Abstract
Telomerase-mediated telomeric DNA synthesis is important for eukaryotic cell immortality. Telomerase adds tracts of short telomeric repeats to DNA substrates using a unique repeat addition form of processivity. It has been proposed that repeat addition processivity is partly regulated by a telomerase reverse transcriptase (TERT)-dependent anchor site; however, anchor site-mediating residues have not been identified in any TERT. We report the characterization of an N-terminal human TERT (hTERT) RNA interaction domain 1 (RID1) mutation that caused telomerase activity defects consistent with disruption of a template-proximal anchor site, including reduced processivity on short telomeric primers and reduced activity on substrates with nontelomeric 5′ sequences, but not on primers with nontelomeric G-rich 5′ sequences. This mutation was located within a subregion of RID1 previously implicated in biological telomerase functions unrelated to catalytic activity (N-DAT domain). Other N-DAT and C-terminal DAT (C-DAT) mutants and a C-terminally tagged hTERT-HA variant were defective in elongating short telomeric primers, and catalytic phenotypes of DAT variants were partially or completely rescued by increasing concentrations of DNA primers. These observations imply that RID1 and the hTERT C terminus contribute to telomerase's affinity for its substrate, and that RID1 may form part of the human telomerase anchor site.
INTRODUCTION
Telomerase is important for telomeric DNA replication in most eukaryotes. The minimal components of in vitro–reconstituted telomerase are the telomerase reverse transcriptase (TERT) and the telomerase RNA (TR), which contains a short template that commonly encodes less than two telomeric DNA repeats (reviewed in Harrington, 2003). Many telomerases add multiple telomeric repeats to a single DNA substrate by repetitive reverse transcription of the TR template (repeat addition processivity; reviewed in Lue, 2004).
It is proposed that telomerase associates specifically with its DNA substrate by two different mechanisms: 1) template-dependent base-pairing with 3′ sequences of DNA substrates; and 2) template-independent interactions with G-rich 5′ substrate sequences that are regulated by a protein-dependent anchor site (Harrington and Greider, 1991; Morin, 1991). Repeat addition processivity requires translocation of the TR template and DNA substrate such that the DNA substrate 3′ end is realigned with template 3′ sequences at the beginning of each round of template reverse transcription. The anchor site is predicted to prevent dissociation of telomerase from its DNA substrate during the translocation step of repeat addition processivity; the anchor site may also facilitate alignment or positioning of primers in the active site and promote the elongation of partially telomeric and nontelomeric substrates (Morin, 1989; Harrington and Greider, 1991; Morin, 1991; Collins and Greider, 1993; Lee and Blackburn, 1993; Melek et al., 1994, 1996; Wang and Blackburn, 1997).
Yeast and ciliate telomerases are thought to interact in a template-independent manner with telomeric DNA at two positions: the template-proximal and template-distal anchor sites (Lue and Peng, 1998; Collins, 1999). The template-proximal anchor site is predicted to interact with primers in a region immediately adjacent to the template-hybridizing nucleotides (up to ∼ nt 12), whereas the template-distal anchor site associates with primer sequences further upstream; primer nucleotides in both of these regions contribute to repeat addition processivity and the affinity of yeast, ciliate, and human telomerases for their DNA substrates, though template-distal anchor site sequences are not essential for repeat addition processivity (Morin, 1989; Collins and Greider, 1993; Lee and Blackburn, 1993; Lue and Peng, 1998; Collins, 1999; Baran et al., 2002; Wallweber et al., 2003). In Euplotes, a TERT-sized protein cross-links to single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) substrates 5′ of the 3′ terminus, suggesting that the anchor site is TERT-dependent (Hammond et al., 1997). However, residues that mediate anchor site function have not been identified in any TERT.
Mutations in telomerase-specific N- and C-terminal sequences of human and Saccharomyces cerevisiae TERTs (hTERT and Est2p, respectively) have been reported to dissociate the biological and catalytic activities of telomerase (N-DAT and C-DAT mutants; Friedman and Cech, 1999; Xia et al., 2000; Armbruster et al., 2001; Banik et al., 2002; Kim et al., 2003). Addition of a hemaglutinin (HA) tag or other extensions to the hTERT C terminus also confers a DAT-like phenotype (Counter et al., 1998; Kim et al., 2003). Though N-DAT mutants exhibit at least 60% of the in vitro telomerase activity of wild-type (WT) telomerase, these variants do not immortalize telomerase-negative cells, and telomere shortening is as extensive as in cells that express no telomerase (Counter et al., 1998; Friedman and Cech, 1999; Xia et al., 2000; Armbruster et al., 2001; Kim et al., 2003). The limited proliferation phenotypes of cells expressing certain hTERT N-DAT variants is partially or fully rescued by fusion of these mutants to the telomere-binding proteins hTRF2 or hPOT1; these observations led to the proposal that some N-DAT sequences may be important for the recruitment and/or activation of telomerase at telomeres (Armbruster et al., 2003, 2004). However, some N-DAT and C-DAT hTERT mutants exhibit primer concentration-dependent catalytic defects when telomerase activity is measured using an entirely nontelomeric primer in the PCR-based TRAP assay; some hTERT DAT mutants also display catalytic defects in direct primer extension telomerase assays performed with a physiological telomeric DNA substrate, though catalytic impairment is not readily apparent when telomerase activity is measured using the TRAP assay and the standard, partially telomeric TRAP primer (Armbruster et al., 2001; Lee et al., 2003). These observations indicate that a number of hTERT DAT residues are catalytically important in the context of low primer concentrations, altered substrate sequences, and more stringent assay conditions.
In hTERT, most DAT sequences are found within RNA interaction domain 1 (RID1), a putative TERT accessory domain that interacts with the hTR pseudoknot/template domain and is essential for repeat addition processivity (Figure 1A; Moriarty et al., 2004). Several groups have proposed that RID1 might constitute the anchor site, based on observations that this domain in Est2p interacts with DNA, and is essential for repeat addition processivity and elongation of entirely nontelomeric primers by human telomerase (Beattie et al., 2000; Xia et al., 2000; Lee et al., 2003; Moriarty et al., 2004). The RID1 domain may function cooperatively with the putative TERT C-terminal thumb, because the isolated Est2p C terminus interacts with DNA, and the hTERT C terminus has been implicated in repeat addition processivity, elongation of nontelomeric primers, and functional and physical interactions with RID1 (Beattie et al., 2001; Hossain et al., 2002; Huard et al., 2003; Lee et al., 2003; Moriarty et al., 2004).
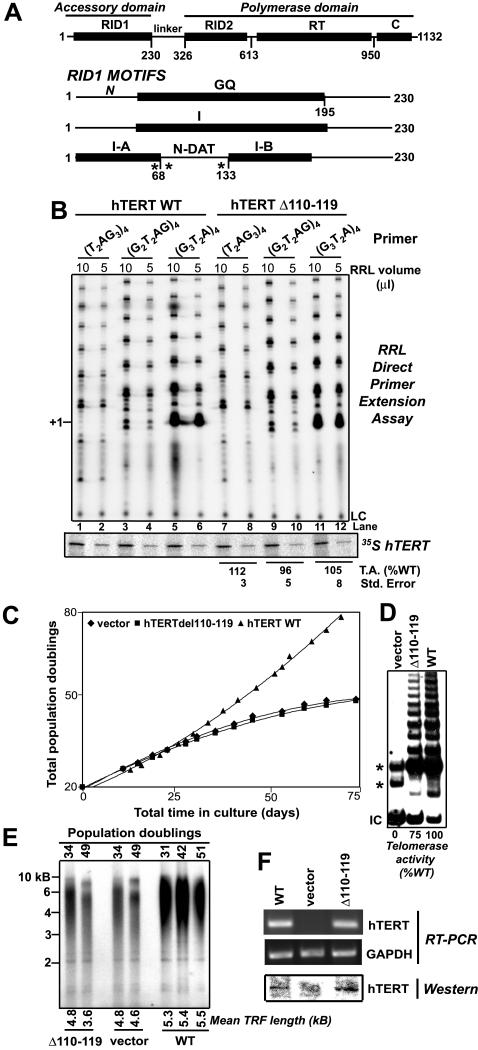
Figure 1.
