Abstract
One component of host defense at mucosal surfaces is epithelium-derived peptides with antimicrobial activity called defensins. We describe in this report the isolation and characterization of a murine homologue of human β-defensin 2 (hBD-2) called mouse β-defensin 3 (mBD-3). The predicted amino acid sequence shows the hallmark features of other known epithelial defensins, including the ordered array of six cysteine residues. Analysis of a genomic clone of mBD-3 revealed two exons separated by a 1.7-kb intron. The mBD-3 gene is localized at the proximal portion of chromosome 8, the site where genes for mouse α- and β-defensins are found. Under basal condition, mBD-3 transcripts were detected at low levels in epithelial cells of surface organs, such as the intestine and lung. After instillation of Pseudomonas aeruginosa PAO1 into mouse airways, mBD-3-specific mRNA was upregulated significantly not only in large airways but also in the small bowel and liver. Recombinant mBD-3 peptide, produced from a baculovirus expression system, showed antimicrobial activity against P. aeruginosa PAO1 (MIC of 8 μg/ml) and Escherichia coli D31 (MIC of 16 μg/ml) in a salt-dependent manner. This study demonstrates that a murine homologue of hBD-2 is present in the respiratory system and other mucosal surfaces. These similarities between murine and human host defense apparatus provide further impetus to evaluate the mouse as a model for studying the human innate host defense system.
Antimicrobial peptides have been found in a wide array of animal species, ranging from insects to lower vertebrates and mammals (11). These peptides contribute to innate host defense against a number of bacterial and fungal pathogens. The β-defensins are small cystine-rich, cationic peptides expressed by various epithelia (8). The first such peptide, called tracheal antimicrobial peptide (TAP), was isolated from cow trachea and is expressed throughout the surface epithelia of the cow lung, where it is believed to contribute to host defense (6). A homologous peptide called lingual antimicrobial peptide (LAP) was subsequently purified from cow tongue and shown to be expressed in multiple epithelia, including those of the lung (22).
A defect of antimicrobial functions has been implicated in the pathogenesis of cystic fibrosis (CF) lung disease (23). Several levels of host defense are present in the human airways, including resident macrophages, mucociliary clearance, and antibacterial proteins such as lysozyme and lactoferrin. More recently, antimicrobial peptides have been isolated and shown to play a role in the host defense apparatus of the human respiratory system. These include human β-defensins 1 and 2 (hBD-1 and -2) (2, 10, 12), which belong to the β-defensin family, and LL-37/hCAP-18, which is a member of the cathelicidins (3). It is not known, however, to what extent these or other molecules are impaired in CF.
Authentic murine models would be of use in the study of antimicrobial substances in pulmonary host defense and in defining their role in the development of CF lung disease. We and others recently isolated a cDNA and the corresponding genomic clone encoding a defensin molecule, called mouse β-defensin 1 (mBD-1) which is homologous to hBD-1 (1, 14, 17). This constitutively expressed peptide is found in multiple epithelia, with highest levels in kidney. Recently, a second mouse β-defensin (mBD-2) was cloned (16). In the present study, we describe the cloning of a mouse cDNA encoding another mouse β-defensin, mBD-3, and analyzed its gene structure, antimicrobial function, expression, and regulation.
MATERIALS AND METHODS
Cloning of mBD-3 cDNA.
We used the β-defensin-specific cysteine pattern to perform a BLAST (basic local alignment search tool) search at the website of the National Center for Biotechnology Information (18a) and found a rat expressed sequence tag clone that showed similarities to β-defensins (Genbank accession no. AA800706). The cDNA sequence of mBD-3 was cloned by using cDNA from mouse lung and primers generated from the cDNA sequence of the putative rat defensin. Reverse transcriptase PCR (RT-PCR) was used to clone the full-length cDNA sequence. Total RNA was isolated from C57BL/6 mouse lung by using Trizol (Gibco BRL) and further purified to poly(A)+ RNA by passage through oligo(dT) columns (Qiagen). Poly(A)+ RNA (approximately 100 ng) was reverse transcribed by using NotI-(dT)18 as a primer (First-Strand cDNA synthesis kit; Pharmacia Biotech), and 10% of the reaction was used for a PCR. Primers that resulted in successful cloning of a candidate cDNA were forward primer (m2-def 1; 5′-CAC GAG GCA CCA GGC TTC AGT C-3′) and reverse primer (m2-def 2; 5′-CAA TGG GAT GAA CAG AAT TTG CTC C-3′). The PCR products were analyzed on a 1.5% agarose gel, and bands between 250 and 500 bp were cut out, purified (QIAquick gel extraction kit; Qiagen), and cloned into pGEM-T (Promega). Inserts were sequenced with an Applied Biosystems model 373 fluorescent DNA sequencer and analyzed for the presence of β-defensin-specific hallmarks.
