Abstract
Background
The β-glucosidases (BGLU) of glycoside hydrolase family 1 hydrolyze the glycosidic bond to release β-D-glucose and related ligands, which are widely involved in important physiological processes in plants. Genome-wide analysis of the BGLU genes in the model crops Arabidopsis thaliana and Oryza sativa revealed that they are functionally diverse. In contrast, the BGLU gene family in Tartary buckwheat remains unclear.
Results
This study identified the FtBGLU gene family based on Tartary buckwheat genomic data and analyzed the biological function of the FtBGLU gene using bioinformatics methods and the expression pattern of the gene using fluorescence quantitative PCR. The results showed that 39 BGLU genes were identified in Tartary buckwheat, which were classified into 10 subfamilies and one unclassified group. They were unevenly distributed on 10 chromosomes, and seven tandem duplication events involving 19 FtBGLU genes were observed, which mainly occurred in subfamily II. Their physicochemical properties are highly variable; however, they have relatively conserved exon-intron structures and high sequence homology in the subfamily, and most of the FtBGLUs contain conserved motifs, among which the expression products FtBGLU1, FtBGLU17, FtBGLU19, FtBGLU21, FtBGLU22, and FtBGLU28 have no β-glucosidase activity. Additionally, we analyzed the tissue expression specificity of 10 FtBGLU genes during Tartary buckwheat growth and development and their expression patterns under adversity stress and hormone treatments. Revealing the important role of the BGLU gene family in Tartary buckwheat growth and development, as well as its response to adversity, provides strong support for further analysis of its regulatory mechanisms and functional applications.
Summary
A total of 39 FtBGLU genes were identified. Bioinformatics analysis of the gene structure, evolutionary relationship, and expression pattern of the Fagopyrum tataricum BGLU gene family establishes a foundation for a better understanding and future research on the Tartary buckwheat BGLU gene family.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05919-3.
Keywords: Fagopyrum tataricum, BGLU gene family, Genome-wide analysis, Evolutionary relationships, Expression patterns
Background
β-glucosidases (BGLUs, EC 3.2.1.21), also known as β-D-glucosidases, are members of the glycoside hydrolase family 1 (GH1). In 1837, Liebig and Wohler first discovered BGLUs, also known as gentian disaccharidases or fiber disaccharidases, in bitter almonds [1, 2]. Since then, BGLUs have been ubiquitous in nature, also present in some bacteria, such as Aspergillus, Xylococcus, and yeast, and exhibit a wide range of functions. The catalytic specificity of the enzyme is demonstrated by its substrate and reaction specificity, leading to more than one glycoside hydrolase family. Currently, 145 glycoside hydrolase families exist in the CAZy database (http://www.CAZy.org/fam/accGH.html), and new hydrolase families are constantly being discovered. Studies have shown that BGLUs can be categorized into the GH3, GH5, GH9, GH10, and GH30 protein families based on their amino acid sequences [3–5]. In addition to binding to glucose on the substrate, BGLU must bind to various glycosides, resulting in different functional specificities. Most BGLUs identified in this study were also members of GH1 and were predominantly of bacterial origin, with fewer members of the glycoside hydrolase family 3 [6].
The reaction mechanisms of enzymes are closely related to their structures. BGLUs belong to the GH-A type and have a classical (β/α)8 barrel structure [3, 5, 6]. The active site contains two carboxylic acid residues that catalyze acid-base and nucleophilic reagents capable of catalyzing the hydrolysis of the non-reducing terminal between two or more carbohydrates or between carbohydrates and non-carbohydrate fractions of the β-1,4-glycosidic bond with the release of β-D-glucose and the corresponding ligand [7]. Bauer et al. have also demonstrated that the two carboxylic acid residues in the active site play a key role in the two-step double substitution reaction mechanism of β-D-glucosidases in catalyzing the hydrolysis of glycosidic bonds [8]. Additionally, BGLUs (GH1) can be connected by a helical linkage at the end of the C-terminal of the extended classical (β/α)8 barrel structure, forming the entrance to the substrate binding pocket [9]. In contrast, the active site of the GH3 enzyme consists of two active structural domains located between the (β/α)8 and (β/α)6 sandwich structures that catalyze a carboxylate residue but are not readily recognized [3, 10].
The biological functions of oligosaccharides and polysaccharides are diverse, and glycosyl hydrolases are involved in many fundamental life processes. For instance, the BGLU gene family is widely involved in important physiological processes in organisms, such as cell wall lignification [11–13], defense responses [14], phytohormone signaling activation [15, 16], secondary metabolism [17], and responses to adversity stresses [18–20]. Studies have shown that the cell wall of plants is a storehouse of carbohydrates, which contain a large number of glucoside residues, indicating that BGLUs can participate in plants cell wall development. Additionally in 1995, Leah et al. have showed that BGLUs are sensitive to oligosaccharides in germinating seedlings [21]. This sensitivity has also been reported by Hrmova et al. [22, 23]. In 1998, Akiyama et al. have confirmed that BGLUs could stabilize the cell wall and degrade oligosaccharides, thereby releasing lignin monomers [24]. Studies have shown that BGLUs (especially lignin monomers, and Os1BGLU1, Os3BGLU8, and Os7BGLU26) can hydrolyze oligosaccharides and cellulosic oligosaccharides, and degrade the cell wall in rice seedlings [25, 26].
β-glucosidases have metabolic functions, releasing glucosyl from metabolic intermediates while metabolizing them to natural products such as cyanohydrin [27], alkaloids [28, 29], and phenylpropanoid [30, 31]. Many phytohormones are glycosylated in plants and can modulate the biological activity by hydrolyzing inactive hormone-glycoside couplings [32]. Some rice BGLUs have been found to hydrolyze gibberellin glucoside [33], and BGLUs have also been found to hydrolyze or stimulate cytokinin glucoside in vitro in maize [34]. Recent, studies have found that AtBGLU18 [35], localized to the endoplasmic reticulum, can dehydrolyze ABA-GE and produce active abscisic acid (ABA), and that AtBGLU10 [36], localized to vesicles, can also act on ABA-GE (ABA-glucose abscisate) to release ABA, thereby accumulating higher ABA levels. Complementation, overexpression, and mutant experiments have demonstrated that overexpression of AtBGLU18 and AtBGLU10 increased ABA levels and showed tolerance to abiotic stresses, especially drought and salt tolerance. OsBGLUs [37] have been found to be induced by drought and low-temperature stress; for example, disruption of chloroplast-localized Os3BGLU6 in rice resulted in an increase in stomatal density and a decrease in ABA content and was responsive to drought and ABA treatments [38]. Not only have the model crops Arabidopsis thaliana and Oryza sativa been studied, but using RNA sequencing technology, the woody plant Populus trichocarpa PtBGLU8 and PtBGLU9 have been found to respond to drought stress through ABA signaling, ZmBGLU1 [39] has been found to increase tolerance to salt stress and is possibly related to its own defense response or phytohormone activation, and SrBGLU16 [40] has been found to be substantially upregulated in response to dark treatment. Collectively, the BGLU gene family exhibits functional biological diversity in networks that regulate plant growth, development, and response to adversity.
Tartary buckwheat (Fagopyrum tataricum L.) is a medicinal crop belonging to the Fagopyrum Mill annual plants [41]. Fagopyrum tataricum is rich in proteins, amino acids, unsaturated fatty acids, and other nutrients, as well as phenolic acids, alkaloids, anthraquinones, triterpenoids, flavonoids, and other biologically active substances that have various health functions [42, 43]. More and more scholars are studying the BGLU gene family, and its functions are continually being explored. This interest has grown, especially after identifying the corresponding functions of the BGLU gene family members in the model crops A. thaliana and O. sativa. Additionally, more plant BGLU gene family members have been identified. For example, 47 AtBGLUs have been identified and classified into 10 subgroups in A. thaliana [44], and 40 OsBGLUs have been identified and classified into eight subgroups in O. sativa [45]. In Brassica rapa [46], Zea mays [39], Stevia rebaudiana [40], Populus trichocarpa [47], Ziziphus jujube [48], Medicago truncatula [49], Dendrobium catenatum [50], and Pyrus bretschneideri Rehd [51], 64 (10 subgroups), 26 (four subgroups), 19 (five subgroups), 44 (four subgroups), 35 (10 subgroups), 51 (seven subgroups), 22 (four subgroups), and 50 (eight subgroups) members of the BGLU gene family were identified, respectively. However, the BGLU gene family in Tartary buckwheat has not been identified, making it difficult to explore the function of its genes. Therefore, this study was based on Tartary buckwheat genomic data, used bioinformatics technology to identify the members of the FtBGLU gene family, and analyzed the physicochemical properties of the members of the gene family, gene structure, evolutionary relationships, chromosomal distribution, cis-acting elements, and multiple sequence comparisons. Real-time fluorescence quantification was also used to analyze the tissue expression specificity of Tartary buckwheat partial BGLUs during the grubbing period, the gene expression patterns of grains and husks during fruit development, and the response mechanism under different hormonal and abiotic stress treatments.
Results
Identification of BGLU gene family members
We identified all possible members of the BGLU family in the Tartary buckwheat genome and 39 FtBGLU genes. Because there is no specific nomenclature, the author named FtBGLU1-FtBGLU39 based on its position on the Tartary buckwheat chromosomes (Chr) (Table S1). The physicochemical properties, including protein length, location, domain, molecular weight (MW), predicted subcellular location, and isoelectric point (pI) of 39 BGLU gene members were analyzed. The 39 BGLU gene members were highly variable in the number of amino acids, with an average of 471. The gene with the least number of amino acids was FtBGLU22, with 173 amino acids, and the gene with the most was FtBGLU12, with 640 amino acids. The MW of the proteins averaged 53.76 kD, mostly in the range of 19.40 kD (FtBGLU22)–72.99 kD (FtBGLU12), which was consistent with the number of amino acids in a positive relationship. The pI of the FtBGLU family averaged 6.34, acidic, and distributed between 4.41 (FtBGLU22)–9.34 (FtBGLU5), and 74.36% of the FtBGLU proteins were enriched with acidic amino acids. All 39 FtBGLU proteins contain a Glyco_hydro_1 conserved structural domain. Based on the results of the subcellular localization prediction, the proteins encoded by the FtBGLU gene were found to be widely distributed. Of these, 23.08% were localized in the endoplasmic reticulum, 17.95% in the cytoplasm, 17.95% in chloroplasts, 12.82% in vacuoles, 12.82% extracellularly, 7.69% in the cytoskeleton, FtBGLU27 in the nucleus, FtBGLU12 in the plasma membrane, and FtBGLU16 in the mitochondria. The physicochemical properties of different members of the FtBGLU gene family are highly variable, and their subcellular localization is high; therefore, members of the FtBGLU gene family are hypothesized to be functionally more diverse and involved in different physiological processes in an organism.
Phylogenetic analysis, classification, and homology analysis of the BGLU gene family
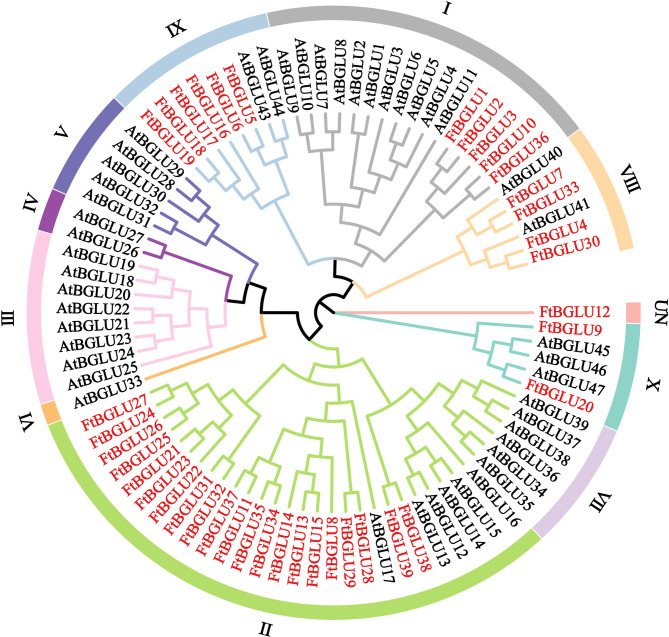
In this study, a phylogenetic tree (bootstrap = 1000) of 39 identified FtBGLU proteins and 47 reported AtBGLU proteins was constructed (Fig. 1, Table S2). The 86 BGLU proteins were categorized into 10 subfamilies and one UN group based on the topology of the phylogenetic tree and the classification of Arabidopsis BGLUs. The FtBGLU proteins were unevenly distributed among the five subfamilies and one unclassified group. Subfamily II contained the most FtBGLU proteins, and the UN group contained only FtBGLU12. Among them, 53.85% (21) of the FtBGLU proteins belonged to subfamily II, followed by six FtBGLU proteins in subfamily IX, five FtBGLU proteins in subfamily I, four FtBGLU proteins in subfamily VIII and two FtBGLU proteins in subfamily VII. The FtBGLU proteins were tightly aggregated (bootstrap support ≥ 80) in subfamilies I, II, and IX, indicating that these proteins have high homology and similar functions. However, subfamilies III, IV, V, VI, and VII in the absence of FtBGLU family members, which is likely to be in the Tartary buckwheat in the process of long-term evolution, and VII occurred in the BGLU gene local duplication, loss, or undifferentiated events. UN FtBGLU12 formed a topology distinct from that of AtBGLU, suggesting that it may represent a new direction in the evolution of Tartary buckwheat diversity.
