Abstract
Molecular switches serve as key regulators of biological systems by acting as one of the crucial driving forces in the initiation of signal transduction pathway cascades. The Ras homolog gene family member A (RhoA) is one of the molecular switches that binds with GTP in order to cycle between an active GTP-bound state and an inactive GDP-bound state. Any aberrance in control over this circuit, particularly due to any perturbation in switching, leads to the development of different pathogenicity. Consequently, the single nucleotide polymorphisms (SNPs) within the RhoA gene, especially deleterious genetic variations, are crucial to study to forecast structural alteration and their functional impacts in light of disease onset. In this comprehensive study, we employed a range of computational tools to screen the deleterious SNPs of RhoA from 207 nonsynonymous SNPs (nsSNPs). By utilizing 7 distinct tools for further analysis, 8 common deleterious SNPs were sorted, among them 5 nsSNPs (V9G, G17E, E40K, A61T, F171L) were found to be in the highly conserved regions, with E40K and A61T at G2 and G3 motif of the GTP-binding domain respectively, indicating potential perturbation in GTP/GDP binding ability of the protein. RhoA-GDP complex interacts with the enzyme phospholipase, specifically PLD1, to regulate different cellular activities. PLD1 is also a crucial regulator of thrombosis and cancer. In that line of focus, our initial structural analysis of Y66H, A61T, G17E, I86N, and I151T mutations of RhoA revealed remarkable decreased hydrophobicity from which we further filtered out G17E and I86N which may have potential impact on the RhoA-GDP-PLD1 complex. Intriguingly, the comparative 250 ns (ns) molecular dynamics (MD) simulation of these two mutated complexes revealed overall structural instability and altered interaction patterns. Therefore, further investigation into these deleterious mutations with in vitro and in vivo studies could lead to the identification of potential biomarkers in terms of different pathogenesis and could also be utilized in personalized therapeutic targets in the long run.
Keywords: RhoA, Molecular switching, Deleterious SNP, Molecular dynamic simulation, RhoA-GDP-PLD1
Highlights
-
•
The hydrophobicity of the Y66H, A61T, G17E, I86N, and I151T mutations in RhoA was observed to decrease remarkably.
-
•
The I86N point mutation significantly reduced the structural stability of the RhoA protein.
-
•
The G17E and I86N nsSNPs were found to have the most detrimental impact on the structure and function of the RhoA protein.
-
•
Wild-type RhoA-GDP-PLD1 complex showed stronger binding affinity than RhoA (G17E/I86N mutants)-GDP-PLD1 complex.
1. Introduction
The Ras homolog gene family member A (RhoA) is one of the key members of the Rho GTPase superfamily. RhoA and other GTPases act as molecular switches that regulate the actin cytoskeleton, cell growth, motility, morphogenesis, differentiation, innate and adaptive immunity, and apoptosis by changing between GTP-bound active and GDP-bound inactive forms (Wheeler and Ridley, 2004; Takai et al., 2001; Vega and Ridley, 2008; Burridge and Wennerberg, 2004). In all eukaryotic organisms, this sort of molecular switching is a fundamental mechanism in respective contexts to cover precise control over processes including survival, proliferation, advancement, defense, diversification, and reproduction (Wittinghofer and Vetter, 2011; Perica et al., 2021). The underlying factor that accounts for such potential is their ambidexterity in switching “ON” and “OFF” in response to various stimuli such as ligand binding, changes in pH or temperature, mechanical stress, and other external as well as internal influences. One specific example of this switching in signal transduction pathways is the Rho family of small GTPases binding to the C-terminal domain of Phospholipase D1 (PLD1), regulating its activity in generating phosphatidic acid. PLD1 is an isoform of Phospholipase which has a crucial role in activating the lipid second messenger phosphatidic acid (Xie et al., 2002; Du et al., 2000).
Rho GTPase activity is determined by Rho guanine nucleotide exchange factors (RhoGEFs) which facilitate the association of GTP, while inactivation is achieved through the action of Rho GTPase activating proteins (RhoGAPs) via the hydrolysis of GTP (Hanna and El-Sibai, 2013). In case of RhoA, when this protein is in its inactive state, guanine nucleotide dissociation inhibitors (GDIs) lock it in the cytosol. Conversely, when activated, RhoA remains associated with the plasma membrane (Leonard et al., 1992).
However, the perturbation of any dynamic switching events leads to a wide range of pathological conditions, such as cancer, cardiovascular disease, and neurological disorders, etc. (Alexanian et al., 2021; McWilliams et al., 2018; Wittinghofer and Waldmann, 2000; Chen et al., 2021). Therefore, the understanding of the complex interplay between molecular switching and its regulatory mechanisms could provide elucidation of the significant fundamental principles behind eukaryotic biology and the development of new strategies for the prevention and treatment of diseases. Recent studies have shown that RhoA can be active at the leading edge of migrating cells, and the cooperation of Rho effector Rhotekin and S100A4 can suppress stress fibre generation to permit RhoA-mediated lamellae formation (O'Connor and Chen, 2013). RhoA also participates in the formation of phagocytic cups mediated by αMβ2 and opsonized particles, whereas β2 integrin controls its activation and recruitment (Wiedemann et al., 2006).
In various cancers, aberrant expression of RhoA plays a role in oncogenic signaling. When cooperating with JAK/STAT3 and NF-κB, it facilitates survival, proliferation, and circumvents apoptosis (O'Halloran et al., 2014; Jaffe and Hall, 2005). Comprehending the frequency and impact of genetic variation is crucial for identifying the detrimental associations of RhoA, as they offer insight into an individual's genetic predisposition to diseases, drug responses, and other phenotypic traits.
Single nucleotide polymorphisms (SNPs) represent the most common type of genetic variation in humans (Collins et al., 1998), with approximately 0.1% of the human genome consisting of unique variants that result from random mutations (Forsberg et al., 2000). SNPs are point mutations in DNA sequence that can be classified into two categories: synonymous SNPs which do not alter the sequence of amino acid, whereas non-synonymous SNPs (nsSNPs) encoding modified proteins may cause structural or functional abnormalities (Lander, 1996; Dobson et al., 2006). As RhoA plays a crucial regulatory role in the downstream signaling cascade, nsSNPs within its coding region may negatively impact these functions (Wheeler and Ridley, 2004; Thumkeo et al., 2013). It has been found that hereditary genetic disorders, including various autoimmune diseases, are caused by about half of the genetic variation resulting from nsSNPs (Yue and Moult, 2006; Chasman and Adams, 2001; Cargill et al., 1999; Begovich et al., 2004).
Studies conducted in recent years have established a strong correlation between nsSNPs and the function of RhoA in the case of specific types of malignancies whereas G17V mutation causes functional loss of over fifty percent of angioimmunoblastic T cell lymphomas (Palomero et al., 2014). Regarding the changes in the effector domain of RhoA, Y42C mutant has been reported to contribute to 20% of diffuse-type gastric cancer (Wang et al., 2014; Kakiuchi et al., 2014). Also, high expression of RhoA in the corpus cavernosum (CC) contributes to RhoA-mediated Ca2+ sensitization and the flaccid state of CC (Wang et al., 2002).
Over the years, numerous computational tools have been developed with the aim of screening, mapping, and postulating the impacts of both functional and deleterious SNPs of targeted genes. However, no previous computational study had investigated the functional impacts of nsSNPs on the RhoA protein using molecular docking and molecular dynamics (MD) simulation. Our objective was to identify the most harmful SNPs, focusing on highly conserved regions within the RhoA gene and to explore their consequences on the RhoA protein through the application of different in silico tools. We also sought to assess the functional effects of identified nsSNPs on the interaction between RhoA and PLD1 by MD simulation, aiming to comprehensively confirm and validate the adverse impacts of these deleterious SNPs on the structure and function of the RhoA protein.
2. Methods and materials
2.1. Collection of datasets from various databases
The raw data of the RhoA gene, comprising its SNP rsIDs, chromosomal location, protein accession number, and residue alteration, was sourced from the dbSNP database of NCBI (https://www.ncbi.nlm.nih.gov/projects/SNP/). Missense variants of the SNP rsIDs were extracted for further screening. Subsequently, protein sequence data of RhoA was collected from the UniProtKB database (http://www.uniprot.org/uniprot/). Overall, a thorough approach was adopted to ensure comprehensive and accurate information on the RhoA gene was gathered.
2.2. Identification of most deleterious SNPs
To identify the highly significant deleterious SNPs from the dbSNP database, a comprehensive bioinformatics approach was applied, utilizing eight different tools. The initial screening was done by SIFT (Sorting Intolerant from Tolerant) algorithm followed by assessments with seven additional tools, which include PolyPhen-2 (Polymorphism Phenotyping v2), SNAP2 (Screening for Non-Acceptable Polymorphism), PROVEAN (Protein Variation Effect Analyzer), CADD (Combined Annotation Dependent Depletion), PhD-SNP (Predictor of human Deleterious Single Nucleotide Polymorphisms), PredictSNP2, and SNPs&GO.
SIFT (https://sift.bii.a-star.edu.sg/) helps to identify and differentiate the amino acid substitutions in the functionally important and neutral domains. The feature of sequence homology and physical properties of the amino acids of this tool calculated the probability scores of deleterious (with a score below 0.05) and tolerated (with a score above 0.05) amino acid substitutions, considering their positions and types (Kumar et al., 2009). Furthermore, to predict the effects of amino acid substitutions on protein structure and function through physical and comparative analysis, Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) considers a range of factors, including the physiochemical properties of the amino acid residues, evolutionary pattern, and the known functional sites of the protein. This tool provides feedback as a score ranging from 0.0 (neutral) to 1.0 (deleterious) (Adzhubei et al., 2010). SNAP-2 (https://www.rostlab.org/services/SNAP/) is a classifier developed using a machine learning technique known as a ‘neural network’ to distinguish the deleterious SNPs from the neutral ones, allowing for a more nuanced assessment of the potential impact of amino acid substitutions on protein function. The result presents a robust prediction ranging from ‘strongly neutral (−100)’ to ‘strongly effective (+100)’ (Hecht et al., 2015). PROVEAN (http://provean.jcvi.org/index.php) utilizes a sequence homology-based approach to predict the functional impact of changes in the amino acid sequence of a protein, including SNPs. This tool generates a score that distinguishes deleterious variants from neutral ones: variants with a PROVEAN score at or below a predefined threshold of 2.5 are predicted to have a deleterious effect, while those with a score above the threshold are predicted to have a neutral effect, providing a cutoff for interpretation of results (Choi and Chan, 2015). The integrative framework known as CADD (https://cadd.gs.washington.edu/) scores human single nucleotide variants and short insertions and deletions by contrasting variants that have survived natural selection against simulated mutations. It accomplishes this by incorporating over 60 genomic features into a single metric. Here, a C score ≥10 indicates that the amino acid substitution is among the 10% most deleterious substitutions, while a score ≥20 predicts it to be within the 1% most deleterious. Potentially pathogenic variants are assumed to be at a score of 15 (Rentzsch et al., 2019). As this assumption is arbitrary, other tools are used to validate this prediction. PhD-SNP (https://snps.biofold.org/phd-snp/phd-snp.html) employs evolutionary information to investigate the effect of polymorphism on protein function. It differentiates SNPs associated with diseases from benign point mutations through the Support Vector Machine (SVM) method. A score >0.5 is indicated to be pathogenic, while a score below this threshold is predicted to be benign (Capriotti et al., 2006). PredictSNP2 (https://loschmidt.chemi.muni.cz/predictsnp2/) is a consensus classifier that integrates the six best-performing methods to achieve a more robust and reliable output of the deleterious SNPs. It generates a score that falls within the continuous interval of < −1, +1>, with values in the interval (<-1,0>) indicating a neutral effect, and values in the interval (0, +1>) suggesting a potentially deleterious effect (Bendl et al., 2016). SNPs&GO (https://snps-and-go.biocomp.unibo.it/snps-and-go/) evaluates the pathogenicity of amino acid substitutions based on protein functional annotation. The potential effect of SNPs on protein-coding transcripts or structures is calculated into a score, with higher scores indicating more deleterious effects (Capriotti et al., 2013).
2.3. Identification of the position of missense SNPs in the domain of RhoA
To map the missense SNPs on the conserved domains of RhoA, InterPro (https://www.ebi.ac.uk/interpro/) tool was used (Hunter et al., 2009). Protein sequence in FASTA format was inserted as a query. This tool can utilize databases containing protein families, domains, and functional sites to identify the functional characterization of a protein through the recognition of motifs and domains.
