Abstract
Dynein is a critical mitotic motor whose inhibition causes defects in spindle pole organization and separation, chromosome congression or segregation, and anaphase spindle elongation, but results differ in different systems. We evaluated the functions of the dynein–dynactin complex by using RNA interference (RNAi)-mediated depletion of distinct subunits in Drosophila S2 cells. We observed a striking detachment of centrosomes from spindles, an increase in spindle length, and a loss of spindle pole focus. RNAi depletion of Ncd, another minus-end motor, produced disorganized spindles consisting of multiple disconnected mini-spindles, a different phenotype consistent with distinct pathways of spindle pole organization. Two candidate dynein-dependent spindle pole organizers also were investigated. RNAi depletion of the abnormal spindle protein, Asp, which localizes to focused poles of control spindles, produced a severe loss of spindle pole focus, whereas depletion of the pole-associated microtubule depolymerase KLP10A increased spindle microtubule density. Depletion of either protein produced long spindles. After RNAi depletion of dynein–dynactin, we observed subtle but significant mislocalization of KLP10A and Asp, suggesting that dynein–dynactin, Asp, and KLP10A have complex interdependent functions in spindle pole focusing and centrosome attachment. These results extend recent findings from Xenopus extracts to Drosophila cultured cells and suggest that common pathways contribute to spindle pole organization and length determination.
INTRODUCTION
It is well established that accurate chromosome segregation is mediated by a protein machine, the spindle, which uses a variety of kinesins, dyneins, and microtubule (MT) polymer ratchets to generate forces associated with spindle assembly and chromosome motility (Karsenti and Vernos, 2001; Scholey et al., 2003; Wadsworth and Khodjakov, 2004). However, the precise roles of different motors and how multiple motors cooperate as ensembles remain unclear, and there remain significant inconsistencies in the proposed roles of specific force generators in different cell types. A case in point is the multifunctional motor protein dynein.
In mammalian cells, dynein activity is required for centrosome separation and chromosome alignment (Vaisberg et al., 1993; Echeverri et al., 1996) and is proposed to control spindle length by transporting the MT-deploymerizing kinesin-13 KLP10A to the spindle poles (Gaetz and Kapoor, 2004; Lawrence et al., 2004). In addition, a large body of evidence suggests that dynein–dynactin forms a complex with the nuclear mitotic apparatus protein NuMA and transports it to spindle poles where it is enriched in a crescent-shaped area and serves to organize MTs into focused spindle poles and to attach centrosomes to spindles (Heald et al., 1996, 1997; Merdes et al., 1996, 2000; Gaglio et al., 1997; Dionne et al., 1999; Quintyne et al., 1999). This activity is thought to cross-link centrosome-associated interpolar microtubules (ipMTs) to ipMTs whose minus ends are detached from centrosomes and lie within the body of the half-spindle (Mastronarde et al., 1993), thereby maintaining spindle pole MTs as an organized continuum.
The inhibition of dynein function in Drosophila has yielded variable results. For example, we previously microinjected dynein monoclonal antibodies and p50-dynamitin into embryos to create a gradient of inhibited dynein function decreasing from the injection site, plausibly mimicking an allelic series of loss-of-function mutants (Sharp et al., 2000a,b). The most severe effect, observed proximal to the injection site, was a zone of spindles displaying failures in spindle pole separation, whereas further away from the injection site we observed defects in chromosome congression and segregation on apparently normal-looking bipolar spindles, and further away still we observed defects in late anaphase B spindle elongation. However, we did not observe any defects in spindle organization and centrosome attachment comparable with those caused by perturbation of the dynein–dynactin–NuMA pathway in vertebrates. Studies of the mitotic functions of dynein in Drosophila mutant embryos revealed defects in early centrosome separation in agreement with our data, but in contrast to our data significant detachment of centrosomes from nuclei and spindle poles was observed (Robinson et al., 1999). Finally, a comprehensive proteomic analysis of multiple mitotic motor function in cultured Drosophila S2 cells yielded minor mitotic defects that were consistent with a delay in the metaphase–anaphase transition after dynein inhibition, but gross defects in chromosome movements and spindle integrity were not observed (Goshima and Vale, 2003).
Here, we have examined the role of dynein in mitosis in cultured Drosophila S2 cells and show that loss of its function leads to defects in spindle pole organization and centrosome attachment comparable with those seen in vertebrate cells, but different from our results by using fly embryos. We show that the RNAi depletion of the abnormal spindle (Asp) protein, which in some respects resembles vertebrate NuMA (Saunders et al., 1997; Carmo-Avides and Glover, 1999; Wakefield et al., 2001) or the spindle pole-associated kinesin-13 KLP10A (Rogers et al., 2004; Goshima and Vale, 2003), also lead to changes in spindle pole morphology. Moreover, the localization of these two proteins is partially disrupted by dynein depletion, suggesting they both require dynein activity for proper localization to spindle poles. Thus, we propose that in S2 cells, dynein–dynactin, Asp, and KLP10A play complex, interdependent roles in spindle pole organization.
MATERIALS AND METHODS
Cell Culture
Drosophila S2 cells were cultured as described previously (Clemens et al., 2000) in Schneider medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin at 27°C.