Fig. 1.
Phylogenetic tree of the relationships between the BGLU proteins of F. tataricum and A. thaliana. I, II, III, IV, V, VI, VII, VIII, IX, X, and UN group represent the different subfamilies
Homology amino acid sequence analysis of FtBGLU genes
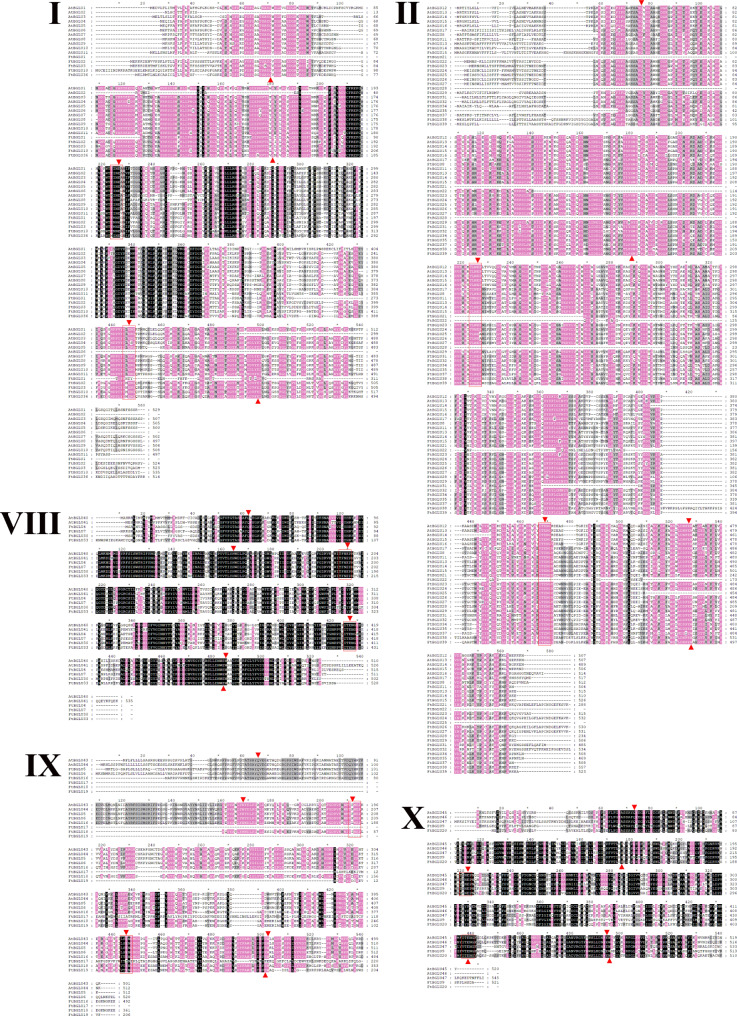
In this study, we analyzed the homology amino acid sequence of FtBGLU and AtBGLU proteins in subfamilies I, II, VIII, IX, and X (Fig. 2). The homology of the BGLU protein in subfamilies I, II, VIII, IX, and X was 58.71%, 55.24%, 70.58%, 56.24%, and 64.24%, respectively. Homology of the BGLU protein in subfamily IX was 56.24%, and that of the BGLU protein in subfamily X was 64.24%. The high homology indicates that the BGLU proteins are relatively conserved. Czjzek et al. identified amino acids critical for glucose binding (Q38, H142, E191, E406, E464, and W465), with E191 and E406 as the key active sites [52]. Based on the multiple sequence comparisons conducted in this study, 38 amino acid sequences (excluding FtBGLU12) generally agreed, with 68.42% of the FtBGLU proteins containing these six important amino acids. Among them, all members of subfamily I lacked W465; FtBGLU21 in subfamily II lacked Q38, H142, and E191 (consistent with FtBGLU28); FtBGLU22 was missing H142, E191, and E406; FtBGLU17 and FtBGLU19 in subfamily IX were missing Q38, H142, and E191; FtBGLU18 was missing Q38; FtBGLU17 with E406 mutated to I406; E464 mutated to K464; and W465 mutated to E465. The similarity of members among the subfamilies suggests that they have similar functions. In addition, 84.21% (32 FtBGLU proteins) contained glutamate residues. However, FtBGLU1, FtBGLU17, FtBGLU19, FtBGLU21, FtBGLU22, and FtBGLU28 were missing or mutated 1/2 glutamates in their conserved motifs, and the N-terminal active site of these six FtBGLUs were defective. In other words, the resulting β-glucosidase may not be active in catalyzing the hydrolysis of the substrate, which can be subsequently verified using enzymatic property assays.
Fig. 2.
Multiple sequence alignments of the BGLU domains of the members of five phylogenetic subfamilies. Black, pink, and gray backgrounds indicate 100%, ≥ 80%, and ≥ 60% amino acid homology, respectively. Red boxes indicate the conserved motifs of β-glucosidase TFNEP and I/VTENG. The red triangles point to six amino acids that are essential for glucose binding (Q38, H142, E191, E406, E464, and W465)
Analysis of BGLU gene structure and conserved motifs of the BGLU protein
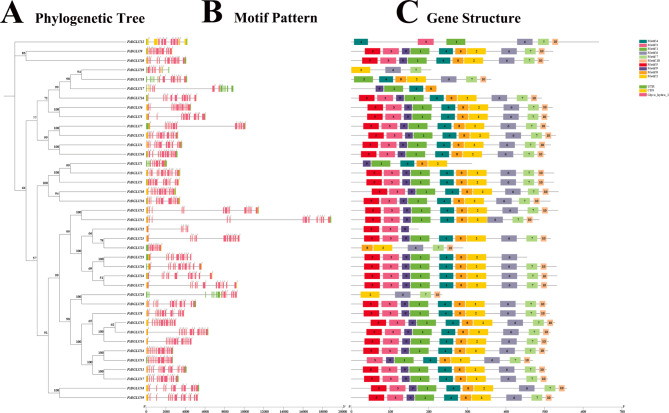
Based on the Tartary buckwheat genome sequence, we constructed a structural map of the buckwheat BGLU gene family (phylogenetic tree + motif pattern + gene structure) and obtained the intron-exon structure of 39 FtBGLUs, which led to a better understanding of the function of the FtBGLU genes (Fig. 3). The number of exons of FtBGLU gene family members varies from 6 to 13, with FtBGLU22 having the fewest exons and 16 FtBGLUs having the most exons. All 39 FtBGLUs had intronic sequences (Table S1). Moreover, 10.26% of FtBGLUs had 11 exons, 33.33% of FtBGLUs had 12 exons, and 41.03% of FtBGLUs had 13 exons.
Fig. 3.
Phylogenetic tree, gene structure, and motif pattern of BGLU genes in F. tataricum. (A) The phylogenetic tree constructed using the neighbor-joining method from the full-length sequence of the FtBGLU protein, with 1000 repetitive sequences per node. (B) Exons and introns are indicated by yellow rectangles and black lines, respectively. The numbers “0”, “1”, and “2” indicate the different stages of CDS in the gene. (C) Green, yellow, and pink rectangles represent the untranslated region (UTR), coding sequence (CDS), and BGLU structural domain (Glyco_hydro_1), respectively
Six genes had 6–10 exons, and the other 33 FtBGLUs had 11–13 exons, consistent with results from soybean [53] and rice [45]. In plants, the closer members of the same subfamily are evolutionarily, and the more homologous they are, the more similar they are likely to be in terms of the structure, pathways involved, and biological processes. Comparative analysis revealed that the homology between individual genes in subfamilies VIII and X was the highest, followed by subfamilies I, II, and IX. Among them, FtBGLU1 had eight exons (subfamily I, 1/5); FtBGLU21 and FtBGLU22 had seven and six exons, respectively (subfamily II, 2/21); and FtBGLU19 had nine exons (subfamily IX, 1/6), suggesting that the presence of these four FtBGLU genes may allow its subfamily to show greater diversity.
We further analyzed 39 FtBGLU proteins in Tartary buckwheat, which have not been previously reported. The conserved motifs of the protein were analyzed, and 10 conserved motifs were selected (named motifs 1–10). The results of the conserved domain analysis (Table S3) showed that motifs 1 and 2 were the longest, containing 50 amino acid residues, followed by motifs 3, 6, 5, 4, and 10, which were the shortest, with only 16 amino acid residues. The 10 motifs were found in most of the FtBGLU family members (30/39), and all of them were in the order of “5, 3, 9, 1, 4, 8, 2, 6, 7, and 10”. A description is a conserved motif of the family that performs a basic function, where a single motif appears in the family at least 31 times. However, FtBGLU1, FtBGLU21, FtBGLU22, FtBGLU28, FtBGLU35, FtBGLU17, FtBGLU18, and FtBGLU19 lacked a complete combination of conserved motifs based on this order (1–7 motifs missing). Motifs 1 and 6 contained the conserved amino acid sequences “TLNEP” and “ITENG”, respectively. The analysis revealed that FtBGLU1 and FtBGLU17 lacked motif 6; FtBGLU21, FtBGLU28, and FtBGLU19 lacked motif 1; and FtBGLU22 lacked motifs 1 and 6, which is consistent with the results of the multiple sequence comparison. This suggests that these six FtBGLUs may not be enzymatically active and may have resulted from incorrect splicing. Moreover, FtBGLU12 of the UN group contained active conserved motifs, but the motifs were not in the same order, suggesting the possibility of a new direction in the evolution of Tartary buckwheat with activity.
Cis-acting element prediction of the FtBGLU gene promoter
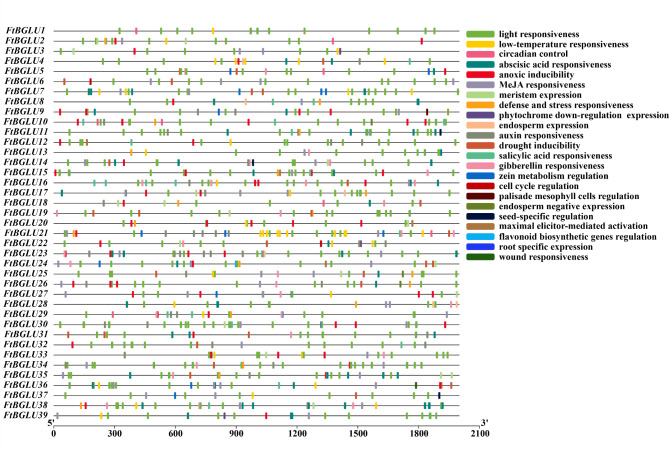
The upstream 2000 bp of the 39 identified FtBGLU genes were used as promoter sequences for cis-acting element analysis using PlantCARE software (Fig. 4, Table S4). A total of 986 components were predicted, with light-responsiveness components being the most abundant at 451 (45.74%). This was followed by hormone-related components (32.05%), including ABA responsiveness (11.36%), methyl jasmonate responsiveness component (11.16%), auxin inducibility component (4.06%), gibberellin (GA) responsiveness element (3.14%), and salicylic acid (SA) responsiveness element (2.33%). There are also components related to environmental stress: anoxic inducibility, low-temperature responsiveness, drought inducibility, defense, and stress responsiveness. Additionally, cis-acting elements include developmentally relevant elements, such as palisade mesophyll cell regulation, root-specific expression, cell cycle regulation, seed-specific regulation, and meristem expression. This indicates that the FtBGLU gene is important for Tartary buckwheat cell growth, normal plant growth and development, and response to adversity stress.
Fig. 4.
