Abstract
Exoenzyme S (ExoS) is an ADP-ribosyltransferase produced and directly translocated into eukaryotic cells by the opportunistic pathogen Pseudomonas aeruginosa. Model systems that allow bacterial translocation of ExoS have found ExoS to have multiple effects on eukaryotic cell function, affecting DNA synthesis, actin cytoskeletal structure, and cell matrix adherence. To understand mechanisms underlying differences observed in cell sensitivities to ExoS, we examined the effects of bacterially translocated ExoS on multiple human epithelial cell lines. Of the cell lines examined, confluent normal kidney (NK) epithelial cells were most resistant to ExoS, while tumor-derived cell lines were highly sensitive to ExoS. Analysis of the mechanisms of resistance indicated that cell association as well as an intrinsic resistance to morphological alterations were associated with increased resistance to ExoS. Conversely, increased sensitivity to ExoS appeared to be linked to epithelial cell growth, with tumor cells capable of undergoing non-contact-inhibited, anchorage-independent growth all being sensitive to ExoS, and NK cells becoming sensitive to ExoS when subconfluent and growing. Consistent with the possibility that growth-related, actin-based structures are involved in sensitivity to ExoS, scanning electron microscopy revealed cellular extensions from sensitive, growing cells to bacteria, which were not readily evident in resistant cells. In all studies, the severity of effects of ExoS on cell function directly correlated with the degree of Ras modification, indicating that sensitivity to ExoS in some manner related to the efficiency of ExoS translocation and its ADP-ribosylation of Ras. Our results suggest that factors expressed by growing epithelial cells are required for the bacterial contact-dependent translocation of ExoS; as normal epithelial cells differentiate into polarized confluent monolayers, expression of these factors is altered, and cells in turn become more resistant to the effects of ExoS.
Exoenzyme S, an ADP-ribosyltransferase produced by Pseudomonas aeruginosa, contributes to the virulence of this opportunistic pathogen by causing increased tissue damage and bacterial dissemination (17, 21, 25, 37). ExoS is translocated into eukaryotic cells by the bacterially mediated type III secretory process (38). Studies of the effects of ExoS on cell function have therefore required the development of model systems which enable the bacterial translocation of ExoS yet allow the effects of ExoS to be differentiated from those of other Pseudomonas factors. The use of such model systems has linked ExoS production to complex effects on eukaryotic cell function, including inhibition of DNA synthesis, alterations in actin cytoskeletal structure, and interference with cell-matrix adherence (12, 28, 29). Alterations in multiple eukaryotic cell processes are therefore likely to contribute to the increased tissue damage associated with ExoS.
The multiple and diverse effects of ExoS on cell function can, to some extent, be explained by its multidomain structure. Residues within the amino-terminal half are required for export and the aggregation properties of ExoS, while the ADP-ribosyltransferase (ADPRT) activity of ExoS is included within the carboxy-terminal 222 amino acids of the protein (18, 38). Consistent with the multifunctional structure of ExoS, effects on cell rounding have been associated with an enzymatically inactive E381A mutant form of ExoS (12). Diversity in the cellular effects of ExoS may also relate to Ras functioning as an in vivo substrate of ExoS (24) and the ability of Ras to modulate multiple signal transduction pathways. In this regard, recent studies have linked the ADP-ribosylation of Ras by ExoS to the disruption of Ras-Raf-mediated signal transduction pathways (14, 36), providing a cellular mechanism to explain inhibitory effects of ExoS on DNA synthesis and PC12 cell differentiation (13, 29). A current understanding of cell signaling pathways also links Ras activation to the regulation of actin cytoskeletal structure, via Rac, Cdc42, and Rho (15).
Our laboratory has used a bacterial-eukaryotic coculture model system that compares the effects of ExoS-producing strain 388 and the isogenic non-ExoS-producing strain 388ΔexoS (388ΔS) to differentiate the effects of ExoS on eukaryotic cell function. During these studies, it became evident that ExoS could exert similar effects on DNA synthesis and morphology of different cell types (29). It was also observed that different cell lines exhibited different levels of sensitivity to ExoS. Studies described in this report further investigate the cytotoxic effects of ExoS by comparing the effects of ExoS on different human epithelial lines and examining processes underlying differential sensitivities to ExoS. Intercellular association as well as intrinsic cellular properties were found to contribute to the resistance of normal kidney (NK) epithelial cells to bacterially translocated ExoS. Conversely, sensitivity to ExoS appeared to be linked to epithelial cell growth, with rapidly growing tumor cell lines and growing subconfluent NK cells showing increased sensitivity to the effects of ExoS. The results are consistent with factors expressed by growing epithelial cells being required for the bacterial contact-dependent translocation of ExoS and the possibility that these factors become down-regulated or redistributed and less accessible as epithelial cells differentiate into confluent polarized monolayers.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. aeruginosa 388, 388ΔS, 388exs1∷Tn1, and 388popD∷Tc* were all kindly provided by Dara Frank, Medical College of Wisconsin, Milwaukee, Wis. Parental strain 388 and construction of the 388ΔS mutant have been previously described (3, 22). The 388ΔS mutant strain fails to produce 49-kDa ExoS due to allelic exchange of the majority of the structural gene with a tetracycline gene cartridge. ExoS complementation studies of strain 388ΔS confirmed that differences in the effects of this strain and parental strain 388 on eukaryotic cell function directly relate to ExoS production (28). Strains 388popD∷Tc* (35) and 388exs1∷Tn1 (38), discussed in scanning electron microscopy (SEM) studies, lack production of the type III PopD translocation and PscC secretion proteins, respectively.
Bacteria were induced for ExoS production in TSBD-N medium and prepared for bacterial-eukaryotic coculture studies as previously described (29), except when ExoS induction by eukaryotic cell contact was examined. In these experiments bacteria were grown in a non-ExoS-inducing medium, unchelated Trypticase soy broth containing 1% glycerol and 100 mM monosodium glutamate (TSB). Also for these experiments, bacteria were grown for 12 h, instead of the usual 18 h, before being added to eukaryotic cell monolayers.
Eukaryotic cell culture.
Cell lines and their specific growth media are described in Table 1. All cells were maintained at 37°C in 5% CO2–95% air, and all media and media components were purchased from Gibco-BRL (Gaithersburg, Md.). Cell lines obtained from the American Type Culture Collection (ATCC) were cultured and passaged as specified by ATCC. Primary cultures of human NK proximal tubule (NK77 and NK97612) cells were obtained from the tissue culture bank in the Department of Pathology at the Medical University of South Carolina, Charleston. NK cells were maintained as previously described (7) and cultured in vessels coated first with bovine collagen type 1 (Collaborative Biomedical, Lexington, Mass.) and then with fetal bovine serum (FBS). Culture growth medium was replaced every 2 to 3 days, and cells were passaged weekly at a subculture ratio of 1:2 or 1:3. All experiments with NK cells were performed from passages 4 to 9. CFT1 and CFT1-LCFSN cell lines were a kind gift from Gerald Pier, Harvard Medical School, Boston, Mass., and were cultured as previously described (27, 39). Medium was replaced every 2 to 3 days, and cells were passaged weekly as described for NK cells. For comparisons of NK and HT-29 cell induction of the ExoS regulon, HT-29 cells were adapted to the 50% high-glucose Dulbecco’s modified Eagle’s medium (DMEM)–50% Ham’s F-12 (DME/F12) used to culture NK cells. For bacterial-eukaryotic cell coculture experiments, all except NK cell monolayers were detached with 0.25% trypsin–1 mM EDTA (trypsin-EDTA; Gibco-BRL), resuspended in their respective culture medium, counted, diluted to the appropriate density, and seeded in 25-cm2 flasks, 35-mm-diameter dishes, or 48- or 6-well plates (Costar). NK cells were similarly detached with trypsin-EDTA, but following detachment, trypsin was neutralized with an equal volume of FBS, and cells were collected, centrifuged for 5 min at 600 × g, resuspended in growth medium, and seeded as described above on collagen-coated surfaces. All cells were allowed to grow for 2 to 3 days prior to addition of bacteria.
