Abstract
In the current study, we introduced a unique method for identifying and segmenting oral squamous cell carcinoma (OSCC) nuclei, concentrating on those predicted to have significant CAF-1/p60 protein expression. Our suggested model uses the StarDist architecture, a deep-learning framework designed for biomedical image segmentation tasks. The training dataset comprises painstakingly annotated masks created from tissue sections previously stained with hematoxylin and eosin (H&E) and then restained with immunohistochemistry (IHC) for p60 protein. Our algorithm uses subtle morphological and colorimetric H&E cellular characteristics to predict CAF-1/p60 IHC expression in OSCC nuclei. The StarDist-based architecture performs exceptionally well in localizing and segmenting H&E nuclei, previously identified by IHC-based ground truth. In summary, our innovative approach harnesses deep learning and multimodal information to advance the automated analysis of OSCC nuclei exhibiting specific protein expression patterns. This methodology holds promise for expediting accurate pathological assessment and gaining deeper insights into the role of CAF-1/p60 protein within the context of oral cancer progression.
Keywords: OSCC, CAF/p60, Deep learning, Stardist, Segmentation, Detection, Classification, QuPath, H&E, IHC
Introduction
CAF-1/p60 is a chromatin assembly factor-1 (CAF-1) complex subunit involved in DNA replication and repair.1,2 CAF-1/p60 expression levels have been reported to be of prognostic value in oral squamous cell carcinoma (OSCC), as well as in other human solid tumors.1,3, 4, 5, 6, 7, 8 High levels of CAF-1/p60 in OSCC tissues are associated with poor survival, tumor recurrence, and lymph node metastasis.4,7 Moreover, CAF-1/p60 can be detected in the peripheral blood of OSCC patients, suggesting a role for CAF-1/p60 as a potential soluble biomarker of OSCC. The detection and quantification of CAF-1/p60 in serum may provide useful information for diagnosing, staging, and monitoring OSCC patients.9 The expression and localization of CAF-1/p60 in OSCCs and normal oral mucosa tissue samples can be examined by immunohistochemistry (IHC).10 Deep learning models have been increasingly used to predict IHC staining from hematoxylin and eosin (H&E)-stained tissue slides.11,12 IHC staining is a widely used technique to assess the expression of various biomarkers in tissue samples, such as estrogen receptors in breast cancer.13 Despite being relatively affordable, the technique is still an extra cost/step that could be avoided. Furthermore, IHC staining is often subjective and prone to inter- and intra-observer variability.14,15 Therefore, developing computational methods to infer IHC staining from H&E staining can significantly benefit clinical diagnosis and research. One of the promising approaches to achieving this goal is to use deep learning models that can learn complex nonlinear mappings between H&E and IHC images. Deep learning models can leverage large-scale datasets of co-registered H&E and IHC slides to train end-to-end image translation networks to generate realistic and accurate IHC images from H&E images.11 For example, de Haan et al.16 proposed a supervised learning-based framework to transform H&E-stained tissues into special stains, such as Masson's trichrome, periodic acid-Schiff, and Jones silver stain, using kidney needle core biopsy tissue sections. They showed that their method improved the diagnosis of several non-neoplastic kidney diseases and achieved comparable quality to histochemically stained slides. Ghahremani et al. presented a multitask deep learning framework, DeepLIIF, to infer multiplex immunofluorescence (mpIF) staining for IHC image quantification from H&E-stained slides.17
Machine learning approaches have been proposed to predict positivity to proliferation markers, such as Ki-67.18 Martino et al. showed a machine learning-based analysis to predict the IHC labeling index for the cell proliferation marker Ki67/MIB1 on cancer tissues using morphometrical features extracted from H&E-stained tumor tissue samples. The authors provided a fast and quantitative method of identifying the proliferating compartment of the tumor on H&E slides.18 Martino et al. also proposed a deep learning approach for the same task based on a generative network.11 These studies demonstrate the potential of machine learning-based models to predict IHC staining from H&E staining, which can facilitate faster, cheaper, and more objective analysis of tissue samples. However, there are still several challenges and limitations that need to be addressed, such as the scarcity of co-registered H&E and IHC datasets,19 the variability of staining protocols and quality across different laboratories,20 the generalization of models to unseen tissue types and markers,21 and the validation of model performance by expert pathologists.22
We herein present an approach aimed at predicting CAF-1/p60-positive OSCC nuclei based on subtle morphological and colorimetric H&E cellular characteristics by a deep learning model trained on H&E-masks pairs with segmentation masks obtained from IHC specific to detect CAF-1/p60 expression on tissue slides.
Our model is based on StarDist23,24 and was constructed by making whole slide image (WSI) pairs first stained with H&E and restained with anti-CAF-1/P60 IHC. Future research directions include expanding the scope of this model approach to more tissue types and markers, improving the quality and availability of co-registered H&E and IHC datasets, and validating the clinical utility of deep learning models by expert pathologists.