Double-stranded RNA (dsRNA) Preparation
Expression plasmids encoding the cDNA of the dynein IC (CG18000), p50-dynamitin (CG8269), and KLP10A (CG1453) were acquired from the Drosophila Genome Resource Center (Bloomington, IN), and Ncd-pET expression constructs were provided by Dr. L.S.B. Goldstein (Howard Hughes Medical Institute, University of California–San Diego, San Diego, CA). Genomic coding sequences for dynein heavy chain and Asp were used as templates to PCR amplify corresponding 600- to 900-residue regions (individual primer sequences may be found in Table S1). Each primer used in the PCR contained a 5′ T7 polymerase binding site (TAATACGACTCACTATAGGG). dsRNA was produced by in vitro transcription by using Megascript kits (Ambion, Austin, TX) according to the methods of Clemens et al., (2000) with minor modifications. S2 cells were treated twice with 50 μg of dsRNA for 4 d (added at days 1 and 3) to deplete dynein heavy chain (DHC), dynein intermediate chain (DIC), p50, and Asp or with 30 μg of dsRNA for 4 d to deplete KLP10A and Ncd. In comparison, Goshima and Vale (2003) used a single treatment of 1–5 μg of dsRNA for 7 d. The effect of dsRNA treatment on cell cultures was determined by immunoblotting, and its effect on individual cells was determined by immunofluorescence to be confident that the target protein was gone. Immunofluorescence microscopy of mitotic morphology was performed on aliquots of the same dsRNA treated as used for immunoblotting, and the reported phenotypes were consistent among at least 90% of mitotic cells in each case.
Antibodies and Western Blotting
A polyclonal Ncd antibody was prepared by immunizing rabbits with the glutathione S-transferase (GST)-Ncd tail (∼200-amino acid) protein. Antibodies were purified on Affigel protein A columns (Bio-Rad, Hercules, CA), acid eluted, neutralized in Tris buffer, dialyzed into PB buffer (10 mM NaPO4, pH 7.4, 1 mM MgCl2, 1 mM EGTA, and 1 mM dithiothreitol), and concentrated. This antibody was highly specific on immunoblots (Figure S1). DHC was detected with a mouse monoclonal antibody as described previously (Sharp et al., 2000a,b). Asp was detected with a rabbit polyclonal anti-Asp antibody provided by D. M. Glover (Department of Genetics, University of Cambridge, Cambridge, United Kingdom). KLP10A was detected with a rabbit polyclonal anti-KLP10A antibody (Rogers et al., 2004) provided by D. J. Sharp (Department of Physiology and Biophysics, Albert Einstein College of Medicine, Bronx, NY). Other antibodies used were rabbit anti-phospho-histone H3 antibody (Upstate Biotechnology, Lake Placid, NY), mouse anti-DIC antibody 74.1 (Chemicon International, Temeculca, CA), mouse anti-p50 dynamitin antibody (BD Biosciences PharMingen, San Diego, CA), and mouse anti-γ-tubulin and anti-tubulin DM1α (Sigma-Aldrich, St. Louis, MO). dsRNAi-treated or control lysates were prepared with 150 μl of cell lysis buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 1% NP-40, and protease inhibitors). Samples were run on 7.5–10% SDS-PAGE gels. Protein concentrations were determined using the Bradford protein assay.
Immunofluorescence Microscopy
After dsRNA treatment, S2 cells were plated onto coverslips previously coated with 0.5 mg/ml concanavalin A for 2 h and fixed either with 0.5% glutaraldehyde (EM Scientific, Gibbstown, NJ), 3% formaldehyde (EM Scientific), and 1 mg/ml saponin in BRB80 buffer at room temperature for 10 min or with 90% methanol, 10% formaldehyde at –20°C for 10 min. Cells were permeabilized with phosphate-buffered saline (PBS) + 0.1% Triton X-100 (Rogers et al., 2002). For blocking, coverslips were incubated at room temperature with PBS + 0.1% Triton X-100 + 0.3% bovine serum albumin (PBST) for 1 h. Blocked slides were incubated with diluted primary antibodies (anti-DHC, anti-DIC, anti-p50, anti-Asp, 1:50; anti-Ncd, anti-KLP10A, 1:1000; anti-H3, 10 μg/ml; anti-γ-tubulin and DM1α, 1:200) at room temperature for 2 h and then washed three times for 10 min in PBST. Fluorescent secondary antibodies donkey anti-rabbit Cy5, donkey anti-mouse Cy5, donkey anti-mouse Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at a final dilution of 1:500. DNA was stained with propidium iodide (1 μg/ml in PBS) for 20 min. Slides were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and visualized on an Olympus microscope equipped with an UltraView spinning disk confocal head (PerkinElmer Life and Analytical Sciences, Boston, MA) and a 100× 1.35-numerical aperture objective.
Data Analysis
Analysis was performed using MetaMorph (Universal Imaging, West Chester, PA). Images are shown as three-dimensional projections generated from stacks of six to eight sections (0.5 μm in thickness). The pole–pole distance is the length of a line drawn from one spindle pole to the other. Student's t test for unpaired data was performed using SigmaPlot (Systat Software, Point Richmond, CA). Line scans were generated from the average intensity across a 10-pixel-wide line drawn from one spindle pole to the other.
RESULTS
RNAi-mediated Depletion of Dynein–Dynactin Causes Defects in Spindle Morphology and Chromosome Positioning
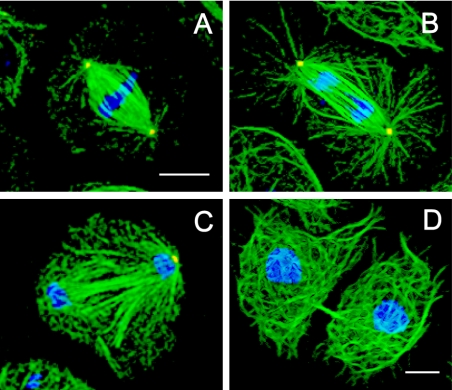
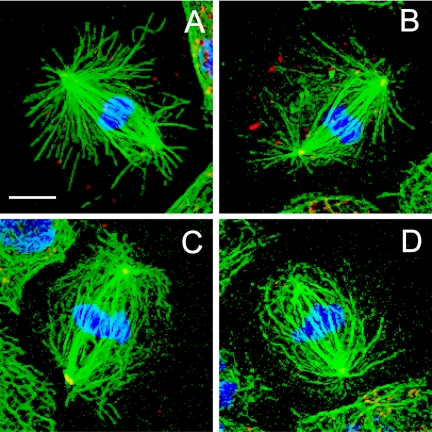
Immunofluorescence microscopy of control mitotic S2 cells typically revealed robust mitotic spindles (Figure 1) containing attached poles with distinct foci of γ-tubulin and radial arrays of astral MTs (Figure 1, A and B). As noted by others, S2 cell spindles display significant variability in their size, morphology, and number of microtubule organizing centers (Goshima and Vale, 2003; Maiato et al., 2004), far more than the multiple, relatively uniform spindles present in syncytial embryos (Sharp et al., 2000a). A consistent feature of S2 cell spindles is a region of decreased fluorescent tubulin intensity located between the spindles and the centrosomes, which is also present in published images of living mitotic S2 cells (Figure S2) and is therefore not a fixation artifact. This plausibly reflects a situation in which the minus ends of most spindle MTs are not anchored in the centrosome but instead have their minus ends free (Mastronarde et al., 1993). This would require a minus-end MT bundling activity to keep the S2 cell spindle focused at the poles, and a mechanism for attaching centrosomes to spindles by cross-linking centrosome-anchored MTs to free spindle MTs.
