Abstract
Microtubule (MT)-based motor proteins, kinesins and dyneins, play important roles in multiple cellular processes including cell division. In this study, we describe the generation and use of an Escherichia coli RNase III-prepared human kinesin/dynein esiRNA library to systematically analyze the functions of all human kinesin/dynein MT motor proteins. Our results indicate that at least 12 kinesins are involved in mitosis and cytokinesis. Eg5 (a member of the kinesin-5 family), Kif2A (a member of the kinesin-13 family), and KifC1 (a member of the kinesin-14 family) are crucial for spindle formation; KifC1, MCAK (a member of the kinesin-13 family), CENP-E (a member of the kinesin-7 family), Kif14 (a member of the kinesin-3 family), Kif18 (a member of the kinesin-8 family), and Kid (a member of the kinesin-10 family) are required for chromosome congression and alignment; Kif4A and Kif4B (members of the kinesin-4 family) have roles in anaphase spindle dynamics; and Kif4A, Kif4B, MKLP1, and MKLP2 (members of the kinesin-6 family) are essential for cytokinesis. Using immunofluorescence analysis, time-lapse microscopy, and rescue experiments, we investigate the roles of these 12 kinesins in detail.
INTRODUCTION
The mitotic spindle, a unique cellular apparatus assembled at the beginning of mitosis, plays a pivotal role in regulating mitosis and cytokinesis. Although tubulins are the most abundant protein in the mitotic spindle, many additional proteins are involved in regulating its formation and function. Most prominent among these are members of the kinesin and dynein families of MT-based motor proteins (Mandelkow and Mandelkow, 2002; Vale, 2003), which generate directional movement along microtubules. The kinesin proteins share a conserved, ∼340 amino acid, motor domain that utilizes ATP to fuel their movement along MTs. Outside the motor domain, kinesins also contain different “stalk” and “tail” domains that mediate oligomerization, regulation of motor activity, and interactions with their specific cargos (Vale, 2003).
The roles of kinesins in mitosis and cytokinesis have been extensively studied in Saccharomyces cerevisiae, in which there are six kinesin genes belonging to five subfamilies. Gene disruption experiments have demonstrated that five of these kinesins play essential roles in regulating the formation, orientation, and elongation of the mitotic spindle and the segregation of chromosomes in mitosis (Hildebrandt and Hoyt, 2000). In addition, one cytoplasmic dynein motor-containing polypeptide, the dynein heavy chain (DHC), has also been implicated in mitotic spindle function (Hildebrandt and Hoyt, 2000). Using RNAi techniques, Goshima and Vale recently examined the mitotic functions of cytoplasmic dynein and all 25 fly kinesins in cultured Drosophila cells (Goshima and Vale, 2003). They found that four kinesins are needed for bipolar spindle assembly and four are crucial for metaphase chromosome alignment. Dynein plays a role in the metaphase-to-anaphase transition, and one kinesin is required for cytokinesis.
The complete sequencing of the human genome allowed the identification of more than 40 human kinesins (Miki et al., 2001; Vale, 2003). Several studies show that several numbers of kinesins are crucial for spindle assembly and function, chromosome segregation, mitotic checkpoint control, and cytokinesis (Blangy et al., 1995; Schaar et al., 1997; Mountain et al., 1999; Levesque and Compton, 2001; McEwen et al., 2001; Neef et al., 2003; Andrews et al., 2004; Ganem and Compton, 2004; Gruneberg et al., 2004; Kurasawa et al., 2004; Matuliene and Kuriyama, 2004; Mazumdar et al., 2004; Mishima et al., 2004; Zhu and Jiang, 2005). Small-molecule inhibitors of specific kinesins perturb mitotic spindle dynamics and arrest cells in mitosis and cytokinesis (Mayer et al., 1999; Sakowicz et al., 2004). A systematic functional analysis of human motor proteins and their potential roles in mitosis and cytokinesis has not been performed. In this study, we report the generation of an Escherichia coli RNase III–prepared human kinesin/dynein esiRNA library for systematically analyzing the functions of all human kinesin/dynein MT motor proteins in mitosis and cytokinesis.
MATERIALS AND METHODS
Generation of Human Kinesin/Dynein esiRNA Library
We used the E. coli RNase III method (Yang et al., 2002) with some modifications to generate esiRNAs specific for all human kinesin/dynein MT motor proteins. Previous studies reported the identification of 45 kinesins and 1 DHC in humans (Miki et al., 2001). After carefully analyzing the cDNA sequences of these reported kinesins, we found that 41 sequences represent unique human kinesin cDNAs whose corresponding genes can be identified in the human genome. The other 4 were either redundant with other human kinesin sequences or from other species. Therefore, we focused our study on the 41 human kinesins and 1 DHC. The Supplementary Table S1 shows the names of these 41 human kinesins, their GenBank accession and recent standardized nomenclature (Lawrence et al., 2004).
Genomic structures (exons, 5′ and 3′ UTRs) of DHC and 41 human kinesin genes were determined using BLAST to compare the appropriate cDNA sequences (Supplementary Table S1) to human genome sequences. We selected 3′ UTRs (∼400 bp) of these genes as DNA templates for preparing esiRNAs. In brief, ∼400 base pairs 3′ UTR DNA fragments of human kinesins and DHC were amplified by PCR from sheared genomic DNA of normal human foreskin fibroblasts using sequence-specific 5′ and 3′ primers containing T7 promoter sequence at the 5′ end (Supplementary Table S1). As a control, a fragment of the luciferase coding region was amplified from a luciferase cDNA (Supplementary Table S1). The PCR products were subjected to in vitro transcription to produce dsRNAs using an in vitro T7 transcription kit (Ambion, Austin, TX). dsRNAs were then treated with DNase I, precipitated with 7.5 M LiCl, washed with 70% (vol/vol) ethanol, dried, and resuspended in water at 1 μg/μl. dsRNAs, 50 μg, were digested with 2.5 μg of GST-RNase III at 37°C for 3 h. The reactions were stopped by addition of 20 mM EDTA, and products were purified through QIAquick spin columns (Qiagen, Chatsworth, CA). esiRNAs were precipitated by ethanol and dissolved in water. The concentrations of esiRNAs were determined by measuring absorbance at UV260. The quality of esiRNAs was also evaluated by electrophoresis on 4% low-melting agarose gels.
There were several reasons that we chose these sequences as DNA templates for preparing esiRNAs. First, the templates could be directly amplified from human genomic DNA by PCR because the 3′ UTR usually comprises part of the last exon of a gene. This approach allowed us to amplify DNA template for any gene of interest, even when an appropriate cDNA was not available. Second, esiRNA generated from 3′ UTRs may provide more specific gene silencing than esiRNA generated from coding regions. Because 3′ UTRs of different genes display very little sequence similarity, it is unlikely that esiRNAs generated from 3′ UTRs will cross-react with other mRNA transcripts. Third, because the esiRNAs generated only target the 3′ UTR, their effects can be rescued by ectopically expressing the target protein using a cDNA lacking the native 3′ UTR. Thus, this approach allows one to easily confirm the specificity of esiRNA effects (see Figure 7).
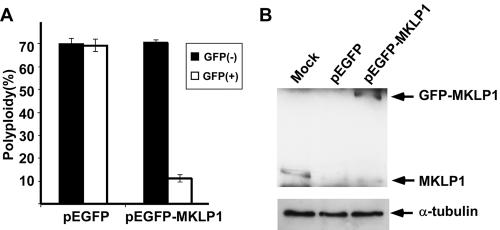
Figure 7.
Rescue of cytokinetic defects in MKLP1 esiRNA-treated cells by ectopic expression of GFP-MKLP1. (A) HeLa cells were transfected with pEGFP or pEGFP-MKLP1 plasmid. Twelve hours later, cells were transfected with 100 nM MKLP1 esiRNA. Two days after esiRNA transfection, cells were fixed and stained with DAPI. The percentage of polyploid cells was scored using a fluorescence microscope. Histograms show the percentages of polyploidy in pEGFP- or pEGFP-MKLP1–untransfected cells (GFP negative) and pEGFP- or pEGFP-MKLP1–transfected cells (GFP positive). (B) HeLa cells were transfected with plasmids and esiRNA as in A. Two days posttransfection, cell lysates were subjected to SDS-PAGE, transferred to PVDF membrane, and immunoblotted with anti-MKLP1 antibodies or anti-α-tubulin.
To determine if the human kinesin/dynein esiRNAs generated could effectively silence expression of their targets, HeLa cells were transfected with individual human kinesin/dynein esiRNAs, and 2 d after transfection RNA was isolated from transfected cells. Quantitative RT-PCR analysis revealed that the DHC esiRNA and 27 of the kinesin esiRNAs significantly ablated mRNA expression (>50%) of their corresponding gene (Supplementary Table S2). Another 5 kinesin esiRNAs moderately ablated target expression (>30% but <50% of ablation). The remaining 8 kinesin esiRNAs did not significantly affect target expression (<10% of ablation). However, the cycles of qRT-PCR required in 6 of these samples were quite high (>25 cycles), suggesting that these genes are weakly expressed in HeLa cells. Thus, the results demonstrate that the majority of our kinesin/dynein esiRNAs effectively silenced expression of their targets.