Methods
Tissue slide preparation
We built our dataset with nine WSIs of OSCCs and a tissue microarray (TMA) with 55 OSCC cores. All the tissue samples came from the archives of the Pathology Unit of “Federico II” University of Naples. TMA was obtained as previously described.10,18 We finally used tumor tissue samples from 64 patients. Tissue WSI and TMA slides were stained with standard H&E staining and then scanned using a Leica Aperio AT2 scanner with a 40× magnifier (pixel size: 0.25 μm). After slide scanning, we use a hot plate and forceps to remove a coverslip from a slide. The slides were then soaked two times in a xylene bath to remove any remaining adhesive and rehydrated to decrease ethanol concentrations and remove the eosin stain. After destaining, the slides were rinsed in tap water, allowed to dry in the fume hood, and immunostained with the antibody anti-CAF-1/p60. IHC staining was performed on a Ventana Benchmark Ultra (Ventana Medical Systems Inc., Tucson, AZ, USA) using the rabbit monoclonal antibody anti-CAF-1/p60 (clone ab8133 AbCam, Cambridge, UK) as described in Morra et al.2 Briefly, sections were heated at 55 °C for 60 min, deparaffinized, and processed for antigen retrieval by microwaving in 1% sodium citrate buffer, pH 6.0. Non-specific binding was blocked with 1.5% non-immune mouse serum (1:20; Dakopatts, Hamburg, Germany), and endogenous peroxidase and alkaline phosphatase activities were quenched with dual endogenous enzyme block (0.5% H2O2 in methanol and detergent). Sections were then incubated with the primary antibody followed by the secondary antibody, and the reaction was detected using 3,3′-diaminobenzidine (Vector Laboratories, Burlingame, CA, USA); nuclei were counterstained with Mayer's hematoxylin. The new IHC-stained slides were then digitized using a Leica Aperio AT2 scanner with a 40× magnifier (pixel size: 0.25 μm). The positivity for CAF-1/p60 was evaluated on immunostained samples on the basis of the presence of DAB in a binary mode (positive/negative) for each tumor cell considered. The nine WSIs were used as a training/validation set, and the 55 TMA cores were used as a test set. The agreement between primary and secondary IHC in the quality control process was performed by Pearson correlation test in SPSS software (IBM Corp. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY, USA).
Dataset preparation
The model described was generated using tiles extracted from full-resolution images exported from the previously prepared and digitized WSI. We superimposed the anti-p60 IHCs images on the corresponding H&E-stained images by SIFT registration in ImageJ (v. 1.54f).25 The aligned pairs were re-imported in QuPath26 project and used to generate the dataset. Annotations were generated using QuPath cell detection algorithm.26 Our project was carried out in the QuPath (v. 0.4.3) version. We subsequently classified the detected objects based on DAB intensity. Following the visual check by two expert pathologists (SS and FM), the whole set of objects was exported by IHC in .geojson format and reimported on H&E images. We finally used a tiling script to export 256 × 256 px H&E tiles and the relative label image. Below is the groovy script used for the cell detection, the classification based on the intensity of staining with DAB, and the subsequent cleaning of the detections to obtain a mask that only highlights positive cells. We compiled groovy scripts modifying the ones available on the QuPath documentation web page [“https://qupath.readthedocs.io/en/stable/“last access: 10/09/2023) The full script is available in Supplementary 2 subsection of Supplementary material [Supplementary Code 1].
The Groovy script we provided is a sequence of commands for the QuPath software used for digital pathology image analysis.
‘runPlugin(‘qupath.imagej.detect.cells. WatershedCellDetection’, …)‘: This command runs the Watershed Cell Detection algorithm on an image. The parameters inside ‘…‘control the algorithm's behavior. For example, ‘“sigma”:6.0′ sets the sigma parameter of the Gaussian filter used in the algorithm, and ‘“minArea”:100.0′ and ‘“maxArea”:1000.0′ set the minimum and maximum allowed cell areas.
‘setDetectionIntensityClassifications(“Nucleus: DAB OD mean”, 3)‘: This command classifies detected cells into three intensity classes based on the mean optical density (OD) of DAB staining in the nucleus.
‘selectObjectsByClassification(“Negative”)‘: This command selects all cells that were classified as “Negative” in the previous step.
‘clearSelectedObjects(true)‘: This command deletes all currently selected objects.
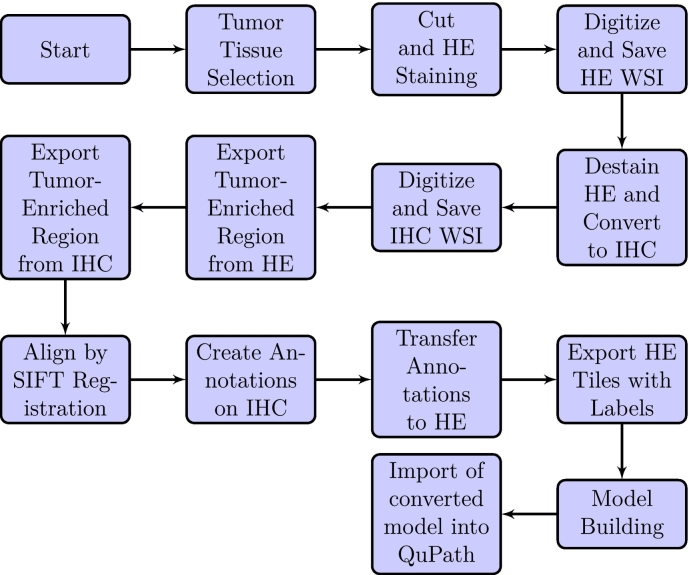
‘clearSelectedObjects()‘: This command clears the selection, leaving no objects selected. Fig. 1 illustrates the dataset preparation flowchart.
Fig. 1.
Dataset preparation flowchart. To generate the masks used to train the StarDist-based model, we used IHC as the ground truth. This was possible due to the precise coupling of IHC and H&E staining, aided by the fact that each sample was the same section stained both ways. We generated the Train/Validation sets with a np.random.RandomState function, randomly assigning images to the Train and the Validation set, respectively. We also managed to convert the model into a portable format and import it into QuPath.
