Abstract
Lyme disease begins at the site of a tick bite, producing a primary infection with spread of the organism to secondary sites occurring early in the course of infection. A major outer surface protein expressed by the spirochete early in infection is outer surface protein C (OspC). In Borrelia burgdorferi sensu stricto, OspC is highly variable. Based on sequence divergence, alleles of ospC can be divided into 21 major groups. To assess whether strain differences defined by ospC group are linked to invasiveness and pathogenicity, we compared the frequency distributions of major ospC groups from ticks, from the primary erythema migrans skin lesion, and from secondary sites, principally from blood and spinal fluid. The frequency distribution of ospC groups from ticks is significantly different from that from primary sites, which in turn is significantly different from that from secondary sites. The major groups A, B, I, and K had higher frequencies in the primary sites than in ticks and were the only groups found in secondary sites. We define three categories of major ospC groups: one that is common in ticks but very rarely if ever causes human disease, a second that causes only local infection at the tick bite site, and a third that causes systemic disease. The finding that all systemic B. burgdorferi sensu stricto infections are associated with four ospC groups has importance in the diagnosis, treatment, and prevention of Lyme disease.
Lyme disease is a progressive multisystem disorder and is the most common vector-borne disease in both North America and Europe. This disease was first described as a focus of pediatric arthritis patients in Old Lyme, Conn. (34). A few years later, the association of this syndrome with the bite of the deer tick, Ixodes scapularis, led to the identification of Borrelia burgdorferi as the causative agent (4). As culture isolation of the bacterium from clinical and field samples became more efficient, Baranton and colleagues (1) described three pathogenic genospecies, B. burgdorferi sensu stricto, Borrelia afzelii, and Borrelia garinii. These are part of a species complex, B. burgdorferi sensu lato, which consists of at least 10 different genospecies (25, 26, 38). All three pathogenic species are found in Europe, but in North America, B. burgdorferi sensu stricto is the only pathogenic genospecies. Each of the three pathogenic genospecies is associated with distinct clinical manifestations (39). This implies that differences in genospecies, and perhaps even strains, play an important role in the wide array of clinical manifestations observed in Lyme disease.
Outer surface protein A (OspA) is the major outer surface protein expressed when B. burgdorferi resides in ticks. As an infected tick begins to feed on a mammal, the synthesis of OspA is repressed (7) and the synthesis of OspC is induced (30). Thus, in early infection, OspC is the major outer membrane protein expressed by B. burgdorferi (10, 24). Even though OspC has been demonstrated to have limited surface exposure (6, 19), OspC is a potent immunogen. Immunization with OspC is protective against tick-transmitted Borrelia infection (12). However, the protection is limited to the immunizing OspC allele, as challenge with heterologous isolates results in infection (27).
OspC is very diverse (16). Wang et al. (40) found 13 ospC alleles of B. burgdorferi sensu stricto within a small tick population, while Livey et al. (17) found 34 alleles in 76 B. burgdorferi sensu lato isolates. Wang et al. (40) defined major ospC groups by using the observation that ospC alleles are either very similar (less than 2% sequence divergence) or very different (greater than 8% sequence divergence), with most having greater than 14% sequence divergence. Using these parameters and including all available sequences from GenBank plus the alleles from the tick population, 19 major ospC groups, groups A through S (Table 1), were defined and strains were associated with each group listed in Wang et al. (40). Strain B31 belongs to group A, strain HB19 belongs to group I, strain N40 belongs to group E, and strain 297 is a mixture of groups I and K. In the present study, we describe two new groups (groups T and U).
TABLE 1.
Alignment of major ospC groups with ospC alleles identified by SSCP analysis
| Major ospC group | ospC allele | GenBank accession no.a | No. of the indicated ospC group in isolates fromb:
|
||
|---|---|---|---|---|---|
| Ticks | Human skin | Secondary sites of human infection | |||
| A | 1 | AF029860 | 17 | 23 | 21 |
| B | 2 | AF029861 | 17 | 19 | 4 |
| C | 3 | AF029862 | 11 | 3 | 0 |
| D | 4 | AF029863 | 10 | 1 | 0 |
| E | 5, 7 | AF029864 | 6 | 1 | 0 |
| F | 6 | AF029865 | 9 | 0 | 0 |
| G | 8 | AF029867 | 5 | 7 | 0 |
| H | 9 | AF029868 | 7 | 6 | 0 |
| I | 10 | AF029869 | 1 | 9 | 3 |
| J | 11, 18 | AF029870 | 3 | 7 | 0 |
| K | 12, 13 | AF029871 | 6 | 32 | 16 |
| L | L42899 | 2 | 0 | 0 | |
| M | 14 | U01892 | 1 | 3 | 0 |
| N | 15 | L42899 | 1 | 3 | 0 |
| O | X84778 | 0 | 1 | 0 | |
| Pc | U91796 | 1 | 0 | 0 | |
| Qc | U91790 | 1 | 0 | 0 | |
| Rc | U91791 | 2 | 0 | 0 | |
| Sc | U91793 | 1 | 0 | 0 | |
| T | 16 | AF065143 | 0 | 1 | 0 |
| U | 17 | AF065144 | 0 | 2 | 0 |
A single GenBank sequence for each type is given as an example. The complete list of sequences is given in reference 40.
The number of each major ospC group observed. This includes both SSCP data and data from the literature, including GenBank.
B. burgdorferi sensu stricto groups P through S are found only in Europe. Groups R and S are excluded from the analysis because nearly identical ospC alleles are found in B. afzelii and B. garinii, showing that these groups were recently created by cross-species transfer.