TABLE 1.
Characteristics of cell lines examined in ExoS coculture studies
| Cell line | Cell type | Basal medium | Characteristics | Source |
|---|---|---|---|---|
| NK77 | Primary kidney epithelial | DME/F12a | Normal | MUSCd |
| NK97612 | Primary kidney epithelial | DME/F12a | Normal | MUSC |
| CFT1 | Tracheal epithelial | Ham’s F-12b | CFTR ΔF508 mutation | Gerald Piere |
| CFT1-LCFSN | Trachael epithelial | Ham’s F-12b | CFT1 with normal CFTR | Gerald Pier |
| HT29 | Colon carcinoma | McCoy’s 5Ac | Normal Ras | ATCC HTB 38 |
| LNCaP | Prostate carcinoma | RPMI 1640c | Normal Ras | ATCC CRL 1740 |
| T24 | Bladder carcinoma | McCoy’s 5Ac | Mutant H-Ras (G12V) | ATCC HTB 4 |
| SW480 | Colon carcinoma | McCoy’s 5Ac | Mutant K-Ras (G12V) | ATCC CCL 228 |
| SK-N-SH | Neuroblastoma | DMEMc | Mutant N-Ras (Q61K) | ATCC HTB 11 |
Supplemented with 5 μg of insulin, 5 μg of transferrin, 5 μg of selenium, 36 ng of hydrocortisone, 4 pg of triiodothyronine, and 10 ng of epidermal growth factor per ml.
Supplemented with 10 μg of insulin per ml, 1 μM hydrocortisone, 3.75 μg of endothelial cell growth supplement per ml, 25 ng of epidermal growth factor per ml, 30 nM triiodothyronine, 5 μg of transferrin per ml, and 10 ng of cholera toxin per ml.
Contains 10% FBS.
MUSC, tissue culture bank, Department of Pathology and Laboratory Medicine, Medical University of South Carolina.
e Brigham & Women’s Hospital, Harvard Medical School.
Examination of bacterial effects on HT-29 and NK cells.
In all experiments, control and strain 388 or strain 388ΔS cells were identically treated except that in controls no bacteria were added to the coculture medium, and in bacterially treated monolayers the indicated concentration of the respective strain was added to the coculture medium. The multiplicity of infection ranged from 10 to 100 bacteria per eukaryotic cell, depending on eukaryotic cell density.
(i) Quantification of DNA synthesis in eukaryotic cells following exposure to bacteria.
Following a 3- or 4-h coculture period, bacteria were removed and monolayers were washed in medium containing gentamicin (200 μg/ml) and ciprofloxacin (100 μg/ml) (antibiotic medium) to limit further bacterial growth. Cells were then analyzed for [3H]thymidine incorporation after a 20-h pulse in antibiotic medium as previously described (29).
(ii) Immunoprecipitation, detection, and quantification of Ras proteins.
Ras was immunoprecipitated from cells after coculture with bacteria, using monoclonal Y13-259 Ras antibody (ATCC), rabbit anti-rat immunoglobulin G (Sigma Chemical Co., St. Louis, Mo.), and protein G-Sepharose (Sigma), as previously described (24). Proteins were resolved on sodium dodecyl sulfate (SDS)–15% polyacrylamide gels and electroblotted, and Ras immunoblots were developed by using pan-Ras-OP22 antibody (Oncogene Research, Cambridge, Mass.) followed by peroxidase-conjugated goat anti-mouse antibody (Sigma) and visualized by enhanced chemiluminescence (Amersham Life Sciences, Arlington Heights, Ill.). Ras modification was quantified by densitometric analysis using NIH Image version 1.60 software. The areas of intensity of modified and unmodified Ras were determined, and modified Ras is expressed as a percentage of total Ras.
(iii) Examination of cell morphology.
Cells were grown and cocultured with bacteria in 48-well plates. Cell morphology was assessed by phase-contrast microscopy following a 3-h coculture period, either immediately after removal of bacteria at 3 h or after the addition of antibiotic medium and further incubation for 24 h.
Quantification of bacteria associated with eukaryotic cells.
Bacterial association was determined by using an adaptation of the procedure of Fleiszig et al. (9). NK and HT-29 cells were seeded at 6 × 105 cells/ml (0.5 ml/well) in 48-well plates and grown until confluent. Bacteria were prepared for coculture as previously described (29) and suspended at 108 CFU/ml in coculture medium; then 0.5 ml was added to each tissue culture well, and the plates were incubated for 3 h. Bacteria were removed and monolayers were washed three times with phosphate-buffered saline (PBS) to remove unassociated bacteria. Cells were detached by scraping and lysed by adding to each well 1 ml of PBS containing 0.25% Triton X-100. Lysates were thoroughly mixed; then aliquots were diluted in PBS containing 1% bovine serum albumin and plated to quantify viable bacteria. Bacterial numbers in the original inoculum were determined, and bacteria associated with cells were reported as percentage of inoculum.
Assay of ExoS activity in coculture medium.
ExoS ADPRT activity was quantified by using the in vitro substrate soybean trypsin inhibitor as previously described (29) except that reactions were modified to detect lower levels of activity. For this, the final concentration of [14C]NAD was raised to 5 μM, the 14-3-3 protein cofactor (FAS) concentration was adjusted to 100 nM, and 20 μl of culture medium was added to reactions, which were allowed to proceed for 1 h. Incorporation of radiolabel was measured by liquid scintillation counting and reported as picomoles of ADP-ribose transferred to substrate per milliliter of coculture medium.
Disruption of intercellular junctions in confluent monolayer cell cultures.
NK and HT-29 cells grown to confluence in 6- or 48-well plates were washed once with PBS free of calcium and then incubated with PBS containing 1 mM EDTA, or as a control PBS supplemented with 0.9 mM calcium, until all EDTA-treated cells were rounded (10 to 15 min). PBS was then removed, cells were cocultured with bacteria for 3 h, and [3H]thymidine uptake assays or Ras immunoprecipitation was performed as described above.
SEM.
P. aeruginosa 388 (108 CFU/ml) was cultured for 3 h with subconfluent HT-29, LNCaP, or NK cell monolayers or confluent NK cell monolayers which were grown on 12-mm-diameter glass coverslips. Bacteria were removed, and cells were washed twice with McCoy’s SA-bovine serum albumin fixed in situ in 2% gluteraldehyde in 0.1 M cacodylate buffer (pH 7.4), rinsed, postfixed in 1% osmium in 0.1 M cacodylate buffer, and then dehydrated in a graded series of ethanol, and treated with hexamethyldisilane. After drying, coverslips were coated with gold and examined in a JEOL JSEM-LV5410 scanning electron microscope.
Statistical analysis.
Statistical analysis was performed with StatView (Abacus Concepts, Inc.).
