91], [33], and an ability catalytically oxidise DTT at rates comparable to EcDsbA (Fig. 4A and B). However, FtDsbA1 lacks the hallmarks of the canonical oxidases, such as hydrophobic surface properties for unfolded substrate interaction and stabilisation or a cis-proline loop for binding and positioning the substrate protein near the CXXC centre for catalysis. The highly atypical structure and modifications to the active site likely limits FtDsbA1’s substrate specificity as shown by this protein being unable to interact with and catalyse thiol oxidation in a F. tularensis substrate and reference E. coli DsbA substrates both in vitro and in cell-based assays.
5. Significance and conclusions
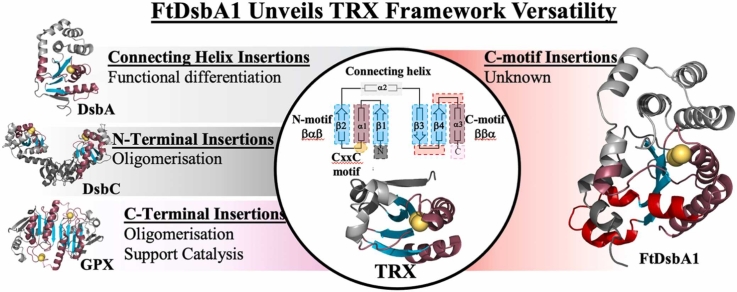
In conclusion, the present study underscores the remarkable adaptability and functional diversity of thioredoxin (TRX) fold proteins, which are essential across all forms of life for their roles in redox regulation, protein folding, and defence against oxidative stress [79], [20], [24], [38], [4]. By incorporating new variations into an ancestral fold through structural modifications, such as extensions, insertions and recombination of domains, these proteins have evolved diverse functions (Fig. 5). Our findings on FtDsbA1 emphasize the extreme levels of structural and functional structural malleability of TRX fold proteins, demonstrating that this fold supports insertions within its core modules to fine-tune protein functionality and specificity. This new knowledge will not only enhance the precision of structural predictions, particularly for TRX proteins, but will also have significant implications for biomedical research and drug discovery, give the multifaceted roles of TRX-like proteins in cellular processes, redox signalling pathways, and disease states [20], [24], [4].
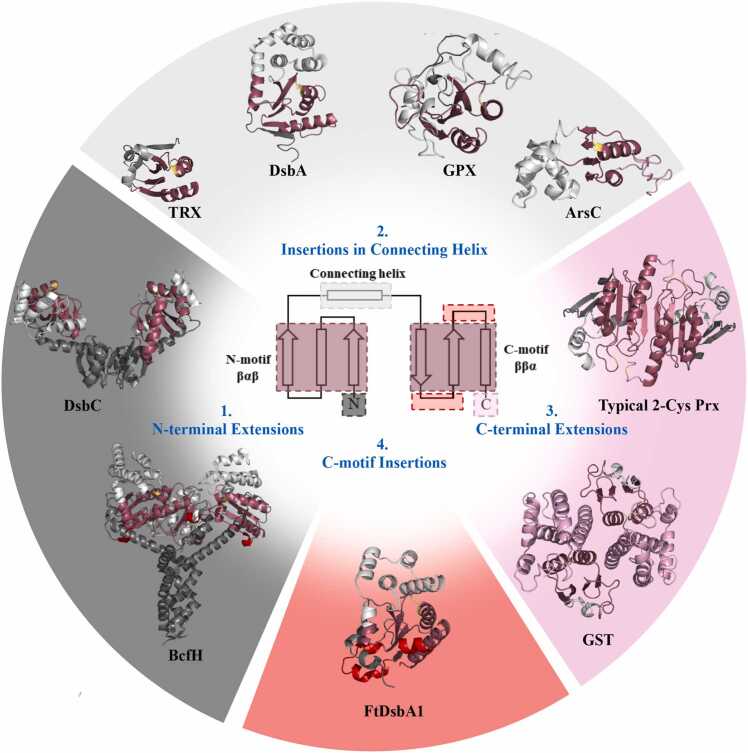
Fig. 5.
Modifications within the TRX-fold lead to functional diversity.