There is evidence that ospC has been transferred between strains (7a, 17) and even between genospecies (40). This is not true of Borrelia chromosomal genes (8, 20). We find that ospA and ospC alleles in B. burgdorferi sensu stricto are almost completely linked (40). This suggests that once an ospC allele has been transferred into a particular background, there is little or no selection for another similar recombinant. Thus each major ospC group represents a clonal population descended from a single recombination. We call each major ospC group a clone and use the terms group and clone interchangeably.
In this study, we compared the frequency distributions of these major groups from ticks with those from human skin and those from skin with those isolated from secondary sites of human infection. While many groups found in ticks are found in primary skin lesions, the frequency distributions are significantly different. All groups are found more or less commonly in ticks but in the primary skin lesions, only four groups are commonly found, and others are found only rarely or not at all. More importantly, only four ospC groups are found in secondary sites.
MATERIALS AND METHODS
Borrelia strains.
B. burgdorferi strains (n = 140) were isolated from primary erythema migrans lesions, blood, or cerebrospinal fluid (CSF) of patients seen at the Lyme Disease Center at Stony Brook, N.Y., the Lyme Disease Diagnostic Center at New York Medical College, Valhalla, N.Y., or the private practices of the two collaborating physicians on the eastern end of Long Island or were obtained from the Centers for Disease Control. All patients met the Centers for Disease Control surveillance definition for Lyme disease (5). Patients were asked to read and sign informed consent statements. Isolates from skin specimens, blood, and CSF were obtained by standard techniques (2, 3, 41). Punch biopsies were taken from the advancing border of the erythema migrans lesion and incubated in BSK-H medium (Sigma, St. Louis, Mo.) at 34°C to create a culture. We have shown previously that there is little culture bias as determined by direct analysis of biopsy tissue compared to culture isolates (31). This differs from the isolation of B. burgdorferi from unfed ticks (22). In addition, 22 B. burgdorferi sensu stricto ospC sequences were retrieved from GenBank. The tick data used was either from GenBank or from the study of Wang et al. (40).
DNA isolation.
For isolation of genomic DNA, log-phase cells were harvested by centrifugation (Eppendorf model 5415 C) at 10,000 rpm for 30 min at 4°C. The bacterial pellet was resuspended in Tris-saline buffer (10 mM Tris [pH 7.5], 150 mM NaCl), pelleted, and resuspended in TNE (10 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA). Freshly prepared lysozyme (20 mg/ml in TNE), sodium dodecyl sulfate (10%), and proteinase K (20 mg/ml) were then added, and the mixture was incubated at 50°C for 1 h prior to RNase treatment. DNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in TE buffer.
PCR.
The ospC gene was amplified by PCR as described previously (40). A 340-bp fragment of the 5′ end of ospC, suitable in size to be studied by single-stranded conformational polymorphism (SSCP) analysis, was amplified by using forward primer 5′-AAA GAA TAC ATT AAG TGC GAT ATT-3′ and reverse primer 5′-CAA TCC ACT TAA TTT TTG TGT TAT TAT-3′. The 3′ end of ospC, 314 bp in size, was amplified by using forward primer 5′-TTG TTA GCA GGA GCT TAT GCA ATA TC-3′ and reverse primer 5′-GGG CTT GTA AGC TCT TTA ACT G-3′.
Amplification was processed in 50 μl of a solution containing Perkin-Elmer Cetus 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl), 2.5 mM MgCl2, deoxynucleoside triphosphates at 0.2 mM per nucleotide, 2.5 U of Taq polymerase (Perkin-Elmer Cetus), and 0.5 M (each) primer. The amplification reaction was carried out for 40 cycles in a DNA thermal-cycler (PTC-100; MJ Research, Inc., Watertown, Mass.) with an amplification profile of denaturation at 95°C for 40 s, annealing at 54°C for 35 s, and extension at 72°C for 1 min, after an initial denaturation step at 96°C for 2 min. Negative controls were included in each experiment to control for contamination.
Cold SSCP analysis.
Although a large number of molecular techniques are available to characterize genetic variation, we chose cold SSCP analysis based on its exquisitely high detection rate of DNA polymorphisms and point mutations at a variety of positions in DNA fragments (23). Single point mutations have been detected in fragments up to 800 bp long (21). However, there is evidence that the ability of SSCP analysis to detect mutations begins to decline significantly as PCR fragments approach 400 bp in size (14). To achieve high efficiency of detection of nucleotide polymorphism, the length of the PCR products used in our study was 340 bp from the 5′ half and 314 bp from the 3′ half of ospC.
Amplified ospC gene fragments from all 140 strains were analyzed for genetic variations by the cold SSCP protocol described by Hongyo et al. (15). Briefly, 5 to 15 μl of the PCR product was added to a mixture containing 4 μl of 5× TBE Ficoll sample buffer (NOVEX, San Diego, Calif.) and 0.4 μl of 1 M methylmercury hydroxide (Alfa Aesaer, Ward Hill, Mass.). The amount of the PCR product used for the SSCP analysis was estimated after visualizing the PCR product on an agarose gel with ethidium bromide. The sample mixture was heated to 95°C for 4 min and then plunged into ice prior to loading the entire 20 μl into the gel sample well. The sharpest bands were observed when the sample was applied to a precast 20% TBE gel (NOVEX). Electrophoresis was performed by using a temperature-controlled electrophoresis system (ThermoFlow ETC unit; NOVEX) with 1.25× TBE running buffer. SSCP runs were conducted at a constant temperature of 8°C for 17 h at 240 V to reveal discernable mobility shifts. Gels were stained with 0.5 μl of ethidium bromide per ml in 1× TBE buffer for 25 min and destained in distilled water for 30 min. Stained bands were viewed with a UV (340-nm) staining box. Samples that showed more than two SSCP bands were reamplified to determine whether the bands found were real alleles or the product of PCR artifacts. Side-by-side SSCP analysis was performed to detect even slight shifts in electrophoretic mobility.