2.4. Analyzing the effect of the nsSNPs on protein stability
The mutation causes changes in protein stability, and to predict these impacts, I-Mutant 3.0 (http://gpcr.biocomp.unibo.it/∼emidio/I-Mutant3.0/I-MutantDDG_Help.html) was employed that is based on the database of ProTherm, which is currently the most comprehensive database of experimental data on protein mutations (Capriotti et al., 2005). This web-based server's reliability index (RI) feature provides values ranging from 0 to 10, with higher values indicating greater reliability.
2.5. Identification of missense SNPs in evolutionarily conserved regions of RhoA
The presence of missense SNPs on highly conserved regions of amino acid residues within RhoA indicates the possibility of significant alteration to the expression of RhoA. Identification of such conserved positions was carried out by the ConSurf web server (https://consurf.tau.ac.il/consurf_index.php) which determines the rate of the conservation of amino acid positions (Ashkenazy et al., 2010). This server employs phylogenetic relation analysis using the homologous sequences to find these conserved regions. The Bayesian method classifies conservation scores into three categories: variable, intermediate, and conserved, with scores of 1–4, 5–6, and 7–9, respectively. The prediction of conserved patterns and structural and functional amino acids, as well as the determination of a conservation score and coloring scheme, was accomplished by inputting the protein FASTA sequence of RhoA. High-risk missense SNPs of RhoA were used for further analyses.
2.6. Prediction of the structural effect of missense SNPs on RhoA protein
In order to evaluate the impact of missense SNPs on RhoA protein structure, two sophisticated and distinct tools were used, including HOPE (https://www3.cmbi.umcn.nl/hope/) and Missense 3D (http://missense3d.bc.ic.ac.uk/missense3d/) (Venselaar et al., 2010; Khanna et al., 2021). HOPE is a web server that identifies the structural effects of the point mutations in a protein sequence. P61586 (UniProt ID of RhoA) and the filtered SNPs were used individually as the input. HOPE utilizes a range of data sources, including WHAT IF Web, distributed annotation system (DAS) servers, and UniProt database to observe the impact of substitution on the 3D structure of the RhoA protein. Missense 3D detects structural changes of the RhoA due to amino acid substitution, leading to the characterization of missense variants by coordinating the 3D structure of proteins collected from experimental models stored in the RCSB database. Swiss-PDB Viewer (v4.1.0) (https://spdbv.vital-it.ch/) is a stand-alone software associated with missense 3D, was used to generate mutated models of the proteins for the corresponding amino acid substitutions (Waterhouse et al., 2018). With the help of the Mutation Tool, a feature of Swiss-PDB Viewer, which is a rotamer library of mutated models, can be browsed, and the best rotamer of the mutated model can be selected. The comparison between wild-type and mutated protein structure was analyzed through TM-align (https://seq2fun.dcmb.med.umich.edu//TM-align/). When the protein structure alignment algorithm runs, it computes the template modeling score (TM-score) and RMSD of both models. TM-score ranges from 0 to 1, where a better match between the structures is indicated by a higher value. The distance between the alpha carbon backbone of the model is measured by the RMSD, and a higher value means that there is greater variation in their structure (Zhang and Skolnick, 2005).
2.7. 3D protein modeling of missense SNPs
To generate a complete 3D model of the mutated protein, AlphaFold3 was used. AlphaFold3 uses a neural networking approach that produces an experimentally close or near resolution of protein structure, as it has revolutionized protein structure prediction for their accuracy and reliability (Abramson et al., 2024).
The resulting 3D protein models were then validated through the PROCHECK tool which performs the Ramachandran plot analysis for accuracy and quality. The geometry of individual residue compared to that of the entire structure is examined by this tool (Laskowski et al., 1993, 1996). The overall model quality is validated by ERRAT based on the statistical relationship of nonbonded interactions between various types of atoms, using typical atomic interactions (Morris et al., 1992; Laskowski et al., 2012). The high standard resolution of structure gives ERRAT score of around 95% or more and for lower resolutions (2.5–3 A) the average overall quality factor is around 91% (Zobayer and Aowlad Hossain, 2018).
2.8. Identification of RhoA interacting proteins
The study of protein-protein interactions provides understanding of how a particular protein interacts with other proteins in various biological pathways, and what the nature of this interaction is. Proteins that have direct interaction with RhoA protein were identified by employing the STRING (https://string-db.org/) tool, and the interrelationships between them were ascertained (Snel et al., 2000; Szklarczyk et al., 2021, 2023). To evaluate the functional impact, certain key proteins were chosen using the KEGG pathway (https://www.genome.jp/kegg/pathway.html), and further analysis was conducted to determine whether their interaction with RhoA protein was altered by the effect of missense SNPs (Kanehisa and Goto, 2000; Ogata et al., 1999; Kanehisa et al., 2017).
2.9. Molecular docking approach to observe the functional effect
Protein-ligand-protein docking analysis was performed to observe the functional effect of this PLD1 activation. The structure of the PLD1 protein was obtained from the RCSB PDB database (https://www.rcsb.org/) (PDB ID: 1FTN) (Berman et al., 2000). The 3D structure of the I86N and G17E point mutation was also obtained using AlphaFold3 to observe the changes in the interaction with PLD1. The structure of the GDP molecule was collected from PubChem (https://pubchem.ncbi.nlm.nih.gov/) (PubChem CID: 5957) and optimization of its energy was done using Avogadro software (Kim et al., 2023; Hanwell et al., 2012). Proteins and ligand structure preparation and visualization were analyzed using Biovia Discovery Studio. The molecular docking of the RhoA-GDP-PLD1 complex was performed using the HADDOCK (High Ambiguity Driven protein-protein Docking) web server (https://wenmr.science.uu.nl/haddock2.4/), which is an information-driven flexible docking approach for modeling biomolecular complexes (Honorato et al., 2021; Van Zundert et al., 2016). All default parameters of HADDOCK were used during the submission of the input data. The active site of PLD1 and RhoA was selected based on a previous study to enhance the accuracy of the docking analysis (Bowling et al., 2020). The docking result and the binding interaction between protein-ligand and receptor proteins were visualized and analyzed by UCSF ChimeraX and PyMol visualization tool (Pettersen et al., 2021; Goddard et al., 2018).
2.10. Molecular dynamics simulation analysis
2.10.1. Preparation of simulation system
The GROMACS-2023 version in the Ubuntu 24.04.1 LTS system was utilized in conducting molecular dynamics simulations of protein-ligand-protein docking complexes in this study (Van Der Spoel et al., 2005; Abraham et al., 2015). The most updated version of the Charm36-jul2021 force field, widely used for biomolecules, was employed (Brooks et al., 2009; Croitoru et al., 2021). The RhoA-GDP-PLD1, a biomolecular complex, was the subject of the study. In this study, the recommended TIP3P_CHARM CHARM-modified TIP3P water model was utilized (Harrach and Drossel, 2014). A cubic-type box system was defined with a box diameter of 13.605 nm and a center of 7.083 7.083 7.083 nm. The system consisted of 374,507 atoms in 119,245 residues and had a total volume of 3800.27 nm3 and a density of 997.133 g/L. The acid dissociation constant value (pKa) for amino acids was also calculated using Ewald summation. To neutralize the charge of the positively charged system, 10 CLA ions were added in place of 10 random solvent molecules using the “genion” tool, and the pH was set to 7. During the equilibration period, the system temperature was raised from 0 K to 300 K. A stable conformation was obtained by performing energy minimization using the steepest descent method for 5000 steps. The energy-minimized systems were then subjected to equilibration for 200 ps in both NPT (constant number of particles, pressure, and temperature) and NVT (constant number of particles, volume, and temperature) ensembles. MD simulation was carried out for 250 ns using the particle-mesh Ewald (PME) method to describe short-range electrostatic interactions within a cut-off distance of 1.2 nm. Throughout the simulation, constant pressure and Berendsen thermostat were sustained, and a multiple timestep algorithm was employed to establish a time step interval of 2.00 fs (Essmann et al., 1998).
2.10.2. Trajectory analysis of MD simulations
Structural trajectories were calculated for Protein-Ligand-Protein systems using the trjconv tool. The proteins and GDP molecule within the cubical box were recentered using the trjconv tool of GROMACS. Values such as van der Waals interaction, electrostatic energy, and interaction energy were calculated using the gmx energy tool of GROMACS. Finally, the structural changes of the native and mutant proteins were calculated through RMSD, RMSF, Rg, and SASA analysis using the “gmx rms”, “gmx rmsf”, “gmx gyrate” and “gmx sasa” tools. The binding free energy of the complexes was determined using the Molecular Mechanics Generalized Born Surface Area (MMGBSA) and Molecular Mechanics Poisson Boltzmann Surface Area (MMPBSA) approaches using the MMPBSA. py module, analyzing 1000 snapshots taken from the final 5 ns of the NPT-MD trajectories (Miller et al., 2012). In standard MM/PBSA, the binding free energy (ΔGbind) between the proteins is calculated as (Genheden and Ryde, 2015; Sun et al., 2018; Rastelli et al., 2010)
| ΔGbind = ΔH − TΔS ≈ ΔEMM + ΔGsol – TΔS | (1) |
| ΔEMM = ΔEele + ΔEvdw | (2) |
| ΔGsol = ΔGPB + ΔGSA | (3) |
The binding free energy (ΔGbind) of the complexes can be broken down into three components including, gas-phase interaction energy (ΔEMM), desolvation free energy (ΔGsol), and conformational entropy (−TΔS). The gas-phase interaction energy (ΔEMM) consists of electrostatic (ΔEele) and van der Waals (ΔEvdw) terms. The desolvation free energy (ΔGsol) is composed of nonpolar (ΔGSA) and polar (ΔGPB) contributions. The nonpolar contribution (ΔGSA) is calculated using the solvent accessible surface area (SASA), with ΔGSA = γ⋅ SASA + b, where γ is the surface tension (2.27 kJ mol−1 nm−2) and b is a constant (3.85 kJ mol−1). The polar contribution is evaluated using the Poisson−Boltzmann (PB) model. The conformational entropy (−TΔS) is calculated by a computationally efficient interaction entropy method. The electrostatic energy (ΔEele) between proteins is calculated using the following equation:
| (4) |
Where qi represents the charge of atom i in one protein, qj denotes the charge of atom j in the other protein, rij is the distance between these atoms, ϵ0 is the dielectric constant in a vacuum, and ϵin is the relative dielectric constant of the solute (proteins), usually set to 2.0 unless otherwise specified (Levin, 2002).
2.10.3. Statistical analysis
All the corresponding plots were generated using the XMGRACE and GnuPlot tools (Turner et al., 2005; Williams and Kelley, 2011). The statistical analysis of the MD trajectories was performed using GraphPad Prism v 8.0 (GraphPad Software, San Diego, CA, USA) software. Unpaired two-tailed Student's t-tests were performed, and p values of <0.0001 were considered highly significant for comparing the simulation results.
3. Results
3.1. Retrieval of SNPs
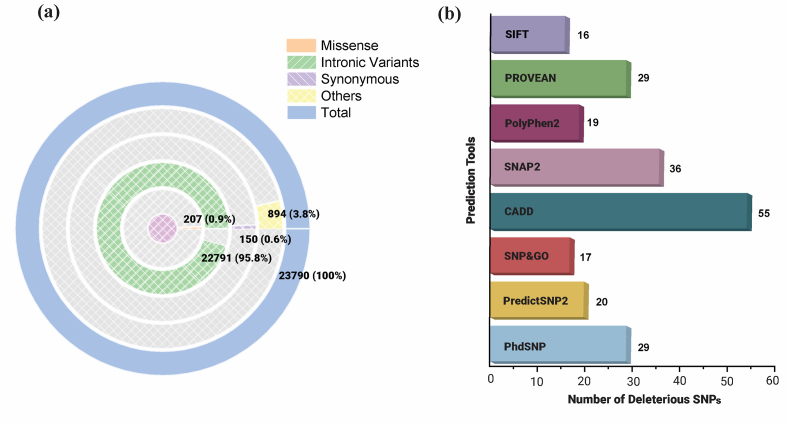
A total of 23,790 SNPs of the human RhoA gene were retrieved from the NCBI dbSNP database (see Fig. 1). Among them, 22,791 SNPs were found to be located in the intronic region, while 207 SNPs were identified as missense, and 150 SNPs were synonymous. Fig. 2(a) indicates that most SNPs are located in the intronic region (95.8%), followed by others (3.76%), then missense SNPs (0.87%) and coding synonymous SNPs (0.63%). Further analysis was carried out on 207 nsSNPs, as they result in altered amino acids due to changes in the coding region.
