Abstract
ADARs (adenosine deaminases that act on RNA) are RNA-editing enzymes that convert adenosines to inosines in structured or double-stranded RNAs. Expression and intracellular distribution of ADAR1 is controlled by a plethora of mechanisms suggesting that enzyme activity has to be tightly regulated. Mammalian ADAR1 is a shuttling protein, whereas Xenopus ADAR1 is exclusively nuclear. In oocytes, Xenopus ADAR1 associates with most nascent transcripts but is strongly enriched at a specific site on chromosome 3, termed the special loop. Enrichment at this site requires the presence of RNAs but is independent of ongoing transcription. Here we show that RNAs transcribed elsewhere in the genome accumulate at the special loop even in the absence of transcription. In situ hybridization experiments, however, indicate the absence of known editing substrates from this site. In the absence of transcription also other RNA binding and processing factors accumulate at the special loop, suggesting that ADAR1 is stored or assembled at the special loop in an RNA-containing complex. Nuclear injection of RNAs providing binding sites for ADAR1 dissociates the enzyme from the special loop, supporting the notion that the special loop represents a site where ADAR1 is stored, possibly for later use during development.
INTRODUCTION
ADARs (adenosine deaminases that act on RNA) are a group of enzymes that catalyze the hydrolytic deamination of adenosines into inosines in structured or double-stranded RNAs (Bass, 2002). As inosines are interpreted as guanosines during translation, this RNA-editing event can lead to codon exchanges in mRNAs. ADAR activity was first described in Xenopus oocytes but seems to exist in all metazoan tissues (Bass and Weintraub, 1987; Wagner et al., 1990). Consistently, cDNAs encoding ADAR proteins have been cloned from different organisms ranging from Caenorhabditis elegans to man, including Xenopus (Bass, 2002).
Three ADAR members, termed ADAR1, ADAR2, and ADAR3 have been characterized in mammals, whereas in most other vertebrates and invertebrates only one or two ADAR variants are known. All ADAR proteins have a highly conserved catalytic domain at their C-terminal ends and one to three double-stranded RNA-binding domains (dsRBDs) located N-terminally of the catalytic domain. The amino-terminal ends of ADARs are highly variable. Although ADAR1 has a long amino-terminus containing a Z-DNA binding domain, ADAR2 and ADAR3 proteins lack this region altogether (Keegan et al., 2004).
ADARs can edit RNAs both in a specific and nonspecific manner, depending on the substrate. During specific editing, only a few selected adenosines are modified, whereas up to 50% of adenosines are affected in hyperedited substrates. Hyperediting frequently occurs in viral RNAs, whereas specific editing is predominantly found in endogenous substrates (Bass, 2002).
The pre-mRNAs encoding the serotonin HT-2c receptor and those encoding subunits of the glutamate-gated ion channel family are probably the best-studied mammalian substrates so far. In both cases, editing sites are defined by base-paired regions formed between intronic and exonic sequences, thus indicating that editing must occur cotranscriptionally, before the removal of intronic sequences (Dabiri et al., 1996; Burns et al., 1997). Consistently, the enzyme can be found in the nucleus associated with nascent transcripts on transcriptionally active chromosomes (Eckmann and Jantsch, 1999). By far the most abundant class of editing substrates, however, represent repetitive elements located in noncoding regions of mRNAs. The functional significance of editing in these mobile elements remains enigmatic at this point (Athanasiadis et al., 2004; Kim et al., 2004; Levanon et al., 2004).
Different mechanisms controlling ADAR activity have been described in various organisms, suggesting that ADAR activity has to be tightly controlled. In mammals, for instance, ADAR1 expression is stimulated by interferon (Patterson and Samuel, 1995). Moreover, mammalian ADAR1 is a shuttling protein and its nuclear accumulation is regulated in an RNA-binding dependent manner (Strehblow et al., 2002). Within the nucleus, ADARs can be found sequestered in nucleoli (Desterro et al., 2003; Sansam et al., 2003). In addition, ADARs act as dimers and it seems possible that heterodimer formation between different ADAR versions can control enzyme activity and substrate specificity (Cho et al., 2003; Gallo et al., 2003). Also, ADAR3 appears enzymatically inactive and might modulate the editing activity of ADAR1 and -2 by competitive binding to editing sites or by forming heterodimers with other ADAR members (Chen et al., 2000). In Drosophila, finally, ADAR can edit its own pre-mRNA, thus giving rise to an enzymatically less active version of the protein (Palladino et al., 2000), whereas a similar self-editing event leads to alternative splicing of ADAR2 in rodents (Rueter et al., 1999; Palladino et al., 2000).
In Xenopus, ADAR1 is the only ADAR member isolated so far and this protein is constitutively nuclear (Hough and Bass, 1997; Eckmann et al., 2001). In oocytes, the protein is found associated with the nascent RNP matrix on transcriptionally active lampbrush chromosomes. Additionally, ADAR1 can be found highly enriched at an atypical chromosomal site, termed the “special loop” (Eckmann and Jantsch, 1999). This special loop is located on chromosome 3 and represents a so called “double loop bridge,” where the two sister chromatids are separated, giving rise to two separated DNA threads. The localization of ADAR1 to the special loop is not sensitive to transcriptional inhibitors, hence suggesting that this site is transcriptionally silent. However, treatment of chromosomes with RNAses removes ADAR1 from the special loop, indicating that an RNA is required to tether ADAR1 or other proteins associated with it to the special loop (Eckmann and Jantsch, 1999).
In this study, we investigate the mechanism and nature of RNA accumulation at the special loop that is required to tether ADAR1 to this site. RNA injection experiments with an artificial ADAR1 binding site can titrate ADAR1 off the special loop, suggesting that ADAR1 is stored or assembled with other RNA processing factors at this site. We therefore suggest that the special loop represents an assembly or storage site of ADAR1 that may contribute to regulate the intranuclear concentration of ADAR1 in Xenopus.
