Abstract
Grain copper (Cu) concentrations represent a qualitative trait mainly controlled by genetic factors, which may differ between wheat varieties from the Sichuan Basin of China and other areas. However, the differences are poorly understood. Here, we investigated the grain Cu concentration in a remaining heterozygous line population derived from a multiparental recombinant inbred line. The grain Cu concentration varied from 4.25 to 13.44 mg/kg and 3.32 to 7.74 mg/kg over a two-year investigation, and the broad-sense heritability was 0.67. Bulked-segregation analysis revealed three quantitative trait loci on chromosomes 2A (QGr_Cu_Conc-2A), 2B (QGr_Cu_Conc-2B), and 4D (QGr_Cu_Conc-4D). QGr_Cu_Conc-2B is a novel locus, which was further narrowed between KASP-52.32 and KASP-56.57 with an interval of 52.32–56.57 Mb, explaining 17.10% of the phenotypic variation; its potential candidate gene was TraesCS2B03G0196500, encoding a chloroplast thylakoid lumen protein. KASP-52.32 successfully genotyped two common wheat populations, and the grain Cu concentration of CC genotype varieties was significantly higher than that of TT genotype varieties. Meanwhile, the concentrations of chlorophyll and the expression levels of three TaZIP8 and two TaZIP9 in flag leaves were higher in plants with high grain Cu concentration than in plants with low grain Cu concentration. These results provide guidance for understanding the genetic mechanisms underlying grain Cu concentration and may aid in wheat breeding.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11132-1.
Keywords: Wheat, Remaining heterozygous line, Grain copper concentration, Quantitative trait loci
Key messages
A novel major QTL (QGr_Cu_Conc-2B) for grain Cu concentrations in common wheat was identified and the marker KASP-52.32 was developed to screen for high and low grain Cu concentrations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11132-1.
Introduction
Copper (Cu) is an essential microelement for the growth and development of all organisms [1, 2]. For example, as a cofactor for numerous proteins, Cu is involved in cell wall metabolism, photosynthetic electron transport, hormonal signalling, and mitochondrial respiration; thus, it ultimately determines plant fertility, seed size, and protein accumulation [2–4]. Cu is deficient in alkaline or organic soils, which account for more than 30% of the world’s arable soils, and Cu deficiency reduces crop fertility, grain yield, and Cu accumulation in crop grains [4, 5]. The human dietary intake of Cu is mainly derived from crop grains. The average daily intake of Cu for adults over 18 age is 0.62 mg; for kindergarten is appropriate reduction; for pregnant women is 0.72mg; and for lactating women is 1.12 mg [6]. Deficient and excess Cu conditions can lead to a variety of disorders in the human body, such as anaemia, reduced immunity, and bone dysplasia [7]. Therefore, the maintenance of appropriate Cu concentrations in crops is important for food safety and human health.
Common wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is one of the most important food crops worldwide [8]. The Cu concentration in wheat grains not only determines the quality but also potentially influences the safety of wheat crops. However, the grain concentration depends on the uptake of Cu and its redistribution from the shoots as they senesce [5]. The redistribution of Cu to wheat grains is also dependent on the Cu concentration in the soil solution [5]. Grain Cu concentrations vary among wheat varieties, and represent a quantitative trait that is predominantly determined by genetic factors [9–13].
To reveal the genetic factors underlying the grain Cu concentration of wheat, several quantitative trait loci (QTL) have been identified [9–13]. Briefly, two major QTL for grain Cu concentration located on chromosomes 2A and 5A were detected in 243 Chinese wheat cultivars collected from the Yellow and Huai Valley of China, and they explained 10.1–32.6% of the phenotypic variation in grain concentration [5]. Moreover, four QTL were identified on chromosomes 1D, 6A, 6B, and 7D in a biparental population, and they explained 8.66%, 7.55%, 8.21%, and 4.71% of the phenotypic variation in grain Cu concentration, respectively [5]. Using a recombinant inbred line (RIL) derived from a cross between durum wheat (T. turgidum L. var. durum, 2n = 4x = 28, AABB) and wild emmer (T. turgidum L. ssp. dicoccoides, 2n = 4x = 28, AABB), Peleg et al. detected ten major QTL underlying grain Cu concentrations in 1A, 2A, 4A, 5A, 6A, 7A, 7B, 3B, 4B, 6B, and 7B, which explained 1–13% of the phenotypic variation [10]. Ma et al. used 372 Chinese local varieties and 71 exotic varieties to identify three major loci underlying grain Cu concentration, and they were located on chromosomes 1D, 5A, and 5D and explained 0.48–2.89% of the phenotypic variation [13]. One QTL located on chromosome 5B was detected in 330 wheat lines from the CIMMYT Biofortification Breeding Program [9]. In addition, several non-major QTL or loci underlying grain Cu concentrations in wheat have been identified in different wheat varieties located on chromosomes 1A, 1B, 1D, 2A, 2D, 3A, 3B, 3D, 4A, 4B, 5A, 5B, 5D, 6A, 6B, 6D, 7A, 7B, and 7D [9, 12–15]. These results suggest that the genetic factors underlying the grain Cu concentration of wheat differ depending on the wheat cultivar and most detected QTL explain less than 10% of the phenotypic variation.
Because it has the warmest temperatures, shortest sunshine hours, and lowest latitude, the Sichuan Basin is an important and region-specific wheat-producing region in China [16]. Most wheat varieties in the Sichuan Basin are spring-type wheat, which require little or no vernalisation. Thus, the genetic factors underlying grain Cu concentration in wheat varieties in the Sichuan Basin are different from those identified in previous studies; moreover, these genetic factors are poorly understood. Therefore, in this study, we investigated the grain Cu concentration in a remaining heterozygous line (RHL) population derived from a multiparental hybrid RIL. Bulked-segregant analysis sequencing (BSA-Seq) and RNA-sequencing (RNA-Seq) were performed to screen the potential loci underlying grain Cu concentration. Kompetitive allele-specific PCR (KASP) markers were exploited and used to fine-map the loci. The aim of this study was to identify novel QTL for grain Cu concentration and develop a breeder-friendly KASP marker for use in breeding, which will provide guidance for revealing the genetic mechanism of grain Cu concentration and aid in wheat breeding.
Materials and methods
Plant materials
The remaining heterozygous line (RHL) was derived from a multiparental hybrid RIL (F5). The parents were dwarf Polish wheat (DPW, T. polonicum L., 2n = 4x = 28. AABB), Jianyangailanmai (JAM, T. turgidum L., 2n = 4x = 28. AABB), the breeding line K1041, and three modern wheat cultivars: Xikemai11 (XK11), Shumai133 (SM133), and Chuanmai64 (CM64). DPW, originated from Turpan, Xinjiang, China, has several valuable traits, including elongated kernel, dwarf, and low grain cadmium concentration [17–19]. JAM, a landrace in Sichuan, processes a dwarfing gene Rht22 [20]. Both DPW and JAM are valuable germplasms for common wheat improvement. The pedigree of this RIL was that the F2 plants of DPW × JAM were continuously hybridised with XK11 and SM133, and their F3 plants were crossed with the F3 plants of K1041 × CM64.