DNA sequencing.
The ospC genes of representatives of each mobility class were reamplified. Double-stranded PCR fragments were purified by agarose gel electrophoresis and subjected to automated DNA sequencing using fluorescent dideoxy terminator chemistry and the forward and reverse primers originally used for PCR amplification.
Statistical analysis.
Chi-square analysis of contingency tables was performed. This tests for significant difference in frequency distributions. The tables were 2 by n where n is the number of major ospC groups that were distinguished. The average expected number in each element of the table must be approximately six or greater for an unbiased test (42). This means that the number of observations should be greater than six times 2n. When the expected average number was less than six, the major ospC groups with the lowest number in the sample were combined until the number of observations was approximately equal to or greater than 12n.
RESULTS
ospC mobility classes in human B. burgdorferi isolates.
We initiated our analysis of ospC sequence diversity by accumulating a large collection of isolates of B. burgdorferi sensu stricto (n = 132) from patient samples of skin, blood, and CSF (Table 2). Each was propagated in vitro and used as a source of DNA for analysis. The ospC genotype of each strain was determined by cold SSCP analysis of the 5′ end (340 bp) of the gene and was later confirmed by SSCP analysis of the 3′ end (314 bp) of ospC. In all B. burgdorferi isolates, the genetic variation at the 5′ end of the gene corresponded to the variation at the 3′ end. At least two representatives of each SSCP mobility class were subsequently sequenced. The sequences of the same mobility classes were identical in all samples and each mobility class had a unique sequence. Therefore, the sensitivity and specificity of SSCP analysis were 100%. This result confirms the findings of previous studies, which also utilized SSCP analysis to determine genetic variability in B. burgdorferi (13, 40). Each SSCP mobility class was designated an allele. Wang et al. (40) recently described 13 ospC alleles. In this study, we present an additional five ospC mobility classes, OC14 to -18. OC14 has the same ospC sequence as the ospC in strain 2591 (GenBank accession no. U01892) and OC15 has the same sequence as the ospC in strain 26815 (accession no. L42897).
TABLE 2.
B. burgdorferi sensu stricto isolates classified according to geographic region, source, and major ospC group
| Geographical source of isolates | Biological source (no. of isolates) | Major ospC groupa (no. observed) |
|---|---|---|
| New York (Westchester County) | Skin (62) | A (8), B (10), D (1), E (1), G (6), H (4), I (4), J (6), K (18), M (2), U (2) |
| Blood (14) | A (5), B (2), I (1), K (6) | |
| CSF (1) | B (1) | |
| New York (eastern Long Island) | Skin (40) | A (10), B (6), C (2), G (1), I (4), K (12), N (3), O (1), T (1) |
| Blood (1) | A (1) | |
| CSF (10) | A (3), K (7) | |
| Connecticut | Skin (2) | C (1), K (1) |
| Blood (3) | A (1), I (1), K (1) | |
| CSF (3) | I (1), K (2) | |
| California | Skin (4) | A (1), H (1), I (1), M (1) |
| Blood (2) | A (2) | |
| CSF (1) | A (1) | |
| Arkansas | Blood (1) | A (1) |
| Wisconsin | Skin (1) | J (1) |
| Pennsylvania | Skin (1) | H (1) |
| CSF (1) | K (1) | |
| Texas | Skin (1) | A (1) |
| Michigan | Skin (2) | B (1), K (1) |
| Europe (Austria, Denmark, France, Germany, Italy) | Skin (5) | A (3), B (2) |
| Blood (2) | A (2) | |
| CSF (4) | A (4) | |
| Synovial fluid (1) | A (1) |
A major ospC group is defined as a set of sequences with more than 98% similarity within each group and less than 92% sequence similarity between groups.
Nucleotide sequence accession numbers.
OC16, OC17, and OC18 are alleles that have not been previously described in the literature. The GenBank accession numbers for these ospC alleles are AF065143, AF065144, and AF097915, respectively.
Multiple infections.
Of the 132 primary isolates from patients with Lyme disease in this study, most contained only a single strain. Seven skin isolates and one CSF isolate contained two different strains as determined by SSCP analysis, thus giving a total of 140 different strains. The ospC allele pairs found in multiply infected erythema migrans biopsy specimens were OC1 and OC12, OC1 and OC14, OC2 and OC3 (two sets), OC2 and OC12 (two sets), and OC8 and OC18. CSF isolate NY940657 contained ospC alleles OC1 and OC12. For CSF isolate 297, which was isolated in Connecticut, there are two ospC sequences published in GenBank, L42893 (analogous to OC10) and U08284 (analogous to OC12). The pairwise difference of ospC sequences of both strains is 16.4%, suggesting central nervous system infection with two different strains in this isolate. Overall, 5.5% of the isolates in our collection contained two strains. Wang et al. (40) found that 50% of ticks collected on the eastern end of Long Island were infected with multiple strains. This suggests that exposure to multiple strains in a single tick bite is common and raises the possibility that different strains are differentially pathogenic.