Figure 1.
Spindle morphology in control S2 cells. Cells plated on concanavalin A displayed a well spread morphology, allowing discrimination between the different stages of mitosis: immunofluorescence localization of α-tubulin (green), γ-tubulin (red), and DNA (blue) at metaphase (A), anaphase (B), telophase (C), and cytokinesis (D). Bar, 10 μm.
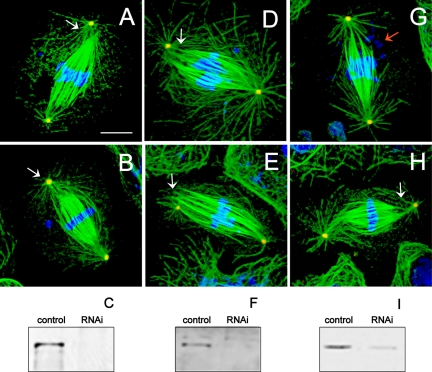
Mitotic S2 cells treated with dsRNA to deplete specific components of the dynein–dynactin complex, namely, DHC, DIC, or p50-dynamitin subunit, however, displayed a striking detachment of centrosomes from spindles and a loss of spindle pole focusing (Figure 2). This centrosome detachment leads to an increase in pole–pole spacing, but we also observed an increase in spindle length as determined by measurement of fluorescent tubulin distribution (Table 1). This phenotype, which differs significantly from those observed in our studies in dynein-inhibited embryos, was observed consistently, and no significant differences were observed after RNAi depletion of all the dynein–dynactin subunits examined.
Figure 2.
Defects in spindle pole organization in dynein–dynactin-depleted cells. Immunofluorescence localization of α-tubulin (green), γ-tubulin (red), and DNA (blue). Cells depleted of DHC (A and B), DIC (D and E), and p50 dynamitin (G and H) displayed longer than normal metaphase spindles with unfocused microtubules at spindle poles and loosely attached or completely detached centrosomes (white arrows). Some spindles displayed defects in chromosome alignment at the metaphase plate and apparent chromosome fragmentation (red arrow). Bar, 10 μm. Immunoblots of S2 cell cultures probed with anti-DHC (C), anti-DIC (F), and anti-p50 dynamitin (I) antibodies show 97, 82, and 80% depletion of protein, respectively (confirmed by densitometry) after dsRNA treatment. All lanes loaded with 50 μg of protein.
Table 1.
Mitotic indices, pole–pole distance, and microtubule length in dynein-KLP10A–and Asp-depleted S2 cells
| Pole—pole distancea | Spindle lengthb | Mitotic indexc | Prophase | Metaphase | Anaphase | Telophase | |
|---|---|---|---|---|---|---|---|
| Control | 9.0 ± 1.3 | 7.3 ± 1.0 | 3.6 | 23 | 50 | 10 | 14 |
| DHC RNAi | 14.0 ± 2.7* | 11.0 ± 3.0* | 10 | 14 | 74 | 12 | 0.5 |
| DIC RNAi | 16.0 ± 3.0* | 10.5 ± 2.0* | 12 | 6 | 89 | 4 | 0.5 |
| p50 RNAi | 13.0 ± 2.0* | 10.5 ± 1.5* | 9 | 16 | 79 | 4 | 0.6 |
| Asp RNAi | 14.0 ± 3.0* | 12.3 ± 2.0* | 8 | 9 | 67 | 4 | 10 |
| KLP10A RNAi | 11.0 ± 2.0* | 10.4 ± 1.5* |
Indicates statistically significant difference from control (P < 0.001) by using Student's t-test. All data are mean ± SD.
Pole—pole distance is expressed as micrometers and is the distance between centrosomes stained with anti-γ-tubulin antibody. Control, 87; DHC, 50; DIC, 52; p50, 30; Asp, 61; and KLP10A, 30 spindles.
Spindle length is expressed as micrometers and is the length of the spindle without centrosomes. Control, 24; DHC, 25; DIC, 21; p50, 17; Asp, 20; and KLP10A, 28 spindles.
Mitotic index and cell cycle stages were determined by scoring all mitotic cells stained with anti-phospho-H3 antibody and are represented as the percentage of mitotic cells in the total population. Control, 2855; DHC RNAi, 926; DIC RNAi, 1677; p50 RNAi, 2008; and Asp, 1026 cells.
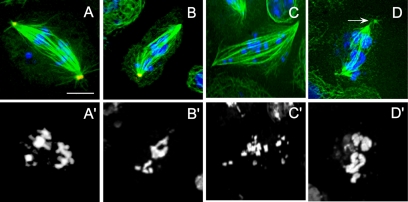
RNAi depletion of dynein–dynactin subunits also was associated with apparent defects in chromosome alignment on spindles (Figure 3), but whether this is due to a direct mechanical or regulatory role for dynein in chromosome motility (Sharp et al., 2000b; Basto et al., 2004) or is instead due to an indirect effect of spindle pole disorganization cannot be determined in fixed cells. This is in contrast to our previous work on living Drosophila embryos, where we observed defects in chromosome congression and segregation on absolutely normal-looking bipolar spindles, at least at the level of light microscopy of spindles in living embryos (Sharp et al., 2000b).