Cloning of the genomic sequence.
A mouse genomic library constructed in FIX II vector (Stratagene) was screened by standard procedures with a probe generated by PCR using mBD-3 cDNA as the template and gene-specific primers. Positive clones were purified, and genomic DNA was isolated and subcloned (19). The clones were sequenced, and the data obtained were analyzed to determine the structure of the mBD-3 gene.
Analysis of chromosomal localization by fluorescence in situ hybridization (FISH).
Two gene-specific primers were used to amplify a sequence spanning the region including parts of exon 1 and the intron (Genome Walker kit; Clontech). The PCR product was used as a probe to screen a mouse genomic bacterial artificial chromosome (BAC) library (GenomeSystems, Inc.). The DNA of positive clones was labeled with digoxigenin-dUTP by nick translation and hybridized to normal mouse metaphase chromosomes. The chromosome of interest was identified by analysis of the 4′,6-diamidino-2-phenylindole banding pattern and finally by cohybridization of the mBD-3-specific probe with a probe specific for the telomeric region of chromosome 8 (GenomeSystems).
Dot blot of poly(A)+ RNA.
[32P]dCTP random primer-labeled probes of mBD-3 and ubiquitin cDNA were hybridized separately to a nylon filter with dotted mRNAs from different mouse organs (Mouse RNA Master Blot; Clontech). After washes at high-stringency conditions, the signals were quantified with a PhosphorImager 445 SI (Molecular Dynamics). The expression signals for mBD-3 were normalized to the signal for hybridization to the housekeeping gene ubiquitin.
RT-PCR and induction of expression by bacterial infection.
For stimulation of mBD-3 expression, Pseudomonas aeruginosa PAO1 (kindly provided by Lisa Saiman, Columbia University, N.Y.) was used in a mouse pneumonia model (9). When needed for an experiment, frozen bacteria were thawed and cultured overnight on a nutrient agar plate. The bacteria in a single colony arbitrarily selected from colonies on the plate were cultured overnight in a nutrient broth medium and used for the experiment. The number of CFU of the organisms was determined by quantitative cultivation on nutrient agar plates. Bacteria (50 μl containing 106 CFU) were inoculated into the nostrils of anesthetized mice in vertical position, allowing for aspiration of the inoculum. After 24 h, the animals were killed and RT-PCR was used to detect levels of mBD-3 mRNA in several tissues. Poly(A)+ RNA was isolated from mouse tissues (heart, trachea/lung, kidney, small and large intestine, and liver) and reverse transcribed as described above. The following primers were used for PCR: forward primer (m2-def 3; 5′-CTC TTT GCA TTT CTC CTG GTG CTG CTG-3′) and reverse primer (m2-def 4; 5′-CAT CTT CAT GGA GGA GCA AAT TCT G-3′). The predicted size of the PCR product was 273 bp. The reverse transcriptase was omitted in the negative control, whereas an RT-PCR with primers specific for mouse glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was used as a positive control. The PCR products were analyzed on a 1.5% agarose gel, photographed, and blotted onto nitrocellulose by using standard procedures. To confirm that the immobilized PCR products represent mBD-3 cDNA, a [32P]dCTP random primer-labeled probe of mBD-3 was used to hybridize with the blots.
In situ hybridization.