Four main sites within the thioredoxin-core fold have been identified that can tolerate structural modifications, which in turn lead to functional diversity between members of the TRX-like protein superfamily. 1. N-terminal extensions. Long extensions before the first β-strand of the βαβ motif commonly results in oligomerisation. For example, dimerization in TRX-proteins such as DsbC and DsbG (EcDsbC shown (PDB ID 1EEJ [53]) occurs via interactions between N-terminal β sheets. Conversely, trimeric TRX-like proteins like BcfH contain a long N-terminal α helical extension that allow homo-trimerization (PDB ID 7JVE [83]). Additionally, monomeric TRX and DsbA homologues feature short N-terminal extensions that fold to their characteristic 5-stranded β sheet core scaffold (E. coli TRX (PDB ID 2TRX [39]) and EcDsbA (PDB ID 1FVK [22]) are shown). 2. Insertions within the TRX connecting helix region. The region spanning the two TRX- motifs; βαβ–ββα, is highly susceptible to modification and can tolerate large insertions without compromising the integrity of the TRX core fold. A large insertion encompassing five alpha helices can be found in DsbA homologues [50] while in bacterial arsenate reductases (E. coli ArsC is shown (PDB ID 1I9D [51])) the topology of the connecting helix insertion comprises only either one or three alpha helices. Additionally, glutathione peroxidases (GPX) [61] (bovine GPX1 is shown (PDB ID 1GP1 [16]) utilise a varied insertion, although universally sharing one helix followed by a series of α-helices and β-strands. 3. C-terminal modifications. The C-terminal region following the TRX-core fold is subject to modification with diverse outcomes, although, like the N-terminus, often is used to facilitate oligomerisation. In some examples of typical 2-Cys peroxiredoxins (Prx) (murine thioredoxin peroxidase 2 is shown (PDB ID 1QQ2 [27])) an inter-protomer disulphide bond is formed between cysteines located at the C-terminal tip of α1 in the TRX-core fold and the C-terminus which contributes to stabilising the dimeric structure and is used in catalysis. Similarly, the glutathione S transferase proteins (GST) (murine glutathione S-transferase is shown (PDB ID 6GST [90])) also utilise their C-terminus, which contains a bundle of six helices, to facilitate dimerisation and catalysis [1]. 4. C-motif insertions. Significant modifications within the core TRX motifs themselves are rare, with examples limited to a helical turn bridging β5 and α7 in many Class I DsbA homologues [54] and a helical turn spanning β4 and β5 in both BcfH (PDB ID 7JVE [83]) and WpDsbA1 (PDB ID 3F4R [41]). The FtDsbA1 structure presented in this work represents one of the only proteins yet described which features significant modification inside the core of the TRX-motif. A more detailed analysis of the structural differences between TRX-like proteins can be found in the supplementary material (Supplementary Figure A6).
CRediT authorship contribution statement
Begona Heras: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition, Formal analysis, Conceptualization. Jason J Paxman: Writing – review & editing, Supervision, Methodology, Investigation, Formal analysis. Makrina Totsika: Writing – review & editing, Supervision. Lilian Hor: Writing – review & editing, Investigation. Taylor Cunliffe: Writing – review & editing, Investigation. Yaoqin Hong: Writing – review & editing, Investigation, Formal analysis. Stephanie Penning: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. The authors would like to acknowledge the La Trobe University Bioimaging Platform. This work was supported by the Australian Research Council (ARC) project grants (DP180102987, DP190101613, DP210100673), and a National Health and Medical Research Council (NHMRC) project grant (GNT1143638; GNT1144046).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.11.034.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Armstrong R.N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem Res Toxicol. 1997;10(1):2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson H.J., Babbitt P.C. An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput Biol. 2009;5(10) doi: 10.1371/journal.pcbi.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardwell J.C., Lee J.O., Jander G., Martin N., Belin D., Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci. 1993;90(3):1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell J.C.A., McGovern K., Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67(3):581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 6.Biran S., Gat Y., Fass D. The Eps1p protein disulfide isomerase conserves classic thioredoxin superfamily amino acid motifs but not their functional geometries. PLOS ONE. 2014;9(12) doi: 10.1371/journal.pone.0113431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bocian-Ostrzycka K.M., Grzeszczuk M.J., Banaś A.M., Jastrząb K., Pisarczyk K., Kolarzyk A., et al. Vol. 7. Original Research,; 2016. Engineering of helicobacter pylori dimeric oxidoreductase DsbK (HP0231) (Frontiers in Microbiology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouwman C.W., Kohli M., Killoran A., Touchie G.A., Kadner R.J., Martin N.L. Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J Bacteriol. 2003;185(3):991–1000. doi: 10.1128/jb.185.3.991-1000.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centre for Disease Control. (2018a, 04/04/2018). Bioterrorism Agents/ Diseases〈https://emergency.cdc.gov/agent/agentlist-category.asp〉. Retrieved 17/08 from 〈https://emergency.cdc.gov/agent/agentlist-category.asp〉.