Two natural common wheat populations were used for marker validation. One natural common wheat population with 134 varieties was primarily collected from the Sichuan and Henan wheat-producing regions of China (NCWSH_China). Another population with 101 varieties was collected from Eurasia between 30°N and 45°N latitude (named NCWE30°N−45°N) and divided into two subgroups based on genetic structure analysis [21]. Most of the varieties collected from Europe and selected germplasms from Asia were classified into subgroup 1, particularly the Bulgarian and Portuguese germplasms, and a portion of the germplasms from Asia and the Middle East were clustered into subgroup 2. Both subgroups exhibited high levels of genetic diversity.
Field experiments
The parents and 103 plants derived from the RHL were grown during the 2022–2023 wheat growing season at the Wenjiang experimental field of Sichuan Agricultural University (2023WJ_RHL). A total of 243 plants generated from 2023WJ_RHL and the parents were grown during the 2023–2024 wheat growing season at the Wenjiang experimental field of Sichuan Agricultural University (2024WJ_RHL). The average of soil Cu concentration of the Wenjiang field was 32.95 mg/kg. The grains of NCWSH_China were sampled during the 2021–2022 wheat growing season at Wenjiang (2022WJ_NCWSH_China). Grains of NCWE30°N−45°N were sampled during the 2018–2019 wheat growing season in the experimental fields at Wenjiang (2019WJ_NCWE30°N−45°N) and Chongzhou (2019CZ_NCWE30°N−45°N) at Sichuan Agricultural University.
All parents and RHLs were planted in a completely randomised block design, and the field was managed using conventional management. Twenty grains of each parent and RHL were planted in rows that were 2 m long and 30 cm apart.
Measurement of chlorophyll concentration and yield traits
At the filling stage, the chlorophyll and carotenoid concentrations of the leaves were measured from high-grain and low-grain Cu concentration plants, as described by Warren [22]. The pigment concentration was calculated according to the method described by Lichtenthaler [23]. At maturity, the yield traits, including grain number per plant, thousand grain weight, grain length, grain width, grain circumference, and grain area, were measured by automatic seed analysis and thousand grain weight meter system (SC-G2; Wseen, Hang Zhou, China).
Determination of grain and leaf Cu concentration
The grain Cu concentrations of 2023WJ_RHL, 2024WJ_RHL, 2022WJ_NCWSH_China, 2019WJ_NCWE30°N−45°N, and 2019CZ_NCWE30°N−45°N and the leaf Cu concentration of 2023WJ_RHL were determined according to the method of high-temperature humidification digestion [24]. Briefly, the leaves and grains were dried to a constant weight and powdered. Each sample (0.20 g) was digested in a mixture of acid (HClO4-HNO3, v/v = 1/4) and diluted by 1% nitric acid and filtered. All samples were loaded into an inductively coupled plasma mass spectrometer (Nexlon2000, PerkinElmer, Waltham, MA, USA) for Cu determination. A certified standard substance (wheat flour [GBW 10011]; National Standard Research Centre of China) was used for quality assurance.
BSA-Seq analysis
To detect the genetic factors underlying grain Cu concentrations, BSA-Seq analysis was performed at Tiansheng Future Biotechnology Company, Chengdu, China. Briefly, pools of plants with grain high- and low-Cu concentration were prepared using equal amounts of genomic DNA from 2023WJ_RHL, and each pool contained 30 plants. Genomic DNA was extracted from each pool using the CTAB method [25]. A total of 4.5 mg of genomic DNA from each pool was fragmented to 300 bp using a Bioruptor UCD-200 sonicator (Diagenode, Denville, USA). Libraries were constructed using Illumina HiSeq Kapa Hyper DNA Library Preparation Kit (Illumina, Shang Hai, China). Sequencing was performed on an Illumina HiSeq Nova platform (Illumina, Shang Hai, China), and each pool was sequenced at a 30 × depth.
The raw sequenced data were screened and filtered to obtain high-quality clean reads using BWA software [26]. Inserted fragments, single nucleotide polymorphism (SNP) variants, and small insertion and deletion (InDel) sites were detected using Picard (http://sourceforge.net/projects/picard/), GATK, and Snp Eff software, respectively [27, 28]. Genome-wide association analysis of grain Cu concentrations was performed using SNP variant density enrichment maps and the G' value statistic algorithm [29].
KASP marker exploitation and QTL analysis
To confirm the QTL for grain Cu concentrations, KASP markers were designed according to the filtered SNP information obtained from BSA-Seq [30], and listed in STable 2. KASP analyses of 2023WJ_RHL and 2024WJ_RHL for QTL mapping and in the natural common wheat population for validation was conducted as described by Liu et al.[30]. Linkage mapping was performed using JoinMap 4.0, and QTL were mapped using IciMapping 4.2.
Candidate gene prediction
Candidate genes within the physical region of the QTL were analysed using WheatOmics 1.0 (http://202.194.139.32/) and then clustered by GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) [31]. To further predict the candidate QTL genes for grain Cu concentration, extremely high- and low-Cu plants from 2023WJ_RHL were used for RNA sequencing (RNA-Seq). RNA-Seq analysis was performed at Novo Gene Bioinformation Technology Co., Ltd. (Beijing, China) [20]. Metal transporter genes with log2 (fold change) > 1 or < −1 and Padj value < 0.005 were considered differentially expressed genes (DEGs). DEGs and/or genes with SNP variant(s) within the QTL were recommended.
Gene amplification
Total RNA was extracted using a Plant RNA Kit (Omega Bio-Tek, Norcross, GA, USA). cDNA was synthesised using an M-MLV First Strand cDNA Synthesis Kit (Invitrogen). PCR primers (STable 7) used for the amplification of the genomic and coding sequences (CDS) of the candidate genes were designed from the Chinese Spring reference genome sequence and used for candidate gene amplification and detection [18].
Data analysis
The broad-sense heritability (H2) was calculated using SAS software (8.0, SAS Institute, USA) [32]. Statistical analyses were performed using SPSS software (version 20.0; IBM Japan Ltd., Tokyo, Japan). Independent samples t-test was used to test for significant differences between two samples at a significance level of P < 0.05 (*), P < 0.01 (**) or P < 0.001 (***). Pearson’s correlation analysis (two-tailed) was used to analyse the correlations between grain Cu concentrations and leaf Cu concentrations, agronomic traits, and yield traits. All graphs were constructed using Origin software (version 64.0; OriginLab USA Ltd., Northampton, USA).