The cis-acting element of the promoter region (upstream 2000 bp) of 39 FtBGLU genes in F. tataricum
Chromosomal distribution of FtBGLU genes and gene duplication events
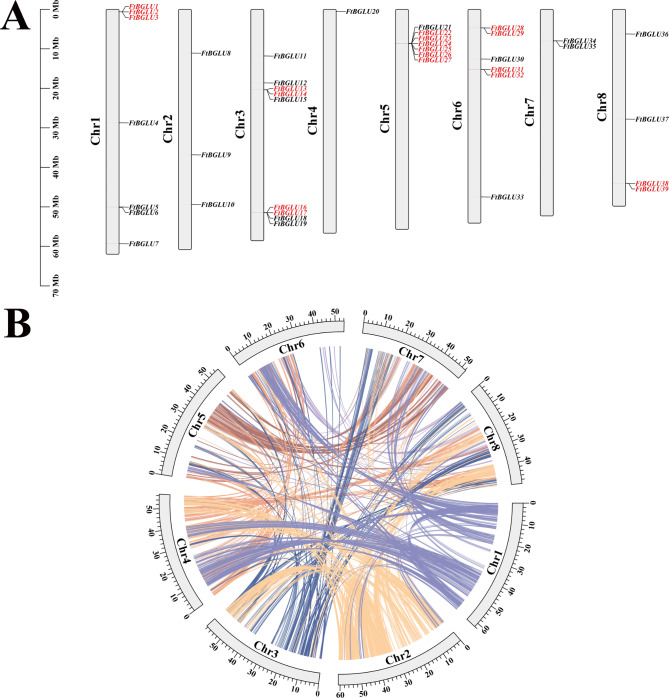
Studies have shown that there are many repetitive sequences in the genome and that both gymnosperms and angiosperms have undergone genome duplication events, leading to an increase in the number of genes in plants [54, 55]. Gene and fragment duplication events are important tools for gene amplification, and exploring the possibility of duplication events in gene families is important to advance the study of gene functions [56, 57]. In this study, we mapped the physical location of FtBGLU genes based on the latest genomic database of Tartary buckwheat (Fig. 5A). Notably, 39 FtBGLUs were unevenly distributed on eight Chr, with the most genes distributed on Chr3 (9, 23.08%), containing some members of subfamily IX (4/6) and subfamily II (4/21) as well as FtBLU12 from the UN group, followed by Chr5 (7, 17.95%), Chr1 (7, 17.95%), and Chr6 (6, 15.38%). Additionally, Chr8 contained four FtBGLUs, Chr2 contained three FtBGLUs, Chr7 contained two FtBGLUs, and Chr4 contained only one gene (FtBGLU20); that is, Chr contained the least number of genes. FtBGLU genes of the same subfamily are also randomly distributed on these eight chromosomes, especially subfamily II, which has the largest number of members. The random distribution on the chromosomes is more obvious, with no obvious preference, and most of the FtBGLU genes are distributed at the ends of the chromosomes and are tightly clustered, as in A. thaliana [32].
Fig. 5.
Schematic diagram of the chromosomal distribution and interchromosomal relationships of the F. tataricum BGLU genes. (A) Vertical bars represent the chromosomes of F. tataricum; each chromosome is labeled with a chromosome number on the left side of the chromosome; the scale on the left represents chromosome length. (B) Colored lines indicate all homologous blocks in the F. tataricum genome; chromosome numbers are labeled at the bottom of each chromosome
Two or more chromosomal regions within 200 kb of the same genomic region are referred to as tandem duplication events [58]. In this study, we found seven tandem duplication events on Chr1, Chr3, Chr5, Chr6, and Chr8 involving 19 (48.72%) FtBGLU genes (subfamilies I, II, and IX) (Fig. 5A, Table S5). The genes involved in one of the tandem duplication events found on Chr1 (FtBGLU1, FtBGLU2, and FtBGLU3) belonged to subfamily I, and the genes involved in one of the tandem duplication events found on Chr3 (FtBGLU16 and FtBGLU17) belonged to subfamily IX. Additionally, one tandem repetition event on Chr3 (FtBGLU13 and FtBGLU14), one tandem repetition event on Chr5 (FtBGLU22, FtBGLU23, FtBGLU24, FtBGLU25, FtBGLU26, and FtBGLU27), two tandem repetition events on Chr6 (FtBGLU28 and FtBGLU29, FtBGLU31 and FtBGLU32), and one tandem duplication event on Chr8 (FtBGLU38 and FtBGLU39) involve basal genes that belong to subfamily II. Duplication events occurred in almost half of the FtBGLU genes, with 73.68% of the members belonging to subfamily II; particularly, one duplication event on Chr5 involved six genes. This suggests a localized duplication event in the FtBGLU gene family, most likely a tandem duplication due to unequal hybridization. Gene fragment duplication analysis showed (Fig. 5B) that no genes involved in fragment duplication events were identified in the FtBGLU gene family.
Synteny analyses of FtBGLU genes
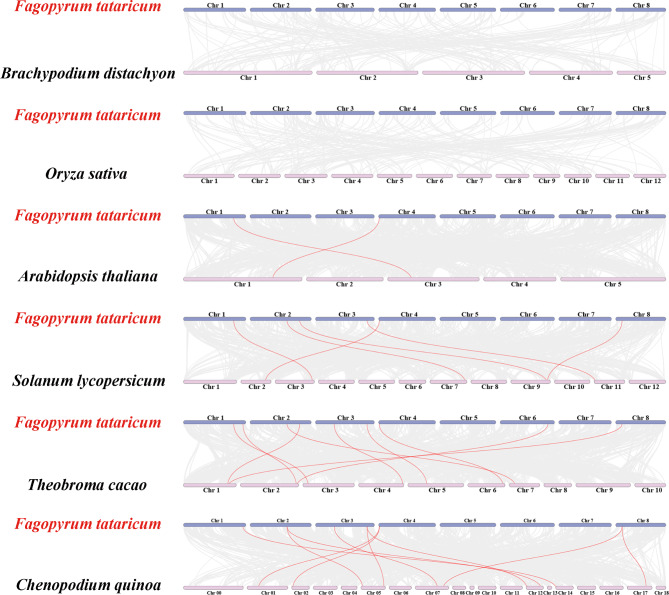
To further infer the kinship evolution of FtBGLUs, this study analyzed the interspecific covariance between Tartary buckwheat and monocotyledon/dicotyledonous and finally constructed an interspecific covariance map between Tartary buckwheat and two monocotyledons (B. distachyon and O. sativa) and four dicotyledons (A. thaliana, C. quinoa, T. cacao, and S. lycopersicum) (Fig. 6, Table S6). The results showed that FtBGLU genes covaried only with those of dicotyledonous plants and not with those of monocotyledonous plants. The FtBGLU genes had the highest number of homologous pairs with T. cacao (10), followed by C. quinoa (9), and the lowest number of homologous pairs with A. thaliana (2). Notably, FtBGLU20 shared a common line of homologous genes with all four dicotyledons. Co-linear homologs were found between FtBGLU6, FtBGLU9, FtBGLU16, and FtBGLU36 and the three dicotyledonous plants. FtBGLU9, FtBGLU16, FtBGLU20, and FtBGL36 had two co-linear homologs in C. quinoa. These five genes were hypothesized to be the most primitive in the FtBGLU gene family and may have played an important role in family evolution. In contrast, FtBGLU7, FtBGLU10, FtBGLU12, and FtBGLU33 covaried with only one or two dicotyledons, suggesting that these four genes may have formed gradually and at a more primitive time after the independent differentiation of dicotyledons. The relatively primitive FtBGLU genes described above belong to the subfamilies I, VIII, IX, X, and UN, whereas subfamily II, which contains the largest number of FtBGLU members, does not. This indirectly indicates the scientific validity of the classification and suggests that some FtBGLU genes in the FtBGLU gene family may have arisen as a result of tandem duplication events, which are the main driving force in the long-term evolution of Tartary buckwheat.
Fig. 6.
Synteny analyses of the BGLU genes between F. tataricum and six representative plants (B. distachyon, O. sativa, A. thaliana, C. quinoa, T. cacao, and S. lycopersicum). The gray lines on the background indicate the collinear blocks in the genomes of F. tataricum and other plants. Red lines highlight syntenic F. tataricum BGLU gene pairs
Analysis of the expression pattern of FtBGLU genes under different adversity stresses
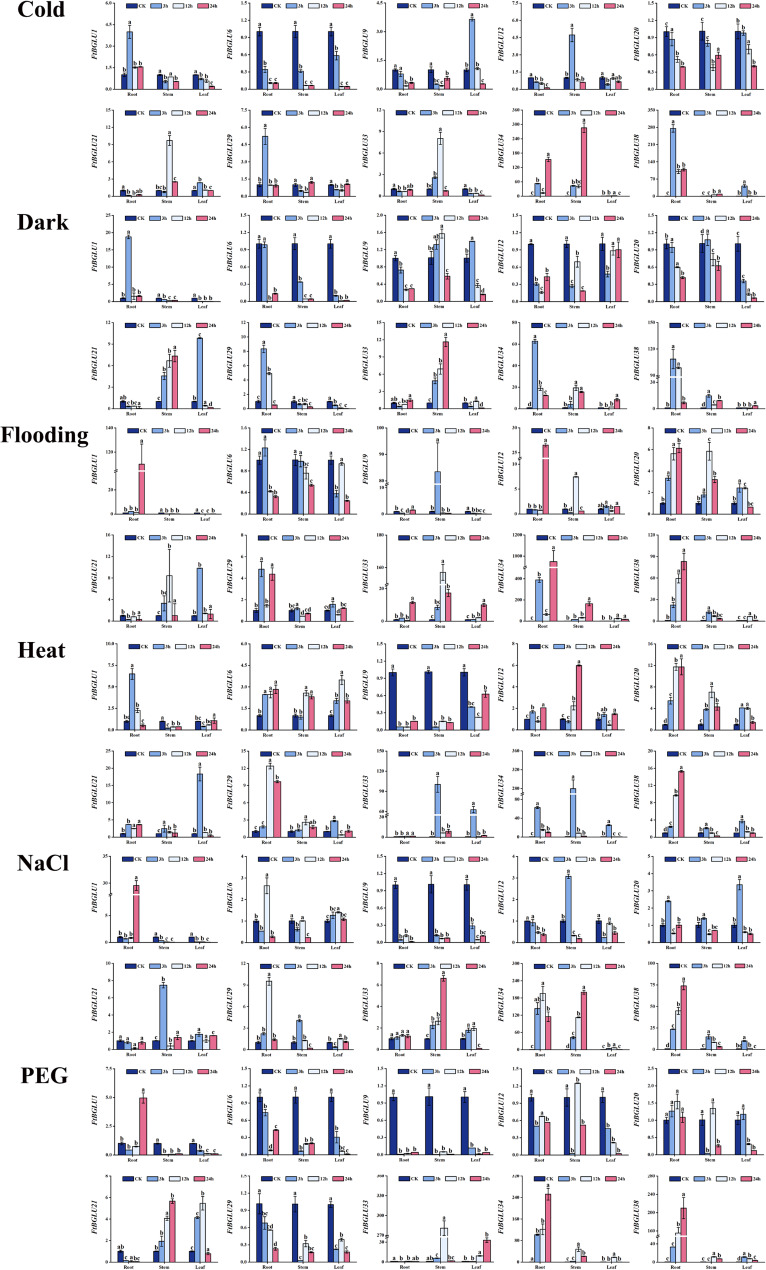
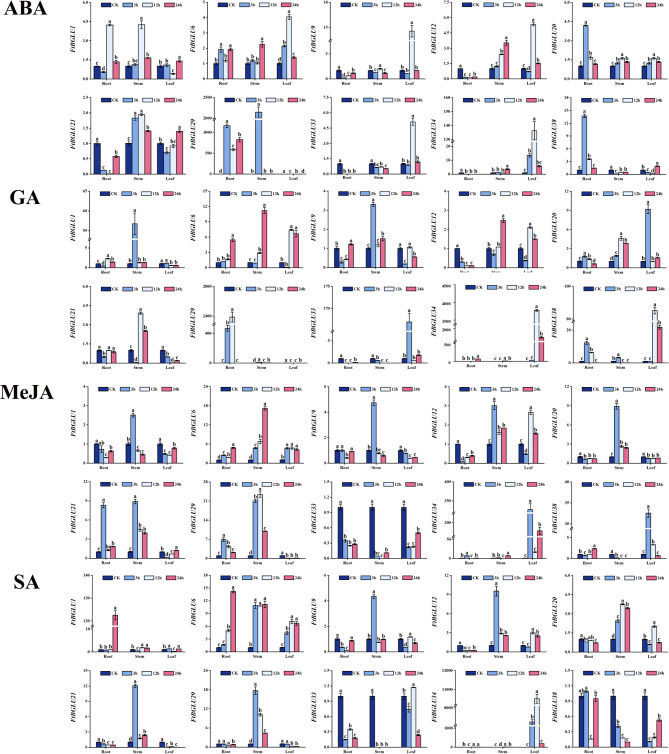
To investigate whether FtBGLU genes play a regulatory role in abiotic stresses, the expression patterns of 10 FtBGLU genes randomly selected under six abiotic stresses (cold, heat, dark, NaCl, flooding, and polyethylene glycol (PEG)) were explored in this study. The expression of 10 genes was analyzed using quantitative PCR technology (quantitative real-time polymerase chain reaction (qRT-PCR)) in the roots, stems, and leaves under each treatment (Fig. 7). FtBGLU34 (subfamily II) and FtBGLU38 (subfamily II) were significantly induced under all six stressors with diverse expression patterns. For example, under flooding stress, the expression of FtBGLU34 in roots, stems, and leaves showed a tendency to decrease and then increase, reaching a peak at 24 h. Under NaCl stress, the expression in roots showed a tendency to increase and then decrease, reaching a peak at 12 h, whereas the expression in stems reached a peak at 24 h. The expression of FtBGLU34 in roots tended to decrease, reaching a peak at 12 h; the expression in stems reached a peak at 24 h. Roots, stems, and leaves responded to dark stress and peaked at 3, 12, and 24 h, respectively. Another example is that the expression of FtBGLU38 under heat, PEG, flooding, and NaCl stress treatments peaked at 24 h in the roots, however, the increase in expression was not evident in the stems and leaves. Under cold stress, expression was induced in both roots and leaves, whereas the amount of change in the stems was insignificant. In contrast, under dark stress, expression was significantly increased in the roots, stems, and leaves. Second, FtBGLU33 was significantly expressed only in the heat, PEG, and flooding treatments, and expression induction occurred mainly in the stems and leaves. Finally, the relative expression of FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, and FtBGLU29 was lower than or equal to that of the CK under the six abiotic stresses.