Effect of an N-DAT mutation on hTERT-mediated immortalization, telomere length maintenance, and in vitro telomerase activity. (A) Top panel: schematic depicting hTERT regions: RNA interaction domains 1 and 2 (RID1 and RID2), reverse transcriptase (RT) motifs, and C terminus (C); hTERT accessory domain: RID1; hTERT polymerase domain: RID2 + RT + C (Moriarty et al., 2004). Bottom panels: RID1 sequence motifs and functional regions previously identified in hTERT and Est2p, mapped onto hTERT. The GQ motif was identified by alignment of sequences conserved in Est2p and other TERTs and overlaps with Est2p hypomutable Region I (Friedman and Cech, 1999; Xia et al., 2000; Armbruster et al., 2001). I-A, I-B: hTERT RID1 sequences required for telomerase catalytic function (Armbruster et al., 2001). N-DAT: sequences previously implicated in telomere length maintenance, but not in vitro telomerase activity (Armbruster et al., 2001). Asterisks indicate the positions in hTERT of Est2p RID1 mutants that exhibit a DAT phenotype (Friedman and Cech, 1999; Xia et al., 2000). (B) Elongation of 24-nt telomeric primers by rabbit reticulocyte lysate (RRL)-reconstituted wild-type (WT) and N-DAT mutant (Δ110–119) telomerases, as detected by a direct primer extension assay. Top panel: telomerase-mediated elongation of DNA primers containing different permutations of the telomeric sequence. +1 indicates the position of products resulting from the addition of one nucleotide to a 24-nt substrate. Telomerase products lower than the + 1 position arise from telomerase-associated nucleolytic primer cleavage (Huard and Autexier, 2004; Oulton and Harrington, 2004). LC, loading control. Middle panel: expression of 35S-labeled hTERT in RRL samples. Bottom: hTERTΔ110–119 T.A. values were expressed relative to activity values for WT hTERT reactions performed using the same RRL volume and DNA substrate. Relative T.A. values were similar for reactions performed with 5 or 10 μl RRL-reconstituted telomerase. Average T.A. values were calculated from three independent experiments. (C–F) Proliferative lifespan (C), telomerase activity (D), telomere length (E), and expression of hTERT mRNA, and protein (F) in polyclonal HA5 cell lines stably expressing vector alone, hTERTΔ110–119, or wild-type (WT) hTERT. Telomerase activity and hTERT protein and mRNA expression were examined in cell lines at population doubling (PD) 42–55. (D) Telomerase activity (T.A.) detected using the TRAP assay. The positions of PCR primer dimers (*) and internal controls (IC) are indicated.
We examined the catalytic phenotypes of a highly active hTERT RID1 DAT mutant and several less active N-DAT and C-DAT variants using a direct primer extension telomerase assay and DNA substrates that varied in length and 5′ sequence. We investigated whether increasing concentrations of DNA substrate could rescue the catalytic defects of DAT mutants on these primers. Our results implicate RID1 and C-terminal hTERT sequences in telomerase's affinity for its DNA substrate, and indicate that residues in RID1 contribute to proximal anchor site-type functions in human telomerase.
MATERIALS AND METHODS
T7 Promoter-driven RRL Expression Constructs
pET28b-hTERT WT, D868N (Bachand and Autexier, 1999), Δ70–79, Δ110–119, Δ150–159, Δ508–517 (Moriarty et al. 2002b), and Δ1123–1132 (Huard et al., 2003) plasmids were previously described; pCR3.1-Flag-hTERT1–250 was previously described (Moriarty et al., 2004); pcDNA3.1-HA-hEST1A, pcDNA3.1-HA-hEST1B, pcDNA-N-MYC-TRF2, and hPOT1-expressing pPB320A constructs were provided by Drs. L. Harrington, T. DeLange, and T. Cech, respectively (Baumann and Cech, 2001; Snow et al., 2003). The T7 reporter-driven TRF2 construct was generated in the lab of T. DeLange and has not been previously described. The pCI-Neo-hTERT-HA construct, provided by Dr. R. Weinberg, was subcloned into the EcoRI/SalI sites of a pcDNA3 vector. Construct identity was confirmed by restriction digest and in vitro expression of 35S-labeled hTERT-HA.
Retroviral Constructs
Retroviral constructs expressing a GFP reporter gene and hTERT (WT or Δ110–119) were created by EcoRI/NotI digestion of pET28b-hTERT plasmids and ligation of hTERT-encoding DNA into an EcoRI/NotI-digested pBMN-IRES-EGFP retroviral vector provided by Dr. G. P. Nolan. Infectious retro virus-containing media collected from Phoenix packaging cells (G. P. Nolan) transiently transfected with pBMN-IRES-GFP-hTERT plasmids were passed through a 0.2-μm filter and stored at –80°C.
Stable Cell Lines
The hTERT-negative hTR-positive SV40-transformed human embryonic kidney cell line HA5 (provided by Dr. S. Bacchetti) was infected at population doubling 20 with packaging cell line media containing retroviruses expressing GFP reporter alone (vector) or GFP reporter gene and WT or Δ110–119 hTERT (WT and Δ110–119 hTERT, respectively). Hexadimethrine bromide (5 μg/ml; polybrene; Sigma, St. Louis, MO) was added to media to promote infection. GFP-expressing infected cells were selected by fluorescence-activated cell sorting with a Becton-Dickinson FACScan machine. Stable cell lines were grown in the presence of 5% CO2, in MEM alpha medium (GIBCO, Rockville, MD) supplemented with 5% heat-inactivated fetal bovine serum (GIBCO) and 1× antibiotic/antimycotic (GIBCO; 100 μg/ml penicillin, 100 μg/ml streptomycin, 250 ng/ml amphotericin B). Population-doubling number of polyclonal stable cell lines was calculated from cell numbers counted at each passage.
TRAP Assays
TRAP assays were performed using 1 μg CHAPS cell extracts, as previously described (Moriarty et al. 2002a, 2002b).
Immunoblotting
Immunoblotting was performed with 50 μg CHAPS cell extracts and αhTERT antibody (Moriarty et al. 2002b). The hTERT peptide sequence used to generate αhTERT (Moriarty et al. 2002b) is the same as the sequence used to generate αTP2 hTERT antibody (Harrington et al., 1997). A typographical error in the latter manuscript erroneously suggests that this peptide is different (L. Harrington, personal communication).