Various mouse tissues (lung, small intestine, and liver before and after administration of bacteria) were embedded in OCT (Tissue-Tek; Miles Laboratories), cryosectioned (6-μm sections), mounted on slides, and fixed in 4% paraformaldehyde in phosphate-buffered saline (pH 7.4, 4 h, 4°C). Following dehydration, sections were treated with proteinase K (10 μg/ml; 30 min, 30°C), fixed in 4% paraformaldehyde in phosphate-buffered saline, treated with acetic anhydride, and dehydrated through ethanol. Prehybridization was performed for 4 h at 54°C in 10 mM Tris (pH 8.0)–50% formamide–2.5× Denhardt’s solution–0.6 M NaCl–1 mM EDTA–0.1% sodium dodecyl sulfate–500 μg of tRNA per ml–10 mM dithiothreitol. RNase control sections were treated with RNase A (200 μg/ml) for 1 h at 37°C before the prehybridization step. Sections were hybridized in the prehybridization solution (16 h, 54°C), using digoxigenin-labeled antisense or sense probes synthesized by in vitro transcription of full-length mBD-1 and mBD-3 cDNA. Sections were incubated with antibodies against digoxigenin conjugated with alkaline phosphatase followed by a solution of nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate. After washes, the sections were covered with mounting medium.
Production of recombinant mBD-3 and testing of antimicrobial activity.
mBD-3 peptide was produced by a recombinant baculovirus system as described for hBD-2 (2). In short, the full-length mBD-3 cDNA was prepared by using gene-specific primers in standard PCR and ligated with the transfer vector pBAC-1 (Novagen Inc.). The recombinant transfer plasmids were cotransfected with the linearized baculovirus DNA (BacVector-3000; Novagen) into Spodoptera frugiperda Sf9 cells, and recombinant viral plaques were purified individually and amplified. Sf9 cells were grown in serum-free medium (SF-900; GIBCO BRL) in suspension (27°C, 110 rpm). Cells were infected with recombinant virus, and medium was collected 72 h after infection by centrifugation (Syncom Corp.), adjusted to pH 5.8, and chromatographed on a cation-exchange column (2.5 by 10 cm, carboxymethylcellulose) equilibrated with ammonium acetate (32 mM, pH 5.8). After washing with ammonium acetate, a one-step elution was performed with 50 ml of 0.8 M NaCl in 32 mM ammonium acetate–20% acetonitrile. The substances were further purified on a 0.5- by 25-cm Dynamax-300Å C18 reverse-phase high-pressure liquid chromatography column (Rainin Instrument Co.) using a linear gradient of acetonitrile with 0.1% trifluoroacetic acid. Fractions were dried, resuspended in 50 μl of distilled water, and tested for antimicrobial activity in agarose diffusion assays (see below). Purified peptides were characterized by mass spectrometry (Voyager BioSpectrometry workstation; PerSeptive Biosystems), capillary zone electrophoresis (270A-HT capillary electrophoresis system; Applied Biosystems), and amino acid composition analysis (Protein and Carbohydrate Structure Facility, University of Michigan). To screen chromatography fractions for antibacterial activity, 2 μl of each fraction was applied on the top of 0.7% agarose containing Escherichia coli D31 (5 × 108 CFU/10 ml) and incubated for 12 h at 37°C. Fractions with antimicrobial activity were determined visually. The isolated peptide was used in antibacterial microdilution assays against E. coli D31 and P. aeruginosa PAO1 (2). In parallel, hBD-2 and magainin I were tested as positive controls. For MIC testing, twofold serial dilutions of peptides were prepared in half-strength Mueller-Hinton broth. Inocula of approximately 105 CFU of bacteria growing in log phase were added to each well. After 24 h of incubation (37°C), bacterial growth was determined by visual analysis and determination of the optical density at 595 nm.
Nucleotide sequence accession numbers.
The sequences shown in Fig. 1A and 2B have been assigned GenBank accession no. AF092929 and AF093245, respectively.
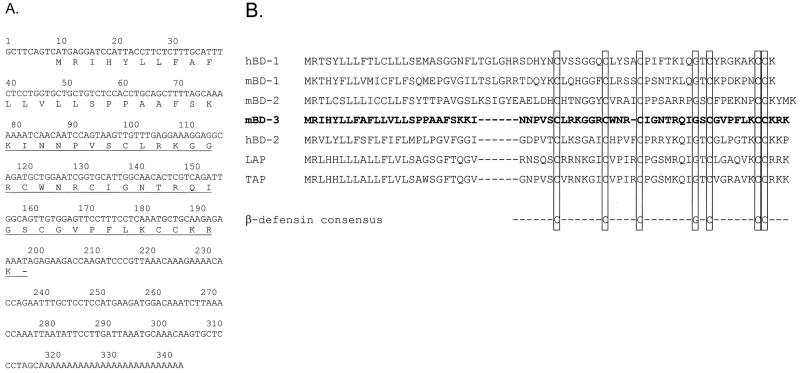
FIG. 1.