Plasmids, Antibodies, and Reagents
A full-length human MLKP1 cDNA (IMAGE EST clone 4830855) was subcloned into SalI-SalI sites of the mammalian expression vector pEGFP-C1 to generate pEGFP-MKLP1. pEYFP-tubulin, and pECFP-H2B plasmids were generated as described previously (Zhu and Jiang, 2005). Affinity-purified rabbit polyclonal anti-INCENP antibodies were generated as described (Zhu et al., 2005). Rabbit polyclonal anti-MKLP1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and sheep polyclonal anti-MKLP2 antibodies were a gift from Dr. Francis A. Barr (Max-Planck-Institute of Biochemistry, Germany). Rabbit polyclonal anti-pericentrin antibodies, mouse monoclonal anti-α-tubulin antibody, and human CREST serum (ADA) were purchased from AbCam (Cambridge, United Kingdom), Sigma-Aldrich (St. Louis, MO), and Cortex Biochem (San Leandro, CA), respectively. All secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA); Monastrol, nocodazole, and taxol were purchased from A.G. Scientific (San Diego, CA), and Sigma-Aldrich, respectively.
Cell Culture, Transfection, Immunoblotting, and Immunostaining Analyses
HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). HeLa cell transfections and monastrol, nocodazole, or taxol treatments were performed as described previously (Kapoor et al., 2000; Carvalho et al., 2003; Zhu and Jiang, 2005). Twenty-four to 72 h after transfection or 12 h after drug treatment, cells were harvested or fixed for immunoblotting or immunofluorescence analysis. For immunoblotting analysis, cells were harvested and lysed in 1% Nonidet P-40 buffer (Jiang et al., 1998). Cell lysates with equal amounts of total protein were subjected to SDS-PAGE, transferred to PVDF membrane and then immunoblotted with corresponding antibodies. For immunofluorescence analysis, cells grown on glass coverslips were fixed in phosphate-buffered saline (PBS) solution containing 3% formaldehyde and 2% sucrose at room temperature for 15 min. After permeabilization and blocking in PBS containing 0.4% Triton X-100 and 10% goat serum, cells were incubated with primary antibodies (α-INCENP 1:500; α-pericentrin 1:500; CREST serum 1:1000; α-MKLP1 1:100; α-MKLP2 1:100; α-tubulin 1:10000) overnight at 4°C. After washing, cells were costained with secondary antibodies (1:200 dilution) and DAPI (0.5 μg/ml) for 1 h at room temperature. After five washes, cells were mounted with FluoroGuard (Bio-Rad, Richmond, CA) and photographed using a Leica DM IRE2 fluorescence microscope (Deerfield, IL). For calcium pretreatment, cells were permeabilized in Pipes buffer (100 mM Pipes, pH 6.8, 1 mM MgCl2, 0.1 mM CaCl2, and 0.1% Triton X-100) for 90 s and then fixed in the same buffer supplemented with 4% formaldehyde (Kapoor et al., 2000).
Time-lapse Microscopy
HeLa cells grown on 35-mm glass-bottom microwell dishes (MatTek, Ashland, MA) were transfected with 0.5 μg of pEYFP-tubulin and pECFP-H2B plasmids together with 100 nM esiRNA using Oligofectamine (Invitrogen, Carlsbad, CA). Twenty-four hours after transfection, cells were cultured with CO2-independent medium (GIBCO, Rockville, MD) containing 10% FBS overnight. Dishes were then covered with mineral oil (Sigma) and transferred to a heated stage (37°C) on a Zeiss Axiovert 100M microscope (Thornwood, NY). Phase-contrast and fluorescence images of live cells were collected at 2-min intervals for 9–10 h and processed using the Slidebook 3.0 software (Intelligent Imaging Innovations, Denver, CO).
Quantitative RT-PCR
Total RNAs were extracted from kinesin/dynein esiRNA-transfected cells using TRIZOL reagent (Invitrogen). cDNAs were synthesized by reverse transcription and quantitative RT-PCRs (qRT-PCRs) were performed using specific kinesin/dynein primers (Supplementary Table S2). The amounts of amplified DNA and the threshold cycles (Ct) of each sample were normalized with housekeeping genes, β-actin and GAPDH (Supplementary Table S2). The percentage of ablation of mRNA expression by esiRNA was calculated assuming that PCR amplification efficiency was 1.9.
RESULTS
Identification of Human Kinesin/Dynein Motor Proteins Involved in Mitosis and Cytokinesis
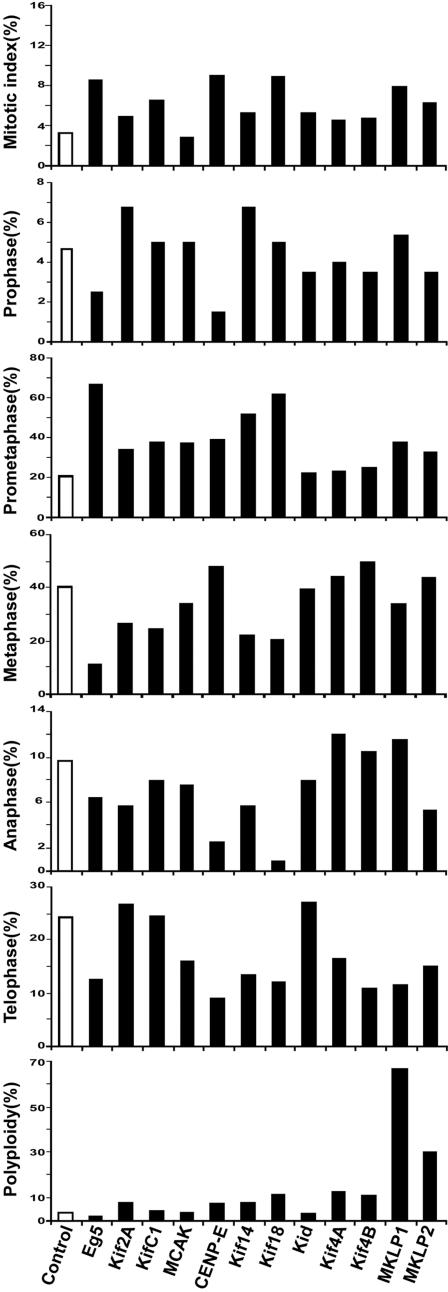
We generated human kinesin/dynein 3′ UTR esiRNA library (for details see Materials and Methods) and screened the library to identify kinesin and dynein motors involved in mitosis and cytokinesis. HeLa cells transfected with kinesin/dynein esiRNAs were fixed and costained with DAPI and anti-α-tubulin antibody to monitor DNA and the mitotic spindle, respectively. Cell morphology, mitotic index, and the percentage of cells at different stages of mitosis and cytokinesis were determined using a fluorescence microscope. Cells transfected with control esiRNA (luciferase) did not show discernible changes in cell morphology, mitotic index, or the distribution of cells in various stages of mitosis and cytokinesis (Figures 1 and 2). By contrast, 12 of the kinesin esiRNAs caused marked but distinct mitotic and cytokinetic defects (Figures 1 and 2). These included Eg5 (a member of the kinesin-5 family), Kif2A and MCAK (members of the kinesin-13 family), KifC1 (a member of the kinesin-14 family), CENP-E (a member of the kinesin-7 family), Kif14 (a member of the kinesin-3 family), Kif18 (a member of the kinesin-8 family), Kid (a member of the kinesin-10 family), Kif4A and Kif4B (members of the kinesin-4 family), and MKLP1 and MKLP2 (members of the kinesin-6 family; Lawrence et al., 2004). qRT-PCR confirmed that all 12 esiRNAs were able to ablate the expression of their corresponding mRNA (Supplementary Table S2). No obvious mitotic and cytokinetic abnormalities were detected in cells transfected with the other 29 kinesin esiRNAs or the DHC esiRNA. We found that the human kinesins were mainly involved in four critical mitotic and cytokinetic processes: 1) spindle formation, 2) chromosome congression and alignment, 3) anaphase spindle dynamics, and 4) the completion of cytokinesis, which we describe in detail in the following sections.
Figure 1.
Quantitation of mitotic and cytokinetic defects in HeLa cells treated with kinesin esiRNAs. HeLa cells were transfected with 100 nM of control (luciferase) esiRNA or kinesin esiRNA. Two days posttransfection, cells were fixed and stained with anti-α-tubulin antibody and DAPI. Mitotic index and the percentage of cells at different stages of mitosis and cytokinesis were scored using a fluorescence microscope. For mitotic index and polyploidy analyses, more than 500 mitotic and interphase cells were counted. For mitosis analyses, more than 150 mitotic cells were counted.
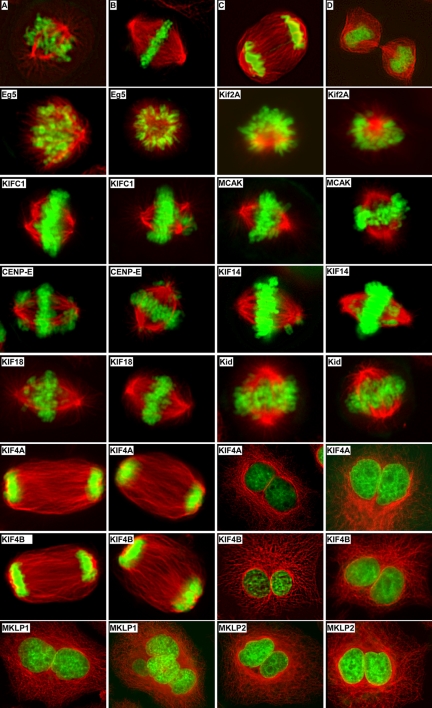
Figure 2.