RT-PCR
Preparation of total cellular RNA and cDNA from stable cell lines using Trizol reagent, Superscript II reverse transcriptase (Invitrogen, Burlington, Ontario, Canada) and pd(N)6 random hexamer (Amersham, Baie D'Urfe, Quebec, Canada) were performed according to manufacturer's instructions. hTERT and GAPDH cDNAs were amplified using 1 ng/μl hTERT (5′-AAGTTCCTGCACTGGCTGATGAG-3′, 5′-TCGTAGTTGAGCACGCTGAACAG-3′) or GAPDH (5′-CGGAGTCAACGGATTTGGTCGTAT-3′, 5′-TGCTAAGCAGTTGGTGGTGCAGGA-3′) specific primers, 200 μM dNTPs and 0.05 U/μl TaqDNA polymerase (Invitrogen). PCR conditions: 94°C 5 min, 31 cycles (94°C 45 s, 57°C 45 s, 72°C 1 min).
Telomere Length Measurement
Two micrograms of RsaI/HinfI-digested genomic DNA were resolved by pulse-field gel electrophoresis on 1% agarose gels (Bryan et al., 1995). In-gel hybridizations and calculations of mean telomere length from blots exposed to phosphorimager cassettes were performed as described (Harley et al., 1990; Bryan et al., 1995).
In Vitro Telomerase Reconstitution
35S-labeled and unlabeled human telomerases were reconstituted in rabbit reticulocyte lysates (RRL), using equal amounts of WT hTR, as previously described (Moriarty et al. 2002b). In complementation assays (Supplementary Figure 6, A and B), 0.75 μg hTERT-encoding DNA was mixed with 0.75 μg DNA encoding the specified proteins in a final RRL reaction volume of 20 μl.
Direct Primer Extension Telomerase Assay
The direct primer extension telomerase assay was performed as described (Huard et al., 2003; Moriarty et al., 2004). Reactions mixtures were supplemented with 1 mM dCTP as this nucleotide is present in the cellular context. All reactions were performed with 20 μl RRL-reconstituted telomerase and 1 μM primer, except where indicated. RRL alone did not mediate nonspecific primer elongation under different assay conditions. This control was performed routinely, though not in every experimental replicate.
Quantification of Repeat Addition Processivity and Telomerase Activity Detected by Direct Primer Extension Assay
Telomerase activity was quantified by measuring the signals of the first six repeats of telomerase products, using gels exposed to phosphorimager cassettes (Moriarty et al., 2004). The signals of products containing more than six telomeric repeats were not measured because nonspecific “shadows” of variable intensities were frequently present on gels above this position, precluding accurate quantification of longer products. Mutant telomerase activity values were expressed relative to activity values for WT hTERT reactions from the same experiment performed with the same RRL volume and DNA substrate. Relative telomerase activity values were similar when reactions were performed with different volumes of RRL-reconstituted enzyme; therefore calculations of average telomerase activity values included data for all sample volumes. Repeat addition processivity values were calculated, as previously described, by measuring the signal intensity of the strongest product in each repeat (corresponding to the last G incorporated before enzyme translocation), normalizing this value to the number of incorporated radiolabeled dGTPs and comparing the normalized values for different repeats to each other in the form of a ratio (Hardy et al., 2001). Repeat addition processivity values were measured between the first and second, and second and third telomere repeats, and both values were included in all statistical calculations. Processivity values were similar over the first three repeats when measured from primer extension assays performed with either 10 or 20 μl RRL-reconstituted telomerase (confirmed by two-way ANOVA tests). Therefore, calculations of average processivity included both of these sets of data. Signal quantification, nonlinear regression analyses and statistical calculations (average, SE, and Student's t-test and two-way ANOVA calculations of statistical significance) were performed using ImageQuant, GraphPad Prism (Amersham/Molecular Dynamics, Sorrento, CA), and Excel (Microsoft, Redmond, WA) software.
Pulse-chase Direct Primer Extension Assay
Pulse-chase telomerase assays were performed as previously described using 20 μl RRL-reconstituted telomerases in a final reaction volume of 40 μl (Moriarty et al., 2004).
RESULTS
A True DAT Mutant?
The direct primer extension telomerase assay reveals at least two classes of catalytically impaired telomerases whose defects cannot be identified using the PCR-based TRAP assay: 1) highly nonprocessive enzymes whose extremely short products cannot be amplified in TRAP reactions; and 2) enzymes that exhibit nearly wild-type levels of telomerase activity in the TRAP assay, but are only weakly active in the direct primer extension assay (Huard et al., 2003; Moriarty et al., 2004). We and others have found that almost all mutations in conserved N- and C-terminal hTERT regions cause severe impairment of telomerase's ability to elongate telomeric primers in the direct primer extension assay (unpublished data; Huard et al., 2003; Lee et al., 2003; Moriarty et al., 2004). One exception was a mutation in the N-DAT region (Δ110–119), which did not impair the ability of in vitro–reconstituted mutant telomerase to elongate 18 or 24 nucleotide (nt) telomeric primers in the direct primer extension assay (Figures 1B and 3). A polyclonal SV40-transformed human embryonic kidney HA5 cell line stably expressing this mutant exhibited proliferation and telomere length maintenance defects similar to a cell line expressing vector alone (Figure 1, C and E), though hTERT protein and mRNA levels were similar to those in cells stably expressing wild-type (WT) hTERT (Figure 1F). Telomerase activity in cells expressing hTERTΔ110–119 was slightly reduced than that in cells expressing WT hTERT, as measured using the TRAP assay and the standard TRAP TS primer (TS-GTT; Figure 1D). The Δ110–119 mutant exhibited WT levels of telomerase activity when expressed in S. cerevisiae, rabbit reticulocyte lysates (RRL), or transiently in HA5 cells (unpublished data; Moriarty et al. 2002b). Therefore, we initially concluded that hTERTΔ110–119 was likely a true DAT mutant that dissociates the in vitro and in vivo activities of telomerase (Armbruster et al., 2001).
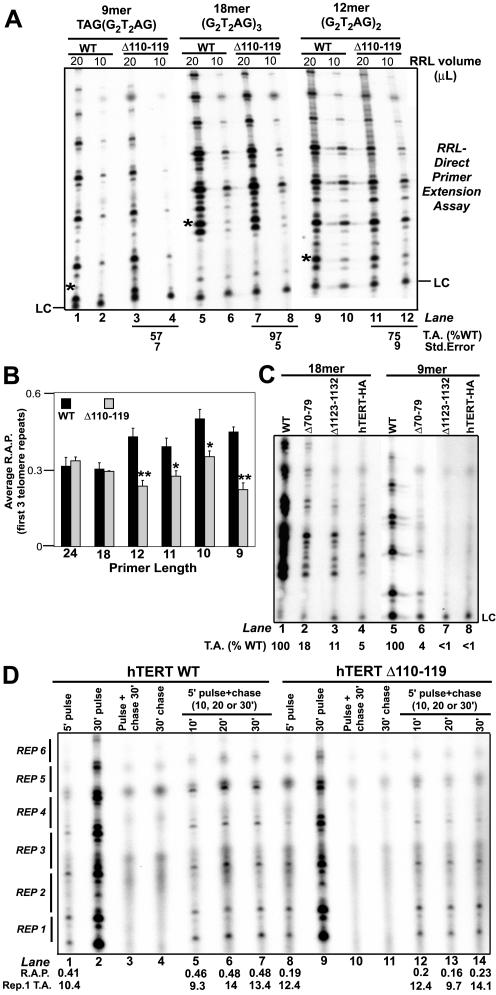
Figure 3.