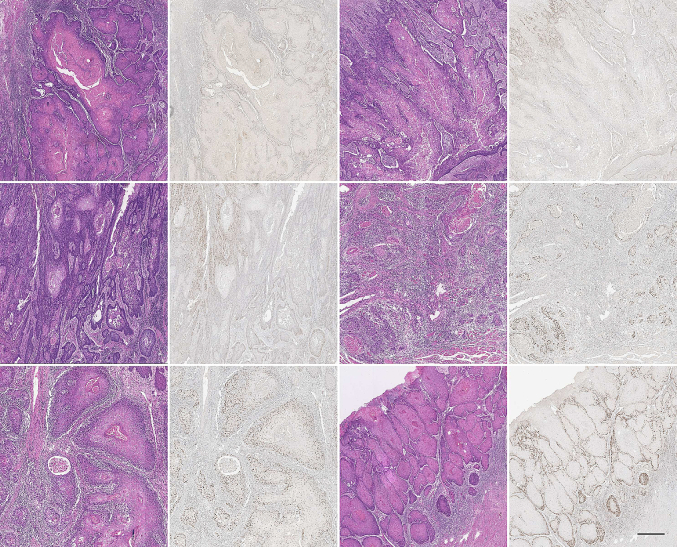
From the paired WSIs (H&E and IHC), we exported tumor-rich areas that we aligned using SIFT registration (Fig. 2). The aligned IHCs were used as ground truth to construct the segmentation masks on H&Es (Fig. 3).
Fig. 2.
Some representative examples showing registered image pairs, H&E, and IHC (Scale bar: 100 μm).
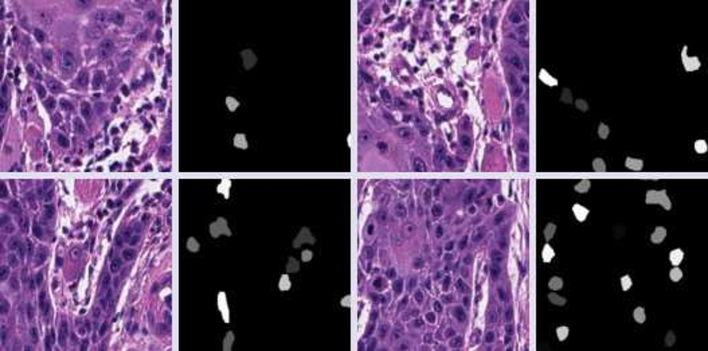
Fig. 3.
An example of tiles and corresponding segmentation masks as exported from QuPath after annotation alignment. The mask represents “CAF-1/p60-positive” nuclei as identified on IHC.
Model training
The StarDist-based model was generated by modifying the scripts distributed on the official page of StarDist [“https://github.com/stardist/stardist“, last access: 04/09/2023)]. StarDist uses a light-weight neural network based on U-Net.27 The StarDist-based model predicts a star-convex polygon for every pixel; for each pixel with index i, j, a regression of the distances has been done to the boundary of the object to which the pixel belongs, along a set of n predefined radial directions with equidistant angles. The model also separately predicts for every pixel whether it is part of an object so that it only considers polygon proposals from pixels with sufficiently high object probability di,j. An over-complete set of candidate polygons is produced for a given input image by training a model to densely predict the distances (r) to the object border along a specific set of rays and object probabilities (d). Non-maximum suppression (NMS) of these candidates yields the final result.23
The model was trained using a Google Colab virtual machine with a V100 GPU 16GB VRAM and 51GB of RAM. All code and statistical analysis have been performed using Python 3.10.
StarDist −0.8.5 csbdeep −0.7.4 configparser −6.0.0.
funcsigs −1.0.2 gputools −0.2.14 mako−1.2.4.
pyopencl −2023.1.2 pytools −2023.1.1 reikna −0.8.0.
Listing 1: Package Version. The list shows the version of specific packages used for model training.
To train our model, we used 9661 256 × 256 px tiles. The dataset has been structured as shown in Table 1. The randomness of the dataset split ensures that the training and test datasets represent the overall distribution of the data.
Table 1.
Dataset random split into training and validation set.
| Number of images | 9661 |
|---|---|
| Training | 8212 |
| Validation | 1449 |
The dataset split strategy was coded by the following (see full code in Supplementary 3):
assert len (X) > 1, " not enough training data "
rng = np . random . RandomState(42)
ind = rng . permutation (len (X))
n_val = max(1, int (round (0.15 ∗ len (ind))))
ind_train, ind_val = ind [: −n_val], ind[−n_val:]
X_val, Y_val = [X[i] for i in ind_val], [Y[i] for i in ind_val]
X_trn, Y_trn = [X[i] for i in ind_train], [Y[i] for i in ind_train]
print (‘number of images: %3d’ % len (X))
print (’ − training: %3d’ % len (X_trn))
print (’ − validation: %3d’ % len (X_val))
This code splits the dataset into training and validation sets. In particular:
’rng = np.random.RandomState(42)’: This line defines a random number generator that may generate numbers based on a seed (42 in this case). The seed ensures that the same random integers are generated each time the code is executed.
’ind = rng.permutation(len(X))’: This line creates a random permutation of indices from the dataset ‘X'.
: This line multiplies the number of validation samples by 15% of the total number of samples in the dataset. The ‘max' function guarantees at least one validation sample.
’’: This line separates the indices into training (‘ind_train’) and validation (‘ind_val’). The final ‘n_val’ indices are utilized for validation, whereas the remainder are used for training.
. This line generates the validation datasets ‘X_val’ and’Y_val’ from the validation indices.
. This line generates the training datasets ‘X_trn’ and’Y_trn’ from the training indices.