RESULTS
Effects of ExoS-producing bacteria on DNA synthesis and Ras modification in different epithelial cell lines.
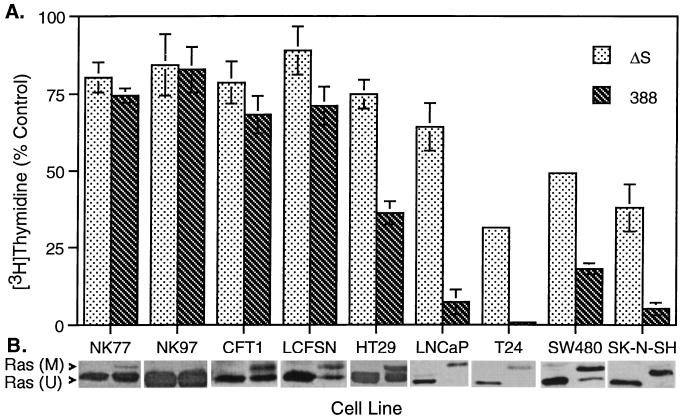
Differences have been observed in the sensitivity of eukaryotic cell lines to the effects of ExoS during coculture studies. To investigate cellular properties that might influence sensitivity to bacterially translocated ExoS, we compared levels of inhibition of DNA synthesis caused by ExoS-producing strain 388 or the non-ExoS-producing strain 388ΔS in a selection of human cell lines described in Table 1. As shown in Fig. 1A, NK epithelial cells were most resistant to the effects of ExoS on DNA synthesis. CFT1-derived tracheal epithelial cells, which express mutant cystic fibrosis transductance regulator (CFTR), appeared slightly more sensitive to the effects of ExoS on DNA synthesis than NK cells. However, LCFSN cells, which express both mutant and normal CFTR, showed a sensitivity to ExoS similar to that of CFT1 cells, suggesting that expression of the mutant form of CFTR associated with cystic fibrosis did not alter sensitivity to ExoS. Tumor-derived cell lines consistently showed a greater sensitivity to ExoS production. As indicated in Fig. 1, and as previously described (36), sensitivity to ExoS did not appear to be affected by the expression of oncogenic forms of Ras by tumor cell lines.
FIG. 1.
Differential sensitivity of human cell lines to ExoS-producing bacteria. (A) Inhibition of DNA synthesis. The indicated cell lines, described in Table 1, were cocultured with 107 CFU of strain 388 or 388ΔS (ΔS) per ml or no bacteria for 3 or 4 h in their respective media. Bacteria were removed and replaced with medium containing 1 μCi of [3H]thymidine per ml and antibiotics to inhibit further bacterial growth. DNA synthesis was assayed after 20 h and is expressed as percent incorporation relative to non-bacterially treated control cells. The mean and SD of assays performed in triplicate are represented. (B) Ras modification. Cells were cocultured as described above, culture supernatants were removed, cells were lysed, and Ras was immunoprecipitated with Ras monoclonal Y13-259 antibody, rabbit anti-rat immunoglobulin G, and protein G-Sepharose. Immunoprecipitates were resolved by SDS–15% polyacrylamide gel electrophoresis. Ras immunoblots were developed using pan-Ras-OP22 antibody and detected by enhanced chemiluminescence. Ras (M) and Ras (U) indicates modified and unmodified Ras, respectively.
Ras has been identified as an in vivo substrate of ExoS, and the ADP-ribosylation of cellular Ras by bacterially translocated ExoS can be recognized by a shift in mobility when examined by SDS-polyacrylamide gel electrophoresis (6, 24). Cell lines showing greater inhibition of DNA synthesis following coculture with ExoS-producing bacteria were also found to have a higher proportion of modified Ras (Fig. 1B). These data indicate that sensitivity of cells to ExoS reflects the accessibility of cellular Ras to ADP-ribosylation by bacterially translocated ExoS.
Relationship between effects of ExoS on DNA synthesis and cellular morphology.
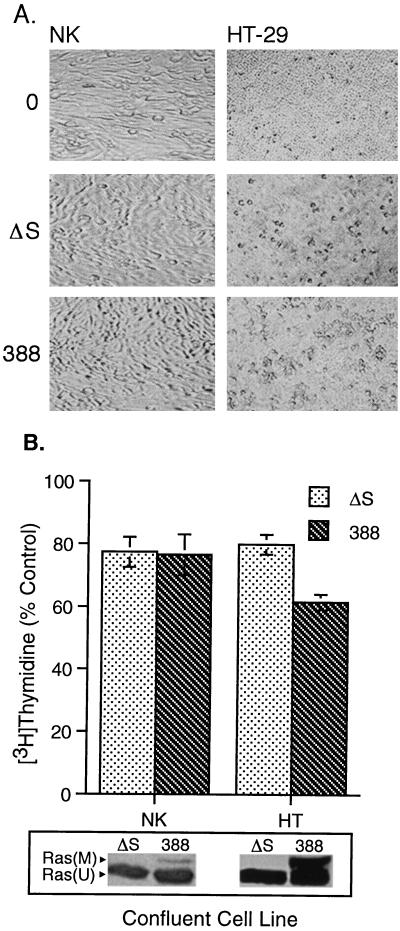
In addition to effects on DNA synthesis and Ras modification, the translocation of ExoS has been associated with cell rounding and the disruption of the actin cytoskeleton (12, 28, 29). To further understand differences in the sensitivities of epithelial cells to ExoS, cells resistant and sensitive to the effects of ExoS on DNA synthesis were compared for ExoS-associated alterations in cellular morphology. Two cell lines were selected for these studies: NK cells, representing an ExoS-resistant cell line, and HT-29 cells, representing an ExoS-sensitive cell line. HT-29 cells were chosen for these studies because their morphology and growth characteristics more closely resembled those of NK cells than other tumor-derived cell lines.
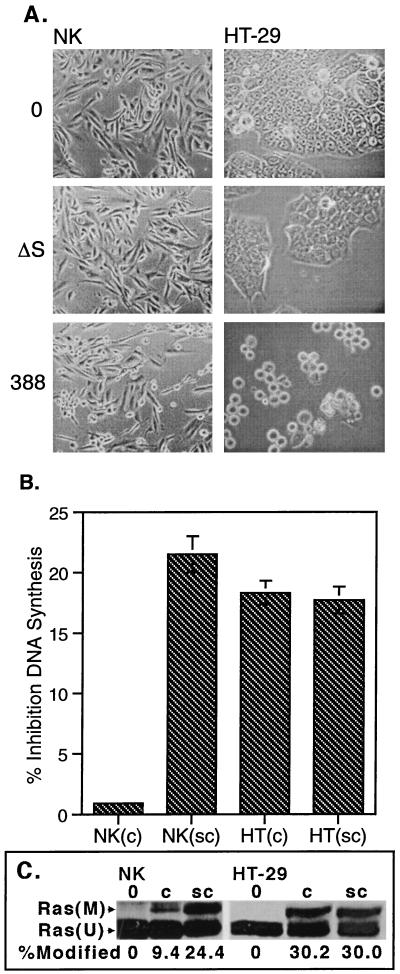
When confluent NK cell morphology was examined by phase-contrast microscopy following removal of 388 or 388ΔS bacteria at 3 h (Fig. 2A) or at 24 h (not shown), no rounding was observed. However, some degree of cell rounding was evident following the coculture of confluent HT-29 with both strains 388 and 388ΔS for 3 h, with more pronounced rounding in 388-treated cells, which persisted for 24 h (not shown). Non-ExoS-associated effects by strain 388ΔS on HT-29 morphology have been previously described and can be differentiated from morphological alterations caused by enzymatically active ExoS based on the ability of cells to recover from morphological alterations caused by strain 388ΔS, but not strain 388, following removal of bacteria (28). Consistent with effects on morphology, inhibition of DNA synthesis was detected in confluent HT-29 cells exposed to ExoS-producing bacteria but not in confluent NK cells (Fig. 2B). The degree of inhibition of HT-29 DNA synthesis, however, was not as great as that shown in Fig. 1, where cells were not examined at confluence. Ras modification was greater in confluent HT-29 cells than in confluent NK cells, again indicating a relationship between cellular sensitivity to ExoS and the efficiency of modification of Ras by ExoS.