cDNA and peptide sequences of mBD-3. (A) Complementary DNA and deduced amino acid sequences of mBD-3. The mature peptide based on an analysis of baculovirus-expressed peptide is underlined; the dash represents the termination codon. (B) Comparison of the putative prepropeptide sequences of mBD-1, mBD-2, mBD-3, hBD-1, hBD-2, TAP, and LAP, all derived from the cDNA sequences.
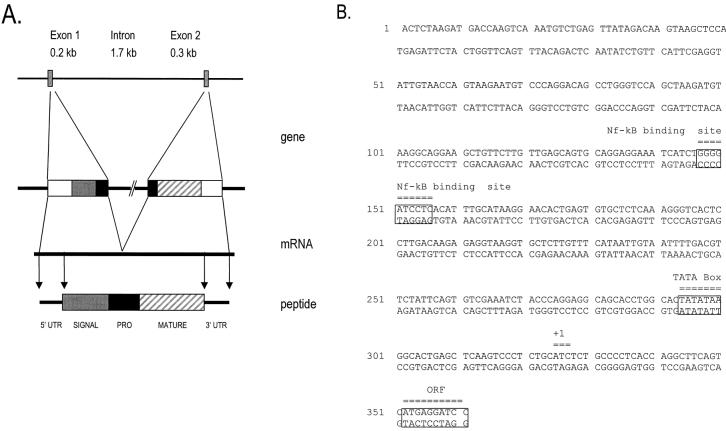
FIG. 2.
Structure of the mBD-3 gene. (A) Schematic drawing of the gene, the cDNA, and the predicted structure of the prepropeptide. The gene is represented schematically, with the following components shown: 5′ untranslated region (5′ UTR), signal sequence, interrupted prosequence (PRO), mature peptide (MATURE), and 3′ UTR. (B) Nucleotide sequence of the mBD-3 promoter, with locations of the TATA box, an NF-κB binding site, the putative start of transcription (+1), and the start of the open reading frame (ORF) indicated.
RESULTS
Cloning of the cDNA and genomic sequence for mBD-3.
Primers based on the sequence of a putative rat defensin found during a BLAST search were used to isolate candidate clones from mouse lung by RT-PCR. One clone homologous to cDNA sequences of β-defensins was obtained and further analyzed (Fig. 1A). The cDNA clone exhibited greater DNA sequence homology to hBD-2 (36.7%) than hBD-1 (25.3%). There was 39.7% amino acid sequence identity to hBD-2. This murine clone was called mouse mBD-3. The predicted peptide of mBD-3 showed the presence of the β-defensin-specific conserved amino acids, including the pattern of six cysteines (Fig. 1B); however, the spacing between the second and third cysteines is reduced by one residue compared to other known β-defensins. The mBD-3 cDNA clone consists of a 192-bp open reading frame encoding a peptide 64 amino acids in length with a putative preprosequence (Fig. 1). Online BLAST searches using the mBD-3 cDNA and amino acid sequences revealed high degrees of homology to sequences of known β-defensins but no novel sequences at the time of the search (March 1999).
A mouse genomic library was screened with PCR-amplified sequences, and several positive clones were isolated and further analyzed by hybridization with radiolabeled probes corresponding to the 5′ and 3′ regions of the mBD-3 cDNA. A sequence spanning the mBD-3 gene was defined. A map of the mBD-3 gene shows the two exons separated by a 1.7-kb intron (Fig. 2A). A TATA box and NF-κB site are located in the 5′ flanking region (Fig. 2B). The exon-intron splice site sequences confirm to the consensus rule (18) (data not shown).
A BAC clone containing the mBD-3 gene was isolated and used as a probe to determine the chromosomal localization of the mBD-3 gene by FISH analysis (GenomeSystems). The mBD-3 probe cohybridized with a probe specific for the telomeric region of chromosome 8. Measurements of 10 metaphase chromosomes localized the mBD-3 specific hybridization signal to the proximal region of chromosome 8, an area that corresponds to band 8A4 (data not shown).