MATERIALS AND METHODS
Preparation of Xenopus Lampbrush Chromosomes
A Xenopus laevis female was anesthetized for ovary removal. Separated oocytes were stored in OR2 medium (Wallace et al., 1973) at 16°C over night. Lampbrush chromosome preparations were made according to Gall et al. (1991).
RNase Digestion of Xenopus Lampbrush Chromosomes
Xenopus lampbrush chromosomes were spread as usual. After centrifugation, chromosome spreads were incubated with one of the following RNAses: RNase A, 1 mg/ml in 0.5 M NaCl; RNase T1, 10.000 U/ml in 0.5 M NaCl; RNase V1, 1 U/10 μl in 0.2 M KCl, 10 mM Tris, 7.5, 50% glycerol. Digestions with RNase A or T1 were incubated at 37°C for 30 min, whereas RNase V1 digestions were incubated at 37°C for 1 h. Subsequently, slides were washed in 1× phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde in PBS for 1 h before immunofluorescence staining.
In Situ Hybridization
Lampbrush chromosome preparations were dehydrated and deparafinized with 100% xylene followed by washes with 100% ethanol and 100% acetone. The preparations were air-dried and stored in vacuo at –70°C.
A 1285-bp cDNA sequence of bFGF was cloned into the BamHI and KpnI sites pBS KS-using forward primer CCCGGTACCTCTTGGCCCAATATAGAG and reverse primer GACTGGATCCTAATAACTACAACACTTAC. RNA-probes were made by in vitro transcription in the presence of fluorescein- or Texas Red–labeled UTP (Molecular Probes, Eugene, OR).
For in situ hybridization, ∼50–75 ng labeled RNA probe was resuspended in 10 μl in situ-mix-5 (40% formamide, 4× SSC, 0.1 M Na,K phosphate, pH 7, 300 μg/μl sonicated salmon sperm DNA, 300 μg/μl tRNA in dH2O). The probe was denatured at 89°C for 7 min, transferred to the chromosome spread, and sealed with a coverslip and rubber cement. Hybridization was carried out over night at 42°C. The slides were washed three times for 5 min in 2× SSC at 37° followed by a final rinse with 1× PBS.
Different antibody-systems were used to amplify the signals. The first antibody was directed against the fluorescent dye incorporated in the RNA-probe. The following antibody-sets were used from Molecular Probes:
1st antibody: rb-α-fluorescein-Alexa 488 diluted 1:62.5 in 2.5%serum/PBST
2nd antibody: gt-α-rb-Alexa 488 diluted 1:125 in 2.5% serum/PBST
1st antibody: gt-α-fluorescein-Alexa 488 diluted 1:100 in 2.5% serum/PBST
2nd antibody: dk-α-gt-Alexa 488 diluted 1:100 in 2.5% serum/PBST
1st antibody: rb-α-Texas-Red diluted 1:100 in 2.5% serum/PBST
2nd antibody: gt-α-rb-Texas-Red diluted 1:250 in 2.5% serum/PBST
Immunofluorescence Staining
Tissue culture cells were grown on coverslips for immunofluorescence staining. The cells were washed three times with PBS and fixed for 3 min with 2% paraformaldehyde, 1× PBS, 0.05% Triton. Three further washing steps with PBS were followed by a 1-min methanol-permeabilization-step before unspecific antibody binding was blocked. LBC spreads were rinsed briefly with 1× PBS before blocking. The anti-xlADAR1 antibody Sat3 (Eckmann and Jantsch, 1999) was used for localization of the special loop. The preparations were fixed and stained with DAPI in antifading buffer. Antibodies used were monoclonal antibody (mAb) Y12, directed against Sm core proteins found on most splicing snRNPs (Lerner et al., 1981); mAb K121, directed against the trimethyl guanosine cap present on most spicing snRNAs (Krainer, 1988); mAb H14, directed against the phosphorylated carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II (Kim et al., 1997); mAb SC35, directed against a non-snRNP factor, SC35, required for spliceosome assembly (Fu and Maniatis, 1990); mAb 10D1, directed against hnRNP A2 (Marcu et al., 2001); and mAb 4D11, directed against hnRNP L (Pinol-Roma et al., 1989). For double-staining experiments secondary FITC- (fluorescein isothiocyanate) or TRITC-labeled antibodies were used.
AMD and alpha Amanitin Treatment of Oocytes
Actinomycin D (AMD; Sigma) was added to oocytes cultured in OR2 to a final concentration of 50 μg/μl AMD over night before LBC preparation. Alpha amanitin (50 nl; 400 μg/ml in dH2O) was injected into oocytes 10 h before chromosome preparation.
Heterokaryon Assays
XlA6 cells were grown on coverslips. Trypsinated HeLa cells were added drop wise. The cells were incubated for 2.5 h at 27°C in L-15 medium supplemented with 15% fetal calf serum. Inhibition of translation was achieved by addition of 20 μg cycloheximide per milliliter of medium and a further incubation period of 30 min. For cell fusions medium was removed and cells were treated with PEG 6000 (1 g/ml in serum-free medium) for 2 min. Two washing steps with PBS were followed by an incubation period of 4 h in cycloheximide containing medium. Fixed cells were stained with species-specific antibodies against putative shuttling proteins. Secondary antibodies were either Alexa 488 or TRITC conjugated.
Probe Preparation
The complementary 25 mer RNA oligonucleotides RNA2 5′ UUCGCCACUGCACCAGCCUGAUUGC and RNA3 5′ GCAAUCAGGCUGGUGCAGUGGCGAA were 5′ end-labeled with 32P using T4 Kinase for 1 h at 37°C. This was followed by two ethanol precipitation steps.