FIG. 2.
Differential effect of ExoS-producing bacteria on confluent NK and HT-29 cell morphology, DNA synthesis, and Ras modification. Confluent and subconfluent monolayers of NK and HT-29 cells were cocultured with 108 CFU of strain 388, strain 388ΔS (ΔS) per ml or no bacteria (0) for 3 h. Bacteria were removed, antibiotic-containing medium was added, and cells were analyzed for alterations in morphology by phase-contrast microscopy at 3 h (A) and differences in DNA synthesis and Ras modification (B), assayed as described for Fig. 1. DNA synthesis results represent the mean and SD of assays performed in triplicate. Modified and unmodified Ras [Ras(M) and Ras(U)] are labeled.
Role of bacterial contact in differential sensitivities of HT-29 and NK to the effects of ExoS.
The observation that NK cells remained more resistant to ExoS than HT-29 cells when both cell types were confluent suggests that intrinsic cellular differences may contribute to their differential sensitivity to ExoS. Expression and translocation of ExoS is triggered via a poorly understood process which is thought to be initiated in vivo by the direct contact of the bacterium with a target cell but can be mimicked in vitro by Ca2+ ion depletion (11, 33). To determine whether differences in the sensitivity of cell lines to effects of ExoS might relate to cell surface properties that affect bacterial association or induction of the ExoS regulon, both parameters were compared in HT-29 and NK cells. When the association of bacteria with HT-29 or NK cells was examined in bacterial plating assays, no significant difference was detected in the percentage of inoculated bacteria associated with confluent HT-29 or NK monolayers following a 3-h exposure to strain 388 (0.13% ± 0.08% and 0.09 ± 0.06% [mean ± standard deviation {SD}] for HT-29 and NK cells, respectively; P = 0.77). However, since NK cells occupy a larger area and produce a more uniform monolayer than the rapidly proliferating HT-29 cells, fewer NK cells were present at confluency, which makes the efficiency of bacterial association with resistant NK cells actually greater than that of HT-29 cells (5:1 and 2:1 bacterium-to-cell ratio for NK and HT-29 cells, respectively). Association of 388ΔS to both cell types was similar to that of strain 388. While these studies indicate that overall differences in bacterial association do not appear to contribute to the increased resistance of NK cells to the effects of ExoS, it cannot be determined from these studies whether the two cell lines differ in the manner of bacterial binding and/or cell surface localization.
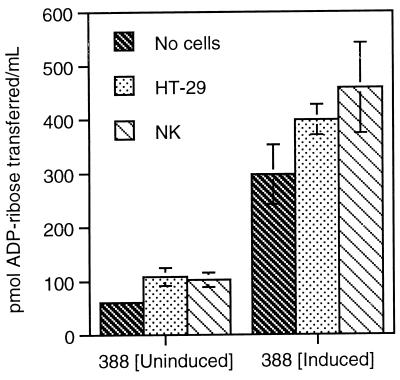
To examine whether differences in the sensitivities of NK and HT-29 cells to ExoS related to their ability to induce the ExoS regulon upon bacterial contact, we assayed ExoS activity in HT-29 or NK coculture supernatants following a 3-h exposure to strain 388. This study takes advantage of our previous finding that low levels of ExoS activity could be detected in the medium following coculture with strain 388 (29). Although it cannot be presumed that levels of ExoS activity in coculture supernatants directly reflect levels of translocated ExoS, the rate of ExoS secretion should provide an accurate index of induction of the ExoS regulon. It was essential that NK and HT-29 cells be adapted to the same medium in these studies, since differences have been observed in the efficiency of ExoS production in different tissue culture media. The medium most suited to the growth of both cell types, DME/F12, contains higher concentrations of cations than the McCoy’s medium normally used in HT-29 cell culture and is associated with less efficient induction of ExoS. As shown in Fig. 3, contact of strain 388 with HT-29 or NK cells resulted in an approximate 1.8-fold increase in ExoS secretion, increasing from 60.5 ± 5 pmol/ml when cultured in the absence of eukaryotic cells to 108 ± 17 and 103 ± 14 pmol/ml in the presence of HT-29 and NK cells, respectively. Levels of ExoS secretion were proportionately increased when strain 388 was preinduced for ExoS production in a low Ca2+ bacterial growth medium (TSBD-N) prior to initiation of coculture studies, with activities increasing from 297 ± 55 pmol/ml in the absence of cells to 399 ± 29 and 458 ± 84 pmol/ml in the presence of HT-29 and NK cells, respectively. No significant differences, however, were observed in the induction of ExoS by HT-29 or NK cells under either of the bacterial culture conditions, indicating that the efficiency of induction of the ExoS regulon was not a factor in the differential sensitivities of the two cell lines to ExoS.
FIG. 3.
Cell contact-dependent secretion of ExoS during coculture with HT-29 or NK cells. Strain 388 was grown in non-ExoS induction medium (TSB) or ExoS induction medium (TSBD-N) for 12 h, and then bacteria (108 CFU/ml) were cocultured with HT-29 or NK cells for 4 h. Coculture supernatants were assayed for ExoS ADPRT activity in reaction mixtures containing 100 μM soybean trypsin inhibitor 100 nM FAS, and 5 μM [14C]NAD in 0.2 M sodium acetate (pH 6.0). Activity is expressed as picomoles of ADP-ribose transferred per milliliter of coculture medium, and the mean and SD of analyses performed in triplicate are represented.
Role of cell junction formation in the resistance of cells to ExoS-producing bacteria.
Susceptibility to P. aeruginosa infection has previously been reported to be influenced by epithelial cell polarity, with the basolateral surface of the cell being more susceptible to P. aeruginosa cytotoxicity than the apical surface (8). Confluent cultures of NK cells can mimic intact epithelial tissue by forming junctional complexes between cells, which can result in cell polarization and the separation of apical and basolateral cell surfaces (4). While HT-29 cells have also been reported to form tight junctions and become polarized under specific growth conditions (30, 34), culture conditions used in this study did not favor HT-29 cell polarization, and cells remained relatively undifferentiated and formed multilayers.