To these 140 strains for which we determined the ospC allele, we added 22 strains of known ospC sequence from GenBank to give a total of 162. The majority of these strains were obtained from either eastern Long Island (n = 51) or Westchester County, N.Y. (n = 77), with the remainder from other areas in the United States (n = 22) and Europe (n = 12) in which the strains are endemic. The isolates were divided into those from sites of the primary infection, erythema migrans skin lesions (n = 118), and secondary sites, where the infection had disseminated (n = 44). This latter group included 20 isolates from CSF, 23 isolates from blood, and 1 isolate from synovial fluid.
Major ospC groups in human B. burgdorferi isolates.
The pairwise differences of ospC sequences within B. burgdorferi sensu stricto fall into two classes. Either a pair is different in less than 2% of its nucleotides or it is different in more than 8% of its nucleotides. Wang et al. (40) defined 19 major ospC groups, designated A to S. The present analysis revealed two additional ospC groups, designated T and U. OC16 represents major group T and OC17 represents major group U (Table 1). The lowest pairwise differences of group T and U from other major ospC groups are 16.1% and 20.5%, respectively.
OC18 is an allele in group J along with OC11. They have 99.6% sequence identity and were both found in skin isolates. OC11 was isolated five times and OC18 was isolated twice from erythema migrans biopsies. Otherwise, all ospC alleles found in this sample were from different major groups.
B. burgdorferi clones are differentially pathogenic.
We wished to show that clones are differentially pathogenic by comparing the frequencies of the various major groups in ticks, in the initial infection in the skin, and in disseminated infections. Our samples are not ideally suited for the comparisons that we wish to make since most of the samples are from southeastern New York state. However, we do not think this is a serious problem because the frequency distributions seem to be similar across the species range.
The strains in GenBank and the literature for which the ospC sequence has been determined are widely sampled from the entire geographic range of the species and were chosen irrespective of whether they were from ticks or humans. They give a small but random sample of the frequencies of the major ospC groups in ticks and humans. The frequency of the major ospC groups from human isolates is significantly different from the frequency found in ticks (Table 3) (17, 18). This is also true locally. Table 4 shows that the frequency distribution of strains from skin from eastern Long Island is significantly different from tick strains collected in the same area.
TABLE 3.
Contingency table for major ospC group data for lesions and ticks as published in GenBanka and the literature
| Isolate source (n) | No. of observations in isolates of major ospC groupb:
|
|||
|---|---|---|---|---|
| A | B | K | Combc | |
| Erythema migrans lesions (22) | 11 | 4 | 4 | 3 |
| Ixodes scapularis ticks (25) | 5 | 5 | 1 | 14 |
GenBank data include all ospC submissions made before February 1998.
The numbers of observations for lesions and for ticks were significantly different (χ2 = 11.1321 with 3 degrees of freedom; P < 0.025).
Combined major groups (Comb) are defined by individual frequencies of 0.025 or less and include groups D, E, F, I, J, L, M, O, Q, and R.
TABLE 4.
Contingency table for major ospC groups retrieved from lesions and ticks collected from eastern Long Island
| Isolate source (n) | No. of observations in isolates of major ospC groupa:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | F | G | H | I | K | Combb | |
| Erythema migrans lesions (46) | 13 | 6 | 2 | 0 | 0 | 1 | 0 | 4 | 16 | 4 |
| Ixodes scapularis ticks (74) | 12 | 12 | 11 | 9 | 6 | 5 | 7 | 1 | 5 | 6 |
The numbers of observations for lesions and for ticks were significantly different (χ2 = 36.3 with 9 degrees of freedom; P < 0.001).
Combined major groups (Comb) are defined by individual frequencies of 0.025 or less and include groups E, J, N, and O.
To support the contention that geography is an insignificant component in the differences in the frequencies of the various groups in ticks and humans, we performed the two following tests. The frequency distributions of human isolates from eastern Long Island and the human isolates from GenBank and the literature are not significantly different (χ2 = 7.07 with 7 degrees of freedom; P > 0.25). Further, the frequency distribution of the tick samples from eastern Long Island is not significantly different from the GenBank and literature sample (χ2 = 10.84 with 8 degrees of freedom; P > 0.25). Our analysis shows that the frequency distribution of alleles in human isolates is significantly different from the distribution in ticks. This difference does not depend upon geography. Thus, we combined the data from diverse locations for the tests below. There is one caveat to the postulate that there is no geographical substructure. When the local and GenBank tick samples are compared without combining any groups, the test is significant (χ2 = 31.94 with 15 degrees of freedom; P < 0.01). The importance of this result is that it shows that not all groups are found at every sampling site. Consequently, there could be a group not present in southeastern New York state which is infectious.
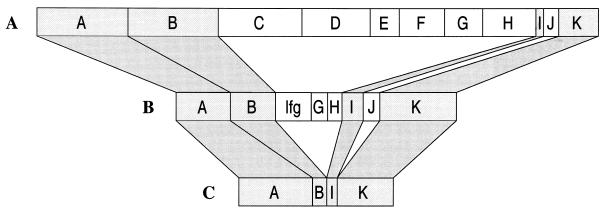
Collective analysis of all ospC groups presented in this study shows that most groups are found in both ticks and humans (Table 1). However, while the different groups are in near-equal frequency in ticks (40), major groups A and K predominate in humans (Fig. 1).
FIG. 1.
The frequency distribution of major ospC groups among B. burgdorferi isolates from eastern Long Island Ixodes scapularis ticks (n = 72) (A), erythema migrans lesions (n = 118) (B), and secondary sites of infection (n = 44) (C). The percentage of group A plus K was 23% in the tick isolates, 47% in the skin isolates, and 84% in the secondary sites. The lengths of the bars in the figure reflect these differences by holding the length of the combined A and K groups constant. In the skin, groups C, D, E, M, N, O, T, and U have been combined since their individual frequencies are 0.025 or less. This combination of groups has been labeled lfg for low-frequency groups. When combined, these groups make up 12.7% of the total number of strains.