Fig. 7.
Relative expression patterns of 10 FtBGLUs (FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, FtBGLU21, FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38) under different stresses (cold, heat, dark, NaCl, flooding, and PEG) at the seedling stage: expression patterns of 10 FtBGLUs at 3, 12, and 24 h in root, stem, and leaf. Values of the column chart are expressed as mean ± SD; the lowercase letters represent significant differences (p < 0.05, Duncan test)
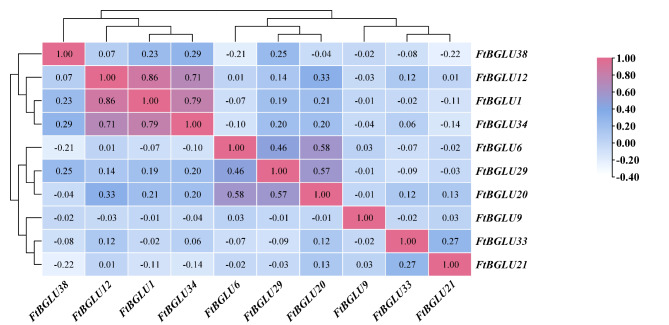
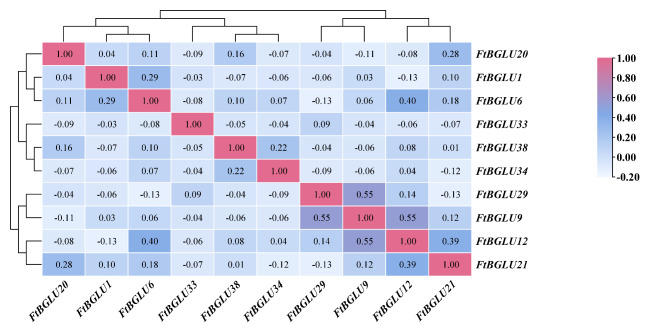
The correlation among the gene expression of these 10 FtBGLUs was also analyzed in this study (Fig. 8). The results revealed that the significant/extremely significant coefficients of correlation between the gene expressions of FtBGLUs were distributed in the range of − 0.21–0.86, with FtBGLU38 being significantly negatively correlated with FtBGLU6 (− 0.21), and FtBGLU12 being highly significantly positively correlated with FtBGLU1 (0.86). These results are consistent with those analyzed above.
Fig. 8.
Correlation analysis of 10 FtBGLUs (FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, FtBGLU21, FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38) under different stresses (cold, heat, dark, NaCl, flooding, and PEG) at the seedling stage. A positive number represents a positive correlation, and a negative number indicates a negative correlation. The right color scale (− 0.40 to 1.00) represents the normalized gene expression correlation
Analysis of the expression pattern of FtBGLU genes under hormone treatment
Cis-acting element analysis of the 2000 bp sequence upstream of the FtBGLU gene family yielded 32.05% hormone-responsive elements. Combined with the corresponding prediction data, this study chose to treat buckwheat with ABA, MeJA, GA, and SA at the seedling stage to analyze the expression patterns of the 10 FtBGLU genes under hormone treatments (Fig. 9). The results showed that the relative expression of FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38 varied significantly under GA treatment and was mainly expressed in the leaves. The remaining FtBGU genes with small but peak changes appeared during the pretreatment period (3 and 12 h). Under MeJA treatment, genes with significant changes in relative expression (FtBGLU34 and FtBGLU38) were expressed in the leaves of Tartary buckwheat, and the peak of expression appeared at 3 h. Genes with non-significant changes in relative expression were mainly found in the stems, which showed a tendency to increase and then decrease, and peak expression appeared at 3 h. Genes with significant changes in relative expression were mainly found in the stems, and peak expression appeared at 3 h. The relative expression of FtBGLU33 and FtBGLU38 was lower than that of the control under SA treatment, and the expression of these genes was suppressed under SA treatment, whereas FtBGLU1 and FtBGLU34 were significantly expressed in roots and leaves, respectively, and the genes that did not show significant changes in their relative expression peaked mainly in stems at 3–12 h. The relative expression of the genes in the roots and leaves was significantly higher than in the control, whereas the relative expression of the genes in the roots and leaves was significantly higher than in the control. Under ABA treatment, FtBGLU34 and FtBGLU29 were significantly expressed in leaves and roots, respectively. Genes with non-significant expression also appeared in leaves and roots, with peaks mainly in leaves at 12 h. Analysis of the correlation results (Fig. 10) revealed that both positive and negative correlations existed between the FtBGLU genes, but only the positive correlations were significant or highly significant, with correlation coefficients distributed between 0.18 and 0.55. The smallest positive and significant correlation coefficients were found between FtBGLU21 and FtBGLU6 (0.18, p < 0.05), and the largest positive and significant correlation coefficients were found between FtBGLU9 and FtBGLU29 (0.55, p < 0.01), followed by FtBGLU12 and FtBGLU9 (0.55, p < 0.01).
Fig. 9.
Relative expression patterns of 10 FtBGLUs (FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, FtBGLU21, FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38) under different hormone treatments (abscisic acid, ABA; methyl jasmonate, MeJA; gibberellin, GA; and salicylic acid, SA) at the seedling stage: expression patterns of 10 FtBGLUs at 3, 12, and 24 h in root, stem, and leaf. Values of the column chart are expressed as mean ± SD; the lowercase letters represent significant differences (p < 0.05, Duncan test)
Fig. 10.
Correlation analysis of 10 FtBGLUs (FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, FtBGLU21, FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38) under different hormone treatments (abscisic acid, ABA; methyl jasmonate, MeJA; gibberellin, GA; and salicylic acid, SA) at the seedling stage. A positive number represents a positive correlation, and a negative number indicates a negative correlation. The right color scale (− 0.20 to 1.00) represents the normalized gene expression correlation
Tissue expression specificity analysis of FtBGLU genes
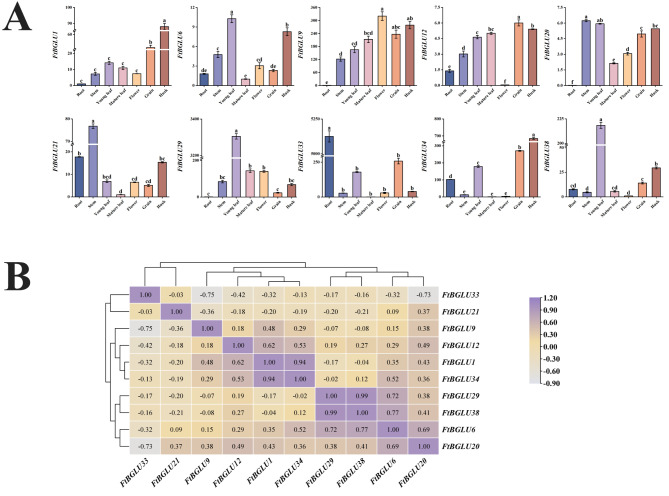
To further understand the role of FtBGLU genes in the development of Tartary buckwheat, we analyzed the expression patterns in roots, stems, young leaves, mature leaves, flowers, fruits, and husks during the mid-period of grain filling. We found that the expression patterns of different genes varied during the middle stage of grain filling, and tissue-specific expression also represented differences in regulatory mechanisms to some extent (Fig. 11A). Overall, the relative expression levels of FtBGLU6, FtBGLU12, and FtBGLU20 were low in all tissues, whereas the relative expression levels of FtBGLU1 and FtBGLU21 were low in most tissues. Secondly, the relative expression of most FtBGLU genes was high in young leaves, grains, and husks. The highest relative expression of FtBGLU29 was observed in young leaves, followed by FtBGLU38, FtBGLU33, FtBGLU34, and FtBGLU9 (The relative expression of FtBGLU29 was approximately 15, 18.4, 18.5, and 20 times higher than them, respectively). High relative expression of FtBGLU34, FtBGLU33, and FtBGLU9 was found in grains, and FtBGLU34 and FtBGLU9 in husks. This indicates that in the middle stage of grain filling, nutrients are mainly present in young leaves, grains, and husks, and FtBGLU33, FtBGLU34, and FtBGLU9 may play a role. Finally, the relative expression of FtBGLU33 was the highest among the 10 FtBGLU genes in root but hardly expressed in mature leaf. The specificity of the expression of individual genes in tissues during plant growth and development facilitates the organs to perform their respective roles. Moreover, correlation analysis showed (Fig. 11B) that FtBGLU33 was negatively correlated with the remaining nine FtBGLU genes. Furthermore, FtBGLU33 exhibited a highly significant negative correlation (p < 0.01) with FtBGLU9 and FtBGLU20, whereas FtBGLU38 had the largest correlation coefficient (0.99) and a highly significant positive correlation with FtBGLU29.
Fig. 11.
Relative expression patterns and correlation analysis of 10 FtBGLUs (FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, FtBGLU21, FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38). (A) Expression patterns of 10 FtBGLUs at the mid-grain-filling stage in root, stem, young leaf, mature leaf, flower, grain, and husk. Values of the column chart are expressed as mean ± SD; the lowercase letters represent significant differences (p < 0.05, Duncan test). (B) Correlation hierarchical cluster analysis between their expression in different tissues at the mid-grain-filling stage. A positive number represents a positive correlation, and a negative number indicates a negative correlation. The right color scale (− 0.90 to 1.20) represents the normalized gene expression correlation
Expression patterns of FtBGLUs in grain-filling stages
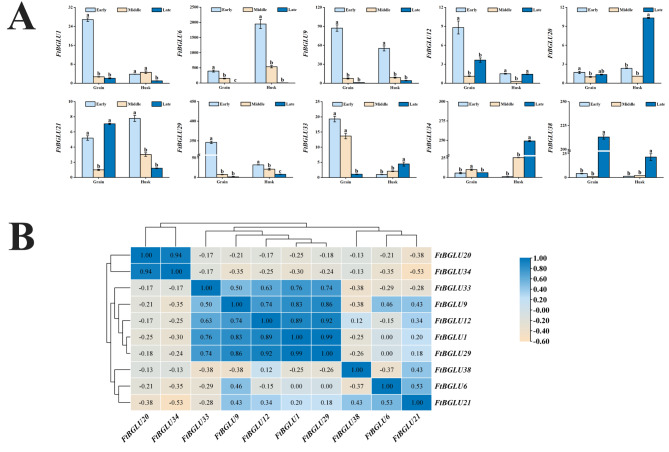
Grain-filling is often a critical period for determining the yield and quality of cereal crops. The above tissue-specific expression pattern in the middle stage of grain filling provides a perspective for understanding the role of the BGLU family in Tartary buckwheat development and further elucidating its mechanism of action. This study explored the expression patterns of FtBGLU in grains and husks during grain filling (early, middle, and late) (Fig. 12A). The relative expression levels of FtBGLU12, FtBGLU20, and FtBGLU21 were lower than those of the other seven genes. FtBGLU6, FtBGLU29, and FtBGLU9 were significantly expressed in both the grains and husks during grain filling (early). The highest relative expression of FtBGLU38 was found in grain filling (late) grains, and the highest relative expression of FtBGLU34 was found in filling (late) husks. Correlation analysis yielded (Fig. 12B) that FtBGLU29 was significantly positively correlated with FtBGLU1 with the largest correlation coefficient (0.99, p < 0.01) and with FtBGLU9 (0.86, p < 0.01). FtBGLU34 was significantly and positively correlated with FtBGLU20 (0.94, p < 0.01), whereas it was negatively and not significantly correlated with the other eight FtBGLU genes. FtBGLU38 was positively correlated with FtBGLU21 and negatively and non-significantly correlated with the other eight FtBGLU genes.