Results
Yield traits
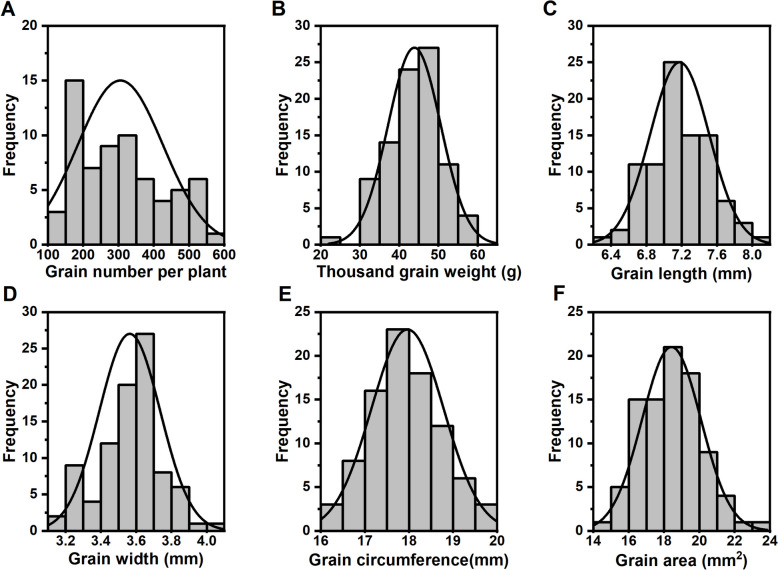
For 2023WJ_RHL, the grain number per plant, thousand grain weight, grain width, grain length, grain circumference, and grain area exhibited continuous variations from 140 to 555, 23.08 to 59.81 g, 3.27 to 3.68 mm, 6.34 to 8.06 mm, 16.01 to 19.92 mm, and 14.88 to 21.62 mm2, with mean values of 305, 43.88 g, 3.56 mm, 7.17 mm, 17.97 mm, and 18.42 mm2, respectively (Fig. 1A-F).
Fig. 1.
Frequency distribution of yield traits in 2023WJ_RHL population. The frequency distribution of grain number per plant (A), thousand kernel weigh (B), grain length (C), grain width (D), grain circumference (E), and grain area (F) in 2023WJ_RHL population
Cu concentration of grains and leaves
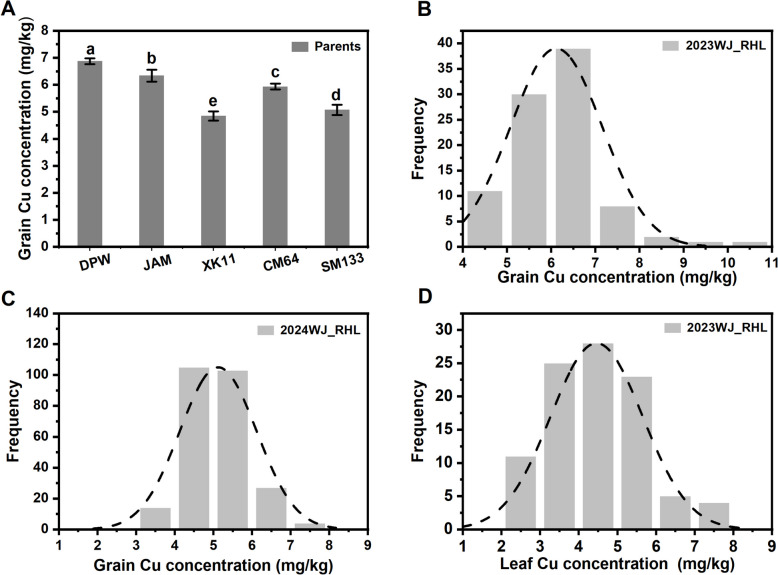
As K1041 is a genetically unstable intermediate material, and its grain Cu concentration was not measured. Grain Cu concentrations of other parents, including DPW, TPW, CM64, SM133, and XK11, showed significantly different average values of 6.87 mg/kg, 6.33 mg/kg, 5.90 mg/kg, 5.07 mg/kg, and 4.84 mg/kg, respectively (Fig. 2A).
Fig. 2.
Grain Cu concentration of parents and RHL populations. A: Grain Cu concentration of parents; B, C: frequency distribution of grain Cu concentration in 2023WJ_RHL and 2024WJ_RHL, respectively; D: frequency distribution of leaf Cu concentration in 2023WJ_RHL
The grain Cu concentrations of the 2023WJ_RHL and 2024WJ_RHL populations exhibited continuous variations, ranging from 4.25 to 13.44 mg/kg (mean value of 6.19 mg/kg) (Fig. 2B) and 3.32 to 7.74 mg/kg (mean value of 5.08 mg/kg), respectively (Fig. 2C). The H2 of the RHL populations was 0.67. These results indicate that the grain Cu concentration in these RHLs is a quantitative trait that is mainly controlled by genetic factors.
Because the grain Cu concentration in wheat is associated with redistribution from leaves as they senesce [5], we also investigated the leaf Cu concentration of 2023WJ_RHL, which varied from 2.05 to 7.92 mg/kg (mean value of 4.46 mg/kg) (Fig. 2D). Correlation analysis revealed that the grain Cu concentration was significantly positively correlated with the leaf Cu concentration in 2023WJ_RHL; however, the correlation coefficient was very low at 0.25. We also analysed the relationship between grain Cu concentrations and tiller numbers and yield traits (including thousand kernel weight, grain width, grain length, grain area, and grain number per plant); however, these traits were not correlated (STable. 1).
BSA-Seq
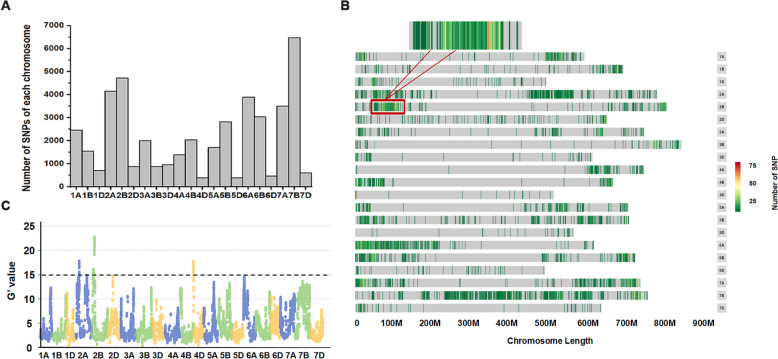
BSA-Seq identified 80,671 SNPs and 3,668 indels. After quality control, 45,001 high-quality SNPs and 1,557 Indels were obtained. In the present study, we used SNPs to exploit KASP markers for QTL screening and validation. All 45,001 SNPs were distributed across all 21 wheat chromosomes, ranging from 386 to 6,478. The largest number of SNPs was found on chromosome 7B (6,478), followed by chromosomes 2B (4,719) and 2A (4,155), and the least number was found on chromosomes 4D (386), 5D (386), and 6D (469) (Fig. 3A; Table 1). SNPs were not uniformly distributed on the chromosomes (except for 7B) but rather showed a centralized distribution on several chromosome fragments (Fig. 3B). For example, out of 4,719 SNPs on chromosome 2B, 2,196 SNPs were distributed within the interval of 49.00–112.0 Mb; 1,102 SNPs out of 4,155 SNPs on chromosome 2A were distributed within the interval of 46.00–91.50 Mb; and 262 SNPs out of 386 SNPs on chromosome 4D were distributed within the interval of 0.50–6.50 Mb (Table 2).