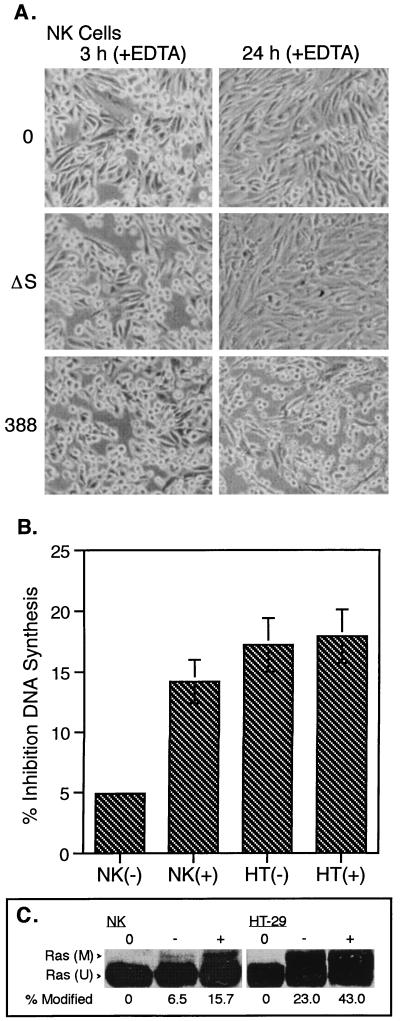
To examine the role of intercellular junctions in the resistance of NK cells to ExoS, junctional complexes of confluent NK and HT-29 cell monolayers were disrupted prior to the addition of bacteria by depleting cell cultures of extracellular calcium. Treatment of cells with 1 mM EDTA, a cation-chelating agent, has been previously reported to disrupt junctional integrity of epithelial cells (16, 31). Consistent with confluent NK cells being affected in a similar manner by this treatment, NK cells were found to round up and separate from each other within 10 min of exposure to EDTA. Figure 4A shows the effects of strains 388 and 388ΔS on the morphology of EDTA-pretreated NK cells. While cell rounding and decreased intercellular association persisted in bacterially treated as well as control monolayers following removal of bacteria at 3 h, strain 388-treated NK cells remained rounded for 24 h. This compared with little lasting effect of EDTA treatment on the morphology of 388ΔS and non-bacterially treated control NK cells after 24 h. Treatment of HT-29 cells with EDTA was found to cause cell rounding more rapid than that observed for NK cells, and cells tended to detach from the growth surface, suggesting that both cell-matrix adhesion and cell-cell junctional complexes had been disrupted. As with NK cells, exposure to strain 388 in EDTA-pretreated HT-29 cells resulted in more long-term cell rounding than observed for strain 388ΔS or control HT-29 cells (not shown).
FIG. 4.
Effects of EDTA treatment on the sensitivity of NK and HT-29 cells to ExoS. NK or HT-29 cells were cultured to confluence and treated with 1 mM EDTA in PBS for ∼10 min, until cells became rounded (+), or with PBS containing 0.9 mM CaCl2 (−). Cells were then cocultured with 108 CFU of strain 388 or 388ΔS (ΔS) per ml or no bacteria (0) for 3 h. (A) NK cells were examined by phase-contrast microscopy for alterations in morphology at 3 and 24 h. (B) NK and HT-29 cells were assayed for DNA synthesis as described for Fig. 1. Results are expressed as percent inhibition of DNA synthesis of 388-treated cells relative to 388ΔS-treated cells, with the mean and SD of assays performed in triplicate or quadruplicate represented. (C) The efficiency of Ras modification was determined by immunoblot analysis as described for Fig. 1 and quantified by densitometric analysis using NIH Image version 1.60 software. The areas of intensity of modified [(M)] and unmodified [(U)] Ras were calculated, and modified Ras is expressed as a percentage of total Ras. 0 represents Ras immunoprecipitated from non-bacterially treated cells.
Consistent with the increased sensitivity of EDTA-treated NK cells to morphological alterations caused by strain 388, pretreatment with EDTA also increased the sensitivity of NK cells to the differential effects of ExoS on DNA synthesis (Fig. 4B). EDTA treatment alone caused no significant alteration of NK cell DNA synthesis in non-bacterially treated or 388ΔS-treated cultures and did not enhance inhibition of DNA synthesis by strain 388 in subconfluent cultures (data not shown). In contrast to NK cells, no relative increase in differential inhibition of DNA synthesis by strain 388 was detected when HT-29 cells were pretreated with EDTA (Fig. 4B). The increased sensitivity of EDTA-treated NK cells to ExoS was associated with increased Ras modification (Fig. 4C). The ability of disruption of cellular junctions by EDTA to alter the resistance of NK, but not HT-29, to ExoS again indicates that the two cell lines differ in sensitivity to ExoS. The observation that the enhanced sensitivity of NK cells to ExoS is coordinated with increased ADP-ribosylation of cellular Ras provides evidence that these cellular differences relate in some manner to the efficiency in which ExoS can translocate across the membrane to modify Ras.
Factors contributing to the sensitivity of epithelial cells to the effects of ExoS-producing bacteria.
While intercellular associations were found to contribute to the resistance of NK cells to the effects of ExoS, the mechanism for the general increased sensitivity of HT-29 and other tumor cells to bacterially translocated ExoS remained unclear. A factor that differentiates HT-29 and other tumor cell lines from resistant confluent NK cells, and could potentially contribute to an increased sensitivity to ExoS, is an increased growth rate. Normal epithelial cell growth is dependent on and regulated by anchorage and cell association processes. Tumor-derived epithelial cell lines differ from normal epithelial cells in the ability to undergo anchorage-independent growth. Consistent with the possibility that factors associated with cell growth processes influence sensitivity to ExoS is the observed increased sensitivity to ExoS in (i) junctionally disrupted confluent NK cells which are in the process of reestablishing intercellular junctions and (ii) tumor cell lines, which can continue to grow in the absence contact-dependent growth signals.
To examine how cell growth-related factors might contribute to sensitivity to bacterially translocated ExoS, attempts were made to stimulate the growth of confluent NK cells. Normal and serum-starved confluent NK cells, however, proved resilient to growth factor and phorbol ester growth-stimulating agents, precluding this approach for analyzing the effect of growth on NK cell sensitivity to ExoS. An alternative approach chosen was to compare the ExoS sensitivity of subconfluent, growing NK cells to that of confluent NK cells, as well as subconfluent HT-29 cells. While no differential effects on NK cell morphology were apparent in subconfluent NK cells after removal of bacteria at 3 h (not shown), some cell rounding was evident after 24 h in cells exposed to strain 388 (Fig. 5A). This differed from the previously observed lack of effect of strain 388 on confluent NK cell morphology at 3 or 24 h (Fig. 2A). When subconfluent HT-29 cells were treated in a similar manner, both strains 388 and 388ΔS caused cell rounding at 3 h, with cell rounding persisting in 388-treated cells for 24 h (Fig. 5A), as previously observed for confluent HT-29 cells. Rounding of 388-treated subconfluent HT-29 cells was notably more severe than that of subconfluent NK cells at 24 h. The sensitivity of subconfluent NK cells to the effects of ExoS was reflected in DNA synthesis analyses, where differential inhibition of DNA synthesis in subconfluent NK cells was found to increase to levels comparable to that of HT-29 cells (Fig. 5B). In contrast, the relative difference in DNA synthesis in HT-29 cells caused by strains 388 and 388ΔS was unaffected by the degree of confluency. The increased sensitivity of subconfluent NK cells to effects of ExoS on morphology and DNA synthesis corresponded with increased Ras modification (Fig. 5C). The data indicate that NK cells become sensitive to ExoS when subconfluent and growing. The observation that HT-29 cells do not show a similar enhanced sensitivity to effects of ExoS on DNA synthesis at subconfluency again highlights intrinsic differences between the two cell lines relative to their sensitivity to ExoS. These differences are also exemplified in the degree of cell rounding caused by both strains 388 and 388ΔS in HT-29 cells, which was not evident in NK cells.