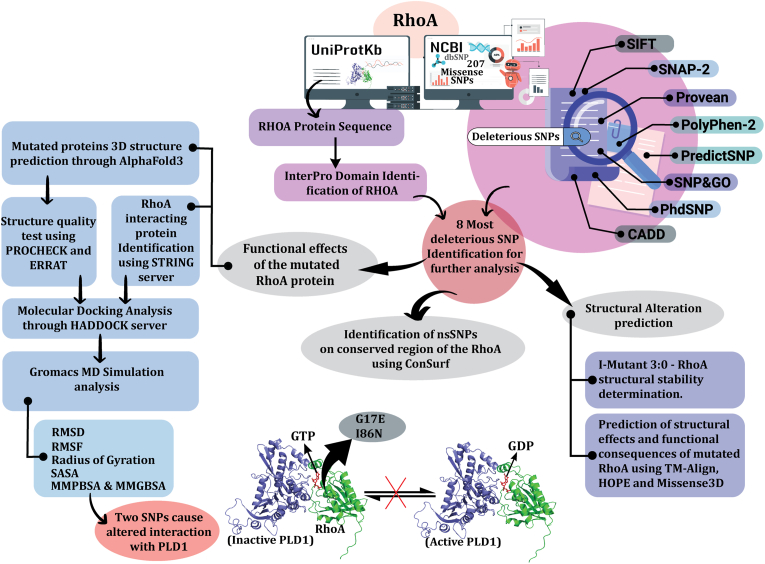
Fig. 1.
Schematic representation of the most deleterious SNP identification of RhoA gene and their subsequent effect analysis.
Fig. 2.
In silico-based identification of deleterious SNPs in the RhoA gene. (a) A circular diagram highlighting the various types of SNPs in RhoA. (b) The total number of missense SNPs and predicted deleterious SNPs by various algorithms are described in the bar plot.
3.2. Identification of the most deleterious SNPs
SNPs of the RhoA retrieved from the dbSNP database were subjected to a comprehensive in silico analysis through a variety of tools such as SIFT, Polyphen-2, SNAP2, PROVEAN, Predict SNP, SNP&GO, and PhD-SNP. Initial screening output obtained by SIFT demonstrated the 65 nsSNPs as either tolerated or deleterious while remaining SNPs out of 207 remained undetected. Among these 65 nsSNPs, SIFT classified 16 nsSNPs as deleterious. To validate the SIFT filtering, the other 8 computational algorithms described above were employed, resulting in the identification of 9 nsSNPs that were predicted to have the most deleterious effects, as shown in Table 1. Out of 8 computational tools, CADD predicted the maximum number of deleterious SNPs, which was 55 followed by SNAP2, with a count of 36, as shown in Fig. 2(b).
Table 1.
List of 9 deleterious nsSNPs in RhoA gene, as identified by eight different in silico tools.
| SNP IDs | AA Changes | SIFT | CADD | PROVEAN | PolyPhen 2 | SNAP2 | SNP & GO | PredictSNP | PhdSNP |
|---|---|---|---|---|---|---|---|---|---|
| rs1057519951 | E40K | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs1057519954 | Y42S | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs1553631976 | Y66H | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs1575647051 | A61T | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs11552761 | G17E | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs112304179 | F171L | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs1381401434 | I86N | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs1465894043 | I151T | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
| rs1575653732 | V9G | Damaging | Deleterious | Deleterious | Probably Damaging | Effect | Disease | Effect | Disease |
3.3. Identification of nsSNPs in the domain of RhoA
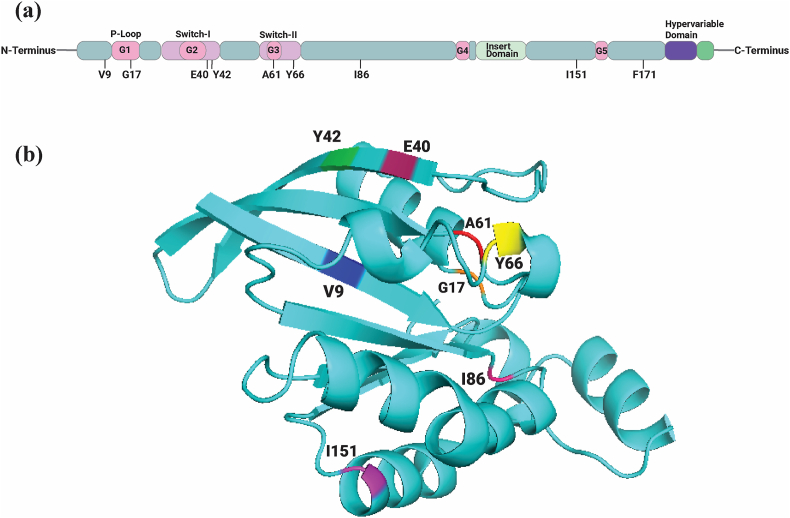
A domain identification tool named InterPro was used to predict the domains and active sites of a protein through the functional analysis of protein families. It predicted one functional domain of RhoA, which is the Small GTP Binding Domain (5–159) and revealed that 8 out of 9 nsSNPs are positioned in this domain. An illustrated physical mapping of these SNPs in Fig. 3(a) shows that G17E is positioned in the p-loop. Besides, E40K and Y42S are positioned in the switch 1 and A61T and Y66H in the switch 2. Positions of these SNPs are marked into a 3D protein cartoon structure as shown in Fig. 3(b).
Fig. 3.
In the human RhoA crystal structure, the structural domains and motifs present in RhoA are highlighted in a representative three-dimensional structure (a), and the identified variant position for 8 nsSNPs is labeled in a cartoon representation (b).
3.4. Analyzing the effect of the nsSNPs on protein stability
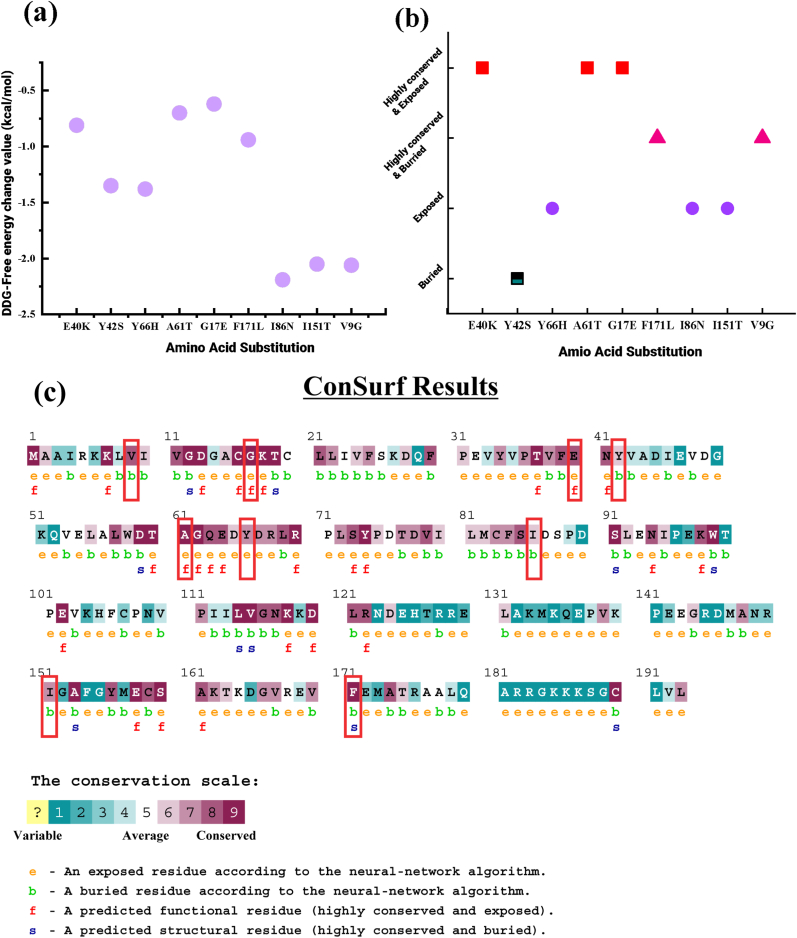
The structural stability of the protein is essential for the functioning of a protein. I-Mutant 3.0 was used to predict the extent of alteration in structural stability in terms of RI and free energy change value (ΔΔG). This tool revealed that structural stability is reduced for each of the 9 deleterious nsSNPs. It also predicted that I86N point mutation causes a maximum decrease in structural stability and ΔΔG value is −2.19 kcal/mol. Y42S, Y66H, I151T, and V9G polymorphisms also cause higher structural instability as shown in Supplementary Table 1. A statistical comparison among 9 SNPs has been shown in Fig. 4(a).
Fig. 4.
Effect of structural stability and evolutionary conservation analysis of protein residues (a) The effects of nsSNPs on protein stability predicted by I-MUTANT 3.0 based on structural stability in terms of RI and free energy change value (DDG) are plotted. ConSurf results are highlighted in red (b) and revealed that 5 out of 9 nsSNPs are highly conserved which is plotted in (c).
3.5. Evolutionary conservation analysis
ConSurf web server was used to determine the conservancy of amino acids of native RhoA protein. The putative functional and structural amino acids are recognized, and their evolutionary conservation profile is identified by the ConSurf server using the Bayesian method. The results revealed structural and functional residues of the 9 high-risk nsSNPs of RhoA protein using evolutionary conservation and solvent accessibility. This tool predicted that Y66, I86, and I151 residues are exposed, whereas the Y42 residue is buried. On the other hand, residues E40, G17, and A61 are highly conserved, functional and exposed, while F171 and V9 are highly conserved, structural, and buried as illustrated in Fig. 4(b).
3.6. Prediction of structural effects
The effect of amino acid substitution on the physical and chemical properties, hydrophobicity, spatial structure, and function of the protein was revealed by HOPE, shown in Table 2. The mutant residues E40K, A61T, G17E, F171L, and I86N were reported to be larger than the wild-type, while the mutant residues Y42S, Y66H, I151T, and V9G were smaller than the wild-type, according to the result of HOPE. Furthermore, the hydrophobicity of the mutant residues A61T, G17E, I86N, I151T, and V9G was found to be decreased. When there is a difference in hydrophobicity and size between wild-type and mutant residues, it can disrupt hydrogen bonds with nearby residues, affecting the protein structure.
Table 2.
The effect of amino acid substitution on the physical and chemical properties, hydrophobicity, spatial structure, and function of protein predicted by Project Hope.
| Mutations | Changes of size | Changes of charge | Changes of hydrophobicity | Interpretation |
|---|---|---|---|---|
| E40K | M > W | Negative > Positive | The mutation occurs within a segment of residues annotated in UniProt as an effector region motif. The differences in amino acid properties are likely to disrupt the motif and impair its function. The mutation at this position introduces a larger residue, which can disrupt multimeric interactions. The mutant residue is near a highly conserved position. Mutation of this residue, which is located on the protein's surface, can disrupt interactions with other molecules or protein subunits. | |
| Y42S | M < W | At position 59, the wild-type residue forms a hydrogen bond with Aspartic Acid. Due to the size difference between the wild-type and mutant residue, the mutant residue cannot form the same hydrogen bond as the original wild-type residue. The mutation occurs within a segment of residues annotated in UniProt as an effector region motif. The differences in amino acid properties are likely to disrupt the motif and impair its function. The mutation is close to a highly conserved position. | ||
| Y66H | M < W | Decreased | Both the wild-type and mutant residues have different hydrophilicity. The mutation will eliminate hydrophobic interactions in the protein's core. | |
| A61T | M > W | Decreased | The 3D structure reveals that the residue of the wild-type protein interacts with a ligand labeled GSP. The difference in properties between the wild-type and the mutant can readily result in the loss of ligand interactions. Because ligand binding is frequently essential to the protein's function, this mutation may impair this function. These differences in properties between the wild-type and mutant residues can readily result in the loss of interactions with the nucleotide (“GTP”). This can directly affect the protein's function. | |
| G17E | M > W | Neutral > Negative | Decreased | The residue of the wild-type is glycine, which is the most flexible of all residues. This flexibility may be required for the protein to perform its function. The mutation of this wild-type's residue is glycine; glycine can render this function ineffective. The mutated residue is not in contact with a metal; however, one of the adjacent residues is in contact with a metal and may be impacted by the mutation in its vicinity. The 3D structure reveals that the residue of the wild-type protein interacts with a ligand labeled GSP. The difference in properties between the wild-type and the mutant can readily result in the loss of ligand interactions. Because ligand binding is frequently essential to the protein's function, this mutation may impair this function. These differences in properties between the wild-type and mutant residues can readily result in the loss of interactions with the nucleotide (“GTP”). This can directly affect the protein's function. |
| F171L | M > W | Only this type of residue was detected at this position. Mutation of a residue that is 100% conserved is typically detrimental to the protein. The mutant residue is more diminutive than its wild-type counterpart. The mutation will result in a vacant space in the protein's core. | ||
| I86N | M > W | Decreased | The residue of the wild-type was concealed in the center of the protein. The mutant residue's size makes it likely to not fit. | |
| I151T | M < W | Decreased | Both the wild-type and mutant residues have different hydrophobicity. The mutation will eliminate hydrophobic interactions in the protein's core. | |
| V9G | M < W | Decreased | This position is changed to glycine by the mutation. Glycine is extremely flexible and can disrupt the protein's required rigidity at this position. The mutation will eliminate hydrophobic interactions in the protein's core. |
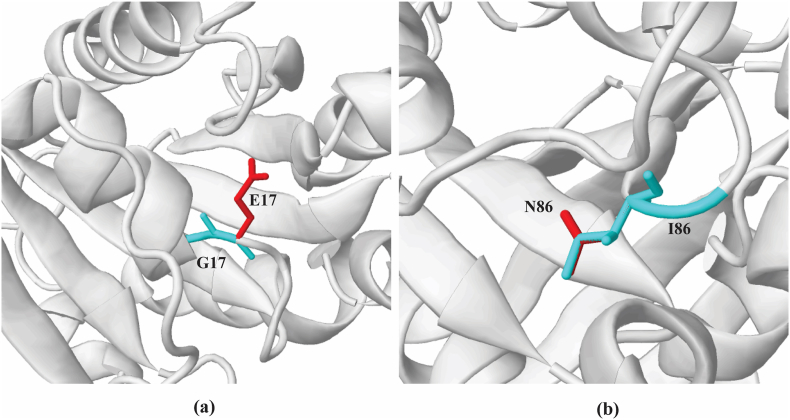
The appearance of a disallowed phi/psi alert by the G17E substitution was revealed by the Missense3D tool. The phi/psi angles were determined to be in the favored region for the wild-type residue but in the outlier region for the mutant residue. This substitution also resulted in the replacement of the originally located glycine in a bending curvature, as shown in Fig. 5(a). The I86N substitute replaced a buried hydrophobic residue (ILE, RSA 0.0%) with a hydrophilic residue (ASN, RSA 0.0%) as shown in Fig. 5(b). No structural damage was predicted by this tool for other substitutions. After energy minimization, the total energy of the wild-type structure was found to be −3.13 × 105 kcal mol−1. Following energy minimization, the total energy of G17E (−3.22 × 105 kcal mol−1) and I86N (−3.18 × 105 kcal mol−1) were significantly decreased, as demonstrated by molecular dynamic simulation results.