For intermolecular basepairing the labeled, and washed probes were resuspended in 1 mM MgCl2 and incubated at 95°C for 3 min. Subsequently probes were slowly cooled to room temperature and precipitated by ethanol.
Oocyte Injection
For brUTP incorporation studies 50 nl of a 5 mM brUTP solution were injected into the cytoplasm of oocytes. These cells were then incubated for different time periods in OR2 at 16°C before AMD-treatment. For nuclear injections of RNAs, oocytes were centrifuged, animal side up, for 30 min at 3000 rpm under OR2, held in place by a synthetic grid. Thirty picomols of single- or double-stranded RNA were injected into the nuclei that were well visible in the dark background of the animal hemisphere. The efficiency of nuclear injection was verified by scintillation counting of hand-isolated nuclei and cytoplasms. Oocytes were stored at 16°C in OR-2 before chromosome preparation.
RESULTS
RNAs Accumulate at the Special Loop with Slow Kinetics
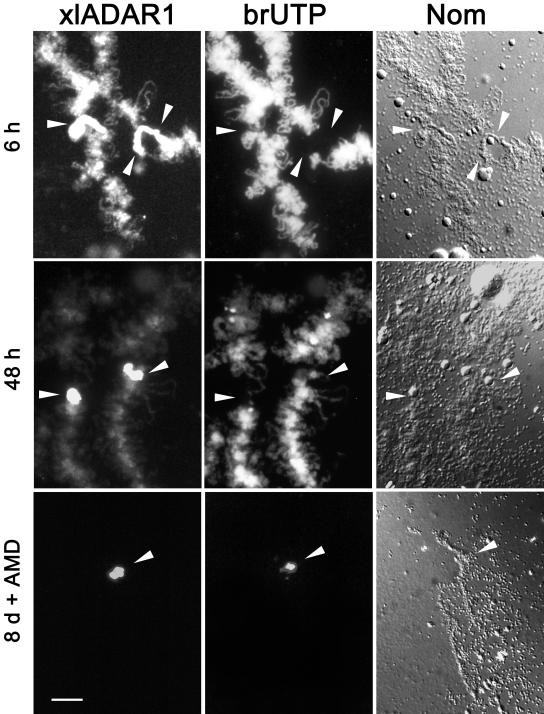
Previous experiments have shown that ADAR1 localization to the special loop does not require ongoing transcription, thus suggesting that this site is transcriptionally silent (Eckmann and Jantsch, 1999). Consistent with this finding, phosphorylated pol II cannot be detected at the special loop (Eckmann and Jantsch, 1999). On the other hand, RNase-treatment of lampbrush chromosomes removes ADAR1 from this site, suggesting that an RNA is required to tether ADAR1 to this site. Interestingly, treatment with both single- and double-strand–specific nucleases leads to removal of ADAR1 from the special loop, suggesting that the RNA(s) located at this site contain both single- and double-stranded regions (unpublished data). To clarify whether the special loop is indeed transcriptionally silent and to test whether RNA could be detected at this site, we decided to label de novo synthesized RNA by injection of 5-bromo UTP (brUTP) into oocytes. Incorporation of these modified nucleotides was subsequently followed over time using fluorescently labeled anti-brUTP antibodies. The earliest time points, taken ∼15 min after injection, showed homogeneous incorporation of brUTP along the majority of all visible loops and in the extrachromosomal nucleoli. This labeling slightly increased over the next couple of hours and remained visible throughout the entire experiment for up to 8 d (Figure 1). No labeling could, however, be detected at the special loop for the first 48 h. After 3 d, minimal brUTP staining was visible at the special loop. Nonetheless, this staining was camouflaged by the intense labeling of regular, transcriptionally active loops. To eliminate the labeling of regular loops and to allow a better observation of brUTP incorporation at the special loop, oocytes were treated with actinomycin D (AMD) 8 d after brUTP injection. This treatment removed all nascent transcripts from the chromosomes and revealed clear brUTP accumulation at the special loop (Figure 1). Similarly, treatment of oocytes with alpha amanitin also allowed observation of minor amounts of brUTP-labeled RNA at the special loop (unpublished data).
Figure 1.
RNA turnover at the special loop. To detect newly synthesized RNA, oocytes were injected with brUTP. At several time points after injection, chromosome spreads were stained with anti-ADAR1 (ADAR1) or brUTP (brUTP) antibodies, respectively. Nomarski optics image of the same region (NOM). Arrowheads mark the position of the special loop on bivalent 3. Six hours (6h) after brUTP injection the majority of regular loops shows massive incorporation of brUTP, whereas the special loop highlighted with the ADAR1 antibody shows no brUTP. Forty-eight hours (48 h) after brUTP injection the special loop appears unlabeled when compared with regular loops. (8d +AMD) Eight days after injection followed by 8 h of AMD treatment transcription at regular loops has ceased, allowing the detection of brUTP-labeled RNAs at the special loop. Scale bar, 10 μm
The Special Loop, a Dedicated Editing Site?
The observed labeling behavior is compatible with a movement of transcripts from regular loops toward the special loop and their subsequent anchoring at this site. Although the nature of the RNAs tethered at the special loop was not clear from these experiments, it appeared conceivable that substrates for editing by ADAR1 synthesized elsewhere in the genome are transported to the special loop to undergo editing at this site. In Xenopus, the mRNA encoding basic fibroblast growth factor (bFGF) is the only substrate proven to be edited by an ADAR-like activity (Kimelman and Kirschner, 1989; Saccomanno and Bass, 1999). An antisense transcript that is complementary to the bFGF mRNA is synthesized from a promoter located in the 3′ region of the gene. Sense and antisense transcripts hybridize over an ∼1000-base pair-long region and subsequently become edited (Kimelman and Kirschner, 1989).