FIG. 5.
Effects of ExoS on subconfluent NK and HT-29 cells. Subconfluent (sc) or confluent (c) NK or HT-29 cell monolayers were cocultured with 108 CFU of strain 388 or 388ΔS (ΔS) per ml or no bacteria (0) for 3 h. (A) Subconfluent NK and HT-29 cells were examined by phase-contrast microscopy for alterations in morphology at 24 h. (B) NK and HT-29 cells were assayed for DNA synthesis as described for Fig. 1. Results are expressed as percent inhibition of DNA synthesis of 388-treated cells relative to 388ΔS-treated cells, with the mean and SD of assays performed in triplicate or quadruplicate represented. (C) The efficiency of Ras modification was determined by immunoblot analysis as described for Fig. 1 and quantified as described for Fig. 4. 0 represents Ras immunoprecipitated from non-bacterially treated cells.
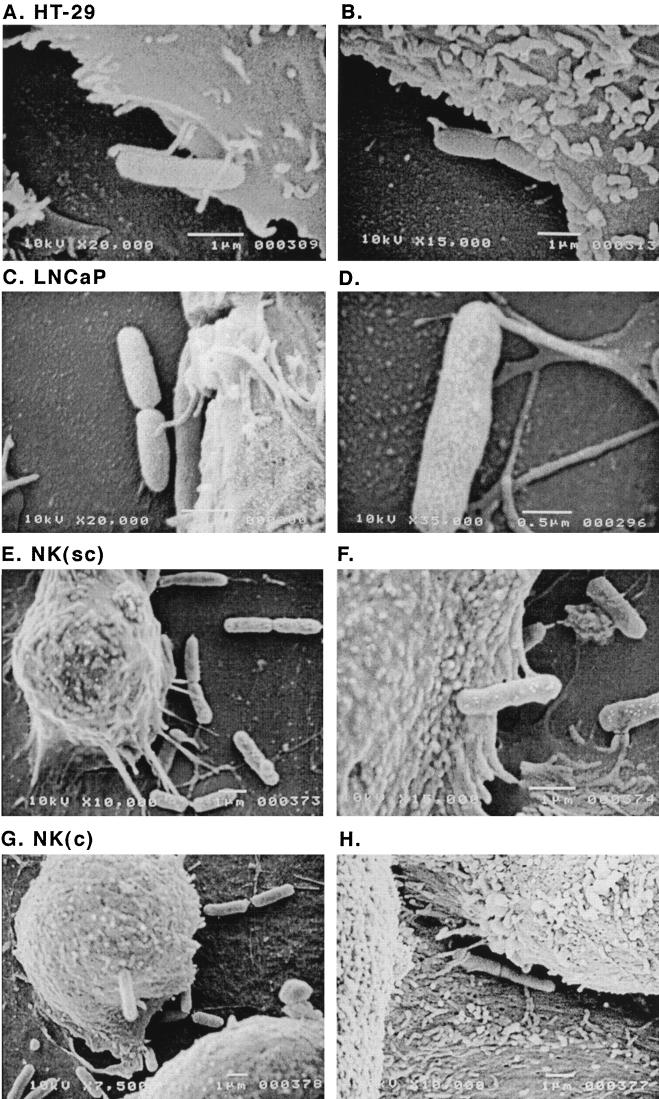
Further visual understanding of cellular factors associated with sensitivity of cell lines to ExoS was gained through SEM studies. When the interaction of strain 388 with the ExoS-sensitive HT-29, LNCaP, and subconfluent NK cells was compared with that of resistant confluent NK cells by SEM, we observed extensions between sensitive cell lines and bacteria which were not as readily apparent in resistant cells. As shown in Fig. 6A to D, elongated microvilli were found to extend from tumor-derived HT-29 and LNCaP cells to the bacteria. Extensions from subconfluent NK cells to 388 bacteria were also readily apparent; however, the appearance of these connections differed from that of HT-29 and LNCaP cells in their adhesive nature and seemingly more secure connection with the bacterium (Fig. 6E and F). In comparison, extensions between resistant confluent NK cells and bacteria were sparse, with only a few connections evident on the lateral aspect of cells (Fig. 6G and H). The appearance of cellular extensions did not relate to ExoS production, nor to the type III secretory/translocation processes of P. aeruginosa, since similar cellular connections were found to extend to strains 388ΔS, 388exs1∷Tn1, the PscC type III secretion mutant, and 388popD∷Tc*, the PopD type III translocation mutant (not shown). The increased frequency of these extensions from cells more sensitive to the effects of ExoS is consistent with the possibility that qualitative differences in cell-bacterium contact may influence the efficiency of ExoS translocation.
FIG. 6.
Scanning electron micrographs of HT-29, LNCaP, and NK cells cocultured with ExoS-producing bacteria. Strain 388 (108 CFU/ml) was cocultured for 3 h with HT-29 or LNCaP cells or subconfluent or confluent NK cell monolayers grown on 12-mm-diameter glass coverslips. Cells were washed, fixed, then coated with gold, and examined in a JEOL JSEM-LV5410 scanning electron microscope. Representative images are shown, with the magnification of each image indicated. (A and B) HT-29 cell images show microvilli extending and then coming in closer contact with the bacterium. (C and D) LNCaP images show similar microvilli extensions to bacteria. (E and F) Subconfluent NK cell images show the basal extension of connections with the bacteria. (G and H) Confluent NK cell images show the general lack of extensions to the bacteria, with the exception of few connections between cells.
DISCUSSION
The requirement of bacterial contact for the translocation of ExoS has posed difficulties in defining ExoS-specific effects on eukaryotic cell function. The development of P. aeruginosa or Yersinia model systems, which allow the effects of ExoS to be differentiated from those of other bacterial factors, confirmed that ExoS is toxic to cultured cells and identified multiple effects on cell function (12, 28, 29). ExoS production was found to cause inhibition of DNA synthesis in both fibroblastic and epithelial cell lines (29), and studies of the HeLa epithelial cell line found ExoS to interrupt actin polymerization, resulting in cell rounding (12). Subsequent studies on the HT-29 epithelial cell line identified additional effects of ExoS on cell matrix adherence and microvillus effacement (28). While it became evident from these studies that bacterially translocated ExoS can exert similar effects on different cell types, the mechanism of the different ExoS cytotoxic effects, as well as an understanding of differential sensitivities of cell lines to the effects of ExoS, remained unknown.
When the effects of ExoS production by P. aeruginosa on different human epithelial cell lines were compared in bacterial-eukaryotic coculture studies, variations in sensitivities to ExoS were observed. Of the cell lines examined, confluent NK cells appeared most resistant to the effects of ExoS on DNA synthesis. These results are consistent with the opportunistic nature of P. aeruginosa infections and the general resistance of healthy epithelial barriers to P. aeruginosa infection. While the CFT1 cell line, which expresses the mutant CFTR, appeared more sensitive to ExoS-specific effects on DNA synthesis and cell morphology (unpublished observation) than confluent NK cells, this sensitivity was not altered by the expression of normal CFTR. This finding suggested that effects of ExoS on cell function were not enhanced by the expression of mutant CFTR protein. Tumor-derived cell lines were consistently highly sensitive to the effects of ExoS, with 10 of 10 cell lines of carcinoma, teratocarcinoma, or neuroblastoma origin examined to date showing a 50% or greater inhibition of DNA synthesis by ExoS. In all cell lines examined, effects of ExoS on DNA synthesis closely correlated with the efficiency of ADP-ribosylation of cellular Ras by ExoS.