Fig. 3.
BSA-Seq analysis. A: Number of SNPs of each chromosome; B: SNP enrichment diagram; C: G’ value statistics, and chromosomes with G’ value greater than 15 were taken as reference
Table 1.
Number of SNPs on all chromosomes of BSA-Seq
| Number | Chromosomes | Number of SNP variants |
|---|---|---|
| 1 | 1A | 2462 |
| 2 | 1B | 1552 |
| 3 | 1D | 703 |
| 4 | 2A | 4155 |
| 5 | 2B | 4719 |
| 6 | 2D | 877 |
| 7 | 3A | 2002 |
| 8 | 3B | 880 |
| 9 | 3D | 956 |
| 10 | 4A | 1390 |
| 11 | 4B | 2032 |
| 12 | 4D | 386 |
| 13 | 5A | 1707 |
| 14 | 5B | 2818 |
| 15 | 5D | 386 |
| 16 | 6A | 3889 |
| 17 | 6B | 3036 |
| 18 | 6D | 469 |
| 19 | 7A | 3504 |
| 20 | 7B | 6478 |
| 21 | 7D | 600 |
Table 2.
Candidate region statistics for SNP locus association analysis
| Chromosome | SNP enrichment diagram | G’ value | Recommended interval (Mb) | |
|---|---|---|---|---|
| Interval (Mb) | SNP number | Interval (Mb) | ||
| 2A | 46.00–91.50 | 1,102 | 158.74–166.37 | 46.00–166.37 |
| 2B | 49.00–112.00 | 2,196 | 67.29–145.97 | 49.00–145.97 |
| 4D | 0.50–6.50 | 262 | 0.60–8.20 | 0.50–8.20 |
Based on a G' value algorithm with a value of more than 15, three candidate regions correlated with grain Cu concentrations were detected on chromosome 2A (158.74–166.37 Mb), 2B (67.29–145.97 Mb), and 4D (0.60–8.20 Mb) (Fig. 3C; Table 2). By combining the results recommended by the SNP numbers and G' value algorithm, three candidate intervals located on chromosome 2A (46.00–166.37 Mb), 2B (49.00–145.97 Mb), and 4D (0.50–8.20 Mb) were recommended for governing grain Cu concentrations (Table 2). These intervals were individually named QGr_Cu_Conc-2A, QGr_Cu_Conc-2B, and QGr_Cu_Conc-4D. Since QGr_Cu_Conc-2B presented the highest G' value and largest number of SNPs, it was further confirmed and narrowed using the 2023WJ_RHL and 2024WJ_RHL populations.
QGr_Cu_Conc_2B fine mapping
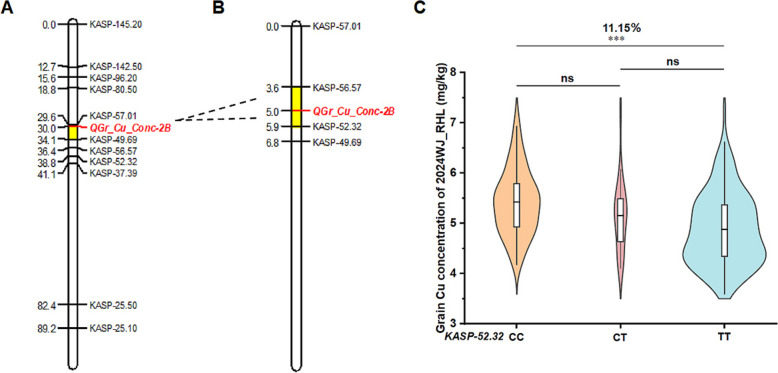
To validate and further narrow down the interval of QGr_Cu_Conc_2B, 40 pairs of KASP markers were designed based on the SNP information. 11 pairs of markers were successfully used to genotype 103 plants of the 2023WJ_RHL population and constructed a genetic map (Fig. 4A). The genetic map spanned 89.17 cM, and the order of the markers on the genetic map was highly consistent with that in the wheat physical map; however, three markers (KASP-49.69, KASP-56.57 and KASP-52.32) showed disorder (Fig. 4A). QTL screening revealed that QGr_Cu_Conc_2B was mapped between markers KASP-49.69 and KASP-57.00 in the 2023WJ_RHL population, and it had a limit of detection (LOD) score of 6.20 and explained 28.07% of the phenotypic variation (Fig. 4A; Table 3). In addition, a QTL governing leaf Cu concentration was mapped between the markers KASP-80.50 and KASP-96.20 with an LOD value of 2.56, and it explained 11.89% of the phenotypic variation (Fig. 4A; Table 3).
Fig. 4.
Fine mapping of QGr_Cu_Conc-2B. A: QTL of grain and leaf concentration in 2023WJ_RHL. B: QTL of grain concentration in 2024WJ_RHL. C: comparison of the significance of the 2024WJ_RHL population based on marker KASP-52.32. Asterisk indicates statistically significant differences ***P < 0.001 by independent samples t-test, and ns indicates no significant difference between the two groups
Table 3.