Comparison of the frequencies of the various groups in primary and secondary sites.
We further investigated the pattern of pathogenicity of the various clones by comparing their frequencies in the primary site of infection, the skin, with their frequencies in secondary sites. Only four major groups (A, B, I, and K) were found in both the skin and secondary sites (Fig. 1B and C). All other major groups were found only in the skin. When all groups with three or fewer isolates are combined to give the lfg (Fig. 1B), a 2-by-8 contingency test comparing the frequency distributions of skin isolates versus those of secondary site isolates indicates significance at P < 0.005 (Table 5). When no groups are combined, a 2-by-15 contingency test is still significant (χ2 = 24.07 with 14 degrees of freedom; P < 0.05). The distribution of strains from primary and secondary sites leads us to postulate that only major groups A, B, I, and K cause disseminated disease. We will refer to these clones as invasive and to other clones as noninvasive.
TABLE 5.
Contingency table for major ospC groups retrieved from lesions and from disseminated infection from all sources
| Isolate source (n) | No. of observations in isolates of major ospC groupa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| A | B | G | H | I | J | K | Combb | |
| Erythema migrans lesions (118) | 23 | 19 | 7 | 6 | 9 | 7 | 32 | 16 |
| Disseminated infections (44) | 21 | 4 | 0 | 0 | 3 | 0 | 16 | 0 |
Numbers of isolates from lesions and from disseminated infections were significantly different (χ2 = 23.6 with 7 degrees of freedom; P < 0.005).
Combined major groups (Comb) are defined by individual frequencies of 0.025 or less and include groups C, D, E, M, N, O, T, and U.
The above test showed only that the frequency distributions are significantly different, not that only four clones cause disseminated disease. We can support this contention by comparing the likelihood of the hypothesis that by chance we simply didn’t find other clones in the sample with the hypothesis that only these four clones cause disseminated disease. If we assume that each infection was caused by a single strain, then the likelihood ratio is a ratio of two binomial samplings. What is the probability that of 44 samples there are only the four clones? The frequency of the four clones in the skin is 0.7034, so the probability ratio of the two hypotheses is 0.703444/144. This means the first hypothesis is 1.4 × 10−68 less likely than the second.
However, we know that the other clones are less pathogenic than the four disseminating clones. The relative infectivity (R) can be estimated from the following equation:
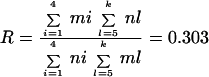 |
where i is groups A, B, I, and K, l is all other groups, m is the number of each group in ticks, and n is the number of each group in skin (primary infection). Assuming the same relative infectivity from skin to disseminated infection, then 0.303 = (83/35)[x/(44 − x)], where x is the number of expected strains which do not belong to the four disseminating clones which are found disseminated; if x = 6.4, the expected frequency of disseminated strains is (44 − 6.4)/44, or 0.8536. The likelihood of this hypothesis compared to the hypothesis that there are only four disseminating clones is 0.0009. Thus the hypothesis that there are only four disseminating clones is significantly more likely, supporting the conclusion that there are only four disseminating groups, A, B, I, and K.
DISCUSSION
Four invasive groups.
The difference in the frequencies of the three pathogenic genospecies in ticks and human infection has led to the hypothesis that the different genospecies are differentially pathogenic (25, 39). Here we report that the different clones of B. burgdorferi sensu stricto, defined by ospC groups, are differentially pathogenic. Some groups very rarely if ever cause human disease, e.g., ospC groups D, E, F, and L. Some groups cause a local infection at the tick bite site but not systemic disease, e.g., ospC groups G, H, J, and T. Finally, there are some groups which are responsible for systemic disease; these are ospC groups A, B, I, and K. Our findings indicate that all systemic B. burgdorferi sensu stricto infections in humans are caused by strains in these four ospC groups.
The data of Steere et al. (33) are consistent with the results of this study. They found that 20% of untreated erythema migrans lesions cleared spontaneously without causing any systemic complications (33). This is not significantly different (P = 0.25 for a 2-by-2 contingency test with double dichotomy) from the percentage of noninvasive strains found in the skin, suggesting that the erythema migrans lesions that clear spontaneously are caused by noninvasive clones.
There is extensive genetic and antigenic diversity of OspC in all three pathogenic genospecies of B. burgdorferi sensu lato (17, 18, 25, 36, 40). In this study, we show that ospC alleles are linked to both infectivity and invasiveness and that invasiveness is confined to a small number of ospC clones. One might expect that systemic disease in the other pathogenic genospecies is also caused by a small number of clones. Thus, ospC is a good marker for human pathogenicity and perhaps its determinator. These findings have important implications not only for our understanding of the pathogenesis of this disease but for its diagnosis and prevention.
Possibility of other invasive clones.
Although we feel that our findings are broadly applicable, two factors, the limited geographic distribution of most of the isolates studied and the source of the invasive isolates, primarily blood and CSF, leave open the possibility that additional invasive groups may be defined. This possibility in no way diminishes the overall importance of the finding that only a limited subset of the ospC groups are associated with invasive disease.
Spirochetemia is a transient phenomenon but is presumably key in seeding secondary skin sites, the heart, joints, and nervous system, where these Borrelia organisms cause the secondary and tertiary clinical manifestations of Lyme disease. All four invasive groups were found in isolates from blood and CSF. The one joint isolate belonged to group A. However, it can be inferred that groups not found in the blood will not be found in the joints, since most if not all dissemination of Borrelia to secondary sites is via blood.