Fig. 5.
Structural alteration of the wild-type residue by the mutant G17E (a) and I86N (b) illustrated by Missense 3D. The wild-type residue is presented as cyan, and the mutant residue is shown in red.
Structural comparison between wild-type and mutants through the TM align reveals the total Root mean square deviation (RMSD) value with the superimposed native protein was determined to be 0.23 Å for mutant G17E and 0.16 Å for mutant I86N. Thus, G17E mutant displayed greater deviation.
3.7. 3D protein modeling of missense SNPs
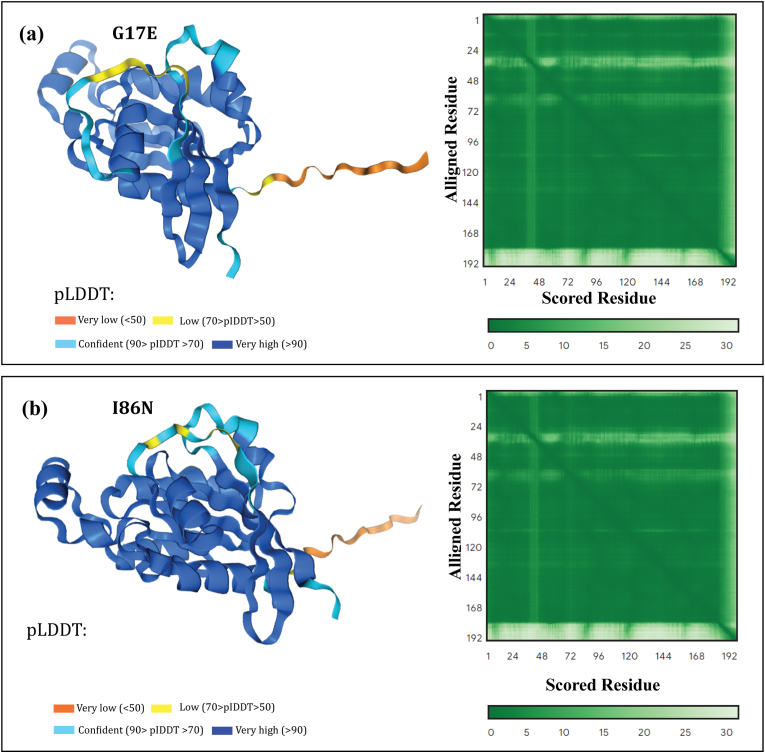
AlphaFold3 is an automated system that provides structure predictions with near-experimental precision. The outputs include three distinct confidence metrics with 3D structures. The first metric is a per-residue confidence metric termed predicted Local Distance Difference Test (pLDDT). It colors residues based on the region's precision, with confidence scores ranging from 0 to 100. A pLDDT greater than 90 predicts with high accuracy and is suitable for predicting vital information such as the correct domain. A pLDDT between 70 and 90 provides a decent prediction of the backbone. The regions between pLDDT 50 and pLDDT 70 must be approached with caution. AlphaFold3 predicts that pLDDT50 will be unstructured and have a ribbon-like appearance when isolated. Consequently, this prediction yields the loops and domains depicted in Fig. 6. The models of the predicted proteins have very high confidence score across lion's share of the structures, making the structures highly reliable.
Fig. 6.
3D protein modeling with AlphaFold3. AlphaFold3 predicted mutated structure of RhoA (a) G17E, and (b) I86N. The lower pLDDT score (the orange and yellow region) indicates disordered region. Both models here have very high and confidence scores across lion's share of the structures, making the structures highly reliable. The lower the PAE value, the better defined the residue's relative position is assumed to be, however when the color changes to light green from dark green, the PAE score rises, and their relative positions are anticipated with less confidence. The predicted template modeling (pTM) score for G17E is 0.89 and for I86N is 0.9. A score above 0.5 predicts that the folds of the model is possibly similar to the true structure.
Predicted Aligned Error (PAE) is another metric that evaluates the confidence in the relative position of the pair residues. The output is an interactive 2D graphic with the expected position error represented in residue x and the true structure aligned on residue y. The lower the score, the more certain AlphaFold3 is that these residues are packed together in a structure as seen in Fig. 6. PAE plot with most of the region with dark green color predicts the model as accurate. The last metric is predicted template modeling (pTM) value, which confirms the accuracy of the folds of the models with a score higher than 0.5.
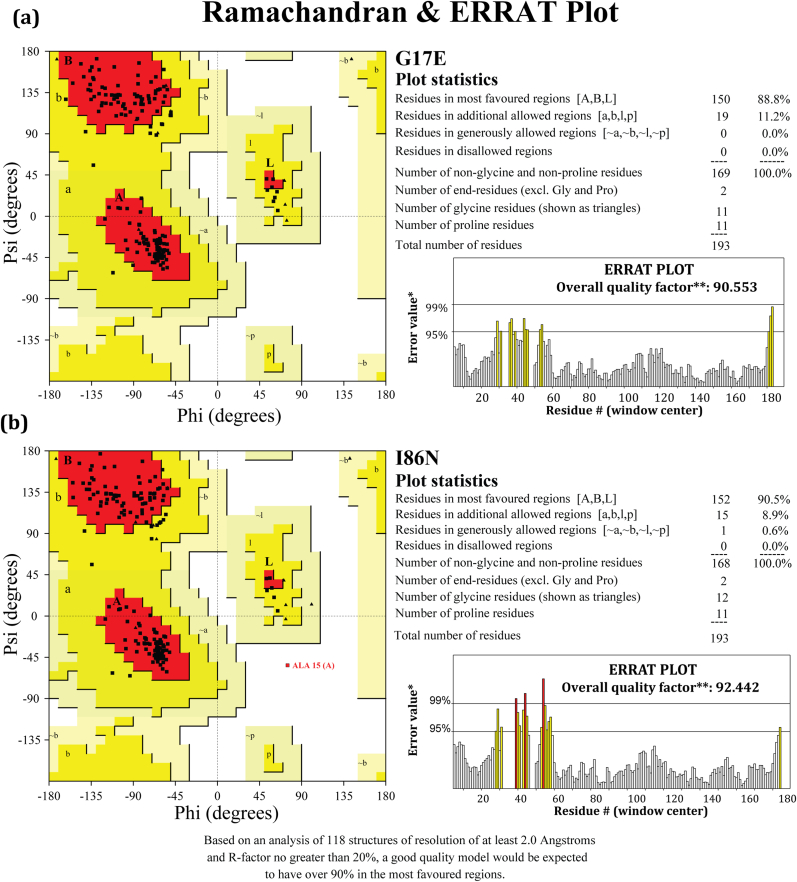
The stereochemical quality of the generated 3D protein was evaluated by PROCHECK which examines various factors such as the overall G-factor, chi1-chi2 plots, phi-psi angles, and sidechain parameters, while ERRAT validated the quality of the model based on non-bonding interactions in proteins. Based on these criteria, the generated structures were found to be acceptable, leading to further analysis. The G17E and I86N mutant protein structures had 88.8% and 90.3% of their Ramachandran plot residues in the most favored region and ERRAT quality factor of 90.553 and 92.442, respectively, as shown in Fig. 7(a) and (b).
Fig. 7.
Predicted structure validation. Ramachandran and ERRAT plots were generated using SAVES server for the mutated RhoA proteins G17E and I86N. The number of residues in the most favored regions are 150 (88.8%) for G17E and 152 (90.5%) for I86N, with no residues found in the disallowed regions in both cases. The overall quality factor for the RhoA G17E and I86N in the ERRAT plot is 90.553 and 92.442, respectively.
3.8. Identification of RhoA interacting proteins
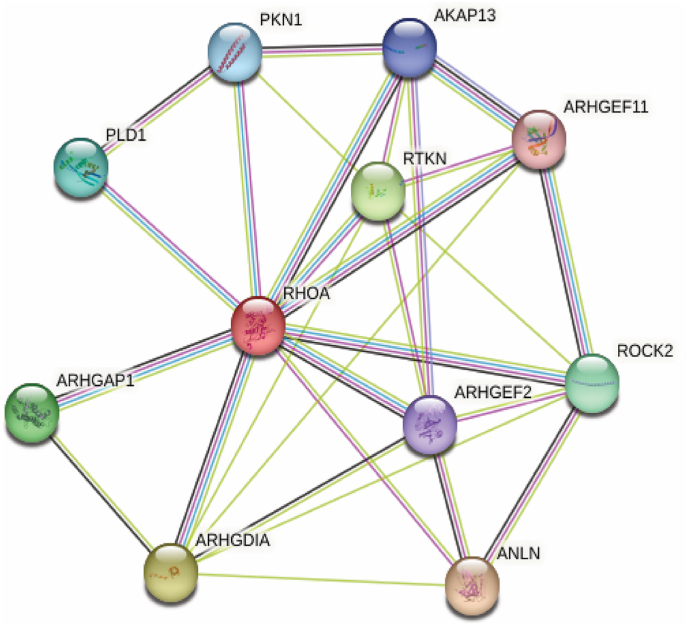
STRING discloses the interaction of RhoA with 10 distinct proteins, as shown in Fig. 8. These proteins are Phospholipase D1 (PLD1), Protein kinase N1 (PKN1), A-kinase anchor protein 13 (AKAP13), Rho guanine nucleotide exchange factor 2 (ARHGEF2), Rho guanine nucleotide exchange factor 11 (ARHGEF11), Rho-associated coiled-coil containing protein kinase 2 (ROCK2), Anillin actin-binding protein (ANLN), Rho GDP Dissociation Inhibitor Alpha (ARHGDIA), Rho GTPase-activating protein 1 (ARHGAP1), Receptor tyrosine kinases (RTKN). RhoA interacts with ROCK2 in the leukocyte trans-endothelial migration pathway, wnt signaling pathway, oxytocin signaling pathway, axon guidance, adherens junction, tight junction, and platelet activation pathway, according to the KEGG pathways database. The effect of RhoA mutations on PLD1 activation at the RhoA-PLD1 binding interface has been investigated further in this study.
Fig. 8.
The STRING web server generates proteins that interact with RhoA proteins in various biological pathways.