The code assures that the training and validation sets are chosen randomly and that the same split is used each time the code is run. This is important for evaluating the performance of various models on the same dataset split.
The model is based on StarDist2D and has been trained for 800 epochs at 100 steps per epoch.
Dataset Matching was obtained with the matching function in stardist.matching module.
matching(y_true, y_pred, thresh = 0.5, criterion=’iou’, report_matches = False): calculate detection/instance segmentation metrics between ground truth and predicted label images. “iou” stands for “intersection over union”.
The following metrics are implemented: ‘fp’ (false positive), ‘tp’ (true positive), ‘fn’ (false negative), ‘precision’, ‘recall’, ‘accuracy’, ‘f1’, ‘criterion’, ‘thresh’, ‘n_true’, ‘n_pred’, ‘mean_true_score’ (the mean intersection over unions (IoUs) of matched true positives but normalized by the total number of Ground Truth objects), ‘mean_matched_score’ (the mean IoUs of matched true positives), ‘panoptic_quality’.28
A full list of abbreviations is in Table 2.
Table 2.
List of abbreviations used in the article.
| CAF | Chromatin Associated Factor |
| DDR | DNA Damage Response |
| fn | False negative |
| fp | False positive |
| H&E | Hematoxylin and Eosin |
| HR | Homologous Recombination |
| IHC | Immunohistochemistry |
| IoU | Intersection over Union |
| NMS | Non-maximum suppression |
| OSCC | Oral squamous cell carcinoma |
| PARP | Poly (ADP-ribose) polymerase |
| SIFT | Scale-invariant feature transform |
| TMA | Tissue micro array |
| WSI | Whole slide image |
QuPath integration of the generated model
Following model training, we converted it into portable formats. The Keras H5 generated model was converted into portable formats, such as .pb and ImageJ-compatible zip formats. We used the QuPath StarDist extension and the Fiji StarDist extension23,26 to test the model. Model conversion has been performed in an Anaconda virtual environment using TensorFlow 1.15 and tfonnx.
We used the StarDist groovy script for QuPath to infer the model on the test images. (Fig. 4).
Fig. 4.
Representative high-resolution tif images exported from WSI. The red overlays show the predicted “CAF-1/p60-positive” nuclei on H&E-stained images (third row) and the ground truth, i.e., “CAF-1/p60-positive” nuclei assessed by IHC and detected by the “positive cell detection” QuPath script (fourth row). The first and second rows show the original H&E and corresponding IHC, respectively. (Scale bar: 100 μm). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Results
Quality check of IHC obtained from H&E slides
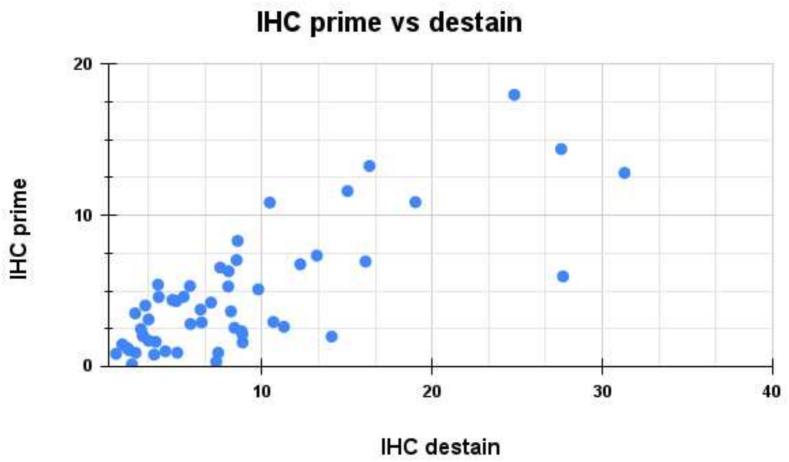
Before collecting patches necessary to train and validate our model, we tested the quality of the “converted” anti-CAF-1/p60 IHC, comparing it with “primary” IHC obtained by staining a tissue section directly. The test was carried out on a TMA tissue block sections (Fig. 5). Agreement between primary and secondary IHC was performed in QuPath by classification of positive tumor cells. The data were plotted in a scatter plot and analyzed with a Pearson correlation test, which returned a value of 0.77 (Fig. 6). Considering that the comparison was carried out between two IHCs on two different sections, in light of the visual analysis of the sample (Fig. 5), we considered the secondary IHC valid.
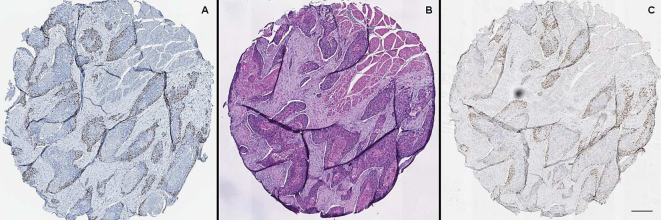
Fig. 5.
The figure shows a representative core out of the 55 that made up the TMA used both for destain and for primary immunostaining: (A) Primary immunostaining with anti-CAF-1/p60. (B) H&E staining on a tissue section different from A. (C) IHC performed on the same section as in B, following destaining. The comparison between the two IHCs shows that the immunostaining of the restained section is comparable to that of the primitive one (Scale bar: 250 μm).
Fig. 6.
The scatter plot compares the quantification of the percentage of tumor cells positive for the marker between the primary IHC and the IHC secondary to destaining. The Pearson correlation test returned a value of 0.77 (Axis values indicates the % of positive tumor cells).