QTL controlling the grain and leaf Cu concentration
| Trait | Genetic position (cM) | Left Marker | Right Marker | LOD | PVE (%) |
|---|---|---|---|---|---|
| Grain Cu concentration in 2023WJ_RHL | 30.00 | KASP-49.69 | KASP-57.01 | 6.20 | 28.07 |
| Grain Cu concentration in 2024WJ_RHL | 5.00 | KASP-52.32 | KASP-56.57 | 10.03 | 17.10 |
| Leaf Cu concentration in 2023WJ_RHL | 16.00 | KASP-80.50 | KASP-96.20 | 2.56 | 11.89 |
We found that the disorders among the markers KASP-49.69, KASP-56.57, and KASP-52.32 probably resulted from the lower number (103) of plants in the 2023WJ_RHL population. To confirm and narrow down the candidate region of QGr_Cu_Conc_2B, 243 plants (2024WJ_RHL) were generated from the 2023WJ_RHL population. Four pairs of KASP markers, i.e. KASP-49.69, KASP-56.57, KASP-52.32, and KASP-57.00, were used to genotype 2024WJ_RHL population. The marker order was KASP-49.69, KASP-52.32, KASP-56.57, and KASP-57.00, which is the same as the physical order (Fig. 4B). Meanwhile, QGr_Cu_Conc_2B was accurately mapped between markers KASP-52.32 and KASP-56.57 in the 2024WJ_RHL population (Fig. 4B). Compared to the reference Chinese Spring gene (RefSeq v2.1), its physical interval was 52.32–56.57 Mb and LOD value was 10.03, and it explained 17.10% of the phenotypic variation (Table 3). According to the genotype of the tightly linked marker KASP-52.32, most plants in the 2024WJ_RHL population were classified into two groups with genotypes CC (82) and TT (145), and the remaining 16 plants had heterozygous genotypes (CT). The average grain Cu concentration of the CC genotype group was 5.42 mg/kg, which was extremely significantly higher (11.15%) than that of the TT genotype group (4.88 mg/kg); moreover, the grain Cu concentration of the CT genotype group was not significantly different from either CC or TT genotypes (Fig. 4C).
Validation of KASP-52.32 in two natural common wheat populations
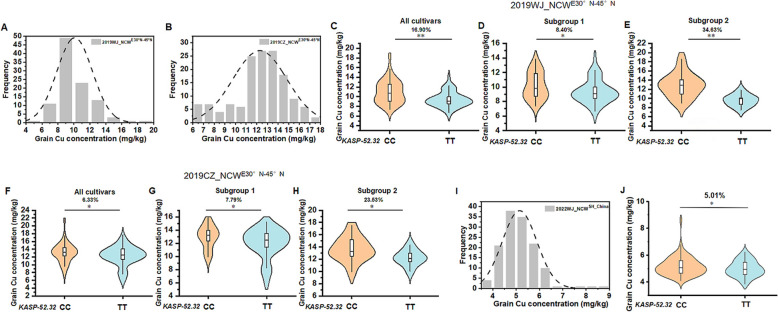
Because the KASP marker KASP52.32 was tightly linked with QGr_Cu_Conc_2B in the 2024WJ_RHL population, it was further validated using two natural common wheat populations. Mineral analysis found that grain Cu concentrations of NCWE30°N−45°N showed continuous variations from 5.97 to 18.50 mg/kg (mean value of 10.33 mg/kg) in 2019WJ_NCWE30°N−45°N and from 6.00 to 17.67 mg/kg (mean value of 12.23 mg/kg) in 2019CZ_NCWE30°N−45°N populations, respectively (Fig. 5A and B). In both environments, the varieties collected from Georgia and Syria had higher grain Cu concentrations while those collected from Greece had lower grain Cu concentrations. Based on the genotypes of the KASP marker KASP-52.32, the average grain Cu concentration of the varieties with the CC genotype was significantly higher than that of the varieties with the TT genotype (Fig. 5C and F). At the subgroup level, similar results were found, with the average grain Cu concentration of varieties with the CC genotype significantly higher than that of varieties with the TT genotype in 2019WJ_NCWE30°N−45°N (Fig. 5D and E) and 2019CZ_NCWE30°N−45°N (Fig. 5G and H). The 2022WJ_ NCWSH_China population also showed continuous variations in grain Cu concentrations (Fig. 5I), and the average grain Cu concentration was higher in varieties with the CC genotype than the TT genotype (Fig. 5J). However, although parents of the RHL populations, including DPW, TPW, CM64, SM133, and XK11, were CC genotypes, their grain Cu concentrations were significantly different, suggesting that the TT genotype may have originated from the intermediate material K1041. These results confirm that the KASP marker KASP-52.32 is tightly linked to grain Cu concentration and could be used for molecular marker-assisted selection in common wheat.
Fig. 5.
Validation of KASP marker KASP-52.32 in NCWE30°N−45°N and NCWSH−China populations. A and B: Frequency distribution of grain Cu concentration separately in 2019WJ_NCWE30°N−45°N (A) and 2019CZ_NCWE30°N−45°N (B); C, D and E: comparison of the significance in the 2019WJ_NCWE30°N−45°N population (C) and its subgroups (D and E) based on the marker KASP-52.32; F, G and H: comparison of the significance in the 2019CZ_NCWE30°N−45°N population and its subgroups based on the marker KASP-52.32; I: frequency distribution of grain Cu concentration in 2022WJ_NCWSH_China; J: comparison of the significance in the 2022WJ_NCWSH_China population. Asterisk indicates statistically significant differences *P < 0.05 and **P < 0.01 by independent samples t-test
Candidate genes
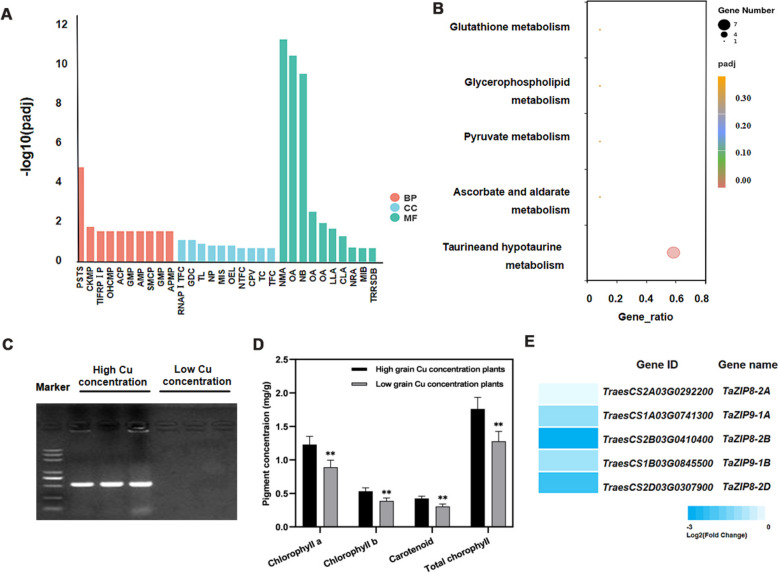
According to the reference genome of Chinese Spring, there were 80 annotated genes in the physical interval of QGr_Cu_Conc_2B but no genes encoding a metal transporter. Among these 80 candidate genes, 47 genes are high confidence gens (STable 6). GO analysis revealed that these genes were enriched in enzyme activities, cellular metabolism, signal transduction, transcription factors, and chloroplast anabolic pathways (Fig. 6A). KEGG analysis revealed that these genes were mainly involved in acid metabolism (Fig. 6B). RNA-Seq analysis revealed that five genes within the interval of QGr_Cu_Conc_2B, including TraesCS2B03G0188400, TraesCS2B03G0188300, TraesCS2B03G0192300, TraesCS2B03G0196200, and TraesCS2B03G0196500, had SNP between plants with grain high- and low-Cu concentrations. They individually encode a UDP-glucoronosyl and UDP-glucosyl transferase, NADP-dependent alkenal double-bond reductase, disease resistance protein, vacuolar protein sorting-associated protein 54, and chloroplast thylakoid lumenal protein. Gene cloning of TraesCS2B03G0196500 using leaf cDNA revealed that TraesCS2B03G0196500 could be cloned from plants with grain high-Cu concentration but not from those with grain low-Cu concentration (Fig. 6C). The concentrations of chlorophyll a, chlorophyll b, carotenoids and total chlorophyll of leaves were higher in high grain Cu concentration plants than in low grain Cu concentration plants (Fig. 6D). Meanwhile, RNA-Seq revealed that only five metal transporter genes in flag leaves showed differential expression between a high grain Cu concentration plant and a low grain Cu concentration plant; the expression levels of three TaZIP8 and two TaZIP9 were higher in high grain Cu concentration plant than in low grain Cu concentration plant (Fig. 6E).