3.9. Molecular docking analysis
Protein-ligand-protein docking analysis demonstrated that the mutant RhoA-GDP complex binds to the PLD1 in a deviated way compared to the native RhoA-GDP complex. HADDOCK generated the top 10 clusters as a result. The top cluster is the most reliable according to HADDOCK, as its Z-score indicates the deviation from the mean in terms of score (with a more negative value being an indicator of better result). Top structures were taken to analyze the binding energy of the protein complex using the Prodigy web server shown in Supplementary Fig. 1A(a), B(a), and C(a). The binding affinity (ΔG) and dissociation constant (Kd) predicted values for wild-type RhoA and PLD1 are −8.3 kcal/mol and 7.5 × E−07 M respectively at 25.0 °C, which differ from those of other point mutations. The binding affinity (ΔG) and dissociation constant (Kd) predicted values for G17E and PLD1 are −9.0 kcal/mol and 2.3 × E−07 M respectively at 25.0 °C. For I86N the binding affinity (ΔG) and dissociation constant (Kd) predicted values are −7.4 kcal/mol and 3.9 × E−06 M, respectively at 25.0 °C. Visualization of the protein-protein interaction was done using the PyMol visualization package. Interacting residues of wild-type RhoA and PLD1, Asp67–Lys559 and Glu97-Phe577, form H-bond. Conversely, interacting residues of mutated RhoA and PLD1 are different from the wild-type shown in Supplementary Figure 1 A(b), B(b), and C(b). Interactions between wild-type RhoA-GDP-PLD1 complex residues may have changed due to the structural changes caused by point mutation. Molecular dynamics simulation analysis was performed using these docking complexes to understand more of this functional alteration.
3.10. Molecular dynamics simulation analysis
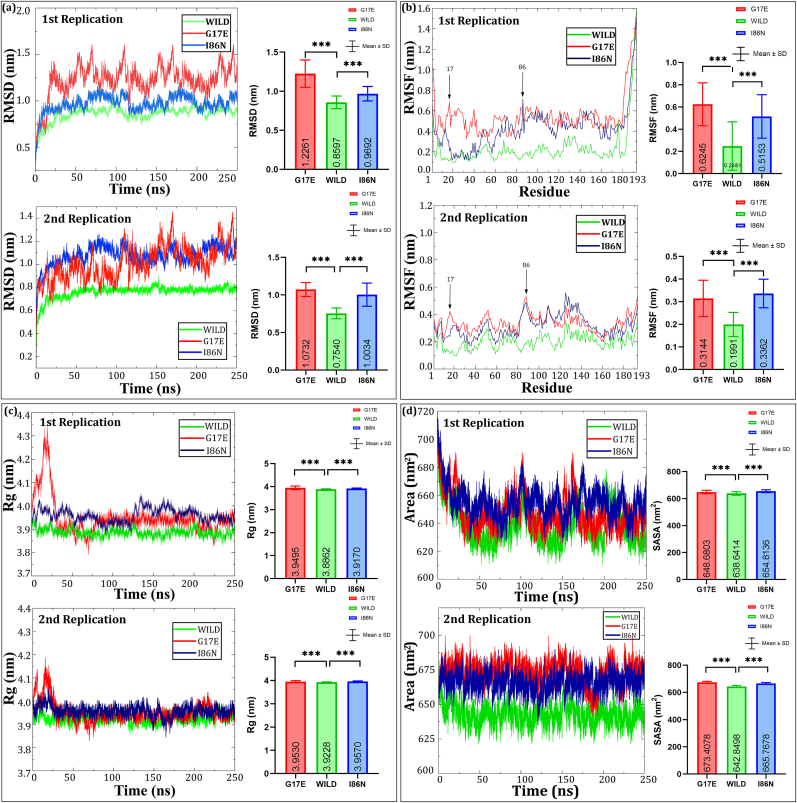
To investigate the deviation of wild-type and mutant proteins in physiological environments, 250 ns MD simulations were performed. Root Mean Square Deviation (RMSD) values between RhoA and PLD1 were calculated. RMSD values of the wild-type RhoA and mutants G17E and I86N differ significantly. The average RMSD values of mutated G17E (mean ± SD, 1.226 ± 0.1740 nm (Rep 1), 1.073 ± 0.09216 nm (Rep 2)) and I86N (mean ± SD, 0.9692 ± 0.09268 nm (Rep 1), 1.003 ± 0.1541 nm (Rep 2)) are greater than the wild-type RhoA (mean ± SD, 0.8597 ± 0.08096 nm (Rep 1), 0.7540 ± 0.07110 nm (Rep 2)), see Fig. 9(a). Throughout the simulation time, the value of RMSD for the mutant G17E fluctuated largely in ∼0.45 nm range in both replication which indicates the interaction instability between RhoA and PLD1.
Fig. 9.
Molecular dynamics analysis of wild-type and mutant (G17E and I86N) RhoA proteins. The Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of Gyration (Rg), and Solvent Accessible Surface Area (SASA) plots are shown for two replicates. (Top left) RMSD plots reveal stability differences among wild-type, G17E, and I86N mutants over 250 ns, with G17E showing higher deviation, indicating reduced stability. (Top right) RMSF plots highlight residue flexibility, with G17E and I86N showing more fluctuation, particularly around residues 60–90 and 180–193. (Bottom left) Rg plots display the compactness of the proteins, with G17E being more expanded. (Bottom right) SASA plots indicate solvent exposure, with the G17E mutant showing the largest surface area, further suggesting a looser structure. Statistical significance is indicated with asterisks.
In addition, RMSF (Root-Mean-Square Fluctuation) value analysis reveals that residues in the C terminal region fluctuate significantly differently between wild-type and mutant structures after 250ns MD simulation. The mean ± SD RMSF values, as compared to wild-type (0.2481 ± 0.2180 nm), were found significantly higher in G17E (0.6245 ± 0.1932 nm) and I86N (0.5153 ± 0.1961 nm) variants in replication 1. Whereas in replication 2, the values are 0.1991 ± 0.0532 nm, 0.3144 ± 0.0805 nm, and 0.3362 ± 0.0632 nm for the wild-type, G17E, and I86N, respectively, as shown in Fig. 9(b). The RMSF plot also indicates that residues 11–27, 78–100 and 112–130 are more flexible than the remaining residues in the wild-type protein. Entire mutated G17E and I86N proteins exhibit flexibility (Fig. 9(b)). Furthermore, in the mutant G17E, the fluctuation of residues (11–27) in the C-terminal flanking tail is greater, with values ranging from 0.4928 to 0.6753 nm, and in the mutant I86N, the fluctuation of residues (78–100) with values ranging from 0.4587 to 0.7173 nm, when compared to that of the native type residues (115–135) ranging from 0.1342 to 0.3851 nm.
The Rg (radius of gyration) study of wild-type and mutant structures (Fig. 9(c)) revealed that the G17E and I86N mutants had greater Mean ± SD of Rg values (3.950 ± 0.07629 nm) and (3.917 ± 0.02298 nm) during the simulation time scale as compared to the wild-type RhoA (3.886 ± 0.01696 nm) in replication 1. Similar type of Rg values trends is also showed in second replication. As a result, mutant G17E and I86N may have greater flexibility. Mutant G17E appeared to vary its Rg value following the start of the simulation to 30ns, while Mutant I86N appeared to deviate throughout the simulation, which could be the explanation for its partial protein folding.
According to Fig. 9(d), the analysis of solvent-accessible surface area (SASA) indicated that the mutants G17E and I86N had statistically significantly higher SASA values (648.7 ± 13.29 nm2 and 654.8 ± 10.84 nm2 respectively) than the wild-type (638.6 ± 13.35 nm2) in replication 1. Though the curves in replication 2 showed less fluctuation than replication 1, the deviation pattern from wild type remained similar. Since a higher SASA value indicates protein expansion, it is possible to conclude that the wild-type protein is more stable than the G17E and I86N mutant proteins. The reason for a greater change in the SASA value of G17E and I86N compared to the wild-type may be due to the effect of amino acid substitution on the size and other properties of the protein surface.
3.11. SNPs disrupts the interaction network between RhoA and PLD1
The binding free energy GB computed using MMGBSA and PB through the MMPBSA protocols has distinguished the affinities of WILD_PLD1, G17E_PLD1, and I86N_PLD1, as shown in Table 3. WILD_PLD1 exhibits a ΔGB of −70.41 kcal/mol, whereas G17E_PLD1 and I86N_PLD1 have a ΔGB of −46.12 kcal/mol and 58.66 kcal/mol, respectively. The energy difference between the complexes is more distinct in MMGBSA calculations, with ΔPB values of −81.53 kcal/mol for WILD_PLD1, -65.65 kcal/mol for G17E_PLD1, and similarly, −68.34 kcal/mol for I86N_PLD1. In all three complexes, the favorable contributions to binding stemmed from van der Waals (vdW) forces and the nonpolar segment of the solvation free energy, contrasting with the unfavorable total electrostatic contributions (EEL + EGB and EEL + EPB). Point mutations due to SNPs at residues G17E and I86N disrupted all binding energy components, with a greater loss in vdW interactions, as shown in Table 3.
Table 3.
The binding affinities derived from MMGBSA and MMPBSA for RhoA wild-type and mutant (G17E and I86N) PLD1 complexes were computed. EEL represents the molecular mechanics electrostatic energy, vdW refers to van der Waals energy, while the polar and non-polar terms are captured by EGB/EPB (electrostatic solvation) and ESURF/ENPOLAR (non-polar solvation) for MMGBSA and MMPBSA, respectively. These components highlight the energetic contributions that distinguish the binding interactions among the wild-type and mutated complexes.
| ΔEbinding (kcal/mol) | WILD | G17E | I86N |
|---|---|---|---|
| vdW | −237.22 | −227.81 | −231.25 |
| EEL | −146.01 | −135.82 | −140.05 |
| EGB | 287.86 | 223.04 | 246.74 |
| ΔGB | −70.41 | −46.12 | −58.66 |
| ESURF | −11.11 | −9.78 | −9.23 |
| EPB | 280 | 227 | 234 |
| ΔPB | −81.53 | −65.65 | −68.34 |
| ENPOLAR | −24.73 | −20.45 | −21.77 |
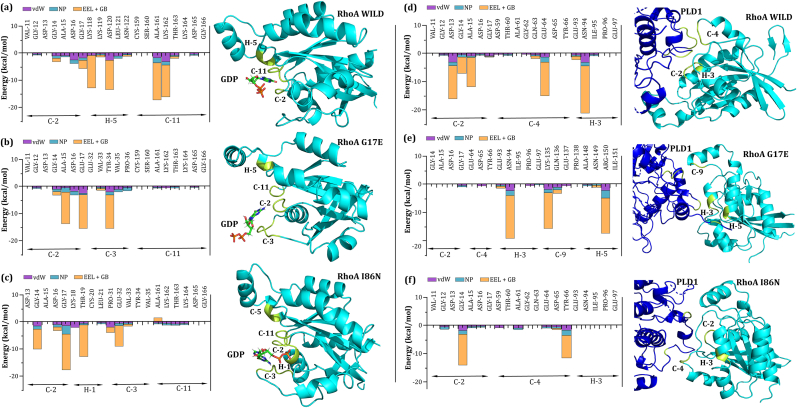
The interaction free energy (IE) of residues was determined by breaking down the binding free energy into van der Waals (vdW) forces, non-polar contributions to solvation free energy (NP), and the combined effects of electrostatic interactions (EEL) and electrostatic contributions to solvation free energy (GB). This per residue decomposition analysis was conducted to identify key residues involved in binding affinity, focusing on residues within a 4 Å proximity between RhoA and GDP, as well as RhoA and PLD1. In addition, different interaction patterns of RhoA complexed with GDP and PLD1 were observed as illustrated in Fig. 10. The residues of WILD RhoA from helix 5 (H-5), coil 2 (C-2), and coil 11 (C-11) maintained close contact with the GDP molecules, exhibiting interaction energies of 38.32 kcal/mol, 17.08 kcal/mol, and 43.82 kcal/mol, respectively. Whereas residues of C-2 and C-3 are in close proximity to GDP molecules for G17E mutated protein with interaction energies of 42.34 kcal/mol and 22 kcal/mol, respectively. For I86N C-2, C-3 and H-1 are in close contact with GDP with a binding energy of 33 kcal/mol, 19 kcal/mol and 16 kcal/mol, respectively. Overall, the involvement of the residues in RhoA-GDP interaction are different in Wild-type and mutated proteins as shown in Fig. 11.
Fig. 10.
Interaction free energy analysis of (a, b, c) Wild-type RhoA, G17E, and I86N mutant residues interacting with GDP, and (d, e, f) Wild-type RhoA, G17E, and I86N mutant residues interacting with PLD1 within a 4 Å binding vicinity. The left panels show the comparison of interaction free energy components (vdW, NP, EEL + GB) for RhoA-GDP complexes, and the right panels display the same for RhoA-PLD1 complexes after 250 ns of molecular dynamics simulations. The most significant residues contributing to the binding free energy in both complexes are highlighted in yellow within the structural representations.