Expression of mBD-3 in mouse tissues and regulation of its expression.
The distribution of mBD-3 expression was evaluated by dot blot hybridization using a commercially available filter that contains RNA from a variety of mouse tissues. The resulting hybridization signal was quantified on a PhosphoImager and normalized to the expression of ubiquitin. Figure 3A presents the relative expression of mBD-3 in all organs that showed a signal above background; muscle is an example of low or no expression. Highest expression was seen in salivary glands, pancreas, and reproductive organs.
FIG. 3.
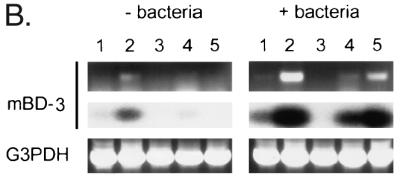
Analysis of mBD-3 expression by dot blot analysis and RT-PCR. (A) Measurement of mBD-3 expression by dot blot analysis. A filter dotted with mRNA from a number of mouse tissues was hybridized to an mBD-3 probe. Signals were quantified with a PhosphorImager system and normalized to expression of the housekeeping gene ubiquitin. Data are expressed as relative hybridization signals. Only those tissues which demonstrated a significant signal over background are presented, except for skeletal muscle, which is an example of an organ with low signal. The experiment was repeated on three occasions with virtually identical results. (B) Detection of mBD-3 expression was measured before and after intratracheal injection of P. aeruginosa PAO1 in various mouse tissues by RT-PCR. Poly(A)+ RNA was isolated from mouse tissues and reverse transcribed, and the cDNAs were amplified by using mBD-3-specific primers. A single 270-bp band visualized directly by staining with ethidium bromide was generated by the amplification (upper panel). The PCR products were blotted onto nitrocellulose filter and hybridized with radiolabeled mBD-3 cDNA (middle panel). The mRNA coding for G3PDH was amplified by using gene-specific primers (lower panel). Lanes: 1, heart; 2, lung/trachea; 3, kidney; 4, small bowel; and 5, liver.
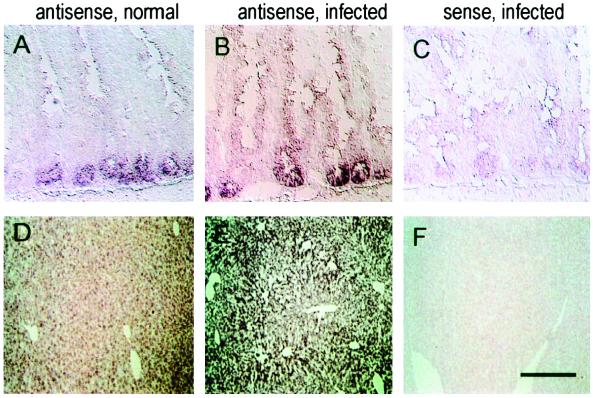
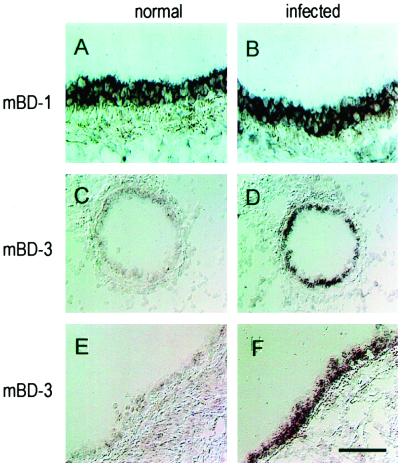
Selected tissues were analyzed by in situ hybridization with an antisense probe to mBD-3 labeled with digoxigenin and detected with alkaline phosphatase-conjugated antibodies (Fig. 4 and 5). In unstimulated animals, mBD-3 RNA was found in epithelial cells of the small bowel (crypts) (Fig. 4A) and the liver (diffusely in all hepatocytes) (Fig. 4D). Expression was barely detectable in the epithelia of the proximal, larger cartilaginous (Fig. 5E), and distal cartilaginous and noncartilaginous (Fig. 5C) native airways, in contrast with high level of mBD-1 RNA in epithelial cells of proximal airway (Fig. 5A). The specificity of this assay was confirmed in serial sections hybridized with the sense probe or RNase-pretreated sections hybridized with the antisense probe (data not shown).