Population genetics analysis.
This paper relies on population genetic analysis, which is an unusual approach to the study of infectious disease. Thus, we would like to point out the strengths and weaknesses of this analysis. Normally, model organisms are used as substitutes for experiments on humans. However, this substitution works only as long as the properties of the model organism and of humans are the same for the studied phenomena. In this study, the human immune system plays a critical role which is expected to be different from the immune response in model organisms, particularly the mouse. Humans are accidental and usually dead-end hosts, while the mouse is a critical host reservoir. Consequently, one cannot do direct experiments but must reach an inferential conclusion from survey data. The field of population genetics has developed sound procedures for reaching conclusions from survey data. However, this analysis still relies on correlation and does not explain cause.
Another major assumption in the analysis is that we are dealing with clones and using ospC as a marker for the clones. If ospC is the determinator of the observed differences in pathogenesis, this assumption is unimportant. However, if other genes are involved, then the assumption that there are clones with the various alleles of different genes linked in clone complexes is important. Ryan et al. (29) have challenged the clone concept. They discovered an interesting phenomenon. In the mixed culture of a major ospC group L strain and a group G strain, the group L strain is selected in the mammal host and the group G strain is selected in the tick. These strains are different not only at the ospC locus but also at ospA and in their plasmid profile. In their discussion, Ryan et al. reject this explanation, instead suggesting that the genome of one strain contains this diversity—two ospC alleles, two ospC alleles, and plasmid content rearranged depending upon the environment. Their reasons for rejecting the mixed culture explanation are as follows. (i) The culture was colony purified before the experiment started. However, the plating efficiency was so low that the colonies that grew up could be mixed colonies where both strains were required for growth. Thus, one need not assume that the culture is a single clone arising from a single cell. (ii) Passage of the rarer strain in the mammal to the tick is unlikely. However, cultures were started using multiple ticks and 10 ticks were used to passage the Borrelia to a new mammalian host. Thus, even if all infected ticks do not pick up the rarer strain, which is then selected in the infected ticks, both the culture and the mammal are likely to contain this strain. Also, we have shown that different clones are not picked up randomly but are likely to be picked up together (28). It is clear that the ospA and ospC alleles are tightly linked even though they are on different plasmids (40). Thus, if the invasiveness is caused by allelic variation at another locus, this variation is likely to be tightly linked to the ospC variation.
OspC as a virulence factor.
OspC could be the important virulence factor that determines the differences in pathogenicity between clones. Using immunoglobulin M antibodies from patients with neuroborreliosis, Mathiesen et al. (19) recently showed that the C-terminal region of OspC is exposed on the surface of the spirochete. The C-terminal polyproline II-like helix, which is conserved in all B. burgdorferi strains, and its adjacent residues are likely to be important for host-parasite interactions. OspC is not required for growth in culture and maintenance of the cp26 plasmid (37).
Practical implications.
Our results have important implications for the development of vaccines and serum-based diagnostic tests for Lyme disease. Two independent trials of first-generation vaccines for the prevention of Lyme disease recently studied the efficacy and safety of a vaccine that is based on recombinant OspA (32, 35). However, a vaccine that consists of recombinant OspA may require frequent booster immunizations, since high antibody titers are necessary to eliminate B. burgdorferi in the tick vector (9). Natural infection with B. burgdorferi does not elicit antibody response to this outer membrane protein, as it does to OspC. Our findings suggest that all systemic B. burgdorferi sensu stricto infections in humans are associated with four ospC groups. Because vaccines against OspC are known to be protective, but have been limited by the diversity of OspC (27), our finding could lead to a highly protective vaccine. We propose that a vaccine that includes these four forms of OspC would be a second level of protection against disseminated infection of the B. burgdorferi spirochete. SSCP analysis may provide a rapid and powerful tool to monitor vaccine efficacy by detecting rare or new invasive ospC groups.
New diagnostic assays based on major ospC groups A, B, I, and K could identify those at risk for progressive illness. A number of investigators have used OspC as a serodiagnostic antigen for early Lyme disease (10, 11, 24). In these tests, the use of OspC as a diagnostic antigen gave highly specific but not sensitive results. However, these studies included only one B. burgdorferi strain and therefore only one type of OspC. Routine tests for the diagnosis of Lyme disease also use a single-strain protocol and therefore a single ospC allele for detection of antibody to the spirochete. Given that OspC proteins are antigenically variable (27), individuals infected with one strain may produce an antibody response that is not reactive with an OspC protein from a different major group. We therefore predict that antibody detection using antigen preparations incorporating a proper mix of invasive clones of B. burgdorferi will be much more sensitive than the present single-strain protocols.
ACKNOWLEDGMENTS
This work was supported by grants from the State of New York (10505-4-3R-97) and the National Institutes of Health (AIAR 37256). G.S. was supported by an Erwin Schroedinger research fellowship of the Austrian Science Fund (J1543-MED).
We gratefully thank D. Qiu, D. S. Dunkin, M. Randesi, J. Medalle, B. Lade, L. Butler, M. L. Hayes, and P. Munoz for technical assistance; J. Kieleczawa for assistance with sequencing PCR samples; K. Ginther-Davis (Centers for Disease Control), I. Schwartz (New York Medical College), B. W. Berger, P. K. Coyle, R. Kahlish, and A. Natarro (all from SUNY at Stony Brook) for providing strains used in this study; B. Rannala for discussions about the statistical tests; and R. Gasser for thoughtful comments on the manuscript.