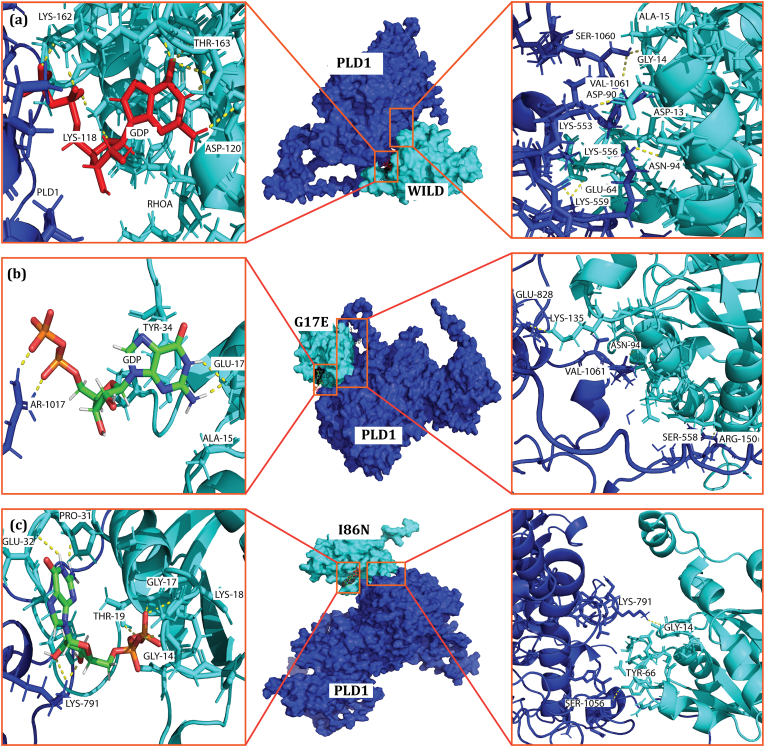
Fig. 11.
Structural representation of interactions between wild-type and mutant (G17E and I86N) RhoA proteins with GDP and PLD1. Panels (a), (b), and (c) show the detailed interactions of key residues within the binding sites of wild-type, G17E, and I86N RhoA with GDP (left) and PLD1 (right), respectively. Significant residues involved in the interactions are labeled, and hydrogen bonds are shown as yellow dashed lines. The surface representation highlights the interaction interface between RhoA (cyan) and PLD1 (blue), with zoomed-in views emphasizing critical residues contributing to the binding between these proteins.
Among the wild RhoA residues in close contact, C-2, C-4, and H-3 significantly contribute to binding affinity by interacting favorably with PLD1 via ASP-13, GLY-14, ALA-15, GLU-64, and ALA-94, with interaction energies of 12.55 kcal/mol (A15-S1060), 7.31 kcal/mol (G14-V1061), 21.47 kcal/mol (N94-K556), 14.7 (E64-K559) and 15.64 kcal/mol (D13-K553). The number of interacting residues altered and reduced in case of G17E and I86N point mutation as shown in Fig. 11(e and f), respectively. Apart from energetic profile, single point mutation G17E has dynamically changed the conformation of the RhoA as C-2 was observed to have moved further away from the PLD. Interacting residues and their binding patterns alteration are also more clearly represented in the last frame of the 250 ns simulation, see Fig. 11.
4. Discussion
Single base pair variation, among the various types of polymorphism in the genome contributes significantly to the development and progression of different types of diseases, including cancer. When this alteration of base pair occurs within a protein coding sequence, it may lead to disrupted function of that protein. Moreover, in silico analysis of deleterious SNPs from huge datasets has been increasingly important in recent years due to the occurrence of damaging SNPs in many oncogenes (George Priya Doss et al., 2013; Kamaraj et al., 2013). Therefore, it is important to identify the effects of deleterious SNPs of every crucial gene and their association with various diseases. As the RhoA protein is involved in the regulation of diverse cellular functions, its altered function can be found in various pathological processes including tumorigenesis, autoimmune disease, and atherosclerosis. Specifically, aberrant expression of this protein has been found in several cancers, such as lung, breast, liver, head, and neck cancer, also in colorectal, ovarian, and testicular carcinomas (Vega and Ridley, 2008; Wiedemann et al., 2006; Wang et al., 2002; Shimokawa et al., 2016; Sakata-Yanagimoto et al., 2014). Furthermore, according to the C-Bioportal database, the G17E mutant of RhoA has been previously associated with breast cancer. The structural and functional properties of RhoA protein can be affected by the changes in the amino acid sequence in the form of SNPs at the genomic level. In this regard, this study intended to find the most deleterious SNPs and their impact on the structure and function of RhoA protein through multiple computational tools (including SIFT, PolyPhen 2, SNAP 2, PROVEAN, CADD, PhD-SNP, Predict SNP2, SNPs&GO). These tools utilized their individual prediction scores to give an integrated result of 9 highly deleterious SNPs from 23,790 SNPs extracted from the dbSNP database. Although, many previous studies have used few tools to evaluate the SNPs, for the comprehensive analysis and filtration of the SNPs according to their harmful impact we used a wide range of tools (Saih et al., 2021; Mustafa et al., 2020).
Proteins have structural and functional units called domains that help them perform a specific function (Basu et al., 2009). The InterPro tool helped to determine whether the nsSNPs have occupied any of the functionally important domains of the protein. Previously, a computational study of high-risk SNPs in the human CHK2 gene responsible for hereditary breast cancer predicted the position of nsSNPs in different protein domains (Badgujar et al., 2019). Our study revealed the presence of 8 out of these 9 nsSNPs in the small GTP binding domain, showing the possibility of disruption of its binding to GTP/GDP. Thus, a deleterious mutation in the domain could significantly impair the function of protein in signal transduction.
Alterations in protein stability can lead to changes in their structure, which, in turn, may distort their function and interactions with other biological molecules (Shoichet et al., 1995; Marquet et al., 2022). I-Mutant predicts the consequence of the presence of these nsSNPs on the stability of the protein, providing valuable insights into the potential consequences of genetic mutation on protein structure and function. Previously, to screen and evaluate the deleterious SNPs in the APOE gene of Alzheimer's disease, a study used the I-Mutant tool (Masoodi et al., 2012). In the present study, the results of the assessment with I-Mutant demonstrated that all the 9 nsSNPs have been associated with decreased protein stability, which can lead to degradation or misfolding of the protein. This may, in turn, lead to loss of protein function in interaction with other molecules within the biological system.
Conserved regions of amino acids of a protein are paramount for the function of the protein, and any missense mutation in this region interrupts the function of the protein. Prior studies have shown the significance of evolutionary information in detecting mutations related to the disease (Zaidem et al., 2019). ConSurf helped us to assess the degree of conservation of the amino acids throughout the evolution. A recent study used ConSurf to identify the conserved SNPs of the NRP1 gene associated with SARS-COV-2 (Özkan Oktay et al., 2023). Our study showed that five nsSNPs (V9G, G17E E40K, A61T, F171L) are found in the highly conserved regions. Interestingly, E40K and A61T are located on the G2 and G3 motifs of the GTP-binding domain respectively. These two motifs are present in switch 1 and switch 2 and cause conformational change after binding to the GTP/GDP. Thus, mutations in these regions will prevent the switches from altering their conformation, which in turn hampers the proper functioning of this domain by inhibiting RhoA's ability to switch between its active and inactive states.
When mutations are introduced in the protein, charge-mass and other physiochemical properties of the amino acids could be modified that could lead to a change in the structure of the protein. Therefore, a detailed analysis of these changes is crucial for understanding the functional impact of genetic variation on protein activity. To this end, Project HOPE and Missense 3D were used to investigate the impact of these nsSNPs on the structure. A previous study characterized both coding and non-coding SNPs in the human Dectin-1 gene, identifying those with a high risk of pathogenicity associated with fungal infections. To predict the structural impact of these SNPs, the study utilized these two tools (Al-nakhle and Khateb, 2023). In our study, Y66H, A61T, G17E, I86N, and I151T mutations were observed to reduce hydrophobicity. Mutation at position 40, which replaces glycine with lysine, alters charge and may disrupt interactions with neighboring residues due to its surface location. This larger lysine variant may interfere with interactions with other biomolecules. Furthermore, the substitution of tyrosine with serine at position 42 could disrupt hydrogen bonding. G17E introduces a negative charge, potentially causing repulsion with nearby residues and affecting interactions with other biomolecules.
The amino acid sequences of the mutated proteins were transformed into 3D structure using AlphaFold3 (which is AI-based). AlphaFold3 was previously used to predict the structure of STK19 protein (Li et al., 2024). In this study, the Alphafold3 predicted 3D structures of mutant RhoA. The predicted models were highly accurate with pLDDT, PAE plot and pTM score proving the predicted structures to be close to the accurate structure. G17E and I86N mutant protein structures exhibited 88.8% and 90.3% of their residues in the most favored regions in Ramachandran plot, respectively, with overall quality factors of 90.553 and 92.442 in ERRAT plot.
Proteins that interact with RhoA were identified employing the widely used tool STRING, and their interrelationships were determined to gain insight into underlying protein-protein interactions. In a previous study, the STRING database was used to perform protein-protein interaction (PPI) network analysis of the selected SNP-containing gene with prognostic value in gastric cancer (Li et al., 2021). PPI information was obtained from the STRING database in this study. Additionally, to assess the functional impact of these interactions, ten key proteins involved in signaling pathways intersecting with RhoA were selected using the KEGG pathway database. As we screened for the proteins that interact with RhoA, we came across various proteins including PLD1 protein (shown in Fig. 8) which has function in a wide variety of cellular processes (Huang and Frohman, 2007).
PLD is a protein that has been investigated thoroughly over the years and has shown crucial role in the regulation of cell survival, intracellular membrane vesicle trafficking, and cell proliferation (Cockcroft and De Matteis, 2001). However, alteration of this protein and its product phosphatidic acid (PA) has been involved in a wide range of diseases such as inflammation, diabetes, phagocytosis, and also involved in metastatic spread of cancer. Among the two isoforms of PLD (PLD1 and PLD2), PLD1 is regulated by the RhoA. The ADP-ribosylation factor (ARF) and protein kinase C (PKC) and RhoA act synergistically to activate the PLD1 (Du et al., 2000; Hammond et al., 1997). Any alteration in any of the regulatory proteins may have implications for the defective activity of the PLD1. However, the level of impact because of the alteration of a specific regulatory protein such as RhoA and its degree of harmfulness in specific tissue is yet to be discovered.
In order to observe the functional anomalies of mutated RhoA, protein-ligand-protein docking analysis was performed, and the activation of PLD1 by a GTP-dependent RhoA family protein was investigated in this study. The PLD1 binds to the mutant RhoA-GDP complex structures slightly differently than the native RhoA-GDP structure, as demonstrated by protein-ligand-protein docking analysis. Studies have shown that mutant protein has reduced binding affinity when compared to the wild-type (Havranek and Islam, 2021). The binding affinity and dissociation constant predicted values for G17E and I86N with PLD1 are different at 25.0 °C than wild-type RhoA and PLD1. The docked protein-protein interaction was visualized using the PyMol visualization package, showing different interacting residues between wild-type and mutated RhoA-GDP-PLD1 complex structures (Supplementary Fig. 1), potentially due to structural changes caused by point mutation.
Previous research has shown that activated PLD1 is responsible for enhancing cell survival when confronted with apoptotic signals (Kiss et al., 1999). Another study has demonstrated the crucial role of PLD1 in facilitating the insulin-stimulated GLUT4 fusion with the plasma membrane, which, in turn, enhances glucose uptake (Huang et al., 2005). Furthermore, in pancreatic β-cells, PLD1 plays a role in mediating insulin secretion in response to glucose stimulation. These observations suggest that impaired PLD1 function could potentially be linked to Type II diabetes (Hughes et al., 2004). Interestingly, PLD1 is located within a recognized genomic region associated with Type II diabetes, although the specific disease-related gene within this locus has yet to be identified (Huang and Frohman, 2007). Phospholipase D1 inhibition is linked to upregulation of ICAT, which blocks colorectal cancer growth when hyperactivated by Wnt/β-Catenin and PI3K/Akt signaling (Kang et al., 2017). Inhibition of phospholipase D1 induces immunogenic cell death and potentiates cancer immunotherapy in colorectal cancer (Hwang et al., 2022).