Figure 3.
Defects in anaphase chromosome positioning in dynein-depleted S2 cells. DHC (A, A′, B, and B′), DIC (C and C′), and p50 dynamitin (D and D′) RNAi-treated cells showed lagging and/or scattered chromosomes during anaphase in addition to disorganized spindle microtubules and poles (arrow). Immunofluorescent localization of α-tubulin (green), γ-tubulin (red), and DNA (blue). Bar, 10 μm.
The defects observed in spindle organization in the dynein–dynactin-depleted S2 cells was accompanied by a 1.5-fold increase in pole–pole distance relative to controls and an ∼2.7-fold elevation of the mitotic index, based on anti-phosphohistone-H3 staining (Table 1). This is consistent with previous results (Goshima and Vale, 2003).
RNAi-mediated Depletion of Ncd, Asp, and KLP10A Also Causes Defects in Spindle Pole Morphology
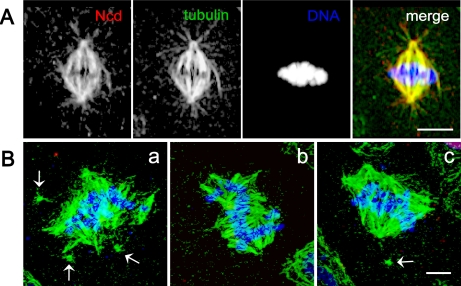
In addition to dynein, Drosophila contains another well characterized minus-end–directed mitotic motor, Ncd, a member of the kinesin-14 family (Lawrence et al., 2004). In Drosophila female meiosis Ncd has been reported to transport mini-spindles protein to the spindle where it associates with D-TACC to stabilize spindle poles (Cullen and Ohkura, 2001). By immunofluorescence with an anti-Ncd–specific antibody (Figure S1), we find that this protein extensively colocalizes with tubulin throughout the mitotic spindle (Figure 4A). We find that RNAi-mediated depletion of Ncd (Figure S1) produces significant defects in spindle morphology (Figure 4B) but that the phenotypes observed were obviously different from those seen after dynein–dynactin depletion. Instead of a single spindle as observed in controls, Ncd depletion seemed to result in the formation of multiple distinct mini-spindles, each associated with a distinct chromatin mass that were not properly connected. This is consistent with the idea that Ncd participates in a distinct spindle assembly pathway from that involving dynein (Lee et al., 2001; Cullen and Ohkura, 2001).
Figure 4.
Spindle morphology and Ncd localization in control and Ncd-depleted S2 cells. (A) During metaphase, Ncd is localized all over the spindle microtubules in a pattern indistinguishable from tubulin. (B) Three different examples (a–c) of dsRNA-treated, Ncd-depleted cells, which displayed wide multipolar spindles giving the impression of two to three fused spindles whose pole–pole axes are oriented in different directions. In some cases, the centrosomes are detached from the spindle poles (arrows). Immunofluorescent localization of Ncd (red), α-tubulin (green), and DNA (blue). Yellow indicates high overlap between tubulin and Ncd. Note the greatly reduced amount of Ncd staining after RNAi treatment (a–c) compared with controls. Bar, 5 μm.
In frog extracts, the dynein–dynactin–NuMA complex is proposed to transport the MT depolymerase Kif2a, a member of the kinesin-13 family (Lawrence et al., 2004), to the spindle poles where it depolymerizes MT minus ends, thereby controlling spindle length (Gaetz and Kapoor, 2004). To determine whether dynein might play a similar role in S2 cells, we initially RNAi-depleted KLP10A a spindle pole-associated kinesin-13 (Rogers et al., 2004) and compared the resulting spindle phenotype to that produced by dynein–dynactin depletion. When KLP10A was depleted as described in Materials and Methods, S2 cells displayed an aberrant bipolar spindle morphology (∼90% cells examined) characterized by ultralong astral and spindle MTs (Figure 5 and Table 1), although we observed monopolar spindles (∼80%) when RNAi treatment was prolonged to 7 d, in agreement with a previous study (Rogers et al., 2004). The abnormal bipolar spindles display some differences from those observed after dynein RNAi in which enlarged asters, for example, are not observed (Figure 5).
Figure 5.
Spindle morphology in KLP10A-depleted S2 cells. Depletion of KLP10A by using RNAi induced an apparent increase in the extent of polymerization of spindle microtubules increasing their apparent density and abundance. Four different examples of spindles (A–D) with long astral MTs. Bar, 10 μm.
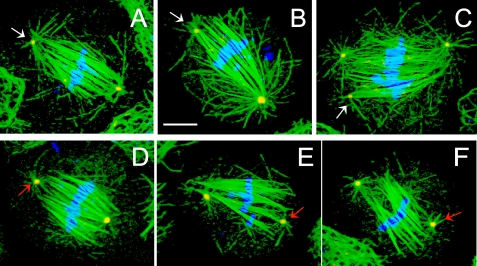
In vertebrates, it has been proposed that the dynein–dynactin-dependent transport of NuMA is required for the formation or maintenance of spindle poles (Merdes et al., 2000; Zeng, 2000). Although the Drosophila genome contains no true NuMA homolog, it does contain a protein with similar properties, the “abnormal spindle” or Asp protein (Saunders et al., 1997). For example, Asp and NuMA share similar sizes (∼200 kDa), high α helical content, similar sequence motifs (e.g., p34cdc-2 sites and calponin homology domains), MT-binding activity, and localization to spindle poles (Compton et al., 1992; Yang et al., 1992; Saunders et al., 1997; Novatchkova and Eisenhaber, 2002). Therefore, we investigated whether depletion of Asp by using RNAi also leads to mitotic defects in S2 cells, and again we observed a loss of spindle pole organization, longer spindles than controls, detached or loosely attached centrosomes, and elevated mitotic index as observed after dynein–dynactin depletion (Figure 6 and Table 1). Based on visual inspection, we find that in Asp-depleted cells, the loss of focusing of spindle poles seems more severe, and the detachment of centrosomes seems less severe, than in dynein–dynactin-depleted cells.