N-DAT, C-DAT, and hTERT-HA variants exhibit elongation defects on telomeric DNA substrates shortened at the 5′ end. WT, N-DAT (hTERTΔ70–79, Δ110–119), C-DAT (hTERTΔ1123–1132), and hTERT-HA mutant telomerase activities on short telomeric DNA substrates. Primers contained identical 3′ sequences and progressively shortened 5′ ends. Primer sequences: 24mer (G2T2AG)4, 18mer (G2T2AG)3, 12mer (G2T2AG)2, 11mer GT2AG(G2T2AG), 10mer T2AG(G2T2AG), 9mer TAG(G2T2AG). In A and C, the telomerase activity (T.A.) values of hTERT mutants expressed relative to WT telomerase are indicated below each panel. LC, loading control. (A) Autoradiograph showing elongation of three representative telomeric primers of different lengths by WT and Δ110–119 telomerases. Asterisks indicate the positions of products resulting from the addition of one nucleotide to primer substrates. (B) Graphical summary of the repeat addition processivity (R.A.P.) of WT and Δ110–119 telomerases on short primers. Average R.A.P. values are expressed in arbitrary units, and SE bars for each value are indicated; R.A.P. calculations are described in detail in Materials and Methods. Average R.A.P. values were calculated from four independent experiments, each of which included direct primer extension assays performed with 10 and 20 μl RRL-reconstituted telomerase; therefore, statistical calculations for each primer include R.A.P. data from 8 samples. Δ110–119 mean processivity values that differed significantly from WT mean processivity values are indicated by *0.05 < p < 0.01 or **p < 0.01. (C) Autoradiograph depicting elongation of 18- and 9-nt telomeric primers by Δ70–79, Δ1123–1132, and hTERT-HA telomerases. (D) Pulse-chase time course experiments. Pulse primer: 1 μM biotinylated 9mer TAG(G2T2AG). Chase primer: 150 μM nonbiotinylated (TTAGGG)3. Control 1 (lanes 3 and 10): pulse and chase primers added simultaneously to demonstrate the efficiency of chase conditions. Control 2 (lanes 4 and 11): reactions performed with chase primer alone to demonstrate that nonbiotinylated chase primer elongation products are not purified by streptavidin beads. Repeat addition processivity (R.A.P.) values and first repeat telomerase activity values (Rep. 1 T.A.) for individual samples (expressed in arbitrary units) are indicated at the bottom of each lane.
Catalytic Defects of an N-DAT Mutant on DNA Substrates with Nontelomeric 5′ Sequences
Altering the sequence of telomeric DNA substrates affects the catalytic activity of endogenous telomerases from diverse organisms, implying that telomerase has a sequence-specific affinity for its substrate (reviewed in Collins, 1999; Harrington, 2003; Lue, 2004). We tested the activity of in vitro-reconstituted WT and Δ110–119 telomerases using primers containing nontelomeric sequences and found that the N-DAT mutant exhibited catalytic defects on partially telomeric substrates, especially those with nontelomeric 5′ sequences (Figure 2 and Supplementary Figure 5). Catalytic defects could be detected visually by comparing the abundance of longer products synthesized by WT and mutant telomerases; these observations were confirmed by measurement of telomerase activity values for different primers in multiple independent experiments (Figure 2 and Supplementary Figure 5). The expression levels of WT, Δ110–119, and other mutant hTERTs analyzed in this study varied by <10% in multiple independent experiments, indicating that the catalytic defects of hTERT mutants were not caused by differences in protein expression.
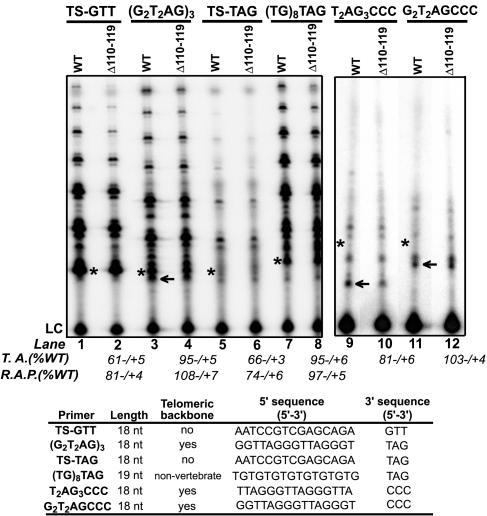
Figure 2.
An N-DAT mutant exhibits elongation defects on DNA substrates with nontelomeric 5′ sequences. Elongation of a telomeric substrate (G2T2AG)3 and DNA primers containing: 1) a nontelomeric `TS' 5′ backbone and telomeric 3′ sequences (TS-GTT and TS-TAG); and 2) a telomeric 5′ backbone and nontelomeric 3′ sequences (T2AG3CCC and G2T2GCCC) by RRL-reconstituted wild-type and N-DAT mutant (Δ110–119) telomerases, as detected by a direct primer extension assay. The (TG)8TAG primer 5′ backbone is composed of G-rich nonvertebrate telomeric sequences. All primers were 18–19 nt in length. Average relative telomerase activity (T.A.) and repeat addition processivity (R.A.P.) values calculated from four independent experiments are indicated at bottom. LC, loading control. Asterisks indicate the positions of products resulting from the addition of one nucleotide to each DNA substrate. Arrows indicate the shortest cleavage-derived elongation products visible on these gels.
Primers with 5′ “backbones” consisting of vertebrate telomeric or G-rich sequences were efficiently elongated by the N-DAT mutant telomerase (Figure 2: (G2T2AG)3 and (TG)8TAG)); however, this mutant elongated TS-type primers containing entirely nontelomeric 5′ sequences and a telomeric 3′ end less efficiently than WT telomerase (Figure 2: TS-GTT, TS-TAG). The elongation defect of the Δ110–119 mutant on TS-type primers was similar for three distinct primers predicted to hybridize with the hTR template at different positions (TS-GTT, TS-TAG, and TS-GGG; Figure 2; unpublished data), suggesting that this N-DAT mutation may not affect formation of the primer/template hybrid. A primer containing an entirely telomeric 5′ backbone and three nontelomeric residues at the primer 3′ end was elongated with similar efficiency by WT and Δ110–119 telomerases (Figure 2: G2T2AGCCC), supporting the hypothesis that the N-DAT mutant's catalytic defect was primarily dependent on 5′ substrate sequences and not on hybridization with the hTR template. DAT mutant-mediated elongation of a second primer consisting of a different permutation of the telomeric 5′ backbone, and the same nontelomeric 3′ residues, was 20% less efficient than that of WT enzyme (Figure 2: T2AG3CCC). Though both of these primers were the same length, the cleavage-derived elongation products generated with these substrates were shorter for T2AG3CCC than for G2T2AGCCC, implying that the T2AG3CCC primer was more extensively cleaved before elongation (Figure 2). Similarly, when we examined the elongation of two 19-nt partially telomeric substrates containing a single internal telomeric repeat cassette at different positions, we found that the substrate that generated shorter cleavage-derived products was elongated less efficiently by Δ110–119 telomerase (Supplementary Figure 5, lanes 7–8 and 11–12). These observations suggested that differences in elongation efficiency might be due to primer length (see Figure 3). The DAT mutation did not appear to affect cleavage of substrates before elongation, because products shorter than the input primer were similar to those generated by WT enzyme (Figure 2 and Supplementary Figure 5). The DAT mutant's activity on a telomeric substrate containing a single nucleotide 3′ mismatch insertion was also slightly reduced compared with WT enzyme, but this inhibition was less pronounced than the defects observed for primers with nontelomeric 5′ backbones (compare Supplementary Figure 5, lanes 3 and 4 with Figure 2, lanes 2 and 6).