Model building
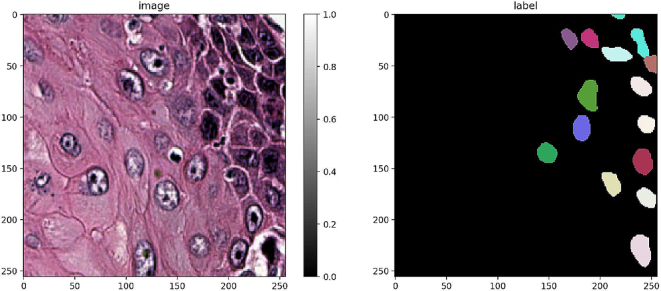
9661 tiles 256 × 256 px in size were generated together with the corresponding segmentation masks. Then, the dataset was loaded into the Google Colab virtual machine runtime in a compiled notebook for model training. Fig. 7 shows a representative tile with the corresponding label.
Fig. 7.
An example of tile image with corresponding mask.
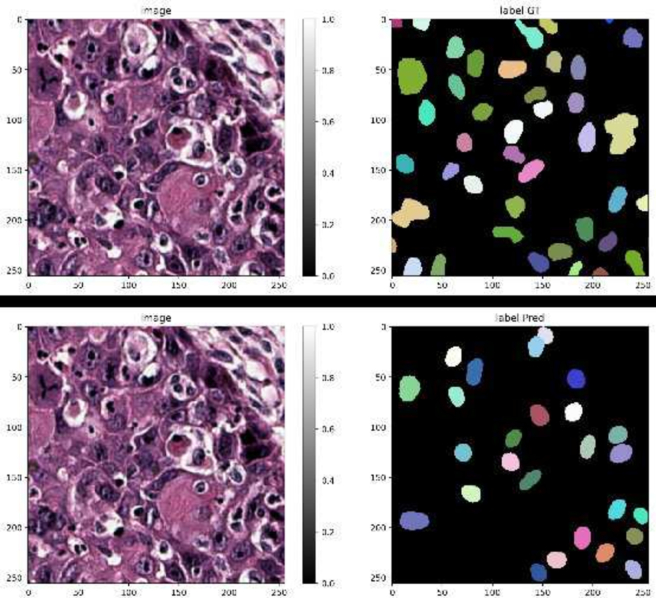
We randomly split the dataset into train and validation sets (8212 and 1449 images, respectively). We trained the model for 800 epochs at 100 steps per epoch. All the configuration parameters are listed in the Supplementary 4 subsection of Supplementary Materials. Fig. 8 shows the prediction label on a representative tile, compared to the corresponding ground truth label image (Fig. 8).
Fig. 8.
Prediction label compared to the ground truth on a representative tile.
Following threshold optimization, the model ended up with the following optimized values: prob_thresh = 0.354425, nms_thresh = 0.5.
The dataset matching values for the calculated threshold were obtained based on the IoU parameter. Results are shown in detail in Supplementary Table S1.
Prediction on TMA test set
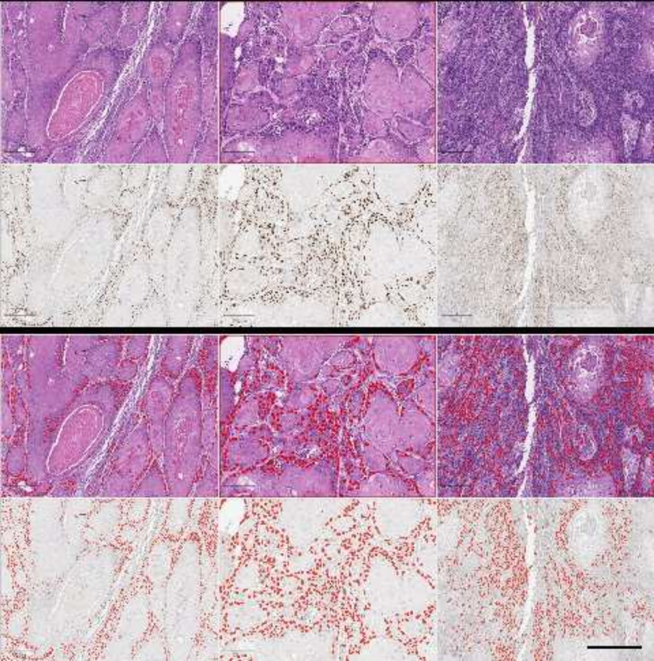
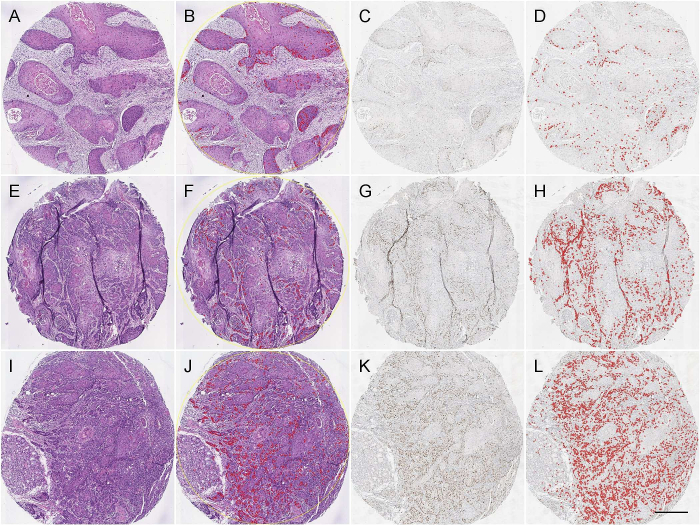
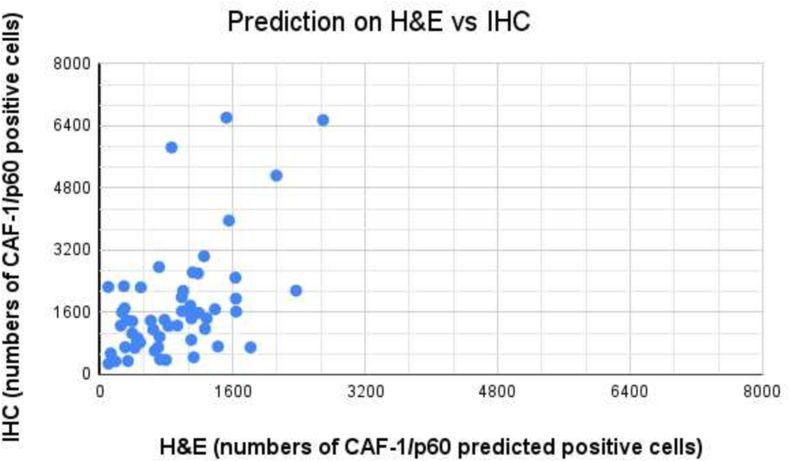
To predict the CAF-1/p60 positivity on H&E-stained WSI, we tested our model on the OSCC TMA to compare the prediction to the IHC restained section. The paired t-test of the two ranges (55 TMA cores, H&E vs p60IHC) returned a p-value <0.0001. Fig. 9 shows the visual results of the prediction. Following the model run in QuPath, we exported TMA measurements on both H&E and corresponding IHC. The correlation analysis is shown in Fig. 10. We demonstrated that a detection and segmentation model, based on StarDist2D, can easily be deployed on WSI in a user-friendly manner to predict CAF-1/p60 tissue marker expression directly on H&E-stained tissue slides.
Fig. 9.
Three representative TMA cores, out of 55, to show the result of p60 positivity prediction on H&E stained WSI compared with IHC. Detection was performed in QuPath: (A, E, I), H&E-stained TMA cores. (C, G, K), CAF-1/p60 IHC on the same cores as in A, E, I, respectively. (B, F, J), Overlay on H&E pointing predicted positive nuclei as determined by the model. (D, H, L), overlay of IHC showing p60 positive cells as determined by cell detection/classification QuPath script described in materials and methods (Scale bar: 250 μm).
Fig. 10.
Scatter plot showing the concordance between p60 positive cell detections on IHC and p60 positivity prediction on H&E. Pearson test value was 0.6. The paired t-test returned a p-value <0.0001. Each dot in the graph is a core of the TMA.
Discussion