Figure 6.
Defects in spindle organization in Asp-depleted S2 cells. Asp RNAi-treated cells displayed longer spindles than controls with detached centrosomes (D, E, F, red arrows) and unfocused, disorganized spindle poles. In some cells, centrosomes remain loosely attached to the spindle (A, B, and C; white arrows) with apparent fragmentation of γ-tubulin foci (C; white arrow). Chromosomes are frequently misaligned (C and E). Immunofluorescent localization of α-tubulin (green), γ-tubulin (red), and DNA (blue). Bar, 10 μm.
RNAi-mediated Depletion of Dynein Causes the Partial Mislocalization of Asp and KLP10A at the Spindle Poles
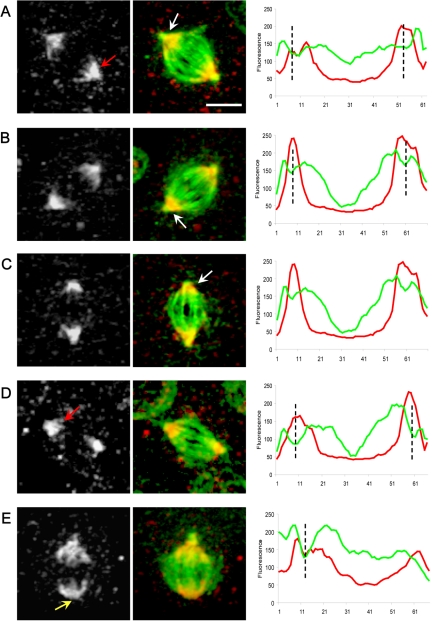
To investigate whether dynein–dynactin might play roles in the localization of Asp and KLP10A to spindle poles, we localized these proteins by immunofluorescence microscopy in dynein-depleted and control S2 cells. In control cells, both proteins were concentrated at the spindle poles (Figures 7 and 8). By densitometry of the fluorescent images along the pole–pole axis, Asp staining was seen throughout the polar regions of the half-spindles giving rise to “crescent-shaped” staining seen in the micrographs with little staining of the centrosomes themselves, and there was relatively intense staining of the “transition zone,” the region of low tubulin density lying between the ends of the half-spindles and the centrosomes (Figures 7 and S2). KLP10A also was concentrated around the poles, but unlike Asp, it seemed highly concentrated on centrosomes (Figure 8). Moreover, scans along the pole–pole axis reveal a decline in the intensity of KLP10A staining in the central region and peaks of fluorescence in the equator were observed, due to the presence of KLP10A on the kinetochores (Figure 8).
Figure 7.
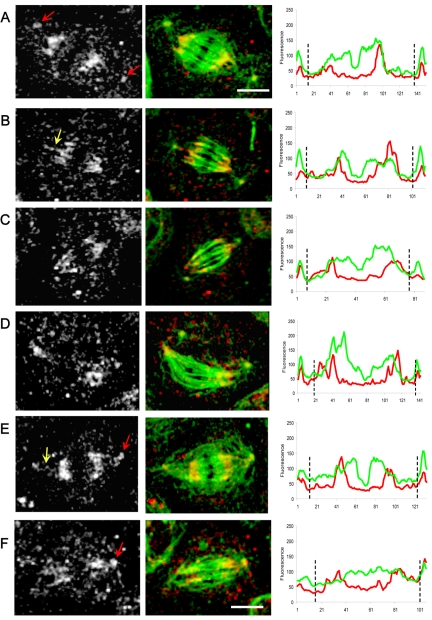
Localization and linescan quantitation of Asp in control S2 cell spindles. During metaphase, Asp is concentrated at the spindle poles (A–E white-gray panels; red arrows) becoming more diffuse toward the central spindle region. (B and C) Asp accumulates in the transition zone between the spindle and the centrosomes (white arrows). Spindles lacking a centrosome at one pole show a normal localization of Asp (E; yellow arrow). Bar, 5 μm. Corresponding linescans of fluorescence intensity (tubulin, green; Asp, red) along the pole-to-pole axis of spindles from A to E show a clear increase in the intensity of Asp staining in the transition zones between the centrosomes and spindle poles (position indicated by black dotted lines).
Figure 8.
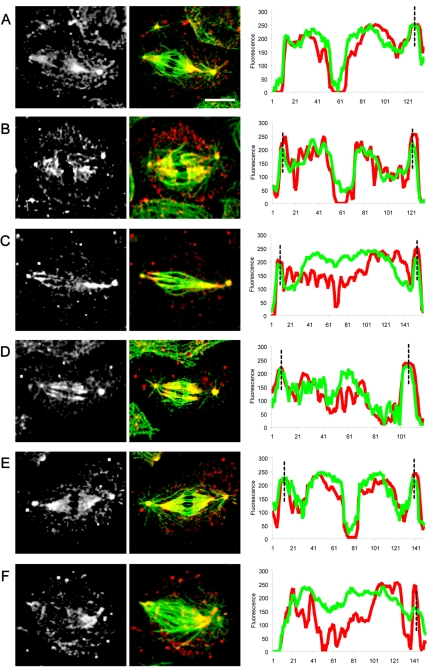
Localization and linescan quantitation of KLP10A in control S2 cell spindles. KLP10A localizes at the centrosomes, spindle poles (A–E white-gray; red arrows) and on the kinetochores (white arrows). Bar, 10 μm. Corresponding linescans of fluorescence intensity (tubulin, green; KLP10A, red) along the pole-to-pole axis of spindles from A to E show KLP10A staining at centrosomes (black dotted lines), and the peaks in the central region of the spindle correspond to KLP10A on the kinetochores.