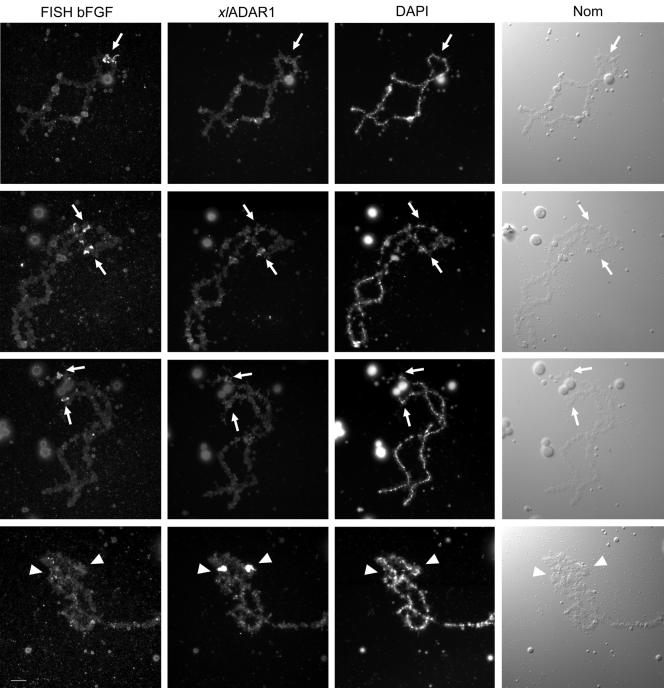
To test whether bFGF can be found at the special loop, we performed fluorescent RNA in situ hybridization experiments. As bFGF is transcribed in both directions, labeled sense and antisense RNA probes were generated. To ensure the specificity of the in situ hybridization, sense and antisense RNA probes of the RNA encoding Xenopus laevis RNA-binding protein A (XlrbpA), were used as negative and positive controls, respectively. The positive control always gave a unique signal on chromosome 3 (unpublished data), whereas no signal could be detected with the XlrbpA sense probe. Both the sense and antisense bFGF-probes could always be detected at the same three chromosomal sites. As Xenopus laevis is pseudotetraploid, it is possible that more than one transcriptionally active gene exists in the genome. Signals could be observed at two different telomeric loci as well as at a single subtelomeric locus (Figure 2). The simultaneous immunodetection of the endogenous ADAR1 protein allowed the identification of the special loop. No accumulation of bFGF sense or antisense RNA could be detected at the special loop in these experiments. Vice versa, no ADAR1 enrichment was detectable at sites of bFGF transcription. These results suggest that the special loop is not a chromosomal site specialized for editing of bFGF mRNA. Whether other substrate RNAs are guided to the special loop for editing remains to be determined as such substrates become identified.
Figure 2.
Fluorescence in situ hybridization (FISH) reveals the absence of bFGF transcripts from the special loop. bFGF was detected by FISH with a fluorescein-labeled antisense probe (FISH bFGF), and the special loop was detected by subsequent immunostaining with anti-ADAR1 antiserum (xlADAR1). bFGF RNA could be consistently detected on three different chromosomes (arrows, top three rows), which were not further identified, whereas bFGF RNA was absent from the special loop (arrowhead, bottom row). The faint signals detected in the anti-ADAR immunostaining at the positions corresponding to the FISH signals (arrows) result from the cross-reactivity of the antibody-based signal amplification system used for FISH. Chromosomal images stained with DAPI and visualized by Nomarski optics are shown for comparison. Scale bar, 10 μm.
Accumulation of RNA-processing Factors at the Special Loop
We previously showed that factors involved in pre-mRNA splicing are present at the special loop (Eckmann and Jantsch, 1999). Immunostaining with antibodies raised against the m3GpppA-caps of snRNAs, the SR protein SC35 and the Sm core proteins found on most splicing snRNPs, revealed that some of these proteins are enriched at the special loop (Eckmann and Jantsch, 1999). Because splicing factors are found associated with nascent pol II transcripts but not with pol I or pol III transcripts, these findings suggested that the RNAs accumulating at the special loop are transcribed by RNA polymerase II.
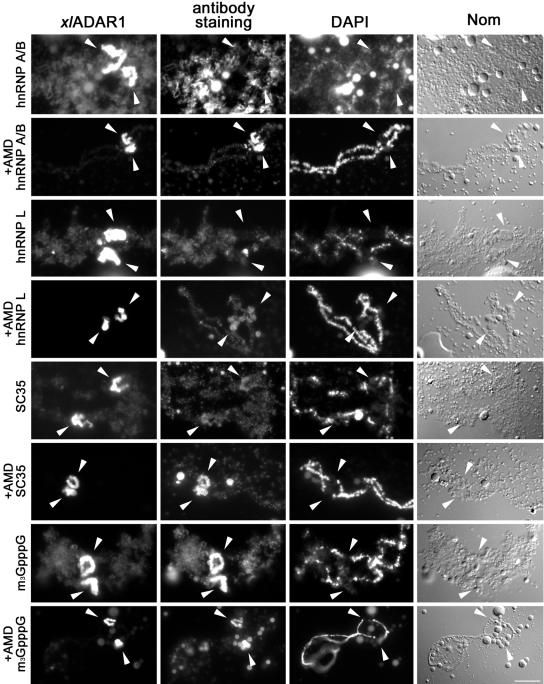
To further investigate whether the RNAs present at the special loop show the protein composition of normal polymerase II transcripts, we tested the special loop for the presence of hnRNPs. These proteins cotranscriptionally associate with pol II transcripts and are essential for their proper processing and export to the cytoplasm. hnRNPs A/B, for instance, are shuttling proteins involved in splicing and nuclear export of RNAs. hnRNP L, on the other hand, is a nuclear protein exclusively involved in RNA-processing (Pinol-Roma et al., 1989). Using antibodies 10D1 and mAb 4D11 raised against hnRNPs A/B (Marcu et al., 2001) and hnRNP L, respectively, we could detect both proteins along regular chromosome loops, whereas neither protein was present at the special loop (Figure 3). The special loop and thus the RNAs located at this site therefore seem to have a different protein composition than regular loops: splicing factors are present at this site, whereas hnRNPs and pol II are absent. Taken together, the absence of hnRNPs from the special loop under normal conditions as well as the previously reported absence of pol II from this site suggests that RNAs localized to the special loop are either not synthesized by pol II or not destined for export into the cytoplasm (see Discussion).