Typical abnormal mitotic and cytokinetic phenotype(s) observed in HeLa cells treated with kinesin esiRNAs. HeLa cells were transfected with kinesin esiRNAs as in Figure 1. Two days posttransfection, cells were fixed, stained with anti-α-tubulin antibody (red) and DAPI (green) and photographed using a fluorescence microscope. Cells transfected with control (luciferase) esiRNA undergo normal mitosis and cytokinesis (A, prometaphase; B, metaphase; C, anaphase; and D, telophase). Cells transfected with Eg5 or Kif2A esiRNA exhibit monopolar spindles. Cells transfected with KifC1 show multipolar spindles and misaligned chromosomes. Cells transfected with MCAK, CENP-E, Kif14, Kif18, or Kid show misaligned chromosomes. Cells transfected with Kif4A or Kif4B show abnormal anaphase spindle morphology and binucleated cells. Cell transfected with MKLP1 or MKLP2 show binucleated/multinucleated cells.
Eg5, Kif2A, and KifC1 Are Essential for Bipolar Spindle Formation
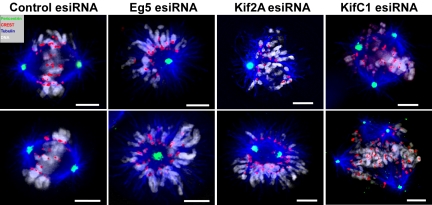
Transfection of HeLa cells with Eg5, Kif2A, or KifC1 esiRNA resulted in perturbation of bipolar spindle formation (Figures 1 and 2). We next immunolocalized pericentrin (a marker of centrosomes), CREST (a marker of centromeres), α-tubulin (mitotic spindles), and DNA in cells transfected with Eg5, Kif2A, or KifC1 esiRNAs and analyzed more than 100 mitotic cells in each transfection. In parallel, we assessed the effects of Eg5, Kif2A, or KifC1 depletion by time-lapse microscopy. To visualize chromosomes and mitotic spindles, we simultaneously transfected cells with mammalian expression plasmids expressing human Histone H2B fused with enhanced cyan fluorescent protein (ECFP) and human α-tubulin fused with an enhanced yellow fluorescent protein (EYFP). Cells transfected with control esiRNA established normal bipolar spindles with bioriented chromosomes at or near the spindle equator in early mitosis (Figure 3). Time-lapse fluorescence and phase-contrast Video images of control cells showed that EYFP-tubulin and ECFP-H2B localized to cytoplasmic cytoskeleton arrays and nuclei in interphase (unpublished data) and then localized to the mitotic spindle and chromosomes during mitosis, respectively. Like untransfected cells, control cells completed mitosis and cytokinesis normally (Supplementary Video 1).
Figure 3.
Eg5, Kif2A, and KifC1 are essential for bipolar spindle formation. HeLa cells were transfected with 100 nM of control, Eg5, Kif2A, or KifC1 esiRNA. Two days posttransfection, cells were fixed and stained with anti-pericentrin antibodies (green), ADA (red), anti-α-tubulin antibody (blue), and DAPI (DNA, white). Two representative cells in each experiment are shown. Bar, 5 μm.
In contrast, the majority of cells transfected with Eg5 esiRNA displayed monopolar spindles with two duplicated, but only slightly separated, centrosomes at the center of the monoasters (Figure 3). Chromosomes in these cells were attached to the radial arrays of MTs emanating from monopolar spindles, and formed “J” or “V” shapes with syntelic orientation (Figure 3). Time-lapse images of cells cotransfected with EYFP-tubulin, ECFP-H2B and Eg5 esiRNA revealed monopolar spindles and inhibition of chromosome segregation in these cells (Supplementary Video 2). Similar results were obtained in HeLa cells treated with the Eg5 inhibitor, monastrol (Supplementary Video 3). These results indicate that depletion of Eg5 inhibits centrosome separation and formation of the bipolar mitotic spindle, thus precluding chromosome segregation.
Cells transfected with Kif2A esiRNA showed initial separation of the two centrosomes at prophase (see Supplementary Video 4). However, the bipolar spindles often collapsed and the poles drew back to form monopolar spindles or asymmetric bipolar spindles with one-half larger than the other as cells entered prometaphase (Figure 3 and Supplementary Video 4). Unlike Eg5 esiRNA- or monastrol-treated cells, the duplicated centrosomes remained separated at the center of monoasters in most Kif2A esiRNA-treated cells (compare pericentrin staining of Eg5 RNAi and Kif2A RNAi cells in Figure 3). Chromosomes with syntelic orientation were also detected in Kif2A esiRNA-treated cells. Previous studies showed that Kif2A has MT-depolymerizing activity in the absence of motile function (Desai et al., 1999). However, we did not observe significant increases in spindle MT length or density in Kif2A esiRNA-treated cells. As our work was in progress, another study showed that Kif2A specifically localizes to centrosomes and spindle poles during mitosis and that depleting Kif2A by siRNA in human U2OS cells leads to the formation of monopolar spindles and a block in cell cycle progression (Ganem and Compton, 2004).
Cells transfected with KifC1 esiRNA revealed multiple pericentrin foci upon staining, indicating the formation of multipolar spindles (Figure 3 and Supplementary Video 5). Although some chromosomes were aligned in the middle of each spindle, misaligned chromosomes were clearly observed in early mitotic cells (Figure 3 and Supplementary Video 5). The abnormal, multipolar spindles clearly affected chromosome congression and alignment. Despite these abnormalities, cells transfected with KifC1 esiRNA eventually entered anaphase with more than three chromosome masses that may have been captured and segregated by multiple polar spindles (Supplementary Video 5). When compared with control cells, we also observed a higher frequency of interphase cells with several small nuclei in KifC1 esiRNA-treated cells, which might be the consequence of nuclear envelope reformation around each separate chromosome mass (unpublished data).
KifC1, MCAK, CENP-E, Kif14, Kif18, and Kid Regulate Chromosome Congression and Alignment
Depletion of KifC1, MCAK, CENP-E, Kif14, Kif18, or Kid resulted in abnormal chromosome congression and alignment during mitosis (Figures 1 and 2). The effects of depleting KifC1 were discussed above (see Figure 3 and Supplementary Video 5). Depletion of MCAK, another human kinesin that has MT-depolymerizing activity in the absence of motile function, resulted in a block of the prometaphase-to-metaphase transition with misaligned chromosomes (Figures 1 and 2). Previous studies showed that ablation of MCAK by immunodepletion or by siRNA in Xenopus egg extracts or cultured Drosophila cells affected spindle MT dynamic instability, resulting in longer spindle MTs with higher density (Walczak et al., 1996; Goshima and Vale, 2003). We did not observe abnormal mitotic spindle formation in MCAK esiRNA-treated HeLa cells, but frequently observed maloriented and misaligned chromosomes (Figure 2). Similar phenotypes were observed in MCAK-depleted mammalian cells in several other studies (Maney et al., 1998; Cassimeris and Morabito, 2004; Kline-Smith et al., 2004). Because MCAK localizes to kinetochores in mammalian cells during mitosis, it was suggested that MCAK is involved in regulating the dynamics of MT plus ends at kinetochores (Kline-Smith et al., 2004).
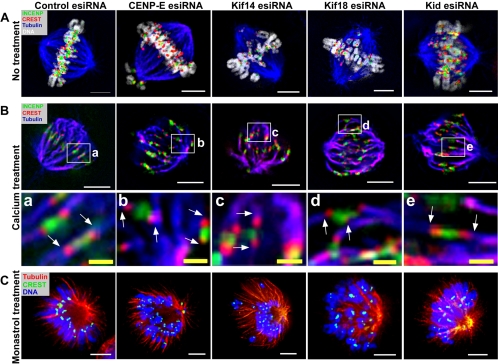
Previous studies indicated that kinesins are involved in regulating the attachment of spindle MTs to kinetochores, generating the polar ejection force that pushes chromosomes away from spindle poles, or monitoring tension developed across kinetochore pairs—all crucial processes that are necessary for chromosome congression and alignment (Kapoor and Compton, 2002; Cleveland et al., 2003). To determine whether CENP-E, Kif14, Kif18, or Kid are involved in regulating the attachment of spindle MTs to kinetochores (k-fibers), we immunolocalized centromeres and spindles in esiRNA-transfected cells using antibodies against INCENP (a marker of inner centromere), ADA (CREST), and α-tubulin (Figure 4A). We preferentially disassembled most of the labile, nonkinetochore-associated MTs by briefly exposing the cells to Ca2+ before fixation (Kapoor et al., 2000) and then stained the cells with the appropriate antibodies (Figure 4B). We also examined whether depleting CENP-E, Kif14, Kif18, or Kid affected the polar ejection force, which is generated on chromosome arms by motor proteins (e.g., chromokinesins) and plays a critical role in antagonizing poleward kinetochore forces (Levesque and Compton, 2001). To monitor the polar ejection force, we treated esiRNA-transfected cells with monastrol and examined the positions of chromosomes on monopolar spindles (Mayer et al., 1999; Levesque and Compton, 2001). In the absence of biorientation, the only force acting to push chromosomes away is the polar ejection force (Levesque and Compton, 2001; Figure 4C). Finally, we determined whether depleting these kinesins perturbed tension across the kinetochore pairs of sister chromatids. We measured interkinetochore distance, an accurate reflection of tension (Waters et al., 1996), in ∼100 kinetochore pairs at the same focal plane (visualized by staining INCENP in the middle of centromeres and CREST at either side of INCENP; Figure 5).
Figure 4.