When mechanisms of resistance to ExoS were examined in resistant NK epithelial cells and the sensitive HT-29 carcinoma cells, neither differences in the efficiency of general bacterial association nor the eukaryotic cell contact-mediated induction of the ExoS regulon appeared to account for the differences in sensitivity of the two cell lines to ExoS. In comparison, intercellular contact did contribute to the resistance of NK cells to ExoS, with EDTA-disrupted confluent NK monolayers showing increased sensitivity to the effects of ExoS on DNA synthesis and morphology. Consistent with the less efficient polarization and formation of intercellular junctions of confluent HT-29 cells, EDTA treatment did not alter the sensitivity of HT-29 cells to ExoS. These results are in agreement with previous studies of canine kidney epithelial (MDCK) cells which found differentiated MDCK cells to be resistant to cytotoxic effects of ExoS-producing P. aeruginosa (9). Collectively the data suggest that cell sensitivity to bacterially translocated ExoS requires the expression of specific eukaryotic cell factors which become altered as epithelial cells differentiate into confluent polarized monolayers. The direct association of sensitivity to ExoS with the efficiency of Ras modification also supports that intrinsic differences in NK and HT-29 cells to ExoS is, in some part, due to differences in the efficiency of internalization of enzymatically active ExoS.
The possibility that differences other than those related to polarization might exist between the two cell lines in their sensitivity to ExoS was also indicated in morphological analyses. Regardless of the degree of cell confluency, more severe cell rounding was detected in HT-29 cells exposed to strain 388 than in NK cells. The increased cell rounding observed in HT-29 cells suggests that their cytoskeletal structure is more sensitive to the effects of ExoS than that of NK cells. Transformed cells differ from normal epithelial cells in their ability to respond to nonadhesive growth factor signals, which are mediated by motile, non-actin-linked adhesion complexes (15, 23). Growth of normal epithelial cells, in comparison, maintains an anchorage-dependent growth factor requirement, mediated by stable, actin-linked adhesion junctions (2). One possible explanation for the differential sensitivity of HT-29 and NK to morphological alterations could therefore be that non-actin-linked adhesion complexes are more sensitive to the effects of ExoS than those that are actin linked. It also remains possible that more severe effects of ExoS on cell morphology are coordinated in some manner with short-term, non-ExoS-associated morphological alterations caused by strain 388ΔS, which are apparent in coculture studies with HT-29 cells but not NK cells.
While cell confluency and intercellular junction formation contributed to the resistance of NK cells to the effects of ExoS, the molecular basis of the increased sensitivity of tumor cell lines to ExoS remains unexplained. Knowing that Ras is modified by ExoS in vivo, it was initially speculated that the increased sensitivity of tumor cells to ExoS related to their rapid proliferative rate and the corresponding increased effects of ExoS on proliferative signals mediated by activated Ras. Contrary to this premise, no biochemical or functional data which support a relationship between the activational state of Ras and the increased sensitivity to bacterially translocated ExoS have yet been obtained. GDP-bound, GTP-bound, and oncogenic Ras have been found to be modified by ExoS in an identical manner in vitro (1a, 36), and no differences have been observed in the modification of normal and oncogenic Ras in vivo (36). Also, as evident in Fig. 1, bacterially translocated ExoS showed no enhanced inhibition of DNA synthesis relative to the expression of normal or oncogenic Ras by tumor cell lines.
A possible clue as to cellular differences that might contribute to an increased sensitivity to ExoS was revealed from SEM. In examining the interaction between eukaryotic cells and P. aeruginosa, extensions were found to protrude from host cells and contact bacteria. These extensions were more numerous in subconfluent NK and tumor-derived HT-29 or LNCaP cell lines sensitive to ExoS compared to resistant confluent NK cells. In this regard, it is notable that while no quantitative differences in P. aeruginosa interaction with HT-29 or NK cells were detected in cell association assays, qualitative differences in the association of cells with bacteria became apparent by SEM. These results are consistent with studies of type III secretory processes of other bacteria which find cell surface receptors mediating type III translocation to differ from those involved in general bacterial adherence (5, 32). Studies are in progress to identify differences in cell receptor expression or adherence-mediated signaling events between ExoS-sensitive and -resistant cells to help understand the molecular basis of the enhanced sensitivity to ExoS.
Manipulation of eukaryotic cytoskeletal structures is becoming a common theme in bacterial mechanisms of pathogenesis. Among the best-characterized manipulations are the actin pedestal formation by enteropathogenic Escherichia coli (19), membrane ruffling by salmonellae and Shigella flexneri (1, 10), and the polymerized actin tail that propels Listeria monocytogenes to adjacent cells (20). While each bacterium appears to use a slightly different cytoskeletal manipulation strategy, the microvilli or filopodium-like structures attracted to P. aeruginosa most closely resemble the cellular structures associated with Legionella pneumophila (26). Although the induction of these cellular structures does not appear to be directly related to ExoS or the type III secretory system, their correlation with the effects of ExoS on cell function suggests that this structural interaction may be used by P. aeruginosa to facilitate the type III-mediated translocation of ExoS.
In conclusion, these studies provide the first direct comparison of the differential effects of bacterially translocated ExoS on human epithelial cell lines and identify cell culture conditions that can influence the cytotoxic effects of ExoS. Cellular properties affecting sensitivity to ExoS appear similar to those affecting sensitivity to P. aeruginosa, in general. Confluent normal epithelial monolayers appeared most resistant to the effects of ExoS, while subconfluent or disrupted monolayers showed increased sensitivity. Tumor cells and normal epithelial cells involved in cell growth were found to be most sensitive to the effects of ExoS, and this may relate to the ability of P. aeruginosa to utilize cellular growth processes in facilitating the translocation of ExoS.
ACKNOWLEDGMENTS
We thank Dara Frank for help with these studies, Gerald Pier for the CFT1 cell lines, and James Yankaskas for helpful advice on maintaining the CFT1 cell lines. We also thank Carol Moskos for assistance with the SEM studies.
This work was supported by NIH grant AI41694.
REFERENCES
- 1.Adam T, Arpin M, Prevost M C, Gounon P, Sansonetti P J. Cytoskeletal rearrangements and the functional role of T-plastin during entry of Shigella flexneri into HeLa cells. J Cell Biol. 1995;129:367–381. doi: 10.1083/jcb.129.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Barbieri, J. Personal communication.