Fig. 12.
Relative expression patterns and correlation analysis of 10 FtBGLUs (FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, FtBGLU21, FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38) in grain and husk at different grain-filling stages. (A) Expression patterns of 10 FtBGLUs in the grain and husk during early, middle, and late grain-filling stages. Values of the column chart are expressed as mean ± SD; the lowercase letters represent significant differences (p < 0.05, Duncan test). (B) Correlation analysis between their expression in the grain and husk during the grain-filling stage. A positive number represents a positive correlation, and a negative number indicates a negative correlation. The right color scale (− 0.60 to 1.00) represents the normalized gene expression correlation
Discussion
Physicochemical properties and phylogenetic analysis of the FtBGLU protein
β-glucosidase is a multifunctional enzyme with special substrate hydrolysis properties that binds to non-reducing glucose residues at the end of oligosaccharide chains to cleave glycosidic bonds to hydrolyze cellobiose and other poly-oligosaccharide molecules to produce glucose. With the rapid development of bioinformatics, the biological functions of BGLU in different plants have gradually been identified. This is the first study to systematically analyze the BGLU gene family in the Tartary buckwheat in a more systematic manner in terms of bioinformatics and gene expression patterns. Based on the genomic data, a bioinformatics approach was used to identify that the FtBGLU family contains 39 FtBGLUs. The number of members was similar to that of BGLU members in the model crops A. thaliana [44] and O. sativa [45], suggesting that there were no excessive gene loss events. Whereas these 39 FtBGLU proteins showed large differences in physicochemical properties, the longest FtBGLU protein was FtBGLU12, which contained 640 amino acids, while the shortest FtBGLU protein (FtBGLU22) contained only 173 amino acids. The MW was distributed between 19.40 kD (FtBGLU22) − 72.99 kD (FtBGLU12). The relative MWs of β-glucosidases from different sources vary markedly but generally remain in the range of 40–300 kD [59, 60]. In this study, 15.4% of the FtBGLU proteins identified had a relative MW lower than 40 kD, indicating that buckwheat in the long-term evolutionary process of adapting to the environment caused by the degree of differentiation was inconsistent, resulting in a large number of differences between the FtBGLU proteins. The theoretical isoelectric points of most β-glucosidases are in the range of 3–6 [61]. FtBGLU proteins have an average pI of 6.34 and are rich in acidic amino acids, meaning that most β-glucosidases are acidic proteases. A few β-glucosidases with isoelectric points exist in the basic range [62] (10 FtBGLU proteins). Next, the subcellular localization of FtBGLU proteins was predicted, and it was found that FtBGLU proteins are widely distributed in the endoplasmic reticulum (9), cytoplasm (7), chloroplast (7), extracellular space (5), vacuole (5) cytoskeleton (3), nucleus (1), plasma membrane (1), and mitochondria (1). Access to physiological substrates for catalytic reactions in these organelles indirectly reflects the diverse functions of the BGLU gene family in Tartary buckwheat.
Based on the constructed phylogenetic tree analysis of AtBGLU and FtBGLU proteins, it was concluded that at least one member of FtBGLU proteins was present in subfamilies II (21), IX (6), I (5), VIII (4), and X (2), except for subfamilies II, IV, V, VI, and VII. These results are in close agreement with the grouping of Z. mays [39], S. rebaudiana [40], P. tomentosa [47], M. truncatula [49], and D. catenatum [50], although some variability has been observed in the model crops. Members of subfamily II far exceeded those of other subfamilies, comprising approximately 50% of the FtBGLU protein. This suggests that the FtBGLU family did not undergo complex differentiation during evolution.
Hydrolysis of the β-D-glycosidic bond in BGLUs involves a two-step process (glycosylation and de-glycosylation) that requires the simultaneous involvement of two glutamate residues, E191 (an acid/base catalyst) and E406 (a nucleophilic reagent) and has to be present simultaneously in the conserved motifs TFNEP and I/VTENG, respectively, of all BGLUs [52, 63–66]. In this study, AtBGLUs were subjected to multiple sequence comparisons with FtBGLUs, and FtBGLUs were found to have conserved structural domains as well as high sequence homology during evolution. Not all identified FtBGLU proteins had glycoside hydrolase activity, with FtBGLU1, FtBGLU17, FtBGLU19, FtBGLU21, FtBGLU22, and FtBGLU28 lacking or having mutated glutamate residues in the conserved motifs. This suggests that all 32 FtBGLUs found in the genome produce active BGLU [45, 67], whereas the BGLU produced by the remaining FtBGLU genes may be inactive, which remains to be verified. Apart from the active amino acid residues, Q38, H142, E464, and W465 are also important amino acid binding sites. Some members of subfamilies I, II, and IX contain deletions or mutations in important amino acid residues. This leads to diversification of BGLU proteins and affects their functional differentiation. The motifs of most FtBGLU (30/39) proteins follow the sequence “5, 3, 9, 1, 4, 8, 2, 6, 7, and 10”, i.e., they are conserved families. The conserved motifs TFNEP and I/VTENG containing active glutamate residues were present in motifs 1 and 6, respectively, and similarly FtBGLU1, FtBGLU17, FtBGLU21, FtBGLU28, FtBGLU19, and FtBGLU22 all lacked one or two motifs with active glutamate residues, in agreement with the results of the multiple sequence comparisons. This indicates that these six FtBGLU genes could not produce active BGLU.
Although the physicochemical properties of the members of the FtBGLU gene family vary widely, each subgroup has a relatively conserved exon-intron structure. In the BGLU family, the 13-exon pattern (13 exons-12 introns) is the most common structure of the ancestral gene, and the appearance of other structural patterns leads to the appearance of other gene structures [68]. Introns also play an important role in the evolution of species because they can increase the length of genes and the frequency of intergenic recombination, which has a regulatory role [69] We identified the exon-intron structure of 39 FtBGLU genes based on the annotation file of the buckwheat genome, and the number of introns was distributed in the range of 5–12, with a pattern of 13 exons being the most common, which is consistent with the fact that most of the crops are expressed, e.g., (A) thaliana [44], O. sativa [45], G. max [57], and (B) rapa [46].
Additionally, UN FtBGLU12 (UN group) has an active conserved motif, but the motifs are not aligned in the same order as all members of the other subfamilies. This suggests that FtBGLU12 may be a new direction for the evolution of the FtBGLU family and demonstrates the correctness and validity of cluster grouping of BGLU proteins.
Analysis of conserved structures and evolutionary relationships of FtBGLU genes
Currently, the number of the same gene family in plants varies among species, which may be related to the natural differentiation of species during evolution, gene duplication events, the frequency of gene recombination, and genome size [70–74]. In contrast, this differentiation is mostly ascribed to gene duplication events; the size of the quantitative gap is related to the magnitude of the degree of gene duplication [75]. The analysis of gene duplication provides a theoretical basis for a better study of the origin of new genes in gene families, formation of families, and diversity of gene expression levels [75, 76].
To further understand the evolutionary relationships among FtBGLU family members, we analyzed the chromosomal localization and gene duplication events of FtBGLU genes. In this study, 39 FtBGLU genes were named according to their locations on the Chrs, and the 39 FtBGLU genes were found to be unevenly distributed on eight chromosomes, and most of them were distributed at the ends of the chromosomes. Tandem duplication events occurred in nearly half the genes in the FtBGLU family, with one tandem duplication event involving five FtBGLU genes. Additionally, most genes that underwent tandem duplication belonged to subfamily II. No fragment duplication events occurred in the members of the FtBGLU gene family. To analyze the kinship of FtBGLU family members in more depth, this study constructed a covariance map was constructed between Tartary buckwheat and two monocotyledons (B. distachyon and O. sativa), and four dicotyledons (A. thaliana, C. quinoa, T. cacao, and S. lycopersicum). The results showed that the FtBGLU family only covaried with dicotyledons and introduced FtBGLU20 (subfamily X), FtBGLU6 (subfamily IX), FtBGLU9 (subfamily X), FtBGLU16 (subfamily IX), and FtBGLU36 (subfamily I) as the most primitive genes in the FtBGLU gene family. In summary, the FtBGLU gene family is closely related to dicotyledonous plants and has evolved to increase the number of BGLU gene family members mainly via tandem duplication events. Members of subfamily II may be derived from subfamilies X and IX, and the members of subfamily I may have undergone extensive gene duplication. Members of subfamily II may be transcriptionally translated into BGLU proteins that play functionally distinct roles, and functional redundancy may occur to some extent.
Expression pattern analysis of FtBGLU genes
GH1 is recognized for its BGLU activity and is largely involved in abiotic and biotic stress responses, growth, and development in plants [7, 45, 77–79]. Most members of subfamilies b, c, d, f, and i are resistant to stress [80]. In conclusion, the predictions of the cis-acting element revealed that in addition to the light-responsive element, the FtBGLU genes also contain a hormone-responsive element, an environmental stress-related element, and a growth and development-related element. Promoters and enhancers of cis-acting elements participate in the regulation of gene expression through the binding of trans-acting factors that regulate the activity of target genes. Therefore, 10 FtBGLU genes were selected from the subfamily containing FtBGLU genes to preliminarily investigate the expression patterns of the FtBGLU gene family during growth, development, and adversity stress. Furthermore, the expression of individual genes was tissue-specific, and the relative expression of young leaves, grains, and husks was high, indicating that nutrients were mainly concentrated in these three tissues during the irrigation period. The relative expression of FtBGLU6, FtBGLU12, and FtBGLU20 was generally low in tissues, whereas FtBGLU29, FtBGLU38, FtBGLU33, FtBGLU34, and FtBGLU9 were highly expressed in young leaves, grains, and husks. Further analysis of the expression patterns of FtBGLU genes in grains and husks during grain filling (early, middle, and late) revealed that FtBGLU6, FtBGLU29, and FtBGLU9 were significantly expressed during grain filling (early). FtBGLU38 and FtBGLU34 were most highy expressed in the grains and husks, respectively, at the late grain-filling stage. Correlation analysis revealed complex interactions among FtBGLUs, suggesting that FtBGLU genes have a complex network regulatory mechanism during the grain filling stage of Tartary buckwheat.
Next, the expression patterns of these 10 FtBGLU genes were investigated under six abiotic stressors: cold, heat, dark, NaCl, flooding, and PEG. Some genes were found to respond to the full range of stress mechanisms and have different expression patterns; however, others responded to only some stresses or were insensitive to adversity stress. FtBGLU34 and FtBGLU38 were significantly induced under all six stressors with diverse expression patterns. For example, the relative expression of FtBGLU34 in the roots, stems, and leaves showed a decreasing and then increasing trend under flooding stress, and the expression in the roots under NaCl stress showed an increasing and then decreasing trend. FtBGLU33 was significantly expressed in the stems and leaves under heat, PEG, and flooding stresses. In contrast, FtBGLU1, FtBGLU6, FtBGLU9, FtBGLU12, FtBGLU20, and FtBGLU29 were either inhibited or unresponsive to these six stressors.