CENP-E, Kif14, Kif18, and Kid are involved in regulating chromosome congression and alignment. (A) HeLa cells were transfected with 100 nM of control, CENP-E, Kif14, Kif18, or Kid esiRNA. Two days posttransfection, cells were fixed and stained with anti-INCENP antibodies (green), ADA (red), anti-α-tubulin antibody (blue), and DAPI (white). A representative metaphase cell from each experiment is shown. Bar, 5 μm. (B) HeLa cells were transfected with esiRNAs as in A. Two days posttransfection, cells were permeabilized, fixed, and then stained with anti-INCENP antibodies (green), ADA (red), and anti-α-tubulin antibody (blue). Top: a representative cell from each experiment. Bottom (a-e): higher magnification views of the images in the top panel. White bar, 5 μm and yellow bar, 1 μm. (C) HeLa cells were transfected with esiRNAs as in A. Thirty-six hours posttransfection, cells were incubated with 100 μM monastrol for an additional 12 h. Then cells were fixed and stained with ADA (green), anti-α-tubulin antibody (red), and DAPI (blue). A representative cell with a monopolar spindle is shown for each experiment. Bar, 5 μm.
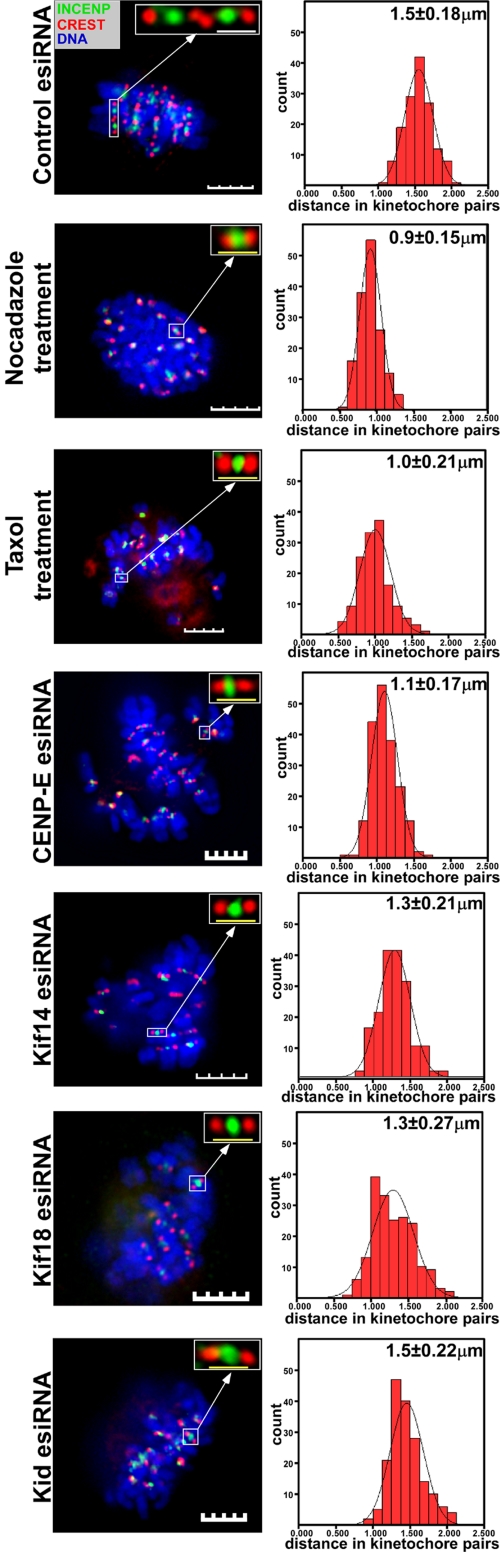
Figure 5.
Analysis of interkinetochore distance on sister chromatids. HeLa cells were transfected with 100 nM of control, CENP-E, Kif14, Kif18, or Kid esiRNA as in Figure 4 or treated with 100 ng/ml nocadazole or 33 nM taxol. Two days posttransfection or 12 h after drug treatment, cells were fixed and immunostained with anti-INCENP antibody (green), ADA (red), and DAPI (blue). Left panel shows a representative immunostained cell from each experiment. Kinetochore pairs on sister chromatids were observed by central INCENP staining and CREST signal at either side in esiRNA- or drug-treated cells (top right corner images; scale bar, 1.25 μm). Interkinetochore distances were measured using Image J software (http://rsb.info.nih.gov/ij/). More than 100 kinetochore pairs were measured in each experiment, and the average interkinetochore distance was calculated using the SPSS 11.5 program (histograms, right panel). White bar, 5 μm and yellow bar, 1.25 μm.
Cells transfected with control esiRNA displayed normal bipolar spindles with biorientated chromosomes aligned perfectly at the metaphase plate (Figure 4A). Kinetochore pairs on sister chromatids were attached with bipolar spindle MTs (Figure 4B). The average interkinetochore distance in these cells was 1.5 ± 0.18 μm, significantly higher than in cells treated with a MT-depolymerizing agent, nocodazole (0.9 ± 0.15 μm), or a MT-stabilizing agent, taxol (1.0 ± 0.21 μm), in which kinetochore pairs were presumably under no tension (Figure 5). Treatment of control esiRNA-transfected cells with monastrol resulted in the formation of monopolar spindles (Figure 4C), with chromosomes arranged around them in a ring. The chromosomes' kinetochore regions were pulled poleward and their arms pushed outward, presumably by the antagonistic actions of poleward kinetochore forces and the polar ejection force (Figure 4C).
Cells transfected with CENP-E esiRNA formed normal bipolar spindles, but the majority were blocked at prometaphase and metaphase (Figures 1, 2, and 4). Although many chromosomes became bioriented and aligned at the metaphase plate, a few were positioned close to spindle poles or at intermediate positions between the metaphase plate and spindle poles (Figures 2 and 4A). Analysis of k-fibers in these cells revealed that sister kinetochore pairs that aligned at the metaphase plate were attached to spindle MTs, whereas kinetochore pairs on sister chromatids that misaligned close to spindle poles were not (Figure 4B). Kinetochore pairs on sister chromatids that misaligned between the metaphase plate and spindle poles showed diminished interactions with spindle MTs (Figure 4B). Stable spindle MT-kinetochore attachment is required for generating poleward kinetochore forces, and indeed, the average interkinetochore distance of misaligned chromosomes in CENP-E esiRNA-treated cells was significantly shorter (1.1 ± 17 μm) than that of aligned kinetochores in control esiRNA-treated cells. This indicated that misaligned kinetochore pairs in CENP-E esiRNA-treated cells were under much less tension. In CENP-E esiRNA-transfected cells treated with monastrol, some chromosomes were pushed away from the chromosome ring around monopolar spindles (Figure 4C). Because the antagonistic actions of poleward kinetochore forces and the polar ejection force largely determine the position of chromosomes on monopolar spindles, this result indicates that CENP-E is not required for generating the polar ejection force (Figure 4C). Consistent with the immunofluorescence analyses, time-lapse microscopy revealed that cells transfected with CENP-E esiRNA displayed a long delay in the metaphase-to-anaphase transition with misaligned chromosomes (Supplementary Video 6). Misaligned chromosomes close to spindle poles halted in spindle pole areas for long periods of time. Taken together, these results indicate that depleting CENP-E perturbs the stability of spindle MT-kinetochore attachments, which in turn affects the tension across kinetochore pairs that is necessary for chromosome congression and alignment.
Depleting Kif14 or Kif18 had no affect on the formation of bipolar mitotic spindles, but greatly perturbed chromosome congression and alignment (Figures 2 and 4 and Supplementary Videos 7 and 8). Almost all Kif14 or Kif18 esiRNA-treated metaphase-like cells showed misaligned chromosomes between the metaphase plate and spindle poles. In Kif14 esiRNA cells, chromosomes that congressed to the metaphase plate showed mal-oriented features (Figure 4A). Chromosomes in Kif18-treated cells also tried to congress, but could not align stably at the metaphase plate and also appeared mal-oriented. Time-lapse microscopy of Kif14 esiRNA-treated cells showed that the majority of chromosomes in these cells initially move to the metaphase plate, but cannot stably remain there (Supplementary Video 7). Almost all of the chromosomes moved back and forth between the metaphase plate and spindle poles, delaying the metaphase-to-anaphase transition by many hours. Kif18 esiRNA-treated cells failed to congress their chromosomes, but showed similar chromosome movements between the metaphase plate and spindle poles (Supplementary Video 8). These cells also showed longer metaphase-to-anaphase delays, but eventually entered anaphase and failed to complete cytokinesis. Analysis of k-fibers in Kif14 or Kif18 esiRNA-treated cells revealed that all kinetochore pairs were attached to spindle MTs, including those that congressed to the metaphase plate or those that misaligned between the metaphase plate and spindle poles (Figure 4B). However, the interkinetochore distance in Kif14 (1.3 ± 0.21 μm) and Kif18 (1.3 ± 0.27 μm) esiRNA-treated cells was shorter than in control esiRNA-treated cells (1.5 ± 0.18 μm, Figure 5). These results indicate that kinetochore pairs in Kif14 and Kif18 esiRNA-treated cells were under less tension, perhaps because kinetochore-MT dynamics were perturbed and poleward kinetochore forces were weakened. Interestingly, treatment of Kif14 or Kif18 esiRNA-transfected cells with monastrol resulted in distinct phenotypes. In Kif14-depleted, monastrol-treated cells, some chromosomes were noticeably pushed away from the others around monopolar spindles, similar to what was observed in CENP-E esiRNA-treated cells (Figure 4C). Thus, unlike CENP-E, Kif14 was not essential for the establishment and/or maintenance of kinetochore-MTs attachment, but it was required for regulating the poleward kinetochore forces that affect tension across kinetochore pairs and regulate chromosome congression and alignment. Kif18 esiRNA-transfected cells treated with monastrol exhibited monopolar spindles but showed atypical arrangements of chromosomes around the spindles with no obvious rings observed (Figure 4C). Instead, some chromosomes were pushed away from the center of the spindles and others were pulled closer. These results indicate that depletion of Kif18 greatly affected the balance between poleward kinetochore forces and the polar ejection force required for maintaining the position of chromosomes around monopolar spindles. Together, our results demonstrate that Kif18 is not involved in regulating the attachment of spindle MTs to kinetochores, but is critical for judging tension developed across kinetochore pairs. Kif18 may be also involved in regulating the polar ejection force necessary for chromosome congression and alignment.