Chromatin remodeling proteins are crucial for DNA replication and repair and the growth and development of several human malignant tumors. They also play a crucial part in genome maintenance activities. During the DNA synthesis phase of the cell cycle, the “histone chaperone” CAF-1 transports newly produced H3/H4 dimers to the replicative fork.29 The p48 component of the heterotrimeric protein CAF-1 works with the Retinoblastoma protein (Rb),30 whereas the p150 and p60 subunits are engaged in DNA replication and repair activities. It is noteworthy that during the past 20 years, p60 has been shown to have a significant role in maintaining the proliferative activity of cells in several malignancies.10,31, 32, 33
We have already demonstrated the validity of the CAF-1/p60 subunit as a prognosis marker for OSCC. Further, we showed that the combined assessment of the p60 and p150 subunits may be particularly useful in stratifying the prognostic classes of OSCC.2 Additionally, we demonstrated that CAF-1/p60 and p150 subunits are involved in HR-DDR, suggesting the potential to induce a radiosensitizing synthetic lethality mechanism by administering PARP inhibitors to tumor cells that have been pharmacologically inhibited for CAF-1/p60 and p150 in OSCC patients with a poorer prognosis.2
Although CAF1 p60 can be measured in the serum of patients with OSCC,9 the best approach to date to evaluate its expression is to test its presence by IHC on sections of neoplastic tissue.7,10
The IHC technique is a valuable and widely used tool in biomedical research and diagnostics to detect and localize specific antigens in the context of intact tissue samples. However, IHC also has some limitations that need to be considered, such as variability in tissue fixation and processing, antigen retrieval methods, antibody quality and specificity, detection systems, and interpretation and quantification of results. Therefore, optimizing and standardizing the IHC techniques for each application and using appropriate controls and validation methods is essential.34
Numerous approaches have been described that address the problem of making predictions on the mutational status or expression of specific tumor cell markers by analyzing H&E images at different levels.35 For our model, we chose to work with a single-cell segmentation model. Our approach was aided by conversion techniques from basic H&E staining to IHC. IHC has proved to be a convenient way to make annotations22,36 but, as we know, the immunostained slide, although very similar, is never pixel-wise aligned to the corresponding H&E counterparts, and annotation process still requires a lot of effort. In our workflow, by converting a former H&E-stained tissue to IHC, we can provide a pixel-wide aligned IHC-to-H&E image. We previously described this technique18 that we have now improved by using registration techniques. We optimized the workflow by combining several image processing tools, from QuPath to ImageJ to Python coding. We obtained a reliable single-cell ground truth, demonstrating that the IHC staining we made by destaining a H&E is comparable to primary IHC.