Figure 3.
RNA processing factors accumulate at the special loop in the absence of transcription. Chromosomal spreads were stained with antibodies against the shuttling hnRNP A/B, the nonshuttling hnRNP L, the nonshuttling SR-protein SC35, or the trimethyl G (3mG) cap found on most splicing snRNPs, both in the presence and absence (AMD) of transcription. xlADAR1 remains at the special loop after AMD treatment (arrowheads), whereas regular loops are stripped of their RNP matrix and fuse with the chromosomal axis (see Nomarski image of AMD-treated chromosomes). HnRNPs A/B and L are absent from the special loop under normal conditions but show strong (A/B) and moderate (L) enrichment at this site upon AMD treatment. SC35, in contrast, is present but not enriched at the special loop under normal conditions but strongly accumulates there when transcription is inhibited. 3mG, indicative of snRNPs, is always enriched at the special loop and thus behaves like ADAR1. Scale bar, 10 μm
Protein Composition at the Special Loop Changes after Inhibition of Transcription
After transcriptional inhibition by AMD, chromosomes shrink and are stripped of their regular loops. Staining for antigens such as ADAR1, hnRNPs, or snRNPs is diminished at regular loops. Interestingly, ADAR1 was shown to remain at the special loop even in the absence of transcription (Eckmann and Jantsch, 1999).
As brUTP accumulation at the special loop could be detected best in the absence of transcription, we wondered whether a similar treatment might allow the detection of minor amounts of hnRNPs at this site. Indeed, hnRNPs A/B, which were normally absent from the special loop were detectable and even accumulated at this site in the absence of transcription, whereas hnRNP L was only moderately enriched. Similarly, the SR protein SC35 showed strong accumulation at the special loop in the absence of transcription, whereas the 3mG antigen of snRNAs was strongly enriched at the special loop both in the presence and absence of transcription (Figure 3).
Given that hnRNPs A/B accumulate at the special loop together with brUTP-labeled RNA in the absence of transcription, it seems possible that at least some RNAs that are transcribed by polymerase II move to this site when transcription is inhibited. Alternatively, hnRNP A/B proteins could accumulate at this site by themselves when transcription is inhibited, possibly by binding to RNAs that are already present at this site.
Xenopus ADAR1 Is an Exclusively Nuclear Protein
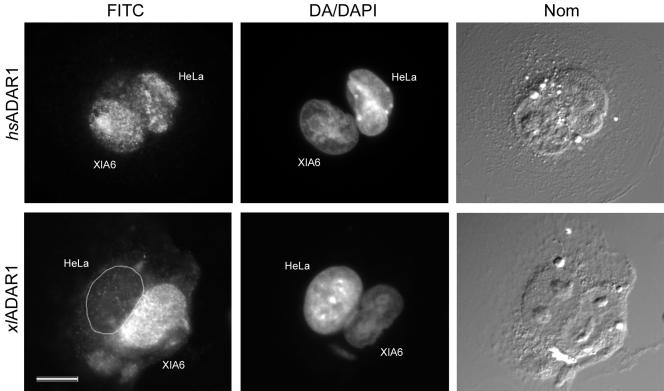
Our results led us to speculate that the special loop might represents a site where ADAR1 is assembled into macromolecular RNP complexes, modified, or stored as a means to regulate the intranuclear concentration of the enzyme. In mammals ADAR1 is a shuttling protein that moves to the cytoplasm when transcription is inhibited which can also be seen as a mechanism to control the intranuclear enzyme concentration. We had shown previously that Xenopus ADAR1, in contrast to its mammalian homologue remains exclusively nuclear even when transcription is inhibited (Eckmann et al., 2001). Although this fact suggests that Xenopus ADAR1 is an exclusively nuclear enzyme, lack of AMD responsiveness cannot be seen as a definitive proof for a lack of shuttling. Therefore, to address this point more clearly, we performed heterokaryon assays of Xenopus XlA6 cells fused to human HeLa cells and determined the shuttling capability of both human and Xenopus ADAR1 proteins. In these assays human ADAR1 could clearly be found in the Xenopus nuclei, whereas the Xenopus enzyme was confined to Xenopus nuclei, thus verifying that human but not Xenopus ADAR1 is a shuttling protein and that Xenopus ADAR1 remains nuclear at all times (Figure 4). This finding might also reflect the different signal sequences for nuclear import and possibly export found in Xenopus and human ADAR1, respectively (Strehblow et al., 2002). Interestingly, in heterokaryons we also observed a slight enrichment of human ADAR1 in nucleoli of Xenopus nuclei, resembling the previously reported accumulation of human ADAR1 in these structures (Desterro et al., 2003; Yang et al., 2003).
Figure 4.