The expression of FtBGLUs was differed at different time points and under different hormone treatments. FtBGLU34 was significantly induced in leaves following ABA, MeJA, GA, and SA treatments. Under the GA treatment, FtBGLU29, FtBGLU33, FtBGLU34, and FtBGLU38 were significantly upregulated in the leaves. FtBGLU34 and FtBGLU38 were also significantly induced in the leaves under the MeJA treatment. SA treatment inhibited the expression of FtBGLU33 and FtBGLU38. In contrast, FtBGLU1 and FtBGLU34 were induced in roots and leaves. Additionally, FtBGLU34 was significantly expressed in the leaves, and FtBGLU29 in roots under ABA treatment. No significant expression of FtBGLU genes was observed in any of the stems under hormone treatment, suggesting that under hormone stress, members of the FtBGLU gene family exhibit a high degree of tissue specificity and are mainly dependent on the leaves and roots for their responses. Combined with correlation analysis data, the diverse expression patterns of FtBGLU gene family members during growth and development, abiotic stress, and hormone treatments have laid a theoretical foundation for understanding the regulatory role of FtBGLU genes. The functional BGLU proteins found in A. thaliana improved drought resistance, salt tolerance, and frost protection (AtBGLU10, AtBGLU2, AtBGLU18, AtBGLU20, AtBGLU21, AtBGLU23, AtBGLU33, and others) [81–86]. BGLU proteins that are tolerant to abiotic stresses have been divided into several subfamilies. This indicates that more BGLU proteins have physiological functions. Both FtBGLU34 and FtBGLU38 that were significantly induced in growth and development, abiotic stress, and hormone-treated adversity, as well as FtBGLU29, which was most pronounced under ABA treatment, were distributed in the late-formed subfamily II. They release ABA for accumulation by transferring ABA-GE, which in turn is transported to the mechanism of response to growth, development, and stress. This also indirectly suggests that BGLU gene members of subfamily II may have similar functions.
Conclusions
This study was based on systematic research at the whole-genome level, discovering the FtBGLU gene family and conducting a series of analyses and verifications. Thirty-nine FtBGLU genes were identified and categorized into 10 subfamilies and one UN group. Subfamily II did not contain the most primitive genes; however, it maintained several FtBGLU gene families through tandem duplication events. Inactive FtBGLU protein was not present in subfamily II. This suggests that the genes of this subfamily may have specific functional requirements and selection pressures, eliminating inactive proteins during the evolutionary process, thus ensuring that the genes of this subfamily have specific and effective biological functions in the process of Tartary buckwheat growth, development, and adaptation to the environment. Members of the FtBGLU gene family vary widely in their physicochemical properties; however, a high degree of homology in gene structure and conserved motifs was observed. The genes were unevenly distributed across 10 chromosomes and were more closely related to dicotyledons. Additionally, FtBGLU34 and FtBGLU38, as BGLU genes were significantly induced in Tartary buckwheat, and FtBGLU29 was most significantly expressed under ABA treatment, which facilitated the maintenance of normal growth and development under adverse conditions by regulating the composition of the cell wall and secondary metabolites, such as the synthesis and degradation of physiological processes, thus enhancing plant resilience. This study lays a theoretical foundation for exploring the mechanisms of regulation during plant growth and development by identifying and analyzing the biological functions of the FtBGLU gene family.
With the advancement and improvement of modern biotechnology, the results of this study can provide new theoretical support and strategies for improving plant stress resistance and yield, as well new ideas and methods for Tartary buckwheat breeding.
Materials and methods
Identification and analysis of FtBGLU gene family members
The whole-genome sequences of Tartary buckwheat and annotation files were downloaded from the website (http://www.mbkbase.org/Pinku1/). First, two BLASTp methods (score ≥ 100, e ≤ 1e10) by Altschul et al. [87] and Liu et al. [88] were used to retrieve all sequences of Tartary buckwheat for possible identification of BGLU proteins. The Hidden Markov Model (HMM) file corresponding to the BGLU structural domain (PF00232) was downloaded (Pfam Protein Family Database [89]: http://pfam.sanger.ac.uk/). Secondly, the FtBGLU protein sequences were compared using the HMM model in HMMER 3.0 (http://plants.ensembl.org/hmmer/index.html; the cutoff value was set to 0.01) [90]. Third, the presence or absence of BGLU core gene sequences was verified using PFAM and the SMART program (http://smart.embl-heidelberg.de/smart/set_mode.cgiGENOMIC=11) to determine the presence or absence of BGLU core gene sequences [91, 92]. Finally, merged integration was performed to obtain the final FtBGLU genes. Additionally, basic characterization of FtBGLU gene family members was obtained from the ExPasy website (http://web.expasy.org/protparam/), including location, domain, MW, protein length, predicted subcellular location, and theoretical pI.
FtBGLU gene structure, conserved motifs, and multiple sequence comparison
First, the coding sequence of the FtBGLU genes were compared with the corresponding genomic DNA sequences using TBtools-ll (Toolbox for Biologists) v2.119 software. Next, the BGLU protein motifs were characterized using the MEME server (http://meme-suite.org/tools/meme) [93] with the parameters set to 10 motifs and 6–200 residues [88, 93, 94]. The exon-intron structure of the FtBGLUs was displayed using the Gene Structure Display Server (http://GSDS.cbi.pku.edu.cn) [65]. Finally, a gene structure diagram of the phylogenetic tree, motif pattern, and gene structure was constructed.
Multiple sequence comparisons of BGLU proteins of different subfamilies from F. tataricum and A. thaliana were performed using MEGA 11 software based on the default parameters of ClustalW [92]. After saving the FASTA files of multiple sequence alignments for each subfamily, the files were put into GeneDoc software to present the conserved structural domains of the sequences. Finally, the sequence homology values of the subfamilies were determined using DNAMAN software.
Chromosomal distribution and gene duplication of FtBGLU genes
Gene duplication events were analyzed using the Multiple Covariance Scanning Toolkit (MCScan X) [95] with default parameters. We used a Dual Synteny Plotter (https://github.com/ CJ-Chen/TBtools) function of TBtools to co-vary the FtBGLU genes with two monocotyledonous (B. distachyon and O. sativa) and four dicotyledonous species (A. thaliana, C. quinoa, T. cacao, and S. lycopersicum) that were analyzed for homology. Non-synonymous (ka) and synonymous (ks) values were calculated for each duplicate BGLU gene using Ka/Ks-Calculator 2.0 [96].
Phylogenetic analysis and classification of the FtBGLU gene family and cis-acting element prediction
Based on the classification of AtBGLU proteins, all identified FtBGLU and AtBGLU proteins were used to construct a phylogenetic tree with a bootstrap value of 1000 using the Jukes-Cantor model in MEGA 7.0, and Genetic R11 was assigned using the BLOSUM62 cost matrix. The cis-acting elements present in the promoter sequence (Upstream 2000 bp) of the FtBGLU gene family were predicted using the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Plant material, growth conditions, and abiotic stresses
The Tartary buckwheat variety selected for this experiment was “Chuanqiao-2”, which was provided by the Alpine Crops Research Station of Xichang Institute of Agricultural Science, Sichuan Province, China (102.20 E, 27.96 N). Uniform sized, pest-free “Chuanqiao-2” seeds were selected after strict sterilization and careful rinsing. The seeds were sown in two layers of sterile, washed textured filter paper (diameter, 9 cm) covered with Petri dishes (diameter, 90 mm). Thereafter, the Petri dishes were placed in a thermostatic incubator and subjected to 16 and 8 h day and night cycles at a temperature of 25 ± 1 °C and 20 ± 1 °C, respectively, with relative humidity maintained at 75%. Petri dishes were incubated for seven days until germination. Seedlings with uniform growth potential were transplanted into pots (25.5 cm in diameter and 17.5 cm in height) with autoclaved mixed nutrient soil (soil: substrate = 1:1), with three seedlings per pot. The pots were incubated under the same conditions. Six abiotic stress treatments and four hormonal treatments were applied to Tartary buckwheat during the three-leaf period. These included cold (4 °C), heat (40 °C), dark (complete shade), flooding (whole plant), PEG (6000: 25% w/v), NaCl: (150 mmol·L− 1), SA (100 µmol·L− 1), MeJA (100 µ µmol·L− 1), GA (100 µmol·L− 1), and ABA (100 µmol·L− 1) treatments, each containing five replicates. Three tissues (root, stem, and leaf) were collected at 0, 3, 12, and 24 h, respectively, and then the tissues were rapidly placed in liquid nitrogen and stored at − 80 °C. Furthermore, the roots, stems, young leaves, mature leaves, flowers, grains, and husks were extracted during the grain-filling stage. The grain and husk were collected during the early, middle, and late grain-filling periods, placed equally rapidly in liquid nitrogen, and stored at − 80 °C.
Total RNA extraction and cDNA synthesis
In this study, total RNA was extracted from the samples according to the instructions of the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek, Inc, USA), and RNA integrity was examined using 1% agarose gel electrophoresis. The concentration and quality of the RNA were determined using an ultra-micro spectrophotometer (Beijing Kaiao Technology Development Co., Ltd., China). cDNA was synthesized using a HiScript II Q RT SuperMix for qPCR Kit (Vazyme Biotech Co., Ltd., China) according to the manufacturer’s instructions.
qRT-PCR analysis
qRT-PCR can accurately measure gene expression, and reference genes can effectively correct the differences caused by RNA initiation and transcription efficiency, so it is widely used, but in fact, there is no ideal reference genes [97]. Li et al. [98] verified the expression stability of 7 candidate reference genes in Tartary Buckwheat and found that FtH3, the most commonly used reference gene in Tartary buckwheat standardization, was a stable gene under abiotic stress. The stress involved in this experiment is abiotic stress, so we chose FtH3 gene as the internal reference gene.
qPCR-specific primers for the FtBGLU genes were designed using Primer Premier software (version 5.0; Premier Corporation, Vancouver, British Columbia, Canada), while and FtH3 [98] was used as the internal reference gene (Table S7). The qRT-PCR method was performed using ChamQ Universal SYBR qPCR Master Mix Kit (Vazyme Biotech Co., Ltd, China) and Fluorescence Quantification Kit, 20 µL system. Detection was performed using a CFX96 Real-Time System instrument (BIO-RAD, USA), and then the relative expression of FtBGLU genes was calculated using the 2−ΔΔCt formula [99]. Three biological and technical replicates were used in the experiment.
Statistical analysis
We performed analysis of variance (ANOVA) (p < 0.05) and multiple comparisons (Duncan) on the data of relative expression using IBM SPSS 26.0 software, and the results are expressed as mean ± standard deviation (mean ± SD). Histograms were plotted using Origin 8.0 software. All experiments were performed in triplicates.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Table S1. List of the 39 FtBGLU genes identified in this study
Supplementary Material 2: Table S2. BGLU information in Arabidopsis thaliana.
Supplementary Material 3: Table S3. Analysis and distribution of conserved motifs in FtBGLU proteins.
Supplementary Material 4: Table S4. Cis-regulatory elements in the promoter region of FtBGLU genes.
Supplementary Material 5: Table S5. The tandem duplication events of FtBGLU genes.
Supplementary Material 6: Table S6. Synteny analyses of the BGLU genes between F. tataricum and representative plants (A. thaliana, C. quinoa, T. cacao and S. lycopersicum).
Supplementary Material 7: Table S7. Primer sequences for qPCR in this study.
Supplementary Material 8: Table S8. Relative expression of FtBGLUs in abiotic stress, grain filling, and different grain filling stages.
Acknowledgements
We express our gratitude to the College of Agronomy at Guizhou University in Guiyang, China, for supplying the essential experimental facilities and materials required for this research. Furthermore, we appreciate the valuable discussions and technical support provided by our colleagues.
Abbreviations
- BGLU
β-glucosidase
- GH1
Glycoside hydrolase family 1
- FtBGLU
Fagopyrum tataricum BGLU
- AtBGLU
Arabidopsis thaliana BGLU
- OsBGLU
Oryza sativa BGLU
- PtBGLU
Populus trichocarpa BGLU
- SrBGLU
Stevia rebaudiana BGLU
- ZmBGLU
Zea mays BGLU
- qRT-PCR
Quantitative real-time polymerase chain reaction
- ABA
Abscisic acid
- GA
Gibberellin
- MeJA
Methyl jasmonate
- SA
Salicylic acid
- UN
unclassified group
Author contributions
H. Y., X. Y., and J. R. conceived and designed the study. X. Y., W. W., and C. M. performed the experiments. H. Y., X. Y., and A. H. performed data analysis and wrote the manuscript. S. Y. and J. R. edited and drafted the manuscript. All the authors contributed to the manuscript and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of China, Grant/Award Numbers: (32312051, 32160669, 32161143005); Research and Integrated Application of Key Technologies for Green and High Yield in Mountainous Characteristic Agriculture, Grant/Award Number: [Guida Lingjun Hezi (2023) 07], and Construction of Scientific and Technological Innovation Talent Team for High-Quality and High-Efficiency Mechanization of Speciality Mixed Grains in Guizhou Province, Grant/Award Number: [Qiankehe Platform Talent-BQW (2024) 009].
Data availability
The Fagopyrum tataricum genome sequence information was obtained from the MKBbase website (https://www.mbkbase.org/Pinku1/). The Fagopyrum tataricum (Chuanqiao-2) used in the experiment was supplied by the Alpine Crops Research Station of the Xichang Institute of Agricultural Science, Sichuan Province, China.