Depleting Kid by esiRNA did not affect the formation of bipolar mitotic spindles, but perturbed chromosome alignment (Figures 2 and 4 and Supplementary Video 9). Again, chromosomes in Kid esiRNA-transfected metaphase-like cells attempted to congress to the metaphase plate, but could not sharply align there. Time-lapse microscopy indicated that these cells display a delay in the metaphase-to-anaphase transition with chromosomes that move back and forth near the metaphase plate, a phenotype that was different from those observed in Kif14 and Kif18 esiRNA-treated cells (Supplementary Video 9). Despite these defects, Kid esiRNA cells eventually entered anaphase, although chromosome segregation appeared highly abnormal. Some sister chromatids pulled to the opposite poles of the spindle, whereas others lagged behind. Examination of k-fibers in these cells showed that all kinetochore pairs were attached to spindle MTs (Figure 4B). The average interkinetochore distance was very similar to that in control cells (1.5 ± 0.22 mm vs. 1.5 ± 0.18 μm, Figure 5). However, Kid esiRNA-transfected cells treated with monastrol showed chromosomes that had collapsed to the center of the monopolar spindles (Figure 4C). These results indicate that Kid is not involved in regulating the establishment and/or maintenance of kinetochore-MT attachment or in judging tension developed across kinetochore pairs, but is primarily involved in regulating the polar ejection force.
Essential Roles of Kif4A and Kif4B in Anaphase Spindle Dynamics and Cytokinesis
We also uncovered functional roles for Kif4A and Kif4B, two closely related kinesins, in regulating anaphase spindle dynamics and cytokinesis. Depleting Kif4A or Kif4B caused indistinguishable defects in anaphase spindle morphology and cytokinesis (Figures 1 and 2). In both cases, we observed highly elongated anaphase spindles with separating chromosomes extremely close to the spindle poles. Binucleated/multinucleated cell populations were significantly higher in Kif4A or Kif4B esiRNA-treated cells (∼11–13%) than in control cells (∼3.5%). Time-lapse microscopy (Supplementary Video 10) indicated that cells transfected with Kif4A esiRNA did not show obvious abnormalities in bipolar spindle formation and chromosome congression/segregation in early mitosis. However, as these cells elongated their anaphase spindles, separating chromosomes moved extremely close to the spindle poles. Although the cleavage furrow formed and ingressed in these cells, the elongated mitotic spindle often twisted around, and ultimately these cells failed to complete cytokinesis and became binucleated. Taken together, our results indicate that Kif4A and Kif4B play essential roles in regulating anaphase spindle dynamics and the completion of cytokinesis.
Distinct roles of Human MKLP1 and MKLP2 in Cytokinesis
The depletion of MKLP1 or MKLP2 also caused cytokinesis failures. More than 30–70% of MKLP1 and MKLP2 esiRNA-treated cells became bi/multinucleated (Figures 1 and 2). MKLP1 orthologues also have functional roles in cytokinesis (Chen et al., 2002; Mishima et al., 2002; Goshima and Vale, 2003; Matuliene and Kuriyama, 2004). MKLP-2 (rabkinesin-6/RAB6-KIFL) was originally identified as a Rab6 GTPase-binding protein that is involved in protein transport at the Golgi apparatus (Echard et al., 1998). However, recent studies concluded that MKLP2 is a mitotic spindle midzone/midbody-associated protein that plays a critical role in regulating cytokinesis (Hill et al., 2000; Fontijn et al., 2001; Neef et al., 2003). MKLP1 and MKLP2 share significant sequence homology and are grouped in the kinesin-6 family (Miki et al., 2001; Lawrence et al., 2004). A third member of this family, MPP1, has also been implicated in cytokinesis (Abaza et al., 2003). However, we did not detect any significant cytokinesis defects in MPP1 esiRNA-treated cells even though qRT-PCR showed significant ablation of MPP1 mRNA expression in these cells (Supplementary Tables S1 and S2 and unpublished data). Because MKLP1 and MKLP2 are closely related but not functionally redundant, we decided to investigate their functions in more detail.
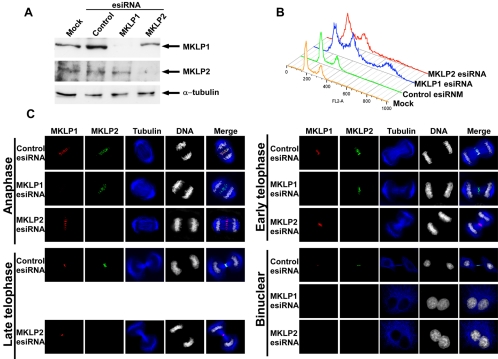
We first examined the expression levels of MKLP1 and MKLP2 proteins in control, MKLP1 or MKLP2 esiRNA-transfected cells by immunoblotting analysis (Figure 6A). Consistent with qRT-PCR results (Supplementary Table S2), transfection of MKLP1 or MKLP2 esiRNA, but not control esiRNA, effectively ablated the expression of MKLP1 or MKLP2 protein in HeLa cells. However, transfection of MKLP1 esiRNA did not cause a substantial reduction of MKLP2 protein and vice versa. No reduction in α-tubulin expression was detected in any of the esiRNA-treated cells. Thus, our MKLP1 and MKLP2 esiRNAs could specifically and effectively ablate the expression of the corresponding genes without affecting the expression of closely related genes, confirming the specificities of these 3′ UTR esiRNAs.
Figure 6.
Effects of depleting MKLP1 or MKLP2 by esiRNA in HeLa cells. (A) HeLa cells were mock-transfected (H2O) or transfected with 100 nM of control (luciferase), MKLP1 or MKLP2 esiRNA. Two days posttransfection, cells lysates were subjected to SDS-PAGE, transferred to PVDF membrane and then immunoblotted with anti-MKLP1, anti-MKLP2, or anti-α-tubulin antibodies. (B) HeLa cells were transfected with esiRNA as in A. Two days posttransfection, cells were fixed and stained with propidium iodide (PI). DNA content was measured by flow cytometry. (C) HeLa cells were transfected with esiRNA as in A. Two days posttransfection, cells were fixed and stained with anti-MKLP1 antibodies (red), anti-MKLP2 antibodies (green), anti-α-tubulin antibody (blue), and DAPI (white).
Cell cycle parameters of MKLP1 or MKLP2 esiRNA-treated cells were examined by fluorescence-activated cell sorting (FACS) analysis. Cells treated with MKLP1 or MKLP2 esiRNA for 48 h accumulated significant 4N (and greater than 4N) DNA content populations when compared with control cells (Figure 6B). These results were consistent with the cell counting data represented in Figure 1, further demonstrating that ablation of MKLP1 or MKLP2 expression perturbed mitotic/cytokinetic progression in HeLa cells.
Cells transfected with control, MKLP1 or MKLP2 esiRNA were stained with anti-MKLP1 and -MKLP2 antibodies and costained with anti-α-tubulin antibody and DAPI. In control cells, MKLP1 and MKLP2 proteins colocalized to the spindle poles and the spindle in early mitosis (unpublished data), and the spindle midzone in anaphase and early telophase (Figure 6C). During late telophase and the final stage of cytokinesis, MKLP1 was concentrated on the center of the midbody, whereas MKLP2 was concentrated on the borders of the midbody (Figure 6C).
MKLP1 levels were greatly reduced in the majority of cells transfected with MKLP1 esiRNA. Cells expressing undetectable MKLP1 showed no obvious abnormalities in chromosome segregation, spindle/spindle midzone assembly, or cleavage furrow formation/ingression before early telophase (Figure 6C). However, the proper formation of the midbody and the completion of cytokinesis were severely inhibited (Figure 6C). MKLP1-depleted cells were rarely detected in late telophase, and accumulations of bi/multinucleated cells were observed (Figure 6C). Association of MKLP2 with the mitotic spindle and the spindle midzone was not affected in MKLP1-depleted cells, indicating that MKLP2 is not dependent on MKLP1 for its localization.
MKLP2 levels were greatly reduced in the majority of cells transfected with MKLP2 esiRNA. Like MKLP1-depleted cells, cells expressing undetectable MKLP2 showed no obvious abnormalities in chromosome segregation, spindle/spindle midzone assembly, or cleavage furrow formation and ingression (Figure 6C). However, MKLP2-depleted cells could be observed in late telophase. Although MKLP2-depleted cells progressed to late telophase, the final step of cytokinesis could not be completed, resulting in the formation of binucleated/multinucleated cells (Figure 6C). The subcellular localization of MKLP1 was not affected in MKLP2-depleted cells.