Human ADAR1 is a shuttling protein, whereas Xenopus ADAR1 is not. Heterokaryons of Xenopus XlA6 and HeLa cells were produced by PEG fusion. The HeLa nuclei were identified by distamycin A+DAPI (DA/DAPI) staining, which results in DAPI-bright heterochromatin spots, whereas the Xenopus nuclei are stained homogeneously. Species-specific antibodies against human ADAR1 (top row) and Xenopus ADAR1 (bottom row) were used to detect the respective proteins in the heterokaryons. Human ADAR1 shuttles out of the human nucleus and can be found in the Xenopus nucleus (hsADAR1), whereas Xenopus ADAR1 remains in the Xenopus nucleus (xlADAR1). Whole cells are visualized by Nomarski optics (NOM). Scale bar, 10 μm.
Nuclear Injection of Double-stranded RNA Titrates ADAR1 off the Special Loop
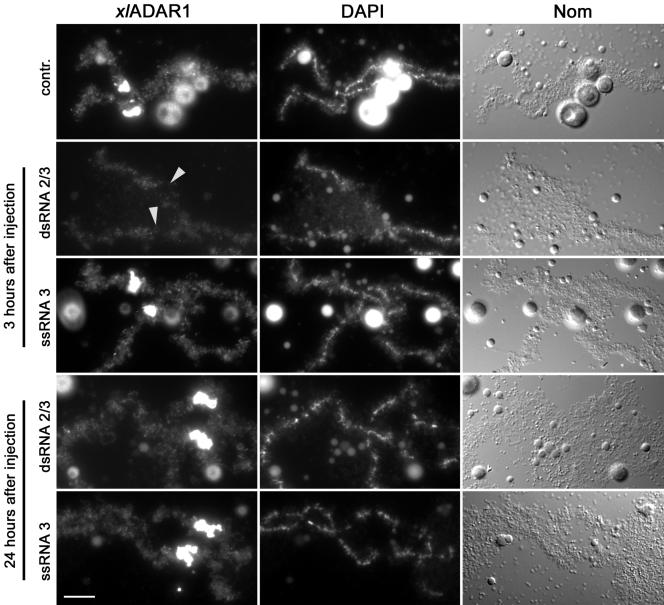
If the special loop represents a site where ADAR1 is stored, possibly for later use during embryogenesis or as a means to regulate the intranuclear concentration of the enzyme, it should be possible to titrate ADAR1 off the special loop by injection of RNAs containing binding sites for ADAR1 into oocyte nuclei. We thus decided to inject synthetic single- or double-stranded RNA into the nuclei of Xenopus oocytes. The RNAs were 25 nucleotides in length and designed in a way that each single-stranded RNA oligo was unable to form a double-stranded structure long enough to serve as a binding site for a dsRBD. On annealing with a complementary strand, however, the formed double-stranded RNA should allow ADAR1 binding to occur. In these experiments, injection of single-stranded RNA-oligos had no effect on chromosome morphology or ADAR1 localization to lampbrush chromosomes. Injection of the double-stranded RNA oligo, instead, led to efficient removal of ADAR1 from the special loop and to a minor extent from regular loops (Figure 5). Removal of ADAR1 was surprisingly slow in these experiments. Although almost no effect could be observed for up to 1 h after injection of the double-stranded oligo, the protein was efficiently removed 3 h after injection. Twenty-four hours after the initial injection, both regular loops and the special loop were again decorated with ADAR1. To verify that ADAR1 could also be removed from the special loop under more physiological conditions we injected part of the RNA containing the R/G editing site encoding the mouse glutamate receptor subunit B (Lomeli et al., 1994). This heterologous substrate of ADAR1 had been shown to be edited by Xenopus ADAR1 (Hurst et al., 1995). As expected, injection of the RNA derived from the GluRB minigene also led to efficient removal of ADAR1 from the special loop (unpublished data). These experiments demonstrate that ADAR1 located at the special loop under normal conditions can be titrated off this site when excess substrate is available.
Figure 5.
Xenopus ADAR1 can be stripped off the special loop by excess substrate. Injection of double-stranded RNA (dsRNA 2/3) into Xenopus oocyte nuclei preferentially removes ADAR1 from the special loop (arrowheads), whereas injection of single-stranded RNA (ssRNA 3) has no effect on ADAR1 localization. The effect is most prominent 3 h after injection of the oligo. Twenty-four hours after injection ADAR1 redecorates the special loop normally. Bar, 10 μm.
DISCUSSION
ADAR activity has been shown to be highly regulated in different species. Among other mechanisms, the intracellular sequestration of ADARs appears to be a highly efficient way of regulating editing activity at the posttranslational level. In mammals, for example, ADAR1 continuously shuttles between the cytoplasm and the nucleus and seems to accumulate in the cytoplasm under certain conditions (Strehblow et al., 2002). Furthermore, mammalian ADAR1 and ADAR2 have been shown to accumulate in nucleoli but can be relocated to the nucleoplasm upon nuclear injection of additional substrate (Desterro et al., 2003; Sansam et al., 2003).
The Special Loop as a Chromosomal Storage Site?