Declarations
Ethics approval and consent to participate
Plant materials, and collection do not necessitate licensing. The plant materials were maintained in accordance with the institutional guidelines established by the College of Agriculture at Guizhou University. The methodologies employed adhered to the pertinent guidelines and regulations. It should be noted that this study did not involve any human participants or animal experimentation carried out by the authors.
Consent for publication
Not applicable.
Clinical trial number
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oliveira RP, Santos BV, Costal, Henrique MA, Daniel, Baffi MA. Xylanase and β-glucosidase production by aspergillus fumigatus using commercial and lignocellulosic substrates submitted to chemical pre-treatments. Ind Crop Prod. 2017;95:453–9. 10.1016/j.indcrop.2016.10.055. [Google Scholar]
- 2.Gonzalez-pombo P, Farina L, Carrau F, Batista-Viera F, Brena B. A novel extracellular β-glucosidase from Issatchenkia terricola: isolation, immobilization and application for aroma enhancement of white Muscat wine. Process Biochem. 2011;46(1):385–9. 10.1016/j.procbio.2010.07.016. [Google Scholar]
- 3.Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G. Conserved catalytic machinery and the prediction of a common Fold for several families of glycosyl hydrolases. P Natl Acad Sci USA. 1995;92(15):7090–4. 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–8. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Opassiri R, Pomthong B, Akiyama T, Nakphaichit M, Onkoksoong T, Ketudat Cairns M, Ketudat Cairns JR. A stress-induced rice (Oryza sativa L.) beta-glucosidase represents a new subfamily of glycosyl hydrolase family 5 containing a fascin-like domain. Biochem J. 2007;408(2):241–9. 10.1042/BJ20070734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litzinger S, Fischer S, Polzer P, Diederichs K, Welte W, Mayer C. Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique asp-his dyad mechanism. J Biol Chem. 2010;285(46):35675–84. 10.1074/jbc.M110.131037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrissa BA. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1992;280(2):309–16. 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer MW, Kelly RM. The family 1 beta-glucosidases from Pyrococcus furiosus and Agrobacterium faecalis share a common catalytic mechanism. Biochemistry. 1998;37(49):17170–8. 10.1021/bi9814944. [DOI] [PubMed] [Google Scholar]
- 9.Nijikken Y, Tsukada T, Igarashi K, Samejima M, Wakagi T, Shoun H, Fushinobu S. Crystal structure of intracellular family 1 β-glucosidase BGL1A from the basidiomycete Phanerochaete Chrysosporium. FEBS lett. 2007;581(7):1514–20. 10.1016/j.febslet.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins J, Lo Leggio L, Harris G, Pickersgill R. Beta-glucosidase, beta-galactosidase, family a cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold beta/alpha architecture and with two conserved glutamates near the carboxy-terminal ends of beta-strands four and seven. FEBS Lett. 1995;362(3):281–5. 10.1016/0014-5793(95)00252-5. [DOI] [PubMed] [Google Scholar]
- 11.Dharmawardhana D. A beta-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin[J]. Plant Physiol. 1995;107(2):331–9. 10.1104/pp.107.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escamilla-Treviño LL, Chen W, Card ML, Shih MC, Cheng CL, Poulton JE. Arabidopsis thalianaglucosidasesidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry. 2006;67(15):1651–60. 10.1016/j.phytochem.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Zhang Y, Zhao Y, Han WL, Lu JJ, Cheng X, Li HG, Jin Q, Cai YP. Genome-wide comparative analysis of the β-glucosidase family in five rosaceae species and their potential role on lignification of stone cells in Chinese white pear. Peer J. 2019;7(3):46971. 10.21203/rs.3.rs-46971/v1. [Google Scholar]
- 14.Duroux L, Delmotte FM, Lancelin JM, Kéravis G, Jay-Allemand C. Insight into naphthoquinone metabolism: β-glucosidase-catalysed hydrolysis of hydrojuglone β-D-glucopyranoside. Biochem J. 1998;333(2):275–83. 10.1042/bj3330275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ketudat Cairns JR, Mahong B, Baiya S, Jeon JS. β-Glucosidases: Multitasking, moonlighting or simply misunderstood? Plant Sci. 2015;241:246–59. 10.1016/j.plantsci.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Sun H, Xue Y, Lin Y. Enhanced catalytic efficiency in quercetin-4’-glucoside hydrolysis of Thermotoga maritima β-glucosidase a by site-directed mutagenesis. J Agric Food Chem. 2014;62(28):6763–70. 10.1021/jf501932v. [DOI] [PubMed] [Google Scholar]
- 17.Sampedro J, Valdivia ER, Fraga P, Iglesias N, Revilla G, Zarra I. Soluble and membrane-bound b-glucosidases are involved in trimming the xyloglucan backbone1. Plant Physiol. 2017;173:1017–30. 10.1104/pp.16.01713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Chen S, Dong Y, Ren R, Chen D, Chen X. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice. New Phytol. 2020;226(4):1042–54. 10.1111/nph.16416. [DOI] [PubMed] [Google Scholar]
- 19.Kong W, Zhong H, Deng X, Gautam M, Gong Z, Zhang Y, Zhao G, Liu C, Li Y. Evolutionary analysis of GH3 genes in six oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica. Plants. 2019,8(2):30. 10.3390/plants8020030 [DOI] [PMC free article] [PubMed]
- 20.Wang PT, Liu H, Hua HJ, Wang L, Song CP. A vacuole localized β-glucosidase contributes to drought tolerance in Arabidopsis. Chin Sci Bull. 2011;56:3538–46. 10.1007/s11434-011-4802-7. [Google Scholar]
- 21.Leah R, Kigel j, Svedsen l, Mundy J. Biochemical and molecular characterization of a barely seed β-glucosidase. J Biol Chem. 1995,270:15789–15797. 10.1074/jbc.270.26.15789 [DOI] [PubMed]
- 22.Hrmova M, Harvey AJ, Wang J, Shirley NJ, Jones GP, Stone BA, Høj PB, Fincher GB. Barley beta-D-glucan exohydrolases with beta-D-glucosidase activity. Purification, characterization, and determination of primary structure from a cDNA clone. J Biol Chem. 1996;271(9):5277–86. 10.1074/jbc.271.9.5277. [DOI] [PubMed] [Google Scholar]
- 23.Hrmova M, MacGregor EA, Biely P, Stewart RJ, Fincher GB. Substrate binding and catalytic mechanism of a barley beta-D-Glucosidase/(1,4)-beta-D-glucan exohydrolase. J Biol Chem. 1998;273:11134–43. 10.1074/jbc.273.18.11134. [DOI] [PubMed] [Google Scholar]
- 24.Akiyama T, Kaku H, Shibuya N. A cell wall-bound β-glucosidase from germinated rice: purification and properties. Phytochemistry. 1998;48:49–54. 10.1016/s0031-9422(97)01099-6. [DOI] [PubMed] [Google Scholar]
- 25.Opassiri R, Ketudat Caims IR, Akiyama T, Wara-Aswapati O, Svasti J, Esen A. Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 2003;165:627–38. 10.1016/S0168-9452(03)00235-8. [Google Scholar]
- 26.Seshadri S, Akyiyama T, Opassiri R, Kuaprasert B, Ketudat Cairns J. Structural andenzymatic characterization of Os3BGlu6, a rice β-glucosidase hydrolyzing hydrophobicglycosides and (1–3)-and (1–2)-linked disaccharides. Plant Physiol. 2009;151:47–58. 10.1104/pp.109.139436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hösel W, Tober I, Eklund SH, Conn EE. Characterization of β-glucosidases with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench seedlings. Arch Biochem Biophys. 1987;252:152–62. 10.1016/0003-9861(87)90019-1. [DOI] [PubMed] [Google Scholar]
- 28.Barleben L, Panjikar S, Ruppert M, Koepke J, Stöckigt J. Molecular architecture of strictosidine glucosidase: the gateway to the biosynthesis of the monoterpenoid indole alkaloid family. Plant Cell. 2007;19:2886–97. 10.1105/tpc.106.045682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia LQ, Ruppert M, Wang MT, Panjikar S, Lin HL, Rajendran C, Barleben L, Stöckigt. Structures of alkaloid biosynthetic glucosidases decode substrate specificity. ACS Chem Biol. 2012;7:226–34. 10.1021/cb200267w. [DOI] [PubMed] [Google Scholar]
- 30.Ahn YO, Shimizu BI, Sakata K, Gantulga D, Zhou ZH, Bevan DR, Esen A. Scopolin-hydrolyzing β-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 2010;51:132–43. 10.1093/pcp/pcp174. [DOI] [PubMed] [Google Scholar]
- 31.Baba SA, Vishwakarma RA, Ashraf N. Functional characterization of CsBGlu12, a β-glucosidase from Crocus sativus, provides insights into its role in abiotic stress through accumulation of antioxidant flavonols. J Biol Chem. 2017;292:4700–13. 10.1074/jbc.M116.762161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z, Escamilla-Treviño L, Zeng L, Lalgondar M, Bevan D, Winkel B, Mohamed A, Cheng CL, Shih MC, Poulton J, Esen A. Functional genomic analysis of Arabidopsis thaliana Glycoside hydrolase family 1. Plant Mol Biol. 2004;55(3):343–67. 10.1007/s11103-004-0790-1. [DOI] [PubMed] [Google Scholar]
- 33.Schliemann W. Hydrolysis of conjugated gibberellins by β-glucosidases from dwarf rice. Plant Physiol. 1984;116:123–32. 10.1016/S0176-1617(84)80069-3. [DOI] [PubMed] [Google Scholar]
- 34.Brzobohaty B, Moore l, Kristoffersen P, Bako L, Campos N, Schell J, Palme K. Releaseof active cytokinin by a β-glucosidase localized to the maize root meristem. Science. 1993;262:1051–4. 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- 35.Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell. 2006;126:1109–20. 10.1016/j.cell.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Wang PT, Liu H, Hua H, Wang L, Song CP. A vacuole localized β-glucosidase contributes to drought tolerance in Arabidopsis. Chin Sci Bull. 2011;56:3538–46. 10.1007/s11434-011-4802-7. [Google Scholar]
- 37.Su ZF. Bioinformation and expression patterns analysis of GH1β-glucosidases in Arabidopsis and rice[D]. TaiAn: Shandong Agricultural University; 2014. [Google Scholar]
- 38.Wang C, Chen S, Dong Y, Ren R, Chen D, Chen X. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice. New Phytol. 2020;226:1042–54. 10.1111/nph.16416. [DOI] [PubMed] [Google Scholar]
- 39.Gómez-Anduro G, Ceniceros-Ojeda EA, Casados-Vázquez LE, Bencivenni C, Sierra-Beltrán A, Murillo- Amador B, Tiessen A. Genome-wide analysis of the beta-glucosidase gene family in maize (Zea mays L. var B73). Plant Mol Biol. 2011;77(1–2):159–83. 10.1007/s11103-011-9800-2. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y, Zhang T, Xu X, Sun Y, Zhang Y, Hou M, Huang S, Yuan H, Tong H. Identification of GH1 gene family fgt members in Stevia rebaudiana and their expression when grown in darkness. Mol Biol Rep. 2020;47(11):8739–46. 10.1007/s11033-020-05920-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, He M, Fan Y, Zhao H, Gao B, Yang K, Li F, Tang Y, Gao Q, Lin T, Quinet M, Janovská D, Meglič V, Kwiatkowski J, Romanova O, Chrungoo N, Suzuki T, Luthar Z, Germ M, Woo SH, Georgiev MI, Zhou M. Resequencing of global Tartary buckwheat accessions reveals multiple domestication events and key loci associated with agronomic traits. Genome Biol. 2021;22(1):23. 10.1186/s13059-020-02217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Lv Q, Liu A, Wang J, Sun X, Deng J, Chen Q, Wu Q. Comparative metabolomics study of Tartary [Fagopyrum tataricum (L.) Gaertn] and common (Fagopyrum esculentum Moench) buckwheat seeds. Food Chem. 2022;371:131–125. 10.1016/j.foodchem.2021.131125. [DOI] [PubMed] [Google Scholar]
- 43.Sun W, Jin X, Ma Z, Chen H, Liu M. Basic helix-loop-helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): genome-wide identification, phylogeny, evolutionary expansion and expression analyses. Int J Biol Macromol. 2020;155:1478–90. 10.1016/j.ijbiomac.2019.11.126. [DOI] [PubMed] [Google Scholar]
- 44.Xu Z, Escamilla-Trevi˜no L, Zeng L, Lalgondar M, Bevan D, Winkel B, Mohamed A, Cheng C, Shih MC, Poulton J. Functional genomicanalysis of Arabidopsis thaliana Glycoside hydrolase family 1. Plant Mol Biol. 2004;55:343–67. 10.1007/s11103-004-0790-1. [DOI] [PubMed] [Google Scholar]
- 45.Opassiri R, Pomthong B, Onkoksoong T, Akiyama T, Esen A, Ketudat Cairns JR. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 beta-glucosidase. BMC Plant Biol. 2006,6(1):33. 10.1186/1471-2229-6-33 [DOI] [PMC free article] [PubMed]