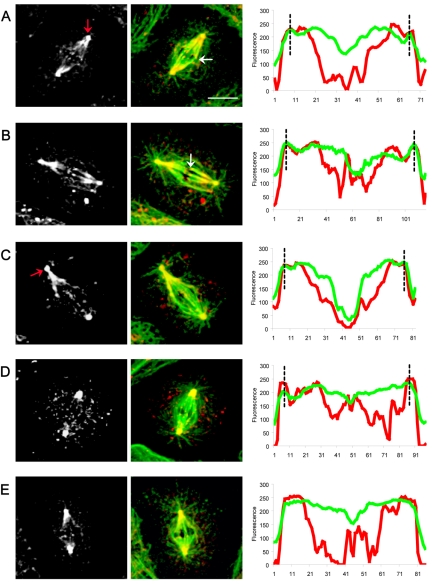
In dynein-depleted spindles, Asp and KLP10A show subtle but significant changes in their distributions in a manner suggesting that dynein activity may normally be required to bias the distribution of the two proteins toward the spindle pole. Immunofluorescence micrographs reveal that Asp and KLP10A both concentrate in the polar regions in a manner superficially resembling controls, but loss of dynein activity produces a partial mislocalization of both proteins (Figures 9 and 10). For example, in dynein-depleted cells, although Asp is relatively concentrated in the poles plausibly accounting for the partial pole-focusing observed in these spindles (Figure 2), there is a specific loss of Asp from the transition regions between the ends of the half-spindles and the centrosomes that is very obvious in the linescans (Figure 9). In dynein-depleted cells, KLP10A staining was high at the poles, as in controls, but moving along the pole–pole axis of the spindle there was an increase in fluorescence intensity at the central region of the spindle, indicating that KLP10A had redistributed throughout the spindle (Figure 10).
Figure 9.
Localization and linescan quantitation of Asp in dynein-depleted S2 cells. After DHC depletion by using RNAi (Figure 2), Asp localization was significantly perturbed. We saw a marked accumulation at the centrosomes (A, E, and F white-gray panels; red arrows) “wispy” fibers of Asp connecting centrosomes to half-spindles (B and E; yellow arrows) and a more diffuse localization of Asp throughout the spindles, including the central spindle (D and F). Bars, 10 μm. Corresponding line scans of fluorescence intensity (tubulin, green; Asp, red) along the pole-to-pole axis of spindles from A to F clearly reveal a loss of Asp from the transition zones between centrosomes and spindles poles, with a corresponding accumulation of the protein at the centrosomes (black dotted lines).
Figure 10.
Localization and linescan quantitation of KLP10A in dynein-depleted S2 cells. After DHC depletion by using RNAi, KLP10A localization was significantly perturbed. KLP10A is all over the spindle (white-gray panels) instead of being concentrated at the poles (compare with Figure 8). Linescan analyses indicate an increase of KLP10A intensity in the central spindle (tubulin, green; KLP10A, red). Bar, 10 μm.
Depletion of dynein–dynactin apparently does not cause the mislocalization of all mitotic proteins that contribute to spindle morphology. For example, we examined the distribution of both Ncd and Aurora-A after dynein RNAi and, although spindle morphology was perturbed as expected, we observed no obvious specific effects of dynein depletion on their localization. For example, in both control and dynein-depleted S2 cells, Ncd colocalized with MTs generally throughout the spindle, whereas Aurora-A localized specifically at the centrosomes and on a few cytoplasmic puncta (Figure S3).
DISCUSSION
These results suggest that in Drosophila S2 cells, dynein–dynactin act together with the NuMA-like protein, Asp, and the MT depolymerase KLP10A to play important roles in the organization of spindle poles. These results are in agreement with data obtained independently by Maiato et al. (2004), whereas Goshima and Vale (2003) observed less severe, albeit significant effects of dynein depletion on S2 cell mitosis in their proteomic survey of mitotic motor function. RNAi depletion of the minus-end–directed kinesin-14 Ncd also gave rise to disorganized spindle poles, but the morphology of the resulting spindles was significantly different, consistent with the idea that it organizes spindle poles via a distinct minus-end–directed transport pathway that targets a complex of two MT-associated proteins, Msps and D-TACC, to spindle poles, where it regulates MT stability and organization (Cullen and Ohkura, 2001; Lee et al., 2001).
Our results lead us to conclude that dynein–dynactin, Asp, and KLP10A cooperate in the organization of spindle poles, providing the required activities of focusing spindle poles and attaching centrosomes to the distal endings of spindles in spindles where the majority of MTs are not directly attached to the centrosomes (Figure S2; Mastronarde et al., 1993). The functional interdependencies of these three spindle proteins, however, seem complex. The simplest interpretation of our data can be summarized as follows. 1) The primary function of the minus-end motor complex dynein–dynactin is to cross-link centrosome-bound MTs to spindle MTs whose minus ends are detached from centrosomes and also to enhance the efficiency of targeting of KLP10A and Asp to the spindle poles (although not being absolutely essential for this). 2) Asp is a MT-binding protein that binds to the minus ends of spindle MTs and cross-links them into focused structures. 3) KLP10A binds to centrosomes and to the minus ends of MTs and depolymerizes MTs at the spindle poles. We propose that the efficiency of Asp and KLP10A localization to the spindle pole regions is enhanced by dynein, but they are capable of localizing there after dynein RNAi, possibly through the action of residual dynein through the action of a second motor with overlapping functions or possibly simply by diffusion and MT minus-end binding.
This explains the observed results of RNAi depletion as follows. 1) After dynein–dynactin depletion, centrosomes are detached from spindles; KLP10A and Asp become diffusely localized on half-spindles, less concentrated on the poles, and severely depleted from the transition zone between the centrosome and half-spindles; the residual Asp can focus the spindle poles from which the centrosomes have detached; and residual or cytosolic KLP10A can depolymerize astral MTs to restrict their length, although not as efficiently as in wild types, so spindle length increases. 2) In the absence of KLP10A, astral and spindle MTs grow too long, but residual Asp can focus the poles and dynein–dynactin can maintain the attachment of centrosomes to spindles. 3) In the absence of Asp, spindle poles become severely unfocused, but residual dynein can maintain the attachment of centrosomes to the disorganized poles, and residual KLP10A can partially regulate spindle length and cytosolic KLP10A can regulate astral MT length.