REFERENCES
- 1.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J-C, Assous M, Grimont P A D. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 2.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 3.Berger B W, Johnson R C, Kodner C, Coleman L. Cultivation of Borrelia burgdorferi from erythema migrans lesions and perilesional skin. J Clin Microbiol. 1992;30:359–361. doi: 10.1128/jcm.30.2.359-361.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease: a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control. Case definitions for infectious conditions under public health surveillance. Morbid Mortal Weekly Rep. 1997;46:20–21. [PubMed] [Google Scholar]
- 6.Cox D L, Akins D R, Bourell K W, Lahidenne P, Norgard M V, Radolf J D. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc Natl Acad Sci USA. 1996;93:7973–7978. doi: 10.1073/pnas.93.15.7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Silva A M, Telford S R, Brunet L R, Barthold S W, Fikrig E. Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J Exp Med. 1996;183:271–275. doi: 10.1084/jem.183.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Dykhuizen, D. E. Unpublished results.
- 8.Dykhuizen D E, Polin D S, Dunn J J, Wilske B, Praec-Mursic V, Dattwyler R J, Luft B J. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc Natl Acad Sci USA. 1993;90:10163–10167. doi: 10.1073/pnas.90.21.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fikrig E, Telford S R, Barthold S W, Kantor F S, Spielman A, Flavell R A. Elimination of Borrelia burgdorferi from vector ticks feeding on OspA-immunized mice. Proc Natl Acad Sci USA. 1992;89:5418–5421. doi: 10.1073/pnas.89.12.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung B P, McHugh G L, Leong J M, Steere A C. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of immunoglobulin M response in the serodiagnosis of early infection. Infect Immun. 1994;62:3213–3221. doi: 10.1128/iai.62.8.3213-3221.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerber M A, Shapiro E D, Bell G L, Sampieri A, Padula S. Recombinant outer surface protein C ELISA for the diagnosis of early Lyme disease. J Infect Dis. 1995;171:724–727. doi: 10.1093/infdis/171.3.724. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore R D, Jr, Kappel K J, Dolan A C, Burkot T R, Johnson B J B. Outer surface protein C (OspC), but not P39, is a protective immunogen against a tick-transmitted Borrelia burgdorferi challenge: evidence for a conformational epitope in OspC. Infect Immun. 1996;64:2234–2239. doi: 10.1128/iai.64.6.2234-2239.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman D S, Wang P, Wang I-N, Bosler E, Luft B, Dykhuizen D E. Multiple infections of Ixodes scapularis ticks by Borrelia burgdorferi as revealed by single-strand confirmation polymorphism. J Clin Microbiol. 1996;34:652–656. doi: 10.1128/jcm.34.3.652-656.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- 15.Hongyo T, Buzard G S, Calvert R J, Weghorst C M. “Cold SSCP”: a simple, rapid, and non-radioactive method for optimized single-stranded conformation polymorphism analyses. Nucleic Acids Res. 1993;21:3637–3642. doi: 10.1093/nar/21.16.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jauris-Heipke S, Fuchs R, Motz M, Preac-Mursic V, Schwab E, Soutschek E, Will G, Wilske B. Genetic heterogeneity of the genes coding for the outer surface protein C (OspC) and the flagellin of Borrelia burgdorferi. Med Microbiol Immunol. 1993;182:37–50. doi: 10.1007/BF00195949. [DOI] [PubMed] [Google Scholar]
- 17.Livey I, Gibbs C P, Schuster R, Dorner F. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol Microbiol. 1995;18:257–269. doi: 10.1111/j.1365-2958.1995.mmi_18020257.x. [DOI] [PubMed] [Google Scholar]
- 18.Masuzawa T, Komikado T, Yanaghihara Y. PCR-restriction fragment length polymorphism analysis of the ospC gene for detection of mixed culture and for epidemiological typing of Borrelia burgdorferi sensu stricto. Clin Diagn Lab Immunol. 1997;4:60–63. doi: 10.1128/cdli.4.1.60-63.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathiesen M J, Holm A, Christiansen M, Blom J, Hansen K, Østergaard S, Theisen M. The dominant epitope of Borrelia garinii outer surface protein C recognized by sera from patients with neuroborreliosis has a surface-exposed conserved structural motif. Infect Immun. 1998;66:4073–4079. doi: 10.1128/iai.66.9.4073-4079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maynard Smith J, Smith N H. Detecting recombination from gene trees. Mol Biol Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- 21.Michaud J, Brody L C, Steel G, Fontaine G, Martin L S, Valle D, Mitchell G. Strand-separating conformational polymorphism analysis: efficacy of detection of point mutations in the human ornithine delta-aminotransferase gene. Genomics. 1992;13:389–394. doi: 10.1016/0888-7543(92)90258-t. [DOI] [PubMed] [Google Scholar]