Although mammalian ADAR1 harbors both the sequences required for nuclear export and import and thus shuttles continuously between the nucleus and cytoplasm, we could show here that Xenopus ADAR1 is exclusively nuclear and fails to shuttle. It therefore appears reasonable to assume that other mechanisms regulating the concentration of free nuclear ADAR1 levels exist in Xenopus. In fact, our data demonstrate that ADAR1 (together with other components of the RNA-processing machinery) is stored at a special chromosome loop. Interestingly, the characteristics of this special loop are quite peculiar. Although no apparent transcription occurs at this site, the chromatin appears completely decondensed; at the same time, RNAs are localized at this site that are required to tether ADAR1 to the special loop. Although this behavior seems odd at first sight, similar specialized chromosomal regions have been described in the past. Roth and Gall (1987), for example, have described several proteins that exclusively localize to a subset of chromosomal sites. Similarly, in Drosophila hydei lampbrush chromosomes, some chromosomal loops serve as storage sites for particular proteins (Hulsebos et al., 1984). Last but not least, the nucleolus organizing regions serve as chromosomal sites on which a specialized proteinacious machinery assembles. Most interestingly, NORs and the attached nucleoli not only serve their primary function, namely the transcription of ribosomal RNAs as well as their processing and assembly into ribosomal subunits but also sequester and regulate the activity of several nuclear proteins including human ADAR1 and ADAR2 (Desterro et al., 2003; Yang et al., 2003; Sansam et al., 2003).
Our experiments suggest that RNAs transcribed elsewhere in the genome localize to the special loop and are required for tethering ADAR1 to this site. At this stage, we do not know which type of RNAs are localized there, but considering the absence of polymerase II from this site and the fact that hnRNPs A/B are absent from this site under normal conditions, it is tempting to speculate that the RNAs located there are transcribed elsewhere in the genome and subsequently transported to the special loop. The special loop shows relatively strong staining with an antibody directed against 3mG cap structures that are present on some snRNAs both in the presence and absence of transcription. This suggests that the RNA(s) present at the special loop possess such a cap structure and might even be snRNAs themselves. However, currently we do not know whether such a 3mG-containing RNA would be at the core of the special loop serving as an anchor for additional factors or whether 3mG-containing RNAs (snRNAs) accumulate there independently. Interestingly, localization of ADAR1 to the special loop is sensitive to digestion with both singleand double-strand specific RNAses, which is compatible with the notion that the RNA(s) at the core of thee special loop contain both structured and unfolded regions.
Nonetheless, although many of our findings are compatible with the presence of RNAs transcribed by pol II at the special loop, we cannot exclude that the RNAs there are transcribed by RNA polymerase III.
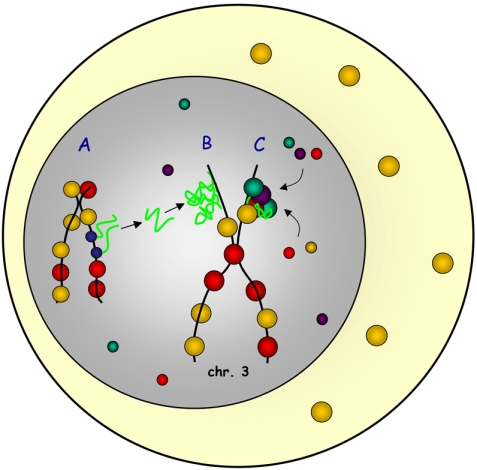
In any case, RNAs have been shown to be involved in the regulation of chromatin states such as X-inactivation, imprinting, or heterochromatin formation (Reinhart and Bartel, 2002; Sleutels et al., 2002; Wutz et al., 2002). It is therefore possible that the RNAs localized to the special loop are essential to define the chromatin activity and protein storage function of this site. Such RNAs might act by controlling the histone code at the special loop on the one hand and by serving as a binding platform for additional RNA-binding proteins on the other (Figure 6).
Figure 6.
The special loop, a chromosomal storage site? RNAs transcribed somewhere in the genome (A) move to the special loop on chromosome 3 where they accumulate (B). RNAs accumulated at the special loop define this site as a storage region for excess RNA-processing proteins and serve as a binding platform to sequester proteins to this site (C).
The Special Loop in Somatic Cells
Our study has primarily focused on the distribution of Xenopus ADAR1 in oocytes. The accumulation of ADAR1 at the special loop in these specialized cells raises the obvious question whether the same structure can also be found in somatic cells. Although we have tried to detect ADAR1 accumulation in somatic cell nuclei, our results are not fully conclusive at this point. If a special loop existed in somatic cells, ADAR1 would be expected to accumulate in two foci corresponding to the two homologous chromosomal sites. Although such foci can be observed in some nuclei of AMD-treated cells, they cannot be found in all nuclei. On average, ADAR1-positive nuclear foci can be found in 30% of all AMD-treated cells, whereas no such foci can be seen in untreated cells (Sallacz and Jantsch, unpublished observation). Although this observation can be taken as an indication for the maintenance of a special loop in somatic cells, it provides no definitive proof thereof. Considering the proposed function of a special loop as a regulator of intranuclear ADAR1 levels, it must be taken into account that ADAR1 levels vary greatly among different tissues. Although oocytes might be highly enriched in ADAR1 protein levels (possibly to serve as a stockpile during early development when high ADAR1 levels might be needed), somatic fibroblasts might have rather low ADAR1 levels. There might thus be little or no need to titrate nuclear ADAR1 levels. It is therefore reasonable to assume that varying intranuclear ADAR1 levels are responsible for the variable association of ADAR1 with intranuclear foci.
Acknowledgments
We thank Andrea Stahlmann for excellent technical assistance, Serafin Pinol-Roma and Gideon Dreyfuss for providing us with antibodies against Xenopus and mammalian hnRNP proteins, and Walter Keller and his lab for providing us with antisera specific for mammalian ADAR1. This work was supported by the Austrian Science Foundation grant SFB 1706.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–01–0016) on April 20, 2005.