Our results with the MT depolymerase KLP10A are consistent with those obtained previously in S2 cells (Goshima and Vale, 2003; Rogers et al., 2004) and also lend support to Gaetz and Kapoor (2004), who propose that dynein localizes the kinesin-13, KIF2A to spindle poles in vertebrates. The results are consistent with the idea that, in the absence of a NuMA homolog in the Drosophila genome, Asp substitutes functionally for NuMA in S2 cells. Concordant with this idea is the observation that Asp and NuMA display a number of similarities, including similar molecular masses (220 kDa for Asp; 240/190 kDa for NuMA), high α helical content with a propensity for coiled-coil formation, calponin-homology domains, p34cdc-2 phosphorylation sites, MT-binding activity, and roles in spindle organization (Compton et al., 1992; Yang et al., 1992; Parry, 1994; Harborth et al., 1995; Saunders et al., 1997; Carmo-Avides and Glover, 1999; Wakefield et al., 2001; Haren and Merdes, 2002). Moreover, NuMA and Asp display similar localizations, being concentrated on prophase centrosomes, forming a crescent-shaped structure at spindle poles and central spindles/midbodies. These molecular properties and localization data are all consistent with the idea that the association of both Asp and NuMA to spindle poles allows them to cross-link MTs, thereby focusing the poles (Merdes et al., 2000, Wakefield et al., 2001).
Recently, Maiato et al. (2004) proposed that “kinetochore-formed kinetochore fibers” in S2 cells become organized at spindle poles by a dynein-dependent mechanism. Our observation that Asp is associated with focused poles lacking a centrosome in control cells (Figure 7E) suggests that dynein may enhance the efficiency of Asp localization to the poles, allowing it to organize such kinetochore MTs as well as other MTs that assemble by centrosome-independent pathways, thereby playing a critical role in the proposed “search-and-transport” mechanism (Maiato et al., 2004; Wadsworth and Khodjakov, 2004).
Our results with dynein depletion differ from the functional proteomic studies of Goshima and Vale (2003). Such proteomic studies are extremely valuable in dissecting the spectrum of functional components that cooperate in a complex system such as the spindle, but the different results observed here underscore the possibility that such high-throughput studies can overlook key functions. Indeed, we suspect that the discrepancy between our results and those of Goshima and Vale (2003) could be due to differences in factors such as the dsRNA concentration used in the RNAi procedure and its time of application (see Materials and Methods). Moreover, as noted by Maiato et al., (2004), RNAi treatment of a population of cells does not affect all cells equally and the treated culture is best considered to be a mixture of cells, some of which are efficiently depleted of the target protein, thereby revealing a loss-of-function phenotype, whereas others retain significant levels of the target protein and thus would phenocopy a “weak hypomorphic” mutant. In our studies, we only scored cells that were efficiently depleted of the target protein by immunofluorescence, so our phenotypic analysis was limited to cells in which RNAi depletion seemed to have been very effective according to the criterion of loss of immunofluorescence staining. It is difficult to apply this criterion in large-scale RNAi studies where the possibility exists that the apparent absence of a phenotype might reflect inefficient depletion of the target protein in the particular cells being scored.
The results reported here differ from our own previous results obtained in dynein- or dynactin-inhibited Drosophila embryos where comparable disorganized spindle poles and detached centrosomes were never observed (Sharp et al., 2000a,b), although we could not rule out subtle effects that would only be seen using, e.g., high-resolution imaging and quantification of spindle pole organization. The present results are instead more concordant with the data obtained by Robinson et al., (1999) who observed the detachment of centrosomes from spindles in dynein mutant embryos. The different results may reflect the different experimental methods used to disrupt dynein function, namely, antibody or p50-dynamitin microinjection (Sharp et al., 2000a,b) versus hypomorphic mutants (Robinson et al., 1999) versus RNAi (this study), but unfortunately technical obstacles prohibit us from testing this directly. For example, our RNAi studies were performed on S2 cells that were depleted of target antigen based on immunofluorescence and were thus likely to reflect a severe loss-of-function phenotype, and it is unclear how we could use this same approach to mimic the hypomorphic alleles studied by Robinson et al., 1999, in which unknown levels of residual mutant protein are presumed to be present. Moreover, our effort to apply RNAi depletion of various mitotic proteins to syncytial embryos has so far been unsuccessful. In addition, a sometimes unappreciated advantage of the large embryos studied by Sharp et al. (2000a,b) is that the injected inhibitors readily diffuse to form a concentration gradient and this in turn produces a corresponding gradient of individual spindle “phenotypes” of decreasing severity, and it would be extremely difficult to mimic these effects by microinjecting antibody and mutant protein inhibitors into small S2 cells. Therefore, we cannot exclude the possibility that differences in experimental techniques contribute to the differences observed.
An alternative possibility, however, is that dynein–dynactin is used to different extents in different cell types. In many cells, including S2 cells, accurate mitosis occurs at a relatively stately pace, and overlapping centrosome-directed and centrosome-independent assembly mechanisms occur together and aspects of each can be readily observed. Drosophila embryonic spindles, on the other hand, are adapted for the much faster mitoses that occur as the mononucleate fertilized egg rapidly divides its nuclei to form the multinuclear syncytial embryo. In embryonic spindles, spindle assembly, chromatid segregation, and spindle elongation occur orders of magnitude faster than in most cell types and to accommodate this “streamlined” mitotic motility, dynein may display specialized functions. For example, it is plausible that, during anaphase A, embryonic dynein not only participates in the spindle assembly checkpoint but also in feeding kinetochore MTs into the kinetochore to facilitate pacman-related depolymerization, thereby enhancing chromatid-to-pole motility (Sharp et al., 2000b; Basto et al., 2004; Rogers et al., 2004). Similarly, we suspect that rapid embryonic mitoses require that efficient centrosome-dependent assembly dominates and thus the influence of other, backup mechanism may only be observed when centrosomes become defective (Megraw et al., 2001). Thus, by comparing the streamlined embryonic spindles to the more stately S2 cell spindles, we may identify fundamental aspects of mitosis that are common to both types of spindle as distinct from functional specializations that reflect adaptations to rapid embryonic mitotic motility.