A molecular dynamics analysis uncovered nsSNPs-induced fluctuations within the kinase, regulatory, and C-terminal regions. Furthermore, the observed RMSD, RMSF, and Rg values suggest instability in the mutant protein structures (Abid et al., 2023). Previously, one study using simulation has shown that mutation in KRAS4B protein disrupts its interaction with GTP, which ultimately hampers its activity in molecular switching (Chen et al., 2024). In this study, molecular dynamics simulations of 250 ns compared native and mutant protein structures using RMSD, RMSF, Rg, and SASA. Mutants G17E and I86N exhibited significantly higher RMSD values compared to wild-type RhoA, indicating structural deviation (Fig. 9(a)). RMSF analysis revealed notable differences in C-terminal fluctuation. Mutant G17E has higher Rg and SASA values, signifying increased flexibility and protein expansion. Additionally, the mutants displayed distinct GDP binding positions, suggesting reduced stability due to amino acid substitutions. As a result, there are altered interaction properties between mutant RhoA and PLD1, which may hamper the proper activation of PLD1. The binding free energy analysis reveals that the wild-type RhoA-PLD1 complex has the strongest affinity, while G17E and I86N mutants show significantly reduced binding energies. Point mutations in G17E and I86N disrupt van der Waals interactions and overall binding patterns, leading to altered conformations and fewer interacting residues. In this study, our computational method anticipates that the identified nsSNPs may be responsible for the improper activation of the PLD1 protein, potentially contributing to various diseases such as diabetes, cancer, and neurological disorders.
5. Conclusion
The human RhoA protein, a Rho family small GTPase, is involved in the regulation of cytoskeleton organization, cell adhesion, cell motility, and gene transcription. Very recently, its impaired function has linked to cancer, cardiovascular, and neurological illnesses. However, this study presents the first comprehensive analysis to map and predict the impact of the most deleterious point mutations which regulate the fate of RhoA, with the validation of molecular dynamics simulation. We reported G17E and I86N as damaging nsSNPs that affect RhoA protein structure and function. In comparison to the structure of the native protein, our molecular dynamics simulation approach revealed a change in the degree of structural deviation in mutant proteins. These mutants exhibit significant variations in the RhoA-GDP-PLD1 complex interaction, which could result in disruption of the activation of the PLD1 protein, thereby impeding cell signaling and membrane trafficking and contributing to the development of numerous diseases. Previously, no other study predicted that the G17E and I86N mutants are linked to any diseases. As a result, it is likely that predisposition to these unreported nsSNPs may cause disease by altering protein activation or efficiency. The outcomes of this study will help future genome association studies to uncover harmful SNPs associated with specific cancer patients and will be essential to develop personalized drugs.
CRediT authorship contribution statement
Mahbub Hasan: Conceptualization, Data curation, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. Md. Nayem Sarker: Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Tazkia Jabin: Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Saifuddin Sarker: Data curation, Methodology, Validation, Writing – review & editing. Shamim Ahmed: Writing – review & editing. Mohammad Abdullah-Al-Shoeb: Project administration, Resources, Supervision, Writing – review & editing. Tanvir Hossain: Conceptualization, Investigation, Project administration, Supervision, Validation, Visualization, Writing – review & editing.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2024.100159.
Contributor Information
Mohammad Abdullah-Al-Shoeb, Email: maashoeb-bmb@sust.edu.
Tanvir Hossain, Email: tanvir-bmb@sust.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abid F., Khan K., Badshah Y., Ashraf N.M., Shabbir M., Hamid A., Afsar T., Almajwal A., Razak S. Non-synonymous SNPs variants of PRKCG and its association with oncogenes predispose to hepatocellular carcinoma. Cancer Cell Int. 2023;23:1–24. doi: 10.1186/S12935-023-02965-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/J.SOFTX.2015.06.001. [DOI] [Google Scholar]
- Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A., Ronneberger O., Willmore L., Ballard A.J., Bambrick J., Bodenstein S.W., Evans D.A., Hung C.C., O'Neill M., Reiman D., Tunyasuvunakool K., Wu Z., Žemgulytė A., Arvaniti E., Beattie C., Bertolli O., Bridgland A., Cherepanov A., Congreve M., Cowen-Rivers A.I., Cowie A., Figurnov M., Fuchs F.B., Gladman H., Jain R., Khan Y.A., Low C.M.R., Perlin K., Potapenko A., Savy P., Singh S., Stecula A., Thillaisundaram A., Tong C., Yakneen S., Zhong E.D., Zielinski M., Žídek A., Bapst V., Kohli P., Jaderberg M., Hassabis D., Jumper J.M. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500. doi: 10.1038/s41586-024-07487-w. 2024 630:8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. 2010 7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-nakhle H.H., Khateb A.M. Comprehensive in silico characterization of the coding and non-coding SNPs in human dectin-1 gene with the potential of high-risk pathogenicity associated with fungal infections. Diagnostics. 2023;13:1785. doi: 10.3390/DIAGNOSTICS13101785/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexanian M., Przytycki P.F., Micheletti R., Padmanabhan A., Ye L., Travers J.G., Gonzalez-Teran B., Silva A.C., Duan Q., Ranade S.S., Felix F., Linares-Saldana R., Li L., Lee C.Y., Sadagopan N., Pelonero A., Huang Y., Andreoletti G., Jain R., McKinsey T.A., Rosenfeld M.G., Gifford C.A., Pollard K.S., Haldar S.M., Srivastava D. A transcriptional switch governs fibroblast activation in heart disease. Nature. 2021;595:438–443. doi: 10.1038/s41586-021-03674-1. 2021 595:7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/NAR/GKQ399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgujar N.V., Tarapara B.V., Shah F.D. Computational analysis of high-risk SNPs in human CHK2 gene responsible for hereditary breast cancer: a functional and structural impact. PLoS One. 2019;14 doi: 10.1371/JOURNAL.PONE.0220711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M.K., Poliakov E., Rogozin I.B. Domain mobility in proteins: functional and evolutionary implications. Briefings Bioinf. 2009;10:205–216. doi: 10.1093/BIB/BBN057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich A.B., Carlton V.E.H., Honigberg L.A., Schrodi S.J., Chokkalingam A.P., Alexander H.C., Ardlie K.G., Huang Q., Smith A.M., Spoerke J.M., Conn M.T., Chang M., Chang S.Y.P., Saiki R.K., Catanese J.J., Leong D.U., Garcia V.E., McAllister L.B., Jeffery D.A., Lee A.T., Batliwalla F., Remmers E., Criswell L.A., Seldin M.F., Kastner D.L., Amos C.I., Sninsky J.J., Gregersen P.K. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendl J., Musil M., Štourač J., Zendulka J., Damborský J., Brezovský J. PredictSNP2: a unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput. Biol. 2016;12 doi: 10.1371/JOURNAL.PCBI.1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/NAR/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling F.Z., Salazar C.M., Bell J.A., Huq T.S., Frohman M.A., Airola M.V. Crystal structure of human PLD1 provides insight into activation by PI(4,5)P2 and RhoA. Nat. Chem. Biol. 2020;16:400–407. doi: 10.1038/s41589-020-0499-8. 2020 16:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Brooks C.L., Mackerell A.D., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A.R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R.W., Post C.B., Pu J.Z., Schaefer M., Tidor B., Venable R.M., Woodcock H.L., Wu X., Yang W., York D.M., Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/JCC.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. Rho and rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/S0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R. I-Mutant 2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/NAR/GKI375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R., Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. doi: 10.1093/BIOINFORMATICS/BTL423. [DOI] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R., Fariselli P., Martelli P.L., Altman R.B., Casadio R. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013;14(Suppl. 3):1–7. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., Patil N., Lane C.R., Lim E.P., Kalyanaraman N., Nemesh J., Ziaugra L., Friedland L., Rolfe A., Warrington J., Lipshutz R., Daley G.Q., Lander E.S. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22:231–238. doi: 10.1038/10290. 1999 22:3. [DOI] [PubMed] [Google Scholar]
- Chasman D., Adams R.M. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol. 2001;307:683–706. doi: 10.1006/JMBI.2001.4510. [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang S., Wang W., Pang L., Zhang Q., Liu X. Mutation-induced impacts on the switch transformations of the GDP-and GTP-bound K-Ras: insights from multiple replica Gaussian accelerated molecular dynamics and free energy analysis. J. Chem. Inf. Model. 2021;61:1954–1969. doi: 10.1021/ACS.JCIM.0C01470. [DOI] [PubMed] [Google Scholar]
- Chen J., Wang J., Yang W., Zhao L., Hu G. Conformations of KRAS4B affected by its partner binding and G12C mutation: insights from GaMD trajectory-image transformation-based deep learning. J. Chem. Inf. Model. 2024;64:6880–6898. doi: 10.1021/ACS.JCIM.4C01174. [DOI] [PubMed] [Google Scholar]
- Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/BIOINFORMATICS/BTV195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S., De Matteis M.A. Inositol lipids as spatial regulators of membrane traffic. J. Membr. Biol. 2001;180:187–194. doi: 10.1007/S002320010069. [DOI] [PubMed] [Google Scholar]
- Collins F.S., Brooks L.D., Chakravarti A. A DNA polymorphism Discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/GR.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Croitoru A., Park S.J., Kumar A., Lee J., Im W., Mackerell A.D., Aleksandrov A. Additive CHARMM36 force field for nonstandard amino acids. J. Chem. Theor. Comput. 2021;17:3554–3570. doi: 10.1021/ACS.JCTC.1C00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson R.J., Munroe P.B., Caulfield M.J., Saqi M.A.S. Predicting deleterious nsSNPs: an analysis of sequence and structural attributes. BMC Bioinf. 2006;7:1–9. doi: 10.1186/1471-2105-7-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G., Altshuller Y.M., Kim Y., Han Jung Min, Ho Ryu Sung, Morris A.J., Frohman M.A. Dual requirement for rho and protein kinase C in direct activation of phospholipase D1 through G protein-coupled receptor signaling. Mol. Biol. Cell. 2000;11:4359–4368. doi: 10.1091/MBC.11.12.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1998;103:8577. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- Forsberg L., De Faire U., Marklund S.L., Andersson P.M., Stegmayr B., Morgenstern R. Phenotype determination of a common pro-leu polymorphism in human glutathione peroxidase 1. Blood Cells Mol. Dis. 2000;26:423–426. doi: 10.1006/BCMD.2000.0325. [DOI] [PubMed] [Google Scholar]
- Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George Priya Doss C., Nagasundaram N., Chakraborty C., Chen L., Zhu H. Extrapolating the effect of deleterious nsSNPs in the binding adaptability of flavopiridol with CDK7 protein: a molecular dynamics approach. Hum. Genom. 2013;7 doi: 10.1186/1479-7364-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/PRO.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Jenco J.M., Nakashima S., Cadwallader K., Gu Q.M., Cook S., Nozawa Y., Prestwich G.D., Frohman M.A., Morris A.J. Characterization of two alternately spliced forms of phospholipase D1: activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and RHO family monomeric GTP-binding proteins and protein kinase C-α. J. Biol. Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- Hanna S., El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cell. Signal. 2013;25:1955–1961. doi: 10.1016/J.CELLSIG.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeerschd T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 2012;4:1–17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrach M.F., Drossel B. Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity. J. Chem. Phys. 2014;140 doi: 10.1063/1.4872239. [DOI] [PubMed] [Google Scholar]
- Havranek B., Islam S.M. Prediction and evaluation of deleterious and disease causing non-synonymous SNPs (nsSNPs) in human NF2 gene responsible for neurofibromatosis type 2 (NF2) J. Biomol. Struct. Dyn. 2021;39:7044–7055. doi: 10.1080/07391102.2020.1805018. [DOI] [PubMed] [Google Scholar]
- Hecht M., Bromberg Y., Rost B. Better prediction of functional effects for sequence variants. BMC Genom. 2015;16:1–12. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honorato R.V., Koukos P.I., Jiménez-García B., Tsaregorodtsev A., Verlato M., Giachetti A., Rosato A., Bonvin A.M.J.J. Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front. Mol. Biosci. 2021;8:708. doi: 10.3389/FMOLB.2021.729513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Frohman M.A. The potential for phospholipase D as a new therapeutic target. Expert Opin. Ther. Targets. 2007;11:707–716. doi: 10.1517/14728222.11.5.707. [DOI] [PubMed] [Google Scholar]