- 10.Centre for Disease Control. (2018b, 13/12/2018). Tularemia〈https://www.cdc.gov/tularemia/〉. 〈https://www.cdc.gov/tularemia/〉.
- 11.Chen, V.B., Arendall, I.I.I.W.B., Headd, J.J., Keedy, D.A., Immormino, R.M., Kapral, G.J. & et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. 66, 12–21. https://doi.org/10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed]
- 12.Delano, W.L. (2002). The PyMOL Molecular Graphics System. 〈https://www.pymol.org〉.
- 13.Dennis D.T., Inglesby T.V., Henderson D.A., Bartlett J.G., Ascher M.S., Eitzen E., et al. Tularemia as a biological weapon: medical and public health management. Jama. 2001;285(21):2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 14.Dutton R.J., Boyd D., Berkmen M., Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105(33):11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66(4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epp O., Ladenstein R., Wendel A. The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur J Biochem. 1983;133(1):51–69. doi: 10.1111/j.1432-1033.1983.tb07429.x. [DOI] [PubMed] [Google Scholar]
- 17.Eschenfeldt W.H., Lucy S., Millard C.S., Joachimiak A., Mark I.D. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol. 2009;498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):282–292. doi: 10.1107/s090744491003982x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furlong E.J., Lo A.W., Kurth F., Premkumar L., Totsika M., Achard M.E.S., et al. A shape-shifting redox foldase contributes to Proteus mirabilis copper resistance. Nat Commun. 2017;8(1) doi: 10.1038/ncomms16065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furniss C., Kaderabkova N., Barker D., Bernal P., Maslova E., Antwi A.A., et al. Breaking antimicrobial resistance by disrupting extracytoplasmic protein folding. elife. 2022 doi: 10.7554/eLife.57974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guddat L.W., Bardwell J.C.A., Martin J.L. Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization. Structure. 1998;6(6):757–767. doi: 10.1016/S0969-2126(98)00077-X. [DOI] [PubMed] [Google Scholar]
- 22.Guddat L.W., Bardwell J.C.A., Glockshuber R., Huber-Wunderlich M., Zander T., Martin J.L. Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci. 1997;6(9):1893–1900. doi: 10.1002/pro.5560060910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heras B., Edeling M.A., Schirra H.J., Raina S., Martin J.L. Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide. Proc Natl Acad Sci USA. 2004;101(24):8876–8881. doi: 10.1073/pnas.0402769101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heras B., Shouldice S.R., Totsika M., Scanlon M.J., Schembri M.A., Martin J.L. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7(3):215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 25.Heras B., Totsika M., Jarrott R., Shouldice S.R., Gunčar G., Achard M.E.S., et al. Structural and functional characterization of three DsbA paralogues from salmonella enterica serovar typhimurium. J Biol Chem. 2010;285(24):18423–18432. doi: 10.1074/jbc.M110.101360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heras B., Totsika M., Peters K.M., Paxman J.J., Gee C.L., Jarrott R.J., et al. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Natl Acad Sci USA. 2014;111(1):457–462. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirotsu S., Abe Y., Okada K., Nagahara N., Hori H., Nishino T., et al. Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product. Proc Natl Acad Sci. 1999;96(22):12333–12338. doi: 10.1073/pnas.96.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hizukuri Y., Yakushi T., Kawagishi I., Homma M. Role of the intramolecular disulfide bond in FlgI, the flagellar P-ring component of Escherichia coli. J Bacteriol. 2006;188(12):4190–4197. doi: 10.1128/jb.01896-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L., Laiho A., Törönen P., Salgado M. DALI shines a light on remote homologs: one hundred discoveries. Protein Sci. 2023;32(1) doi: 10.1002/pro.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254(19):9627–9632. doi: 10.1016/S0021-9258(19)83562-7. [DOI] [PubMed] [Google Scholar]
- 31.Huber-Wunderlich M., Glockshuber R. A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold Des. 1998;3(3):161–171. doi: 10.1016/S1359-0278(98)00024-8. [DOI] [PubMed] [Google Scholar]
- 32.Hutchinson E.G., Thornton J.M. PROMOTIF—A program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5(2):212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inaba K. Disulfide bond formation system in escherichia coli. J Biochem. 2009;146(5):591–597. doi: 10.1093/jb/mvp102. [DOI] [PubMed] [Google Scholar]