FIG. 4.
Detection of transcripts encoding mBD-3 in mouse gastrointestinal organs. (A to C) Small intestine; (D to F) liver. Left column, sections from animals under normal conditions hybridized with an antisense probe; middle column, sections from infected mice with an antisense probe; right column, sections from infected animals hybridized to a sense probe. Bars represent 50 μm (A to C) and 120 μm (D to F).
FIG. 5.
Detection of transcripts encoding mBD-1 and mBD-3 in mouse respiratory organs. Tissue samples were frozen in OCT and evaluated by in situ hybridization using digoxigenin-labeled probes. (A and B) Detection of mBD-1-specific transcripts in large airways; (C and D) mBD-3-specific transcripts in small airways; (E and F) mBD-3-specific transcripts in large airways. Left column, sections from animals under normal conditions; right column, sections from mice infected with P. aeruginosa PAO1. Bar, 35 μm.
The human and cow homologues of mBD-3 (i.e., hBD2 and TAP) are induced in response to both infection and inflammatory cytokines. To analyze the regulation of mBD-3 expression, we infected mice via nasal inoculation of P. aeruginosa PAO1. The mice were killed after 24 h, and their organs were harvested for analysis of mBD-3 mRNA by RT-PCR and in situ hybridization. The results of RT-PCR analysis (Fig. 3B) showed expression of mBD-3 at low levels in lung/trachea (left, lane 2) and bowel (left, lane 4) of unstimulated mice, consistent with the dot blot analysis. Substantially increased expression of mBD-3 was observed in both lung (right, lane 2) and bowel (right, lane 4) following exposure to P. aeruginosa. mBD-3 mRNA was detected in liver after intrapulmonary infection of P. aeruginosa, whereas no expression was seen in this organ before infection (compare lanes 5). G3PDH was again used as a positive control. No bands were detected when reverse transcriptase was omitted (data not shown).
Tissue distribution of mBD-3 mRNA before and after infection was analyzed at the cellular level by in situ hybridization. Whereas under normal conditions, the mBD-3-specific signal was weak in the surface epithelium of the airways (Fig. 5C and E) or the liver parenchyma (Fig. 4D), after lung infection, strong signals were detected in epithelial cells throughout the conducting respiratory epithelium (Fig. 5D and F) and in pericentral hepatocytes (Fig. 4E). In contrast, expression of mBD-1 was detected at equivalent signal intensities in epithelia of normal and infected mice (Fig. 5A and B).
Production of recombinant mBD-3 peptide and analysis of antimicrobial activity.
To analyze the properties of mBD-3, the peptide was produced in a baculovirus expression system and purified by sequential fractionation applying cation-exchange and reverse-phase chromatography as described for hBD-2 (2). Capillary zone electrophoresis, amino acid composition, and mass spectroscopy (data not shown) were used to analyze the primary peaks of antimicrobial activity. These studies revealed a preparation of the mature peptide with a molecular weight of 4,360.3 spanning the 39 COOH-terminal amino acids of mBD-3. This mass indicates the presence of three disulfide bonds.
The MICs of purified mBD-3 were 16 μg/ml for E. coli and 8 μg/ml for P. aeruginosa, which compared favorably to the activity of a magainin peptide (MICs against E. coli D31 of 16 μg/ml and against P. aeruginosa PAO1 of 8 μg/ml) and hBD-2 (MICs against E. coli D31 of 62 μg/ml and against P. aeruginosa PAO1 of 62 μg/ml).
DISCUSSION
We describe in this study the isolation of a third mouse β-defensin, called mBD-3. The cDNA was cloned by using RT-PCR with primers based on the sequence of a putative rat β-defensin found during a BLAST search in GenBank. The putative mature peptide contains six cysteine residues and other conserved amino acids that may have important roles for the conformation and function of β-defensins (25). Recombinant mBD-3 peptide derived from a baculovirus expression system demonstrated bacterial killing activity against different bacteria.