Time-lapse microscopy showed that MKLP1 esiRNA-treated cells progressed through mitosis normally until anaphase (Supplementary Video 11), when their mitotic spindles appeared to be highly aberrant. The formation of the midbody was severely inhibited and separating sister chromosomes moved back into close proximity. Although the formation and ingression of the cleavage furrow was observed in these cells, the furrowing remained incomplete. As chromosomes were decondensed, the cells became binucleated, demonstrating an essential role of MKLP1 in cytokinesis.
MKLP2 esiRNA-treated cells progressed through mitosis normally until late telophase (Supplementary Video 12). In these cells, chromosomes segregated normally and assembled into two nuclei. The cleavage furrows ingressed to form the intercellular bridges between two separating cells, but the cells could not break the intercellular bridges. Instead, the intercellular bridges widened and eventually the two cells fused back together. These results indicate that MKLP2 is essential for cell abscission, the final step of cytokinesis.
Ectopic Expression of MKLP1 Rescues Cytokinesis Defect in MKLP1 esiRNA-treated Cells
The specificity of RNAi-mediated gene silencing has been challenged by several studies (Jackson et al., 2003; Snove and Holen, 2004). To ascertain the specificity of esiRNA effects in our study, we next performed rescue experiments. HeLa cells were cotransfected with MKLP1 esiRNA together with a mammalian expression vector expressing either EGFP (control) or EGFP-tagged human MKLP1 protein, in the latter case using a MKLP1 cDNA lacking the native 3′UTR. MKLP1 esiRNA, cotransfected with either EGFP or EGFP-MKLP1 plasmid, could effectively ablate the expression of endogenous MKLP1 (Figure 7B). However, MKLP1 esiRNA did not affect the expression of the exogenous EGFP-MKLP1 in these cells.
Expression of EGFP-MKLP1 protein, but not EGFP, rescued the cytokinesis defects observed in cells depleted of endogenous MKLP1, and bi/multinucleated cells were dramatically reduced (Figure 7A). Similar rescue experiments were successfully performed in Kif4A esiRNA-treated cells (Zhu and Jiang, 2005). These experiments demonstrate that the defects observed in esiRNA-treated cells were due to specific inhibition of the endogenous gene targeted by the corresponding 3′ UTR esiRNA.
DISCUSSION
Advances in cell imaging and RNAi technology have provided powerful new tools for studying biological processes. In this study, we report the generation of an E. coli RNase III–prepared 3′UTR esiRNA library and the use of this library to systematically analyze the functions of all human kinesin/dynein MT motor proteins. The majority of 3′ UTR kinesin/dynein esiRNAs were able to efficiently and specifically ablate target mRNA and/or protein expression (Supplementary Table S2 and Figure 6A). The specific effects we observed with 3′ UTR esiRNA could be rescued by ectopically expressing the corresponding protein using a cDNA that lacks the native 3′ UTR (Figure 7; Zhu and Jiang, 2005). Our study demonstrates that 3′ UTR esiRNAs can be used to analyze the function of individual genes or gene families, and, because they are cheap and easy to make, they are particularly useful for large-scale functional genomic screens. Indeed, a large-scale screen in human cells using an esiRNA library (though in this case not directed against target 3′ UTRs) was recently described (Kittler et al., 2004).
Our screen revealed that 12 human kinesins are involved in four critical mitotic and cytokinetic processes in HeLa cells. Eg5, Kif2A, and KifC1 are crucial for bipolar mitotic spindle formation; KifC1, MCAK, CENP-E, Kif14, Kif18, and Kid are required for chromosome congression and alignment; Kif4A and Kif4B are involved in regulating anaphase spindle dynamics; and Kif4A, Kif4B, MKLP1, and MKLP2 are essential for cytokinesis. Immunofluorescence and time-lapse microscopy analyses, in which we monitored chromosome congression, alignment, and segregation, and bipolar mitotic spindle formation and dynamics, confirmed our screening results. We showed that depleting Eg5 with esiRNA or inactivating it with a specific inhibitor, monastrol, blocked centrosome separation and bipolar spindle assembly, resulting in the formation of monopolar spindles. Previous studies showed that Eg5, a homotetrameric kinesin, is required for cross-linking, and the sliding apart of, antiparallel spindle MTs that are necessary for establishing bipolar spindles (Kapoor and Mitchison, 2001). Our results are consistent with these findings, demonstrating that the Eg5 kinesin is essential for bipolar spindle formation in human cells.
Depletion of Kif2A, an MT-depolymerizing KinI motor, resulted in the formation of monopolar or asymmetric bipolar spindles. In contrast to cells treated with Eg5 esiRNA or monastrol, time-lapse microscopy indicated that cells depleted of Kif2A initially separated their duplicated centrosomes and attempted to establish bipolar spindles at prophase. However, as cells entered prometaphase, bipolar spindles in these cells collapsed and spindle poles drew back to become monopolar or asymmetric bipolar spindles. Thus, unlike Eg5, Kif2A is not crucial for centrosome separation, but is essential for maintaining spindle bipolarity. Recent studies have shown that Kif2A specifically localizes to centrosomes and spindles poles during mitosis and is required for the disassembly of MT minus ends associated with poleward MT flux (Ganem and Compton, 2004). Because poleward MT flux generates tension at kinetochores that induces addition of tubulin subunits to MT plus ends, it was postulated that disruption of poleward MT flux by Kif2A depletion would relax tension and switch kinetochore MT dynamics from polymerization to depolymerization, thereby drawing spindle poles back together into monopolar spindles (Ganem and Compton, 2004). Our results are consistent with this model and confirm that the regulation of spindle MT dynamics by Kif2A is crucial for spindle bipolarity.
In contrast to previous studies, which reported that microinjection of anti-KifC1 (HSET) antibodies into cultured mammalian cells did not perturb spindle formation (Mountain et al., 1999), we found that depletion of KifC1 by esiRNA caused the formation of multipolar spindles and abnormal chromosome congression and alignment. The simplest explanation for the discrepancy is that transfection of KifC1 esiRNA may ablate KifC1 activity more efficiently than microinjection of KifC1 antibodies. Consistent with our results, depletion of the Drosophila KifC1 ortholog, Ncd, resulted in defects in spindle pole integrity and the formation of multipolar spindles in cultured Drosophila S2 cells (Goshima and Vale, 2003). Because KifC1 is a minus end-directed motor and localizes on mitotic spindles, we speculate that KifC1 moves toward spindle poles and is required for tethering centrosomes and MTs together to form bipolar mitotic spindles.
Our results also indicate that MCAK, CENP-E, Kif14, Kif18, and Kid are involved in regulating chromosome congression and alignment. Depletion of MCAK, an MT-depolymerizng KinI motor, did not grossly affect formation of bipolar mitotic spindles, but perturbed chromosome congression and alignment. Inactivation of MCAK orthologues in Xenopus egg extracts (XKCM1) or cultured Drosophila S2 cells (Klp10A) caused longer mitotic spindle MTs with higher density (Walczak et al., 1996; Goshima and Vale, 2003), and our results suggest that MCAK is not a major mitotic spindle MT destabilizer in mammalian cells. Similar results were reported by others (Maney et al., 1998; Cassimeris and Morabito, 2004; Kline-Smith et al., 2004). MCAK has been shown to colocalize with the Aurora-B kinase to centromeres. Aurora-B is a chromosomal passenger protein that regulates chromosome movement and spindle dynamics, and MCAK is one of its critical downstream targets (Shannon and Salmon, 2002; Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004). Phosphorylation by Aurora-B regulates the localization of MCAK to centromeres and also its MT depolymerase activity. Thus, the function of MCAK at centromeres, regulated by Aurora-B, may fine-tune spindle MT-kinetochore dynamics for guiding chromosome congression and alignment during mitosis in human cells (Gorbsky, 2004).
Consistent with previous studies (Putkey et al., 2002; Weaver et al., 2003; Tanudji et al., 2004), cells depleted of CENP-E exhibit unaligned chromosomes close to spindle poles and misaligned chromosomes positioned between the metaphase plate and spindle poles at metaphase. Kinetochore pairs on misaligned sister chromatids showed no interaction or diminished interaction with spindle MTs. Previous studies demonstrated that CENP-E binds to kinetochores and MTs and stabilizes their interaction, thereby contributing to silencing the mitotic spindle checkpoint (Putkey et al., 2002; Weaver et al., 2003; Tanudji et al., 2004). CENP-E associates with and activates the mitotic checkpoint kinase BubR1, indicating that the spindle checkpoint is sensed through CENP-E kinetochore activity (Chan et al., 1998, 1999; Mao et al., 2003; Weaver et al., 2003). However, the observation that there are few unaligned and/or misaligned chromosomes in CENP-E esiRNA-treated cells suggests that functionally redundant mechanisms may operate to control spindle MT-kinetochore attachment and chromosome congression (Tanudji et al., 2004).