- 34.Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., et al. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127(4):789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 35.Jacob-Dubuisson F., Pinkner J., Xu Z., Striker R., Padmanhaban A., Hultgren S.J. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc Natl Acad Sci. 1994;91(24):11552–11556. doi: 10.1073/pnas.91.24.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66(2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadokura H., Tian H., Zander T., Bardwell J.C.A., Beckwith J. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science. 2004;303(5657):534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 39.Katti S.K., LeMaster D.M., Eklund H. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J Mol Biol. 1990;212(1):167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 40.Kurth F., Rimmer K., Premkumar L., Mohanty B., Duprez W., Halili M.A., et al. Comparative sequence, structure and redox analyses of Klebsiella pneumoniae DsbA show that anti-virulence target DsbA enzymes fall into distinct classes. PLOS ONE. 2013;8(11) doi: 10.1371/journal.pone.0080210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurz M., Iturbe-Ormaetxe I., Jarrott R., Shouldice S.R., Wouters M.A., Frei P., et al. Structural and functional characterization of the oxidoreductase α-DsbA1 from Wolbachia pipientis. Antioxid Redox Signal. 2009;11(7):1485–1500. doi: 10.1089/ars.2008.2420. [DOI] [PubMed] [Google Scholar]
- 42.Lafaye C., Iwema T., Carpentier P., Jullian-Binard C., Kroll J.S., Collet J.-F., et al. Biochemical and structural study of the homologues of the Thiol–Disulfide oxidoreductase DsbA in Neisseria meningitidis. J Mol Biol. 2009;392(4):952–966. doi: 10.1016/j.jmb.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 43.Landeta C., Boyd D., Beckwith J. Disulfide bond formation in prokaryotes. Nat Microbiol. 2018;3(3):270–280. doi: 10.1038/s41564-017-0106-2. [DOI] [PubMed] [Google Scholar]
- 44.Lebowitz J., Lewis M.S., Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11(9):2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K., Dzubeck V., Latshaw L., Schneider J.P. De novo designed peptidic redox potential probe: linking sensitized emission to disulfide bond formation. J Am Chem Soc. 2004;126(42):13616–13617. doi: 10.1021/ja047300r. [DOI] [PubMed] [Google Scholar]
- 46.Leverrier P., Declercq J.P., Denoncin K., Vertommen D., Hiniker A., Cho S.H., et al. Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J Biol Chem. 2011;286(19):16734–16742. doi: 10.1074/jbc.M111.224865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lillig C.H., Holmgren A. Thioredoxin and related molecules--from biology to health and disease. Antioxid Redox Signal. 2007;9(1):25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 48.Madan Babu M., K S. DOLOP- database of bacterial lipoproteins. Bioinformatics. 2002;18(4):641–643. doi: 10.1093/bioinformatics/18.4.641. [DOI] [PubMed] [Google Scholar]
- 49.Martin J.L. Thioredoxin--a fold for all reasons. Structure. 1995;3(3):245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 50.Martin J.L., Bardwell J.C.A., Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365(6445):464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 51.Martin P., DeMel S., Shi J., Gladysheva T., Gatti D.L., Rosen B.P., et al. Insights into the structure, solvation, and mechanism of arsc arsenate reductase, a novel arsenic detoxification enzyme. Structure. 2001;9(11):1071–1081. doi: 10.1016/S0969-2126(01)00672-4. [DOI] [PubMed] [Google Scholar]
- 52.Matthews B.W. Solvent content of protein crystals. J Mol Biol. 1968;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 53.McCarthy A.A., Haebel P.W., Törrönen A., Rybin V., Baker E.N., Metcalf P. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat Struct Biol. 2000;7(3):196–199. doi: 10.1038/73295. [DOI] [PubMed] [Google Scholar]
- 54.McMahon R.M., Premkumar L., Martin J.L. Four structural subclasses of the antivirulence drug target disulfide oxidoreductase DsbA provide a platform for design of subclass-specific inhibitors. Biochim Et Biophys Acta (BBA) - Proteins Proteom. 2014;1844(8):1391–1401. doi: 10.1016/j.bbapap.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 55.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19(6):679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mössner E., Huber-Wunderlich M., Rietsch A., Beckwith J., Glockshuber R., Åslund F. Importance of redox potential for the in vivo function of the cytoplasmic disulfide reductant thioredoxin from escherichia coli. J Biol Chem. 1999;274(36):25254–25259. doi: 10.1074/jbc.274.36.25254. [DOI] [PubMed] [Google Scholar]