Fusion of hTRF2 or hPOT1 to certain N-DAT mutants partially or fully rescues the limited proliferation phenotype of cells expressing these variants, and hPOT1 regulates telomerase activity on telomeric substrates (Armbruster et al., 2003, 2004; Kelleher et al., 2005). The telomerase-associated protein Est1p also stimulates the catalytic function of Candida albicans telomerase on short substrates and primers containing nontelomeric 5′ sequences and can rescue the proliferation defects of S. cerevisiae cells expressing Est2p proteins with mutations in the RID1 region (Friedman et al., 2003; Singh and Lue, 2003). Coexpression of telomere-binding proteins hTRF2 or hPOT1, or the hTERT-interacting human Est1p orthologues hEST1A or hEST1B, did not rescue the N-DAT mutant's TS-TAG substrate elongation defect (Supplementary Figure 6; Snow et al., 2003). The Δ110–119 variant coimmunoprecipitated hEST1A and hEST1B as efficiently as WT hTERT in the presence of hTR (unpublished data). Furthermore, coexpression of a catalytically dead hTERT RT mutant (D868N) or the hTERT RID1 domain did not improve the N-DAT mutant's ability to elongate the TS-TAG primer, though we and others have shown that functionally complementing hTERT molecules can restore processive telomerase activity to other RID1 mutants (Supplementary Figure 6; Beattie et al., 2001; Moriarty et al., 2004). This result suggests that more than one copy of the N-DAT region or RID1 domain may be required for elongation of substrates with nontelomeric 5′ ends or that elongation of partially telomeric substrates may require cis interactions between hTERT's polymerase and RID1 accessory domains (Moriarty et al., 2004).
Substrate Length-dependent Repeat Addition Processivity and Activity Defects of an N-DAT Mutant
Anchor site–type interactions with the 5′ end of DNA substrates are thought to be facilitated by the presence of G-rich sequences. Because the telomerase activity of the Δ110–119 mutant appeared to be dependent on the presence of G-rich 5′ sequences in DNA substrates, we investigated its ability to elongate entirely telomeric primers consisting of identical 3′ sequences and 5′ ends of varying lengths (shortened 5′ ends; Figure 3). Previous studies have shown that endogenous human telomerases elongates 12-nt telomeric primers more efficiently than longer 18-nt telomeric substrates and that the catalytic rate and activity of Tetrahymena and Oxytricha telomerases are enhanced on substrates containing two or fewer telomere repeats (Morin, 1989; Zahler et al., 1991; Lee and Blackburn, 1993). It has been proposed that elongation of such short telomeric substrates is regulated by interaction with a template-proximal anchor site (Lue and Peng, 1998; Collins, 1999). Similar to these previous observations, quantitative analysis of repeat addition processivity within the first three telomere repeats synthesized by human telomerase indicated that WT telomerase was more processive on short (9–12 nt) primers than on longer (18–24 nt) substrates (Figure 3B). Repeat addition processivity values between the first and second, and second and third telomere repeats were similar to each other, and both values were included in all calculations shown in Figure 3.
WT and Δ110–119 telomerases exhibited similar levels of processivity on 24- and 18-nt primers (Figure 3B). However, the Δ110–119 variant did not exhibit the enhanced repeat addition processivity observed for WT telomerase on substrates 12-nt and shorter (Figure 3B), suggesting that this mutation disrupts the activity-stimulating function of the template-proximal anchor site. After the addition of three repeats to short primers, the DAT mutant became as processive as WT telomerase on most short primers (unpublished data), possibly because it elongated longer products more efficiently or because telomerase entered a new mode of elongation after the addition of the first three repeats (Melek et al., 1994); therefore, we did not include processivity values for products longer than three repeats in the data summarized in Figure 3.
Pulse-chase telomerase assays indicated that the Δ110–119 mutant added the first repeat of telomeric DNA to a 9-nt substrate as efficiently as WT enzyme (Figure 3D, compare first repeat telomerase activity values), but synthesized fewer products containing multiple telomere repeats, implying a specific defect in repeat addition processivity on short primers (Figure 3D). As we have previously observed, WT telomerase exhibited quite limited repeat addition processivity over time in pulse-chase time-course assays, because longer products were not detected after the first 5 min of chase conditions (Moriarty et al., 2004). This may indicate that recombinant human telomerase reconstituted in RRL is less processive or less stable than endogenous enzyme or that human telomerase is less processive than previously assumed. Pulse-labeling of telomerase products for fewer than 5 min might have facilitated detection of chased longer products; however, we could not perform pulse-labeling for less than 5 min because of the reduced activity of WT and mutant telomerases on 9-nt primers.
It has previously been demonstrated that Euplotes telomerase forms an anchor site–type cross-link to both ssDNA and dsDNA substrates, implying that telomerase may also interact with double-stranded DNA (Hammond et al., 1997). Furthermore, the isolated RID1 region of Est2p can interact with both ssDNA and dsDNA (Xia et al., 2000). Because the DAT mutant was fully active and processive on telomeric and partially telomeric G-rich primers 18 nt and longer (Figures 1, 2, 3), we hypothesized that extending the 5′ length of short substrates by the addition of duplex sequences might also restore WT levels of activity. Nontelomeric double-stranded substrates with telomeric 3′ overhangs of varying lengths were generated by adapting an oligonucleotide annealing technique previously used to examine the activity of Tetrahymena telomerase on 3′ overhang DNA; a variation of this method has also been used recently to examine the 3′ overhang length requirements of recombinant and endogenous human telomerases in the PCR-based TRAP assay (Supplementary Figure 7; Wang and Blackburn, 1997; Rivera and Blackburn 2004). Using the direct primer extension assay, we found that WT telomerase could elongate dsDNA substrates containing 3′ overhangs as short as 7 nt; interestingly, we also observed that the DAT mutant elongated substrates with telomeric overhangs 12 nt and shorter as efficiently as WT telomerase (Supplementary Figure 7). Similarly, extending the length of 5′ sequences also stimulates the ability of Tetrahymena telomerase to elongate entirely nontelomeric substrates (Wang and Blackburn, 1997). These observations suggest that extending the length of 5′ sequences by adding either ssDNA or dsDNA sequences (Figures 1, 2, 3; Supplementary Figure 7) might engage a potential template-distal anchor site or dsDNA interaction site that could enhance the affinity or stabilize the association of telomerase with suboptimal DNA substrates.
Catalytic Defects of Other N-DAT, C-DAT, and hTERT-HA Variants on Short DNA Substrates
Because the N-DAT mutant characterized in these experiments clearly exhibited catalytic defects on short telomeric primers, we investigated the effect of primer length on the telomerase activities of other DAT-type mutants. A C-terminally HA-tagged hTERT variant (hTERT-HA; Counter et al., 1998), a C-DAT mutant (Δ1123–1132; Banik et al., 2002; Huard et al., 2003) and a second N-DAT mutant (Δ70–79; Moriarty et al. 2002b) elongated 18-nt primers less efficiently than WT enzyme (Figure 3C). All of these variants were expressed at wild-type levels in RRL (unpublished data). The Δ70–79 N-DAT mutant exhibited repeat addition processivity defects in addition to an overall impairment of telomerase activity on 18-nt primers (unpublished data). These putative DAT variants were completely or almost completely inactive on 9-nt primers, indicating that they could not elongate a minimal substrate that was sufficient for WT telomerase function (Figure 3C). The reduced activity of these variants on both 18- and 9-nt telomeric primers prevented us from examining potential anchor site-type catalytic defects using the types of analysis performed for the Δ110–119 mutant (Figure 3B).