Supplementary Material
Acknowledgments
We thank members of the Scholey and Sharp laboratories for discussions, Drs. Steve Rogers and Ron Vale for advice on S2 cell culturing and RNAi, and Greg Rogers for preparing the Ncd antiserum. We are especially grateful to Dr. Patrizia Sommi and Amy Gunnarson for help in the preparation of the manuscript. This work was supported by National Institutes of Health Grant GM-55507 (to J.M.S.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1110) on May 11, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Basto, R., Scaerou, F., Mische, S., Wojcik, C. L., Gomes, R., Hays, T., and Karess, R. (2004). In vivo dynamics of the Rough Deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14, 56–61. [DOI] [PubMed] [Google Scholar]
- Carmo-Avides, M., and Glover, D. M. (1999). Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science 283, 1733–1735. [DOI] [PubMed] [Google Scholar]
- Clemens, J. C., Worby, C. A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B. A., and Dixon, J. E. (2000). Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97, 6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D. A., Szilak, I., and Cleveland, D. W. (1992). Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J. Cell Biol. 116, 1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, C. F., and Ohkura, H. (2001). Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3, 637–642. [DOI] [PubMed] [Google Scholar]
- Dionne, M. A., Howard, L., and Compton, D. A. (1999). NuMA is a component of an insoluble matrix at mitotic spindle poles. Cell Motil. Cytoskeleton 42, 189–203. [DOI] [PubMed] [Google Scholar]
- Echeverri, C. J., Paschal, B. M., Vaughan, K. T., and Vallee, R .B. (1996). Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz, J., and Kapoor, T. M. (2004). Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 166, 465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio, T., Dionne, M. A., and Compton, D. A. (1997). Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 138, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Vale, R. D. (2003). The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth, J., Weber, K., and Osborn, M. (1995). Epitope mapping and direct visualization of the parallel, in-register arrangement of the double-stranded coiled-coil in the NuMA protein. EMBO J. 14, 2447–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., and Merdes, A. (2002). Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci. 115, 1815–1824. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Blank, T., Sandaltzopoulos, R., Becker, P., Hyman, A., and Karsenti, E. (1996). Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420–425. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Habermann, A., Karsenti, E., and Hyman, A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization J. Cell Biol. 138, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti, E., and Vernos, I. (2001). The mitotic spindle: a self-made machine. Science 294, 543–547. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. J., Gergely, F., Geffers, K., Teak-Chew, S., and Raff, J. W. (2001). Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behavior. Nat. Cell Biol. 3, 643–649. [DOI] [PubMed] [Google Scholar]
- Maiato, H., Rieder, C. L., and Khodjakov, A. (2004). Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D. N., McDonald, K. L., Ding, R., and McIntosh, J. R. (1993). Interpolar spindle microtubules in PTK cells.J. Cell Biol. 123, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T. L., Kao, L. R., and Kaufman, T. C. (2001). Zygotic development without functional mitotic centrosomes. Curr. Biol. 11, 116–120. [DOI] [PubMed] [Google Scholar]
- Merdes, A., Ramyar, K., Vechio, J. D., and Cleveland, D. W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458. [DOI] [PubMed] [Google Scholar]
- Merdes, A., Heald, R., Samejima, K., Earnshaw, W. C., and Cleveland, D. W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova, M., and Eisenhaber, F. (2002). A CH domain-containing N terminus in NuMA? Protein Sci. 11, 2281–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, D. A. (1994). NuMA/centrophilin: sequence analysis of the coiled-coil rod domain. Biophys. J. 67, 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne, N. J., Gill, S. R., Eckley, D. M., Crego, C. L., Compton, D. A., and Schroer, T. A. (1999). Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. T., Wojcik, E. J., Sanders, M. A., McGrail, M., and Hays, T. S. (1999). Cytoplasmic dyenin is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S. L., Rogers, G. C., Sharp, D. J., and Vale, R. D. (2002). Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158, 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G. C., Rogers, S. L., Schwimmer, T. A., Ems-McClung, S. C., Walczack, C. E., Vale, R. D., Scholey, J. M., and Sharp, D. J. (2004). Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427, 364–370. [DOI] [PubMed] [Google Scholar]
- Saunders, R.D.C., Avides, M. C., Howard, T., Gonzalez, C., and Glover, D. M. (1997). The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 137, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey, J. M., Brust-Mascher, I., and Mogilner, A. (2003). Cell division. Nature 422, 746–752. [DOI] [PubMed] [Google Scholar]
- Sharp, D. J., Brown, H. M., Kwon, M., Rogers, G. C., Holland, G., and Scholey, J. M. (2000a). Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell 11, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., Rogers, G. C., and Scholey, J. M. (2000b). Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2, 922–930. [DOI] [PubMed] [Google Scholar]
- Vaisberg, E. A., Koonce, M. P., and McIntosh, J. R. (1993). Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 123, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth, P., and Khodjakov, A. (2004). E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol. 14, 413–419. [DOI] [PubMed] [Google Scholar]
- Wakefield, J. G., Bonaccorsi, S., and Gatti, M. (2001). The Drosophila protein Asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 153, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. H., Lambie, E. J., and Snyder, M. (1992). NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. 116, 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, C. (2000). NuMA: a nuclear protein involved in mitotic centrosome function. Microsc. Res. Technique 49, 467–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.