- 57.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D: Biol Crystallogr. 2011;67(4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pace C.N., Hebert E.J., Shaw K.L., Schell D., Both V., Krajcikova D., et al. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa311Edited by P. E. Wright. J Mol Biol. 1998;279(1):271–286. doi: 10.1006/jmbi.1998.1760. [DOI] [PubMed] [Google Scholar]
- 59.Pan J.L., Bardwell J.C. The origami of thioredoxin-like folds. Protein Sci. 2006;15(10):2217–2227. doi: 10.1110/ps.062268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paxman J.J., Borg N.A., Horne J., Thompson P.E., Chin Y., Sharma P., et al. The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes. J Biol Chem. 2009;284(26):17835–17845. doi: 10.1074/jbc.M109.011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pei J., Pan X., Wei G., Hua Y. Research progress of glutathione peroxidase family (GPX) in redoxidation [Review] Front Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1147414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petit G.A., Hong Y., Djoko K.Y., Whitten A.E., Furlong E.J., McCoy A.J., et al. The suppressor of copper sensitivity protein C from Caulobacter crescentus is a trimeric disulfide isomerase that binds copper(I) with subpicomolar affinity. Acta Crystallogr Sect D. 2022;78(3):337–352. doi: 10.1107/S2059798322000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Premkumar L., Heras B., Duprez W., Walden P., Halili M., Kurth F., et al. Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases. Acta Cryst D. 2013;69(10):1981–1994. doi: 10.1107/S0907444913017800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi Y., Grishin N.V. Structural classification of thioredoxin-like fold proteins. Proteins. 2005;58(2):376–388. doi: 10.1002/prot.20329. [DOI] [PubMed] [Google Scholar]
- 65.Qin A., Scott D.W., Mann B.J. Francisella tularensis subsp. tularensis Schu S4 disulfide bond formation protein B, but not an RND-type efflux pump, is required for virulence. Infect Immun. 2008;76(7):3086–3092. doi: 10.1128/iai.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin A., Scott D.W., Thompson J.A., Mann B.J. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun. 2009;77(1):152–161. doi: 10.1128/iai.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qin A., Zhang Y., Clark M.E., Rabideau M.M., Millan Barea L.R., Mann B.J. FipB, an essential virulence factor of Francisella tularensis subsp. tularensis, has dual roles in disulfide bond formation. J Bacteriol. 2014;196(20):3571–3581. doi: 10.1128/jb.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin A., Zhang Y., Clark M.E., Moore E.A., Rabideau M.M., Moreau G.B., et al. Components of the type six secretion system are substrates of Francisella tularensis Schu S4 DsbA-like FipB protein. Virulence. 2016;7(8):882–894. doi: 10.1080/21505594.2016.1168550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quan S., Schneider I., Pan J., Von Hacht A., Bardwell J.C.A. The CXXC motif is more than a redox rheostat. J Biol Chem. 2007;282(39):28823–28833. doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- 70.Read R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr Sect D: Biol Crystallogr. 2001;57(10):1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 71.Ren G., Champion M.M., Huntley J.F. Identification of disulfide bond isomerase substrates reveals bacterial virulence factors. Mol Microbiol. 2014;94(4):926–944. doi: 10.1111/mmi.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ren G., Stephan D., Xu Z., Zheng Y., Tang D., Harrison R.S., et al. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J Biol Chem. 2009;284(15):10150–10159. doi: 10.1074/jbc.M809509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rietsch A., Bessette P., Georgiou G., Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179(21):6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santiago A.E., Mann B.J., Qin A., Cunningham A.L., Cole L.E., Grassel C., et al. Characterization of Francisella tularensis Schu S4 defined mutants as live-attenuated vaccine candidates. Pathog Dis. 2015;73(6):1. doi: 10.1093/femspd/ftv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santos-Martin C., Wang G., Subedi P., Hor L., Totsika M., Paxman J.J., et al. Structural bioinformatic analysis of DsbA proteins and their pathogenicity associated substrates. Comput Struct Biotechnol J. 2021;19:4725–4737. doi: 10.1016/j.csbj.2021.