Depletion of Kif14 or Kif18, two previously uncharacterized human kinesins, resulted in perturbation of chromosome congression and alignment. Immunofluorescence analysis indicated that almost all Kif14 or Kif18 esiRNA-treated metaphase-like cells displayed misaligned chromosomes between the metaphase plate and spindle poles. However, unlike CENP-E esiRNA-treated cells, Kif14 or Kif18 esiRNA-treated cells did not show any unaligned chromosomes close to spindle poles. Consistently, analyses of k-fibers and interkinetochore distance indicated that Kif14 and Kif18 are not essential for the establishment and/or maintenance of kinetochore-MT attachment, but are required for judging tension across kinetochore pairs. Time-lapse microscopy revealed that, although the majority of chromosomes in Kif14 esiRNA-treated cells initially moved to the metaphase plate, they could not remain there and instead migrated back and forth between the metaphase plate and spindle poles. In contrast, chromosomes failed to congress in cells transfected with Kif18 esiRNA, although misaligned chromosomes moved back and forth between the metaphase plate and spindle poles. Depletion of Kif14 or Kif18 caused a prolonged delay of the metaphase-to-anaphase transition, suggesting that the mitotic spindle checkpoint was activated in these cells. Previous studies indicate that the Kif14 Drosophila ortholog, Klp38B, is also required for chromosome segregation (Molina et al., 1997; Ohkura et al., 1997). Klp38B colocalizes with condensed chromosomes and has been speculated to push chromosome arms away from spindle poles, either by coupling chromosome arms to astral MT dynamics or by plus end-directed MT motor activity. We found no evidence that Kif14 is involved in generating the polar ejection force in HeLa cells (Figure 4C). Thus, how Kif14 functions in chromosome congression/alignment and mitotic checkpoint control requires further study.
Database analysis indicates that Kif18 shares significant sequence homology with the Kip3 subfamily of N-terminal positioned motor kinesins, Kip3 (S. cerevisiae), Klp5/6 (Schizosaccharomyces pombe), KipB (Aspergillus nidulans), and Klp67A (Drosophila) (unpublished data and Cottingham and Hoyt, 1997; Garcia et al., 2002; West et al., 2002; Gandhi et al., 2004; Rischitor et al., 2004; Savoian et al., 2004). Klp5/6, KipB, and Klp67A are involved in spindle formation and chromosome segregation during meiosis and/or mitosis (Garcia et al., 2002; West et al., 2002; Gandhi et al., 2004; Rischitor et al., 2004; Savoian et al., 2004). S. pombe Klp5/6 and Drosophila Klp67A localize to kinetochores and the mitotic spindle in early mitosis and concentrate to the spindle midzone/midbody during cytokinesis. Double knockout of klp5+ and klp6+ in S. pombe causes chromosome pairs to move back and forth along the spindle for an extended period before sister chromatid separation (Garcia et al., 2002; West et al., 2002). Mutations in the Klp67A gene in Drosophila caused an increase in MT length, formation of disorganized bipolar spindles, abnormal chromosome congression, alignment and segregation, and failure of cytokinesis (Gandhi et al., 2004; Savoian et al., 2004). Our preliminary results indicate that human Kif18 also localizes to the kinetochores and mitotic spindle during early mitosis and translocates to the spindle midzone/midbody during cytokinesis (C. Zhu and W. Jiang, unpublished observations). Depletion of Kif18 by esiRNA did not cause gross mitotic spindle abnormalities, indicating that Kif18 may not be involved in regulating mitotic spindle formation. Alternatively, such a function for Kif18 may be masked by redundant functions of other proteins. Nevertheless, our results demonstrate that Kif18 is crucial for chromosome congression and alignment and for the metaphase to anaphase transition, indicating that Kif18 is a key determinant involved in chromosome segregation and spindle checkpoint in human cells.
Our immunofluorescence and time-lapse studies indicated that the chromokinesin Kid is important for chromosome movement. Although chromosomes in Kid esiRNA-treated cells attempted to congress to the metaphase plate, they were unable to sharply align and moved back and forth close to the metaphase plate, which caused a delay of the metaphase-to-anaphase transition. As cells entered anaphase, chromosome segregation was highly abnormal. Some sister chromatids moved to the opposite poles of the spindle, but others lagged behind. K-fiber and interkinetochore analyses showed that Kid is not required for the establishment and maintenance of kinetochore-spindle MT attachment or the judgment of tension across kinetochore pairs. However, when Kid esiRNA-transfected cells were treated with monastrol, chromosomes clustered into a mass close to the center of the monopolar spindle. Previous studies postulated that Kid is required for generating the polar ejection force necessary for chromosome congression and alignment (Levesque and Compton, 2001). In Xenopus, chromosome congression is inhibited in the absence of Xkid (Antonio et al., 2000; Funabiki and Murray, 2000). In human cells, microinjection of Kid-specific antibodies inhibited chromosome orientation and congression, and coinjection of Kid- and Eg5-specific antibodies resulted in chromosomes moving very close to the center of monopolar spindles (Levesque and Compton, 2001). Our findings support the idea that that Kid is essential for generating the spindle ejection force in human cells.
Our study also revealed that Kif4A and Kif4B, two closely related kinesins, play essential roles in anaphase spindle dynamics and the completion of cytokinesis. Previous studies indicated that the Drosophila or Xenopus Kif4 orthologues, Klp3A or Klp1, played an essential for spindle MT dynamics and Klp3A is essential for cytokinesis (Walczak et al., 1998; Kwon et al., 2004). Kif4A localizes to condensed chromosomes, mitotic spindles and the spindle midzone/midbody during mitosis and cytokinesis (Lee and Kim, 2004). Depletion of Kif4A in human fibroblasts by siRNA resulted in defective prometaphase organization, chromosome misalignment, spindle defects, chromosome mis-segregation, and cytokinesis failures (Mazumdar et al., 2004). We did not observe significant abnormalities in bipolar spindle formation or chromosome segregation in HeLa cells treated with Kif4A (or Kif4B) esiRNA. However, we found that depletion of Kif4A or Kif4B resulted in the formation of highly elongated anaphase spindles with separating chromosomes extremely close to the spindle poles and also failures in cytokinesis. These results are consistent with reports that Kif4A specifically interacts with the spindle midzone-associated Cdk substrate PRC1, an essential protein required for midzone formation and cytokinesis (Kurasawa et al., 2004; Zhu and Jiang, 2005). Kif4A is required for the translocation of PRC1 to the plus ends of interdigitating spindle MTs during the metaphase-to-anaphase transition, and Cdk phosphorylation of PRC1 controls the timing of PRC1 translocation by Kif4 (Zhu and Jiang, 2005). Because the midzone/midbody is critical for cytokinesis in animal cells, cell cycle–dependent translocation of PRC1 by Kif4 could regulate anaphase spindle dynamics, midzone/midbody formation, and the completion of cytokinesis.
Our analyses showed that MKLP1 and MKLP2, two closely related mitotic spindle midzone/midbody-associated kinesins, play independent and distinct roles in regulating cytokinesis. Neither protein was dependent on the other for its association with the spindle midzone/midbody. MKLP1 is crucial for the midbody formation and early telophase to late telophase transition, whereas MKLP2 is essential for cell abscission, the final step of cytokinesis. Consistent with our findings, MKLP1 and MKLP2 do not interact with each other to form heterodimers in human cells (Neef et al., 2003). MKLP1 is required for the formation of the midbody matrix and MKLP1 and MKLP2 are essential for the completion of cytokinesis (Neef et al., 2003; Matuliene and Kuriyama, 2004).
Taken together, our study presents a detailed analysis of MT-based motor proteins involved in mitosis and cytokinesis in human cells. We identified at least 12 human kinesins that are involved in regulating mitotic spindle formation, chromosome segregation, anaphase spindle dynamics, and cytokinesis in HeLa cells. Among these, we identified two new human kinesins, Kif14 and Kif18, which play essential roles in mitosis and cytokinesis. Because of technical limitations of the gene knockdown approach, we cannot exclude the possibility that other kinesins and/or dynein might also be involved in mitosis and cytokinesis. Indeed, dynein has previously been implicated in spindle formation and mitosis (Echeverri et al., 1996; Goshima and Vale, 2003; Gaetz and Kapoor, 2004). However, our study and others (Kittler et al., 2004) indicate that depletion of dynein by siRNA does not cause any mitotic or cytokinetic abnormalities. There are several explanations for the results we obtained, including 1) dynein does not have a crucial role in mitosis in HeLa cells, 2) depletion of dynein by esiRNA might not be complete, and trace amounts of dynein protein in esiRNA-treated cells may be sufficient to carry out its mitotic functions, or 3) there are functional redundancies among dynein and other kinesin motor proteins. Thus, experiments involving various combinations of kinesin/dynein esiRNA treatments will be required to address this point in the future.