Catalytic Defects of N-DAT and C-DAT Mutants Can Be Partially or Fully Rescued by Increasing Concentrations of DNA Substrate
The presence of a template-independent anchor site in telomerase has been deduced not only from analysis of substrate sequences that influence telomerase activity, but also from the observations that 5′ primer sequences regulate elongation in a concentration-dependent manner (Harrington and Greider, 1991; Collins and Greider, 1993). We therefore examined whether inefficient elongation of telomeric primers by RID1 and C-terminal hTERT variants could be rescued in the presence of increasing concentrations of DNA substrate (Figure 4; unpublished data; see text below). The telomerase activity of the Δ70–79 mutant improved with increasing concentrations of 18-nt substrate, and the elongation defects of the Δ110–119 variant were rescued at higher concentrations of 9-nt substrate (Figure 4, B and C). The primer concentration-dependent catalytic phenotype of these mutants was specific, because increasing DNA concentrations did not restore telomerase activity to a completely inactive RNA interaction domain 2 (RID2) mutant (Δ508–517) defective in the ability to assemble with the telomerase RNA (Figure 4C; Moriarty et al., 2002b, 2004).
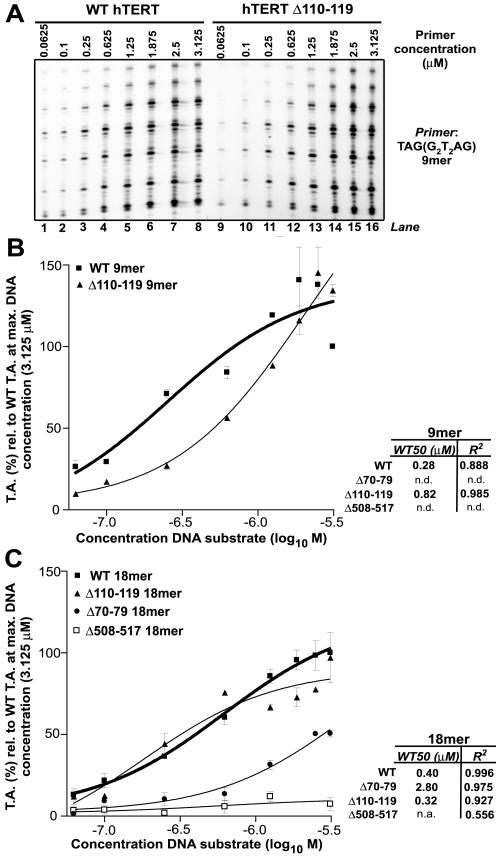
Figure 4.
Increasing concentrations of DNA substrate rescue catalytic defects of N-DAT mutants. Elongation activities of WT, N-DAT (hTERTΔ110–119 and Δ70–79), and RID2 (hTERT Δ508–517) telomerases in the presence of different concentrations of telomeric DNA primers. (A) Representative autoradiograph showing elongation of a 9-nt telomeric substrate by WT and Δ110–119 telomerases at the indicated primer concentrations. (B and C) Graphical summary of the average telomerase activities (T.A.) and SE (SE) values of WT and indicated mutant telomerases at increasing concentrations of 9-nt (B) or 18-nt (C) telomeric primers. The dose-response curves for WT telomerase are indicated by bold lines. At least two independent repeats of each experiment were performed. In each experiment, the T.A. of WT and mutant telomerases at different concentrations of the same primer were expressed relative to the T.A. of WT enzyme at the maximum primer concentration (3.125 μM, –5.5 on the log10 M scale). Therefore, the WT value at 3.125 μM is 100% (SE is 0). R2 values for dose-response curves calculated by nonlinear regression analysis are shown to the right of each graph, as are the primer concentrations at which WT and mutant telomerases were predicted by linear regression analysis to achieve 50% of the maximum measured activity of WT enzyme on the specified substrates (WT50). Note that in B WT T.A. at the maximum 9-mer primer concentration is lower than the maximum observed T.A. because activity decreased at higher primer concentrations; a similar trend was observed for Δ110–119 telomerase on the same primer. Maximal WT T.A. (140% of T.A. at the maximum primer concentration) was observed at 1.875 μM primer (–5.7 on the log10 M scale); therefore, WT50 values for the 9-mer primer were calculated as the primer concentrations at which T.A. was predicted to achieve 70% of WT T.A. The SE values for Δ110–119 T.A. were very small at lower concentrations of 9-mer primer and are not readily discernible on this graph. In C, the SE bar for Δ110–119 T.A. at the maximum primer concentration overlaps with the WT datapoint at the same primer concentration; average WT T.A. at this concentration had an SE of 0. The Δ508–517 RID2 mutant exhibited activity levels only slightly above background at all primer concentrations, and the WT50 concentration was predicted to be infinite (n.a.). n.d., not determined.
We could not calculate Km values for different telomerases and primers because the primer concentrations that would be required to reach saturation for more weakly active telomerases exceeded the binding capacity of the streptavidin magnetic beads used in the telomerase assay. Instead, dose-response curves plotted from these data were used to estimate the primer concentrations required to achieve 50% of maximum WT telomerase activity. These values were similar for WT and Δ110–119 telomerases on 18-nt primers (Figure 4C). In contrast, the Δ110–119 and Δ70–79 mutants required ∼threefold and sevenfold greater concentrations of 9- and 18-mer DNA substrate, respectively, to reach 50% of the maximum telomerase activity of WT enzyme (Figure 4, B and C). An approximately eightfold greater concentration of 18-nt substrate was required for a C-DAT mutant (Δ1123–1132) to achieve 50% of the activity of WT telomerase on the same primer (unpublished data). It has previously been reported that RID1 and the TERT C terminus can physically interact and regulate similar, though not identical catalytic properties, including repeat addition processivity and affinity for the DNA substrate (Beattie et al., 2000; Xia et al., 2000; Peng et al., 2001; Hossain et al., 2002; Huard et al., 2003; Lee et al., 2003; Moriarty et al., 2004). Our observations that hTERT C-terminal variants are profoundly impaired in the ability to elongate short telomeric substrates and exhibit a reduced affinity for DNA substrates suggests that the TERT C-terminus might also contribute to anchor site-type functions, perhaps by cooperative interaction with or regulation of the RID1 domain.
Interestingly, the three- and sevenfold reductions in WT50 telomerase activity values that we observed for deletion 110–119 and deletion 70–79 mutant-mediated elongation of 9- and 18-mer primers, respectively, were comparable to the template-independent reductions in affinity recently measured for human telomerase substrates containing substituted 5′ sequences; this previous study indicates that substitution of telomeric sequences 4–8 nt upstream of the 3′ end with nontelomeric sequences reduces the affinity of endogenous human telomerase for primer DNA by 2.6-fold and that substitution of nt 4–18 reduces affinity by 8.3-fold (Wallweber et al., 2003). Increasing substrate concentrations also improved, but did not fully rescue the activity defects caused by a RID1 mutation outside the N-DAT region (Δ150–159); however, this mutant is entirely nonprocessive, and its predicted WT50 value on 18-nt primers was exceedingly large (unpublished data; Moriarty et al., 2004). Collectively, these observations suggested that the RID1 and C-DAT hTERT regions are important for telomerase's affinity for its DNA substrate and that RID1 contributes to proximal anchor site-type functions in human telomerase.
DISCUSSION
Is RID1 Part of the Human Telomerase Anchor Site?