The chromosomal localization of mBD-3 was analyzed by FISH and mapped to the proximal region of chromosome 8, an area which also contains the Defcr locus, where genes for mouse α- and β-defensins are found (1, 14, 16, 17, 20). This area is homologous to the human chromosome 8p23, where genes for human α- and β-defensins reside (2, 13, 15). These results indicate a close phylogenetic relationship of α- and β-defensins in mice and humans. The development of these two groups of antimicrobial peptides may have taken place during the development of mammalia before rodents and primates separated.
The tissue distribution of mBD-3 expression, initially evaluated by using a commercial dot blot of mouse RNAs, revealed low-level expression in several surface organs, such as lung, salivary glands, and reproductive organs. In situ hybridization further demonstrated expression of the gene in epithelia of some of these organs. In the respiratory tract, expression is low under basal conditions and mainly restricted to surface epithelia of the large airways. Introduction of bacteria directly into the airway resulted in a substantial induction of mBD-3 expression throughout the epithelia of the conducting airways, consistent with bacterium-mediated induction of hBD-2 observed in vitro as well as in vitro and in vivo induction studies with LAP and TAP (5, 12, 21, 22, 24). Interestingly, there was a marked induction of mBD-3 expression in liver following intratracheal infection, suggesting systemic regulation of defensin genes by diffusible molecules. The precise mechanism by which mBD-3 is regulated locally and systemically is unclear. All homologues of this subfamily of β-defensins (i.e., hBD-2, mBD-3, TAP, and LAP) are expressed from a promoter that contains a classic TATA box with an upstream NF-κB site. NF-κB is an important intracellular signal of both innate and acquired immunity triggered by a wide array of inflammatory mediators.
Our study contributes to the evolving story of innate immunity in the mouse. An important question is the role of murine systems in characterizing biology of peptide antibiotics relevant to human disease such as CF. The type 1 β-defensins, such as hBD-1 and mBD-1, show tremendous similarities between humans and mice; they are salt sensitive and active against an array of bacteria (1, 4). Expression of the type 1 β-defensin genes is seen throughout epithelia of many mucosal surfaces, with a predominate site being the urogenital system. The type 2 β-defensin genes, such as hBD-2 and mBD-3, are also expressed throughout epithelia of multiple mucosal surfaces, with relatively higher levels found in the gastrointestinal tract. These genes are significantly upregulated in response to infection and inflammation (2, 6, 12, 22). The recently isolated mBD-2 is interesting in that it shows more homology to type 1 β-defensins although it appears to be regulated by inflammatory mediators (16). Both human and mice contain α- and β-defensin genes located in homologous chromosomal locations. Whereas in humans neutrophils represent a primary site of α-defensin expression, these substances seem to be absent from mouse neutrophils (7).
In summary, we describe a murine homologue of hBD-2 with significant similarities in structure, function, and regulation. Further characterization of the β-defensin genes in the mouse could enhance understanding of innate immunity in health and disease.
ACKNOWLEDGMENTS
The contribution of the Cell Morphology Core of the Institute of Human Gene Therapy was greatly appreciated.