Supplementary Material
Acknowledgments
We thank Drs. Francis A. Barr, Michael Glotzer, Fabienne Pirollet, and Jason Myers for reagents; Dr. Nanxin Li for critical reading of the manuscript; and Ningning Sai for laboratory support. This work was supported by grants from Edward Mallinckrodt, Jr. Foundation, Lisa U. Pardee Foundation, National Science Foundation (NSF-0233997), and National Institutes of Health (GM67859) to W.J.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0167) on April 20, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abaza, A., Soleilhac, J. M., Westendorf, J., Piel, M., Crevel, I., Roux, A., and Pirollet, F. (2003). M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J. Biol. Chem. 278, 27844–27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P. D., Ovechkina, Y., Morrice, N., Wagenbach, M., Duncan, K., Wordeman, L., and Swedlow, J. R. (2004). Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6, 253–268. [DOI] [PubMed] [Google Scholar]
- Antonio, C., Ferby, I., Wilhelm, H., Jones, M., Karsenti, E., Nebreda, A. R., and Vernos, I. (2000). Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102, 425–435. [DOI] [PubMed] [Google Scholar]
- Blangy, A., Lane, H. A., d'Herin, P., Harper, M., Kress, M., and Nigg, E. A. (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Carvalho, A., Carmena, M., Sambade, C., Earnshaw, W. C., and Wheatley, S. P. (2003). Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 116, 2987–2998. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L., and Morabito, J. (2004). TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell 15, 1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G. K., Jablonski, S. A., Sudakin, V., Hittle, J. C., and Yen, T. J. (1999). Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146, 941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G. K., Schaar, B. T., and Yen, T. J. (1998). Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. C., Zhou, Y., and Detrich, H. W., 3rd. (2002). Zebrafish mitotic kinesin-like protein 1 (Mklp1) functions in embryonic cytokinesis. Physiol. Genom. 8, 51–66. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Mao, Y., and Sullivan, K. F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421. [DOI] [PubMed] [Google Scholar]
- Cottingham, F. R., and Hoyt, M. A. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Verma, S., Mitchison, T. J., and Walczak, C. E. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69–78. [DOI] [PubMed] [Google Scholar]
- Echard, A., Jollivet, F., Martinez, O., Lacapere, J. J., Rousselet, A., Janoueix-Lerosey, I., and Goud, B. (1998). Interaction of a Golgi-associated kinesin-like protein with Rab6. Science 279, 580–585. [DOI] [PubMed] [Google Scholar]
- Echeverri, C. J., Paschal, B. M., Vaughan, K. T., and Vallee, R. B. (1996). Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontijn, R. D., Goud, B., Echard, A., Jollivet, F., van Marle, J., Pannekoek, H., and Horrevoets, A. J. (2001). The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol. Cell. Biol. 21, 2944–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., and Murray, A. W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411–424. [DOI] [PubMed] [Google Scholar]
- Gaetz, J., and Kapoor, T. M. (2004). Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 166, 465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, R., Bonaccorsi, S., Wentworth, D., Doxsey, S., Gatti, M., and Pereira, A. (2004). The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell 15, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N. J., and Compton, D. A. (2004). The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 166, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M. A., Koonrugsa, N., and Toda, T. (2002). Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 21, 6015–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky, G. J. (2004). Mitosis: MCAK under the aura of Aurora B. Curr. Biol. 14, R346–R348. [DOI] [PubMed] [Google Scholar]
- Goshima, G., and Vale, R. D. (2003). The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg, U., Neef, R., Honda, R., Nigg, E. A., and Barr, F. A. (2004). Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt, E. R., and Hoyt, M. A. (2000). Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1496, 99–116. [DOI] [PubMed] [Google Scholar]
- Hill, E., Clarke, M., and Barr, F. A. (2000). The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19, 5711–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P. S. (2003). Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Jimenez, G., Wells, N. J., Hope, T. J., Wahl, G. M., Hunter, T., and Fukunaga, R. (1998). PRC1, a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell 2, 877–885. [DOI] [PubMed] [Google Scholar]
- Kapoor, T. M., and Compton, D. A. (2002). Searching for the middle ground: mechanisms of chromosome alignment during mitosis. J. Cell Biol. 157, 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T. M., Mayer, T. U., Coughlin, M. L., and Mitchison, T. J. (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T. M., and Mitchison, T. J. (2001). Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154, 1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler, R. et al. (2004). An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Kline-Smith, S. L., Khodjakov, A., Hergert, P., and Walczak, C. E. (2004). Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell 15, 1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa, Y., Earnshaw, W. C., Mochizuki, Y., Dohmae, N., and Todokoro, K. (2004). Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, M., Morales-Mulia, S., Brust-Mascher, I., Rogers, G. C., Sharp, D. J., and Scholey, J. M. (2004). The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell 15, 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, W., Zhang, X., Kline-Smith, S. L., Rosasco, S. E., Barrett-Wilt, G. A., Shabanowitz, J., Hunt, D. F., Walczak, C. E., and Stukenberg, P. T. (2004). Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14, 273–286. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. J. et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. M., and Kim, W. (2004). Kinesin superfamily protein member 4 (KIF4) is localized to midzone and midbody in dividing cells. Exp. Mol. Med. 36, 93–97. [DOI] [PubMed] [Google Scholar]
- Levesque, A. A., and Compton, D. A. (2001). The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154, 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow, E., and Mandelkow, E. M. (2002). Kinesin motors and disease. Trends Cell Biol. 12, 585–591. [DOI] [PubMed] [Google Scholar]
- Maney, T., Hunter, A. W., Wagenbach, M., and Wordeman, L. (1998). Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y., Abrieu, A., and Cleveland, D. W. (2003). Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell 114, 87–98. [DOI] [PubMed] [Google Scholar]
- Matuliene, J., and Kuriyama, R. (2004). Role of the midbody matrix in cytokinesis: RNAi and genetic rescue analysis of the mammalian motor protein CHO1. Mol. Biol. Cell 15, 3083–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, T. U., Kapoor, T. M., Haggarty, S. J., King, R. W., Schreiber, S. L., and Mitchison, T. J. (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974. [DOI] [PubMed] [Google Scholar]
- Mazumdar, M., Sundareshan, S., and Misteli, T. (2004). Human chromokinesin KIF4A functions in chromosome condensation and segregation. J. Cell Biol. 166, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B. F., Chan, G. K., Zubrowski, B., Savoian, M. S., Sauer, M. T., and Yen, T. J. (2001). CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell 12, 2776–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., Setou, M., Kaneshiro, K., and Hirokawa, N. (2001). All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA 98, 7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima, M., Kaitna, S., and Glotzer, M. (2002). Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2, 41–54. [DOI] [PubMed] [Google Scholar]
- Mishima, M., Pavicic, V., Gruneberg, U., Nigg, E. A., and Glotzer, M. (2004). Cell cycle regulation of central spindle assembly. Nature 430, 908–913. [DOI] [PubMed] [Google Scholar]
- Molina, I., Baars, S., Brill, J. A., Hales, K. G., Fuller, M. T., and Ripoll, P. (1997). A chromatin-associated kinesin-related protein required for normal mitotic chromosome segregation in Drosophila. J. Cell Biol. 139, 1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain, V., Simerly, C., Howard, L., Ando, A., Schatten, G., and Compton, D. A. (1999). The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef, R., Preisinger, C., Sutcliffe, J., Kopajtich, R., Nigg, E. A., Mayer, T. U., and Barr, F. A. (2003). Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi, R., Sapra, T., Howard, J., and Mitchison, T. J. (2004). Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell 15, 2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura, H., Torok, T., Tick, G., Hoheisel, J., Kiss, I., and Glover, D. M. (1997). Mutation of a gene for a Drosophila kinesin-like protein, Klp38B, leads to failure of cytokinesis. J. Cell Sci. 110(Pt 8), 945–954. [DOI] [PubMed] [Google Scholar]
- Putkey, F. R., Cramer, T., Morphew, M. K., Silk, A. D., Johnson, R. S., McIntosh, J. R., and Cleveland, D. W. (2002). Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell 3, 351–365. [DOI] [PubMed] [Google Scholar]
- Rischitor, P. E., Konzack, S., and Fischer, R. (2004). The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot. Cell 3, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakowicz, R. et al. (2004). Antitumor activity of a kinesin inhibitor. Cancer Res. 64, 3276–3280. [DOI] [PubMed] [Google Scholar]
- Savoian, M. S., Gatt, M. K., Riparbelli, M. G., Callaini, G., and Glover, D. M. (2004). Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J. Cell Sci. 117, 3669–3677. [DOI] [PubMed] [Google Scholar]
- Schaar, B. T., Chan, G. K., Maddox, P., Salmon, E. D., and Yen, T. J. (1997). CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, K. B., and Salmon, E. D. (2002). Chromosome dynamics: new light on Aurora B kinase function. Curr. Biol. 12, R458–R460. [DOI] [PubMed] [Google Scholar]
- Snove, O., Jr., and Holen, T. (2004). Many commonly used siRNAs risk off-target activity. Biochem. Biophys. Res. Commun. 319, 256–263. [DOI] [PubMed] [Google Scholar]
- Tanudji, M., Shoemaker, J., L'Italien, L., Russell, L., Chin, G., and Schebye, X. M. (2004). Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell 15, 3771–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale, R. D. (2003). The molecular motor toolbox for intracellular transport. Cell 112, 467–480. [DOI] [PubMed] [Google Scholar]
- Walczak, C. E., Mitchison, T. J., and Desai, A. (1996). XKCM1, a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84, 37–47. [DOI] [PubMed] [Google Scholar]
- Walczak, C. E., Vernos, I., Mitchison, T. J., Karsenti, E., and Heald, R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8, 903–913. [DOI] [PubMed] [Google Scholar]
- Waters, J. C., Skibbens, R. V., and Salmon, E. D. (1996). Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J. Cell Sci. 109(Pt 12), 2823–2831. [DOI] [PubMed] [Google Scholar]
- Weaver, B. A., Bonday, Z. Q., Putkey, F. R., Kops, G. J., Silk, A. D., and Cleveland, D. W. (2003). Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, R. R., Malmstrom, T., and McIntosh, J. R. (2002). Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 115, 931–940. [DOI] [PubMed] [Google Scholar]
- Yang, D., Buchholz, F., Huang, Z., Goga, A., Chen, C. Y., Brodsky, F. M., and Bishop, J. M. (2002). Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 99, 9942–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C., Bossy-Wetzel, E., and Jiang, W. (2005). Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem. J. (in press). [DOI] [PMC free article] [PubMed]
- Zhu, C., and Jiang, W. (2005). Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA 102, 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.