- 22.Norris D E, Johnson B J B, Piesman J, Maupin G O, Clark J L, Black W C. Culturing selects for specific genotypes of Borrelia burgdorferi in an enzootic cycle in Colorado. J Clin Microbiol. 1997;35:2359–2364. doi: 10.1128/jcm.35.9.2359-2364.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padula S J, Dias F, Sampieri A, Craven R B, Ryan R W. Use of recombinant OspC from Borrelia burgdorferi for serodiagnosis of early Lyme disease. J Clin Microbiol. 1994;32:1733–1738. doi: 10.1128/jcm.32.7.1733-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picken R N, Strle F, Picken M M, Ruzic-Sabljic E, Maraspin V, Lotric-Furlan S, Cimperman J. Identification of three species of Borrelia burgdorferi sensu lato (B. burgdorferi sensu stricto, B. garinii, and B. afzelii) among isolates from acrodermatitis chronica atrophicans lesions. J Investig Dermatol. 1998;110:211–214. doi: 10.1046/j.1523-1747.1998.00130.x. [DOI] [PubMed] [Google Scholar]
- 26.Postic D, Assous M V, Grimont P A D, Baranton G. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf 5S-rrl 23S intergenic amplicons. Int J Syst Bacteriol. 1994;44:743–752. doi: 10.1099/00207713-44-4-743. [DOI] [PubMed] [Google Scholar]
- 27.Probert W S, Crawford M, Cadiz R B, LeFebvre R B. Immunization with outer surface protein (Osp) A, but not OspC, provides cross-protection of mice challenged with North American isolates of Borrelia burgdorferi. J Infect Dis. 1997;175:400–405. doi: 10.1093/infdis/175.2.400. [DOI] [PubMed] [Google Scholar]
- 28.Qiu W G, Bosler E M, Campbell J R, Ugine G D, Wang I-N, Luft B J, Dykhuizen D E. A population genetic study of Borrelia burgdorferi sensu stricto from eastern Long Island, New York; suggested frequency-dependent selection, gene flow and host adaptation. Hereditas. 1997;127:203–216. doi: 10.1111/j.1601-5223.1997.00203.x. [DOI] [PubMed] [Google Scholar]
- 29.Ryan J R, Levine J F, Apperson C S, Lubke L, Wirtz R A, Spears P A, Orndorff P E. An experimental chain of infection reveals that distinct Borrelia burgdorferi populations are selected in arthropod and mammalian hosts. Mol Microbiol. 1998;30:365–379. doi: 10.1046/j.1365-2958.1998.01071.x. [DOI] [PubMed] [Google Scholar]
- 30.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seinost, G., W. T. Golde, B. W. Berger, J. J. Dunn, D. Qiu, D. S. Dunkin, D. E. Dykhuizen, B. J. Luft, and R. J. Dattwyler. Infection with multiple Borrelia burgdorferi sensu stricto in patients with Lyme disease. Arch. Derm., in press. [DOI] [PubMed]
- 32.Sigal L H, Zahradnik J M, Lavin P, Patella S J, Bryant G, Haselby R, Hilton E, Kunkel M, Adler-Klein D, Doherty T, Evans J, Malawista S E, Molloy P J, Seidner A L, Sabetta J R, Simon H J, Klempner M S, Mays J, Marks D. A vaccine consisting of recombinant Borrelia burgdorferi outer-surface protein A to prevent Lyme disease. N Engl J Med. 1998;339:216–222. doi: 10.1056/NEJM199807233390402. [DOI] [PubMed] [Google Scholar]
- 33.Steere A C, Schoen R T, Taylor E. The clinical evolution of Lyme arthritis. Ann Int Med. 1987;107:725–731. doi: 10.7326/0003-4819-107-5-725. [DOI] [PubMed] [Google Scholar]
- 34.Steere A C, Malawista S E, Snydman D R, Shope R E, Andiman W A, Ross M R, Steele F M. Lyme arthritis: an epidemic of oligoarticular arthritis in children and adults in three Connecticut communities. Arth Rheum. 1977;20:7–17. doi: 10.1002/art.1780200102. [DOI] [PubMed] [Google Scholar]
- 35.Steere A C, Sikand V K, Meurice F, Parenti D L, Fikrig E, Schoen R T, Nowakowski J, Schmid C H, Laukamp S, Buscarino C, Krause D S. Vaccination against Lyme disease with recombinant Borrelia burgdorferi outer-surface lipoprotein A with adjuvant. N Engl J Med. 1998;339:209–215. doi: 10.1056/NEJM199807233390401. [DOI] [PubMed] [Google Scholar]
- 36.Theisen M, Frederiksen B, Lebech A M, Vuust J, Hansen K. Polymorphism in the OspC gene of Borrelia burgdorferi and immunoreactivity of OspC protein: implication for taxonomy and for use of OspC protein as a diagnostic agent. J Clin Microbiol. 1993;31:2570–2576. doi: 10.1128/jcm.31.10.2570-2576.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilly K, Casjens S, Stevenson B, Bono J L, Samuels D S, Hogan D, Rosa P. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 38.Valsangiacomo C, Balmelli T, Piffaretti J-C. A phylogenetic analysis of Borrelia burgdorferi sensu lato based on sequence information from the hbb gene, coding for a histone-like protein. Int J Syst Bacteriol. 1997;47:1–10. doi: 10.1099/00207713-47-1-1. [DOI] [PubMed] [Google Scholar]
- 39.Van Dam A P, Kuiper H, Vos K, Widjojokusumo A, de Jongh B M, Spanjaard L, Ramselaar A C P, Kramer M D, Dankert J. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin Infect Dis. 1993;17:708–717. doi: 10.1093/clinids/17.4.708. [DOI] [PubMed] [Google Scholar]
- 40.Wang, I.-N., D. E. Dykhuizen, Q. Weigang, J. J. Dunn, E. M. Bosler, and B. J. Luft. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15–30. [DOI] [PMC free article] [PubMed]
- 41.Wormser G P, Nowakowski J, Nadelman R B, Bittker S, Cooper D, Pavia C. Improving the yield of blood cultures for patients with early Lyme disease. J Clin Microbiol. 1998;36:296–298. doi: 10.1128/jcm.36.1.296-298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zar J H. Biostatistical analysis. 3rd ed. Upper Saddle River, N.J: Prentice-Hall International, Inc.; 1996. [Google Scholar]