Fig. 6.
Candidate gene analysis. A: GO pathways of candidate genes at the interval of QGr_Cu_Conc-2B; among them, biological progress (BP) pathways include phosphorelay signal transduction system (PSTS), cellular ketone metabolic process (CKMP), transcription initiation from RNA polymerase I promoter (TIFRPIP), organic hydroxy compound metabolic process (OHCMP), aldehyde catabolic process (ACP), glycerol metabolic process (GMP), alditol metabolic process (AMP), small molecule catabolic process (SMCP), glycerol-3-phosphate metabolic process (GMP) and alditol phosphate metabolic process (APMP); cellular component (CC) pathways include RNA polymerase I transcription factor complex (PIRPITFC), glycerol-3-phosphate dehydrogenase complex (GDC), thylakoid lumen (TL), nucleolar part (NP), mitochondrial intermembrane space (MIS), organelle envelope lumen (OEL), nuclear transcription factor complex (NTFC), cytoplasmic vesicle part (CVP), tethering complex (TC) and transcription factor complex (TFC); Molecular Function (MF) pathways include N,N-dimethylaniline monooxygenase activity (NMA), oxidoreductase activity and acting on paired donors (OA), NADP binding (NB), oxidoreductase activity and acting on the CH-CH group of donors with NAD or NADP as acceptor (OA), oxidoreductase activity, acting on the CH-CH group of donors (OA), lactoylglutathione lyase activity (LLA), carbon–sulfur lyase activity (CLA), nutrient reservoir activity (NRA), manganese ion binding (MIB) and transcription regulatory region sequence-specific DNA binding (TRTSDB); B: KEGG pathways of candidate genes at the interval of QGr_Cu_Conc-2B; C: amplification of TraesCS2B03G0196500 between high and low grain Cu concentration plants; full-length gels are presented in Supplementary Fig. 2; D: chlorophyll content of leaf in the high and low grain Cu concentration; E: different expression of metal transporter genes in flag leaves between the high grain Cu concentration plants and the low grain Cu concentration plants
Discussion
According to the agricultural industry standard (NY 861–2004) released by the Ministry of Agriculture of the People’s Republic of China, the recommended grain Cu concentration for wheat is 3–10 mg/kg [33]. In the present study, we found that the grain Cu concentrations of the 2023WJ_RHL, 2024WJ_RHL, and 2022WJ_NCWSH_China populations were consistent with the recommended range, except for a few lines of 2023WJ_RHL, which had Cu concentrations greater than 10 mg/kg (Fig. 2B, C, and 5I). Most varieties in the NCWE30°N−45°N population collected from Georgia and Syria exhibited higher grain Cu concentrations when grown in the Wenjiang and Chongzhou experimental fields (Fig. 5A and B), although they did not exceed the limit of 20 mg/kg recommended by the United States Environment Protection Agency [34]. Many common wheat varieties in China also have higher grain Cu concentrations; for example, 18.53% of wheat varieties collected from the Yellow and Huai Valleys accumulated more than 10 mg/kg of Cu in their grains [11]. We found different frequencies of grain Cu concentrations between the 2023WJ_RHL and 2024WJ_RHL populations and between the 2019WJ_NCWE30°N−45°N and 2019CZ_NCWE30°N−45°N populations. This result indicates that grain Cu concentrations are influenced by environmental factors, such as soil properties [35, 36], which likely caused certain varieties to accumulate more than 10 mg/kg Cu [11, 13].
Ma et al. [13] found that grain Cu concentrations in common wheat was significantly negatively correlated with thousand grain weight. However, we found no correlation between grain Cu concentrations and yield traits such as thousand kernel weight, grain width, grain length, grain area, or grain number per plant (STable 1). These results indicate that the characteristics of grain Cu concentrations depend on the wheat variety. Because the values of H2 (0.61) for the 2023WJ_RHL and 2024WJ_RHL populations were greater than 0.50, the grain Cu concentration in those RHL populations were mainly controlled by genetic factors.
BSA-Seq has been successfully used to identify candidate intervals for wheat traits, such as plant height and defective kernels [20, 37]. In this study, three candidate intervals underlying grain Cu concentrations of common wheat, QGr_Cu_Conc-2A, QGr_Cu_Conc-2B, and QGr_Cu_Conc-4D, were identified using BSA-Seq (Fig. 3, Table 2). The region of QGr_Cu_Conc-4D (0.50–8.20 Mb) has been identified as a non-major QTL for grain Cu concentration of wheat [12]; and QGr_Cu_Conc-2A (46.00–166.37 Mb) overlapped with one QTL on chromosome 2A was reported [15] and it explained 15.70% of grain Cu concentration variation. QGr_Cu_Conc-2B (49.00–145.97 Mb) was further narrowed to the interval of 52.32–56.57 Mb using KSAP markers KASP-52.32 and KASP-56.57 (Fig. 4B); it explained 17.10% of the phenotypic variation, which was higher than that of other reported QTL explaining the grain Cu concentration variation. Thus, QGr_Cu_Conc-2B is a novel QTL for grain Cu concentrations in wheat, because QTL for grain Cu concentration in wheat have not been previously reported in this interval [9–13, 15]. QGr_Cu_Conc-2B was linked to a QTL for leaf Cu concentrations on chromosome 2B (Fig. 4A), which supports that the grain Cu concentration was positively correlated with the leaf Cu concentration (STable 1).