The presence of a template-independent anchor site in telomerase was originally deduced based on observations that altering the length or G-rich composition of 5′ substrate sequences affects the processive elongation of primers in a concentration-dependent manner (Harrington and Greider, 1991; Morin, 1991; Collins and Greider, 1993). We found that the catalytic activity of an N-DAT RID1 mutant was specifically dependent on the presence of G-rich sequences in the 5′ region of DNA primers and on the length of substrate 5′ sequences. Furthermore, this mutant exhibited specific repeat addition processivity defects on short telomeric primers, and its impaired catalytic phenotype on a short telomeric primer was rescued by increasing DNA substrate concentration. Therefore, we concluded that the catalytic phenotype of the RID1 mutant characterized most extensively in this study is consistent with an anchor site–type defect. We have been unsuccessful in our attempts to analyze hTERT-DNA interactions by more direct, telomerase activity-independent methods that would permit the examination of anchor site function in other N-DAT and RID1 mutants that exhibit catalytic defects on both long and short primers. However, the sensitivity of these mutants to altered substrate concentration and substrate sequence suggests that RID1 sequences in addition to residues 110–119 likely contribute to telomerase's affinity for its DNA substrate (this study; Beattie et al., 2000; Lee et al., 2003). Collectively, these observations suggest that much of RID1 may interact with DNA substrates.
Evidence of a Role for a Template-proximal Anchor Site in Human Telomerase Repeat Addition Processivity
We found that recombinant WT human telomerase, like endogenous human and Oxytricha telomerases, was more processive on telomeric substrates containing two or fewer telomere repeats than on primers consisting of 3 or 4 telomere repeats (Morin, 1989; Zahler et al., 1991). In Tetrahymena telomerase, DNA sequences predicted to interact with the putative template-proximal anchor site also contribute to repeat addition processivity, and the use of short telomeric primers (<12 nt) increases enzyme catalytic rate (Collins and Greider, 1993; Lee and Blackburn, 1993; Baran et al., 2002). Oxytricha telomerase activity is reduced on certain long primers as a result of conformational changes in the substrates themselves; however, Lee et al. have demonstrated that the enhanced catalytic rate of Tetrahymena telomerase on short substrates is unlikely to be due to such conformational changes (Zahler et al., 1991; Lee and Blackburn, 1993). This implies that stimulation of activity may be attributable to telomerase interactions with template-proximal DNA sequences. The hypothesis that up-regulation of telomerase activity on short telomeric substrates is dependent on the enzyme itself is supported by our observation that mutation of hTERT RID1 significantly reduced the enhanced repeat addition processivity of human telomerase on short telomeric substrates but did not affect processive elongation of longer primers. These data also imply that RID1 residues 110–119 likely regulate template-proximal anchor site–type functions in human telomerase. Interestingly, our observation that human telomerase preferentially elongates short telomeric substrates complements recent data indicating that hPOT1 binding to internal sites in telomeric primers also stimulates telomerase processivity, likely by restricting the length of primer sequences available for interaction with telomerase (Lei et al., 2005). Thus, hPOT1 might promote telomere elongation in vivo by regulating the exposure of substrate sequences that can interact with the template-proximal anchor site of telomerase.
hTERT RID1 mutations, including deletion of residues 110–119, impair both hTR interactions and affinity for telomeric substrates, and RID1 associates with the hTR pseudoknot/template domain (this study; Moriarty et al., 2002b, 2004). At present, we cannot determine if hTERT RID1's affinity for the DNA substrate requires the presence of hTR. However, the observations that cross-linking of DNA substrates to the anchor site of Euplotes TERT is dependent on TR and that Est2p RID1 interacts with both TR and DNA suggest that the telomerase anchor site may not be entirely TERT-dependent (Hammond et al., 1997; Xia et al., 2000). DNA sequences predicted to interact with the template-proximal anchor site could regulate conformational changes in the enzyme-substrate complex that facilitate alignment and/or positioning of the template/primer duplex in the catalytic active site, or unwinding of RNA/DNA duplexes during translocation (Lee and Blackburn, 1993; Lue and Peng, 1998; Baran et al., 2002). In ciliates, 5′ substrate sequences regulate default positioning of the template in the active site (Yu and Blackburn, 1991; Melek et al., 1994, 1996; Wang and Blackburn, 1997; Wang et al., 1998). A repeat addition processivity- and activity-stimulating template recognition element (TRE) in the Tetrahymena TR that interacts with TERT also regulates default template positioning and is proposed to form sequence-specific contacts with the DNA substrate and/or other regions of the telomerase ribonucleoprotein (Licht and Collins, 1999; Lai et al., 2001; Miller and Collins, 2002). Thus, hTERT RID1 might mediate repeat addition processivity both by stabilizing telomerase interactions with 5′ substrate sequences and by regulating interactions between the TR template and DNA 3′ end.
Catalytic Phenotypes of DAT Mutants
Data reported here and previously collectively indicate that the C-DAT region and most N-DAT sequences are important for human telomerase catalytic function, especially when telomerase activity is examined with partially telomeric, entirely nontelomeric or short telomeric primers (this study; Armbruster et al., 2001; Huard et al., 2003; Lee et al., 2003). This indicates that the DAT domains are not simply required for recruitment of telomerase to telomeres, but are also important for the affinity and specificity of telomerase for its substrate. We found that the telomerase activity of most DAT-type variants was profoundly reduced on short telomeric primers. Because catalytic impairment on short primers might account for the inability of these mutants to maintain telomere length, it will be important to determine the length of the telomeric substrate accessible to human telomerase in vivo. The effects of N-DAT mutations spanning amino acids 86–91, 98–109, and 128–133 on telomerase activity have not yet been examined by these more stringent activity assays; however, because these sequences are short and interspersed among regions known to be catalytically important, we propose that all N-DAT sequences likely contribute to telomerase catalytic function. This conclusion does not preclude the possibility that N- and C-DAT regions have biological functions in addition to their role in catalysis, as yeast and human N-DAT and C-DAT TERT sequences may interact directly or indirectly with proteins that could recruit or activate telomerase at the telomere, including Est1p, Est3p, and hnRNPs A1 and C1/C2 (Ford et al., 2000; Friedman et al., 2003; Lee et al., 2003; Singh and Lue, 2003).
CONCLUSIONS
RID1 is important for repeat addition processivity, anchor site–type sequence- and length-specific affinity for the DNA substrate, and interactions with the TR pseudoknot/template domain, ssDNA and dsDNA (this study; Xia et al., 2000; Beattie et al., 2001; Lee et al., 2003; Moriarty et al., 2004). Further characterization of this fascinating, telomerase-specific domain will be essential for our understanding of telomerase's unique catalytic properties.
Supplementary Material
Acknowledgments
We thank S. Bacchetti, T. Cech, T. DeLange, L. Harrington, G. P. Nolan, and R. Weinberg for providing reagents; S. Dupuis and J. Sanchez for technical assistance with retroviral constructs and cell sorting; M. A. Cerone for sharing RT-PCR and telomere length measurement techniques; and members of the Autexier laboratory for critical review of the manuscript. We also thank anonymous peer reviewers for criticisms and comments that substantially improved the quality of this article. This work was funded by Canadian Institutes of Health Research grant MOP68844 to C.A., a CIHR doctoral research award and McGill Graduate Studies Fellowship award to T.J.M., and Boehringer-Ingelheim (Canada) Young Investigator and Fonds de la Recherche en Santé du Québec Chercheur-Boursier awards to C.A.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0148) on April 27, 2005.