- 2.Barth A I M, Nathke I S, Nelson W J. Cadherins, catenins and APC protein: interplay between cytoskeletal complexes and signaling pathways. Curr Opin Cell Biol. 1997;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- 3.Bjorn M J, Pavlovskis O R, Thompson M R, Iglewski B H. Production of exoenzyme S during Pseudomonas aeruginosa infections of burned mice. Infect Immun. 1979;24:837–842. doi: 10.1128/iai.24.3.837-842.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn J G, Hazen-Martin D J, Detrisac C J, Sens D A. Electrophysiology and ultrastructure of cultured human proximal tubule cells. Kidney Int. 1988;33:508–516. doi: 10.1038/ki.1988.27. [DOI] [PubMed] [Google Scholar]
- 5.Boyd A P, Sory M-P, Iriarte M, Cornelis G R. Heparin interferes with translocation of Yop proteins into HeLa cells and binds to LcrG, a regulatory component of the Yersinia Yop apparatus. Mol Microbiol. 1998;27:425–436. doi: 10.1046/j.1365-2958.1998.00691.x. [DOI] [PubMed] [Google Scholar]
- 6.Coburn J, Gill D M. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1991;59:4259–4262. doi: 10.1128/iai.59.11.4259-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Detrisac C J, Sens M A, Garvin A J, Spicer S S, Sens D A. Tissue culture of human kidney epithelial cells of proximal tubule origin. Kidney Int. 1984;25:383–390. doi: 10.1038/ki.1984.28. [DOI] [PubMed] [Google Scholar]
- 8.Fleiszig S M J, Evans D J, Do N, Shin S, Mostov K E. Epithelial cell polarity affects susceptibility to Pseudomonas aeruginosa invasion and cytotoxicity. Infect Immun. 1997;65:2861–2867. doi: 10.1128/iai.65.7.2861-2867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleiszig S M J, Wiener-Kronish J P, Miyazaki H, Vallas V, Mostov K E, Kanada D, Sawa T, Yen T S B, Frank D W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis C L, Ryan T A, Jones B D, Smith S J, Falkow S. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature. 1993;364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 11.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 12.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 13.Ganesan A K, Frank D W, Misra R P, Schmidt G, Barbieri J T. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 14.Ganesan, A. K., T. S. Vincent, J. C. Olson, and J. T. Barbieri. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor catalyzed nucleotide exchange. Submitted for publication. [DOI] [PubMed]
- 15.Giancotti F G. Integrin signaling: specificity and control of cell survival and cell cycle progression. Curr Opin Cell Biol. 1997;9:691–700. doi: 10.1016/s0955-0674(97)80123-8. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi K. Role of tight junctions of polarized epithelial MDCK cells in the replication of herpes simplex virus type 1. J Med Virol. 1995;47:323–329. doi: 10.1002/jmv.1890470406. [DOI] [PubMed] [Google Scholar]
- 17.Iglewski B H. Pseudomonas toxins. In: Hardegree M C, Tu A T, editors. Handbook of toxins. Vol. 4. New York, N.Y: Marcel Dekker; 1988. pp. 249–265. [Google Scholar]
- 18.Knight D A, Finck-Barbancon V, Kulich S M, Barbieri J T. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1995;63:3182–3186. doi: 10.1128/iai.63.8.3182-3186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface glycoprotein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 21.Kudoh I, Wiener-Kronish J P, Hashimoto S, Pittet J F, Frank D. Exoproduct secretions of Pseudomonas aeruginosa strains influence severity of alveolar epithelial injury. Am J Physiol Lung Cell Mol Physiol. 1994;267:L551–L556. doi: 10.1152/ajplung.1994.267.5.L551. [DOI] [PubMed] [Google Scholar]
- 22.Kulich S M, Frank D W, Barbieri J T. Expression of recombinant exoenzyme S of Pseudomonas aeruginosa. Infect Immun. 1995;63:1–8. doi: 10.1128/iai.63.1.1-8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lampugnani M G, Dejana E. Interendothelial junctions: structure, signalling and functional roles. Curr Opin Cell Biol. 1997;9:674–682. doi: 10.1016/s0955-0674(97)80121-4. [DOI] [PubMed] [Google Scholar]
- 24.McGuffie E M, Frank D W, Vincent T S, Olson J C. Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1998;66:2607–2613. doi: 10.1128/iai.66.6.2607-2613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicas T I, Frank D W, Stenzel P, Lile J D, Iglewski B H. Role of exoenzyme S in chronic Pseudomonas aeruginosa lung infections. Eur J Clin Microbiol. 1985;4:175–179. doi: 10.1007/BF02013593. [DOI] [PubMed] [Google Scholar]
- 26.Oldman L J, Rogers F G. Adhesion, penetration and intracellular replication of Legionella pneumophila: an in vitro model of pathogenesis. J Gen Microbiol. 1985;131:697–706. doi: 10.1099/00221287-131-4-697. [DOI] [PubMed] [Google Scholar]
- 27.Olsen J C, Johnson L G, Stutts M J, Sarkadi B, Yankaskas J R, Swanstrom R, Boucher R C. Correction of the apical membrane chloride permeability defect in polarized cystic fibrosis airway epithelia following retroviral-mediated gene transfer. Hum Gene Ther. 1992;3:253–266. doi: 10.1089/hum.1992.3.3-253. [DOI] [PubMed] [Google Scholar]
- 28.Olson, J. C., J. E. Fraylick, E. M. McGuffie, K. M. Dolan, T. L. Yahr, D. W. Frank, and T. S. Vincent. Interruption of multiple cellular processes in HT-29 epithelial cells by Pseudomonas aeruginosa exoenzyme S. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 29.Olson J C, McGuffie E M, Frank D W. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect Immun. 1997;65:248–256. doi: 10.1128/iai.65.1.248-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto M, Appay M-D, Simon-Assmann P, Chevalier G, Dracopoli N, Fogh J, Zweibaum A. Enterocyte differentiation of cultured human colon cancer cells by replacement of glucose by galactose in the medium. Biol Cell. 1982;44:193–196. [Google Scholar]
- 31.Rajasekaran A K, Hojo M, Huima T, Rodriguez-Boulan E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J Cell Biol. 1996;132:451–463. doi: 10.1083/jcb.132.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenshine I, Ruschkowski S, Stein M, Reinscheid D J, Mills S D, Finlay B B. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 1996;15:2613–2624. [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson M R, Bjorn M J, Sokol P A, Lile J D, Iglewski B H. Exoenzyme S: an ADP-ribosyltransferase produced by Pseudomonas aeruginosa. In: Smulson M, Sugimura T, editors. Novel ADP-ribosylations of regulatory enzymes and proteins. Amsterdam, The Netherlands: Elsevier; 1980. pp. 425–432. [Google Scholar]
- 34.Trainer D L, Kline T, McCabe F L, Faucette L F, Feild J, Chaikin M, Anzano M, Rieman D, Hoffstein S, Li D-J, Gennaro D, Buscarino C, Lynch M, Poste G, Greig R. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int J Cancer. 1988;41:287–296. doi: 10.1002/ijc.2910410221. [DOI] [PubMed] [Google Scholar]
- 35.Vallis A J, Yahr T L, Barbieri J T, Frank D W. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent, T. S., J. E. Fraylick, E. M. McGuffie, and J. C. Olson. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol. Microbiol., in press. [DOI] [PubMed]
- 37.Woods D E, Sokol P A. Use of transposon mutants to assess the role of exoenzyme S in chronic pulmonary disease due to Pseudomonas aeruginosa. Eur J Clin Microbiol. 1985;4:163–169. doi: 10.1007/BF02013591. [DOI] [PubMed] [Google Scholar]
- 38.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 39.Yankaskas J R, Haizlip J E, Conrad M, Koval D, Lazarowski E, Paradiso A M, Rinehart C A J, Sarkadi B, Schlegel R, Boucher R. Papilloma virus immortalized tracheal epithelial cells retain a well differentiated phenotype. Am J Physiol. 1993;264:C1219–C1230. doi: 10.1152/ajpcell.1993.264.5.C1219. [DOI] [PubMed] [Google Scholar]