This work was supported by the Cystic Fibrosis Foundation, grants NIDDK P30 and NHLBI R01 from the NIH to J.M.W., as well as Genovo, Inc., a biotechnology company that James Wilson founded and holds equity in. Robert Bals was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Bals R, Goldman M J, Wilson J M. Mouse beta-defensin 1 is a salt-sensitive antimicrobial peptide present in epithelia of the lung and urogenital tract. Infect Immun. 1998;66:1225–1232. doi: 10.1128/iai.66.3.1225-1232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bals R, Wang X, Wu Z, Freeman T, Banfa V, Zasloff M, Wilson J M. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Investig. 1998;102:874–880. doi: 10.1172/JCI2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bals R, Wang X, Zasloff M, Wilson J M. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc Natl Acad Sci USA. 1998;95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensch K W, Raida M, Magert H-J, Schulz-Knappe P, Forssmann W-G. hBD-1: a novel β-defensin from human plasma. FEBS Lett. 1995;368:331–335. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 5.Diamond G, Russell J P, Bevins C L. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond G, Zasloff M, Eck H, Brasseur M, Maloy W L, Bevins C L. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci USA. 1991;88:3952–3956. doi: 10.1073/pnas.88.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenhauer P B, Lehrer R I. Mouse neutrophils lack defensins. Infect Immun. 1992;60:3446–3447. doi: 10.1128/iai.60.8.3446-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganz T, Lehrer R I. Defensins. Pharmacol Ther. 1995;66:191–205. doi: 10.1016/0163-7258(94)00076-f. [DOI] [PubMed] [Google Scholar]
- 9.George S E, Kohan M J, Gilmour M I, Taylor M S, Brooks H G, Creason J P, Claxton L D. Pulmonary clearance and inflammatory response in C3H/HeJ mice after intranasal exposure to Pseudomonas spp. Appl Environ Microbiol. 1993;59:3585–3591. doi: 10.1128/aem.59.11.3585-3591.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 11.Hancock R E W. Peptide antibiotics. Lancet. 1997;349:412–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 12.Harder J, Bartels J, Christophers E, Schroeder J-M. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 13.Harder J, Siebert R, Zhang Y, Matthiesen P, Christophers E, Schlegelberger B, Schroeder J M. Mapping of the gene encoding human beta-defensin-2 (DEFB2) to chromosome region 8p22-p23.1. Genomics. 1997;46:472–475. doi: 10.1006/geno.1997.5074. [DOI] [PubMed] [Google Scholar]
- 14.Huttner K M, Kozak C A, Bevins C L. The mouse genome encodes a single homolog of the antimicrobial peptide human beta-defensin 1. FEBS Lett. 1997;413:45–49. doi: 10.1016/s0014-5793(97)00875-2. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Zhao C, Heng H H, Ganz T. The human beta-defensin-1 and alpha-defensins are encoded by adjacent genes: two peptide families with differing disulfide topology share a common ancestry. Genomics. 1997;43:316–20. doi: 10.1006/geno.1997.4801. [DOI] [PubMed] [Google Scholar]
- 16.Morrison G, Davidson D, Dorin J. A novel mouse beta defensin, Defb2, which is upregulated in the airways by lipopolysaccharide. FEBS Lett. 1999;442:112–116. doi: 10.1016/s0014-5793(98)01630-5. [DOI] [PubMed] [Google Scholar]
- 17.Morrison G M, Davidson D J, Kilanowski F M, Borthwick D W, Crook K, Maxwell A I, Govan J R, Dorin J R. Mouse beta defensin-1 is a functional homolog of human beta defensin-1. Mamm Genome. 1998;9:453–457. doi: 10.1007/s003359900795. [DOI] [PubMed] [Google Scholar]
- 18.Mount S. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.National Center for Biotechnology Information. [12 January 1999, posting date.] BLAST search engine. [Online.] http://www.ncbi.nlm.nih.gov./BLAST. [31 December 1998, last date accessed.]
- 19.Nehls M, Messerle M, Sirulnik A, Smith A J, Boehm T. Two large insert vectors, lambda PS and lambda KO, facilitate rapid mapping and targeted disruption of mammalian genes. BioTechniques. 1994;17:770–775. [PubMed] [Google Scholar]
- 20.Ouellette A J, Pravtcheva D, Ruddle F H, James M. Localization of the cryptdin locus on mouse chromosome 8. Genomics. 1989;5:233–239. doi: 10.1016/0888-7543(89)90051-7. [DOI] [PubMed] [Google Scholar]
- 21.Russell J P, Diamond G, Tarver A P, Scanlin T F, Bevins C L. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators lipopolysaccharide and tumor necrosis factor alpha. Infect Immun. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schonwetter B S, Stolzenberg E D, Zasloff M A. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 23.Smith J, Travis S, Greenberg E, Welsh M. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 24.Stolzenberg E D, Anderson G M, Ackermann M R, Whitlock R H, Zasloff M. Epithelial antibiotic induced in states of disease. Proc Natl Acad Sci USA. 1997;94:8686–8690. doi: 10.1073/pnas.94.16.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerman G R, Legault P, Selsted M E, Pardi A. Solution structure of bovine β-defensin-12: the peptide fold of the β-defensins is identical to that of the classical defensins. Biochemistry. 1995;34:13663–13671. doi: 10.1021/bi00041a048. [DOI] [PubMed] [Google Scholar]