Similar to grain Fe and Zn concentrations [38], grain Cu concentrations in wheat is also dependent on redistribution from flag leaves during senescence [5]. In this study, we found that the concentrations of chlorophyll a, chlorophyll b, carotenoids, and total chlorophyll in leaves were higher in plants with high grain Cu concentration than in plants with low grain Cu concentration (Fig. 6D), which indicated the senescence of flag leaves in plants with low grain Cu concentration. Meanwhile, the candidate gene of QGr_Cu_Conc-2B, TraesCS2B03G0196500, which encodes a chloroplast lumenal protein, could be cloned from the cDNA of the flag leaf of grain high-concentration Cu plants, but not from the cDNA of the flag leaf of grain low-concentration Cu plants (Fig. 6C). Chloroplast lumenal proteins play crucial roles in regulating thylakoid biogenesis and the activity and turnover of photosynthetic protein complexes, especially photosystem II, and then mediate leaf senescence [39]; and the thylakoid compartments of plant chloroplasts are a vital destination for Cu [40]. On the other hand, during flag leaf senescence, several metal transporter genes, such as members of the ZIP family, showed downregulation to limit the remobilisation of Fe and Zn from leaves to grains [38, 41]. Here, the expression levels of three TaZIP8 and two TaZIP9 were consisted with chlorophyll concentration, TraesCS2B03G0196500 expression and grain Cu concentration. Their homologous genes OsZIP1 transport Cu and OsZIP9 transport Zn for Zn uptake [42, 43]. Thus, we theoretically understood that the lack of TraesCS2B03G0196500 in flag leaves promoted the senescence to downregulate the expression levels of TaZIP8 and TaZIP9, which probably limited Cu transport from flag leaves to grain via the phloem. However, further studies are required to confirm whether TraesCS2B03G0196500 is the target gene of QGr_Cu_Conc-2B.
Conclusion
In this study, a novel major QTL (QGr_Cu_Conc-2B) within the interval 52.32–56.57 Mb was mapped on chromosome 2B, and it is responsible for grain Cu concentration in wheat. We also used its tightly linked marker KASP-52.32 to successfully track grain Cu concentrations in two other natural common wheat populations. Our results also showed that grain Cu concentration was not correlated with yield traits such as grain number per plant or thousand grain weight. Collectively, our results provide a new guide for understanding the genetic mechanisms of grain Cu concentration in wheat and improving this trait.
Supplementary Information
Acknowledgements
This study was supported by the National Natural Science Foundation of China [Nos. 32301752 and 32272032], the project funded by China Postdoctoral Science Foundation [No 2022M712291], and the State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China [SKL-ZY202204].
Authors’ contributions
YW, YC and YH planned and designed the research. YH performed the experiments. QZ, XL, and DL analyzed the data. YW, YH and YC wrote the paper. YW, YC, YZ, JZ, DW, LS, XF, HK, and HZ discussed the results and commented on the paper. All authors read, revised, and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China [Nos. 32301752 and 32272032], the project funded by China Postdoctoral Science Foundation [No 2022M712291], and the State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China [SKL-ZY202204].
Data availability
The data underlying this article are available in the article and in its online supplementary material.Sequence data that support the findings of this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2024), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA020853) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Declarations
Ethics approval and consent to participate
No license was required for the samples collected in this study. All materials were stored at the Triticeae Research Institute of Sichuan Agricultural University, Chengdu, Sichuan Province, China. Wheat grains were collected in accordance with institutional, national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yi Wang, Email: wangyi@sicau.edu.cn.
Yiran Cheng, Email: chengyiran@sicau.edu.cn.
Reference
- 1.Kumar V, Pandita S, Sidhu G, Sharma A, Khanna K, Kaur P, Bali S, Setia R. Copper bioavailability, uptake, toxicity, and tolerance in plants: A comprehensive review. Chemosphere. 2021;262:127810. [DOI] [PubMed] [Google Scholar]
- 2.Shabbir Z, Sardar A, Shabbir A, Abbas G, Shamshad S, Khalid S, Natasha, Murtaza G, Dumat C, Shahid M. Copper uptake, essentiality, toxicity, detoxification, and risk assessment in soil-plant environment. Chemosphere. 2020;259:127436. [DOI] [PubMed] [Google Scholar]
- 3.Moreira A, Moraes L, Melo T, Heinrichs R, Moretti L. Management of copper for crop production. Adv Agron. 2022;173:257–97. [Google Scholar]
- 4.Sheng H, Jiang Y, Rahmati M, Chia J, Dokuchayeva T, Kavulych Y, Zavodna T, Mendoza P, Huang R, Smieshka L, Miller J, Woll A, Terek O, Romanyuk N, Piñeros M, Zhou Y, Vatamaniuk OK. YSL3-mediated copper distribution is required for fertility, seed size and protein accumulation in Brachypodium. Plant Physiol. 2021;186:655–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garnett TP, Graham RD. Distribution and remobilization of iron and copper in wheat. Ann Botany. 2005;95:817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Expert Committee on Dietary Nutrient Reference Intakes 2023 of the Chinese Nutrition Society. Dietary Reference Intake of Nutrients for Chinese Residents. Acta Nutrimenta Sinica. 2024;45:521–4. [Google Scholar]
- 7.Prohaska JR. Impact of copper deficiency in humans. Ann N Y Acad Sci. 2014;1314:1–5. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Ren W, Han L. Screening and characterisation of wheat germplasm against stem collapse in the southern part of the Huanghuai wheat region. J South Agric. 2023;13:9007. [Google Scholar]
- 9.Cu ST, Guild G, Alison N, Velu G, Singh R, Stangoulis J. Genetic dissection of zinc, iron, copper, manganese and phosphorus in wheat (Triticum aestivum L.) grain and rachis at two developmental stages. Plant Sci. 2020;291:110338. [DOI] [PubMed] [Google Scholar]
- 10.Peleg Z, Cakmak I, Ozturk L, Yazici A, Jun Y, Budak H, Korol AB, Fahima T, Saranga Y. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat × wild emmer wheat RIL population. Theor Appl Genet. 2009;119:353–69. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L, Pan Y, Dong Z, Zheng Y, Liu J, Geng J, Ren Y, Zhang N, Chen F. Investigation and genome-wide association study of grain copper content in Chinese common wheat. Cereal Sci. 2020;95:102991. [Google Scholar]
- 12.Liu Y, Chen Y, Yang Y, Zhang Q, Fu B, Cai J, Guo W, Shi L, Wu J, Chen Y. A thorough screening based on QTLs controlling zinc and copper accumulation in the gran of different wheat genotypes. Environ Sci Pollut Res. 2021;28:15043–54. [DOI] [PubMed] [Google Scholar]