Abbreviations used: TERT, telomerase reverse transcriptase; RID1, RNA interaction domain 1; DAT, dissociates activities of telomerase; TR, telomerase RNA; nt, nucleotide; WT, wild-type; TRAP, telomeric repeat amplification protocol; RRL, rabbit reticulocyte lysate.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Armbruster, B., Banik, S., Guo, C., Smith, A., and Counter, C. (2001). N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21, 7775–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster, B. N., Etheridge, K. T., Broccoli, D., and Counter, C. M. (2003). Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol. Cell. Biol. 23, 3237–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster, B. N., Linardic, C. M., Veldman, T., Bansal, N. P., Downie, D. L., and Counter, C. M. (2004). Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 24, 3552–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachand, F., and Autexier, C. (1999). Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem. 274, 38027–38031. [DOI] [PubMed] [Google Scholar]
- Banik, S.S.R., Guo, C., Smith, A. C., Margolis, S. S., Richardson, D. A., Tirado, C. A., and Counter, C. M. (2002). C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell. Biol. 22, 6234–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran, N., Haviv, Y., Paul, B., and Manor, H. (2002). Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res. 30, 5570–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, P., and Cech, T. R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Beattie, T., Zhou, W., Robinson, M., and Harrington, L. (2000). Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11, 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie, T., Zhou, W., Robinson, M., and Harrington, L. (2001). Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 21, 6151–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, T. M., Englezou, A., Gupta, J., Bacchetti, S., and Reddel, R. R. (1995). Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, K. (1999). Ciliate telomerase biochemistry. Annu. Rev. Biochem. 68, 187–218. [DOI] [PubMed] [Google Scholar]
- Collins, K., and Greider, C. W. (1993). Tetrahymena telomerase catalyzes nucleolytic cleavage and non-processive elongation. Genes Dev. 7, 1364–1376. [DOI] [PubMed] [Google Scholar]
- Counter, C. M., Hahn, W. C., Wei, W., Dickinson Caddle, S., Beijersbergen, R. L., Lansdorp, P. M., Sedivy, J. M., and Weinberg, R. A. (1998). Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA 95, 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, L., Suh, J., Wright, W., and Shay, J. (2000). Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol. Cell. Biol. 20, 9084–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, K. L., and Cech, T. R. (1999). Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13, 2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, K. L., Heit, J. J., Long, D. M., and Cech, T. R. (2003). N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol. Biol. Cell 14, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, P. W., Lively, T. N., and Cech, T. R. (1997). The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 17, 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, C. D., Schultz, C. S., and Collins, K. (2001). Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276, 4863–4871. [DOI] [PubMed] [Google Scholar]
- Harley, C. B., Futcher, A. B., and Greider, C. W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460. [DOI] [PubMed] [Google Scholar]
- Harrington, L. (2003). Biochemical aspects of telomerase function. Cancer Lett. 194, 139–154. [DOI] [PubMed] [Google Scholar]
- Harrington, L., Zhou, W., McPhail, T., Oulton, R., Yeung, D.S.K., Mar, V., Bass, M. B., and Robinson, M. O. (1997). Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 11, 3109–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L. A., and Greider, C. W. (1991). Telomerase primer specificity and chromosome healing. Nature 353, 451–454. [DOI] [PubMed] [Google Scholar]
- Hossain, S., Singh, S. M., and Lue, N. F. (2002). Functional analysis of the C-terminal extension of telomerase reverse transcriptase: a `putative' thumb domain. J. Biol. Chem. 277, 36174–36180. [DOI] [PubMed] [Google Scholar]
- Huard, S., and Autexier, C. (2004). Human telomerase catalyzes nucleolytic primer cleavage. Nucleic Acids Res. 32, 2171–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard, S., Moriarty, T. J., and Autexier, C. (2003). The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31, 4059–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, C., Kurth, I., and Lingner, J. (2005). Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell. Biol. 25, 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Xu, L., and Blackburn, E. H. (2003). Catalytically active human telomerase mutants with allele-specific biological properties. Exp. Cell Res. 288, 277–287. [DOI] [PubMed] [Google Scholar]
- Lai, C., Mitchell, J., and Collins, K. (2001). RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21, 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. S., and Blackburn, E. H. (1993). Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 13, 6586–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. R., Wong, M. Y., and Collins, K. (2003). Human telomerase reverse transcriptase motifs required for elongation of a telomeric substrate. J. Biol. Chem. 278, 52531–52536. [DOI] [PubMed] [Google Scholar]
- Lei, M., Zaug, A. J., Podell, E., and Cech, T. R. (2005). Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 280, 20449–20456. [DOI] [PubMed] [Google Scholar]
- Licht, J. D., and Collins, K. (1999). Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 13, 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue, N. F. (2004). Adding to the ends: what makes telomerase processive and how important is it? BioEssays 26, 955–962. [DOI] [PubMed] [Google Scholar]
- Lue, N. F., and Peng, Y. (1998). Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 26, 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melek, M., Davis, B. T., and Shippen, D. E. (1994). Oligonucleotides complementary to the Oxytricha nova telomerase RNA delineate the template domain and uncover a novel mode of primer utilization. Mol. Cell. Biol. 14, 7827–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melek, M., Greene, E. C., and Shippen, D. E. (1996). Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol. Cell. Biol. 16, 3437–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. C., and Collins, K. (2002). Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. USA 99, 6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty, T. J., Dupuis, S., and Autexier, C. (2002a). Rapid upregulation of human telomerase activity in human leukemia HL-60 cells treated with clinical doses of the DNA-damaging drug etoposide. Leukemia 16, 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty, T. J., Huard, S., Dupuis, S., and Autexier, C. (2002b). Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 22, 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty, T. J., Marie-Egyptienne, D. T., and Autexier, C. (2004). Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 24, 3720–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, G. B. (1989). The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59, 521–529. [DOI] [PubMed] [Google Scholar]
- Morin, G. B. (1991). Recognition of a chromosome truncation site associated with α-thalassaemia by human telomerase. Nature 353, 454–456. [DOI] [PubMed] [Google Scholar]
- Oulton, R., and Harrington, L. (2004). A human telomerase-associated nuclease. Mol. Biol. Cell 15, 3244–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y., Mian, I., and Lue, N. (2001). Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7, 1201–1211. [DOI] [PubMed] [Google Scholar]
- Rivera, M. A., and Blackburn, E. H. (2004). Processive utilization of the human telomerase template: lack of a requirement for template switching. J. Biol. Chem. 279, 53770–53781. [DOI] [PubMed] [Google Scholar]
- Singh, S. M., and Lue, N. F. (2003). Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc. Natl. Acad. Sci. USA 100, 5718–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow, B. E., Erdmann, N., Cruickshank, J., Goldman, H., Gill, R. M., Robinson, M. O., and Harrington, L. (2003). Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13, 698–704. [DOI] [PubMed] [Google Scholar]
- Wallweber, G., Gryaznov, S., Pongracz, K., and Pruzan, R. (2003). Interaction of human telomerase with its primer substrate. Biochemistry 42, 589–600. [DOI] [PubMed] [Google Scholar]
- Wang, H., and Blackburn, E. H. (1997). De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 16, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Gilley, D., and Blackburn, E. H. (1998). A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. 17, 1152–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J., Peng, Y., Mian, I., and Lue, N. (2000). Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20, 5196–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G.-L., and Blackburn, E. H. (1991). Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell 67, 823–832. [DOI] [PubMed] [Google Scholar]
- Zahler, A. M., Williamson, J. R., Cech, T. R., and Prescott, D. M. (1991). Inhibition of telomerase by G-quartet DNA structures. Nature 350, 718–720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.