- 46.Dong X, Jiang Y, Hur Y. Genome-wide analysis of glycoside hydrolase family 1 beta-glucosidase genes in Brassica rapa and their potential role in pollen development. Int J Mol Sci. 2019;20(7):1663. 10.3390/ijms20071663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bian Z, Wang D, Liu Y, Xi Y, Wang X, Meng S. Analysis of Populus glycosyl hydrolase family I members and their potential role in the ABA treatment and drought stress response. Plant Physiol Biochem. 2021;163:178–88. 10.1016/j.plaphy.2021.03.057. [DOI] [PubMed] [Google Scholar]
- 48.Yang M, Ma Y, Si X, Liu X, Geng X, Wen X, Li G, Zhang L, Yang C, Zhang Z. Analysis of the Glycoside Hydrolase Family 1 from wild jujube reveals genes involved in the degradation of Jujuboside A. Genes. 2023;14(6):1135. 10.3390/genes14061135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J, Ma L, Jiang W, Yao Y, Tang Y, Pang Y. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol Biochem. 2021;158:21–33. 10.1016/j.plaphy.2020.11.046. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Zhao M, Zhang X, Deng X, Li J, Wang M. Genome-wide identification and characterization of active ingredients related β-Glucosidases in Dendrobium catenatum. BMC Genomics. 2022;23(1):612. 10.1186/s12864-022-08840-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Zhang Y, Feng X, Peng F, Mazoor MA, Zhang Y, Zhao Y, Han W, Lu J, Cao Y, Cai Y. Analysis of the β-Glucosidase family reveals genes involved in the lignification of Stone cells in Chinese White Pear (Pyrus Bretschneideri Rehd). Front Plant Sci. 2022;13:852001. 10.3389/fpls.2022.852001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Czjzek M, Cicek M, Zamboni V, Bevan DR, Henrissat B, Esen A. The mechanism of substrate (aglycone) specificity in beta-glucosidases is revealed by crystal structures of mutant maize beta-glucosidase-DIMBOA, -DIMBOAGlc, and-dhurrin complexes. Proc Natl Acad Sci U S A. 2000;97(25):13555–60. 10.1073/pnas.97.25.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ke D, Liu Y, Zhang J, Chen Q. Genome-wide identification and expression analysis of BGLU family genes in soybean. J Xinyang Normal Univ (Natural Sci Edition) 2019,32(3):372–8. 10.3969/j.issn.1003-0972.2019.03.006
- 54.de Martin X, Sodaei R, Santpere G. Mechanisms of binding specificity among bHLH transcription factors. Int J Mol Sci. 2021;22(17):9150. 10.3390/ijms22179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48(5):501–16. 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- 56.Moore RC, Purugganan MD. The early stages of duplicate gene evolution. Proc Natl Acad Sci U S A. 2003;100:15682–7. 10.1073/pnas.2535513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu Y, Wu N, Song W, Yin G, Qin Y, Yan Y, Hu Y. Soybean (Glycine max) expansin gene superfamily origins, segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014;14:93. 10.1186/1471-2229-14-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson KA, Koepke JI, Kharodawala M, Lopes JM. A network of yeast basic helix-loop-helix interactions. Nucleic Acids Res. 2000;28(22):4460–6. 10.1093/nar/28.22.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallecha A, Mishra S. Purification and characterization of two β-glucosidases from a thermotolerant yeast Pichia etchellsii. Acta Bioch Bioph Sin. 2003;1649(1):74–84. 10.1016/s1570-9639. [DOI] [PubMed] [Google Scholar]
- 60.Fan HX, Miao LL, Liu Y, Liu HC, Liu ZP. Gene cloning and characterization of a cold-adapted β-glucosidase belonging to glycosyl hydrolase family 1 from psychrotolerant bacterium micrococcus antarcticus. Enzyme Microb Tech. 2011;49(1):94–9. 10.1016/j.enzmictec.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Bylina EJ. Comparison of a beta-glucosidase and a beta-mannosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Purification, characterization, gene cloning, and sequence analysis. J Biol Chem. 1996;271(39):23749–55. 10.1074/jbc.271.39.23749. [DOI] [PubMed] [Google Scholar]
- 62.Ketudat Cairns JR, Esen A. β-Glucosidases. Cell Mol Life Sci. 2010;67(20):3389–405. 10.1007/s00018-010-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aguilar CF, Sanderson I, Moracci M, Ciaramella M, Nucci R, Rossi M, Pearl LH. Crystal structure of the beta-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus: resilience as a key factor in thermostability. J Mol Biol. 1997;271(5):789–802. 10.1006/jmbi.1997.1215. [DOI] [PubMed] [Google Scholar]
- 64.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3(9):853–9. 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 65.Burmeister WP, Cottaz S, Driguez H, Iori R, Palmieri S, Henrissat B. The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure. 1997;5(5):663–75. 10.1016/s0969-2126(97)00221-9. [DOI] [PubMed] [Google Scholar]
- 66.Barrett T, Suresh CG, Tolley SP, Dodson EJ, Hughes MA. The crystal structure of a cyanogenic beta-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure. 1995;3(9):951–60. 10.1016/s0969-2126(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 67.Müllegger J, Jahn M, Chen HM, Warren RA, Withers SG. Engineering of a thioglycoligase: randomized mutagenesis of the acid base residue leads to the identification of improved catalysts. Protein Engin Des Sel. 2005;18:33–40. 10.1093/protein/gzi003. [DOI] [PubMed] [Google Scholar]
- 68.Xu Z, Escamilla-Trevino L, Zeng L, Lalgondar M, Bevan D, Winkel B, Mohamed A, Cheng CL, Shih MC, Poulton J, Esen A. Functional genomic analysis of Arabidopsis thaliana Glycoside hydrolase family1. Plant Mol Biol. 2004;55:343–67. 10.1007/s11103-004-0790-1. [DOI] [PubMed] [Google Scholar]
- 69.Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol. 2010;27(8):1745–9. 10.1093/molbev/msq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ke Q, Tao W, Li T, Pan W, Chen X, Wu X, Nie X, Cui L. Genome-wide identification, evolution and expression analysis of basic Helix-loop-helix (bHLH) Gene Family in Barley (Hordeum vulgare L). Curr Genomics. 2020;21(8):621–44. 10.2174/1389202921999201102165537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141(4):1167–84. 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–7. 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 73.Deschamps S, Zhang Y, Llaca V, Ye L, Sanyal A, King M, May G, Lin H. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat Commun. 2018;9(1):4844. 10.1038/s41467-018-07271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fan Y, Lai D, Yang H, Xue G, He A, Chen L, Feng L, Ruan J, Xiang D, Yan J, Cheng J. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L). BMC Genomics. 2021;22(1):778. 10.1186/s12864-021-08095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raes J, Vandepoele K, Simillion C, Saeys Y, Van de Peer Y. Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct. 2003;3:117–29. 10.1023/A:1022666020026. [PubMed] [Google Scholar]
- 76.Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D. Genome-wide identification of NBS genes in rice revealssignificant expansion of divergent non-TIR NBS genes. Mol Genet Genomics. 2004;271:402–15. 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- 77.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (cazy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–8. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (cazy) in 2013. Nucleic Acids Res. 2014;42:D490–5. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chapelle A, Morreel K, Vanholme R, Le-Bris P, Morin H, Lapierre C, Boerjan W, Jouanin L, Demont-Caulet N. Impact of the absence of stem-specific beta-glucosidases on lignin and monolignols. Plant Physiol. 2012;160:1204–17. 10.1104/pp.112.203364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dong X, Jiang Y, Hur Y. Genome-wide analysis of Glycoside Hydrolase Family β-glucosidase genes in Brassica rapa and their potential role in Pollen Development. Int J Mol Sci. 2019;20(7):1663. 10.3390/ijms20071663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu ZY, Lee KH, Dong T, Jeong JC, Jin JB, Kanno Y, Kim DH, Kim SY, Seo M, Bressan RA, Yun DJ. A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell. 2012;24:2184–99. 10.1105/tpc.112.095935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Costet L, Fritig B, Kauffmann S. Scopoletin expression in elicitor-treated and tobacco mosaic virus-infected tobacco plants. Plant Physiol. 2002;115:228–35. 10.1034/j.1399-3054.2002.1150208.x. [DOI] [PubMed] [Google Scholar]
- 83.Baillieul F, de Ruffray P, Kauffmann S. Molecular cloning and biological activity of alpha-, beta-, and gamma-megaspermin, three elicitins secreted by Phytophthora Megasperma H20. Plant Physiol. 2003;131(1):155–66. 10.1104/pp.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hino F, Okazaki M, Miura Y. Effect of 2, 4-dichlorophenoxyacetic acid on glucosylation of scopoletin to scopolin in tobacco tissue culture. Plant Physiol. 1982;69:810–3. 10.1104/pp.69.4.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahn YO, Shimizu B, Sakata K, Gantulga D, Zhou Z, Bevan DR, Esen A. Scopolin hydrolyzing beta-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 2010;51:132. 10.1093/pcp/pcp174. [DOI] [PubMed] [Google Scholar]
- 86.Fourrier N, Bedard J, Lopez-Juez E, Barbrook A, Bowyer J, Jarvis P, Warren G, Thorlby G. A role for sensitive to freezing2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J. 2008;55:734–45. 10.1111/j.1365-313X.2008.03549.x. [DOI] [PubMed] [Google Scholar]
- 87.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu M, Ma Z, Wang A, Zheng T, Huang L, Sun W, Zhang Y, Jin W, Zhan J, Cai Y, Tang Y, Wu Q, Tang Z, Bu T, Li C, Chen H. Genome-wide investigation of the Auxin Response Factor Gene Family in Tartary Buckwheat (Fagopyrum tataricum). Int J Mol Sci. 2018;19(11):3526. 10.3390/ijms19113526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1). 10.1093/nar/gkx922. D493-D496. [DOI] [PMC free article] [PubMed]
- 92.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinf. 2002. 10.1002/0471250953.bi0203s00. Chap. 2: Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 93.Xie T, Chen C, Li C, Liu J, Liu C, He Y. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics. 2018;19(1):490. 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu M, Ma Z, Sun W, Huang L, Wu Q, Tang Z, Bu T, Li C, Chen H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genomics. 2019;20(1):113. 10.1186/s12864-019-5500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinf. 2010;8(1):77–80. 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Remans T, Keunen E, Bex GJ, Smeets K, Vangronsveld J, Cuypers A. Reliable gene expression analysis by reverse transcription-quantitative PCR: reporting and minimizing the uncertainty in data accuracy. Plant Cell. 2014;26(10):3829–37. 10.1105/tpc.114.130641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li C, Zhao H, Li M, Yao P, Li Q, Zhao X, Wang A, Chen H, Tang Z, Bu T, Wu Q. Validation of reference genes for gene expression studies in tartary buckwheat (Fagopyrum tataricum Gaertn.) Using quantitative real-time PCR. PeerJ. 2019;7:e6522. 10.7717/peerj.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001,25(4):402-408.2013. 10.1006/meth.2001.1262 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Table S1. List of the 39 FtBGLU genes identified in this study
Supplementary Material 2: Table S2. BGLU information in Arabidopsis thaliana.
Supplementary Material 3: Table S3. Analysis and distribution of conserved motifs in FtBGLU proteins.
Supplementary Material 4: Table S4. Cis-regulatory elements in the promoter region of FtBGLU genes.
Supplementary Material 5: Table S5. The tandem duplication events of FtBGLU genes.
Supplementary Material 6: Table S6. Synteny analyses of the BGLU genes between F. tataricum and representative plants (A. thaliana, C. quinoa, T. cacao and S. lycopersicum).
Supplementary Material 7: Table S7. Primer sequences for qPCR in this study.
Supplementary Material 8: Table S8. Relative expression of FtBGLUs in abiotic stress, grain filling, and different grain filling stages.
Data Availability Statement
The Fagopyrum tataricum genome sequence information was obtained from the MKBbase website (https://www.mbkbase.org/Pinku1/). The Fagopyrum tataricum (Chuanqiao-2) used in the experiment was supplied by the Alpine Crops Research Station of the Xichang Institute of Agricultural Science, Sichuan Province, China.