One of the main reasons AI models do not find widespread use in routine is the difficulty of using codes on diagnostic material daily with ease.37 QuPath represents a valid solution for WSI visualization. Over time, the platform has been enriched with extensions that allow for a (relatively) easy implementation of artificial intelligence models.26 The workflow chosen to build a classification/segmentation model capable of predicting positivity to a marker on sections stained with H&E was StarDist because we believe it is a valid approach for building nuclear segmentation models and because we have been able to take advantage of a fairly simple integration flow that will now allow us to implement the model in the clinical routine. As shown in Fig. 9, prediction of CAF-1/p60 positivity on H&E is fairly good and visually corresponds to the immunostain. AI-assisted prediction allows us to categorize tumor samples in low/high expression as we use to do following regular IHC.
Because we are dealing with distinguishing cells in H&E based on subtle differences not appreciable to the naked eye, we did not expect much better metrics than those obtained. Like any AI model applied to pathological anatomy, we believe this model should also be understood as a diagnosis assistance tool to be used by specialists in the field. The model's performance is enormously enhanced if the use, as it should always be done, is entrusted to personnel with specific domain knowledge. However, we are working to design a better strategy to improve metrics and prediction capability (e.g., working on multiclass models).
In conclusion, our work demonstrates that with the right contribution of the wet lab and a workflow designed around the needs of the average pathologist, with the tools that the community of developers makes available to computational pathologists, it is possible to generate predictive models with which we can maximize the information obtainable from H&E stains, all so that pathologists can easily integrate the tool into the diagnostic routine. Predicting the expression of a promising prognostic marker such as CAF-1 p60 directly from H&E-stained sections paves the way to numerous potential applications. Maximizing the information obtainable from H&E could help recover information from exhausted samples, for which there is no possibility of cutting out sections for IHC or making the best use of cryostat sections.
Author contributions statement
Conceptualization, F.M.; methodology, S.V. and G.I.; software, F.M.; validation, D.R., R.M.D.C.; formal analysis, F.M., and A.C.; investigation, S.V.; resources, G.I.; data curation, A.C.; writing—original draft preparation, F.M.; writing—review and editing, S.V. and S.S.; funding, S.S.; supervision, F.M. All authors reviewed the manuscript.
Ethics statement
The study was performed according to the Declaration of Helsinki and in agreement with Italian law for studies based only on retrospective analyses on routine archival FFPE-tissue; a written informed consent from the living patient, following the indication of Italian DLgs No. 196/03 (Codex on Privacy), as modified by UE 2016/679 law of the European Parliament and Commission, was obtained at the time of surgery.38
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
We thank Dr. Filippo Fraggetta for critical reading and helpful discussions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2024.100407.
Appendix A. Supplementary data
Supplementary 1 - Dataset matching values based on the intersection of union (iou) parameter
Supplementary 2 - QuPath Groovy script used for the cell detection, the classification based on the intensity of staining with DAB, and the subsequent cleaning of the detections to obtain a mask that only highlights positive cells.
Supplementary 3 - Python script of the trained model
Supplementary 4 - Model config
References
- 1.Staibano S., Mignogna C., Lo Muzio L., et al. Chromatin assembly factor-1 (CAF-1)-mediated regulation of cell proliferation and DNA repair: a link with the biological behaviour of squamous cell carcinoma of the tongue? Histopathology. 2007;50(7):911–919. doi: 10.1111/j.1365-2559.2007.02698.x. [DOI] [PubMed] [Google Scholar]
- 2.Morra F., Merolla F., Picardi I., et al. Caf-1 subunits levels suggest combined treatments with parp-inhibitors and ionizing radiation in advanced HNSCC. Cancers. 2019;11(10):1582. doi: 10.3390/cancers11101582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staibano S., Mascolo M., Rocco A., et al. The proliferation marker chromatin assembly factor-1 is of clinical value in predicting the biological behaviour of salivary gland tumours. Oncol. Rep. 2011;25(1):13–22. [PubMed] [Google Scholar]
- 4.Staibano S., Mascolo M., Mancini F.P., et al. Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology. 2009;54(5):580–589. doi: 10.1111/j.1365-2559.2009.03266.x. [DOI] [PubMed] [Google Scholar]
- 5.Mascolo M., Travaglino A., Varricchio S., et al. Role of chromatin assembly factor-1/p60 and poly [ADP-ribose] polymerase 1 in mycosis fungoides. Virchows Arch. 2021;478:961–968. doi: 10.1007/s00428-020-02952-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mascolo M., Vecchione M.L., Ilardi G., et al. Overexpression of chromatin assembly factor-1/p60 helps to predict the prognosis of melanoma patients. BMC Cancer. 2010;10:1–14. doi: 10.1186/1471-2407-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mascolo M., Ilardi G., Romano M.F., et al. Overexpression of chromatin assembly factor-1 p60, poly (ADP-ribose) polymerase 1 and nestin predicts metastasizing behaviour of oral cancer. Histopathology. 2012;61(6):1089–1105. doi: 10.1111/j.1365-2559.2012.04313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo D., Travaglino A., Varricchio S., et al. Brit-1 expression and its relationship with parp-1 and caf-1/p60 in cutaneous melanoma. J. Eur. Acad. Dermatol. Venereol. 2021;35(4) doi: 10.1111/jdv.16977. [DOI] [PubMed] [Google Scholar]
- 9.Merolla F., Ilardi G., Di Spigna G., et al. Detection of caf-1/p60 in peripheral blood as a potential biomarker of HNSCC tumors. Oral Oncol. 2021;120 doi: 10.1016/j.oraloncology.2021.105367. [DOI] [PubMed] [Google Scholar]