- Huang P., Altshuller Y.M., Hou J.C., Pessin J.E., Frohman M.A. Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1. Mol. Biol. Cell. 2005;16:2614–2623. doi: 10.1091/MBC.E04-12-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes W.E., Elgundi Z., Huang P., Frohman M.A., Biden T.J. Phospholipase D1 regulates secretagogue-stimulated insulin release in pancreatic beta-cells. J. Biol. Chem. 2004;279:27534–27541. doi: 10.1074/JBC.M403012200. [DOI] [PubMed] [Google Scholar]
- Hunter S., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R.D., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Laugraud A., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., Mcanulla C., McDowall J., Mistry J., Mitchell A., Mulder N., Natale D., Orengo C., Quinn A.F., Selengut J.D., Sigrist C.J.A., Thimma M., Thomas P.D., Valentin F., Wilson D., Wu C.H., Yeats C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/NAR/GKN785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.C., Song D., Lee H., Oh C., Lim S.H., Bae H.J., Kim N.D., Han G., Min D.S. Inhibition of phospholipase D1 induces immunogenic cell death and potentiates cancer immunotherapy in colorectal cancer. Exp. Mol. Med. 2022;54:1563–1576. doi: 10.1038/s12276-022-00853-6. 2022 54:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. vol. 21. 2005. pp. 247–269. (RHO GTPASES: Biochemistry and Biology). [DOI] [PubMed] [Google Scholar]
- Kakiuchi M., Nishizawa T., Ueda H., Gotoh K., Tanaka A., Hayashi A., Yamamoto S., Tatsuno K., Katoh H., Watanabe Y., Ichimura T., Ushiku T., Funahashi S., Tateishi K., Wada I., Shimizu N., Nomura S., Koike K., Seto Y., Fukayama M., Aburatani H., Ishikawa S. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat. Genet. 2014;46:583–587. doi: 10.1038/ng.2984. 2014 46:6. [DOI] [PubMed] [Google Scholar]
- Kamaraj B., Rajendran V., Sethumadhavan R., Purohit R. In-silico screening of cancer associated mutation on PLK1 protein and its structural consequences. J. Mol. Model. 2013;19:5587–5599. doi: 10.1007/S00894-013-2044-0. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/NAR/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/NAR/GKW1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.W., Lee B.H., Suh Y.A., Choi Y.S., Jang S.J., Kim Y.M., Choi K.Y., Min D.S. Phospholipase D1 inhibition linked to upregulation of ICAT blocks colorectal cancer growth hyperactivated by Wnt/β-catenin and PI3K/Akt signaling. Clin. Cancer Res. 2017;23:7340–7350. doi: 10.1158/1078-0432.CCR-17-0749/14725. [DOI] [PubMed] [Google Scholar]
- Khanna T., Hanna G., Sternberg M.J.E., David A. Missense3D-DB web catalogue: an atom-based analysis and repository of 4M human protein-coding genetic variants. Hum. Genet. 2021;140:805–812. doi: 10.1007/S00439-020-02246-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., Zaslavsky L., Zhang J., Bolton E.E. PubChem 2023 update. Nucleic Acids Res. 2023;51:D1373–D1380. doi: 10.1093/NAR/GKAC956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss Z., Petrovics G., Olàh Z., Lehel C., Anderson W.B. Overexpression of protein kinase C-epsilon and its regulatory domains in fibroblasts inhibits phorbol ester-induced phospholipase D activity. Arch. Biochem. Biophys. 1999;363:121–128. doi: 10.1006/ABBI.1998.1066. [DOI] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. 2009 4:7. [DOI] [PubMed] [Google Scholar]
- Lander E.S. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/SCIENCE.274.5287.536. (1979) [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Thornton J.M. PROCHECK: validation of protein-structure coordinates. 2012. 684–687. [DOI]
- Leonard D., Hart M.J., V Platkot J., Eva A., Henzelll W., Evansll T., Cerione R.A. The identification and characterization of a GDP-dissociation inhibitor (GDI) for the CDC42Hs protein. J. Biol. Chem. 1992;267:22860–22868. doi: 10.1016/S0021-9258(18)50026-0. [DOI] [PubMed] [Google Scholar]
- Levin Y. Electrostatic correlations: from plasma to biology. Rep. Prog. Phys. 2002;65:1577. doi: 10.1088/0034-4885/65/11/201. [DOI] [Google Scholar]
- Li H., Guo J., Cheng G., Wei Y., Liu S., Qi Y., Wang G., Xiao R., Qi W., Qiu W. Identification and validation of SNP-containing genes with prognostic value in gastric cancer via integrated bioinformatics analysis. Front. Oncol. 2021;11 doi: 10.3389/FONC.2021.564296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ma X., Wang X., Hu X., Fang S., Jin G., Liu K., Dong Z. Mutations found in cancer patients compromise DNA binding of the winged helix protein STK19. Sci. Rep. 2024;14:1–10. doi: 10.1038/s41598-024-64840-9. 2024 14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet C., Heinzinger M., Olenyi T., Dallago C., Erckert K., Bernhofer M., Nechaev D., Rost B. Embeddings from protein language models predict conservation and variant effects. Hum. Genet. 2022;141:1629–1647. doi: 10.1007/S00439-021-02411-Y/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoodi T.A., Al Shammari S.A., Al-Muammar M.N., Alhamdan A.A. Screening and evaluation of deleterious SNPs in APOE gene of Alzheimer's disease. Neurol Res Int. 2012;2012 doi: 10.1155/2012/480609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams T.G., Barini E., Pohjolan-Pirhonen R., Brooks S.P., Singh F., Burel S., Balk K., Kumar A., Montava-Garriga L., Prescott A.R., Hassoun S.M., Mouton-Liger F., Ball G., Hills R., Knebel A., Ulusoy A., Di Monte D.A., Tamjar J., Antico O., Fears K., Smith L., Brambilla R., Palin E., Valori M., Eerola-Rautio J., Tienari P., Corti O., Dunnett S.B., Ganley I.G., Suomalainen A., Muqit M.M.K. Phosphorylation of Parkin at serine 65 is essential for its activation in vivo. Royal Soc. Open Biol. 2018;8 doi: 10.1098/RSOB.180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 2012;8:3314–3321. doi: 10.1021/CT300418H. [DOI] [PubMed] [Google Scholar]
- Morris A.L., MacArthur M.W., Hutchinson E.G., Thornton J.M. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/PROT.340120407. [DOI] [PubMed] [Google Scholar]
- Mustafa H.A., Albkrye A.M.S., AbdAlla B.M., Khair M.A.M., Abdelwahid N., Elnasri H.A. Computational determination of human PPARG gene: SNPs and prediction of their effect on protein functions of diabetic patients. Clin. Transl. Med. 2020;9:1–10. doi: 10.1186/S40169-020-0258-1. 2020 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/NAR/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan Oktay E., Kaman T., Karasakal Ö.F., Enisoğlu Atalay V. In silico prediction and molecular docking of SNPs in NRP1 gene associated with SARS-COV-2. Biochem. Genet. 2023:1–20. doi: 10.1007/S10528-023-10409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K.L., Chen M. vol. 4. 2013. pp. 141–147. (Dynamic Functions of RhoA in Tumor Cell Migration and Invasion). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran P.D., Blackstock F., Shields N., Holland A., Iles R., Kingsley M., Bernhardt J., Lannin N., Morris M.E., Taylor N.F. vol. 28. 2014. pp. 1159–1171. (Motivational Interviewing to Increase Physical Activity in People with Chronic Health Conditions: a Systematic Review and Meta-Analysis). [DOI] [PubMed] [Google Scholar]
- Palomero T., Couronné L., Khiabanian H., Kim M.Y., Ambesi-Impiombato A., Perez-Garcia A., Carpenter Z., Abate F., Allegretta M., Haydu J.E., Jiang X., Lossos I.S., Nicolas C., Balbin M., Bastard C., Bhagat G., Piris M.A., Campo E., Bernard O.A., Rabadan R., Ferrando A.A. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014;46:166–170. doi: 10.1038/ng.2873. 2014 46:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perica T., Mathy C.J.P., Xu J., Jang G., Zhang Y., Kaake R., Ollikainen N., Braberg H., Swaney D.L., Lambright D.G., Kelly M.J.S., Krogan N.J., Kortemme T. Systems-level effects of allosteric perturbations to a model molecular switch. Nature. 2021;599:152–157. doi: 10.1038/s41586-021-03982-6. 2021 599:7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/PRO.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli G., Del Rio A., Degliesposti G., Sgobba M. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J. Comput. Chem. 2010;31:797–810. doi: 10.1002/JCC.21372. [DOI] [PubMed] [Google Scholar]
- Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/NAR/GKY1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saih A., Baba H., Bouqdayr M., Ghazal H., Hamdi S., Kettani A., Wakrim L. In silico analysis of high-risk missense variants in human ACE2 gene and susceptibility to SARS-CoV-2 infection. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/6685840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata-Yanagimoto M., Enami T., Yoshida K., Shiraishi Y., Ishii R., Miyake Y., Muto H., Tsuyama N., Sato-Otsubo A., Okuno Y., Sakata S., Kamada Y., Nakamoto-Matsubara R., Tran N.B., Izutsu K., Sato Y., Ohta Y., Furuta J., Shimizu S., Komeno T., Sato Y., Ito T., Noguchi M., Noguchi E., Sanada M., Chiba K., Tanaka H., Suzukawa K., Nanmoku T., Hasegawa Y., Nureki O., Miyano S., Nakamura N., Takeuchi K., Ogawa S., Chiba S. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014;46:171–175. doi: 10.1038/ng.2872. 2013 46:2. [DOI] [PubMed] [Google Scholar]
- Shimokawa H., Sunamura S., Satoh K. RhoA/Rho-kinase in the cardiovascular system. Circ. Res. 2016;118:352–366. doi: 10.1161/CIRCRESAHA.115.306532. [DOI] [PubMed] [Google Scholar]
- Shoichet B.K., Baase W.A., Kuroki R., Matthews B.W. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. USA. 1995;92:452–456. doi: 10.1073/PNAS.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/NAR/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Duan L., Chen F., Liu H., Wang Z., Pan P., Zhu F., Zhang J.Z.H., Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys. Chem. Chem. Phys. 2018;20:14450–14460. doi: 10.1039/C7CP07623A. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., Jensen L.J., von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/NAR/GKAA1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., Bork P., Jensen L.J., von Mering C. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/NAR/GKAC1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y., Sasaki T., Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/PHYSREV.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Thumkeo D., Watanabe S., Narumiya S. Physiological roles of Rho and Rho effectors in mammals. Eur. J. Cell Biol. 2013;92:303–315. doi: 10.1016/J.EJCB.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/JCC.20291. [DOI] [PubMed] [Google Scholar]
- Van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., Van Dijk M., De Vries S.J., Bonvin A.M.J.J. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/J.JMB.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Vega F.M., Ridley A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/J.FEBSLET.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Venselaar H., te Beek T.A.H., Kuipers R.K.P., Hekkelman M.L., Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinf. 2010;11:1–10. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Eto M., Steers W.D., Somlyo A.P., Somlyo A.V. RhoA-mediated Ca2+ sensitization in erectile function. J. Biol. Chem. 2002;277:30614–30621. doi: 10.1074/jbc.M204262200. [DOI] [PubMed] [Google Scholar]
- Wang K., Yuen S.T., Xu J., Lee S.P., Yan H.H.N., Shi S.T., Siu H.C., Deng S., Chu K.M., Law S., Chan K.H., Chan A.S.Y., Tsui W.Y., Ho S.L., Chan A.K.W., Man J.L.K., Foglizzo V., Ng M.K., Chan A.S., Ching Y.P., Cheng G.H.W., Xie T., Fernandez J., Li V.S.W., Clevers H., Rejto P.A., Mao M., Leung S.Y. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014;46:573–582. doi: 10.1038/ng.2983. 2014 46:6. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/NAR/GKY427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler A.P., Ridley A.J. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 2004;301:43–49. doi: 10.1016/J.YEXCR.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Wiedemann A., Patel J.C., Lim J., Tsun A., Van Kooyk Y., Caron E. Two distinct cytoplasmic regions of the β2 integrin chain regulate RhoA function during phagocytosis. JCB (J. Cell Biol.) 2006;172:1069–1079. doi: 10.1083/JCB.200508075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A., Vetter I.R. vol. 80. 2011. pp. 943–971. (Structure-Function Relationships of the G Domain, a Canonical Switch Motif). [DOI] [PubMed] [Google Scholar]
- Wittinghofer A., Waldmann H. Ras-A molecular switch involved in tumor formation. Angew Chem. Int. Ed. Engl. 2000;39:4192–4214. doi: 10.1002/1521-3773(20001201)39:23<4192::aid-anie4192>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Xie Z., Ho W.T., Spellman R., Cai S., Exton J.H. Mechanisms of regulation of phospholipase D1 and D2 by the heterotrimeric G proteins G13 and Gq. J. Biol. Chem. 2002;277:11979–11986. doi: 10.1074/jbc.M109751200. [DOI] [PubMed] [Google Scholar]
- Yue P., Moult J. Identification and analysis of deleterious human SNPs. J. Mol. Biol. 2006;356:1263–1274. doi: 10.1016/J.JMB.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Zaidem M.L., Groen S.C., Purugganan M.D. Evolutionary and ecological functional genomics, from lab to the wild. Plant J. 2019;97:40–55. doi: 10.1111/tpj.14167. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2303–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobayer N., Aowlad Hossain A.B.M. In silico characterization and homology modeling of histamine receptors. J. Biol. Sci. 2018;18:178–191. doi: 10.3923/JBS.2018.178.191. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.