References
- Athanasiadis, A., Rich, A., and Maas, S. (2004). Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2, e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L., and Weintraub, H. (1987). A developmentally regulated activity that unwinds RNA duplexes. Cell 48, 607–613. [DOI] [PubMed] [Google Scholar]
- Burns, C. M., Chu, H., Rueter, S. M., Hutchinson, L. K., Canton, H., Sanders-Bush, E., and Emeson, R. B. (1997). Regulation of serotonin-2C receptor G-protein coupling by RNA editing [see comments]. Nature 387, 303–308. [DOI] [PubMed] [Google Scholar]
- Chen, C. X., Cho, D. S., Wang, Q., Lai, F., Carter, K. C., and Nishikura, K. (2000). A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, D. S., Yang, W., Lee, J. T., Shiekhattar, R., Murray, J. M., and Nishikura, K. (2003). Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 278, 17093–17102. [DOI] [PubMed] [Google Scholar]
- Dabiri, G. A., Lai, F., Drakas, R. A., and Nishikura, K. (1996). Editing of the GLuR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J. 15, 34–45. [PMC free article] [PubMed] [Google Scholar]
- Desterro, J. M., Keegan, L. P., Lafarga, M., Berciano, M. T., O'Connell, M., and Carmo-Fonseca, M. (2003). Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 116, 1805–1818. [DOI] [PubMed] [Google Scholar]
- Eckmann, C. R., and Jantsch, M. F. (1999). The RNA-editing enzyme ADAR1 is localized to the nascent ribonucleoprotein matrix on Xenopus lampbrush chromosomes but specifically associates with an atypical loop. J. Cell Biol. 144, 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann, C. R., Neunteufl, A., Pfaffstetter, L., and Jantsch, M. F. (2001). The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol. Biol. Cell 12, 1911–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X. D., and Maniatis, T. (1990). Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343, 437–441. [DOI] [PubMed] [Google Scholar]
- Gall, J. G., Murphy, C., Callan, H. G., and Wu, Z. A. (1991). Lampbrush chromosomes. Methods Cell Biol. 36, 149–166. [PubMed] [Google Scholar]
- Gallo, A., Keegan, L. P., Ring, G. M., and O'Connell, M. A. (2003). An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 22, 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough, R. F., and Bass, B. L. (1997). Analysis of Xenopus dsRNA adenosine deaminase cDNAs reveals similarities to DNA methyltransferases. RNA 3, 356–370. [PMC free article] [PubMed] [Google Scholar]
- Hulsebos, T. J., Hackstein, J. H., and Hennig, W. (1984). Lampbrush loop-specific protein of Drosophila hydei. Proc. Natl. Acad. Sci. USA 81, 3404–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, S. R., Hough, R. F., Aruscavage, P. J., and Bass, B. L. (1995). Deamination of mammalian glutamate receptor RNA by Xenopus dsRNA adenosine deaminase: similarities to in vivo RNA editing. RNA 1, 1051–1060. [PMC free article] [PubMed] [Google Scholar]
- Keegan, L. P., Leroy, A., Sproul, D., and O'Connell, M. A. (2004). Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 5, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. D., Kim, T. T., Walsh, T., Kobayashi, Y., Matise, T. C., Buyske, S., and Gabriel, A. (2004). Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 14, 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E., Du, L., Bregman, D. B., and Warren, S. L. (1997). Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman, D., and Kirschner, M. W. (1989). An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Krainer, A. R. (1988). Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 16, 9415–9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, E. A., Lerner, M. R., Janeway, C. A., Jr., and Steitz, J. A. (1981). Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA 78, 2737–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon, E.Y. et al. (2004). Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J. R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P. H. (1994). Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266, 1709–1713. [DOI] [PubMed] [Google Scholar]
- Marcu, A., Bassit, B., Perez, R., and Pinol-Roma, S. (2001). Heterogeneous nuclear ribonucleoprotein complexes from Xenopus laevis oocytes and somatic cells. Int. J. Dev. Biol. 45, 743–752. [PubMed] [Google Scholar]
- Palladino, M. J., Keegan, L. P., O'Connell, M. A., and Reenan, R. A. (2000). dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing [In Process Citation]. RNA 6, 1004–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, J. B., and Samuel, C. E. (1995). Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma, S., Swanson, M. S., Gall, J. G., and Dreyfuss, G. (1989). A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell Biol. 109, 2575–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B. J., and Bartel, D. P. (2002). Small RNAs correspond to centromere heterochromatic repeats. Science 297, 1831. [DOI] [PubMed] [Google Scholar]
- Roth, M. B., and Gall, J. G. (1987). Monoclonal antibodies that recognize transcription unit proteins on newt lampbrush chromosomes. J. Cell Biol. 105, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter, S. M., Dawson, T. R., and Emeson, R. B. (1999). Regulation of alternative splicing by RNA editing. Nature 399, 75–80. [DOI] [PubMed] [Google Scholar]
- Saccomanno, L., and Bass, B. L. (1999). A minor fraction of basic fibroblast growth factor mRNA is deaminated in Xenopus stage VI and matured oocytes. RNA 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam, C. L., Wells, K. S., and Emeson, R. B. (2003). Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. USA 100, 14018–14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., and Barlow, D. P. (2002). The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. [DOI] [PubMed] [Google Scholar]
- Strehblow, A., Hallegger, M., and Jantsch, M. F. (2002). Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol. Biol. Cell 13, 3822–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, R. W. et al. (1990). Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol. Cell. Biol. 10, 5586–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, R. A., Jared, D. W., Dumont, J. N., and Sega, M. W. (1973). Protein incorporation by isolated amphibian oocytes. 3. Optimum incubation conditions. J. Exp. Zool. 184, 321–333. [DOI] [PubMed] [Google Scholar]
- Wutz, A., Rasmussen, T. P., and Jaenisch, R. (2002). Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174. [DOI] [PubMed] [Google Scholar]
- Yang, J. H., Nie, Y., Zhao, Q., Su, Y., Pypaert, M., Su, H., and Rabinovici, R. (2003). Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J. Biol. Chem. 278, 45833–45842. [DOI] [PubMed] [Google Scholar]