- 13.Ma J, Qi S, Yuan M, Zhao D, Zhang D, Feng J, Wang J, Li W, Song C, Wang T, Zeng Q, Wu J, Han D, Jiang L. A genome-wide association study revealed the genetic variation and candidate genes for grain copper content in bread wheat (Triticum aestivum L). Food Funct. 2022;13:5177. [DOI] [PubMed] [Google Scholar]
- 14.Rahman MM, Azirun SM, Boyce AN. Enhanced accumulation of copper and lead in amaranth (Amaranthus paniculatus), Indian mustard (Brassica juncea) and sunflower (Helianthus annuus). PLoS ONE. 2013;8: e62941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi R, Tong Y, Jiang R, Zhang F, Zou C. Characterization of quantitative trait loci for grain minerals in hexaploid wheat (Triticum aestivum L). J Integr Agric. 2013;12:1512–21. [Google Scholar]
- 16.Li M, Tang Y, Li C, Wu X, Tao X, Liu M. Climate warming causes changes in wheat phenological development that benefit yield in the Sichuan Basin of China. Eur J Agron. 2022;139: 126574. [Google Scholar]
- 17.Chai S, Yao Q, Zhang X, Xiao X, Fan X, Zeng J, Sha L, Kang H, Zhang H, Li J, Zhou Y, Wang Y. The semi-dwarfing gene Rht-dp from dwarf Polish wheat (Triticum polonicum L.) is the Green Revolution gene Rht-B1b. BMC Genomic. 2021;22:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chai S, Yao Q, Liu R, Xiang W, Xiao X, Fan X, Zeng J, Sha L, Kang H, Zhang H, Long D, Wu D, Zhou Y, Wang Y. Identification and validation of a major gene for kernel at the P1 locus in Triticum polonicum. Crop J. 2022;10:387–96. [Google Scholar]
- 19.Cheng Y, Liu R, Yang T, Yang S, Chen J, Huang Y, Long D, Zeng J, Wu D, Kang H, Fan X, Sha L, Zhang H, Zhou Y, Wang Y. Genetic factors of grain cadmium concentration in Polish wheat (Triticum polonicum L). Plant Physiol. 2024;196:979–95. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Bao Y, Yao Q, Long D, Xiao X, Fan X, Kang H, Zeng J, Sha L, Zhang H, Wu D, Zhou Y, Zhou Q, Wang Y, Cheng Y. Fine mapping of the reduced height gene Rht22 in tetraploid wheat landrace Jianyangailanmai (Triticum turgidum L). Theor Appl Genet. 2022;135:3643–60. [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Tan B, Liu H, Zhu W, Xu L, Wang Y, Fan X, Sha L, Zhang H, Zeng J, Wu D, Jiang Y, Hu X, Chen G, Zhou Y, Kang H. Genetic diversity and population structure of Asian and European common wheat accessions based on genotyping by sequencing. Front Genet. 2020;11:580782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren CR. Rapid Measurement of Chlorophylls with a Microplate Reader. J Plant. 2008;31:1321–32. [Google Scholar]
- 23.Lichtenthaler HK. Chlorophylls and carotenoids: pigment photosynthetic biomembranes. Methods Enzymol. 1987;148:350–82. [Google Scholar]
- 24.Chen X, Yang S, Ma J, Huang Y, Wang Y, Zeng J, Li J, Li S, Long D, Xiao X, Sha L, Wu D, Fan X, Kang H, Zhang H, Zhou Y, Cheng Y. Manganese and copper additions differently reduced cadmium uptake and accumulated in dwarf Polish wheat (Triticum polonicum L). J Hazard Mater. 2023;448:130998. [DOI] [PubMed] [Google Scholar]
- 25.Sharp PJ, Kreis M, Shewry PR, Gale MD. Location of β-amylase sequence in wheat and its relatives. Theor Appl Genet. 1988;75:286–90. [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Bur rows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Rudenet DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly MA, DePristo M. The Genome Analysis Toolkit: a MapReduce frame work for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magwene PM, Willis JH, Kelly JK. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput Biol. 2011;7: e1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Mullan D, Zhang A, Liu H, Liu D, Yan G. Identification of KASP markers and putative genes for pre-harvest sprouting resistance in common wheat (Triticum aestivum L). Crop J. 2023;11:549–57. [Google Scholar]
- 31.Siavoshi A, Taghizadeh M, Dookhe E, Piran M. Gene expression profiles and pathway enrichment analysis to identification of differentially expressed gene and signaling pathways in epithelial ovarian cancer based on high-throughput RNA-seq data. Genomics. 2022;114:161–70. [DOI] [PubMed] [Google Scholar]
- 32.Wu D, Sato K, Ma JF. Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol. 2015;208:817–29. [DOI] [PubMed] [Google Scholar]
- 33.Chu H, Mu W, Dang H, Wang T, Sun R, Hou S, Huang T, Huang Q, Shi M, Wang Z. Evaluation on concentration and nutrition of micro-elements in wheat grains in major wheat production regions of China. Acta Agron Sinica. 2022;48:2853–65. [Google Scholar]
- 34.USEPA. Environmental Protection Agency, Region 9, Preliminary remediation goals. 2012. http://www.epa.gov/region9/superfund/prg/.
- 35.Deng D, Wu S, Wu F, Deng H, Wong H. Effects of root anatomy and Fe plaque on arsenic uptake by rice seedlings grown in solution culture. Environ Pollut. 2010;158:2589–95. [DOI] [PubMed] [Google Scholar]
- 36.DiDonato RJ, Roberts LA, Sanderson T, Eisley RB, Walker EL. Arabidopsis Yellow Stripe-Like 2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. 2004;39:403–14. [DOI] [PubMed] [Google Scholar]
- 37.Tang H, Dong H, Guo X, Cheng M, Li M, Chen Q, Yuan Z, Pu Z, Wang J. Identification of candidate gene for the defective kernel phenotype using bulked segregant RNA and exome capture sequencing methods in wheat. Front Plant Sci. 2023;14: 1173861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearce S, Tabbita F, Cantu D, Buffalo V, Avni R, Vazquez-Gross H, Zhao R, Conley CJ, Distelfeld A, Dubcovksy J. Regulation of Zn and Fe transporters by the GPC1 gene during early wheat monocarpic senescence. BMC Plant Biol. 2014;14:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarvi S, Gollan PJ, Aro EM. Understanding the roles of the thylakoid lumen in photosynthesis regulation. Front Plant Sci. 2013;4:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banci L, Bertini I, Ciofi-Baffoni S, Kandias N, Robinson N, Spyroulias G, Su X, Tottey S, Vanarotti M. The delivery of copper for thylakoid import observed by NRM. Proc Natl Acad Sci USA. 2006;103:8320–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wani SH, Gaikwad K, Razzaq A, Samantara K, Kumar M, Govindan V. Improving zinc and iron biofortification in wheat through genomic approaches. Mol Biol Rep. 2022;49:8007–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Feng S, Zhang B, Wang M, Cao H, Rono JK, Chen X, Yang Z. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 2019;19:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang M, Li Y, Liu Z, Tian J, Liang L, Qiu Y, Wang G, Du Q, Cheng D, Cai H, Shi L, Xu F, Lian X. A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J. 2020;103:1695–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.Sequence data that support the findings of this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2024), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA020853) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.