- 10.Mascolo M., Ilardi G., Merolla F., et al. Tissue microarray-based evaluation of chromatin assembly factor-1 (CAF-1)/p60 as tumour prognostic marker. Int. J. Mol. Sci. 2012;13(9):11044–11062. doi: 10.3390/ijms130911044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martino F., Ilardi G., Varricchio S., et al. A deep learning model to predict Ki-67 positivity in oral squamous cell carcinoma. J. Pathol. Inform. 2024;15 doi: 10.1016/j.jpi.2023.100354. https://linkinghub.elsevier.com/retrieve/pii/S2153353923001682 URL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Li X., Zheng A., et al. Predict Ki-67 positive cells in H&E-stained images using deep learning independently from IHC-stained images. Front. Mol. Biosci. 2020;7:183. doi: 10.3389/fmolb.2020.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janowczyk A., Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016;7(1):29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veta M., Van Diest P.J., Willems S.M., et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med. Image Anal. 2015;20(1):237–248. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Caputo A., Fraggetta F., Cretella P., et al. Digital examination of lymph node cytopathology using the Sydney system (Delycyus). An international, multi-institutional study. Cancer Cytopathol. Jul 2023 doi: 10.1002/cncy.22741. [DOI] [PubMed] [Google Scholar]
- 16.de Haan K., Zhang Y., Zuckerman J.E., et al. Deep learning-based transformation of H&E stained tissues into special stains. Nat. Commun. 2021;12(1):1–13. doi: 10.1038/s41467-021-25221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghahremani P., Li Y., Kaufman A., et al. Deep learning-inferred multiplex immunofluorescence for immunohistochemical image quantification. Nat. Mach. Intell. 2022;4(4):401–412. doi: 10.1038/s42256-022-00471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martino F., Varricchio S., Russo D., et al. A machine-learning approach for the assessment of the proliferative compartment of solid tumors on hematoxylin-eosin-stained sections. Cancers. 2020;12(5):1344. doi: 10.3390/cancers12051344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litjens G., Kooi T., Bejnordi B.E., et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Macenko M., Niethammer M., Marron J.S., et al. Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, June 28–July 1, 2009. IEEE; 2009. A method for normalizing histology slides for quantitative analysis; pp. 1107–1110. [DOI] [Google Scholar]
- 21.Tellez D., Balkenhol M., Otte-Höller I., et al. Whole-slide mitosis detection in H&E breast histology using phh3 as a reference to train distilled stain-invariant convolutional networks. IEEE Trans. Med. Imaging. 2018;37(9):2126–2136. doi: 10.1109/TMI.2018.2820199. [DOI] [PubMed] [Google Scholar]
- 22.Saltz J., Gupta R., Hou L., et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23(1):181–193. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt U., Weigert M., Broaddus C., Myers G. Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16–20, 2018, Proceedings, Part II 11. Springer; 2018. Cell detection with star-convex polygons; pp. 265–273. [Google Scholar]
- 24.Weigert M., Schmidt U., Haase R., Sugawara K., Myers G. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. 2020. Star-convex polyhedra for 3D object detection and segmentation in microscopy; pp. 3666–3673. [Google Scholar]
- 25.Lowe D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004;60:91–110. [Google Scholar]
- 26.Bankhead P., Loughrey M.B., Fernández J.A., et al. Qupath: open source software for digital pathology image analysis. Sci. Rep. 2017;7(1):1–7. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronneberger O., Fischer P., Brox T. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 28.Kirillov A., He K., Girshick R., Rother C., Dollár P. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019. Panoptic segmentation; pp. 9404–9413. [Google Scholar]
- 29.Mjelle R., Hegre S.A., Aas P.A., et al. Cell cycle regulation of human dna repair and chromatin remodeling genes. DNA Repair. 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S. Versatile assembler. Nature. 2003;423(6942):815–817. doi: 10.1038/423814a. [DOI] [PubMed] [Google Scholar]
- 31.Taddei A., Roche D., Sibarita J.-B., Turner B.M., Almouzni G. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 1999;147(6):1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polo S.E., Theocharis S.E., Klijanienko J., et al. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 2004;64(7):2371–2381. doi: 10.1158/0008-5472.can-03-2893. [DOI] [PubMed] [Google Scholar]
- 33.Balmain A., Barrett J.C., Moses H., Renan M.J. How many mutations are required for tumorigenesis? Implications from human cancer data. Mol. Carcinog. 1993;7(3):139–146. doi: 10.1002/mc.2940070303. [DOI] [PubMed] [Google Scholar]
- 34.Dabbs D.J. Elsevier Health Sciences; 2021. Diagnostic Immunohistochemistry: Theranostic and Genomic Applications. [Google Scholar]
- 35.Lee K., Lockhart J.H., Xie M., et al. Deep learning of histopathology images at the single cell level. Front. Artif. Intell. 2021;4 doi: 10.3389/frai.2021.754641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He W., Li C., Nie X., et al. Recognition and detection of aero-engine blade damage based on improved cascade mask r-cnn. Appl. Optics. 2021;60(17):5124–5133. doi: 10.1364/AO.423333. [DOI] [PubMed] [Google Scholar]
- 37.Van der Laak J., Litjens G., Ciompi F. Deep learning in histopathology: the path to the clinic. Nat. Med. 2021;27(5):775–784. doi: 10.1038/s41591-021-01343-4. [DOI] [PubMed] [Google Scholar]
- 38.W. M. Association, et al. World medical association declaration of Helsinki: ethical principles for medical research involving human subjects. Jama. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary 1 - Dataset matching values based on the intersection of union (iou) parameter
Supplementary 2 - QuPath Groovy script used for the cell detection, the classification based on the intensity of staining with DAB, and the subsequent cleaning of the detections to obtain a mask that only highlights positive cells.
Supplementary 3 - Python script of the trained model
Supplementary 4 - Model config