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schuck P., Perugini M.A., Gonzales N.R., Howlett G.J., Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: Strategies and application to model systems. Biophys J. 2002;82(2):1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shepherd M., Heras B., Achard M.E.S., King G.J., Argente M.P., Kurth F., et al. Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from salmonella enterica serovar typhimurium. Antioxid Redox Signal. 2013;19(13):1494–1506. doi: 10.1089/ars.2012.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shouldice S.R., Heras B., Walden P.M., Totsika M., Schembri M.A., Martin J.L. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid Redox Signal. 2011;14(9):1729–1760. doi: 10.1089/ars.2010.3344. [DOI] [PubMed] [Google Scholar]
- 79.Silverman P.M., Rosenthal S., Mobach H., Valentine R.C. Two new classes of F-pili mutants of Escherichia coli resistant to infection by the male specific bacteriophage f2. Virology. 1968;36(1):142–146. doi: 10.1016/0042-6822(68)90125-6. [DOI] [PubMed] [Google Scholar]
- 80.Snowden J., Simonsen K.A. StatPearls. StatPearls Publishing LLC,; 2022. Tularemia. [Google Scholar]
- 81.Studier F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 82.Subedi P., Paxman J.J., Wang G., Ukuwela A.A., Xiao Z., Heras B. The Scs disulfide reductase system cooperates with the metallochaperone CueP in Salmonella copper resistance. J Biol Chem. 2019;294(44):15876–15888. doi: 10.1074/jbc.RA119.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subedi P., Paxman J.J., Wang G., Hor L., Hong Y., Verderosa A.D., et al. Salmonella enterica BcfH Is a Trimeric Thioredoxin-Like Bifunctional Enzyme with Both Thiol Oxidase and Disulfide Isomerase Activities. Antioxid Redox Signal. 2021;35(1):21–39. doi: 10.1089/ars.2020.8218. [DOI] [PubMed] [Google Scholar]
- 84.Totsika M., Vagenas D., Paxman J.J., Wang G., Dhouib R., Sharma P., et al. Inhibition of diverse DsbA enzymes in multi-DsbA encoding pathogens. Antioxid Redox Signal. 2018;29(7):653–666. doi: 10.1089/ars.2017.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verderosa A.D., Dhouib R., Hong Y., Anderson T.K., Heras B., Totsika M. A high-throughput cell-based assay pipeline for the preclinical development of bacterial DsbA inhibitors as antivirulence therapeutics. Sci Rep. 2021;11(1):1569. doi: 10.1038/s41598-021-81007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walden P.M., Heras B., Chen K.-E., Halili M.A., Rimmer K., Sharma P., et al. The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production. Acta Crystallogr Sect D. 2012;68(10):1290–1302. doi: 10.1107/S0907444912026388. [DOI] [PubMed] [Google Scholar]
- 88.Walden P.M., Whitten A.E., Premkumar L., Halili M.A., Heras B., King G.J., et al. The atypical thiol-disulfide exchange protein [alpha]-DsbA2 from Wolbachia pipientis is a homotrimeric disulfide isomerase. Acta Crystallogr Sect D. 2019;75(3):283–295. doi: 10.1107/S2059798318018442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang G., Qin J., Verderosa A.D., Hor L., Santos-Martin C., Paxman J.J., et al. A buried water network modulates the activity of the escherichia coli disulphide catalyst DsbA. Antioxidants. 2023;12(2):380. doi: 10.3390/antiox12020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao G., Liu S., Ji X., Johnson W.W., Chen J., Parsons J.F., et al. First-sphere and second-sphere electrostatic effects in the active site of a class mu glutathione transferase. Biochemistry. 1996;35(15):4753–4765. doi: 10.1021/bi960189k. [DOI] [PubMed] [Google Scholar]
- 91.Zapun A., Bardwell J.C.A., Creighton T.E. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry. 1993;32(19):5083–5092. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 92.Zapun A., Missiakas D., Raina S., Creighton T.E. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry. 1995;34(15):5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 93.Zeldin O.B., Gerstel M., Garman E.F. RADDOSE-3D: time- and space-resolved modelling of dose in macromolecular crystallography. J Appl Crystallogr. 2013;46(4):1225–1230. doi: 10.1107/S0021889813011461. [DOI] [Google Scholar]
- 94.Zhao Z., Peng Y., Hao S.-f, Zeng Z.-h, Wang C.-c. Dimerization by domain hybridization bestows chaperone and isomerase activities. J Biol Chem. 2003;278(44):43292–43298. doi: 10.1074/jbc.M306945200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material